Abstract
Transcriptional silencing selectively impedes gene expression. Silencing is often accompanied by replication delay and can be prevented by replicator sequences. Here we report a replicator-binding protein complex involved in the prevention of transcriptional silencing. The protein complex interacts with an essential asymmetric region within the human β-globin Rep-P replicator and includes hnRNP C1/C2, SWI/SNF complex, and MeCP1, which are members of the locus control region (LCR)-associated remodeling complex (LARC). Interaction between LARC and Rep-P prevented transcriptional silencing and replication delay. Transgenes that did not contain the asymmetric LARC-binding region of Rep-P replicated late and exhibited stable silencing that could not be affected by a DNA methylation inhibitor. In contrast, transgenes that contain a mutation of the asymmetric region of Rep-P that could not bind LARC exhibited a silent state that could transiently be reactivated by DNA demethylation. The effect of DNA demethylation was transient, and prolonged exposure to a methylation inhibitor induced distinct, stable, methylation-independent silencing. These observations suggest that the interaction of LARC complex with replicators plays a role in preventing gene silencing and provides support for a novel, epigenetic mechanism of resistance to methylation inhibitors.
INTRODUCTION
Dynamic activation and silencing of gene expression are essential for the execution of cell fate decisions during development and differentiation (24). Silencing of transgenes continues to be a major impediment to gene therapy, and silencing of tumor suppressor genes has been implicated as an important mechanism in tumor progression (38). The combinatorial activity of several chromatin remodeling complexes, including histone modifications and methylation of CpG sequences often accompany gene silencing (20, 44). Silencing is also often accompanied by replication delay, but the relationship between chromatin remodeling, replication timing, and transcriptional silencing is not well understood.
Epigenetic activation and silencing of gene expression were studied extensively at the human β-globin locus. In that locus, the locus control region (LCR) regulates both gene expression and DNA replication (2, 15, 16, 45). Interactions between the LCR and globin promoters confer highly regulated tissue- and developmental stage-specific expression of β-globin-like genes (32, 36, 52). In cells actively expressing β-globin-like genes, the LCR and globin promoters are physically associated (43, 52), and their interaction is mediated by several protein factors, including transcription factors GATA-1, EKLF, NF-E2, CTCF, and the LCR-associated remodeling complex (LARC) (36, 41). Interestingly, the LCR can act not only as a transcriptional enhancer at endogenous and ectopic chromosomal sites (5, 17, 22, 23, 32) but also as an orientation- and context-dependent gene silencer (12, 14).
LCR-mediated silencing is accompanied by changes in replication timing, DNA methylation, and histone modifications (28, 49–51). The LCR is required for DNA methylation at ectopic sites (11). Although DNA methylation is not essential for gene silencing, it confers epigenetic memory and maintains the silenced state (11, 47). DNA sequences that facilitate initiation of DNA replication (replicators) can prevent gene silencing and replication delays (19). The interaction between the LCR and replicators is consistent with the observation that the LCR plays a role in the initiation of DNA replication at the endogenous locus (2). When active replicators are inserted adjacent to LCR-containing transgenes, expression is restored, and transgenes exhibit early replication as well as epigenetic marks of decondensed chromatin (11).
The LCR-associated remodeling complex is a multiprotein complex involved in chromatin remodeling that includes the MeCP1 complex, the SWI/SNF complex, and heterogeneous nuclear ribonucleoprotein (hnRNP) C1/C2. MeCP1 is a protein complex associated with epigenetic mechanisms of transcriptional repression, particularly during development. It includes the methyl-DNA-binding protein MBD2 (9), p66/p68 (8), and the multisubunit Mi2/NuRD complex that contains the nucleosome-stimulated Mi2β ATPase and the histone deacetylases HDAC1 and HDAC2 (10). Examples of the transcriptional repressive activity of MeCP1 include interactions with GATA-1 during erythroid development (46) and establishment of altered epigenetic marks in acute promyelocytic leukemia via interactions with PML-RARα (39). At the human β-globin locus, MeCP1 interacts with the SWI/SNF chromatin remodeling complex and hnRNP C1/C2 to form LARC. LARC interacts with hypersensitive site 2 (HS2) of the LCR and with the β-globin promoter (37).
Our goals were to characterize the relationship between gene silencing and chromatin modifications and to gain insight into the mechanism underlying LCR-mediated transcriptional silencing. We mapped protein-DNA interactions within asymmetric purine:pyrimidine sequences of Rep-P in the human β-globin locus that are essential to the prevention of LCR-mediated silencing (19). We then introduced mutations that prevent those protein-DNA interactions into a transgene cassette carrying the LCR and determined their effect on transgene expression and protein complex recruitment. The results showed that the asymmetric region forms several protein-DNA complexes, including a complex with hnRNP C1/C2, SWI/SNF, and MeCP1, three components of LARC. Deletion of Rep-P resulted in late replication and stable silencing. A mutation that preserved the asymmetric region but eliminated the interaction between the asymmetric region and LARC delayed replication and resulted in silencing that can be transiently prevented by zebularine, an inhibitor of DNA methylation. Long-term demethylation induced resilencing that was independent of DNA methylation and exhibited distinct epigenetic modifications. These data imply that LCR can mediate transcriptional silencing via two distinct mechanisms, one that is affected by methylation and another that is not. These data also imply that the LARC complex plays a role in preventing LCR-mediated gene silencing.
MATERIALS AND METHODS
Cell lines and culture conditions.
We grew human erythroleukemic K562 cells, human T-lymphoblastoid Jurkat cells, and murine erythroleukemia-derived cells at 37°C in a 5% CO2 atmosphere in Dulbecco's modified Eagle medium (catalog no. 10564-011; Invitrogen) supplemented with 10% heat-inactivated fetal calf serum and added 5% penicillin-streptomycin (catalog no. 15140-163; Invitrogen) and 5% Fungizone (catalog no. 15290-018; Invitrogen) as needed.
Transgene cassettes.
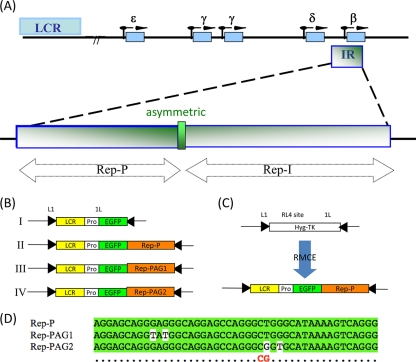
To determine the role of the LCR/LARC/Rep-P interaction in gene silencing or reexpression, we constructed transgene cassettes carrying an enhanced green fluorescent protein (EGFP) gene downstream of the β-globin LCR/promoter region and upstream of the wild-type Rep-P sequence (Fig. 1B, construct II). We also constructed control transgenes that lacked Rep-P (noPAG) (Fig. 1B, construct I) or that contained a mutant form, either Rep-PAG1 (Fig. 1B, construct III) or Rep-PAG2 (Fig. 1B, construct IV). Transgene Rep-PAG1 had two G-to-T substitutions, one at position 10 and one at position 12, and Rep-PAG2 had a T-to-G substitution at position 28 and a G-to-T replacement at position 30 (Fig. 1D).
Fig. 1.
Experimental system and constructs used in this study. (A) Structure of the human β-globin locus with the locations of the two replicators (Rep-P and Rep-I) and the asymmetric purine:pyrimidine (AG) region marked. LCR, locus control region; IR, initiation region. (B) Transgene constructs. LCR, a mini-LCR sequence that includes the DNase I-hypersensitive sites 4, 3, and 2 from the human β-globin locus control region; Pro, human β-globin promoter; EGFP, enhanced green fluorescence protein gene. (C) All transgene constructs flanked by a pair of inverted loxP sites, L1 and 1L, were inserted into a late-replicating RL4 site of the murine chromosome 15 by recombinase-mediated cassette exchange (RMCE). The insertion replaces the hygromycin-thymidine kinase (Hyg-TK) hybrid selection marker. (D) Wild-type and mutant sequences of the AG region of Rep-P used in this study. Nucleotides shown on a white background are mutations. The CG (red) below the sequences indicates the CpG dinucleotide created in the Rep-PAG2 mutation.
We inserted the LCR-Pro-EGFP cassettes into the random locus 4 (RL4) site of MEL cells using Cre recombinase-mediated cassette exchange (RMCE) as described elsewhere (12, 18) and used the PCR to confirm insertion and determine its orientation. We used primer pairs LCRperFW (FW stands for forward) (5′-CAGGAGAAGGGTCAGAGGCTTG-3′) and LCRuniRE (RE stands for reverse) (5′-AGGTCATGGCTATTCTTATTCTCAC-3′) to detect the cassette inserted in permissive orientation and the primer pair LCRnonperFW (nonper stands for nonpermissive) (5′-GGAGCCTATGGAAAAACGCC-3′) and LCRuniRE (5′-AGGTCATGGCTATTCTTATTCTCAC-3′) to detect the cassette inserted in nonpermissive orientation, and we used only cells with insertion in the nonpermissive orientation. For convenience, we refer to all cell lines that carry a transgene cassette at the RL4 site as “RL4 cells.” For chronic treatment, we added zebularine (a gift from Victor E. Marquez) at 100 μM directly to the culture medium, which we changed every 3 or 4 days. We monitored GFP expression by fluorescence-activated cell sorting (FACS) or fluorescence microscopy, counted cells every 3 or 4 days with a Z2 Coulter particle counter (Beckman Coulter Corporation), and calculated cell growth using the mean cell number from two sets of experiments.
Nuclear protein extract preparation.
We washed the harvested cells with phosphate-buffered saline (PBS) and incubated the cells in sucrose buffer containing NP-40 (320 mM sucrose, 10 mM Tris HCl [pH 8.0], 3 mM CaCl2, 2 mM magnesium acetate [MgOAc], 0.1 mM EDTA, 0.5% NP-40, 1 mM dithiothreitol [DTT], 0.5 mM phenylmethylsulfonyl fluoride [PMSF], protease inhibitor cocktail [catalog no. P8340; Sigma], and phosphatase inhibitor cocktail [catalog no. 04906845001; Roche]). We harvested the nuclei by centrifugation, washed, and resuspended them in low-salt buffer (20 mM HEPES [pH 7.9], 20 mM KCl, 0.2 mM EDTA, 25% glycerol [vol/vol], 0.5 mM DTT, 0.5 mM PMSF, protease inhibitor cocktail, and phosphatase inhibitor cocktail) and then added high-salt buffer (20 mM HEPES [pH 7.9], 800 mM KCl, 0.2 mM EDTA, 25% glycerol [vol/vol], 1% NP-40, 0.5 mM DTT, 0.5 mM PMSF, protease inhibitor cocktail, and phosphatase inhibitor cocktail). We incubated the suspension at 4°C for 45 min with rotation, centrifuged it at 14,000 × g for 15 min at 4°C, and determined protein concentrations with a Bio-Rad DC protein assay kit (catalog no. 500-0112; Bio-Rad).
Western blot analysis.
Using nuclear extracts prepared from MEL RL4 cells with EpiQuik nuclear extraction kit I (catalog no. Op-0002-1; Epigenetek), we mixed 25 μg of total protein with 2× Tris-glycine sample buffer (catalog no. LC2676; Invitrogen), heated the mixture in a boiling water bath for 10 min, centrifuged it at 13,200 rpm for 5 min, and analyzed it on 4% Tris-glycine gel (catalog no. EC6058BOX; Invitrogen) at 125 V for 1 h. We used goat anti-DNA methyltransferase 1 (anti-DNMT1) (sc-10221) diluted 1:200, goat anti-DNMT3a (sc-10232) diluted 1:200, goat anti-DNMT3b (sc-10236) diluted 1:200, and mouse anti-proliferating cell nuclear antigen (anti-PCNA) (sc-56) diluted 1:300 as primary antibodies and horseradish peroxidase (HRP)-conjugated donkey anti-goat IgG (sc-2020) diluted 1:2,000 and HRP-conjugated donkey anti-mouse IgG (sc-2314) diluted 1:1,000 as secondary antibodies (all antibodies from Santa Cruz Biotechnology). We used SuperSignal West Pico chemiluminescent substrate (catalog no. 34080; Pierce) for immunodetection and, when necessary, stripped polyvinylidene difluoride (PVDF) membranes with 15 ml of Restore Western blot stripping buffer (catalog no. 21059; Thermo Scientific) at room temperature for 15 min.
Electrophoretic mobility shift assay (EMSA) and supershift assays.
To further characterize the antisilencing function of Rep-P, we examined its protein-binding specificity by performing the electrophoretic mobility shift assay using an oligonucleotide substrate containing the 45-bp asymmetric region (Oligo AG) or a variant in which positions 28 and 30 of the asymmetric region were mutated (Oligo AG2).
We labeled DNA oligonucleotides extracted from logarithmic-growth-phase cells with biotin using a biotin 3′-end DNA labeling kit (catalog no. 89818; Pierce) and concentrated nuclear protein extracts with Microcon YM-10 centrifugal filter units (catalog no. 42406; Millipore). We incubated the labeled oligonucleotides with 20 μg protein for each binding reaction mixture in the binding buffer [10 mM Tris-HCl, 2.5% glycerol, 0.05% NP-40, 25 mM KCl, 1 μg poly(dI·dC), with 1 mM DTT] for 30 min at room temperature and electrophoresed them on a 6% DNA retardation gel (catalog no. EC6365BOX; Invitrogen) on ice for 1.5 h. We transferred the samples to a positively charged nylon membrane and cross-linked them in a UV cross-linker. We used the LightShift chemiluminescent EMSA kit (catalog no. 20148; Pierce) for biotin-labeled DNA detection. For the competition experiments, we added unlabeled oligonucleotides to the nuclear proteins before adding the biotin-labeled oligonucleotides. For the supershift assays, we added antibodies to the prepared nuclear extracts before adding the labeled oligonucleotides. We obtained the antibodies (anti-hnRNP C1/C2 [sc-15386X], anti-Orc4 [sc-19725], anti-TRRAP [sc-5405], and anti-TIP60 [sc-5727]) from Santa Cruz Biotechnology.
In vivo DNA footprinting.
We performed in vivo DNase I genomic footprinting experiments according to a protocol by Drouin et al. (7) with the following modifications. In brief, we washed 2 × 106 K562 cells with 1× PBS, resuspended the cells in solution II (150 mM sucrose, 80 mM KCl, 35 mM HEPES [pH 7.4], 5 mM MgCl2, and 2 mM CaCl2) and permeabilized them with prewarmed (37°C) 0.1% lysolecithin (catalog no. L4129; Sigma) in solution II for 1 to 2 min. We treated the permeabilized cells with 0.125 U of DNase I (catalog no. M0303L; New England BioLabs) for 3 min and stopped the reaction by heating at 76°C for 10 min. We extracted the DNA using the phenol-chloroform method and performed ligation-mediated PCR (LMPCR) with fluorescence-labeled amplification primer.
We performed LMPCR by the methods of Mueller et al. (40) and Paixão et al. (42) with modifications. We performed primer extension by annealing DNase I-treated DNA to 0.5 pmol of the ASYMMET1F extension primer (5′-ACTCCTAAGCCAGTGCCAGA-3′) and incubating the sample at 57°C with 1 U of Vent exopolymerase (catalog no. M0254L; New England BioLabs) to obtain blunt-end products. For blunt-end ligation, we added 30 μl of linker mix containing 4 μM asymmetric double-stranded linker LMPCR1 (5′-GGTGACCCGGGAGATCTGAATTC-3′) and LMPCR2 (5′-GAATTCAGATC-3′) and 1 μl of T4 DNA ligase in 1× ligation buffer (New England BioLabs) to the sample and incubated the sample in buffer overnight at 16°C. We labeled and amplified the ligation products with the ASYMMET2F fluorescence-labeled primer (5′-AGGTACGGCTGTCATCACTTAGACC-3′) at 59°C with 1 U Vent polymerase. We purified amplified products to eliminate salts that can interfere with capillary electrophoresis, ran the samples with GeneScan 500 ROX internal lane size standard (Applied Biosystems) on an automated DNA sequencer, and analyzed the samples using GeneMapper fragment analysis software (Applied Biosystems).
Nascent-strand DNA analysis.
We performed nascent-strand DNA analysis as described previously (3, 55). In brief, DNA was extracted from asynchronous RL4 cells, denatured by boiling for 10 min, incubated on ice for 10 min, and fractionated on a neutral sucrose gradient. We collected 0.5- to 1-kb DNA fractions, treated them with λ exonuclease to remove non-RNA-primed genomic DNA fragments, purified them, and measured DNA concentration using NanoDrop 1000 (Thermo Scientific). We quantified nascent-strand DNA by real-time PCR in an ABI 7900 thermocycler (primers and probes used for real-time PCR are listed in Table 1).
Table 1.
List of primers and probes for real-time PCR
| Primer or probea | Sequence |
|---|---|
| bG59.8 | |
| Fwd primer | TGGAAAAGCAACCCCTGC |
| Rev primer | AACTATGGATCCTTCTCTTGTGTTGG |
| Probeb | GCTGCAGATACCATCATCCTGGCTTCAA |
| LB2 | |
| Fwd primer | TGGGACCCTGCCCTTTTT |
| Rev primer | CGTGACGAAGAGTCAGCT |
| Probeb | TTCTAGTGAGCCTCCGAC |
| Rep-I | |
| Fwd primer | GGTGAAGGCTCATGGCAAGA |
| Rev primer | AAAGGTGCCCTTGAGGTTGTC |
| Probeb | CCTTTAGTGATGGCCTGGCTCACCTG |
| hBG | |
| Fwd primer | TGAGGGTTTGAAGTCCAACTCC |
| Rev primer | GGTCTAAGTGATGACAGCCGTACC |
| Probeb | AAGCCAGTGCCAGAAGAGCCAAGGA |
| mAmy | |
| Fwd primer | TCATATTCTAATCAAGACTAGTGACTTTAGAGC |
| Rev primer | TGCCACAACTACCAATCCTTTT |
| Probe | CAACTTCATTTCACACATGACTTTGCTGAGAAA |
| GFP | |
| Fwd primer | AGCAAAGACCCCAACGAGAA |
| Rev primer | GGCGGCGGTCACGAA |
| Probeb | CGCGATCACATGGTCCTGCTGG |
| mBG | |
| Fwd primer | CCAGCCTCAGTGAGCTCCA |
| Rev primer | CCCATCAGACTCACCCTGAAG |
| Probeb | TGTGACAAGCTGCATGTGGATCCTGA |
| mCh15 | |
| Fwd primer | TCCGTCCCCTTCTCCTCC |
| Rev primer | TTCAGGTTCCATTGCCACG |
| Probeb | CACCATTCACACAGCCCACGAGCA |
| AG | |
| Fwd primer | CAACTCCTAAGCCAGTGCCAGAAG |
| Rev primer | TGCCCTGACTTTTATGCCCAGC |
| Probeb | TCATCACTTAGACCTCACCCTGT |
| HS2 | |
| Fwd primer | TGTGTAACCTTCTAAGCAAACCTTCTG |
| Rev primer | GCCATCTGCCCTGTAAGCATC |
| Probeb | GTTCTCAGCCTAGAGTGATGAC |
The forward (Fwd) and reverse (Rev) primers and probe are shown for Rep-P 5′-end sequence (bG59.8), lamin B2 replicator (LB2), replicator Rep-I in the β-globin locus, human β-globin promoter (hBG), mouse amylase gene (mAmy), enhanced green fluorescent protein (EGFP) gene (GFP), mouse β-globin locus (mBG), murine chromosome 15 (mCh15), the AG region of Rep-P, and hypersensitive site 2 (HS2).
These probes were modified with 6-carboxyfluorescein (FAM) at the 5′ end and with 6-carboxytetramethylrhodamine (TAMRA) at the 3′ end.
Replication timing analyses.
Replication timing analyses were performed as described previously (31, 33) with the following modifications. Cells were labeled with bromodeoxyuridine (BrdU) for 90 min prior to harvesting. Cells from different cell cycle fractions were separated by centrifugation elutriation and verified by FACS. Newly replicated, BrdU-substituted DNA was isolated by immunoprecipitation with anti-BrdU antibodies (catalog no. 555627; BD Pharmingen). DNA from Drosophila S2 cells was added as a control to verify that each fraction contained similar quantities of amplifiable DNA strands. The level of DNA in the samples was determined by real-time PCR on an ABI 7900. Experiments were repeated at least twice for each cell line.
ChIP analysis.
We performed chromatin immunoprecipitation (ChIP) analyses with 1% formaldehyde-fixed K562, Jurkat, and RL4 cells using the Millipore ChIP assay kit (catalog no. 17-295) following the manufacturer's protocol. We purchased normal rabbit IgG (sc-2027), normal mouse IgG, normal goat IgG (sc-2028), antibodies against hnRNP C1/C2 (sc-15386), Brg1 (sc-12520), Mi2β (sc-12541), MBD2 (sc-9397), and MBD3 (sc-9402) from Santa Cruz Biotechnology, and antibodies against acetyl-histone H4 (6-866), acetyl-histone H3 (6-599), dimethyl-histone H3 Lys4 (07-030), and trimethyl-histone H3 Lys9 (07-442) from Upstate/Millipore. We analyzed ChIP samples by real-time PCR in an ABI 7900 thermocycler using the primers and probes listed in Table 1.
Plasmid constructs and site-directed mutagenesis.
The Rep-P sequence and the asymmetric region from the human β-globin locus that we used have been described previously (55, 56). The Rep-PAG1 mutation contains substitutions at positions 10 and 12 of the asymmetric region (G10T and G12T), and the Rep-PAG2 mutation contains substitutions at positions 28 and 30 of the asymmetric region (T28G and G30T). We used the QuikChange II kit (catalog no. 200523; Stratagene) to introduce the mutations into Rep-P and the following primers for site-directed mutagenesis at positions 10 to 12 and 28 to 30 of the asymmetric (AG) region. To introduce the PAG1 mutations, the forward primer was 5′-CTACTCCCAGGAGCAGGTATGGCAGGAGCCAGGGCTG-3′, and the reverse primer was 5′-CAGCCCTGGCTCCTGCCATACCTGCTCCTGGGAGTAG-3′. To introduce the PAG2 mutation, the forward primer was 5′-GAGGGCAGGAGCCAGGGCGGTGCATAAAAGTCAGGGCAG-3′, and the reverse primer was 5′-TGCCCTGACTTTTATGCACCGCCCTGGCTCCTGCCCTC-3′.
Constructs that contained pLCR–Pro–EGFP–Rep-P with the PAG1 or PAG2 mutations were designated PAG1 or PAG2, respectively. We cloned the wild-type and mutant Rep-P sequences into the pLCR-Pro-EGFP vector (13) by standard cloning techniques.
ChIP-3C.
We carried out ChIP-chromosome conformation capture (ChIP-3C) experiments by the method summarized by Simonis et al. (48) and the method of Horike et al. (27) with modifications. Briefly, 1 × 107 K562 cells were fixed with 1% formaldehyde in PBS with 10% fetal bovine serum (FBS) for 10 min. Cells were lysed with 10 ml of lysis buffer (10 mM Tris-HCl [pH 8.0], 10 mM NaCl, 0.2% NP-40) on ice for 10 min and washed with PBS twice. Restriction enzyme digestion was done by incubating the cell lysate in 1× restriction buffer with 0.3% SDS, 2% Triton X-100, and 400 U of HindIII (catalog no. R0104; New England BioLabs) at 37°C overnight. We immunoprecipitated the digested chromatin with antibody against hnRNP C1/C2 (sc-15386X) before proceeding to ligation. Subsequently, the sample was ligated in 7 ml of 1× T4 ligation buffer with 4,000 U of T4 DNA ligase (catalog no. M0202S; New England BioLabs) at 16°C overnight. Ligated DNA sample was reverse cross-linked and extracted by phenol-chloroform. We used nested PCR to confirm the interaction between HS2 and Rep-P. The primers used in this study were as follows: HS2 outer primer (5′-GAACTGCTCATGCTTGGAC-3′), Rep-P outer primer (5′-TGCTGCAGATACCATCATCC-3′), HS2 inner primer (5′-GAGGTGGAGTTTTAGTCAGGTGGTC-3′) (6), and Rep-P inner primer (5′-TGGTCAGAGCCTCAGTTTCA-3′). The PCR products were TA cloned and sequenced.
DNMT1 assay.
We used EpiQuik nuclear extraction kit I (catalog no. OP-0002-1; EpiGenetek) to prepare nuclear extracts and the EpiQuik DNMT1 assay kit (catalog no. P-3011-2; EpiGenetek) to quantify DNMT1 activity.
Bisulfate sequencing of genomic DNA.
We extracted genomic DNA from RL4 cells using a standard protocol and processed it with a Methylamp whole-cell bisulfite modification kit (catalog no. P-1016; Epigentek), following the manufacturer's protocol. We amplified modified DNA samples by nested PCR using outside primer pairs Met-Rep-P_F2-1 (5′-AAGTTGAATTATGGTAGATAAAG-3′) and Met-Rep-P_R2-2 (5′-CAATTAACCCTCACTAAAAAAAAC-3′) and inside primer pairs Met-Rep-P_F2-2 (5′-TTGTAAAGGAGGATGTTTTTAG-3′) and Met-Rep-P_R2-1 (5′-CCTCACTAAAAAAAACAAAAACTAAATACC-3′). We gel purified the PCR products and cloned them into TA cloning vector pCR2.1 using the TOPO TA cloning kit (Invitrogen), and we used an M13 reverse primer for DNA sequencing.
RACE.
We performed 5′ rapid amplification of cDNA ends (5′ RACE) to determine the transcription start site and promoter usage of the EGFP transgene cassette in RL4 cells. We used the FirstChoice RNA ligase-mediated RACE (RLM-RACE) kit (catalog no. AM1700; Ambion) to perform the 5′ RACE following the manufacturer's instructions. In brief, 1 μg of total RNA extracted from GFP-expressing RL4 cells using the RNeasy minikit (catalog no. 74104; Qiagen) was treated with calf intestine alkaline phosphatase (CIP) at 37°C for 1 h. CIP-treated RNA was then treated with tobacco acid pyrophosphatase at 37°C for 1 h. The RNA samples were reverse transcribed into cDNA. Primers used for the nested PCR are as follows: for outer primers, 5′ RACE outer primer (provided by the Ambion kit) and EGFP_R2 (5′-TGCTTCATGTGGTCGGGGTAG-3′); for inner primers, 5′ RACE inner primer (provided by the Ambion kit) and EGFP_R3, (5′-CAGATGAACTTCAGGGTCAG-3′). Both outer and inner PCR products were cloned by using the TA cloning kit (catalog no. 45-0046; Invitrogen) and sequenced.
hnRNP C1/C2 gene silencing.
The short hairpin RNA (shRNA) hnRNP C1/C2 vector was made following Clontech's knockout single vector inducible RNA interference (RNAi) system user manual (6a) using a hairpin-forming sequence (5′-GCGTCCACACTTTGCTAGACAATTCAAGAGATTGTCTAGCAAAGTGTGGACGTTTTTTACGCGT-3′). K562 cells harboring the shRNA vector were exposed to 1 μg/ml doxycycline added to the culture medium each day for 10 days. Gene expression of hnRNP C1/C2 and human β-globin mRNA was measured using real-time PCR. Applied Biosystem's Power SYBR PCR mix was used for each real-time PCR experiment. All mRNA expression values were normalized against the mRNA expression value of human beta-2-microglobulin (hB2M).
FACS analysis.
We used fluorescence-activated cell sorting to monitor the levels of GFP expression of the transgenes. Zebularine-treated and parallel cultured untreated cells were observed using FACS or fluorescence microscopy. Dead cells were gated out on the basis of morphological parameters (forward scatter [FSC] and side scatter [SSC]). The percentage of cells expressing the GFP transgene was estimated using untreated cells as a negative control.
RESULTS
Binding of hnRNP C1/C2, SWI/SNF, and MeCP1 to the Rep-P asymmetric region.
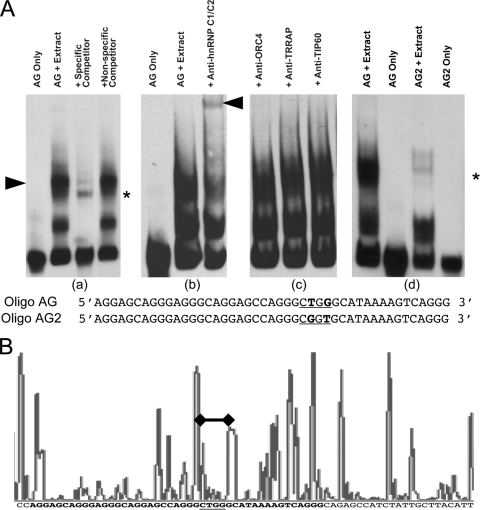
To identify proteins that interact with the essential asymmetric regions, we investigated whether proteins in nuclear extracts can interact with a 45-bp oligonucleotide (Oligo AG) containing the asymmetric sequence. We chose the sequence of Oligo AG to reflect the fact that initiation of DNA replication is prevented when the 45-bp sequences are deleted from Rep-P (56).
When we incubated protein extracts from K562 cells with Oligo AG, we detected two complexes with distinct electrophoretic mobility [Fig. 2A, panel (a)]. Mass spectrometry analysis of proteins in the slower-moving complex revealed the presence of hnRNP C1/C2. Coincubation with antibody to hnRNP C1/C2 specifically supershifted the slowly migrating protein complex [Fig. 2A, panel (b)], but antibodies to other chromatin components did not [Fig. 2A, panel (c)], thus confirming the result.
Fig. 2.
Protein-DNA interactions at the asymmetric purine:pyrimidine region of Rep-P. (A) Electrophoretic mobility shift assays and supershift assays with K562 cell nuclear extract and Oligo AG (wild-type oligonucleotide) or Oligo AG2 (mutant oligonucleotide) with their sequences. The protein-binding sites are underlined, and the bold letters show base alterations. The black arrowhead in panel (a) points to the position of the slower-migrating complex. The black arrowhead in panel (b) indicates the supershift caused by anti-hnRNP C1/C2 antibody. The asterisks in panel (a) and (d) indicate the nonspecific band that cannot be competed with the specific competitor. (B) DNase I footprinting indicating protection at the AG2 site. The figure shows the automated DNA sequence analysis of ligation-mediated PCR-amplified products from DNase I-treated permeabilized asynchronous K562 cells. In the histogram, high-intensity peaks represent DNase I-hypersensitive sequences, whereas no peak or low-intensity peaks represent protected regions. Genomic DNA was used as the standard for sequence alignment. The asymmetric AG region is in bold font. As shown in the figure, the AG2 site in the AG region is protected from DNase, indicating protein binding at the AG2 site (marked by the black bar with solid black diamonds on the histogram and the underlined sequence below).
We next introduced an extensive series of mutations into Oligo AG to identify the DNA sequences required for hnRNP C1/C2 binding. We have identified two nucleotides at positions 28 and 30 that were essential for the binding interaction. When we incubated the extracts with Oligo AG2 [Fig. 2A, panel (d)], we did not detect the complex, indicating that positions 28 to 30 in the oligonucleotide were essential for interaction with the complex. We confirmed this result by in vivo DNase I footprinting of the Rep-P region in K562 cells (Fig. 2B), which showed protection of positions 28 to 30 in the template.
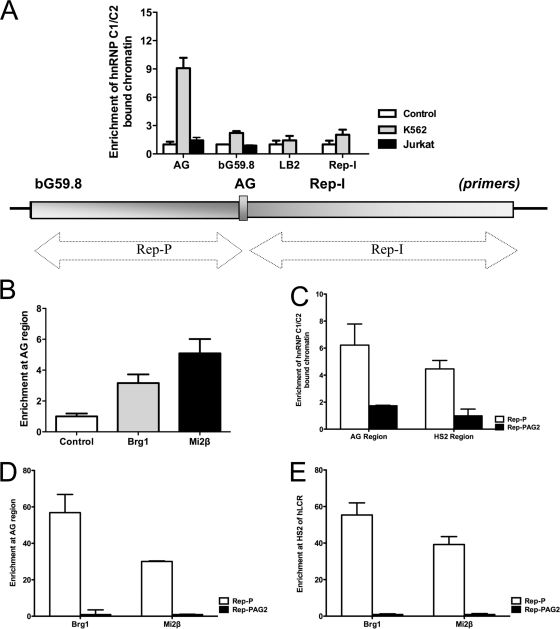
ChIP analysis, which we performed to determine whether other components of the locus control region (LCR)-associated remodeling complex (LARC) besides hnRNP C1/C2 interacted with Rep-P via the asymmetric region, indicated that Brg1 (a component of SWI/SNF) and Mi2β (a component of MeCP1) were enriched at the Rep-P asymmetric region of the endogenous β-globin locus in K562 cells (Fig. 3A and B). We also observed binding of hnRNP C1/C2 to Rep-I, a second replicator adjacent to Rep-P that can prevent gene silencing (19; data not shown). In Jurkat cells, no enrichment of hnRNP C1/C2 at the Rep-P asymmetric region was evident, which is consistent with the absence of gene expression from the globin locus in those cells (Fig. 3A). This is consistent with the fact that K562 cells express γ-globin, whereas Jurkat cells do not. These results suggest that the LARC complex binds the asymmetric region of Rep-P and participates in a tissue-specific LCR/Rep-P interaction and that the interaction may be required for the expression of β-globin-like genes.
Fig. 3.
MeCP1, SWI/SNF, and hnRNP C1/C2 interactions with Oligo AG. (A) ChIP analysis measuring the binding of hnRNP C1/C2 to the asymmetric Rep-P region in K562 cells and Jurkat cells. Primers and probes include the following: LB2, lamin B2 origin; for the globin IR, see the illustration; bG59.8, Rep-P 5′-end sequence; AG, AG region of Rep-P; and Rep-I, another replicator in the β-globin locus. (B) Binding of the AG region by SWI/SNF and MeCP1 components (Brg1 and Mi2β, respectively) in K562 cells. (C) ChIP analysis for hnRNP C1/C2 binding at AG region and hypersensitive site 2 (HS2) region of LCR in RL4 cells carrying wild-type (Rep-P) or mutant (Rep-PAG2) transgene cassettes. (D) ChIP analysis for LCR-associated remodeling complex (LARC) components binding at the AG region in RL4 cells carrying Rep-P or Rep-PAG2 transgene cassettes. (E) ChIP analysis for LARC components binding at the HS2 region of LCR in RL4 cells carrying Rep-P or Rep-PAG2 transgene cassettes. (Primer and probe sets used in this study are listed in Table 1.)
We then investigated whether the sequences required for the interaction of Oligo AG with hnRNP C1/C2 in vitro were also essential for interaction of MeCP1-SWI/SNF-hnRNP C1/C2 with Rep-P in cells. To that end, we introduced a transgene cassette that includes the LCR, an enhanced GFP expression cassette driven by the β-globin promoter and Rep-P variants into the RL4 site in MEL cells. We tested a transgene cassette that did not include Rep-P (Fig. 1B, construct I), a cassette that included an intact Rep-P (Fig. 1B, construct II), a cassette that included a Rep-P variant mutated in nucleotides 10 and 12 (Rep-PAG1, illustrated in Fig. 1B [construct III]), and a cassette that included a Rep-P variant mutated in nucleotides 2 and 30 (Rep-PAG2, illustrated in Fig. 1B [construct IV]). Rep-P PAG2 recapitulated the PAG2 mutation that prevented the interaction of Oligo AG with MeCP1-SWI/SNF-hnRNP C1/C2 complex (Fig. 2); Rep-PAG1 was chosen as a control, because the mutated nucleotides in positions 10 and 12 corresponded to an interaction site detected by genomic footprinting, but a variant of Oligo AG harboring this mutation could interact with MeCP1-SWI/SNF-hnRNP C1/C2 complex (data not shown). After inserting transgene cassettes into the silencing-prone orientation of the RL4 site in MEL cells (Fig. 1C), we selected stably transfected clones and verified their structure and orientation by PCR.
To investigate the binding of hnRNP C1/C2 to Rep-P in vivo, we performed ChIP assays using RL4 cells carrying a Rep-P variant. We found that hnRNP C1/C2-bound chromatin was enriched when transgene cassettes carried Rep-PAG1 or Rep-P, but not Rep-PAG2 (Fig. 3C), recapitulating in vivo the interactions we observed in vitro. Similarly, Brg1- and Mi2β-bound chromatin was enriched in transgene sequences in cells harboring Rep-P but not in cells harboring Rep-PAG2 (Fig. 3D).
LARC was previously shown to bind hypersensitive site 2 (HS2) of the LCR in K562 cells (37). As in the K562 cells, hnRNP C1/C2, Brg1, and Mi2β were also found to bind to the HS2 in the transgene harboring the intact Rep-P (Fig. 3C and E). No hnRNP C1/C2, Brg1, and Mi2β binding was observed in the Rep-PAG2 mutant transgene (Fig. 3C and E). These results suggest that LARC binding to the HS2 region required its binding to the asymmetric region.
Direct interaction between LCR and Rep-P.
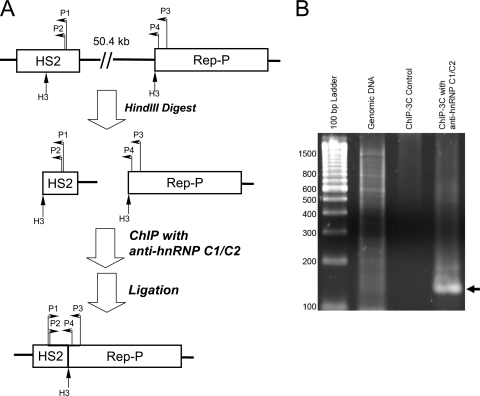
We used ChIP-chromosome conformation capture (ChIP-3C) to investigate whether LARC associates with HS2 sequences that directly interact with Rep-P. K562 cells were fixed with formaldehyde to cross-link DNA-protein complexes. Chromatin was then digested with a restriction enzyme, HindIII, immunoprecipitated with anti-hnRNP C1/C2 antibody, and religated with T4 ligase. The ChIP procedure in the ChIP-3C assay isolated the hnRNP C1/C2-bound chromatin. If two distant cis elements are interacting with each other, the ligation will link the two sequences together (Fig. 4A). Interaction between Rep-P and HS2 was detected by PCR using a primer pair that included a primer from HS2 and another primer from Rep-P (Fig. 4B). The PCR product was cloned into a TA cloning vector and sequenced. The sequencing results showed that both HS2 and Rep-P sequences were present in the PCR product, linking at a HindIII site. These results demonstrated a direct interaction between LCR and Rep-P in chromatin from K562 cells precipitated with antibody against hnRNP C1/C2, consistent with the hypothesis that the interaction between the two elements was mediated through a protein complex containing hnRNP C1/C2.
Fig. 4.
Chromatin immunoprecipitation-chromosome conformation capture (ChIP-3C) experiments demonstrating direct interactions between HS2 and Rep-P in hnRNP C1/C2-containing chromatin. (A) Schematic illustration of the experiment. Fixed K562 cells were lysed and digested with HindIII (H3). Cross-linked chromatin fragments were immunoprecipitated with hnRNP C1/C2 antibody and ligated with T4 ligase. The hnRNP C1/C2-containing protein complexes that bound to both sequences brought remote sequences together. The locations of primers marked on the illustration are as follows: P1, HS2 outer primer; P2, HS2 inner primer; P3, Rep-P outer primer; P4, Rep-P inner primer. All primers are listed in Materials and Methods. (B) An agarose gel showing the nested PCR product of the ChIP-3C sample (black arrow). The PCR product was cloned and sequenced. In the control sample, no antibody was added in the ChIP step. No nested PCR products were generated from the control. K562 genomic DNA was used as a negative control for the experiment. Without HindIII digestion and religation, the nested PCR primers are unable to generate PCR products from the genomic DNA template. The positions of molecular size standards (in base pairs) are shown to the left of the gel.
Interactions between members of the LARC complex and Rep-P prevent gene silencing.
In an earlier study, we found that inclusion of Rep-P in the transgene cassette advances the replication timing and prevents gene silencing (19). To determine whether prevention of gene silencing requires the interaction between the LARC complex and Rep-P, we measured GFP expression in RL4 cells carrying the Rep-PAG1 or Rep-PAG2 cassette. We found that recently isolated RL4 cells carrying the Rep-PAG1 or Rep-PAG2 cassette expressed GFP (Fig. 5A). Cells carrying the Rep-PAG1 cassette continued to express GFP for at least a year (data not shown), but cells carrying the Rep-PAG2 cassette showed no GFP expression after 1 to 2 months (Fig. 5A). This suggests that Rep-PAG1 retains antisilencing activity while Rep-PAG2 does not and that binding of MeCP1-SWI/SNF-hnRNP C1/C2 to Rep-P is required for Rep-P antisilencing activity. Consistent with this suggestion, reduction of hnRNP C1/C2 levels diminished expression of the human β-globin gene (Fig. 5B). This observation is consistent with the idea presented earlier that MeCP1-SWI/SNF-hnRNP C1/C2 promotes interaction between LCR and Rep-P and that the interaction is essential for derepression of the β-globin promoter.
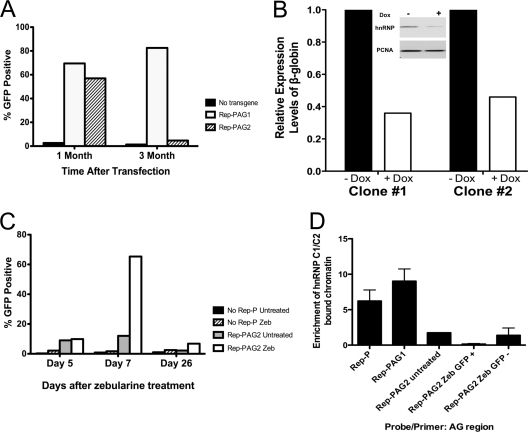
Fig. 5.
hnRNP C1/C2 antisilencing activity. (A) The percentages of GFP-positive cells of RL4 cells at 1 month and 3 months after transfection with the indicated transgene cassette. The GFP expression of RL4 cells was analyzed using FACS. Viable cells were gated on the basis of morphological parameters (forward scatter [FSC] and side scatter [SSC]). RL4 cells without a transgene were treated as a negative control. (B) Partial silencing of hnRNP C1/C2 expression in K562 cells. Cells (two independent clones) harboring a doxycycline-inducible shRNA targeting hnRNP C1/C2 were exposed to doxycycline (+ Dox) for 10 days, and the levels of β-globin mRNA were measured. The relative expression levels were normalized according to the control levels (no doxycycline [− Dox]). (Inset) Levels of hnRNP C1/C2 and PCNA in clone 1 with and without doxycycline. This experiment was repeated in 3 independent clones with similar results; error bars were too small and not visible on the histogram. hnRNP, hnRNP C1/C2; PCNA, proliferating cell nuclear antigen. (C) Outcome of zebularine exposure (the results of 1 representative experiment of 3 experiments is shown). RL4 cells containing the indicated constructs were treated continuously with zebularine (Zeb). The GFP expression of RL4 cells carrying no Rep-P or Rep-PAG2 transgene cassette were analyzed using FACS. Viable cells were gated on the basis of morphological parameters (FSC and SSC). The percentage of cells expressing the GFP transgene was estimated using untreated cells as a negative control. Data points presented are day 5 (before GFP expression), day 7 (expressing GFP), and day 26 (GFP expression resilenced after prolonged zebularine treatment). (D) ChIP analysis of the binding of hnRNP C1/C2 to the indicated constructs before and during exposure to zebularine. GFP +, GFP expression; GFP −, GFP expression resilenced.
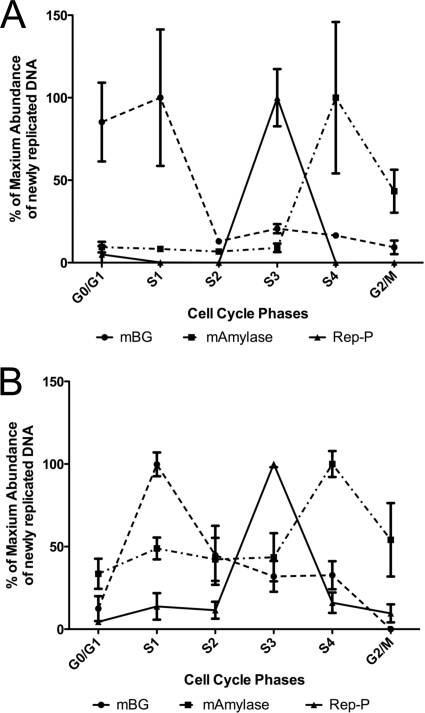
We found that the Rep-PAG2 transgene replicated in mid to late S phase (Fig. 6A). Since wild-type Rep-P replicates early when inserted into the same RL4 site in MEL cells (19), these data suggest that interaction between the LARC complex and the Rep-P asymmetric region is essential for establishing early replication of the transgene and preventing LCR-mediated replication delay.
Fig. 6.
Replication timing of cells transfected with the LCR–Pro–EGFP–Rep-PAG2 cassette before zebularine treatment (A) and after zebularine treatment (B). We quantified newly replicated transgene (Rep-P) DNA and host sequences (mBG and mAmylase) at different cell cycle phases by real-time PCR. mBG, mouse β-globin locus, an early replicating region in mouse MEL cells; mAmylase, mouse amylase gene, a late replicating region; Rep-P, the Rep-PAG2 region of the inserted transgene cassette.
Two modes of silencing distinguished by response to zebularine.
DNA methylation is known to stabilize gene silencing, and our previous studies suggested that transcription of transgenes that were silenced by LCR in the absence of Rep-P could not be reactivated by methylation inhibitors. We have tested whether the same phenomenon occurred in cells that harbor the mutant RepP that cannot bind LARC; we inhibited DNA methylation in cells harboring transgenes containing Rep-PAG2. When grown in the presence of zebularine, a methyltransferase inhibitor, RL4 cells carrying an EGFP cassette lacking Rep-P did not express GFP even after more than 1 month (Fig. 5C and data not shown), while RL4 cells carrying the Rep-PAG2 transgene expressed GFP strongly but transiently. Seven days after zebularine treatment, ∼70% of the Rep-PAG2 cells were GFP positive compared to untreated controls, whereas very few zebularine-treated cells that did not carry Rep-P were GFP positive (Fig. 5C). GFP expression was not accompanied by early replication (Fig. 6B) and was lost upon removal of the drug after 1 week of exposure. In cells chronically exposed to zebularine, GFP expression persisted for only 3 to 4 weeks, and the fraction of cells exhibiting GFP expression after 26 days of exposure to zebularine was similar in cells exposed to zebularine and cells that were not exposed to zebularine (Fig. 5C). These observations suggest that Rep-PAG2 can transiently promote GFP expression in RL4 cells when methylation is inhibited.
We next performed 5′-RACE experiments to determine whether demethylation transiently activated the promoter from which transcription of eGFP originates in the transgene or whether transcription started from another cryptic promoter. Sequencing 10 clones isolated following the 5′-RACE procedure showed that eGFP transcription within the transgene initiated from the original promoter. We also measured the nearby transcript in cells carrying the transgenes to investigate whether zebularine exposure induced a nearby transcript that could cause silencing in the transgene by transcriptional interference (14). We found that resilencing was not accompanied by induction of the nearby transcript in all silenced transgenes (data not shown), suggesting that resilencing was not triggered by the reported nearby transcript.
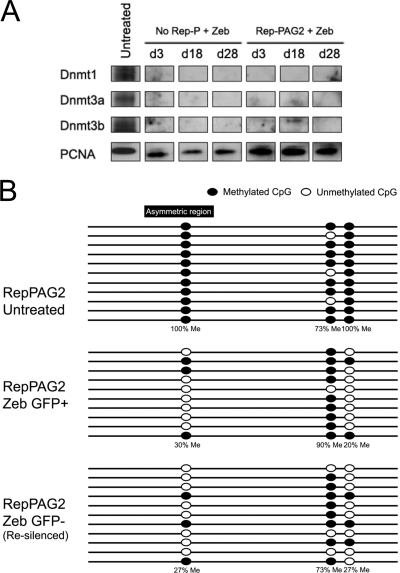
With continuous exposure to zebularine, RL4 cells grew at a slightly decreased rate (data not shown), and Western blot analysis confirmed that the cells had a low level of DNMT1 protein that was retained during GFP derepression and resilencing (Fig. 7A). We also detected low DNMT1 activity in zebularine-treated RL4 cells in an in vitro DNA methylation assay (data not shown), indicating that GFP resilencing was not due to acquired global drug resistance.
Fig. 7.
Effects of zeburaline treatment on Dnmt protein levels, Dnmt1 activity, and CpG methylation. (A) Western blot analysis of Dnmt proteins in RL4 cells that were exposed to zebularine for 3 days (d3), 18 days (d18), or 28 days (d28). RL4 cells carrying an LCR–Pro–EGFP transgene cassette (No Rep-P) and RL4 cells carrying an LCR–Pro–EGFP–Rep-PAG2 transgene cassette (Rep-PAG2) were exposed to zebularine (Zeb). (B) Results of bisulfite sequencing of genomic DNA from RL4 cells exposed to zebularine. Me, methylated.
Bisulfite sequencing showed that the CpG dinucleotide generated by T28G was methylated in the genomic DNA of RL4 cells carrying Rep-PAG2 but was demethylated after exposure to zebularine (Fig. 7B). Although demethylation coincided with derepression of GFP in zebularine-treated RL4/PAG2 cells, it was persistent after continuous exposure to zebularine even though GFP was resilenced after 4 weeks. These results demonstrate that demethylation exhibited a transient effect on gene expression and that in the prolonged absence of methylation, the transcriptional state of the transgene cassette was determined by other mechanisms. These observations support the notion that while cells often use methylation to maintain the silent state, silencing can occur without DNA methylation.
To determine whether MeCP1-SWI/SNF-hnRNP C1/C2 complex binding depended on CpG methylation, we performed ChIP assays using an antibody against hnRNP C1/C2 on RL4/PAG2 cells demethylated with zebularine for various time periods. We observed that hnRNP C1/C2 failed to bind Rep-PAG2 under any condition, even after chronic exposure to zebularine (more than 4 weeks [Fig. 5D]). Thus, hnRNP C1/C2 failed to bind either methylated or demethylated Rep-PAG2, independent of the transcriptional status of the EGFP transgene. These observations suggest that transient activation of gene expression does not require binding of hnRNP C1/C2 to the asymmetric region.
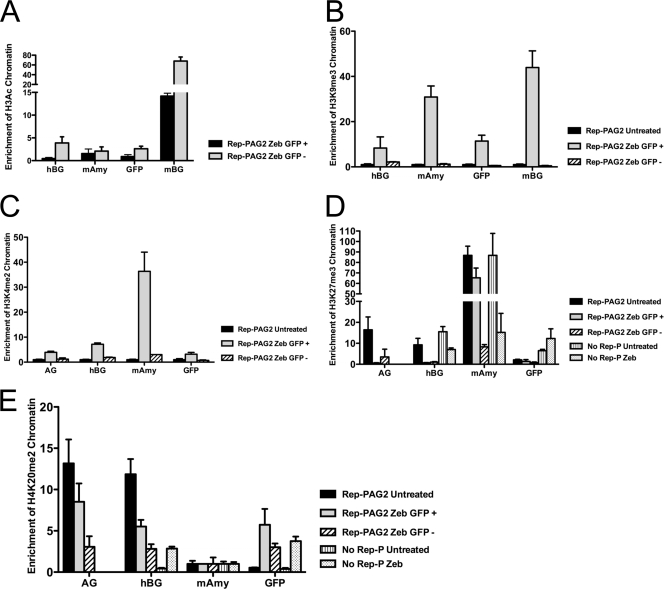
Posttranslational modification of histone H3 and H4 in silenced and reactivated transgenes.
To evaluate whether changes in histone modifications accompanied gene expression in the Rep-P variants, we performed ChIP on Rep-P-containing cassettes in RL4 cells using antibodies against modified histones. The level of acetylated histone H3 in the transgene cassette was similar in RL4/PAG2 cells when GFP expression was derepressed by zebularine (1- to 2-week exposure) or when GFP was resilenced (4- to 5-week exposure) (Fig. 8A). Thus, histone H3 acetylation did not correlate with derepression of GFP in PAG2 cells. Another chromatin marker, trimethylated lysine 9 of histone H3 (H3K9me3), also correlated with activated gene expression but did not correlate with methylation of CpG (Fig. 8B). Similarly, we observed that dimethylation of lysine 4 of histone H3 correlated with activated transgene expression (Fig. 8C). This marker, which typically accompanies nonexpressing genes, is known to serve as an active chromatin marker at the β-globin locus (30, 54).
Fig. 8.
Analysis of histone H3 and H4 modifications in the transgene cassette. ChIP analysis of acetylated histone H3 (H3Ac) (A), histone H3 with trimethylated lysine 9 (H3K9me3) (B), H3K4me2 (C), H3K27me3 (D), and H4K20me3 (E) in RL4 cells carrying the indicated constructs with or without zebularine treatment. The primers and probes used included mAmy (murine amylase), GFP (the EGFP gene), AG (the AG region of Rep-P), mBG (a murine β-globin sequence), and hBG (human β-globin promoter). GFP, AG, and hBG are sequences from the transgene. Zeb GFP +, cells treated for 1 to 2 weeks with zebularine and exhibiting expression of the EGFP marker; Zeb GFP −, cells treated for 4 to 5 weeks with zebularine and exhibiting resilencing. The results of 1 representative experiment of 3 experiments are shown.
ChIP analysis also showed H3K27me3 (H3 histone with trimethylated lysine 27) enrichment in the transgene cassette in RL4/PAG2 cells, but the enrichment was abolished by treatment with zebularine (Fig. 8D). We observed similar enrichment of H4K20me2 at the Rep-PAG2 transgene before but not after treatment with zebularine and not at transgenes lacking Rep-P (Fig. 8E). Interestingly, RL4 cells carrying LCR/EGFP without Rep-P were not H3K27me3 enriched either before or after treatment with zebularine. These data suggest that H3K27me3 and H4K20me2 may be useful markers of methylation-dependent gene silencing.
DISCUSSION
In this study, we found that the protein complex MeCP1-SWI/SNF-hnRNP C1/C2 (LARC) bound the essential asymmetric region of the Rep-P replicator. The interaction between Rep-P and MeCP1-SWI/SNF-hnRNP C1/C2 affected the ability of Rep-P to prevent gene silencing, replication delay, and CpG methylation. These results imply that the binding interaction is required to prevent LCR-mediated, methylation-dependent silencing and is essential for early replication and stable gene expression.
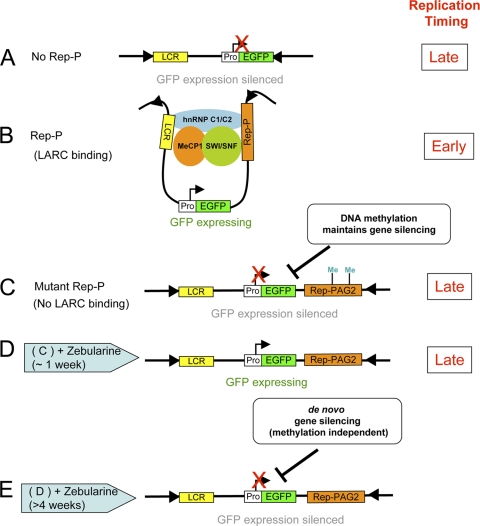
In Fig. 9 we present a model that is consistent with our observations. When inserted into a nonpermissive RL4 site, transgenes that contain LCR but no Rep-P establish a stable silenced state and replicate late (Fig. 9A), but transgenes that contain both LCR and Rep-P activate gene expression and replicate early (Fig. 9B). Persistent transcription and early replication from transgenes that contain Rep-P occur concomitant with the binding of MeCP1-SWI/SNF-hnRNP C1/C2 to the asymmetric region. Transgenes that contain Rep-P in the absence of the MeCP1-SWI/SNF-hnRNP C1/C2 complex binding site are unable to prevent silencing (Fig. 9C); gene expression is prevented, and replication is delayed. Inhibition of DNA methylation by exposure to zebularine can temporarily relieve silencing, and gene expression occurs despite the late replication of the transgene (Fig. 9D), but this is a transient effect, and resilencing can occur via a methylation-independent mechanism (Fig. 9E). In the model, binding of the MeCP1-SWI/SNF-hnRNP C1/C2 complex to Rep-P is required to prevent LCR-mediated silencing and is essential for early replication and stable gene expression.
Fig. 9.
Model for two different gene silencing mechanisms. (A) Transgene cassette without Rep-P cannot establish LCR–Rep-P interaction. The transgene is silenced by a DNA methylation-dependent maintenance gene silencing mechanism. (B) Transgene cassette with wild-type Rep-P. The LCR-AG interaction mediated by the LARC complex prevents gene silencing. (C and D) In the Rep-PAG2 cassette, GFP expression is silenced (C), but after about 1 week of zebularine treatment (D), GFP expression is activated upon DNA demethylation. (E) Gene expression is resilenced by the primary/de novo silencing mechanism after prolonged DNA demethylation.
Two distinct phenotypes are associated with the absence of the MeCP1-SWI/SNF-hnRNP C1/C2 complex–Rep-P interaction. Cells containing a transgene with no Rep-P exhibit late replication and stable silencing that is not affected by DNA demethylation (11, 19). Cells containing a transgene with a mutant Rep-P that cannot bind the MeCP1-SWI/SNF-hnRNP C1/C2 complex exhibit a similar silenced state, but in such cells demethylation can produce a transient relief of silencing and gene expression occurs despite the late replication of the transgene (Fig. 9D). Several weeks later, resilencing can occur via a methylation-independent mechanism (Fig. 9E). Reactivation of gene expression and methylation-independent resilencing are characterized by distinct posttranslational histone modifications. Thus, transgene expression can be repressed by two distinct processes, one involving DNA methylation and another that is methylation independent. Since transgenes with no Rep-P underwent irreversible, zebularine-insensitive silencing and transgenes with the mutant Rep-P that does not bind MeCP1-SWI/SNF-hnRNP C1/C2 underwent reversible, zebularine-sensitive silencing, our observations suggest that another component of Rep-P cooperates with the MeCP1-SWI/SNF-hnRNP C1/C2 complex to establish gene expression patterns.
One possible consequence of DNA methylation is interference with DNA-protein interactions at the methylated sites. In the present study, Rep-PAG2, which carried a novel CpG site created by mutagenesis, did not bind to the MeCP1-SWI/SNF-hnRNP C1/C2 complex, suggesting that methylation might directly inhibit interaction between the LCR and the Rep-P asymmetric region. Zebularine-induced demethylation of Rep-PAG2, however, did not stabilize the binding of hnRNP C1/C2, even though it transiently derepressed transgene expression. These observations imply that CpG methylation is not responsible for the loss of binding of the MeCP1-SWI/SNF-hnRNP C1/C2 complex to the asymmetric region.
DNA methylation can recruit chromatin remodeling factors (4, 53), and we found transient enhancement of the binding of modified histones H3K4me and H3K9me after demethylation of the Rep-P asymmetric region. However, in agreement with previous studies (11, 34, 36), the H3K4me- and H3K9me-enriched pattern was maintained only briefly in cells treated continuously with zebularine. Similarly, although transcriptionally active and early replicating regions of the β-globin locus in erythroid cells typically show strong enrichment for acetylated histone H3 and H4 (11, 12, 19, 21), no such enrichment was evident in nonbinding Rep-P mutant transgenes when eGFP expression was derepressed by zebularine. Taken together, these observations support the notion that histone hyperacetylation is dependent on the context (18) and does not necessarily correlate directly with gene expression and suggest that demethylation did not remodel the transgene chromatin into a euchromatic state.
We observed that wild-type Rep-P was associated with early S-phase replication while a nonbinding mutant of Rep-P that underwent reversible silencing was associated with later replication. These data suggest that the interaction between Rep-P and MeCP1-SWI/SNF-hnRNP C1/C2 is required to establish an early replication pattern and to prevent gene silencing accompanied by DNA methylation.
Early replication is often correlated with active transcription (1, 26, 35), and evidence from whole-genome high-throughput analysis suggests that in metazoans, early replicating regions of the genome tend to contain transcribed genes, whereas late-replicating regions are enriched in nontranscribed heterochromatin (25, 26, 29). In previous studies, we showed that when transgenes undergo silencing, replication delay precedes the loss of expression (19). That is consistent with our present finding that transgenes that did not bind MeCP1-SWI/SNF-hnRNP C1/C2 underwent gene silencing and replicated late. Moreover, reactivation of gene expression by demethylation did not change the timing of transgene replication, emphasizing that transcription can occur from late replicating regions and that DNA methylation is not essential for the maintenance of late replication. The observations reported here, taken together with data from previous studies, suggest that late replication can mark regions that are prone to transcriptional silencing but that replication timing and gene expression are regulated independently.
ACKNOWLEDGMENTS
We thank Victor E. Marquez for providing reagents, for invaluable discussions, and for critically reading the manuscript. We thank Naomi Heilweil, Vidushani Jayalal, Ofer Kimchi, Lucas Chang, Shikha Talwar, and Sitalaximi Thirunavukkarasu for help with plasmid construction and chromatin analyses. We thank Eric E. Bouhassira for helpful advice and critically reading the manuscript and Ofir Hakim for help with the ChIP-3C experiment.
This study was supported by the Intramural Research Program of the Center for Cancer Research, NCI, NIH.
Footnotes
Published ahead of print on 20 June 2011.
REFERENCES
- 1. Aladjem M. I. 2007. Replication in context: dynamic regulation of DNA replication patterns in metazoans. Nat. Rev. Genet. 8:588–600 [DOI] [PubMed] [Google Scholar]
- 2. Aladjem M. I., et al. 1995. Participation of the human beta-globin locus control region in initiation of DNA replication. Science 270:815–819 [DOI] [PubMed] [Google Scholar]
- 3. Aladjem M. I., et al. 2002. Replication initiation patterns in the beta-globin loci of totipotent and differentiated murine cells: evidence for multiple initiation regions. Mol. Cell. Biol. 22:442–452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bird A. 2002. DNA methylation patterns and epigenetic memory. Genes Dev. 16:6–21 [DOI] [PubMed] [Google Scholar]
- 5. van Assendelft G. Blom, Hanscombe O., Grosveld F., Greaves D. R. 1989. The beta-globin dominant control region activates homologous and heterologous promoters in a tissue-specific manner. Cell 56:969–977 [DOI] [PubMed] [Google Scholar]
- 6. Chan P. K., Wai A., Philipsen S., Tan-Un K. C. 2008. 5′HS5 of the human beta-globin locus control region is dispensable for the formation of the beta-globin active chromatin hub. PLoS One 3:e2134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6a. Clontech Laboratories, Inc 2007. Knockout single vector inducible RNAi system user manual. Clontech Laboratories, Inc, Mountain View, CA [Google Scholar]
- 7. Drouin R., Therrien J. P., Angers M., Ouellet S. 2001. In vivo DNA analysis. Methods Mol. Biol. 148:175–219 [DOI] [PubMed] [Google Scholar]
- 8. Feng Q., et al. 2002. Identification and functional characterization of the p66/p68 components of the MeCP1 complex. Mol. Cell. Biol. 22:536–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Feng Q., Zhang Y. 2001. The MeCP1 complex represses transcription through preferential binding, remodeling, and deacetylating methylated nucleosomes. Genes Dev. 15:827–832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Feng Q., Zhang Y. 2003. The NuRD complex: linking histone modification to nucleosome remodeling. Curr. Top. Microbiol. Immunol. 274:269–290 [DOI] [PubMed] [Google Scholar]
- 11. Feng Y. Q., et al. 2006. DNA methylation supports intrinsic epigenetic memory in mammalian cells. PLoS Genet. 2:e65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Feng Y. Q., Lorincz M. C., Fiering S., Greally J. M., Bouhassira E. E. 2001. Position effects are influenced by the orientation of a transgene with respect to flanking chromatin. Mol. Cell. Biol. 21:298–309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Feng Y. Q., et al. 1999. Site-specific chromosomal integration in mammalian cells: highly efficient CRE recombinase-mediated cassette exchange. J. Mol. Biol. 292:779–785 [DOI] [PubMed] [Google Scholar]
- 14. Feng Y. Q., et al. 2005. The human beta-globin locus control region can silence as well as activate gene expression. Mol. Cell. Biol. 25:3864–3874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Forrester W. C., et al. 1990. A deletion of the human beta-globin locus activation region causes a major alteration in chromatin structure and replication across the entire beta-globin locus. Genes Dev. 4:1637–1649 [DOI] [PubMed] [Google Scholar]
- 16. Forrester W. C., Novak U., Gelinas R., Groudine M. 1989. Molecular analysis of the human beta-globin locus activation region. Proc. Natl. Acad. Sci. U. S. A. 86:5439–5443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Fraser P., Grosveld F. 1998. Locus control regions, chromatin activation and transcription. Curr. Opin. Cell Biol. 10:361–365 [DOI] [PubMed] [Google Scholar]
- 18. Fromm G., et al. 2009. Histone hyperacetylation within the beta-globin locus is context-dependent and precedes high-level gene expression. Blood 114:3479–3488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Fu H., et al. 2006. Preventing gene silencing with human replicators. Nat. Biotechnol. 24:572–576 [DOI] [PubMed] [Google Scholar]
- 20. Fuks F. 2005. DNA methylation and histone modifications: teaming up to silence genes. Curr. Opin. Genet. Dev. 15:490–495 [DOI] [PubMed] [Google Scholar]
- 21. Goren A., Tabib A., Hecht M., Cedar H. 2008. DNA replication timing of the human beta-globin domain is controlled by histone modification at the origin. Genes Dev. 22:1319–1324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Grosveld F. 1999. Activation by locus control regions? Curr. Opin. Genet. Dev. 9:152–157 [DOI] [PubMed] [Google Scholar]
- 23. Grosveld F., van Assendelft G. B., Greaves D. R., Kollias G. 1987. Position-independent, high-level expression of the human beta-globin gene in transgenic mice. Cell 51:975–985 [DOI] [PubMed] [Google Scholar]
- 24. Hager G. L., McNally J. G., Misteli T. 2009. Transcription dynamics. Mol. Cell 35:741–753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hansen R. S., et al. 2010. Sequencing newly replicated DNA reveals widespread plasticity in human replication timing. Proc. Natl. Acad. Sci. U. S. A. 107:139–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hiratani I., Takebayashi S., Lu J., Gilbert D. M. 2009. Replication timing and transcriptional control: beyond cause and effect—part II. Curr. Opin. Genet. Dev. 19:142–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Horike S., Cai S., Miyano M., Cheng J. F., Kohwi-Shigematsu T. 2005. Loss of silent-chromatin looping and impaired imprinting of DLX5 in Rett syndrome. Nat. Genet. 37:31–40 [DOI] [PubMed] [Google Scholar]
- 28. Hsu M., Mabaera R., Lowrey C. H., Martin D. I., Fiering S. 2007. CpG hypomethylation in a large domain encompassing the embryonic beta-like globin genes in primitive erythrocytes. Mol. Cell. Biol. 27:5047–5054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Karnani N., Taylor C. M., Malhotra A., Dutta A. 2010. Genomic study of replication initiation in human chromosomes reveals the influence of transcription regulation and chromatin structure on origin selection. Mol. Biol. Cell. 21:393–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kim A., Kiefer C. M., Dean A. 2007. Distinctive signatures of histone methylation in transcribed coding and noncoding human beta-globin sequences. Mol. Cell. Biol. 27:1271–1279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kobayashi T., Rein T., DePamphilis M. L. 1998. Identification of primary initiation sites for DNA replication in the hamster dihydrofolate reductase gene initiation zone. Mol. Cell. Biol. 18:3266–3277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Li Q., Peterson K. R., Fang X., Stamatoyannopoulos G. 2002. Locus control regions. Blood 100:3077–3086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lin C. M., Fu H., Martinovsky M., Bouhassira E., Aladjem M. I. 2003. Dynamic alterations of replication timing in mammalian cells. Curr. Biol. 13:1019–1028 [DOI] [PubMed] [Google Scholar]
- 34. Lindahl Allen M., Koch C. M., Clelland G. K., Dunham I., Antoniou M. 2009. DNA methylation-histone modification relationships across the desmin locus in human primary cells. BMC Mol. Biol. 10:51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Machida Y. J., Hamlin J. L., Dutta A. 2005. Right place, right time, and only once: replication initiation in metazoans. Cell 123:13–24 [DOI] [PubMed] [Google Scholar]
- 36. Mahajan M. C., Karmakar S., Weissman S. M. 2007. Control of beta globin genes. J. Cell. Biochem. 102:801–810 [DOI] [PubMed] [Google Scholar]
- 37. Mahajan M. C., Narlikar G. J., Boyapaty G., Kingston R. E., Weissman S. M. 2005. Heterogeneous nuclear ribonucleoprotein C1/C2, MeCP1, and SWI/SNF form a chromatin remodeling complex at the beta-globin locus control region. Proc. Natl. Acad. Sci. U. S. A. 102:15012–15017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. McGarvey K. M., et al. 2006. Silenced tumor suppressor genes reactivated by DNA demethylation do not return to a fully euchromatic chromatin state. Cancer Res. 66:3541–3549 [DOI] [PubMed] [Google Scholar]
- 39. Morey L., et al. 2008. MBD3, a component of the NuRD complex, facilitates chromatin alteration and deposition of epigenetic marks. Mol. Cell. Biol. 28:5912–5923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Mueller P. R., Wold B., Garrity P. A. 2001. Ligation-mediated PCR for genomic sequencing and footprinting. Current protocols in molecular biology, chapter 15, unit 15.3. John Wiley & Sons, Inc, Hoboken, NJ: [DOI] [PubMed] [Google Scholar]
- 41. Noordermeer D., de Laat W. 2008. Joining the loops: beta-globin gene regulation. IUBMB Life 60:824–833 [DOI] [PubMed] [Google Scholar]
- 42. Paixão S., et al. 2004. Modular structure of the human lamin B2 replicator. Mol. Cell. Biol. 24:2958–2967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Palstra R. J., et al. 2003. The beta-globin nuclear compartment in development and erythroid differentiation. Nat. Genet. 35:190–194 [DOI] [PubMed] [Google Scholar]
- 44. Perissi V., Jepsen K., Glass C. K., Rosenfeld M. G. 2010. Deconstructing repression: evolving models of co-repressor action. Nat. Rev. Genet. 11:109–123 [DOI] [PubMed] [Google Scholar]
- 45. Reik A., et al. 1998. The locus control region is necessary for gene expression in the human beta-globin locus but not the maintenance of an open chromatin structure in erythroid cells. Mol. Cell. Biol. 18:5992–6000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Rodriguez P., et al. 2005. GATA-1 forms distinct activating and repressive complexes in erythroid cells. EMBO J. 24:2354–2366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Schubeler D., et al. 2000. Genomic targeting of methylated DNA: influence of methylation on transcription, replication, chromatin structure, and histone acetylation. Mol. Cell. Biol. 20:9103–9112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Simonis M., Kooren J., de Laat W. 2007. An evaluation of 3C-based methods to capture DNA interactions. Nat. Methods 4:895–901 [DOI] [PubMed] [Google Scholar]
- 49. Singal R., Ferris R., Little J. A., Wang S. Z., Ginder G. D. 1997. Methylation of the minimal promoter of an embryonic globin gene silences transcription in primary erythroid cells. Proc. Natl. Acad. Sci. U. S. A. 94:13724–13729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Singal R., vanWert J. M., Ferdinand L., Jr 2002. Methylation of alpha-type embryonic globin gene alpha pi represses transcription in primary erythroid cells. Blood 100:4217–4222 [DOI] [PubMed] [Google Scholar]
- 51. Singal R., Wang S. Z., Sargent T., Zhu S. Z., Ginder G. D. 2002. Methylation of promoter proximal-transcribed sequences of an embryonic globin gene inhibits transcription in primary erythroid cells and promotes formation of a cell type-specific methyl cytosine binding complex. J. Biol. Chem. 277:1897–1905 [DOI] [PubMed] [Google Scholar]
- 52. Tolhuis B., Palstra R. J., Splinter E., Grosveld F., de Laat W. 2002. Looping and interaction between hypersensitive sites in the active beta-globin locus. Mol. Cell 10:1453–1465 [DOI] [PubMed] [Google Scholar]
- 53. Tost J. 2010. DNA methylation: an introduction to the biology and the disease-associated changes of a promising biomarker. Mol. Biotechnol. 44:71–81 [DOI] [PubMed] [Google Scholar]
- 54. Vakoc C. R., Mandat S. A., Olenchock B. A., Blobel G. A. 2005. Histone H3 lysine 9 methylation and HP1gamma are associated with transcription elongation through mammalian chromatin. Mol. Cell 19:381–391 [DOI] [PubMed] [Google Scholar]
- 55. Wang L., et al. 2004. The human beta-globin replication initiation region consists of two modular independent replicators. Mol. Cell. Biol. 24:3373–3386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Wang L., Lin C. M., Lopreiato J. O., Aladjem M. I. 2006. Cooperative sequence modules determine replication initiation sites at the human beta-globin locus. Hum. Mol. Genet. 15:2613–2622 [DOI] [PubMed] [Google Scholar]