Abstract
Iridoviruses (IV) are nuclear cytoplasmic large DNA viruses that are receiving increasing attention as sublethal pathogens of a range of insects. Invertebrate iridovirus type 9 (IIV-9; Wiseana iridovirus) is a member of the major phylogenetic group of iridoviruses for which there is very limited genomic and proteomic information. The genome is 205,791 bp, has a G+C content of 31%, and contains 191 predicted genes, with approximately 20% of its repeat sequences being located predominantly within coding regions. The repeated sequences include 11 proteins with helix-turn-helix motifs and genes encoding related tandem repeat amino acid sequences. Of the 191 proteins encoded by IIV-9, 108 are most closely related to orthologs in IIV-3 (Chloriridovirus genus), and 114 of the 126 IIV-3 genes have orthologs in IIV-9. In contrast, only 97 of 211 IIV-6 genes have orthologs in IIV-9. There is almost no conservation of gene order between IIV-3, IIV-6, and IIV-9. Phylogenetic analysis using a concatenated sequence of 26 core IV genes confirms that IIV-3 is more closely related to IIV-9 than to IIV-6, despite being from a different genus of the Iridoviridae. An interaction between IIV and small RNA regulatory systems is supported by the prediction of seven putative microRNA (miRNA) sequences combined with XRN exonuclease, RNase III, and double-stranded RNA binding activities encoded on the genome. Proteomic analysis of IIV-9 identified 64 proteins in the virus particle and, when combined with infected cell analysis, confirmed the expression of 94 viral proteins. This study provides the first full-genome and consequent proteomic analysis of group II IIV.
INTRODUCTION
Iridoviruses (IV) are members of the nucleocytoplasmic large DNA viruses (NCLDV) (19). They possess a linear double-stranded DNA (dsDNA) genome with circular permutation and terminal redundancy (6, 13), and replication of the viral genome includes distinct nuclear and cytoplasmic phases (12). The genomes are encapsidated within an icosahedral shell ranging between 120 and 180 nm in diameter and comprised predominantly of a 50-kDa major capsid protein (MCP). The invertebrate iridoviruses (IIV), studied by cryo-electron microscopy, have 2-nm-diameter surface fibrils (23, 42); for invertebrate iridovirus type 6 (IIV-6), these fibrils extend from the 3-fold rotational axis of the 1,460 hexameric capsids found in the virus particle (43). IV are divided into 5 genera (Table 1), with members of three genera infecting poikilothermic vertebrates and members of the Iridovirus and Chloriridovirus genera infecting invertebrates. The Chloriridovirus genus has only one member, IIV-3 (mosquito iridovirus), and the primary defining differences between the Chloriridovirus and Iridovirus genera are particle sizes of approximately 180 and 135 nm, respectively, and the mosquito host range restriction of IIV-3 (4).
Table 1.
Fully sequenced genomes from vertebrate and invertebrate iridoviruses
| Genus and virusa | Genome size (bp) | % G+C | ORFb | Coding density (%) | Protein size range (aa) | GenBank accession no. | Reference |
|---|---|---|---|---|---|---|---|
| Iridovirus | |||||||
| IIV-9 | 205,791 | 31 | 191 | 90 | 50–2,051 | GQ918152 | This study |
| IIV-6 | 212,482 | 29 | 243c | 85 | 40–2,432 | AF303741 | Jakob et al. (20) |
| Chloriridovirus | |||||||
| IIV-3 | 191,132 | 48 | 126 | 68 | 60–1,377 | DQ643392 | Delhon et al. (5) |
| Lymphocystivirus | |||||||
| LCDV-1 | 102,653 | 29 | 110 | 82 | 40–1,199 | L63545 | Tidona and Darai (35) |
| LCDV-C | 186,250 | 27 | 240 | 67 | 40–1,193 | AY380826 | Zhang et al. (46) |
| Ranavirus | |||||||
| TFV | 105,057 | 55 | 105 | 94 | 40–1,294 | AF389451 | He et al. (17) |
| ATV | 106,332 | 54 | 96 | 79 | 32–1,294 | AY150217 | Jancovich et al. (21) |
| FV-3 | 105,903 | 55 | 98 | 80 | 50–1,293 | AY548484 | Tan et al. (34) |
| STIV | 105,890 | 55 | 105 | 80 | 40–1,294 | EU627010 | Huang et al. (18) |
| SGIV | 140,131 | 49 | 162 | 98 | 41–1,268 | AY521625 | Song et al. (32) |
| GIV | 139,793 | 49 | 120 | 83 | 62–1,268 | AY666015 | Tsai et al. (36) |
| Megalocytivirus | |||||||
| ISKNV | 111,362 | 55 | 124 | 93 | 40–1,208 | AF371960 | He et al. (16) |
| RBIV | 112,080 | 53 | 118 | 86 | 50–1,253 | AY532606 | Do et al. (7) |
| RSIV | 112,414 | 53 | 93 | 86–1,309 | BD143114 | Kurita et al. (25) | |
| OSGIV | 112,636 | 54 | 121 | 91 | 40–1,168 | AY894343 | Lu et al. (26) |
IIV-3, invertebrate iridescent virus type 3; IIV-6, invertebrate iridescent virus type 6; LCDV-1, lymphocystis disease virus 1; LCDV-C, lymphocystis disease virus, China strain; TFV, tiger frog virus; ATV, ambystoma tigrinum virus; FV-3, frog virus 3; STIV, soft-shelled turtle iridovirus; SGIV, Singapore grouper iridovirus; GIV, grouper iridovirus; ISKNV, infectious spleen and kidney necrosis virus; RBIV, rock bream iridovirus; RSIV, red sea bream iridovirus; OSGIV, orange spotted grouper iridovirus.
Essentially nonoverlapping ORF encoding a minimum length of 40 to 62 aa.
Revised annotation of Eaton et al. (8).
The vertebrate IV cause disease in fish, amphibians, and reptiles and have received considerable attention due to their effects upon aquaculture. In contrast, the IIV cause predominantly subpathogenic infections, and their consequently limited utility for pest control has meant that less is known about IIV. Of particular importance has been the recent study of Bromenshenk et al. (3) linking colony collapse disorder in honey bees to coinfection with Nosema and an unidentified iridovirus(es). A strong causal relationship was established; however, the identity of the IV was not established, at least in part due to a lack of IIV genomic information. In addition, the refraction of light by assemblies of IIV particles offers new opportunities in materials development (23, 28) that would benefit from more information on the virus particle and its constituents. The roles of viral proteins, such as the surface fiber, in iridescence are unknown, and the proteins and functional activities associated with the virus particle remain to be elucidated. Central to this is the need for information on IIV genomes and the proteomic analysis of the virus particle.
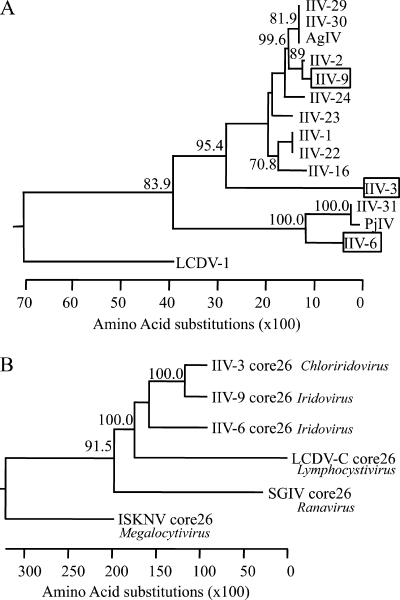
Fourteen iridovirus species have been fully sequenced (Table 1), with multiple members of the Ranavirus, Lymphocystivirus, and Megalocytivirus genera providing a comprehensive coverage of these vertebrate genera of IV. Vertebrate IV genomes range from 105 kbp for tiger frog virus (17) to 186 kbp for lymphocystis disease virus, China strain (LCDV-C) (46). The Ranavirus and Megalocytivirus species have G+C contents of approximately 50%, while the Lymphocystivirus species have G+C contents of less than 30%. There is a consistent lack of genome colinearity between IV except with very closely related isolates, although all IV sequenced to date possess a core cohort of 26 conserved genes (8). In contrast to the vertebrate IV, the only fully sequenced IIV are IIV-6 (Chilo iridovirus [CIV]) (20) and IIV-3 (mosquito chloriridovirus [MIV]) (5). IIV-6 is the type species of the Iridovirus genus, with a genome of 212 kbp and a G+C content of 29%; however, phylogenetic studies show that IIV-6 belongs in a clade distant from that of most iridoviruses (Fig. 1 A) (38). IIV-3, with a genome of 191 kbp and a G+C content of 48%, represents a different genus that may be more closely related to members of the Iridovirus genus than its placement in a separate genus suggests.
Fig. 1.
Phylogenetic trees of iridoviruses. (A) Alignment of the genus Iridovirus based upon a partial major capsid protein sequence as described in the work of Webby and Kalmakoff (38). The Chloriridovirus IIV-3 and the Lymphocystivirus LCDV-1 are included. (B) Alignment of representatives of the five iridovirus genera Chloriridovirus (IIV-3), Iridovirus (IIV-6, IIV-9), Lymphocystivirus (LCDV-1), Ranavirus (ISKNV), and Megalocytivirus (SGIV) based upon a concatenated amino acid sequence of the 26 core iridovirus genes. Bootstrap support from 1,000 iterations is indicated for all branches, with at least 70% support.
To date, only limited sequence information is available from members of the major clade of IIV, defined as group II iridoviruses by Williams and Cory (41), and genome analysis of a member of this clade would provide information on the relationships between disparate IIV. IIV-9 (Wiseana iridovirus [WIV]), a representative of the major clade, was isolated in New Zealand from larvae of the pasture pest Wiseana spp. (Lepidoptera: Hepialidae) (9). The mechanism of transmission of this virus is unknown, though the presence of this virus in damp and cryptic habitats is consistent with many other IIV (40), and suggestions of vector transmission have been made, though not confirmed. Like most invertebrate iridoviruses, IIV-9 replicates in larvae of the greater wax moth Galleria mellonella upon injection, and heavily infected larvae display typical iridescence upon accumulation of paracrystalline arrays of virus particles within infected tissues (9). IIV-9 also replicates in Spodoptera frugiperda (Sf9, Sf21) cells, albeit at the restricted temperature of 21°C. IIV-9 is a member of the major clade of IIV, as determined by partial major capsid protein phylogeny (38).
This study presents the complete genomic sequence of IIV-9 and uses this information for proteomic analysis of IIV-9's encoded proteins in purified virus particles and within infected cells. Analysis of the genome indicates that IIV-9 is more closely related to IIV-3 than to IIV-6 and provides the first complete genome from the major clade of invertebrate iridoviruses.
MATERIALS AND METHODS
IIV-9 purification, DNA extraction, and sequencing.
Sf21 cells were grown in SF900II serum-free medium (Invitrogen, Auckland, New Zealand) and infected with dilutions of a field isolate of IIV-9 that had been passaged repeatedly through G. mellonella larvae. Infected cells were incubated for 5 days at 21°C under an agarose overlay and stained with neutral red. Individual plaques were picked and passaged once in cell culture. One plaque isolate was randomly selected, propagated in G. mellonella, and purified on sucrose gradients as described previously (23). Genomic DNA was extracted by phenol-chloroform extraction (37), and 50 μl (100 ng μl−1) of genomic DNA in deionized water was sequenced using the Roche/454 GS FLX High Throughput Sequencing Service provided by the Department of Anatomy and Structural Biology, University of Otago. All contig junctions were determined by sequencing of available restriction fragment clones or by PCR. Briefly, primers were designed near the termini of contigs and used with primers on adjacent contigs to generate PCR products directly from genomic DNA using the Expand high-fidelity PCR kit (Roche Diagnostics, Auckland, New Zealand). The PCR products were either sequenced directly at the Allan Wilson Sequencing Centre, Palmerston North, New Zealand, on an ABI 3730 automated sequencer or cloned into pGEMTeasy (Promega Corp., Madison, WI) prior to sequencing. Sequence conflicts, long repeats, and long runs of single nucleotides were confirmed by PCR and sequencing of the region in question. All ABI 3730-generated sequences were edited in SeqMan (DNAStar) for sequence quality prior to use.
Sequence analysis.
Newbler Assembler software (454 Life Sciences, Branford, CT) was used to assemble data into unordered and unoriented contigs (default settings). The contigs were exported to the SeqMan program in the Lasergene suite of DNA analysis programs (DNAStar, Madison, WI) and reassembled into a draft alignment using the SeqMan assembler (match size, 12; minimum match percentage, 80%; minimum sequence length, 100; maximum number of added gaps per kb in the contig, 70; maximum number of added gaps per kb in the sequence, 70; maximum register shift difference, 70; last group considered, 2; gap penalty, 0.00; gap length penalty, 0.70). All contigs were aligned to generate a draft alignment with a minimum match percentage of 95%. PCR primers were designed using PrimerDesign (DNAStar). An in silico analysis of the restriction profile of the complete genome was performed using GeneQuest (DNAStar), and results were compared to published restriction profiles of IIV-9 genomic DNA as a confirmation of the assembly profile.
Tandem repeats within the IIV-9 genome were identified using Tandem Repeats Finder (2), with parameters set for match and mismatch and indels equal to 2, 7, and 7, respectively. The minimum alignment score was set at 50, with a maximum period size of 2,000 bases. Direct, inverted, and dyad repeats were identified using GeneQuest (DNAStar) with an unlimited loop size. The minimum period sizes set for direct, inverted, and dyad repeats were 25 bp, 25 and 50 bp, and 16 bp, respectively. Dot plot analysis was performed to identify DNA repeat clusters using MegAlign (DNAStar), with a window size of 50 bp and a 75% match.
Open reading frames (ORF) encoding proteins with a minimum size of 50 amino acids (aa) and that contained a start codon were designated using SeqBuilder (DNAStar). All designated ORF were named with “orf” followed by numbers corresponding to their position and a forward/reverse (right [R]/left [L], respectively) designation to indicate their orientation. ORF that fell completely within a larger ORF were excluded. Heavily overlapping open reading frames where the most likely ORF could not be determined were given the same number but different orientations. All designated IIV-9 open reading frames were exported from SeqBuilder (DNAStar) to EditSeq (DNAStar), and BLASTP analysis of the predicted amino acid sequences was performed for each open reading frame. Amino acid identity to the closest BLASTP match was performed using MegAlign (DNAStar). Analysis of IIV-9 ORF function was performed via the ExPASy Proteomics Server and included InterProScan, SignalP, and PredictProtein. Protein repeats were identified using the XSTREAM prediction server (27), and subsequent alignments of protein repeats were generated using MegAlign.
A phylogenetic tree was constructed based on the alignment of the 26 core gene amino acid sequences found in IIV-9, IIV-6, IIV-3, Singapore grouper iridovirus (SGIV), lymphocystis disease virus 1 (LCDV-1), and infectious spleen and kidney necrosis virus (ISKNV) using MegAlign (DNAStar), with bootstrap trials set at 1,000. All core gene-encoded proteins were combined as one continuous amino acid sequence in the same gene order prior to assembly. This was compared to the partial major capsid protein tree as described in the work of Webby and Kalmakoff (38).
The complete genome was scanned for miRNA coding regions using VMir (14, 33) and possible miRNA coding sequences further analyzed by MiPred (22). All images were generated using Microsoft PowerPoint and/or Adobe Photoshop CS4.
MS analysis.
Purified IIV-9 virions or infected Sf21 cells harvested 24 h postinfection were denatured in SDS-PAGE sample buffer, and proteins were separated on individual 10% SDS-PAGE gels using standard techniques. The gels were stained with Coomassie G250 and protein lanes cut into five (for liquid chromatography coupled with electrospray ionization linear ion trap [LC-ESI LTQ] Orbitrap tandem mass spectrometry [MS/MS] analyses of IIV-9 virions and infected Sf21 cells) or eight (for LC–matrix-assisted laser desorption ionization–tandem time of flight [MALDI TOF/TOF] analysis of IIV-9 virions) equally sized fractions. Fractions were subjected to in-gel protein digestion with trypsin essentially by following the protocol of Shevchenko et al. (30), using a liquid handling robotic workstation (DigestPro MSi; Intavis AG, Cologne, Germany). Each digested fraction was concentrated using a centrifugal vacuum concentrator and reconstituted in a 10-μl aqueous solution of 2% (vol/vol) acetonitrile (ACN) supplemented with either 0.1% (vol/vol) trifluoroacetic acid (TFA) for LC-MALDI TOF/TOF analyses or 0.2% formic acid for LC-ESI LTQ Orbitrap analyses.
Structural proteins from purified IIV-9 virions were analyzed by LC-MALDI TOF/TOF MS and LC-ESI LTQ Orbitrap MS/MS, and proteins from infected Sf21 cells were analyzed by LC-ESI LTQ Orbitrap MS/MS according to the details of methods described in the supplemental material.
Peak lists were processed through the 4000 series Explorer software (Applied Biosystems, MA) for MALDI TOF/TOF data and the Proteome Discoverer 1.1 software (Thermo Scientific, San Jose, CA) for all ESI LTQ Orbitrap data using the software's default settings. All peak lists were then searched with an in-house Mascot server (version 2.1.0; Matrix Science) against an amino acid sequence database combining all predicted and translated IIV-9 ORF and all entries from the NCBI nonredundant sequence database, matching the taxa Lepidoptera and Drosophila melanogaster (downloaded January 2011; 355,290 sequence entries). Mascot search settings allowed for full tryptic peptides with up to 3 missed cleavage sites and variable modifications of carbamidomethyl (C), oxidation (M), and pyroglutamic acid (E, Q). The precursor and fragment mass tolerances were set to ±10 ppm and 0.8 Da for LTQ Orbitrap data and 75 ppm and 0.4 Da for TOF/TOF data. To evaluate the false-discovery rate (FDR), all peak lists were searched against a decoy database using identical search settings. The decoy database was built using the decoy database tool at the Trans-Proteomic Pipeline (TPP; Seattle Proteome Center), comprising the reversed sequence entries of the aforementioned combined database. The FDR was calculated by determining the number of false-positive peptide hits from the decoy search versus the number of peptide identifications from the true search using the same Mascot score as a significance threshold.
Only peptide hits with an individual ion score of >40 (Mascot significance threshold at a P of <0.05) were accepted as significant identifications. This resulted in an FDR of <0.02 for all searches. A significant protein identification required at least two significant peptide hits covering different sequences of the protein. In addition, a protein that was identified by a single peptide-based protein identification in one experiment (IIV-9 particles analyzed by LC-MALDI TOF/TOF or LC-ESI LTQ Orbitrap MS/MS or infected cells analyzed by LC-ESI LTQ Orbitrap MS/MS) that was also confirmed by a different peptide identification covering another sequence stretch in one of the other experiments was considered a significant multipeptide identification.
Nucleotide sequence accession number.
The IIV-9 genome has been deposited in GenBank under accession number GQ918152.
RESULTS AND DISCUSSION
Genome assembly and properties.
Sequencing of the IIV-9 genome using a 454 FLX sequencer generated 20,734 sequences totaling 5,597,884 bases of sequence with 50.4% and 49.6% sequence orientation biases, for an average coverage of 27-fold. The initial Newbler assembly generated 3 large and 10 small contigs that were subsequently assembled into a single contiguous sequence by targeted PCR-based cloning and sequencing. The initial contig boundaries were defined by repeat sequences that the assembler was unable to resolve. The genome was shown to be 205,791 bp in size, with a G+C content of 31% (Table 1). This genome size compares to estimates of 192.5 and 222.6 kbp, as estimated by restriction profiles using standard (37) and pulsed-field gel (39) electrophoresis, respectively. Based upon an estimated 4.7% terminal redundancy in the IIV-9 genome (39), this equates to approximately 9.7 kbp of redundant sequence. Due to the high A+T content in the genome and the challenge of resolving long single base runs using 454 technology, a total of 34 PCR-based clones were generated to resolve 57 potential base conflicts. All base calls were inspected visually and resolved as necessary. In silico restriction endonuclease profiles were compared to experimentally derived restriction endonuclease profiles (37) to confirm correct global assembly of the genomic sequence (data not shown).
Genome analysis identified a range of complex repeat sequences, including tandem, direct, dyad, and inverted repeats. The percentage of repeat sequences in the genome is dependent upon the stringency of parameters employed and ranges from 20 to 23% of the genome. The largest repeat identified is 3.4 copies of a 1,002-bp repeat between nucleotides 69621 and 73024, with a 75% consensus match. Identification of repeat sequences on the genome is illustrated by dot plot analysis (Fig. 2 A). The repeats highlighted in the boxed region of the genome shown in Fig. 2A represented 10 of the contigs generated in the initial sequence assembly.
Fig. 2.
Sequence repeats and IIV-9 versus IIV-3 gene parity plot analysis. (A) The IIV-9 genome was compared against itself by dot plot analysis to identify repeats within the IIV-9 genome. The major clusters of sequence repeats are boxed. (B) The IIV-9 and IIV-3 gene orders were compared by parity plot analysis. Where three or more genes are contiguous in both genomes, regardless of orientation, they have been boxed. Numbers on the x and y axes represent ORF numbers.
IIV-9 ORF and their predicted protein products.
Analysis of the complete genome predicted 191 predominantly nonoverlapping ORF encoding proteins of 50 aa or more in length with an AUG start codon, with the genome displaying a coding density of 90% (Fig. 3). The genome shows a bias in that 63% of genes were oriented in the reverse direction. In conjunction with the genome analysis, we conducted a proteomic analysis, first to confirm expressed ORF and second to establish the first profile of expressed IIV-9 ORF in both isolated virions and infected insect cells. Of the total of 191 ORF, 94 were identified in either isolated virions (64 ORF detected) or infected insect cells (72 ORF detected), with 42 being expressed in both (Table 2; see also Tables S1 to S3 in the supplemental material). The number of expressed proteins in isolated virions roughly correlates with 44 identified proteins in a previous proteomic study of SGIV particles (31). Open reading frames that are discussed in the following paragraph are marked with a superscript “p” if their expression has been confirmed by proteomics of isolated virions or with a superscript “i” if they were confirmed to be protein products in infected insect cells.
Fig. 3.
Open reading frame map of the IIV-9 genome. The 205,791-bp IIV-9 genome is represented as a solid line, and predicted open reading frames are indicated with arrows. Arrows representing genes in the forward (right) direction are stippled, and those in the reverse (left) direction are open. The 26 core IV genes are indicated with bold ORF numbering. Arrows with a bold outline are HTH 7 domain-containing orthologs of orf091L of IIV-3, and those with a broken outline are orthologs of IIV-6 468L. orf061R/L and -139R/L are almost fully overlapping genes facing in opposite directions and have been represented as both R and L forms.
Table 2.
IIV-9 predicted open reading frames
| IIV-9 ORFa | Nucleotide positions | Length (aa) | Best match(es)b |
Predicted motif and/or functione | |||
|---|---|---|---|---|---|---|---|
| IIV protein(s)c | GenBank accession no. | BLASTP score | % aa identityd | ||||
| 001R | 30–1223 | 397 | IIV-3 004R; IIV-6 067R | YP_654576 | 466 | 62.6 | |
| 002R | 1238–1516 | 92 | Signal peptide | ||||
| 003R | 1741–1980 | 79 | |||||
| 004L | 2999–2037 | 320 | IIV-3 005L | YP_654577 | 62 | 31.7 | Signal peptide, RING finger |
| 005R | 3158–4705 | 515 | IIV-3 006R; IIV-6 118L | YP_654578 | 582 | 56.9 | NCLDV membrane protein |
| 006L | 6016–4757 | 419 | IIV-6 468L* | NP_149463 | 285 | 44.9 | Helix-turn-helix 7 motif |
| 007R | 6133–6459 | 108 | IIV-6 248R; PBCV N288R | NP_149711 | 70 | 44.1 | Transmembrane |
| 008R | 6587–7456 | 289 | IIV-6 404L | NP_149867 | 102 | 32.5 | |
| 009Lf | 8013–7519 | 164 | IIV-22 15.9 kDa; IIV-3 15R | P25097 | 134 | 48.6 | 15.9-kDa protein, 5′ MCP gene |
| 010R | 8235–9689 | 484 | IIV-9-MCP; IIV-1 MCP; IIV-3 014L; IIV-6 274L | O39163 | 987 | 100.0 | MCP |
| 011L | 9967–9782 | 61 | |||||
| 012R | 10245–11540 | 431 | MAR 344; IIV-6 273R | NP_149736 | 108 | 47.0 | |
| 013R | 11545–11865 | 106 | Transmembrane | ||||
| 014L | 12780–11935 | 281 | IIV-6 219L; IIV-3 036R,091L | NP_149682 | 239 | 51.8 | |
| 015R | 13053–13316 | 87 | IIV-3 013L | YP_654585 | 84 | 55.8 | Signal peptide |
| 016R | 13458–16700 | 1080 | IIV-3 035R; IIV-6 179R | YP_654607 | 1154 | 51.2 | Tyr protein kinase-like domain |
| 017R | 16743–17156 | 137 | IIV-3 055R; IIV-6 349L | YP_654627 | 148 | 54.1 | TF IIS C-terminal domain |
| 018R | 17457–17726 | 89 | IIV-3 019R | YP_654591 | 55 | 52.8 | Bro-N domain |
| 019L | 19158–17881 | 425 | IIV-3 069L; IIV-6 198R | YP_654641 | 389 | 49.9 | |
| 020R | 19213–20130 | 305 | Yersinia ruckeri chitinase | ZP_04617184 | 352 | 57.2 | Chitinase, family 18 glycohydrolase |
| 021R | 20183–20626 | 147 | IIV-3 057L | YP_654629 | 44 | 25.6 | |
| 022R | 20680–21762 | 360 | IIV-3 056L; IIV-6 287R; MAR 339 | YP_654628 | 301 | 44.7 | Putative phosphodiesterase |
| 023L | 23370–21841 | 509 | IIV-6 380R; IIV-3 010L,011L | NP_149843 | 367 | 45.0 | Serine/threonine protein kinase |
| 024R | 23480–23920 | 146 | IIV-6 293R | NP_149756 | 123 | 44.9 | |
| 025R | 24044–25165 | 373 | IIV-3 012R; IIV-6 302L | YP_654584 | 322 | 47.6 | C2H2 Zn finger |
| 026R | 25236–26552 | 438 | IIV-6 468L*; IIV-3 093 | NP_149463 | 306 | 44.9 | Helix-turn-helix 7 motif |
| 027L | 26619–27086 | 155 | IIV-3 085L; IIV-6 325L | YP_654657 | 188 | 58.4 | Signal peptide |
| 028R | 27029–27229 | 66 | Transmembrane | ||||
| 029R | 27260–28093 | 277 | IIV-3 054L | YP_654626 | 243 | 48.0 | |
| 030Rf | 28156–28518 | 120 | IIV-3 102R; IIV-6 122R | YP_654674 | 119 | 55.0 | |
| 031R | 28627–29910 | 427 | IIV-3 047R; IIV-6 337L | YP_654619 | 446 | 66.3 | Transmembrane |
| 032Lf | 31068–30394 | 224 | IIV-3 021L | YP_654593 | 194 | 55.6 | C3HC4 RING finger/BIR |
| 033L | 31730–31113 | 205 | IIV-3 022L | YP_654594 | 142 | 45.4 | Transmembrane |
| 034L | 32649–31816 | 277 | IIV-3 101R; IIV-6 142R | YP_654673 | 430 | 75.8 | RNase III |
| 035L | 34117–32813 | 434 | IIV-6 468L* | NP_149463 | 295 | 43.8 | Helix-turn-helix 7 motif |
| 036Lf | 34736–34176 | 186 | IIV-3 104L; IIV-6 355R | YP_654676 | 271 | 66.7 | Phosphatase |
| 037R | 34846–35577 | 243 | IIV-3 105R; IIV-6 359L | YP_654677 | 330 | 65.3 | |
| 038L | 37133–35655 | 492 | IIV-6 159L,219L,261R,443R; IIV-3 091L,36R | NP_149622 | 135 | 33.3 | |
| 039L | 38695–37220 | 491 | IIV-6 159L,219L,261R,443R; IIV-3 091L | NP_149622 | 151 | 36.1 | |
| 040R | 38826–40250 | 474 | IIV-3 106R; IIV-6 030L | YP_654678 | 629 | 63.9 | ATP-dependent exo-DNase α subunit |
| 041R | 40283–41143 | 286 | Transmembrane | ||||
| 042L | 41817–41188 | 209 | IIV-3-071L; IIV-6 259R | YP_654643 | 285 | 68.4 | Transmembrane |
| 043R | 42393–42890 | 165 | IIV-3 020R; IIV-6 196R | YP_654592 | 195 | 59.9 | Thioredoxin domain/isomerase |
| 044L | 43528–42923 | 201 | IIV-3 097L; IIV-6 170L; MAR 216 | YP_654669 | 228 | 55.0 | Holliday junction resolvase |
| 045Rf | 43660–44226 | 188 | PBCV N269L | XP_973701 | 137 | 44.4 | Putative dUTPase |
| 046R | 44338–45663 | 441 | IIV-6 468L*; IIV-3 093L | NP_149463 | 294 | 42.0 | Helix-turn-helix 7 motif |
| 047L | 45971–45708 | 87 | Transmembrane | ||||
| 048R | 46162–47862 | 566 | IIV-3 059L; IIV-6 012L | YP_654631 | 729 | 63.9 | XRN 5′–3′ exonuclease |
| 049R | 48016–48504 | 162 | |||||
| 050R | 48602–49111 | 169 | IIV-3 032R | YP_654604 | 43 | 31.2 | |
| 051L | 49744–49310 | 144 | IIV-3 058R, IIV-6 391R | YP_654630 | 200 | 65.3 | |
| 052L | 50273–49851 | 140 | IIV-6 413R; IIV-3 021L | RING/U box motif | |||
| 053R | 50475–51242 | 255 | IIV-3 060L; PBCV A193L | YP_654632 | 254 | 56.0 | Proliferating cell nuclear antigen |
| 054R | 51304–52656 | 450 | IIV-6 468L* | NP_149463 | 303 | 45.4 | Helix-turn-helix 7 motif |
| 055L | 55623–52687 | 978 | IIV-3 087L; IIV-6 022L | YP_654659 | 1258 | 63.5 | DEAD/H motif/putative NTPase |
| 056R | 55805–56563 | 252 | IIV-3 070L; IIV-6 306R | YP_654642 | 240 | 59.7 | SWIB/MDM2 domain |
| 057R | 56642–57964 | 440 | Acanthamoeba polyphaga mimivirus L12 | YP_142366 | 152 | 26.7 | |
| 058L | 59618–58074 | 514 | IIV-3 098L; IIV-6 493L | YP_654670 | 627 | 60.9 | Serine/threonine protein kinase |
| 059R | 59726–61090 | 454 | IIV-3 039R; IIV-6 393L | YP_654611 | 501 | 55.3 | |
| 060Rf | 61210–61602 | 130 | Ixodes scapularis RNA-binding protein; IIV-6 340R | EEC17992 | 53 | 26.7 | dsRNA binding protein |
| 061L | 61781–61599 | 60 | |||||
| 061R | 61662–61832 | 56 | |||||
| 062R | 61866–62225 | 119 | IIV-3 041R; IIV-6 453L | YP_654613 | 152 | 59.7 | Thioredoxin domain |
| 063R | 62315–63559 | 414 | IIV-6 420R* | NP_149883 | 204 | 33.9 | |
| 064Lf | 64363–63656 | 235 | Transmembrane | ||||
| 065R | 64427–66082 | 551 | IIV-3 038R; IIV-6 098R | YP_654610 | 589 | 53.2 | |
| 066L | 66388–66197 | 63 | IIV-3 043R; IIV-6 010R | YP_654615 | 101 | 67.2 | Transmembrane |
| 067L | 69088–66401 | 894 | IIV-3 091L; IIV-6 443R,261R,396L | YP_654663 | 447 | 43.0 | |
| 068L | 75310–69158 | 2050 | IIV-3 091L; IIV-6 443R,261R | YP_654663 | 256 | 33.9 | Transmembrane |
| 069R | 75461–77572 | 703 | IIV-3 074L; IIV-6 268L | YP_654646 | 617 | 50.5 | |
| 070R | 77864–80311 | 815 | IIV-16-RNR; IIV-3 065R; IIV-6 085L | AAY24450.1 | 559 | 71.4 | RNR large-chain precursor/intein |
| 071R | 80408–81727 | 439 | IIV-6 468L*; IIV-3 093L | NP_149463 | 289 | 43.7 | Helix-turn-helix 7 motif |
| 072L | 82690–81764 | 308 | IIV-3 091L,036R,008L; IIV-6 219L,443R | YP_654663 | 114 | 40.0 | |
| 073Lf | 83243–82767 | 158 | IIV-3 042R; IIV-6 136R | YP_654614 | 192 | 59.2 | |
| 074L | 84548–83364 | 394 | IIV-3 079L; IIV-6 282R | YP_654651 | 535 | 69.8 | Poxvirus very late transcription factor |
| 075R | 84711–85385 | 224 | IIV-3 080R | YP_654652 | 181 | 43.8 | NUDIX hydrolase domain |
| 076R | 85649–86833 | 394 | Mimivirus L5, L12, R821, R865, L754, R433 protein | YP_142359 | 151 | 30.3 | Bro domain |
| 077R | 86853–87038 | 61 | |||||
| 078L | 87331–87053 | 92 | Signal peptide | ||||
| 079R | 87349–88107 | 252 | IIV 3088R; IIV-6 075L | YP_654660 | 410 | 77.9 | NTPase domain |
| 080L | 88717–88181 | 178 | Histones | Q27443 | 55 | 17.5 | H4 and H3 histone domains |
| 081R | 88803–89180 | 125 | Pseudoalteromonas haloplanktis TAC125 | YP_339369 | 92 | 48.0 | GIY-YIG endonuclease |
| 082L | 90202–89231 | 323 | IIV-3 044L | YP_654616 | 333 | 51.7 | Protein kinase domain |
| 083R | 90412–90696 | 94 | IIV-3 045R | YP_654617 | 127 | 70.0 | |
| 084R | 90821–92098 | 425 | IIV-6 229L; IIV-3 046R | NP_149692 | 389 | 47.6 | |
| 085R | 92175–92822 | 215 | IIV-6 378R,232R; IIV-3 100L | NP_149841 | 216 | 69.4 | 2-Cys adaptor domain |
| 086L | 93855–92863 | 330 | IIV-3 099R; IIV-6 329R | YP_654671 | 249 | 48.0 | |
| 087R | 94000–95811 | 603 | IIV-3 019R; IIV-6 420R | YP_654591 | 318 | 56.4 | Bro-N domain |
| 088L | 96116–95925 | 63 | Transmembrane | ||||
| 089R | 96461–99868 | 1135 | IIV-3 086L; IIV-6 045L | YP_654658 | 1401 | 62.0 | DNA topoisomerase II |
| 090R | 99858–100511 | 217 | IIV-3 064L | YP_654636 | 88 | 27.2 | |
| 091L | 101269–100583 | 228 | IIV-3 063R; IIV-6 309L | YP_654635 | 170 | 47.6 | |
| 092R | 101417–102835 | 472 | IIV-6 420R*; IIV-3 019R,093L | NP_149883 | 283 | 40.8 | |
| 093L | 103109–102885 | 74 | |||||
| 094L | 104606–103173 | 477 | IIV-3 061R; IIV-6 467R | YP_654633 | 346 | 39.5 | |
| 095R | 104681–105583 | 300 | Bombyx mori thymidylate synthase | XP_001033394 | 322 | 52.8 | Thymidylate synthase |
| 096R | 105651–107114 | 487 | IIV-3 019R,093R; IIV-6 420R* | YP_654591 | 248 | 39.5 | |
| 097R | 107167–108060 | 297 | IIV-3 028R | YP_654600 | 191 | 39.7 | |
| 098R | 108107–108679 | 190 | IIV-3 029R; IIV-6 143R | YP_654601 | 231 | 55.0 | Deoxyribonucleoside kinase |
| 099L | 109129–108725 | 134 | IIV-3 030L | YP_654602 | 109 | 42.3 | |
| 100R | 109185–109688 | 167 | IIV-3 031R: IIV-6 115R | YP_654603 | 84 | 32.4 | |
| 101R | 109843–110592 | 249 | IIV-3 032R | YP_654604 | 139 | 33.5 | |
| 102R | 110660–110956 | 98 | Macaca mulatta regulatory subunit | XP_001097695 | 60 | 35.4 | Protein phosphatase 1C binding |
| 103R | 110998–112287 | 429 | IIV-6 468L*; IIV-3 093L | NP_149463 | 288 | 43.6 | Helix-turn-helix 7 motif |
| 104L | 112901–112320 | 193 | IIV-3 033L: IIV-6 307L | YP_654605 | 222 | 61.5 | Signal peptide, transmembrane |
| 105Rf | 113487–114305 | 272 | IIV-3 034R; IIV-6 077L | YP_654606 | 164 | 38.6 | C3H1 Zinc finger |
| 106R | 114438–116879 | 813 | IIV-3 094L; IIV-6 050L | YP_654666 | 509 | 34.9 | |
| 107R | 117308–117733 | 141 | IIV-3 053L | YP_654625 | 143 | 51.1 | |
| 108L | 118009–117770 | 79 | |||||
| 109R | 118083–119972 | 629 | IIV-3 052L; IIV-6 205R | YP_654624 | 387 | 41.3 | NAD-dependent DNA ligase |
| 110R | 120070–121347 | 425 | IIV-3 007R | YP_654579 | 415 | 52.6 | |
| 111L | 121746–121513 | 77 | Signal peptide | ||||
| 112L | 123866–121758 | 702 | IIV-3 091L; IIV-6 443R,261R,396L,219L,159L | YP_654663 | 226 | 43.1 | |
| 113R | 123945–127328 | 1127 | IIV-3 009R; IIV-6 428L | YP_654581 | 1801 | 77.2 | DNA-dependent RNA Pol subunit 2 |
| 114R | 127384–127998 | 204 | IIV-6 404L | NP_149867 | 238 | 60.5 | |
| 115L | 129575–128067 | 502 | IIV-3 051L; IIV-6 213R | YP_654623 | 141 | 46.3 | |
| 116Rf | 129723–133142 | 1139 | IIV-3 120R; IIV-6 037L | YP_654692 | 1503 | 66.1 | DNA polymerase |
| 117R | 133208–133576 | 122 | IIV-6 049L | NP_149512 | 88 | 41.5 | Transmembrane |
| 118R | 133691–135031 | 446 | IIV-6 468L* | NP_149463 | 277 | 39.7 | Helix-turn-helix 7 motif |
| 119R | 135057–135410 | 117 | |||||
| 120R | 135520–138408 | 962 | IIV-3 121R; IIV-6 184R | YP_654693 | 1342 | 69.2 | Helicase/primase |
| 121R | 138595–138990 | 131 | Amsacta moorei entomopoxvirus AMV-075 | NP_064857 | 107 | 45.8 | |
| 122L | 139310–139029 | 93 | IIV-3 126R | YP_654698 | 91 | 48.4 | Transmembrane |
| 123L | 140261–139410 | 283 | IIV-3 125R | YP_654697 | 271 | 49.5 | |
| 124L | 144216–140356 | 1286 | IIV-6 443R,261R,396L; IIV-3 091L | NP_149906 | 587 | 47.5 | |
| 125L | 144906–144316 | 196 | IIV-3 124R | YP_654696 | 112 | 42.3 | |
| 126R | 144925–145308 | 127 | IIV-3 123L | YP_654695 | 80 | 39.5 | |
| 127L | 145500–145342 | 52 | Transmembrane | ||||
| 128R | 145520–145729 | 69 | IIV-3 117L | YP_654689 | 35 | 34.4 | |
| 129R | 145818–148631 | 936 | IIV-1 L96; IIV-3 084L; IIV-6 232R | P22856 | 927 | 70.0 | OTU-like cysteine protease |
| 130R | 148675–149193 | 172 | IIV-3 083L; IIV-6 358L | YP_654655 | 95 | 34.7 | |
| 131R | 149308–150282 | 324 | IIV-6 420R* | NP_149883 | 179 | 36.5 | |
| 132R | 150254–150514 | 86 | |||||
| 133R | 150722–151150 | 142 | IIV-3 082L | YP_654654 | 82 | 34.5 | |
| 134L | 151813–151349 | 154 | IIV-3 096R; IIV-6 347L | YP_654668 | 108 | 40.5 | ErvI/Alr sulfhydryl oxidase domain |
| 135R | 151908–152948 | 346 | Acyrthosiohon pisum metalloprotein; IIV-3 095L | XP_001945941 | 189 | 38.5 | Matrix metalloproteinase |
| 136L | 154120–153011 | 369 | IIV-3 091L,036R,008L; IIV-6 443R,219L,317L | YP_654663 | 132 | 34.0 | |
| 137L | 154906–154187 | 239 | IIV-3 067L; IIV-6 197R | YP_654639 | 264 | 54.0 | Protein tyrosine phosphatase |
| 138R | 155025–156068 | 347 | IIV-3 078R; IIV-6 244L | YP_654650 | 402 | 56.4 | Phosphodiesterase domain |
| 139L | 156398–156222 | 58 | |||||
| 139R | 156283–156492 | 69 | |||||
| 140L | 157089–156538 | 183 | IIV-3 073R, IIV-6 234R | YP_654645 | 134 | 48.0 | Transmembrane |
| 141R | 157589–158050 | 153 | IIV-3 072L; IIV-6 374R | YP_654644 | 188 | 60.1 | |
| 142R | 158110–159399 | 429 | IIV-6 468L*; IIV-3 093L | NP_149463 | 333 | 48.0 | Homeodomain |
| 143L | 162815–159438 | 1125 | IIV-3 016R; IIV-6 295L | YP_654588 | 1079 | 49.9 | |
| 144R | 162892–164241 | 449 | IIV-6 468L* | NP_149463 | 308 | 45.2 | Homeodomain |
| 145Rf | 164430–165746 | 438 | IIV-6 161L; IIV-3 109L,108L | NP_149624 | 342 | 44.0 | Helicase |
| 146R | 165859–166065 | 68 | IIV-6 212L,211L | NP_149675 | 40 | 39.7 | |
| 147R | 166109–166318 | 69 | IIV-6 388R*; IIV-3 093L | NP_149851 | 43 | 42.9 | |
| 148L | 167577–166534 | 347 | |||||
| 149L | 168222–167611 | 203 | IIV-3 066L; IIV-6 357R | YP_654638 | 102 | 30.9 | Transmembrane |
| 150R | 168376–170697 | 773 | IIV-3 113L; IIV-6 155L,149L | YP_654685 | 809 | 53.7 | |
| 151L | 171154–170834 | 106 | IIV-3 112R; IIV-6 466R | YP_654684 | 101 | 48.1 | Transmembrane |
| 152L | 171684–171214 | 156 | IIV-3 111R, IIV-6 414L | YP_654683 | 203 | 63.7 | NUDIX hydrolase domain |
| 153R | 171761–172324 | 187 | IIV-3 001R; IIV-6 395R | YP_654573 | 104 | 47.3 | |
| 154L | 173033–172515 | 172 | IIV-3 092R; IIV-6 454R | YP_654664 | 206 | 61.9 | RPB5 domain |
| 155L | 173580–173080 | 166 | Burkholderia oklahomensis EO147; MAR 217 | ZP_02357920 | 80 | 31.9 | dNMP kinase |
| 156L | 174825–173689 | 378 | Dictyostelium discoideum AX4 | XP_636066 | 88 | 24.3 | |
| 157R | 175059–175748 | 229 | IIV-3 032R | YP_654604 | 94 | 32.3 | |
| 158R | 175933–176637 | 234 | Ixodes scapularis E3 UBQ ligase | EEC07169 | 48 | 24.7 | RING finger |
| 159R | 176791–177318 | 175 | IIV-3 018L; IIV-6 415R | YP_654590 | 157 | 50.6 | |
| 160R | 177923–179023 | 366 | IIV-3 076L; IIV-6 369L | YP_654648 | 372 | 52.2 | XPG-like protein (excision repair) |
| 161R | 179110–179382 | 90 | |||||
| 162R | 179441–179710 | 89 | |||||
| 163L | 180006–179749 | 85 | |||||
| 164L | 181405–180044 | 453 | A. polyphaga mimivirus L5,L12 | YP_142359 | 176 | 30.6 | Bro-N domain |
| 165R | 181529–185557 | 1342 | IIV-3 090L; IIV-6 176R | YP_654662 | 1964 | 72.3 | DNA-depependent RNA Pol II large subunit |
| 166L | 186055–185801 | 84 | IIV-3 089L | YP_654661 | 44 | 48.8 | |
| 167L | 186230–186060 | 56 | Transmembrane | ||||
| 168R | 186298–187545 | 415 | IIV-6 420R* | NP_149883 | 181 | 33.8 | |
| 169L | 188173–187583 | 196 | IIV-3 068R; IIV-6 401R | YP_654640 | 343 | 84.7 | HMG box |
| 170Rf | 188324–188803 | 159 | Apis mellifera | XP_624869 | 121 | 43.4 | Dual-specificity phosphatase |
| 171R | 188941–189651 | 236 | IIV-3 116R | YP_654688 | 46 | 20.2 | |
| 172L | 190186–189704 | 160 | IIV-3 119R | YP_654691 | 73 | 48.0 | |
| 173R | 190308–190547 | 79 | IIV-6 420R,200R | NP_149883 | 55 | 40.5 | |
| 174L | 190957–190700 | 85 | IIV-3 115R; IIV-6 342R | YP_654687 | 97 | 64.5 | |
| 175R | 191065–191487 | 140 | IIV-3 114L | YP_654686 | 51 | 27.2 | Signal peptide |
| 176R | 191553–192140 | 195 | IIV-3 081L | YP_654653 | 97 | 34.7 | FasI domain |
| 177R | 192245–193678 | 477 | IIV-3 024R; IIV-6 361L,224L | YP_654596 | 539 | 56.3 | Cathepsin |
| 178R | 193729–194130 | 133 | |||||
| 179R | 194134–194400 | 88 | |||||
| 180L | 195002–194424 | 192 | CfDEF NPV 110; Eppo NPV 102 | NP_932719 | 182 | 46.1 | |
| 181R | 195132–196025 | 297 | IIV-3 017R; IIV-6 335L | YP_654589 | 320 | 60.4 | |
| 182R | 196333–197700 | 455 | IIV-6 468L* | NP_149463 | 282 | 40.0 | Helix-turn-helix 7, homeodomain |
| 183R | 197801–198733 | 310 | IIV-3 107R; IIV-6 117L | YP_654679 | 232 | 54.4 | Transmembrane |
| 184L | 199094–198795 | 99 | IIV-3 023R | YP_654595 | 145 | 68.4 | |
| 185R | 199157–199636 | 159 | IIV-3 050L | YP_654622 | 162 | 63.6 | |
| 186L | 202115–199674 | 813 | IIV-3 049R | YP_654621 | 95 | 18.0 | |
| 187R | 202220–203323 | 367 | IIV-3 048L; IIV-6 376L | YP_654620 | 581 | 72.2 | RNR small subunit |
| 188R | 203536–204051 | 171 | IIV-3 025R; IIV-6 111R | YP_654597 | 87 | 33.5 | Transmembrane |
| 189R | 204093–204767 | 224 | IIV-3 026R; IIV-6 350L | YP_654598 | 296 | 67.6 | |
| 190R | 204824–205276 | 150 | IIV- 027R; IIV-6 157L | YP_654599 | 73 | 31.8 | C3HC4 RING finger |
| 191L | 205760–205347 | 137 | |||||
IIV-9 open reading frame number. Proteins identified by the proteomic experiments are indicated by bold ORF numbers for the purified IIV-9 particles and by underlined ORF numbers for infected cells. Note that the protein products of 071R and 182R were identified by the same set of peptides and can therefore not be distinguished unambiguously by the proteomic analysis.
Most closely related gene by BLASTP analysis.
Matching IIV proteins, with the first-listed protein being the most closely related. Non-IIV proteins with a high similarity score to an IIV-9 protein by BLASTP analysis are indicated. NCLDV species abbreviations are PBCV, Paramecium bursaria chlorella virus, and MAR, Marseilles virus. CfDEF NPV, Choristoneura fumiferana nucleopolyhedrovirus; Eppo NPV, Epiphyas postvittana nucleopolyhedrovirus.
Amino acid percent identity for most closely related protein. * indicates similarity to the cluster of proteins encoded by IIV-6 468L and its homologous genes in IIV-6.
TF IIS, transcription factor IIS; BIR, baculovirus inhibitor of apoptosis protein repeat; Bro, baculovirus repeated ORF; NUDIX, nucleoside diphosphate linked.
Protein identified by a single peptide hit.
The genome orientation and gene designation were defined by the start codon of the IIV-9 orf001R ortholog of the first conserved iridovirus core gene in mosquito IIV-3 (orf004R [5]). Four short ORF (orf007, -088, -061, -139) that were represented by dual heavily overlapping ORF in opposite orientations could not be resolved as forward or reverse by bioinformatics analysis alone. orf007 and -088 were subsequently designated orf007Rp and -088Lp based on the identification of their protein products by the proteomic analysis (Table 2; see also Table S1 in the supplemental material). The remaining two ORF could not be resolved, and hence, both overlapping ORF were designated orf061R and -061L or -139R and -139L, respectively.
The majority of IIV ORF have no predicted function. However, a wide range of predicted proteins showed similarity to proteins involved in nucleotide metabolism and DNA replication. These include enzymes required for deoxyribonucleotide synthesis, such as thymidylate synthase (095R), dUTPase (045R), deoxyribonucleoside kinase (098R), and both the large and small subunits of ribonucleotide reductase (070Ri, 187Ri). The last two have been confirmed as expressed proteins in infected cells. There are two putative NUDIX hydrolase proteins (075R and 152L), and these may play an important role in regulating nucleotides in the host cell. Delhon et al. (5) postulated that the IIV-3 ortholog of 075R (IIV-3 080R) might act similarly to the vaccinia virus NUDIX ortholog (with a D10R mutation) and function as a repressor of transcription and translation. The reported presence of an intein in the large subunit of ribonucleotide reductase (orf070Ri) was confirmed (10).
Forty-four genes that could be postulated to have a role in DNA metabolism or DNA replication were identified. These include viral DNA ligase (109R), DNA polymerase (116R), helicase/primase (120R,145R), PCNA (053R), endonuclease (081R), DNA exonuclease (040R), topoisomerase II (089Ri), and phosphodiesterase (022Ri) genes. However, only topoisomerase II (089Ri) and phosphodiesterase (022Ri) have been identified in infected cells. Genes encoding putative chromatin-binding regions, such as SWID/MDM2 (056Rpi) and HMG box domains (169Lpi), are also present. Although it is not known if these affect the host or viral genome structure, the identification of both proteins in isolated virions suggests a possible association with the viral genome.
This study is the first to identify a putative chitinase gene (020Ri) in an IIV. Analysis of this chitinase indicates that it is a member of the family 18 glycohydrolases (exochitinases) and is most closely related to the chitinase of a bacterial pathogen of fish, Yersinia ruckeri (57% identity), and to the chitinases of other bacteria and slime molds. Baculovirus chitinases, along with cathepsin, have been shown to be important in facilitating the release of virus from the host (15). Despite the IIV-9 chitinase displaying less than 20% identity to baculovirus chitinases, the presence of a viral cathepsin (177Rpi) in IIV-9 may reflect similar roles of chitinase and cathepsin, acting in concert to degrade the insect, thereby facilitating viral release and dissemination from the host insect (15). Both enzymes were identified in IIV-9-infected insect cells by our proteomic analysis.
Expressed orf180Lpi also encodes a protein with strong similarity to baculovirus genes, possessing 46% identity and 66% similarity to orf110 of Choristoneura fumiferana nucleopolyhedrovirus (CfDEF NPV). A related gene is also present in IIV-6 (422L), and alignment of CfDEF NPV orf110, Epiphyas postvittana NPV orf102, and the IIV-9 and IIV-6 proteins shows the presence of a highly conserved pan-caspase DEVD cleavage site. It is not known if this exploits caspase activity for processing or if it might regulate apoptosis. A further enzyme identified in IIV-9 is a putative ErvI/augmenter of liver regeneration (ALR) sulfhydryl oxidase (134Lp). This protein is common in large cytoplasmic DNA viruses (29), and in common with poxviruses, this was found in the IIV-9 virus particle. The role of this enzyme activity is unclear but has been postulated to work in concert with glutaredoxin or thioredoxin systems for regulating cytoplasmic disulfide bonds and protein folding. IIV-9 encodes two proteins with putative thioredoxin domains, 043Rp and 062Rpi, both of which were identified in the virus particle.
The repeat sequences identified in the genome are located predominantly in coding regions. This is reflected in the presence of multiple copies of closely related genes on the genome (see Fig. S1 and S3 in the supplemental material). IIV-9 orf006Lpi, -026R, -035L, -046Ri, -054Ri, -071Ri, -103R, -118R, -142R, -144Ri, and -182Ri form one cluster of paralogs, as reflected in amino acid identities ranging from 60 to 84% between the encoded proteins (see Fig. S1 in the supplemental material). With InterProScan, all of these proteins were predicted to contain helix-turn-helix 7 (HTH 7 [Pfam 02796]) motifs and/or the more stable homeodomain motifs that are involved in DNA binding, with a wide spectrum of roles, ranging from transcription regulation to DNA repair (1). These proteins may have a role in the regulation of viral gene expression or viral genome replication, with an array of closely related proteins being involved in sequence-specific fine-tuning of the viral gene cascade. An alternative role could be in the resolution of the branched complexes generated during IV genome replication, although it is not clear why so many copies would be required.
In addition, IIV-9 orf063R, -131R, and -168R (53 to 79% identity) form a less well conserved cluster of proteins (Fig. 3; see also Fig. S2 in the supplemental material) whose genes display some motif conservation to orf092R and -096R (46% identical). No motifs or predicted functions were identified for this cluster of repeated genes, but by BLAST analysis, they were distantly related to the same helix-turn-helix cluster of genes identified above.
Analysis of the protein of orf068Lp, which is indicated by the double-boxed repeat highlighted in Fig. 2A, using the XSTREAM protein tandem repeat finder (27) identified 4.8 copies of a 131-aa repeat at aa 104 to 731 (Fig. 4 A) and an immediately adjacent repeat consisting of 14.4 copies of an 84-aa repeat at aa 714 to 1922 (Fig. 4B). The respective C- and N-terminal flanks of the repeats overlap. The orf067Lpi protein possesses a repeat related to that illustrated in Fig. 4A (Fig. 4C). Both proteins have a high level of predicted β-sheet composition, and both were identified in the virus particle. The high β-sheet structure is similar to what is found in a range of fiber structures, such as bacteriophage fibers and tubulin. BLAST analysis of a single copy of the 131-aa repeat identified a very weak match between a short sequence located around the conserved PDATT motif and bacteriophage fiber proteins (data not shown). However, the location of orf067Lpi and orf068Lp in the particle is unknown, and hence a possible role in the surface fibril cannot be confirmed. An ortholog of this protein is in IIV-3 (091L) and IIV-6 (443R) but is not conserved in vertebrate IV, which would be consistent with the lack of surface fibrils on the vertebrate IV.
Fig. 4.
Tandem protein repeats in the proteins of orf068L and -067L. The repeat regions from aa 104 to 731 (A) and 714 to 1922 (B) of the orf068L protein are shown with residues matching the consensus (shaded). Underlined residues are the same amino acids. (C) The repeat sequence within the orf067L protein is shown aligned with the orf068L protein repeat shown in panel A (boxed underneath). The starred Y's are the same residue (to orient the 068L and 067L repeats).
Relationship to other viruses.
Of the 191 ORF predicted in IIV-9, 108 were most closely related to IIV-3 ORF (Table 2), indicating that IIV-9 is more closely related to the chloriridovirus IIV-3 than to IIV-6. Analysis of IIV-3 shows that 114 of the 126 ORF in IIV-3 (5) were identified as having an ortholog in IIV-9. In contrast, IIV-6 (Iridovirus genus) has 211 ORF, as defined by Eaton et al. (8), of which only 97 have orthologs in IIV-9. A total of 88 ORF are common to all 3 fully sequenced IIV. Interestingly, of the 45 ORF without an ortholog in other IV, 23 encoded proteins that were smaller than 100 amino acids, of which four (002R, 088L, 093L, 111L) were confirmed as expressed proteins by the proteomic analysis. In contrast to the high level of conserved genes between IIV genomes, there is a very low level of conservation in gene order. Comparison to IIV-3 with a gene parity plot (Fig. 2B) indicates only 5 clusters of 3 or more genes, with the largest conservation of gene order and orientation being a cluster of 5 genes represented by IIV-9 097R,101R and IIV-3 028R,032R (Table 2). IIV-9 and IIV-6 genomes possess no more than two genes that are conserved in order in any one cluster.
The 26 core genes previously identified as being conserved in all iridoviruses (8) were identified in IIV-9, and consistently with other genes in the genome, no conservation of gene order was apparent for these conserved genes. Phylogenetic analysis of the coding sequences for all 26 genes collated as a concatenated protein sequence for each of IIV-9, IIV-6, IIV-3, SGIV, LCDV-1, and ISKNV (Fig. 1B) shows the clear separation of the vertebrate and invertebrate IV. In addition, the core set provides strong evidence for the main features of the phylogenetic trees established for the partial MCP sequence (Fig. 1A) (38), with IIV-6 being in a separate clade and IIV-3 being more closely related to the major clade of IIV than its current taxonomic position in a separate genus suggests.
There were 36 NCLDV genes identified in the genome, including nine conserved orthologs found in all NCLDV (19) and seven that were present in all four families but that are missing from some lineages within those families. IIV-9 057R, 076R, and 164L were most closely related to predicted proteins of unknown function from Acanthamoeba polyphaga mimivirus.
miRNA coding prediction.
The analysis of miRNA shows an increasing complexity of viral interactions with this posttranscriptional control system, including the control of host and viral genes. Roles have included the establishment of latency and the avoidance of mammalian immune responses, as well as manipulation of the cellular environment to facilitate replication. Examples of viral miRNA to date have predominantly focused upon viruses that are relatively slow in their replication, such as the herpesviruses, and upon the role of virally encoded miRNA in latency. Because IIV have a nuclear replication phase, are relatively slow growing, and often have a range of sublethal effects upon their host insect, they are potential candidates for encoding miRNA.
Combined analysis for pre-miRNA sequences using VMir and MiPred generated seven possible pre-miRNAs (Table 3). All were located within open reading frames of unknown function, and five had no predicted motifs observable. Three putative pre-miRNAs were identified in the same orientation as the associated ORF, and four were in the opposite orientation. The absence of host cell sequence information makes identification of potential host target genes unfeasible. An XRN exonuclease gene (048R) is predicted on the genome of IIV-9, with orthologs in IIV-3 and IIV-6. This enzyme has a role in the processing of miRNA, in particular, the degradation of mature miRNA (24); hence, even if IIV genomes do not encode miRNAs, there is a strong likelihood that IIV interact with small noncoding RNA systems within the cells that they infect.
Table 3.
Predicted pre-miRNA sequences in the IIV-9 genomec
| miRNAa | Start siteb | Apex nucleotide | Sequence (5′–3′) (length [bp]) | VMir score | % G+C | MiPred MFE | P value | MiPred % conf |
|---|---|---|---|---|---|---|---|---|
| MR171 | 11099 | 1134 | CAAAGUCGACUCUUCCACUCGGAAAUGUGAAUGGUUUCCGAGUAAAAGCUGAAGAAAUGACUUUG (65) | 130 | 42 | −23.6 | 0.001 | 63 |
| MR529 | 31179 | 31212 | AAUGGGGUGUGUGAUGGAAUUGGAUUACCCACCCUUAUUUUAGGGUGGGUAAUUUUGUAUUACACUUUUGA (71) | 181 | 38 | −31 | 0.001 | 75 |
| MR598 | 35790 | 35822 | GGGUUGUGGGAGAGCCAACUGGAUCUACAUAUGUAGAUCCAGCUUGGGAUACAUCUACAGC (61) | 127 | 48 | −27.9 | 0.001 | 66 |
| MD653 | 37793 | 37835 | UGAGUAUUAUCAGCUUUUACCAAAGAUGCUGGAUCUACAAUUAACGUUGGACUAGCACCAGCUGAUAAACUUAAA (75) | 127 | 36 | −21.6 | 0.001 | 63 |
| MR1469 | 89360 | 89395 | UUUGGUAUUAUGUUGCACUUUUUAACCUUGAUGAAAUAUCCUUUUUUACGAGAAGAAGAAGUGCUCGAGUAUCA (74) | 150 | 32 | −20.2 | 0.005 | 63 |
| MD2135 | 128453 | 128488 | GGUGAUGUAAUCUGUGGAAUAACUCUGUUUAGUUUUUUUAGACAUUUGUUCAACAAAGAUUAAAUCGUCACCGU (74) | 141 | 34 | −22.9 | 0.001 | 69 |
| MR2250 | 135129 | 135164 | CGCCAUUAUAAUCAUUUUUAUAGUGGAUAGACUCCAAACUAUCAUUGUUCAAAUGAUUAUAAGAACCGG (69) | 148 | 30 | −18.6 | 0.001 | 64 |
R indicates that pre-miRNA is derived from the reverse genomic strand, and D indicates the direct genomic strand. The number refers to the pre-miRNA identified from the initial screen of the entire genomic sequence by VMir.
First nucleotide position of the pre-miRNA in the IIV-9 genome.
The minimum free energy (MFE), P value, and percent confidence (conf) were determined by MiPred.
The presence of an RNase III gene (orf034L) that has also been identified in IIV-3, IIV-6, and vertebrate IV (45), and a dsRNA binding protein (060R; IIV-6 340R), supports a role for noncoding RNA in IV replication. RNase III was identified in purified virus particles and infected cells by our proteomic experiments (Table 2; see also Tables S1 and S3 in the supplemental material) and was previously identified in particles of SGIV (31), confirming that this protein is produced and, hence, likely to have a role in IV replication. miRNA has also been predicted for soft-shelled turtle iridovirus (STIV) (18), and the presence of miRNAs has recently been confirmed for SGIV (44).
Conclusions.
IIV-9 is a member of the major IIV (group II) clade, and the complete genome provides insight into the relationships within and between IIV genera. The apparent close relationship to IIV-3, a virus from a separate genus, and the more distant relationship to IIV-6 have been confirmed through full-genome analysis. The genome encodes a wide range of proteins for which there is no functional prediction, and many of these are found in the complex virus particle. The presence of paralog proteins on the genome is a major contributor to the high incidence of repeat sequences associated with the genome, unlike with IIV-3, where the repeats are more likely to be in noncoding regions (5). The clustering of repeats within predominantly the β-sheet proteins suggests that these proteins may form filamentous structures that are associated with the virus particle and, as such, are candidates for the surface fibrils identified on IIV-9 particles. As for other IV, a large number of proteins are predicted to be involved in nucleotide regulation and genome replication, consistent with a life cycle that includes DNA replication in both the cytoplasm and nucleus and the branched concatemeric replication strategy of IV, which requires resolution of complex genome structures (11).
Supplementary Material
ACKNOWLEDGMENT
This research was supported by the University of Otago.
Footnotes
Supplemental material for this article may be found at http://jvi.asm.org/.
Published ahead of print on 1 June 2011.
REFERENCES
- 1. Aravind L., Anantharaman V., Balaji S., Babu M. M., Iyer L. M. 2005. The many faces of the helix-turn-helix domain: transcription regulation and beyond. FEMS Microbiol. Rev. 29:231–262 [DOI] [PubMed] [Google Scholar]
- 2. Benson G. 1999. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 27:573–580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bromenshenk J. J., et al. 2010. Iridovirus and microsporidian linked to honey bee colony decline. PLoS One 5:e13181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chinchar V. G., et al. 2005. Iridoviridae, p. 145–162 In Fauquet C. M., Mayo M. A., Maniloff J., Desselberger U., Ball L. A. (ed.), Virus taxonomy. Eighth report of the International Committee on Taxonomy of Viruses. Elsevier Academic Press, San Diego, CA [Google Scholar]
- 5. Delhon G., et al. 2006. Genome of invertebrate iridescent virus type 3 (mosquito iridescent virus). J. Virol. 80:8439–8449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Delius H., Darai G., Flugel R. M. 1984. DNA analysis of insect iridescent virus 6: evidence for circular permutation and terminal redundancy. J. Virol. 49:609–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Do J. W., et al. 2004. Complete genomic DNA sequence of rock bream iridovirus. Virology 325:351–363 [DOI] [PubMed] [Google Scholar]
- 8. Eaton H. E., et al. 2007. Comparative genomic analysis of the family Iridoviridae: re-annotating and defining the core set of iridovirus genes. Virol. J. 4:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Fowler M., Robertson J. S. 1972. Iridescent virus-infection in field populations of Wiseana-Cervinata Lepidoptera-Hepialidae) and Witlesia sp. (Lepidoptera-Pyralidae) in New Zealand. J. Invertebr. Pathol. 19:154–155 [Google Scholar]
- 10. Goodwin T. J., Butler M. I., Poulter R. T. 2006. Multiple, non-allelic, intein-coding sequences in eukaryotic RNA polymerase genes. BMC Biol. 4:38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Goorha R. 1982. Frog virus 3 DNA replication occurs in two stages. J. Virol. 43:519–528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Goorha R., Murti G., Granoff A., Tirey R. 1978. Macromolecular synthesis in cells infected by frog virus 3. VIII. The nucleus is a site of frog virus 3 DNA and RNA synthesis. Virology 84:32–50 [DOI] [PubMed] [Google Scholar]
- 13. Goorha R., Murti K. G. 1982. The genome of frog virus 3, an animal DNA virus, is circularly permuted and terminally redundant. Proc. Natl. Acad. Sci. U. S. A. 79:248–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Grundhoff A., Sullivan C. S., Ganem D. 2006. A combined computational and microarray-based approach identifies novel microRNAs encoded by human gamma-herpesviruses. RNA 12:733–750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hawtin R. E., et al. 1997. Liquefaction of Autographa californica nucleopolyhedrovirus-infected insects is dependent on the integrity of virus-encoded chitinase and cathepsin genes. Virology 238:243–253 [DOI] [PubMed] [Google Scholar]
- 16. He J. G., et al. 2001. Complete genome analysis of the mandarin fish infectious spleen and kidney necrosis iridovirus. Virology 291:126–139 [DOI] [PubMed] [Google Scholar]
- 17. He J. G., et al. 2002. Sequence analysis of the complete genome of an iridovirus isolated from the tiger frog. Virology 292:185–197 [DOI] [PubMed] [Google Scholar]
- 18. Huang Y., et al. 2009. Complete sequence determination of a novel reptile iridovirus isolated from soft-shelled turtle and evolutionary analysis of Iridoviridae. BMC Genomics 10:224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Iyer L. M., Aravind L., Koonin E. V. 2001. Common origin of four diverse families of large eukaryotic DNA viruses. J. Virol. 75:11720–11734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Jakob N. J., Muller K., Bahr U., Darai G. 2001. Analysis of the first complete DNA sequence of an invertebrate iridovirus: coding strategy of the genome of Chilo iridescent virus. Virology 286:182–196 [DOI] [PubMed] [Google Scholar]
- 21. Jancovich J. K., et al. 2003. Genomic sequence of a ranavirus (family Iridoviridae) associated with salamander mortalities in North America. Virology 316:90–103 [DOI] [PubMed] [Google Scholar]
- 22. Jiang P., et al. 2007. MiPred: classification of real and pseudo microRNA precursors using random forest prediction model with combined features. Nucleic Acids Res. 35:W339–W344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Juhl S., et al. 2006. Assembly of Wiseana iridovirus: viruses for colloidal photonic crystals. Adv. Funct. Mater. 16:1086–1094 [Google Scholar]
- 24. Kim Y. K., Heo I., Kim V. N. 2010. Modifications of small RNAs and their associated proteins. Cell 143:703–709 [DOI] [PubMed] [Google Scholar]
- 25. Kurita J., Nakajima K., Hirono I., Aoki T. 2002. Complete genome sequencing of Red Sea bream iridovirus (RSIV). Fish. Sci. 68:1113–1115 [Google Scholar]
- 26. Lu L., et al. 2005. Complete genome sequence analysis of an iridovirus isolated from the orange-spotted grouper, Epinephelus coioides. Virology 339:81–100 [DOI] [PubMed] [Google Scholar]
- 27. Newman A. M., Cooper J. B. 2007. XSTREAM: a practical algorithm for identification and architecture modeling of tandem repeats in protein sequences. BMC Bioinformatics 8:382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Radloff C., Vaia R. A., Brunton J., Bouwer G. T., Ward V. K. 2005. Metal nanoshell assembly on a virus bioscaffold. Nano Lett. 5:1187–1191 [DOI] [PubMed] [Google Scholar]
- 29. Senkevich T. G., White C. L., Koonin E. V., Moss B. 2000. A viral member of the ERV1/ALR protein family participates in a cytoplasmic pathway of disulfide bond formation. Proc. Natl. Acad. Sci. U. S. A. 97:12068–12073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Shevchenko A., et al. 1996. Linking genome and proteome by mass spectrometry: large-scale identification of yeast proteins from two dimensional gels. Proc. Natl. Acad. Sci. U. S. A. 93:14440–14445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Song W., Lin Q., Joshi S. B., Lim T. K., Hew C. L. 2006. Proteomic studies of the Singapore grouper iridovirus. Mol. Cell. Proteomics 5:256–264 [DOI] [PubMed] [Google Scholar]
- 32. Song W. J., et al. 2004. Functional genomics analysis of Singapore grouper iridovirus: complete sequence determination and proteomic analysis. J. Virol. 78:12576–12590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sullivan C. S., Grundhoff A. 2007. Identification of viral microRNAs. Methods Enzymol. 427:3–23 [DOI] [PubMed] [Google Scholar]
- 34. Tan W. G., Barkman T. J., Chinchar V. G., Essani K. 2004. Comparative genomic analyses of frog virus 3, type species of the genus Ranavirus (family Iridoviridae). Virology 323:70–84 [DOI] [PubMed] [Google Scholar]
- 35. Tidona C. A., Darai G. 1997. The complete DNA sequence of lymphocystis disease virus. Virology 230:207–216 [DOI] [PubMed] [Google Scholar]
- 36. Tsai C. T., et al. 2005. Complete genome sequence of the grouper iridovirus and comparison of genomic organization with those of other iridoviruses. J. Virol. 79:2010–2023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ward V. K., Kalmakoff J. 1987. Physical mapping of the DNA genome of insect iridescent virus type 9 from Wiseana spp. larvae. Virology 160:507–510 [DOI] [PubMed] [Google Scholar]
- 38. Webby R., Kalmakoff J. 1998. Sequence comparison of the major capsid protein gene from 18 diverse iridoviruses. Arch. Virol. 143:1949–1966 [DOI] [PubMed] [Google Scholar]
- 39. Webby R. J., Kalmakoff J. 1999. Comparison of the major capsid protein genes, terminal redundancies, and DNA-DNA homologies of two New Zealand iridoviruses. Virus Res. 59:179–189 [DOI] [PubMed] [Google Scholar]
- 40. Williams T. 2008. Natural invertebrate hosts of iridoviruses (Iridoviridae). Neotrop. Entomol. 37:615–632 [DOI] [PubMed] [Google Scholar]
- 41. Williams T., Cory J. S. 1994. Proposals for a new classification of iridescent viruses. J. Gen. Virol. 75:1291–1301 [DOI] [PubMed] [Google Scholar]
- 42. Yan X., et al. 2000. Structure and assembly of large lipid-containing dsDNA viruses. Nat. Struct. Biol. 7:101–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Yan X., et al. 2009. The capsid proteins of a large, icosahedral dsDNA virus. J. Mol. Biol. 385:1287–1299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Yan Y., et al. 2011. Identification of a novel marine fish virus, Singapore grouper iridovirus-encoded microRNAs expressed in grouper cells by Solexa sequencing. PLoS One 6:e19148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Zenke K., Kim K. H. 2008. Functional characterization of the RNase III gene of rock bream iridovirus. Arch. Virol. 153:1651–1656 [DOI] [PubMed] [Google Scholar]
- 46. Zhang Q. Y., Xiao F., Xie J., Li Z. Q., Gui J. F. 2004. Complete genome sequence of lymphocystis disease virus isolated from China. J. Virol. 78:6982–6994 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.