Abstract
We have identified in a skin swab sample from a healthy donor a new virus that we have named human gyrovirus (HGyV) because of its similarity to the chicken anemia virus (CAV), the only previously known member of the Gyrovirus genus. In particular, this virus encodes a homolog of the CAV apoptin, a protein that selectively induces apoptosis in cancer cells. By PCR screening, HGyV was found in 5 of 115 other nonlesional skin specimens but in 0 of 92 bronchoalveolar lavages or nasopharyngeal aspirates and in 0 of 92 fecal samples.
TEXT
The chicken anemia virus (CAV) is highly contagious and causes severe anemia, hemorrhage, and depletion of lymphoid tissue through the destruction of bone marrow progenitor cells in young chickens. CAV has been the only known species of the Gyrovirus genus, which is part of the family Circoviridae. CAV is a nonenveloped virus, with an icosahedral capsid 19 to 27 nm in diameter surrounding a single-stranded (ss) circular genomic DNA (8). In a skin swab taken from a healthy person, we discovered a new human virus that we have designated human gyrovirus (HGyV) because of its homology with the chicken anemia virus.
DNA was extracted with an automatic EasyMag apparatus (bioMérieux, Marcy l'Etoile, France) and amplified using the bacteriophage phi29 polymerase-based multiple-displacement amplification (MDA) assay and random primers. The reaction was performed using a Repli-g Midi kit (Qiagen, Courtaboeuf, France), according to the manufacturer's instructions. Sequencing was conducted using an Illumina HighSeq 2000 sequencer. High-molecular-weight DNA resulting from isothermal amplification was fragmented into 200- to 350-nucleotide (nt) fragments, to which adapters were ligated. A large number (7,588,712) of reads of 100 nt each were obtained from the sample. Illumina sequences were first sorted by a subtractive database comparison procedure as previously described (2). In short, the whole human genome sequence (NCBI build 37.1/assembly hg19) was scanned with SOAPaligner (http://soap.genomics.org.cn). A very restrictive BLASTN study was also performed to eliminate additional host reads. The CLC Genomics Workbench program was used for de novo assembly. The comparison of the results of analysis of the single reads and contigs to known genomic and taxonomic data was performed using dedicated specialized viral and generalist databases maintained locally (i.e., the entire NCBI nucleotide database or only the extracted viral part). The aforementioned databases were scanned using BLASTN and BLASTX. We used Paracel BLAST software, which distributes the BLAST tasks among multiple processors. Taxonomic assignments were based on the lowest common ancestor from the best hits among reads with a significant e-value (generally 10-e4). Among different contigs harboring a size compatible with a viral genome, a contig of 2,315 nt assembled from 436 reads was more deeply analyzed, as it disclosed similarities to CAV. Although CAV from chickens with severe anemia was described and isolated more than 30 years ago (10), no virus of this genus is currently known in humans, and this finding was thus unexpected.
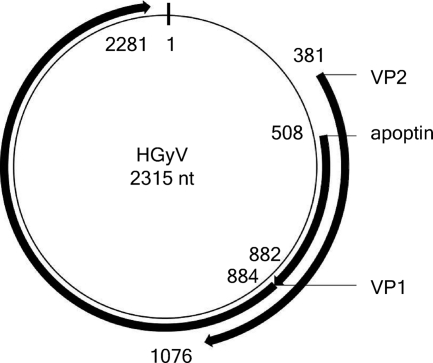
Based on the sequence of this contig, we defined a set of primers allowing amplification of the entire genome (see Table S1 in the supplemental material) and then confirmed the sequence by the Sanger method. The resulting sequence of the virus we have named HGyV (Human Gyrovirus) (accession number FR823283), along with its principal features, is presented in Fig. S1 in the supplemental material. The CAV genome consists of a single molecule of circular, single-stranded negative-sense DNA that forms a closed circle. The complete genome of CAV is 2,290 to 2,320 nucleotides long and contains three partially overlapping open reading frames encoding three proteins, VP1, VP2, and VP3. The genome of HGyV is 2,315 nt long and displays similar characteristics (Fig. 1; see also Fig. S1 in the supplemental material). Alignment of the whole genome of HGyV with that of CAV is depicted in Fig. S1 in the supplemental material. The overall nucleotide identity between the two genomes is low: in the region of maximal identity, between nt 100 and 700 (CAV accession number M55918), the overall identity is around 70%.
Fig. 1.
Map of HGyV (accession number FR823283) and principal features. Principal features are designated by homology with cognate features in the annotated sequence of CAV (accession number M55918).
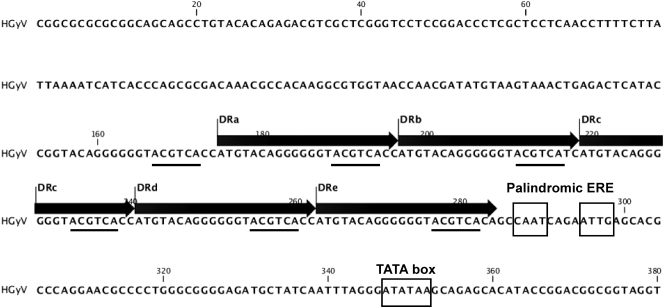
The genome of HGyV presents an overall organization similar to that of CAV. It contains a 5′ region displaying general features of the CAV promoter-enhancer (6) (see Fig. 2). In particular, the 5′ region of HGyV comprises repeated elements of 22 nt (ATGT….TCA C/T C/A), designated direct repeat a (DRa) to DRe. The DR elements of HGyV and CAV contain the sequences ACGTCA and AGCTCA, respectively, which both differ by a single nucleotide from the estrogen-responsive element (ERE) half-site (A)GGTCA. In HGyV, there is also a less-conserved repetition just upstream of DRa (C/A G/T GTACAGGGGGGTACGTCA T/C C/A at positions nt 153 to 174; note that this sequence also contains the ACGTCA sequence [not shown in Fig. 2]). In HGyV, these DRs are repeated without intervening sequences. In CAV, 4 to 5 (depending on the strain) copies of these repeats are present and are separated by linkers of 15 nt between the first two or three elements and the last two. While we did not find any evidence of the presence of the ERE found in the vicinity of nt 50 in CAV, we identified a palindromic sequence (CAATCAGAATTG) downstream from DRe that shows a certain similarity to it. Indeed, while EREs generally consist of pentamers or hexamers such as (A)GGTCA separated by a 2- to 3-nt insert, this consensus sequence has been shown to occur in a wide range of variations (1).
Fig. 2.
Nucleotide sequence of the promoter region of HGyV and principal features (see text). Direct repeats (DRa to DRe) of 22 nt are shown with putative estrogen-responsive elements (ACGTCA; underlined). The palindromic CAAT CAGA ATTG putative estrogen-responsive element and the TATA box are depicted.
Like CAV, the HGyV genome contains three partially overlapping open reading frames encoding different proteins. We have used the names of the CAV counterparts to identify those open reading frames (see details in Table S2 in the supplemental material). The CAV VP1 is a 51-kDa capsid protein that also contains motifs for rolling-circle replication in the C-terminal region. Given the distance from the other ss DNA viruses, phylogenetic analysis was impossible. CAV VP3, a nonstructural protein, is also called apoptin. The alignment of HGyV apoptin with CAV apoptin is shown in Fig. 3, together with the domains of CAV apoptin that have been shown to be functionally important (see review in reference 4). The overall identity of HGyV apoptin with CAV apoptin is relatively low (31%).
Fig. 3.
Comparison of the primary structures of CAV and HGyV apoptin. HGyV apoptin (H-apoptin) was aligned with its CAV homolog (A-apoptin) (accession number P54094) by the use of the CLC Genomics Workbench program. Principal domains of the CAV apoptin protein are shown (LRS, leucine-rich domain; NLS1 and NLS2, nuclear localization signals; NES, putative nuclear exportation signal). The amino acid Thr in position 108 of CAV apoptin is depicted by an arrow. The phosphorylation site at amino acid Thr-111 of HGyV apoptin predicted by the Netphos program (www.cbs.dtu.dk/services/NetPhos/) is also shown.
The CAV apoptin alone can induce apoptosis in a broad range of cancer cells but not in nontransformed or primary cells (4). It induces apoptosis by a mechanism implicating the mitochondrial (intrinsic) pathway and is thus independent of the death receptor (extrinsic) pathway. As shown in Fig. 3, the so-called N-terminal leucine-rich stretch (LRS), characterized by a high content of hydrophobic aliphatic leucine or isoleucine residues, seems conserved. This region interacts with different cellular proteins that seem important for its function (summarized in reference 4). A bipartite nuclear localization signal (NLS), comprising motifs NLS1 and NLS2, also seems to be conserved, together with the putative nuclear exportation signal (NES). Of note, the Netphos 2.0 server predicts a phosphorylation site at position 111 of HGyV apoptin, which is located between NES and NLS2 and seems homologous to position Thr-108 of CAV apoptin. In fact, nuclear accumulation of CAV apoptin, which is important for its proapoptotic activity, is dependent on the phosphorylation of Thr-108. This phosphorylation is mediated by a kinase (cyclin-dependent kinase 2) that can be constitutively active in tumors and transformed cells but not in normal cells (5). Apoptin expression is able to induce cell cycle arrest and apoptosis in p53 null cells (4) and is a very attractive candidate for use in cancer therapy. Nevertheless, CAV apoptin is derived from a virus restricted to birds. It can therefore be anticipated that HGyV apoptin would be more potent in mammals than its avian counterpart.
Since this new virus was first identified in a skin swab sample, we have investigated HGyV prevalence in a set of 115 nonlesional skin specimens sampled by cutaneous swabbing from volunteer subjects by a nested-PCR assay targeting the VP1 gene (see Table S3 in the supplemental material). Consent was obtained for collection of human samples according to French regulations. Subjects included 35 patients hospitalized or attending outpatient clinics at the dermatology unit of the Montpellier university hospital, 20 immunocompromized patients (including 10 HIV-1-infected patients and 10 renal transplant recipients), and 60 healthy control subjects enrolled within the Montpellier university hospital staff. Since respiratory and fecal-oral routes of transmission could be involved in the epidemiology of the virus, we also tested additional sets of human samples, including 46 bronchoalveolar lavage (BAL) fluid samples from intensive care unit patients and 46 nasopharyngeal aspirates plus 92 fecal samples from children attending the pediatric unit of the Montpellier university hospital. Caution was exercised to limit the risk of cross-contamination. HGyV was detected in five additional skin samples, and specificity was confirmed by sequencing the amplicons. Four of these HGyV-positive samples were recovered from healthy volunteers and one from an HIV-positive patient. We failed to detect HGyV sequences in both respiratory and stool samples. Our results show that HGyV is a putative member of the human skin virome detected in nearly 5% of individuals and is reminiscent of its close relative CAV, which is transmitted by contaminated feathers (3).
The importance of HGyV could be 2-fold. First, the potential association of HGyV with disease needs to be determined. Also, the description of a HGyV apoptin provides a promising lead for development of biotherapy for cancer in human and animals (7). It may also serve as a tool to decipher pathways of apoptosis in cancer cells. We are currently exploring these different aspects of HGyV biology.
Nucleotide sequence accession number.
The sequence of HGyV has been deposited in the NCBI database under accession number FR823283.
Supplementary Material
Acknowledgments
The platform “Genotyping of Pathogens and Public Health” is supported in part by the Institut de Veille Sanitaire (Saint-Maurice, France). This study was partly supported by grants from both region Ile de France and the Programme Hospitalier de Recherche Clinique of the Montpellier University Hospital (AOI 2008, Protocole UF8425).
Footnotes
Supplemental material for this article may be found at http://jvi.asm.org/.
Published ahead of print on 1 June 2011.
REFERENCES
- 1. Aumais J. P., Lee H. S., DeGannes C., Horsford J., White J. H. 1996. Function of directly repeated half-sites as response elements for steroid hormone receptors. J. Biol. Chem. 271:12568–12577 [DOI] [PubMed] [Google Scholar]
- 2. Cheval J., et al. Evaluation of high-throughput sequencing for identifying known and unknown viruses in biological samples. J. Clin. Microbiol., in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Davidson I., et al. 2008. The contribution of feathers in the spread of chicken anemia virus. Virus Res. 132:152–159 [DOI] [PubMed] [Google Scholar]
- 4. Los M., et al. 2009. Apoptin, a tumor-selective killer. Biochim. Biophys. Acta 1793:1335–1342 [DOI] [PubMed] [Google Scholar]
- 5. Maddika S., et al. 2009. Unscheduled Akt-triggered activation of cyclin-dependent kinase 2 as a key effector mechanism of apoptin's anticancer toxicity. Mol. Cell. Biol. 29:1235–1248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Miller M. M., Jarosinski K. W., Schat K. A. 2005. Positive and negative regulation of chicken anemia virus transcription. J. Virol. 79:2859–2868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Noteborn M. H. M. 2009. Proteins selectively killing tumor cells. Eur. J. Pharmacol. 625:165–173 [DOI] [PubMed] [Google Scholar]
- 8. Schat K. A. 2009. Chicken anemia virus. Curr. Top. Microbiol. Immunol. 331:151–183 [DOI] [PubMed] [Google Scholar]
- 9. Reference deleted.
- 10. Yuasa N., Taniguchi T., Yoshida I. 1979. Isolation and some characteristics of an agent inducing anemia in chicks. Avian Dis. 23:366–385 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.