Abstract
The flap domain of multisubunit RNA polymerases (RNAPs), also called the wall, forms one side of the RNA exit channel. In bacterial RNAP, the mobile part of the flap is called the flap tip and makes essential contacts with initiation and elongation factors. Cocrystal structures suggest that the orthologous part of eukaryotic RNAPII, called the flap loop, contacts transcription factor IIB (TFIIB), but the function of the flap loop has not been assessed. We constructed and tested a deletion of the flap loop in human RNAPII (subunit RPB2 Δ873-884) that removes the flap loop interaction interface with TFIIB. Genome-wide analysis of the distribution of the RNAPII with the flap loop deletion expressed in a human embryonic kidney cell line (HEK 293) revealed no effect of the flap loop on global transcription initiation, RNAPII occupancy within genes, or the efficiency of promoter escape and productive elongation. In vitro, the flap loop deletion had no effect on promoter binding, abortive initiation or promoter escape, TFIIS-stimulated transcript cleavage, or inhibition of transcript elongation by the complex of negative elongation factor (NELF) and 5,6-dichloro-1-β-d-ribofuranosylbenzimidazole (DRB) sensitivity-inducing factor (DSIF). A modest effect on transcript elongation and pausing was suppressed by TFIIF. Although similar to the flap tip of bacterial RNAP, the RNAPII flap loop is not equivalently essential.
INTRODUCTION
Multisubunit RNA polymerases (RNAP) are responsible for genomic transcription of protein-coding genes in all organisms from bacteria to humans. These RNAP exhibit strong conservation in the core subunit sequence, 3-dimensional structure, and protein-nucleic acid contacts (1, 7–9, 24, 25, 56). Despite this conservation, the extent to which evolution has produced functional divergence, especially in surface-located motifs that bear a superficial resemblance to each other, is generally unknown. An excellent case in point is the flap tip in the second-largest subunit of RNAP (β in bacteria; RPB2 in eukaryotes). In prokaryotes, the β flap tip (corresponding to the flap loop on the wall domain in eukaryotic RPB2) is capped by a hydrophobic alpha helix (the flap tip helix) located near the mouth of the RNA exit channel. The flap tip helix helps recruit different regulators during the initiation and elongation phases of transcription (e.g., σ and NusA, respectively) (17–19, 28, 35, 46, 47).
Interaction of the flap tip helix with σ region 4 is essential for transcription initiation at the major class of bacterial promoters that depend on consensus −10 and −35 promoter elements. Deletion of the flap tip helix (ΔFTH) in Escherichia coli RNAP (Δ887-897 in the E. coli β subunit, corresponding to Δ873-884 in human RPB2) blocks initiation at −35-dependent promoters but not at extended −10 promoters (17, 28, 46). Interaction of the flap tip helix with σ region 4 positions σ region 4.2 for contact with the −35 promoter element (17, 28, 35). Although the interaction of the flap tip with region 4 of the σ factor is essential for initiation, this interaction also hinders promoter escape (35).
In E. coli RNAP, the flap tip also plays roles in transcript elongation. It is required for pause enhancement by nascent RNA hairpins at pause sites (46, 47). In addition, the flap tip helix is required for enhancement of pausing or termination by the elongation factor NusA (19, 46, 47). Thus, the flap domain in E. coli RNAP plays a significant role via direct protein-protein interactions in transcription initiation, as an indirect modulator of active-site properties in pausing, and in protein factor-mediated control of transcriptional pausing at the his pause signal.
A recent X-ray cocrystal structure of yeast RNAPII with transcription factor IIB (TFIIB) reveals flap loop and flap loop helix structures in RPB2 that are similar in structure and location to the flap tip and flap tip helix in bacterial RNAP (26) (see Fig. 1A). However, the eukaryotic flap loop exhibits only limited sequence similarity to the bacterial flap tip. In the TFIIB-RNAPII structure, the flap loop helix contacts the N-terminal ribbon domain of TFIIB, which may be analogous to the interaction of region 4 of σ with the flap tip helix of bacterial RNAP (26). The TFIIB reader segment lies C-terminal of the TFIIB ribbon domain and appears to make key contacts with a promoter element involved in start site selection (26, 31). These similarities between the flap tip and flap loop of bacterial and eukaryotic RNAP, respectively, raise the possibility that the flap loop could play key roles in transcription initiation and promoter escape by eukaryotic RNAPII.
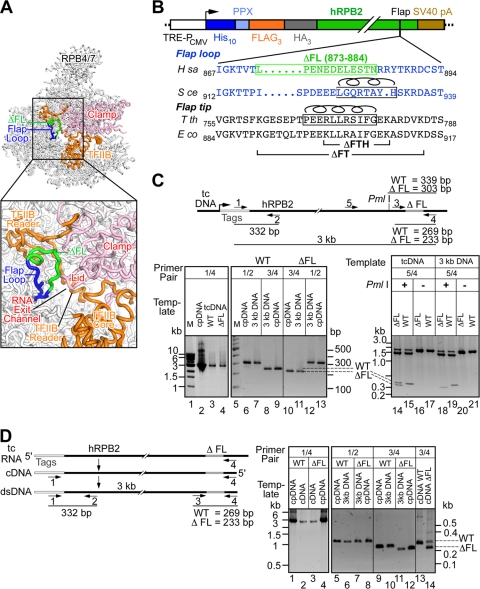
Fig. 1.
Expression of wild-type and ΔFL ettRNAPII. (A) Structure of Saccharomyces cerevisiae RNAPII in complex with TFIIB (Protein Data Bank identification code [PDB ID] 3k1f) (26) with the flap loop (blue/green) enlarged in the inset. The flap loop deletion is shown in green. (B) Schematic diagram of the epitope-tagged human RPB2 gene. The sequence alignment of the human flap loop (blue) is shown with the deletion boxed in green. The flap loop in S. cerevisiae RNAPII is shown in blue, and the flap loop helix is indicated by a helix above the sequence. The Thermus thermophilus and E. coli flap tip sequences are shown in black, with the flap tip helix boxed and indicated by a helix above the sequence. The locations of the E. coli flap tip (ΔFT) and flap tip helix (ΔFTH) deletions studied previously (43, 44) are indicated. SV40, simian virus 40; H sa, Homo sapiens; S ce, S. cerevisiae; T th, T. thermophilus; E co, E. coli. (C) The epitope tag is genetically linked to the flap loop deletion in the genome. The structure of the integrated transgene, the locations of the primers, and the sizes of the amplicons are shown schematically. Genomic DNA was first amplified using primer set 1/4 (lanes 1 to 4) from the control plasmid DNA (cpDNA) or from genomic DNA from the transgenic cell line (tcDNA), and the gel-purified 3-kb fragment was used as a template for the second round of PCR using primer set 1/2 (lanes 6, 7, 12, and 13), 3/4 (lanes 8 to 11), or 5/4 (lanes 14 to 21). The WT or ΔFL plasmids used for transfection served as positive-control templates (cpDNA). Samples were separated on a 1% (lanes 1 to 4) or a 1.5% (lanes 5 to 21) agarose gel. (D) The mRNA that codes for the epitope tags is linked to the flap loop deletion. The structure of mRNA coding for the transgene, the locations of the primers, and the sizes of the amplicons are shown schematically. cDNA was synthesized from total RNA using primer 4, and a 3-kb DNA fragment was PCR amplified from the cDNA or control plasmids (cpDNA) using primer set 1/4 (lanes 1 to 4). The gel-purified 3-kb amplicon was then used as a template for a second round of PCR using primer set 1/2 (lanes 5 to 8) or 3/4 (lanes 9 to 14). The WT or ΔFL plasmids used for transfection served as positive-control templates (cpDNA). Samples were separated on a 1% (lanes 1 to 4) or a 1.5% (lanes 5 to 14) agarose gel.
Transcript elongation in eukaryotes is regulated by multiple protein factors that affect transcript elongation by RNAPII either positively or negatively but whose precise contacts with RNAPII are unknown (e.g., the positive factor TFIIF and the negative factor DSIF/NELF, which is composed of negative elongation factor [NELF] and 5,6-dichloro-1-β-d-ribofuranosylbenzimidazole [DRB] sensitivity-inducing factor [DSIF]) (15, 22, 38, 42, 49, 52, 53, 55). Recent studies using cross-linking–mass spectrometry (4) and protein footprinting (14) suggest that the TFIIF dimerization domain contacts the lobe domain of RNAPII close to RPB9 and place the Tfg2/RAP30 subunit of TFIIF near the wall domain of RNAPII. This mapping raises the possibility that Tfg2/RAP30, which bears some similarity to bacterial σ, contacts the flap loop, a contact analogous to that of the bacterial elongation factor NusA with the flap tip. NELF, in complex with DSIF, induces promoter-proximal transcriptional pausing (38, 42, 49, 52), in part by interacting with the emerging nascent RNA (51). The flap loop, which is located at the mouth of the RNA exit channel, could facilitate the interaction of NELF with the emerging transcript.
During transcript elongation, at certain sequences on the DNA, RNAPII reverse translocates, or backtracks, along RNA and DNA chains with the 3′ end of the RNA dislodged from the active site and located in the secondary channel (20, 36, 37). Interaction of the exiting RNA with the flap loop located immediately outside the RNA exit channel might provide a physical barrier to backtracking and might thus play a key role in maintaining the active state of the enzyme.
Based on these similarities, we sought to test whether the flap loop of human RNAPII plays a significant role in transcription by RNAPII. To study the role of the flap loop in vivo and in vitro, we constructed and studied the effects of a deletion of the flap loop (ΔFL; RPB2 Δ873-884). We used immunoprecipitation (IP) of formaldehyde-cross-linked DNA with monoclonal antibodies (MAbs) specific for the mutant RNAPII followed by hybridization to promoter region tiling arrays (chromatin IP [ChIP]-chip) to study the activity of RNAPII lacking the flap loop in human cell lines. We also purified the mutant human RNAPII from the same stable cell lines and used in vitro transcription assays to test for essential roles of the flap loop in either transcription initiation or elongation.
MATERIALS AND METHODS
Cell line and proteins.
The HEK 293 Tet-ON cell line and the pTRE2hyg expression vector were purchased from Clontech (Mountain View, CA). Fast protein liquid chromatography (FPLC)-purified nucleoside triphosphates (NTPs) were from GE Healthcare (Piscataway, NJ). DNA and RNA oligonucleotides were from IDT (Coralville, IA). Anti-Flag M2 agarose and anti-Flag M2 monoclonal antibodies were from Sigma (St. Louis, MO). Anti-RPB1 8WG16 monoclonal antibodies were a generous gift from Richard Burgess, University of Wisconsin—Madison. TFIIF, DSIF/NELF, and TFIIS were purified as described previously (42, 44, 54).
Stable cell lines for conditional expression of the hRPB2 subunit and purification of epitope-tagged, transgenic RNAPII (ettRNAPII).
The wild-type (WT) human RPB2 coding sequence was PCR amplified from the cDNA carried in a plasmid (a gift from M. Vignernon, Strasbourg, France) using a forward primer carrying MluI and AscI sites at its 5′ end followed by the coding sequence of RPB2 and a reverse primer carrying a NotI site at its 5′ end. The PCR product was restricted with MluI and NotI and was cloned between the same sites in pTRE2-hyg. The Kozak sequence, followed by N-terminal epitope tags (a His10 tag, a precision protease [PPX] recognition site, three copies of a Flag tag, and three copies of a hemagglutinin [HA] tag) (for the sequence, see Fig. S1 in the supplemental material), was cloned in frame upstream of the ATG codon of RPB2 between the MluI and AscI sites by ligating oligonucleotides coding for the epitope tag sequence. This resulted in the deletion of the AscI site immediately downstream of the HA tag and the introduction of a new, unique AscI site immediately upstream of the HA tag to create the pJG012 expression plasmid (see Table S1 in the supplemental material). Plasmid pJG012 was then used to create the flap loop deletion (RPB2 Δ873-884) by an oligonucleotide-directed QuikChange (Stratagene, Santa Clara, CA) procedure, resulting in plasmid pJG019.
HEK 293 Tet-ON cells were grown as monolayers in Dulbecco's modified Eagle medium (DMEM; Sigma, St. Louis, MO) supplemented with Tet-free fetal bovine serum (FBS; 10%; Sigma, St. Louis, MO) and sodium bicarbonate (0.375%; Sigma, St. Louis, MO) and were cultured at 37°C in the presence of 5% CO2. Cells were transfected with the expression plasmids by using Lipofectamine according to the manufacturer's instructions (Takara Mirus Bio, Madison, WI). Cells were selected with hygromycin (200 μg/ml), and a stable pool of cells that conditionally expressed the epitope-tagged transgenic RPB2 subunit upon induction with doxycycline (2 μg/ml) was established. The ettRNAPII was partially purified from cell lysates by using anti-Flag M2 agarose (Sigma, St. Louis, MO) as described previously (50). Except for the experiments for which results are presented in Fig. 7 and 10 (see below), all experiments were performed using these cell lines or this partially purified protein.
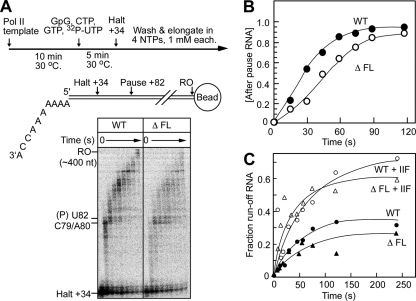
Fig. 7.
The flap loop modestly affects transcript elongation but not TFIIF-accelerated elongation. (A) Reaction scheme and tailed-template structure, with halt and pause sites indicated. The time course of transcript elongation from halted +34 complexes was determined on the tailed template as described in Materials and Methods. Aliquots were removed at the indicated times, and RNA products were separated on a 10% denaturing polyacrylamide gel. The positions of the halted complex RNA and pause RNA (U82) are indicated next to the gel. RO, runoff. (B) The RNA in each lane was quantitated, and the fraction of the RNA longer than the U82 pause RNA was plotted as a function of reaction time. (C) Transcription elongation reactions were performed as in panel A in the absence or presence of TFIIF (10 nM). The fraction of runoff RNA was quantitated and plotted as a function of reaction time.
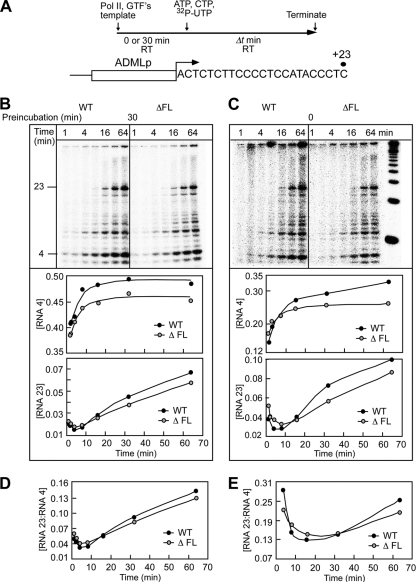
Fig. 10.
The flap loop is not required for transcription initiation or promoter clearance in vitro. (A) Schematic representation of preinitiation complex formation and transcription initiation from the adenovirus major late promoter (ADMLp) by wild-type and ΔFL ettRNAPII. (B) Preinitiation complexes were assembled using wild-type or ΔFL ettRNAPII with a 30-min preincubation at 30°C. Transcription was initiated by the addition of NTPs for the indicated times, and RNA products were separated on a 20% denaturing polyacrylamide gel. The fractions of 4-mer and 23-mer RNAs were quantitated and plotted as a function of reaction time. (C) Preinitiation complexes were assembled using wild-type or ΔFL ettRNAPII without preincubation at 30°C. Transcription was initiated by the addition of NTPs; an aliquot was removed after the indicated time intervals; and RNA products were separated on a 20% denaturing polyacrylamide gel. The fractions of 4-mer and 23-mer RNAs were quantitated and plotted as a function of reaction time. (D and E) Ratio of the 23-mer to the 4-mer synthesized with a 30-min preincubation (D) or with no preincubation (E).
An independently derived plasmid encoding the wild-type or ΔFL RPB2 subunit was constructed with an N-terminal TAP tag and was purified from stable cell lines as described previously (23). The results of experiments performed using TAP-tagged, transgenic RNAPII purified from these strains are presented in Fig. 7 and 10.
Chromatin immunoprecipitation and hybridization to microarrays.
ChIP assays were performed essentially as described elsewhere (33). Cells were grown on 10-cm2 dishes, cross-linked by the addition of formaldehyde to 1% and incubation at room temperature for 15 min, washed twice with phosphate-buffered saline (PBS), scraped off the plates, and centrifuged in 15-ml conical tubes for 10 min at 1,000 × g. The pelleted cells were transferred to a 1.5-ml microcentrifuge tube in 550 μl buffer A (50 mM Tris-HCl [pH 7.5], 10% glycerol, 1 mM EDTA, 1 mM EGTA, 150 mM NaCl, and 1% Triton X-100) and sonicated (with a microtip sonicator at 60% output) 8 times on ice in pulses of 30 s each. Cell debris was removed by centrifugation at 20,800 × g for 10 min at 4°C; the sample was then precleared by the addition of 30 μl Pansorbin (fixed protein A-bearing Staphylococcus aureus cells; Calbiochem), and incubation continued at 4°C for 1 h. RNAPII was immunoprecipitated with anti-Flag antibody M2 or with monoclonal antibody 8WG16 (45). The antibody (2 to 5 μg) was added to the precleared sample, which was then incubated overnight at 4°C with mixing. Antibody complexes were recovered by binding to 30 μl Pansorbin and centrifugation for 4 min at 20,800 × g; they were then washed sequentially with buffer A, buffer B (50 mM Tris-HCl [pH 7.5], 750 mM KCl, 1 mM EDTA, 10% glycerol, and 1% Triton X-100), and buffer C (20 mM Tris-HCl [pH 8.0], 100 mM KCl, 10 mM β-mercaptoethanol, 0.2 mM EDTA, 20% glycerol, and 0.1% Triton X-100). Captured complexes were released by incubation for 20 min at room temperature in elution buffer (10% sodium dodecyl sulfate [SDS], 100 mM NaHCO3) and recovery of supernatants after centrifugation at room temperature for 10 min at 20,800 × g. The Pansorbin pellet was washed an additional time, and the supernatant from the first wash was combined with that from the second. Cross-links were reversed by overnight incubation at 65°C, and the released DNA was then purified using a QIAquick PCR purification column. The IP DNA sample and an input DNA sample (recovered after similar treatment but without immunoprecipitation) were amplified by linker-mediated PCR as described in the protocol from NimbleGen. The amplified DNA was then labeled with Cy3 (input DNA) or Cy5 (IP DNA) dye, hybridized to a NimbleGen high-density 2.1M promoter tiling array using a Maui hybridization apparatus (BioMicro Systems), and quantified using a Axon 4000B scanner (Molecular Devices) by following the protocol from NimbleGen. All of the microarray data have been deposited in the Gene Expression Omnibus (GEO) database.
ChIP-chip data analysis.
Eight ChIP data sets were generated as log2(IP/input) fluorescence intensities using anti-Flag IP of wild-type RPB2-Flag cells in biological triplicate, anti-Flag IP of ΔFL RPB2-Flag cells in biological triplicate, and IP with antibody 8WG16 against the RPB1 C-terminal domains (CTD) from both wild-type and ΔFL cells in biological duplicate. The four 8WG16 IP data sets were combined to measure the distribution of the RPB1 signal. Data were analyzed using the statistical program R (Bioconductor) and publicly available packages (40). For the comparison of RNAPII distributions in wild-type and ΔFL RPB2 cells, replicate data sets were quantile normalized using the “normalizeQuantiles” function in the affyPLM package (2), averaged at each probe position to generate single values for each target, and then quantile normalized against each other to allow comparison (see Fig. 3). The R program CMARRT (27) was used to identify regions of significant difference between the data sets, with the false discovery rate set to 0.05 (see Fig. 6). For each region of significant difference identified by CMARRT (CMARRT region) with at least one transcription start site (TSS), the absolute distance to the closest TSS either upstream or downstream was determined. If a CMARRT region did not contain at least one TSS, it was omitted from the analysis.
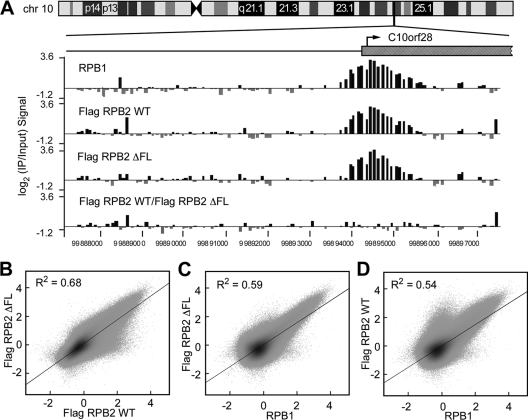
Fig. 3.
Wild-type and ΔFL ettRNAPII are transcriptionally active in vivo. (A) Map of chromosome 10 (chr 10) showing the location of the randomly selected C10orf28 gene. Log2 ratios of IP to input signals from RPB1, wild-type ettRPB2, and ΔFL ettRPB2 samples, and the ratio of wild-type ettRPB2 to ΔFL ettRPB2 at the orf28 gene, are shown. (B to D) Scatter plots of RPB1, Flag RPB2 WT, and Flag RPB2 ΔFL signals. The signal for each probe is plotted against those of the other probes.
Fig. 6.
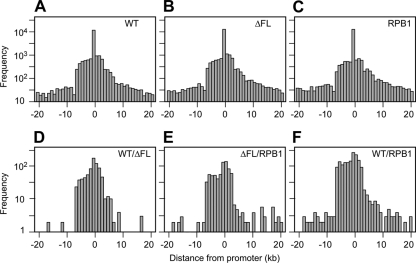
Distribution of ChIP signals or CMARRT regions relative to TSSs. The regions tiled in the array were classified into subsets, eliminating regions of <10 kb and >500 kb. The distance of the nearest TSS was calculated for each of the IPs and was plotted as a function of distance upstream or downstream from the nearest promoter. If a promoter was present within a CMARRT region, then the distance from the promoter to the region was assigned as zero. The total number of signals used for each condition to generate each plot is presented in Table S2 in the supplemental material. The nearest promoter was identified as described in Materials and Methods. (A) WT; (B) ΔFL; (C) RPB1; (D) WT/ΔFL ratio; (E) ΔFL/RPB1 ratio; (F) WT/RPB1 ratio.
RNAPII occupancy relative to promoters.
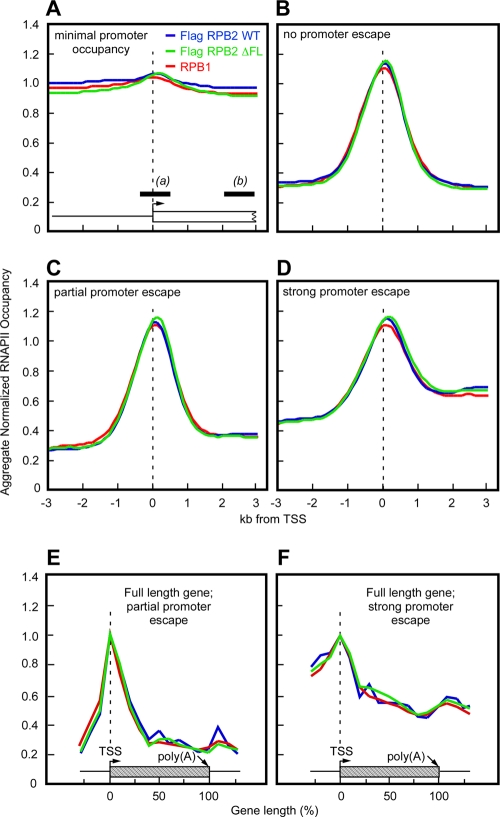
Relative to each TSS, the probe values at two regions, a TSS/promoter region (from bp −500 to +500) and a gene region (from bp +2250 to +3250), were calculated based on averages. Any TSS that lacked probes in promoter or gene regions and any TSS that lacked a RefLink name (http://genome.csdb.cn/cgi-bin/hgTables) were omitted from the analysis. The traveling ratio (TR) was then calculated as the ratio of the signal within the gene to the signal at the promoter. TSSs were then binned as follows. Bin 1 included TSSs with minimal promoter occupancy, where the promoter signal for wild-type IP was <2 (see Fig. 4A), and strong promoter occupancy, where the promoter signal for wild-type IP was ≥2. The cases with strong promoter occupancy were then further divided as follows: (i) no promoter escape, where the signal within the gene for wild-type IP was <1.5 (bin 2) (see Fig. 4B), (ii) partial promoter escape, where the signal within the gene for wild-type IP was ≥1.5 and the traveling ratio was <0.49 (bin 3) (see Fig. 4C), and (iii) strong promoter escape, where the signal within the gene for wild-type IP was ≥1.5 and the traveling ratio was >0.49 and <1 (bin 4) (see Fig. 4D). The signals for each gene were normalized to the signal at the TSS. We then calculated a sliding average for each gene using a 500-bp window every 125 bp from −3 kb to +3 kb relative to the TSS, and we averaged the signals at each position to generate a normalized, aggregate RNAPII occupancy profile for each bin (see Fig. 4).
Fig. 4.
Wild-type and ΔFL RNAPII behave similarly at all promoters in vivo. Promoters were classified into subsets based on the wild-type RNAPII ChIP signals in a 1-kb promoter region centered at the transcription start site (TSS) (promoter signal, designated a in panel A) and in a 1-kb intragenic region between 2 and 3 kb downstream of the promoter (signal within gene, designated b in panel A). The TR was calculated as b/a separately for each TSS on the array. The IP/input signals for each class were averaged and plotted as a function of distance from the TSS. The hatched boxes in panels E and F represent a theoretical promoter and gene extending to 3 kb. Each panel contains averaged ChIP signals for wild-type ettRNAPII (blue) (anti-Flag MAb), ΔFL ettRNAPII (green) (anti-Flag MAb), and all RNAPII (red) (anti-RPB1 CTD MAb). (A) Minimal or no promoter occupancy by RNAPII (promoter signal for wild-type ettRNAPII, <2). (B) Little or no promoter escape (signal within gene for wild-type ettRNAPII, <1.5). (C) Partial promoter escape (for wild-type ettRNAPII, the signal within the gene was ≥1.5 and the TR was <0.49). (D) Strong promoter escape (for wild-type ettRNAPII, the signal within the gene was ≥1.5 and the TR was ≥0.49 and <1). (E and F) Full ChIP signal profiles for genes in ENCODE regions that exhibited partial promoter escape (for wild-type ettRNAPII, the signal within the gene was ≥1.5 and the TR was <0.49) (E) and strong promoter escape (for wild-type ettRNAPII, the signal within the gene was ≥1.5 and the TR was ≥0.49 and <1) (F). See Materials and Methods for a description of the averaging method.
We further analyzed the subsets of bins 3 and 4 that were present in ENCODE regions by dividing each primary transcript into 10 intervals corresponding to adjacent 10% fractions of the transcript length. Average values within each interval were determined, and the average values for each interval of all ENCODE transcripts in the bin were calculated and used to create aggregate profiles (see Fig. 4E and F).
Tailed-template elongation assay.
The tailed-template transcript elongation assay was performed as described previously (37).
EC reconstitution and transcript elongation.
Elongating complexes (ECs) (2.5 nM) were reconstituted on a nucleic acid scaffold containing template DNA (2.5 nM), 5′-32P-labeled RNA oligonucleotides (2.5 nM) (see Fig. 2, 8, and 9; see also Table S1 in the supplemental material), and RNAPII (3 nM), followed by the addition of nontemplate DNA oligonucleotide (3 nM) in elongation buffer as described previously (29). Reconstituted ECs were incubated with ATP and GTP (2.5 μM each) at 30°C for 10 min in order to form halted A24 ECs.
Fig. 2.
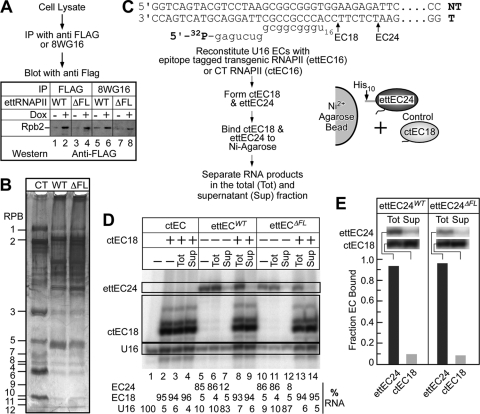
Conditional expression, partial purification, and in vitro transcriptional activities of wild-type and ΔFL ettRNAPII. (A) Conditional expression of wild-type and ΔFL ettRNAPII in HEK 293 cells. Cell lysates from cultures induced with doxycycline (2 μg/ml) were immunoprecipitated with either anti-Flag antibody M2 (lanes 1 to 4) or RPB1-specific 8WG16 antibodies (lanes 5 to 8). The immunoprecipitated proteins were fractionated on an SDS–4 to 12% polyacrylamide gel and were detected by immunoblotting. Anti-Flag antibodies were used to detect the expression of the ettRPB2 subunit and its assembly into RNAPII. (B) Partial purification of wild-type and ΔFL ettRNAPII. Flag-tagged wild-type and ΔFL RNAPII were affinity purified on anti-Flag M2 agarose, fractionated on an SDS–4 to 16% polyacrylamide gel, and stained with silver. Purified calf thymus (CT) RNAPII served as a marker. The positions of the 12 subunits of RNAPII are indicated. (C) Sequences of the DNA and RNA oligonucleotides used to reconstitute ECs. The positions of the halted EC18 and EC24 are indicated. Transcription elongation complex reconstitution, EC18 and EC24 formation, and immobilization of ECs on Ni2+ agarose beads are shown schematically. (D) Purified wild-type and ΔFL ettRNAPII are transcriptionally active and are not contaminated with endogenous wild-type RNAPII. Reconstituted wild-type EC16 and ΔFL ettRNAPII EC16 were elongated in the presence of ATP and GTP to make G24 ECs (ettEC24). Control ctEC18 was made using purified CT RNAPII and was mixed with either wild-type or ΔFL ettRNAPII. The complexes were then immobilized on Ni2+ agarose beads and were separated by centrifugation, and the RNA in the total (Tot) and supernatant (Sup) fractions was separated on a 20% denaturing polyacrylamide gel. (E) The fractions of ettEC24 and ctEC18 bound to Ni2+ agarose beads were determined, establishing that >90% of the transcriptional activity in the ettRNAPII preparations was derived from the tagged enzymes and not from native, untagged RNAPII, which could potentially contaminate the preparations.
Fig. 8.
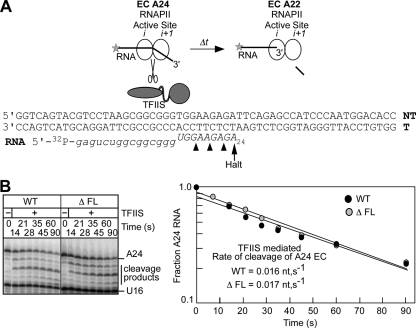
The flap loop does not inhibit backtracking. (A) Reaction scheme for TFIIS-stimulated transcript cleavage. The sequence of the nucleic acid scaffold used to reconstitute ECs is shown with the position of the halted A24 EC indicated. (B) A24 complexes made in the presence of ATP and GTP were incubated with apyrase to convert NTPs to NDPs. The ECs were then incubated with TFIIS; aliquots were removed at the times indicated; and the RNA products were separated on a 20% denaturing polyacrylamide gel. The fraction of remaining A24 RNA is plotted as a function of reaction time.
Fig. 9.
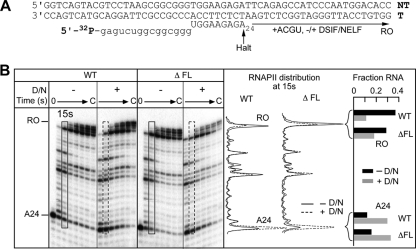
The flap loop does not contribute to DSIF/NELF-mediated inhibition of transcript elongation. (A) Sequence of the nucleic acid scaffold used to reconstitute ECs. The positions of the halted A24 EC and the runoff transcript (RO) are indicated. (B) (Left) A24 ECs made with wild-type or ΔFL ettRNAPII were elongated in the absence or presence of DSIF/NELF (D/N) and in the presence of all 4 NTPs (10 μM each). Aliquots were removed at predetermined time intervals, and RNA was separated on a 15% denaturing polyacrylamide gel. (Center) Densitometric plot of the distribution of RNA species in the lanes representing the 15-s time point in the absence (solid lines) or presence (dashed lines) of DSIF/NELF. (Right) Bar graph showing the fractions of the 24-mer and runoff RNAs for WT or ΔFL ettRNAPII in the absence (solid bars) or presence (shaded bars) of DSIF/NELF.
To exclude the possibility that contaminating endogenous wild-type RNAPII would copurify with the deletion enzyme and be responsible for transcription in vitro, we first reconstituted ettRNAPII and formed ettEC24 with 5′-32P-labeled RNA. The labeled wild-type and mutant ettECs were then immobilized on Ni2+-nitrilotriacetic acid (NTA) agarose and were separated into bound and supernatant fractions, and the RNAs in the total and supernatant fractions were resolved on a 20% denaturing polyacrylamide gel. To control for adventitious binding of endogenous RNAPII to the Ni2+-NTA agarose or to ettRNAPII, ECs containing calf thymus (CT) RNAPII (18-mer RNA) and ECs containing ettRNAPII (24-mer RNA) were mixed, incubated with Ni2+-NTA agarose beads, and separated into the pellet and supernatant fractions. The 32P-labeled RNAs in the total and supernatant fractions were then separated on a 20% denaturing polyacrylamide gel and were quantified by phosphorimaging.
DSIF/NELF-mediated inhibition of transcript elongation.
The halted A24 complexes were incubated in the absence or presence of DSIF/NELF (9 and 6 nM, respectively) at 30°C for 5 min before the addition of all four NTPs (250 μM each). Aliquots of the reaction mixture were removed at the times indicated (see Fig. 9), and reactions were terminated with an equal volume of 2× stop mixture (29). The RNA products were separated on a 17.5% polyacrylamide–8 M urea gel in 0.5× Tris-borate-EDTA buffer and were exposed to a phosphorimager screen.
Transcript cleavage reaction.
A24 ECs were prepared as described above and were incubated with apyrase (50 mU/20-μl reaction mixture) at 30°C for 5 min in order to convert NTPs into nucleoside diphosphates (NDPs). TFIIS was then added to the reaction mixture to a final concentration of 1 nM; aliquots were removed; the reactions were terminated by the addition of 2× stop buffer at the times indicated; and the RNA products were separated as described above. The remaining A24 RNA was calculated as a fraction of the total RNA in each lane see (Fig. 8), and this value was plotted as a function of reaction time.
Transcription initiation.
Transcription initiation reactions were performed as described previously (30). Following preincubation of RNAPII and the template with general transcription factors (TATA-binding protein [TBP], TFIIB, TFIIE, TFIIF, and TFIIH) for 30 min at room temperature, the reaction was initiated by the addition of a nucleotide mixture (100 μM ATP, 100 μM CTP, 5 μCi of 0.08 μM [32P]UTP, 3 mM EGTA, 12.5 mM MgCl2, 1 U/μl RNase inhibitor) and was allowed to proceed for various time intervals. Reactions were terminated by the addition of stop solution (200 μl; 0.1 M sodium acetate, 0.5% SDS, 2 mM EDTA, 100 μg/ml tRNA). The RNA was extracted with phenol-chloroform and was precipitated with ethanol, and transcripts were analyzed on an 18% polyacrylamide–8 M urea denaturing gel in 0.5× Tris-borate-EDTA buffer.
All in vitro transcript initiation, elongation, and transcript cleavage experiments were performed at least twice. The results presented are averages from two independent experiments, and the individual values were within 10 to 15% of each other.
Microarray data accession numbers.
All microarray data have been deposited in the GEO public database (http://www.ncbi.nlm.nih.gov/projects/geo/) with the following accession numbers: GSE29734 (RPB2 flap loop), GSM737207 (Flag_Rpb2), GSM737208 (Flag_Δ_flap_Rpb2), and GSM737209 (8wg16_Rpb2).
RESULTS
Expression and assembly into human RNAPII in vivo of RPB2 lacking the flap loop.
To test the role of the flap loop in transcription, we deleted RPB2 amino acid residues 873 to 884 in human RNAPII (ΔFL) (Fig. 1A and B). The deletion removed the entire interaction interface of the flap loop with TFIIB. In our engineered constructs, the RPB2 subunit carried at its N terminus a His10 tag, a precision protease (PPX) recognition site, three copies of a Flag epitope tag, and three copies of a hemagglutinin (HA) epitope tag (Fig. 1B; see also Fig. S1 in the supplemental material). The expression of the epitope-tagged RPB2 relied on a Tet-regulated cytomegalovirus (CMV) promoter and was inducible by the addition of doxycycline. The expression plasmids carrying either the wild-type or the mutant RPB2 subunit were stably transfected into HEK 293 Tet-ON cells for conditional expression (see Materials and Methods). We first tested to verify that the epitope tag remained linked to the flap loop deletion in the inserted DNA and in mRNA. We then used the tags to track the ΔFL RPB in ettRNAPII.
To verify that the epitope tag DNA was linked only to transgenic RPB2 in the transformed cell lines, we PCR amplified a 3-kb region that included the epitope tags and RPB2 residues 873 to 884 from genomic DNA isolated from the transformed cell lines (Fig. 1C, primers 1 and 4 in the schematic and in lanes 2 to 4 of the gel). The 3-kb PCR product was gel purified, and the epitope tags and flap deletion were detected by PCR with specific primers (primers 1 and 2 for the tags [Fig. 1C, lanes 5 to 7, 12, and 13] and primers 3 and 4 for the flap loop [Fig. 1C, lanes 8 to 11]). As a further test, we PCR amplified a 1.8-kb fragment from the ΔFL genomic DNA using primers that targeted both endogenous (untagged, wild-type) RPB2 and transgenic RPB2 (primers 5 and 4 [Fig. 1C, schematic]). Restriction digestion of the 1.8-kb fragment with PmlI yielded a 339-bp fragment from endogenous RPB2 and a 303-bp fragment from transgenic ΔFL RPB2 (Fig. 1C, compare lanes 14 and 15). However, no 339-bp fragment was detected after digestion of the 1.8-kb fragment when it was amplified from the purified, tag-specific 3-kb DNA fragment (Fig. 1C, primers 5 and 4 in the schematic; also compare lanes 14 and 18). This finding confirmed that the tags were genetically linked only to ΔFL RPB2 in the transformed cell line.
To detect ΔFL RPB2 expression, we performed reverse transcription-PCR (RT-PCR) on RNA isolated from wild-type or flap deletion cell lines after induction with doxycycline (Fig. 1D, primer pair 3/4 in the schematic). The cDNA was initially amplified using tag-specific primers to generate the 3-kb fragment that spans the tags and the flap loop deletion (Fig. 1D, primers 1 and 4 in the schematic and lanes 1 to 4 in the gel). The 3-kb fragment was then purified and analyzed as described above for genomic DNA (Fig. 1D). This yielded fragments from the epitope tag (332 bp) or the flap region (269 and 233 bp for the wild type and deletant, respectively) (Fig. 1D, lanes 5 to 12). Detection of the 233-bp fragment confirmed the linkage of the epitope tags to ΔFL RPB2 in the mRNA. Importantly, we detected only the 233-bp fragment specific for ΔFL RPB2 when using the purified, tag-specific 3-kb fragment as a template, but we detected both the 269- and 233-bp fragments when using cDNA as a template (Fig. 1D, lanes 13 and 14). We conclude that the mRNA encoding the epitope tag also encodes the flap loop deletion.
To detect incorporation of the mutant RPB2 into RNAPII, we immunoprecipitated the Flag-tagged RPB2 from crude cell lysates and then analyzed the immunoprecipitate by Western blotting using the same antibodies. The recombinant wild-type and ΔFL RPB2 subunits were conditionally expressed by doxycycline treatment (Fig. 2A, lanes 1 to 4). To determine if the mutant RPB2 subunit was assembled into RNAPII, we first immunoprecipitated crude cell lysates with 8WG16 antibodies that recognize the RPB1 CTD (45) and then used Western blotting of the immunoprecipitate with anti-Flag M2 antibodies to detect the Flag-tagged RPB2 subunit. The transgenic Flag-tagged wild-type RPB2 subunit and the ΔFL RPB2 subunit coimmunoprecipitated with RPB1, verifying that ΔFL RPB2 was assembled into RNAPII in vivo (Fig. 2A, lanes 5 to 8).
RNAPII containing the Flag-tagged wild-type or ΔFL RPB2 subunit was then partially purified using an anti-Flag M2 agarose affinity matrix and was fractionated on a 4 to 16% SDS-polyacrylamide gel (PAG) to determine the presence of the other 11 subunits of RNAPII. Silver staining of the gel indicated that the partially purified enzyme has all 12 subunits of RNAPII (Fig. 2B), indicative of complete assembly of the recombinant Flag-tagged RPB2 subunit into RNAPII.
Purified wild-type and ΔFL ettRNAPII enzymes are active in vitro.
To test the activities of the ettRNAPII enzymes, we first reconstituted ECs (29) on nucleic acid scaffolds containing 5′-32P-labeled 16-mer RNA and then elongated the RNA to position A24 in the presence of ATP and GTP (Fig. 2C and D). More than 80% of the RNA elongated to position A24 (Fig. 2D, lanes 5, 6, 10, and 11) establishing that the ettRNAPII was transcriptionally active in vitro. To verify that the radiolabeled RNA in the A24 ECs was indeed associated with ettRNAPII, the transgenic enzyme was immobilized on Ni-NTA agarose beads via its His10 tag, and the reaction product was separated into supernatant and pellet. If the elongation products were associated with ettRNAPII, then the labeled RNA would be in the pellet. If endogenous RNAPII copurified with the transgenic enzyme, it would yield labeled A24 RNA in the supernatant. Less than 10% of the labeled RNA was observed in the supernatant (Fig. 2D, compare lanes 6 and 7 and lanes 11 and 12). As a control for adventitious binding of RNAPII to Ni-NTA or to ettRNAPII, we synthesized G18 ECs using calf thymus RNAPII, mixed them with the A24 ettECs, and incubated the mixture with Ni2+-NTA (Fig. 2C and D). Virtually all of the G18 ECs remained in the supernatant fraction (Fig. 2D, compare lanes 8 and 9 and lanes 13 and 14), establishing that binding to the agarose beads was specific for ettRNAPII and that RNA synthesis detected as labeled RNA24 was attributable to ettRNAPII. We conclude that the ettRPB2 subunit was expressed in transformed cells and was assembled into active RNAPII that can be purified from lysates using the epitope tags.
ΔFL RNAPII initiated and transcribed normally in vivo.
We next used ChIP-chip to test the abilities of the recombinant wild-type and ΔFL RNAPII to transcribe genes in vivo. IP was performed using either anti-Flag antibodies recognizing Flag-tagged RPB2 or 8WG16 antibodies recognizing the C-terminal repeats of the RPB1 subunit of RNAPII. The immunoprecipitated DNA was labeled with Cy5, mixed with Cy3-labeled input DNA, and hybridized to a high-density 2.1M promoter array (NimbleGen) spanning the regions from kb −7 to 3 of ∼60,000 annotated human promoters in addition to all of the ENCODE regions (16). Both wild-type and ΔFL ettRNAPII bound, initiated, and transcribed similarly to nontagged wild-type RNAPII, as evidenced by the detection of ΔFL RNAPII at known transcription start sites (TSSs) and within the genes (e.g., C10orf28 [Fig. 3A]). To assess whether global occupancy was altered by the flap loop deletion in vivo, we plotted log2(IP/input) signals for all probes from wild-type and ΔFL RPB2 subunits against each other and also against the signal from RPB1 immunoprecipitated using 8WG16 antibodies (Fig. 3B to D). The signals from wild-type or ΔFL ettRNAPII at promoter regions were indistinguishable from each other and from the signals from control, untagged RNAPII. We conclude that wild-type and ΔFL RNAPII behave similarly at most, if not all, promoters represented on the array.
To examine ΔFL RNAPII further, we performed two additional analyses. First, we categorized the promoter regions into four classes based on the ratio of the signal within the transcribed region to the signal at the promoter (called the traveling ratio) (Fig. 4). We then calculated average occupancy profiles for each class and each RNAPII (ΔFL or wild type RPB2 and RPB1) (Fig. 4). No significant difference in occupancy for ΔFL RPB2 was evident in any of the four classes. Thus, promoter escape by ΔFL RNAPII was indistinguishable from promoter escape by wild-type RNAPII. Additionally, no differences were observed in the occupancy of wild-type and ΔFL ettRNAPII across the full length of the subset of genes present in the ENCODE regions for which significant RNAPII signals were detectable (Fig. 4E and F).
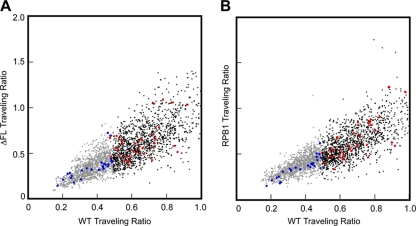
To ensure that we did not overlook significant differences in small numbers of genes, we also calculated scatter plots of the traveling ratios for ΔFL versus wild-type signals and for wild-type versus RPB1 signals (Fig. 5). No differences were evident either for all promoters on the array or for the genes in the ENCODE regions, arguing strongly that ΔFL RNAPII and wild-type RNAPII exhibit indistinguishable occupancy profiles. Finally, to detect any differences that might appear by use of an unbiased approach, we searched for regions in which any difference in the signal was statistically significant using the CMARRT algorithm (27) (see Materials and Methods) (Fig. 6A to C). Although we identified 850 regions where the wild-type signal was significantly greater than the ΔFL signal (Fig. 6D), these regions were not evident in a comparison of the ΔFL signal to the RPB1 signal (see Table S2 in the supplemental material). Further, these 850 regions showed the same distribution relative to TSSs as 966 regions in which the ΔFL signal was greater than the RPB1 signal (Fig. 6E) and 1,988 regions in which the wild-type ettRPB2 signal was greater than the RPB1 signal (Fig. 6F). The fact that we detected more regions of difference between wild-type ettRPB2 and RPB1 than in the wild-type–ΔFL and ΔFL-RPB1 comparisons and that we detected few or no significant regions in the RPB1–ΔFL and ΔFL–wild-type comparisons (see Table S2 in the supplemental material) suggests that the differences detected between the ΔFL and wild-type signals simply reflect noise in the data, to be expected with the 0.05 false discovery rate used for CMARRT analysis. Thus, we conclude that the flap loop has no significant effects on promoter occupancy, initiation, or elongation within genes.
Fig. 5.
Comparison of traveling ratios for wild-type and ΔFL RNAPII on genes with partial or strong promoter escape. (A) ΔFL ettRNAPII versus wild-type ettRNAPII. Data are color coded gray for partial promoter escape (TR, <0.49), black for strong promoter escape (TR, >0.49), and blue (TR, <0.49) or red (TR, >0.49) for genes in ENCODE regions. (B) RPB1 signals versus wild-type ettRNAPII signals. The color scheme is the same as that in panel A.
A modest effect of the flap loop deletion on transcript elongation in vitro is suppressed by TFIIF.
Although no effect of the flap loop on transcript elongation in vivo was observed, it is possible that an intrinsic defect could be masked by transcription regulators. To test whether the purified ΔFL RNAPII exhibited any elongation defects, we used a tailed template that codes for the well-characterized HIV-1 pause and that allowed us to form initially halted ECs with a 34-nucleotide (nt) RNA (37). The halted ECs were washed in transcription buffer and were elongated in the presence of all 4 NTPs at 1 mM each (Fig. 7A). Deletion of the flap loop had only a modest effect on elongation by RNAPII. Although a new pause site appeared ∼2 nt upstream of the HIV-1 pause signal, there was no significant difference in pausing at the HIV-1 pause site (Fig. 7A). Quantitative analysis revealed that the rate of accumulation of the RNA beyond the HIV-1 pause band was delayed by a factor of ∼1.5 in ΔFL RNAPII relative to the wild-type enzyme (Fig. 7B). Simple explanations for this modest effect of deletion of the flap loop could be increased backtracking or increased resistance to translocation. Ordinarily, the emerging RNA passes near the flap loop (Fig. 1A). The deletion could remove contacts that assist translocation or prevent backtracking. Conversely, an altered flap loop in the deletant could interfere with translocation or increase backtracking.
This effect of the flap loop on elongation was largely suppressed by the addition of TFIIF to the assay mixture (Fig. 7C). This result establishes that the flap loop is not required for enhancement of the elongation rate by TFIIF. It also suggests that the flap loop affects elongation by affecting a step that is targeted by TFIIF. Like bacterial NusG (21), TFIIF appears to favor forward translocation (55). Thus, TFIIF may suppress effects of the flap loop deletion either on pause-free elongation or on backtracking by favoring forward translocation.
The flap loop has little or no effect on backtracking.
If the slightly slower elongation by ΔFL RNAPII is a consequence of increased backtracking, then the ΔFL enzyme should be more sensitive than the wild type to TFIIS-stimulated transcript cleavage. We tested this possibility by comparing the sensitivities of wild-type and ΔFL enzymes to TFIIS-stimulated transcript hydrolysis in an isolated EC with a 24-nt RNA (Fig. 8A). We first reconstituted ECs using nucleic acid scaffolds containing a 5′-32P-labeled 16-mer RNA. The ECs were then elongated to A24 in the presence of ATP and GTP. Before the incubation of A24 ECs with TFIIS, we added apyrase to convert NTPs to NDPs and to prevent the reextension of cleaved complexes by reaction with NTPs (32). The A24 ECs were then incubated with TFIIS. The reaction products were fractionated on a denaturing polyacrylamide gel and were quantitated, and the fraction of remaining 24-mer RNA was plotted as a function of time (Fig. 8B). We observed no significant difference between the rate of TFIIS-stimulated transcript cleavage by the ΔFL enzyme and that of wild-type RNAPII, suggesting that the flap loop does not affect backtracking. Thus, the modest effect on transcript elongation might be caused by inhibition of forward translocation during on-pathway RNA synthesis.
The flap loop is dispensable for DSIF/NELF-mediated inhibition of transcript elongation by RNAPII.
DSIF/NELF is thought to interact with the exiting RNA and to require at least 18 nt of RNA in the EC to promote pausing by RNAPII (38, 51). Since the flap loop is located at the mouth of the RNA exit channel, we reasoned that the flap loop might play a role in the interaction of DSIF/NELF either with the RNA or with RNAPII and might influence the ability of DSIF/NELF to promote pausing by RNAPII. To test this hypothesis, we used the reconstituted ECs with the labeled 24-mer RNA and elongated them in the absence or presence of DSIF/NELF. The reactions were performed at subsaturating concentrations of DSIF/NELF (9 and 6 nM, respectively) to ensure that an effect of the flap loop on DSIF/NELF binding would be detected. DSIF/NELF was able to enhance pausing by the WT or ΔFL RNAPII equivalently (Fig. 9 A and B), showing that the flap loop is dispensable for DSIF/NELF function.
ΔFL RNAPII is not defective in promoter binding, abortive initiation, or promoter escape from the adenovirus major late promoter in vitro.
To confirm the ability of ΔFL RNAPII to initiate normally in vivo (Fig. 4), we tested initiation in vitro at the adenovirus major late promoter. The promoter DNA was first preincubated with RNAPII and general transcription factors (TBP, TFIIB, TFIIF, TFIIE, and TFIIH [reviewed in references 39, 41, and 43]); then NTPs were added to initiate transcription; and finally samples were removed to a quench solution at predetermined times (Fig. 10 A). This yielded abortive (4-mer) and productive (9- to 23-mer) transcripts, which were quantitated as the fraction of each species generated as a function of time. After preincubation of the enzyme with promoter DNA for 30 min, we observed little if any difference in the ability of the ΔFL enzyme to bind the promoter and initiate transcription (Fig. 10B). The wild-type and ΔFL ettRNAPII synthesized similar amounts of abortive products (4-mer), indicating that the rate of synthesis of the abortive products was not altered by the deletion (Fig. 10B). Similarly, we observed no significant difference in the amount of productive transcripts (23-mer).
To verify that the flap loop deletion did not affect the rate of preinitiation complex formation, we also performed the assay without preincubation of the enzyme with DNA. Again, we observed no significant difference in the rates of accumulation of abortive or productive transcripts synthesized by the wild-type and ΔFL enzymes (compare Fig. 10B and C). Thus, deletion of the flap loop causes no defect in either the rate of preinitiation complex assembly or the rate of initiation in vitro. To test if the ΔFL ettRNAPII was defective in promoter escape, we compared the ratio of abortive to productive transcripts. We observed no difference in the ratio between the wild-type and the ΔFL enzyme with or without a 30-min preincubation time (Fig. 10D and E). Taken together, these results indicate that the flap loop of human RNAPII plays no significant role in promoter binding, abortive initiation, or promoter escape in vitro.
DISCUSSION
We have conducted a comprehensive evaluation of the contribution of the flap loop of human RNAPII to enzyme function, including both genome-scale in vivo tests and detailed in vitro mechanistic tests. Our overall conclusion, that the flap loop plays no significant role in gene transcription by human RNAPII, is surprising given the multiple important roles played by the orthologous part of bacterial RNAP. Nonetheless, our findings have several important implications for the study of RNAPII structure and function.
The flap loop of human RNAPII appears dispensable for TFIIB function.
Although the TFIIB linker, core, and ribbon domains make contacts with RNAPII in locations similar to those of the contacts made by σ70 regions 2, 3, and 4 (26, 31), our results demonstrate conclusively that the contact between the flap loop and the TFIIB ribbon is not important for transcription. This is in stark contrast to the essentiality of the flap tip-σ70 region 4 contact for initiation at most promoters (17, 28, 46). Although the contrasting importance of these seemingly orthologous contacts between the bacterial and eukaryotic systems is surprising at first, consideration of the TFIIB-RNAPII cocrystal structures offers explanations for this difference.
First, the contact itself does not appear to be especially strong. A significant segment of the TFIIB ribbon indeed lies near the flap loop in the cocrystal structure (TFIIB residues 13 to 32) (26). However, few specific contacts are made. The carbonyl O of TFIIB Pro32 appears to H-bond to Nε of Gln 927 on the flap loop, and a salt bridge is formed between TFIIB Arg13 and Glu923 on the flap loop, but otherwise most of this segment of TFIIB either contacts other parts of RNAPII (e.g., the clamp) or makes no contacts with RNAPII. Much more significant contacts of the TFIIB ribbon domain are made with the RNAPII dock domain (26). C-terminal of the TFIIB ribbon, the TFIIB reader domain lies within the RNA exit channel, much as σ70 region 3.2 does in bacterial RNAP (Fig. 1A). The reader is held in place by the lid domain and does not appear to depend on the ribbon domain to maintain its positioning. Thus, loss of the limited contacts between the flap loop and the TFIIB ribbon would not appear to alter the binding of TFIIB to the RNAPII surface significantly. In contrast, loss of the flap tip-σ70 region 4 contact would untether region 4 from the bacterial RNAP surface.
Second, the potential involvement of the flap loop in upstream promoter contacts differs dramatically between eukaryotic RNAPII and bacterial RNAP. In bacterial RNAP, the flap tip anchors σ region 4 to RNAP, and σ region 4 in turn makes the key upstream promoter contact with the −35 promoter DNA element (34). In eukaryotic RNAPII, upstream promoter contacts are made by the TFIIB core cyclin domains (26, 31) and by TBP. The N-terminal core cyclin domain binds the RNAPII protrusion, similarly to the σ region 3 contact with bacterial RNAP (34, 48). The C-terminal core cyclin domain contacts DNA upstream of the TBP contact with DNA, but neither of these DNA contacts depends on the TFIIB ribbon-RNAPII flap loop contact. Thus, whereas the bacterial flap tip-σ contact is crucial for positioning a σ-promoter DNA contact, the eukaryotic flap loop-TFIIB contact is not involved in promoter contacts.
Less comparison of the involvement of the bacterial flap tip and eukaryotic flap loop in contacts with elongation factors is possible, because much less in known about elongation factor contacts with RNAP in general. Although eukaryotic factors, such as TFIIF and DSIF/NELF, could contact nascent RNA in the vicinity of the flap loop, there is no close eukaryotic ortholog of the bacterial elongation factor known to depend on a flap tip contact, NusA. Thus, the apparent absence of a role of the flap loop in the function of eukaryotic elongation regulators poses no obvious contradiction to expectations of conservation between bacteria and eukaryotes.
Compromised elongation might alter cotranscriptional events.
Deletion of the flap loop resulted in a decrease in the elongation rate by a factor of ∼1.5, but with no significant effects on factor-induced regulation of transcript elongation in vitro. However, a modest decrease in the elongation rate might be sufficient to compromise the association with elongating RNAPII of factors involved in other cotranscriptional events. For instance, a single amino acid substitution (R749H) in the rim helix of the large subunit of RNAPII slows elongation by a factor of ∼2 in vitro (6) and significantly impairs cotranscriptional splicing of RNA in vivo (11). If transcript elongation is not universally suppressed by elongation factors, a reduced elongation rate in vivo caused by deletion of the flap loop might also contribute to impaired splicing events or other cotranscriptional events, which would not necessarily have been detected in our assays.
The flap tip has undergone evolutionary specialization.
Evolutionary divergence of RNAP is inversely correlated with distance from the catalytic center and is greatest on the enzyme surface (10, 12). In this sense, it is not surprising that the flap tip should differ significantly between bacterial and human enzymes. However, the essential functions of the bacterial flap tip, positioning a σ factor domain for promoter recognition and a NusA domain to modulate nascent RNA effects, are each involved in core enzymatic activities shared by all RNAP, namely, initiation, elongation, and termination of transcription. If anything, mediation of these activities involves a larger number of accessory proteins for eukaryotic RNAPII than for bacterial RNAP. Thus, one might expect that a binding site as conveniently positioned as the flap tip would be even more heavily used in eukaryotes. Instead, it appears either to have acquired no necessary interactions or to have lost whatever function might have been present in the last common ancestor during evolutionary divergence.
One possible explanation of this puzzle lies in the much greater complexity of eukaryotic regulation. Broadly speaking, regulation of eukaryotic RNAP, and especially of metazoan RNAPII, relies on assemblies of large complexes of regulatory proteins that may contact the enzyme at multiple locations. This is most evident in the large number of regulators that traffic on and off the RPB1 C-terminal repeat domain (3, 13) but is reinforced by the significant number of regulators that do not appear to contact the CTD (e.g., TFIIB, TFIIF, TFIIS, and hepatitis virus δ antigen). Thus, eukaryotic regulation, which appears to have accumulated complexity via a multiplicity of components and contacts, may not have evolved a use for the flap tip. In contrast, the regulation of bacterial RNAP involves fewer regulators and a relatively precise set of contacts (e.g., promoter contacts on successive turns of the DNA duplex near the enzyme). The need for economy of contacts during the evolution of transcriptional regulation in bacteria may have led to functional specialization of the flap tip to contact both initiation and elongation factors. Meanwhile, the expansion of contacts among multiple regulators and further from the enzyme surface during the evolution of transcriptional regulation in eukaryotes may have allowed the flap tip to be bypassed, even though it would have been a suitable option.
Human ettRNAPII can be assayed in vivo and in vitro.
Most structure/function studies of RNAP employing mutant enzymes have been conducted using bacterial or yeast RNAP. However, the regulation of metazoan transcription exhibits several features, such as extensive promoter-proximal pausing and regulation by DSIF/NELF, that lack mechanistic analogs in yeast or bacteria. Thus, an approach that allows the generation of mutant human RNAPII enzymes and the characterization of their activities in vivo and in vitro offers a significant advantage for the dissection of transcriptional regulatory mechanisms. The use of epitope tags on RNAPII subunits of interest to follow mutant RNAPII in vivo and to purify enzymes for in vitro studies offers a powerful approach to understanding the mechanisms of transcriptional regulation in mammalian cells (5, 23, 30; also this study). However, we have found that experiments with many mutant RPB1 or RPB2 genes yield cell lines that rapidly lose the ability to express the proteins of interest. This likely reflects the deleterious effects of low levels of the mutant proteins made even in the absence of induction. Thus, especially for alterations that seriously compromise RNAPII function, improved methods are needed. Methods that incorporate tighter regulation and that precisely control transgene location, either by targeted integration or by maintenance on an episome, are likely to allow easier exploitation of the approach we describe here.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by grants from the NIH (GM72795) to R.L. and from the Canadian Institutes for Health Research to B.C.
Footnotes
Supplemental material for this article may be found at http://mcb.asm.org/.
Published ahead of print on 13 June 2011.
REFERENCES
- 1. Boeger H., et al. 2005. Structural basis of eukaryotic gene transcription. FEBS Lett. 579:899–903 [DOI] [PubMed] [Google Scholar]
- 2. Bolstad B. M., Irizarry R. A., Astrand M., Speed T. P. 2003. A comparison of normalization methods for high density oligonucleotide array data based on bias and variance. Bioinformatics 19:185–193 [DOI] [PubMed] [Google Scholar]
- 3. Buratowski S. 2009. Progression through the RNA polymerase II CTD cycle. Mol. Cell 36:541–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chen Z. A., et al. 2010. Architecture of the RNA polymerase II-TFIIF complex revealed by cross-linking and mass spectrometry. EMBO J. 29:717–726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Coulombe B., Langelier M. F. 2005. Functional dissection of the catalytic mechanism of mammalian RNA polymerase II. Biochem. Cell Biol. 83:497–504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Coulter D. E., Greenleaf A. L. 1985. A mutation in the largest subunit of RNA polymerase II alters RNA chain elongation in vitro. J. Biol. Chem. 260:13190–13198 [PubMed] [Google Scholar]
- 7. Cramer P. 2006. Recent structural studies of RNA polymerases II and III. Biochem. Soc. Trans. 34:1058–1061 [DOI] [PubMed] [Google Scholar]
- 8. Cramer P., et al. 2008. Structure of eukaryotic RNA polymerases. Annu. Rev. Biophys. 37:337–352 [DOI] [PubMed] [Google Scholar]
- 9. Cramer P., et al. 2000. Architecture of RNA polymerase II and implications for the transcription mechanism. Science 288:640–649 [DOI] [PubMed] [Google Scholar]
- 10. Cramer P., Bushnell D. A., Kornberg R. D. 2001. Structural basis of transcription: RNA polymerase II at 2.8 angstrom resolution. Science 292:1863–1876 [DOI] [PubMed] [Google Scholar]
- 11. de la Mata M., et al. 2003. A slow RNA polymerase II affects alternative splicing in vivo. Mol. Cell 12:525–532 [DOI] [PubMed] [Google Scholar]
- 12. Ebright R. H. 2000. RNA polymerase: structural similarities between bacterial RNA polymerase and eukaryotic RNA polymerase II. J. Mol. Biol. 304:687–698 [DOI] [PubMed] [Google Scholar]
- 13. Egloff S., Murphy S. 2008. Cracking the RNA polymerase II CTD code. Trends Genet. 24:280–288 [DOI] [PubMed] [Google Scholar]
- 14. Eichner J., Chen H. T., Warfield L., Hahn S. 2010. Position of the general transcription factor TFIIF within the RNA polymerase II transcription preinitiation complex. EMBO J. 29:706–716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Elmendorf B. J., Shilatifard A., Yan Q., Conaway J. W., Conaway R. C. 2001. Transcription factors TFIIF, ELL, and Elongin negatively regulate SII-induced nascent transcript cleavage by non-arrested RNA polymerase II elongation intermediates. J. Biol. Chem. 276:23109–23114 [DOI] [PubMed] [Google Scholar]
- 16. ENCODE Project Consortium 2004. The ENCODE (ENCyclopedia Of DNA Elements) Project. Science 306:636–640 [DOI] [PubMed] [Google Scholar]
- 17. Geszvain K., Gruber T. M., Mooney R. A., Gross C. A., Landick R. 2004. A hydrophobic patch on the flap-tip helix of E. coli RNA polymerase mediates σ70 region 4 function. J. Mol. Biol. 343:569–587 [DOI] [PubMed] [Google Scholar]
- 18. Gruber T. M., et al. 2001. Binding of the initiation factor σ70 to core RNA polymerase is a multistep process. Mol. Cell 8:21–31 [DOI] [PubMed] [Google Scholar]
- 19. Ha K. S., Toulokhonov I., Vassylyev D. G., Landick R. 2010. The NusA N-terminal domain is necessary and sufficient for enhancement of transcriptional pausing via interaction with the RNA exit channel of RNA polymerase. J. Mol. Biol. 401:708–725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hawley D. K., Wiest D. K., Holtz M. S., Wang D. 1993. Transcriptional pausing, arrest, and readthrough at the adenovirus major late attenuation site. Cell. Mol. Biol. Res. 39:339–348 [PubMed] [Google Scholar]
- 21. Herbert K. M., et al. 2010. E. coli NusG inhibits backtracking and accelerates pause-free transcription by promoting forward translocation of RNA polymerase. J. Mol. Biol. 399:17–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Izban M. G., Luse D. S. 1992. Factor-stimulated RNA polymerase II transcribes at physiological elongation rates on naked DNA but very poorly on chromatin templates. J. Biol. Chem. 267:13647–13655 [PubMed] [Google Scholar]
- 23. Jeronimo C., et al. 2004. RPAP1, a novel human RNA polymerase II-associated protein affinity purified with recombinant wild-type and mutated polymerase subunits. Mol. Cell. Biol. 24:7043–7058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kornberg R. D. 1998. Mechanism and regulation of yeast RNA polymerase II transcription. Cold Spring Harb. Symp. Quant. Biol. 63:229–232 [DOI] [PubMed] [Google Scholar]
- 25. Kornberg R. D. 2007. The molecular basis of eukaryotic transcription. Proc. Natl. Acad. Sci. U. S. A. 104:12955–12961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kostrewa D., et al. 2009. RNA polymerase II-TFIIB structure and mechanism of transcription initiation. Nature 462:323–330 [DOI] [PubMed] [Google Scholar]
- 27. Kuan P. F., Chun H., Keles S. 2008. CMARRT: a tool for the analysis of ChIP-chip data from tiling arrays by incorporating the correlation structure. Pac. Symp. Biocomput. 13:515–526 [PMC free article] [PubMed] [Google Scholar]
- 28. Kuznedelov K., et al. 2002. A role for interaction of the RNA polymerase flap domain with the sigma subunit in promoter recognition. Science 295:855–857 [DOI] [PubMed] [Google Scholar]
- 29. Kyzer S., Ha K. S., Landick R., Palangat M. 2007. Direct versus limited-step reconstitution reveals key features of an RNA hairpin-stabilized paused transcription complex. J. Biol. Chem. 282:19020–19028 [DOI] [PubMed] [Google Scholar]
- 30. Langelier M. F., et al. 2005. The highly conserved glutamic acid 791 of Rpb2 is involved in the binding of NTP and Mg(B) in the active center of human RNA polymerase II. Nucleic Acids Res. 33:2629–2639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Liu X., Bushnell D. A., Wang D., Calero G., Kornberg R. D. 2010. Structure of an RNA polymerase II-TFIIB complex and the transcription initiation mechanism. Science 327:206–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Molnar J., Lorand L. 1961. Studies on apyrases. Arch. Biochem. Biophys. 93:353–363 [DOI] [PubMed] [Google Scholar]
- 33. Ng H. H., Robert F., Young R. A., Struhl K. 2003. Targeted recruitment of Set1 histone methylase by elongating Pol II provides a localized mark and memory of recent transcriptional activity. Mol. Cell 11:709–719 [DOI] [PubMed] [Google Scholar]
- 34. Nickels B. E., Dove S. L., Murakami K. S., Darst S. A., Hochschild A. 2002. Protein-protein and protein-DNA interactions of σ70 region 4 involved in transcription activation by λcI. J. Mol. Biol. 324:17–34 [DOI] [PubMed] [Google Scholar]
- 35. Nickels B. E., et al. 2005. The interaction between σ70 and the beta-flap of Escherichia coli RNA polymerase inhibits extension of nascent RNA during early elongation. Proc. Natl. Acad. Sci. U. S. A. 102:4488–4493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Palangat M., Landick R. 2001. Roles of RNA:DNA hybrid stability, RNA structure, and active site conformation in pausing by human RNA polymerase II. J. Mol. Biol. 311:265–282 [DOI] [PubMed] [Google Scholar]
- 37. Palangat M., Meier T. I., Keene R. G., Landick R. 1998. Transcriptional pausing at +62 of the HIV-1 nascent RNA modulates formation of the TAR RNA structure. Mol. Cell 1:1033–1042 [DOI] [PubMed] [Google Scholar]
- 38. Palangat M., Renner D. B., Price D. H., Landick R. 2005. A negative elongation factor for human RNA polymerase II inhibits the anti-arrest transcript-cleavage factor TFIIS. Proc. Natl. Acad. Sci. U. S. A. 102:15036–15041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ranish J. A., Hahn S. 1996. Transcription: basal factors and activation. Curr. Opin. Genet. Dev. 6:151–158 [DOI] [PubMed] [Google Scholar]
- 40. R Development Core Team 2009. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria: http://www.R-project.org [Google Scholar]
- 41. Reinberg D., et al. 1998. The RNA polymerase II general transcription factors: past, present, and future. Cold Spring Harb. Symp. Quant. Biol. 63:83–103 [DOI] [PubMed] [Google Scholar]
- 42. Renner D. B., Yamaguchi Y., Wada T., Handa H., Price D. H. 2001. A highly purified RNA polymerase II elongation control system. J. Biol. Chem. 276:42601–42609 [DOI] [PubMed] [Google Scholar]
- 43. Roeder R. G. 1996. The role of general initiation factors in transcription by RNA polymerase II. Trends Biochem. Sci. 21:327–335 [PubMed] [Google Scholar]
- 44. Tan S., Conaway R. C., Conaway J. W. 1994. A bacteriophage vector suitable for site-directed mutagenesis and high-level expression of multisubunit proteins in E. coli. Biotechniques 16:824–826, 828 [PubMed] [Google Scholar]
- 45. Thompson N. E., Aronson D. B., Burgess R. R. 1990. Purification of eukaryotic RNA polymerase II by immunoaffinity chromatography. Elution of active enzyme with protein stabilizing agents from a polyol-responsive monoclonal antibody. J. Biol. Chem. 265:7069–7077 [PubMed] [Google Scholar]
- 46. Toulokhonov I., Artsimovitch I., Landick R. 2001. Allosteric control of RNA polymerase by a site that contacts nascent RNA hairpins. Science 292:730–733 [DOI] [PubMed] [Google Scholar]
- 47. Toulokhonov I., Landick R. 2003. The flap domain is required for pause RNA hairpin inhibition of catalysis by RNA polymerase and can modulate intrinsic termination. Mol. Cell 12:1125–1136 [DOI] [PubMed] [Google Scholar]
- 48. Vassylyev D. G., et al. 2002. Crystal structure of a bacterial RNA polymerase holoenzyme at 2.6 Å resolution. Nature 417:712–719 [DOI] [PubMed] [Google Scholar]
- 49. Wu C. H., et al. 2003. NELF and DSIF cause promoter proximal pausing on the hsp70 promoter in Drosophila. Genes Dev. 17:1402–1414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Wu S. Y., Chiang C. M. 1998. Properties of PC4 and an RNA polymerase II complex in directing activated and basal transcription in vitro. J. Biol. Chem. 273:12492–12498 [DOI] [PubMed] [Google Scholar]
- 51. Yamaguchi Y., Inukai N., Narita T., Wada T., Handa H. 2002. Evidence that negative elongation factor represses transcription elongation through binding to a DRB sensitivity-inducing factor/RNA polymerase II complex and RNA. Mol. Cell. Biol. 22:2918–2927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Yamaguchi Y., et al. 1999. NELF, a multisubunit complex containing RD, cooperates with DSIF to repress RNA polymerase II elongation. Cell 97:41–51 [DOI] [PubMed] [Google Scholar]
- 53. Yan Q., Moreland R. J., Conaway J. W., Conaway R. C. 1999. Dual roles for transcription factor IIF in promoter escape by RNA polymerase II. J. Biol. Chem. 274:35668–35675 [DOI] [PubMed] [Google Scholar]
- 54. Yoo O. J., et al. 1991. Cloning, expression and characterization of the human transcription elongation factor, TFIIS. Nucleic Acids Res. 19:1073–1079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Zhang C., Burton Z. F. 2004. Transcription factors IIF and IIS and nucleoside triphosphate substrates as dynamic probes of the human RNA polymerase II mechanism. J. Mol. Biol. 342:1085–1099 [DOI] [PubMed] [Google Scholar]
- 56. Zhang G., et al. 1999. Crystal structure of Thermus aquaticus core RNA polymerase at 3.3 Å resolution. Cell 98:811–824 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.