Abstract
The cap-dependent endonuclease activity of the influenza virus RNA-dependent RNA polymerase cleaves host mRNAs to produce capped RNA fragments for primers to initiate viral mRNA synthesis. The influenza A virus (FluA) cap-dependent endonuclease preferentially recognizes the cap1 structure (m7GpppNm). However, little is known about the substrate specificity of the influenza B virus (FluB) endonuclease. Here, we determined the substrate specificity of the FluB polymerase using purified viral RNPs and 32P-labeled polyribonucleotides containing a variety of cap structures (m7GpppGm, m7GpppG, and GpppG). We found that the FluA polymerase cleaves m7G-capped RNAs preferentially. In contrast, the FluB polymerase could efficiently cleave not only m7G-capped RNAs but also unmethylated GpppG-RNAs. To identify a key amino acid(s) related to the cap recognition specificity of the PB2 subunit, the transcription activity of FluB polymerases containing mutated cap-binding domains was examined by use of a minireplicon assay system. In the case of FluA PB2, Phe323, His357, and Phe404, which stack the m7GTP, and Glu361 and Lys376, which make hydrogen bonds with a guanine base, were essential for the transcription activity. In contrast, in the case of FluB PB2, the stacking interaction of Trp359 with a guanine base and putative hydrogen bonds using Gln325 and Glu363 were enough for the transcription activity. Taking these results together with the result for the cap-binding activity, we propose that the cap recognition pocket of FluB PB2 does not have the specificity for m7G-cap structures and thus is more flexible to accept various cap structures than FluA PB2.
INTRODUCTION
Influenza A virus (FluA) and influenza B virus (FluB) belong to the family of Orthomyxoviridae. The genomes of FluA and FluB are composed of a set of eight segments of RNA (vRNA) of negative polarity. vRNA is complexed with nucleoprotein (NP) and associated with the RNA polymerase to form viral ribonucleoprotein (vRNP) complexes. vRNP is an essential unit for both transcription and replication (9). In transcription, the RNA polymerase catalyzes not only RNA polymerization and polyadenylation of mRNA but also cleavage of host mRNAs to generate capped RNA fragments. The RNA polymerase is composed of one molecule each of three viral proteins, PB1, PB2, and PA. PB1 plays central roles in both RNA polymerase assembly (27, 31) and RNA polymerization (6). It contains the conserved motifs characteristic of RNA-dependent RNA polymerases and is directly involved in RNA chain elongation (1, 2). It binds to 5′- and 3′-terminal sequences of vRNA and cRNA (cRNA to vRNA), which are conserved in all segments and act as cis-acting elements for the viral RNA synthesis. PB2 is required for transcription and binds to the cap structures of host mRNAs. Recently, the structural features of the cap-binding site in FluA PB2 and the FluA PB1-PB2 contact site have been determined by functional studies and crystallography (12, 31). PA is involved in not only virus genome replication but also transcription as an endonuclease for generation of primers for RNA synthesis (8, 10, 13, 19, 36). It is also reported that PA is important for the polymerase assembly (19). The structure of the PB1-PA contact site has also been determined crystallographically (14, 27).
The FluA polymerase exhibits a cap-dependent endonuclease activity, which cleaves host mRNAs to produce capped RNA fragments with lengths of 11 to 13 nucleotides (nt). The resulting capped RNA fragment serves as a primer to initiate viral mRNA synthesis. It is well known that in the case of the FluA polymerase, eukaryotic mRNAs containing m7G(5′)ppp(5′)Nm (cap1) and m7G(5′)ppp(5′)NmN′m (cap2) structures stimulate in vitro viral RNA transcription strongly (4, 5, 29). Removal of m7G of the cap from mRNA eliminates the priming activity, and naturally occurring uncapped mRNAs do not prime transcription (5, 29). In addition, the presence of methyl groups in the cap is required for the priming activity; reovirus mRNAs with 5′-terminal GpppG are inactive as primers (3). It has also been demonstrated that each of the two methyl groups in the cap1 structure, the 7-methyl residue of guanine and the 2′-O-methyl on the ribose of guanosine, strongly influences the capped RNA-primed transcription activity (4).
Biochemical and structural studies revealed the functional structures of the cap-binding proteins, including FluA PB2 (12), human translation initiation factor 4E (eIF4E) (33, 34), human nuclear cap-binding protein 20 (CBP20) (23), and vaccinia virus (nucleoside-2′-O-)-methyltransferase (VP39) (16). The overall structures of these four cap-binding proteins differ widely due to their evolutionarily unrelated origins, but the cap-binding pockets form a common structure and preferentially bind to the 7-methylated cap structure. These cap-binding proteins hardly bind to the unmethylated cap structure.
Most of our knowledge on the transcription mechanism of the influenza virus genome has been derived from studies on the FluA polymerase, whereas little is known about the FluB polymerase. It is reported that α-amanitin, a potent inhibitor for the host cell RNA polymerase II, inhibits influenza virus transcription, suggesting that eukaryotic mRNAs containing the cap structure are essential for influenza virus transcription (21). Using α-amanitin, we found that the growth of FluB is more sensitive to the amount of cellular mRNA than that of FluA (data not shown). To elucidate the transcription initiation mechanism of the FluB polymerase, we tried to determine the specificity of cap recognition by the FluB polymerase. First, we compared the substrate specificities of FluA and FluB polymerases using purified vRNPs and various capped RNA substrates (m7GpppGm-, m7GpppG-, and GpppG-RNA) and found that the FluB polymerase efficiently cleaves not only m7G-capped RNAs but also unmethylated GpppG-RNA, whereas the FluA polymerase cleaves m7G-capped RNAs specifically. We then tried to identify key amino acids related to the cap recognition of FluB PB2. In order to examine the transcription activity using mutated PB2 proteins, we utilized FluA and FluB minireplicon assay systems using a virus polymerase-dependent reporter gene (17, 35). The minireplicon system has been utilized for a number of functional analyses of cis-acting elements with the viral genome and trans-acting viral factors (10, 35). The reporter gene contains a coding region flanked by each viral 5′ and 3′ untranslated region (UTR), which function as promoters, and therefore mimics an influenza virus genomic segment. Using this assay system, we identified the important amino acids required for the cap recognition by the FluB polymerase by referencing functionally important amino acids in the FluA polymerase (12).
Based on the findings using the assay systems, we propose that the FluB polymerase possesses a novel cap recognition mechanism, which is different not only from the FluA polymerase but also from well-known cap-binding proteins. These findings could be important to develop novel anti-influenza virus drugs targeting the cap recognition and cleavage reaction.
MATERIALS AND METHODS
Biological materials.
Monolayer cultures of 293T and MDCK cells were maintained at 37°C in Dulbecco's modified Eagle medium (DMEM) and minimal essential medium (MEM) (Nissui), respectively, supplemented with 10% fetal calf serum (Cell Culture Technologies). Influenza virus A/Panama/2007/99 (A/PA/99) and B/Shanghai/361/2002 (B/SH/02) were kindly supplied by Y. Suzuki and T. Gotanda (Kitasato Institute, Research Center for Biologicals, Saitama, Japan). Vaccinia virus capping enzyme and recombinant human mRNA (guanine-7-)methyltransferase (rhMTase) were prepared according to a previously described procedure (28).
Cloning of cDNAs for viral RNA polymerase subunits and nucleoprotein cDNA.
For construction of mammalian expression vectors for influenza virus polymerase subunits (PB1, PB2, and PA) and nucleoprotein (NP), cDNAs corresponding to the full-length PB1, PB2 with a FLAG tag at its C terminus (PB2cFLAG), PA, and NP were amplified by reverse transcription-PCR (RT-PCR) from vRNAs of influenza virus A/PA/99 and B/SH/02 as templates using the following sets of phosphorylated primers (see Table S1 in the supplemental material): A-PB1-FOR and A-PB1-REV for FluA-PB1, A-PB2-FOR and A-PB2-cFLAG-REV for FluA-PB2cFLAG, A-PA-FOR and A-PA-REV for FluA-PA, A-NP-FOR and A-NP-REV for FluA-NP, B-PB1-FOR and B-PB1-REV for FluB-PB1, B-PB2-FOR and B-PB2-cFLAG-REV for FluB-PB2cFLAG, B-PA-FOR and B-PA-REV for FluB-PA, and B-NP-FOR and B-NP-REV for FluB-NP. The PCR products were then cloned into the EcoRV site of pCAGGS-P7 (7), resulting in construction of pCAGGS-Panama-PB1, pCAGGS-Panama-PB2-cFLAG, pCAGGS-Panama-PA, pCAGGS-Panama-NP, pCAGGS-Shanghai-PB1, pCAGGS-Shanghai-PB2-cFLAG, pCAGGS-Shanghai-PA, and pCAGGS-Shanghai-NP. cDNAs for PB2 mutants were prepared by site-directed mutagenesis using the primer sets for FluA-PB2-cFLAG and FluB-PB2-cFLAG and mutant primer sets (see Table S2 in the supplemental material). The PB2 mutant genes have been fully sequenced by standard methods (35).
Preparation of influenza virus vRNP.
To prepare vRNP, we first treated purified influenza virus virions at 30°C for 60 min with a disruption buffer consisting of 50 mM Tris-HCl (pH 8.0), 100 mM KCl, 5 mM MgCl2, 1 mM dithiothreitol (DTT), 5% glycerol, 2% Triton X-100, and 2% lysolecithin according to a method described previously (32). The sample was then directly subjected to centrifugation on a 30 to 60% (wt/vol) linear gradient of glycerol on a 70% (wt/vol) glycerol cushion in 50 mM Tris-HCl (pH 8.0) and 150 mM NaCl in a Beckman MLS-50 rotor with adapters at 163,000 × gAV for 3 h at 4°C. Fractionation was carried out from the top of the gradient. Fractions containing vRNP were pooled and then used for in vitro endonuclease and elongation assays.
Preparation of various RNA substrates.
Triphosphate-ended RNA with the 33-nucleotide sequence 5′-GAAAAAAAAAAAAAAAAAAAAAAAAAAAAUAAA-3′, designated pppG-RNA, was synthesized by using T7 RNA polymerase (Amersham Biosciences) and a synthetic DNA template. The protocol was previously described (30). Briefly, to prepare the template for the T7 RNA polymerase, the oligonucleotide T7P (5′-TAATACGACTCACTATA-3′), corresponding to the T7 promoter (−17 to −1), was annealed to the template oligonucleotide T7-polyA-R1 (5′-TTTATTTTTTTTTTTTTTTTTTTTTTTTTTTTCTATAGTGAGTCGTATTA-3′, where the underlined sequence is complementary to the T7 promoter [−17 to −1]). After the transcription reaction, the transcription mixture was treated with DNase I (Roche Applied Science). RNA was then extracted with phenol-chloroform, ethanol precipitated, and used as a capping substrate. To synthesize m7G[32P]pppGm-RNA and G[32P]pppG-RNA, 50 pmol of pppG-RNA was incubated at 37°C for 2 h in the presence of 8 μM [α-32P]GTP (800 cpm/fmol) and an appropriate amount of purified vaccinia virus capping enzyme, which has guanylyltransferase, guanine-7-methyltransferase, and ribose-2′-O-methyl-transferase activities, in a reaction mixture (50 μl) containing 50 mM Tris-HCl (pH 7.9), 2 mM MgCl2, 40 mM NaCl, and 20 mM DTT in the presence or absence of 150 μM S-adenosyl-l-methionine (AdoMet). After the reaction, capped RNA was extracted with phenol-chloroform, ethanol precipitated, and dissolved in H2O. To synthesize m7G[32P]pppG-RNA, 0.4 pmol of G[32P]pppG-RNA was incubated at 30°C for 20 min with 15 ng/μl of rhMTase in a reaction mixture (20 μl) containing 25 mM Tris-HCl (pH 7.9), 0.5 mM DTT, 0.1 mg/ml bovine serum albumin (BSA), and 50 μM AdoMet. The RNA was extracted with phenol-chloroform, ethanol precipitated, and dissolved in H2O. To confirm the cap structure on the synthesized RNA, the cap structure of the synthesized 32P-capped RNA was liberated by digestion with nuclease P1 (Wako) (28). The reaction product was analyzed by thin-layer chromatography (TLC) on a polyethyleneimine (PEI)-cellulose plate (PEI-CEL UV254; Macherey-Nagel) with 0.65 M LiCl and visualized by autoradiography.
In vitro capped RNA cleavage and RNA elongation reactions.
The determination of Flu cap-dependent endonuclease activity and the subsequent RNA elongation reaction were carried out in a reaction mixture (25 μl) containing 50 mM Tris-HCl (pH 7.9), 0.1 mM ammonium acetate, 5 mM MgCl2, 2.5 mM DTT, 0.1% Nonidet P-40, 8 U of RNasin, 3 to 5 fmol of each 32P-capped RNA (800 cpm/fmol), and an appropriate amount of purified vRNPs without or with ATP, UTP, GTP, or CTP. The reaction mixture was incubated at 30°C for 2 h, and then RNA products were extracted with phenol-chloroform and ethanol precipitated. The RNA products denatured with formamide were electrophoresed in a 20% acrylamide gel containing 8 M urea. After electrophoresis, the gel was dried, and RNAs were visualized by autoradiography. The amount of synthesized RNA was measured with a liquid scintillation counter (LS6000IC; Beckman). The endonuclease activity was represented as a ratio of the amount of cleaved RNAs to that of total capped RNAs, and the RNA elongation efficiency was represented as a ratio of the amount of transcripts to that of total capped RNAs.
Cap-binding assay.
UV cross-linking was carried out to measure the cap-binding activity of viral RNA polymerases. A reaction mixture (12 μl) containing 50 mM Tris-HCl (pH 7.9), 0.1 mM ammonium acetate, 5 mM MgCl2, 2.5 mM DTT, 250 fmol of uncapped RNA substrate, 50 fmol of each 32P-capped RNA (∼800 cpm/fmol), and an appropriate amount of purified vRNPs was incubated for 30 min on ice and then irradiated on ice for 10 min with 254-nm UV light (FUNA-UV-Linker FS-1500 [Funakoshi, Japan]) with 0.2 mg/ml of heparin. The 32P-labeled products were digested with nuclease P1, analyzed by 6% SDS-PAGE, and detected by autoradiography.
Minireplicon assay.
Two plasmid vectors carrying a reporter gene (an artificial influenza virus genome containing the firefly luciferase gene of negative polarity, which is synthesized in cells by the human DNA-dependent RNA polymerase I [Pol I]), were constructed as described previously (35). A fragment containing the luciferase gene sandwiched by 5′- and 3′-terminal sequences of FluA/PA/99 and FluB/SH/02 segment 8 was amplified by PCR with specific primers 5′-GTAGTAGAAACAAGGGTGTTTTTTACTCGAGATCTTACAATTTGGACTTTCCGCCCTT-3′ and 5′-GATCCGTCTCCGGGAGCAAAAGCAGGGTGACAAAGACATAATGCATATGGAAGACGCCAAAAACATAAAGAAAGG-3′ for FluA/PA/99 and 5′-TATTCGTCTCAGGGAGCAGAAGCAGAGGATTTGTTTAGTCACTGGCAAACGGAAAAAAATGGAAGACGCCAAAAACATAAAG-3′ and 5′-ATATCGTCTCGTATTAGTAGTAACAAGAGGATTTTTATTTTAAATTTACAATTTGGACTTTCCGCC-3′ for FluB/SH/02, using pGV-B (the promoterless luciferase reporter vector; Toyo Inc.) as a template. The amplified PCR products were digested with BsmBI and cloned into pHH21 containing the promoter region of the human rRNA gene (24, 25), which had been digested with BsmBI. The constructed plasmids were designated pHH-A-vNS-Luc and pHH-B-vNS-Luc, in which the luciferase gene in reverse orientation sandwiched with 23- and 26-nucleotide 5′- and 3′-terminal sequences of the FluA/PA/99 segment 8 or 30 and 44-nucleotide 5′- and 3′-terminal sequences of the FluB/SH/02 segment 8, respectively, is placed under the control of the human Pol I promoter. 293T cells were transfected with plasmids for the expression of the FluA minireplicon (pCAGGS-Panama-PB1, pCAGGS-Panama-PB2-cFLAG, pCAGGS-Panama-PA, pCAGGS-Panama-NP, and pHH-A-vNS-Luc) or FluB minireplicon (pCAGGS-Shanghai-PB1, pCAGGS-Shanghai-PB2-cFLAG, pCAGGS-Shanghai-PA, pCAGGS-Shanghai-NP, and pHH-B-vNS-Luc). A plasmid for the expression of Renilla luciferase driven by the simian virus 40 (SV40) promoter was used as an internal control for the dual-luciferase assay. As a negative control, 293T cells were transfected with the same plasmids, except for the omission of the PB2 expression plasmid. After transfection, the cells were incubated at 37°C for 24 h, and then the luciferase activity was determined using commercially available reagents (Promega) according to the manufacturer's protocol. The relative luminescence intensity was measured with a luminometer for 20 s. To measure the levels of accumulation of viral mRNA, cRNA, and vRNA, quantitative RT-PCR was performed. Total RNA was extracted from transfected cells and then reverse transcribed with either (i) oligo(dT)20 for synthesizing cDNA from viral mRNA, (ii) 5′-ATATCGTCTCGTATTAGTAGTAACAAGAGCATT-3′, which is complementary to the 3′ portion of cRNA of the reporter gene, for synthesizing cDNA from cRNA, or (iii) 5′-TCCATCACGGTTTTGGAATGTTTACTACAC-3′, which is complementary to vRNA, for synthesizing cDNA from vRNA of the reporter gene. These single-stranded cDNAs were subjected to real-time quantitative PCR analyses (Thermal Cycler Dice real-time system TP800; TaKaRa) with SYBR Premix Ex Taq (TaKaRa) and two specific primers, 5′-TCCATCACGGTTTTGGAATGTTTACTACAC-3′, corresponding to the firefly luciferase mRNA between nucleotide sequence positions 728 and 757, and 5′-GTGCGCCCCCAGAAGCAATTTC-3′, which is complementary to the firefly luciferase mRNA between nucleotide sequence positions 931 and 952. Renilla luciferase mRNA was also amplified with two specific primers, 5′-GCAGCATATCTTGAACCATTC-3′, corresponding to the Renilla luciferase mRNA between nucleotide sequence positions 598 and 618, and 5′-CATCACTTGCACGTAGATAAG-3′, which is complementary to the Renilla luciferase mRNA between nucleotide sequence positions 725 and 745. The relative amounts of mRNA, cRNA, and vRNA were calculated by using the second-derivative maximum method and normalized to the amount of Renilla luciferase mRNA. The ratio of the amounts of mRNA and cRNA relative to vRNA is shown.
Detection of capped RNA coprecipitated with the viral RNA polymerase.
293T cells were transfected with plasmids for the expression of the FluB viral proteins, PB1, FLAG-tagged PB2 (wild-type or mutated PB2), and PA. At 24 h posttransfection, cells were resuspended in a lysis buffer (20 mM Tris-HCl [pH 7.9], 100 mM NaCl, 30 mM KCl, and 0.1% Nonidet P-40). The RNA polymerase complex composed of PB1, FLAG-tagged PB2, and PA was purified by incubation with anti-FLAG M2 agarose (Sigma) at 4°C for 3 h and eluted with an elution buffer (50 mM Tris-HCl [pH 7.9], 100 mM ammonium acetate, 5 mM MgCl2, and 10% [vol/vol] glycerol) containing 0.1 mg/ml FLAG peptide (Sigma). RNAs which interact with the viral RNA polymerase was extracted from recombinant RNA polymerase complexes (100 ng of PB1 equivalents) with phenol-chloroform and ethanol precipitated with 20 μg of carrier tRNA. After treatment with calf intestinal alkaline phosphatase (CIAP), which removes free phosphate groups, periodate oxidation under mild conditions followed by β-elimination with aniline was carried out to remove 5′-terminal m7G from capped RNA, generating RNA with 5′-triphosphate, which is the substrate for vaccinia virus capping enzyme, as described previously (4, 11). The RNA was then recapped using vaccinia virus capping enzyme with [α-32P]GTP as described in the previous section. To measure the amount of 32P-labeled capped RNA, the RNA was digested with tobacco acid pyrophosphatase (TAP) (Sigma) at 37°C for 1 h in a buffer containing 50 mM sodium acetate (pH 5.5), 5 mM EDTA, and 10 mM 2-mercaptoethanol. The reaction product was analyzed by thin-layer chromatography on a PEI-cellulose plate as described above, and the amount of [32P]m7Gp was measured with a liquid scintillation counter.
RESULTS
In vitro capped RNA cleavage reaction and subsequent RNA elongation reaction.
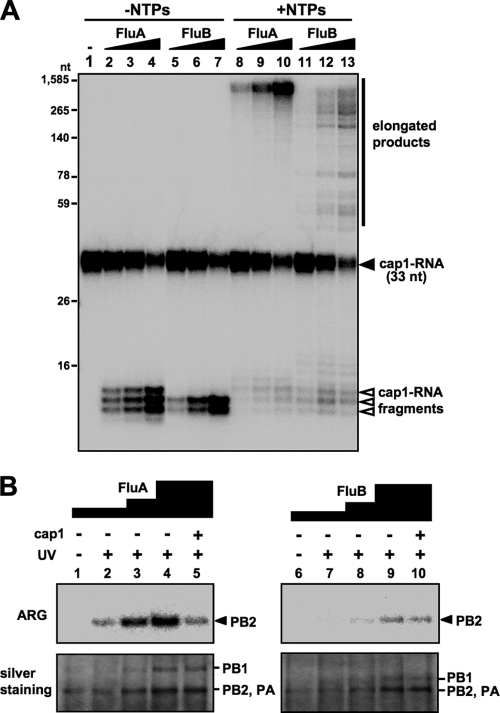
The FluA polymerase requires the cap1 structure (m7GpppNm) stringently for transcription (4). In contrast, little is known about the requirement for the cap structure of the FluB polymerase. Thus, we first examined the efficiency of the capped RNA cleavage reaction and subsequent RNA elongation reaction by FluA and FluB polymerases using cap1-RNA (m7GpppGm-RNA). The cap1-RNA labeled with 32P in the cap structure was incubated with purified vRNP (see Fig. S1A in the supplemental material) in the absence or presence of nucleoside triphosphates (NTPs) (Fig. 1A). RNA products were analyzed by 15% PAGE containing 8 M urea. FluA and FluB polymerases cleaved the cap1-RNA and produced 11- to 13-nucleotide and 11- to 12-nucleotide RNAs, respectively, in the absence of NTPs (Fig. 1A, lanes 2 to 7), indicating that the endonuclease activity of FluB is different from that of FluA in the distance of cleavage site from the cap structure. This cleavage pattern was observed commonly among FluA strains and among FluB strains (see Fig. S1B, lanes 2 to 6, in the supplemental material). The cleaved RNA products were elongated in the presence of NTPs in a dose-dependent manner (Fig. 1A, lanes 8 to 13), but the elongation efficiency of the FluB polymerase was lower than that of the FluA polymerase. We also confirmed that these elongated products contain full-length transcripts from 8 segments and are partially polyadenylated (see Fig. S2 in the supplemental material). To investigate the cap-binding activity of the polymerases, UV cross-linking assays were carried out (Fig. 1B). Cap1-RNA specifically bound to PB2 in both the FluA and FluB polymerases, although the cap-binding activity of FluB PB2 is less (∼25%) than that of FluA PB2. These results suggest that the FluA and FluB polymerases are different in their binding to RNA containing the cap1 structure and in their cleavage modes.
Fig. 1.
In vitro capped RNA cleavage, RNA elongation and cap-binding reactions. (A) Dose dependency of in vitro capped RNA cleavage and subsequent RNA elongation by vRNP. In vitro capped RNA cleavage and RNA elongation reactions were performed with 20, 40, and 80 ng of FluA (lanes 2 to 4 and 8 to 10) and FluB (lanes 5 to 7 and 11 to 13) vRNP using 2 fmol of m7GpppGm-RNA. Capped RNA cleavage was performed in the absence of NTPs (lanes 2 to 7), while RNA elongation was performed in the presence of NTPs (lanes 8 to 13). Synthesized RNA products were analyzed by 15% PAGE containing 8 M urea. (B) Interaction of vRNP with the cap1 structure. UV cross-linking was performed using 50, 100, and 200 ng of FluA (lanes 1 to 5) and FluB (lanes 6 to 10) vRNPs with (lanes 2 to 5 and 7 to 10) or without (lanes 1 and 6) UV irradiation at 254 nm. Competition experiments were performed in the presence of 100 pmol of m7GpppGm analogue (lanes 5 and 10). Upper panels show autoradiography (ARG), while lower panels show silver staining patterns.
Specificity of recognition of cap structures by Flu polymerases.
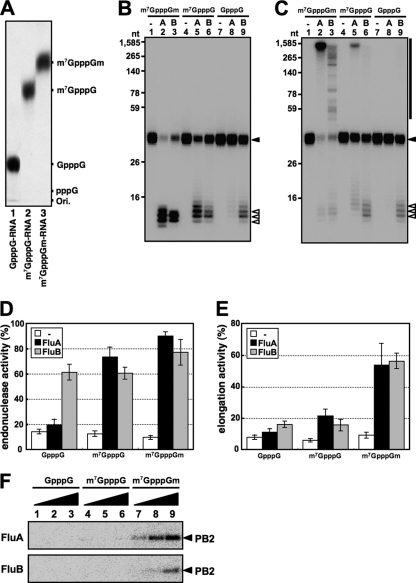
To investigate the specificity of recognition of cap structures by FluA and FluB polymerases, we carried out similar experiments using RNA primers containing various cap structures. To this end, we prepared 32P-labeled RNAs containing differently methylated cap structures, such as m7GpppGm, m7GpppG, and GpppG, as described in Materials and Methods. After preparation, we analyzed the terminal cap structure using nuclease-digested samples (see Materials and Methods) and thin-layer chromatography on a PEI-cellulose plate. As shown in Fig. 2A, we confirmed that each RNA had the expected cap structure. Using these RNAs as substrates, we carried out in vitro capped RNA cleavage and subsequent RNA elongation reactions with FluA or FluB vRNPs. As expected, FluA vRNP specifically cleaved both m7GpppGm-RNA and m7GpppG-RNA, although the latter was less efficiently cleaved (Fig. 2B, lanes 2, 5, and 8, and D). The m7GpppGm-RNA fragments were most successfully elongated into viral mRNAs (Fig. 2C, lane 2, and E). In contrast, FluB vRNP could cleave GpppG-RNA efficiently in addition to the m7GpppGm-RNA and m7GpppG-RNA (Fig. 2B, lanes 3, 6, and 9, and D). It is noteworthy that m7GpppGm-RNA fragments also served as an efficient primer for chain elongation, as is the case for the FluA polymerase (Fig. 2C, lane 3, and E). Moreover, we carried out UV cross-linking assays using RNA primers containing various cap structures (Fig. 2F). Interestingly, the cap-binding activity was detected just using m7GpppGm-RNA with both FluA and FluB vRNPs. These results indicate that the guanine-7-methyl residue is a key for stable cap binding of both FluA and FluB polymerases. It is also indicated that the cap-binding activity is strictly related to the elongation efficiency but not to the cleavage reaction. It is presently unknown why the binding of GpppG and m7GpppG was not detected under the conditions employed, while m7GpppG-RNA was recognized and cleaved by both FluA and FluB polymerases and GpppG-RNA was by the FluB polymerase. Since m7GpppG-RNA and GpppG-RNA were not effective for elongation, the cleavage of these cap structures would be abortive for transcription, possibly due to improper recognition.
Fig. 2.
Specificity of recognition of cap structures by Flu polymerases. (A) Analysis of 5′-terminal cap structures of RNAs. T7 RNA polymerase-synthesized RNAs were treated with nuclease P1 and analyzed by TLC (PEI-CEL, 0.65 M LiCl), and radioactive nucleotides were detected by autoradiography. (B and C) In vitro capped RNA cleavage (B) and RNA elongation (C) reactions were performed with 600 ng of FluA (lanes 2, 5, and 8) or FluB (lanes 3, 6, and 9) vRNP using 2 fmol of variously methylated capped RNAs (m7GpppGm-RNA, lanes 1 to 3; m7GpppG-RNA, lanes 4 to 6; GpppG-RNA, lanes 7 to 9). RNA products were analyzed by 15% PAGE containing 8 M urea. The input capped RNAs (33 nt), the cleaved capped RNA products, and the elongated products are indicated as a closed triangle, open triangles, and a black bar, respectively, at the right. (D and E) Ratios of cleaved RNA products (D) and RNA transcripts (E) to total input primer RNAs. (F) Cap-binding activity for variously methylated capped RNAs. UV cross-linking was performed using 50, 100, and 200 ng of FluA (upper panel) and FluB (lower panel) vRNP and 50 fmol of variously methylated capped RNAs (GpppG-RNA, lanes 1 to 3; m7GpppG-RNA, lanes 4 to 6; m7GpppGm-RNA, lanes 7 to 9).
Identification of key amino acids involved in the cap recognition specificity of the PB2 subunit of the FluB polymerase.
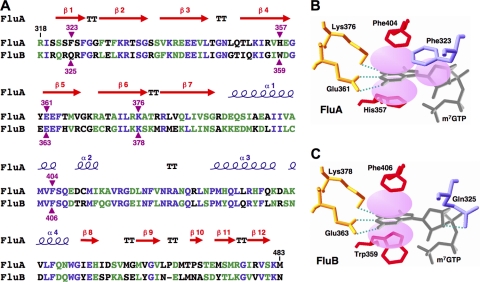
To clarify the cap recognition mechanism, we focused our structure-related functional studies on the interaction between the cap1 structure and the PB2 subunit, which has the cap-binding domain. It is quite likely that amino acid residues essential for cap binding are conserved between FluA and FluB (Fig. 3A). Three-dimensional (3D) structural studies (12) revealed that in the FluA PB2 cap-binding domain (Fig. 3B), Phe404 and His357 sandwich the methylated guanine and Phe323 stacks on the ribose of m7GTP. Glu361 makes hydrogen bonds with the N1 and N2 positions of guanine, and Lys376 also makes a hydrogen bond with position O6 of guanine. Computer-associated modeling could make the FluB PB2 cap-binding domain fit on the FluA PB2 cap-binding domain (Fig. 3C). In the model of the FluB cap-binding domain, 2 amino acids, Gln325 and Trp359, are different from Phe323 and His357 of the FluA cap-binding domain, respectively.
Fig. 3.
Structure of the PB2 cap-binding domain. (A) Sequence alignment of the PB2 cap-binding domains of FluA (A/Panama/2007/99) and FluB (B/Shanghai/361/2002). The secondary structure of FluA is displayed over the sequence alignment. Blue letters and green letters show identical residues and similar residues, respectively. Purple triangles indicate the residues in contact with the cap analogue m7GTP. (B and C) Model of m7GTP interaction with the cap-binding domains of FluA PB2 (B) (10) and FluB PB2 (C) drawn by computer-associated calculation, with putative hydrogen bonds shown as green dotted lines.
To determine key amino acids related to the cap recognition specificity, the transcription activity was measured using a minireplicon assay system. In this assay system, we have used a transient-transfection system with a viral genome, in which the coding region for a viral gene is replaced with a luciferase reporter gene while cis-acting regulatory regions (24) remain intact (35). The cellular RNA polymerase I produces a negative-sense luciferase RNA sandwiched with viral terminal sequences. Luciferase mRNA is synthesized by transcription of the negative-sense RNA with the viral RNA polymerase and NP and subjected to translation. This system has been used to measure the transcription activity of the Flu polymerase (20, 22).
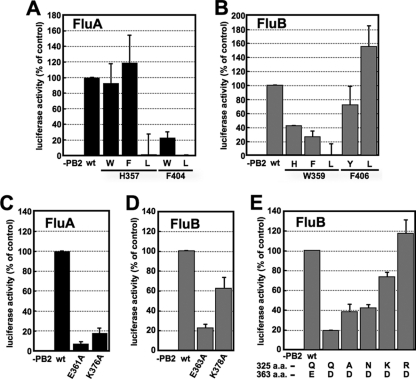
In the case of FluA PB2, His357, with which methylated guanine is stacked, could be replaced by other aromatic residues such as Trp and Phe, while Phe404, which is also involved in stacking methylated guanine, could not be (Fig. 4A). Leu could not substitute for either His357 and Phe404. On the other hand, in the case of FluB PB2, Trp359 could be replaced with other aromatic residues (but with less efficiency than for the FluA polymerase), but Phe406 could be replaced with hydrophobic residues such as Tyr and Leu (Fig. 4B). To confirm the importance of the hydrogen bonds with methylated guanine, Glu361 and Lys376 in FluA PB2 and Glu363 and Lys378 in FluB PB2 were replaced with alanine (Ala). Ala substitutions in FluA PB2 abolished the transcription activity, while Ala substitution for Lys378 of FluB PB2 caused only a small decrease in the transcription activity (Fig. 4C and D). These results suggest that the stacking interaction of His357 and Phe404 and the hydrogen bonds of Glu361 and Lys376 with methylated guanine are essential for cap recognition by the FluA polymerase. This is in good agreement with a previous report (12). In contrast, it is suggested that the stacking interaction of Trp359 and the hydrogen bonds of Glu363 with methylated guanine are sufficient for cap recognition by the FluB polymerase. These results indicate that the mechanism for recognition of methylated guanine by the FluB polymerase could be different from that for the FluA polymerase. It is also speculated that the cap-binding pocket of the FluB polymerase may be more flexible or less stringent than that of the FluA polymerase in recognition of various cap structures, since Phe406 of FluB PB2 is changeable with other amino acids.
Fig. 4.
Transcription activities of PB2 mutants in a minireplicon system. (A and B) Effects of mutations of m7GTP stacking residues in FluA (A) and FluB (B) PB2 on transcription activity. (C and D) Effects of mutations in residues involved in hydrogen bonds with the guanine residue of m7GTP in FluA (C) and FluB (D) PB2 on transcription activity. (E) Effect of mutations in Gln325 with an Asp mutation at position 363 in FluB PB2 on transcription activity. The firefly luciferase activity was normalized to Renilla luciferase activity. The results are averages and standard deviations (SD) from four independent experiments.
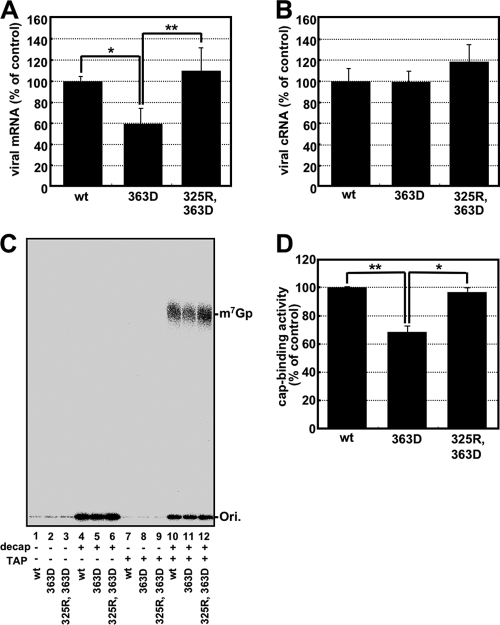
Phe323 in FluA PB2 stacks on the ribose of m7GTP and was essential for cap recognition (see Fig. S3 in the supplemental material) (12). However, it is likely that Gln325 in FluB PB2, which is located in the same position of Phe323 in FluA PB2, makes a hydrogen bond with the ribose of m7GTP. We speculated that FluB PB2 recognizes the cap structure in a flexible pocket as discussed above, so that the hydrogen bonds made by Gln325 and Glu363 could be more crucial for cap binding than those in FluA PB2. In addition, there could be an appropriate amino acid in the amino acid combination between amino acid positions 325 and 363 in FluB PB2 in order to keep the flexible pocket. To confirm this prediction, the transcription activities of mutants with substitutions at position 325 were examined in the presence of the Asp363 mutant (Fig. 4E). The transcription activity of the Asp363 single mutant was reduced to 20% of the wild-type level, possibly because of a longer distance between Asp363 and guanine residues for hydrogen bonds (Fig. 4E; see Fig. S4B in the supplemental material). Interestingly, Lys and Arg mutations but not Ala and Asn mutations at position 325 could rescue the transcription activity of Asp363 (Fig. 4E). We also examined the effect of an Asp363 single mutation and an Arg325-Asp363 double mutation on the transcription and replication processes and the cap-binding activity (Fig. 5). According to the levels of accumulation of mRNA (Fig. 5A) and cRNA (Fig. 5B), the level of reporter expression (Fig. 4E) is well correlated with the transcription but not the replication activities. To examine the cap-binding activity in vivo, capped RNAs that could interact with the viral RNA polymerase were coprecipitated from cells expressing the recombinant RNA polymerase, and the cap structure was detected by recapping of RNA which had been CIAP treated and then decapped (β-eliminated) (Fig. 5C). We could detect the [32P]m7Gp labeled by [α-32P]GTP and vaccinia virus capping enzyme, depending on TAP digestion. In contrast, uncapped RNA treated with CIAP was poorly labeled by this protocol. These results indicate that this recapping method is suitable for the detection of capped RNA specifically. Using this method, we found that the cap-binding activities of these mutants (Fig. 5D) are well correlated with these transcription activities (Fig. 4E) and the mRNA accumulation levels (Fig. 5A). These results indicate that the Arg at position 325 in FluB PB2 supports cap recognition when Glu363 is replaced with Asp363.
Fig. 5.
Suppression mutation in transcription and cap-binding activities for the FluB PB2-363D mutant. (A and B) The levels of accumulation of viral mRNA (A) and cRNA (B) were measured by qPCR. (C) Cap-binding activities of mutants. Coprecipitated capped RNAs with 100 ng of recombinant RNA polymerase complexes (wild type [wt], lanes 1, 4, 7, and 10; 363D mutant, lanes 2, 5, 8, and 11; 325R-363D double mutant, lanes 3, 6, 9, and 12) were recapped before (lanes 1 to 3 and 7 to 9) and after (lanes 4 to 6 and 10 to 12) decapping by β-elimination. Recapped RNAs were treated without (lanes 1 to 6) or with (lanes 7 to 12) tobacco acid pyrophosphatase (TAP) and analyzed by TLC (PEI-CEL, 0.65 M LiCl), and radioactive nucleotides were determined by autoradiography. (D) The radioactivity of [32P]m7Gp of TAP-treated products which were recapped after decapping was counted with a liquid scintillation counter. The cap-binding activity is represented as a ratio to the amount of [32P]m7Gp derived from the wild type. These results are averages and SD from three independent experiments, and the level of significance was determined by Student's t test (unpaired) (*, P < 0.0025; **, P < 0.0005).
DISCUSSION
Most of our knowledge on the transcription mechanism of the influenza virus genome has been derived from studies on FluA, while little has been demonstrated for FluB. This is also the case for studies on the enzymatic aspects of these viral RNA polymerases. Each of the two methyl groups in the cap1 structure, the 7-methyl residue of the guanine base and the 2′-O-methyl residue on the ribose of the penultimate base, strongly influences the transcription activity of the FluA polymerase (4). Recently, the structure of the PB2 cap-binding domain of the FluA polymerase with m7GTP has been clarified (12). Based on these reports, we tried to identify the specificity of cap recognition and characterize key amino acids for cap recognition of the FluB polymerase.
First, we compared the efficiencies of capped RNA cleavage and subsequent RNA elongation reactions of the FluA polymerase with those of the FluB polymerase using cap1-RNA. As expected, the FluA polymerase exhibited efficient endonuclease activity, elongation activity, and cap-binding affinity. The pattern of cleavage of cap1-RNA by the FluB polymerase was different from that by the FluA polymerase (Fig. 1A; see Fig. S1B in the supplemental material), and the RNA elongation and cap-binding activities of the FluB polymerase were lower than those of the FluA polymerase (Fig. 1A and B). These results indicate that the cap binding and cleavage mechanism of the FluB polymerase are different from those of the FluA polymerase.
We then examined the specificity of recognition of cap structures by the FluB polymerase compared with that by the FluA polymerase. Using various methylated capped RNAs, we performed capped RNA cleavage and RNA elongation assays (Fig. 2). The FluA polymerase cleaved RNA containing m7G specifically, while the FluB polymerase could cleave GpppG-RNA as well as RNA containing m7G. Both the FluA and FluB polymerases elongated and bound to the cap structure efficiently only in the case of m7GpppGm-RNA compared with other capped RNAs (Fig. 2C, 2E, and 2F). Based on these results, we propose that the FluA polymerase recognizes strictly the guanine-7-methyl residue in the cleavage reaction and that the FluB polymerase recognizes only the cap core structure (GpppX), which may result in its weak cap1-binding activity. In addition, these results suggest that the ribose 2′-O-methyl residue and/or the guanine-7-methyl residue may be responsible for the elongation reaction by both FluA and FluB polymerases, because cap binding and efficient elongation could not be observed except for m7GpppGm-RNA.
To elucidate the mechanism of cap recognition by the FluB polymerase, we studied the PB2 subunit, which has the cap-binding domain. Recently, the 3D structure of the FluA PB2 cap-binding domain was revealed (12). Amino acid residues essential for cap binding were identified and found to be conserved between FluA and FluB polymerases (Fig. 3A). In the FluA PB2 cap-binding domain (Fig. 3B), the methylated guanine base is sandwiched with His357 and Phe404, and Phe323 stacks on the ribose of m7GTP. Glu361 makes hydrogen bonds with the N1 and N2 positions of guanine, and Lys376 also makes hydrogen bonds with the O6 position of guanine. Based on the structure of the FluA PB2 cap-binding domain, a model of the FluB PB2 cap-binding domain was postulated (Fig. 3C). Five amino acids which contact the guanine-7-methyl residue are highlighted. Minireplicon assays showed that Trp359 in FluB PB2 is crucial for possible stacking interaction with a methylated guanine base without sandwiching with Phe406 (Fig. 4B). Moreover, the hydrogen bond made by Lys378 to the O6 position of guanine seemed not to be essential for cap recognition (Fig. 4D). These results suggest that the FluB polymerase recognizes the cap structure in a manner different from the FluA polymerase. We illustrated a new proposed computer-associated model for cap recognition by FluB PB2 (see Fig. S4A in the supplemental material), although the 3D structure of the FluB PB2 cap-binding domain has not been determined. The overall structures of four cap-binding proteins, FluA PB2 (12), eIF4E (33, 34), CBP20 (23), and VP39 (16), differ each other widely due to their evolutionarily unrelated origins, but the cap-binding pockets are essentially quite similar (see Fig. S5 in the supplemental material), although there are some differences in details. In addition to the two aromatic amino acids, an acidic residue is directed toward the pocket to accommodate the positively charged π-ring system of the methylated guanine. These amino acids provide high specificity for the recognition of m7GTP and exhibit low affinity for nonmethylated cap analogues (>100-fold difference in affinity compared with N7-methylated ones) (15, 18, 26). Compared with these well-known cap-binding proteins, the cap-binding pocket of FluB PB2 contains only one aromatic amino acid, Trp359. This feature may cause the low affinity of FluB PB2 for the cap1 structure (Fig. 1B) and the recognition of nonmethylated capped RNA (GpppG-RNA) (Fig. 2) compared with FluA PB2.
In the case of FluA PB2, the stacking interaction of Phe323 with the ribose of m7GTP is also essential for cap recognition. However, Gln325 of FluB PB2 seems to make a hydrogen bond with the ribose of m7GTP instead of a stacking interaction. To examine our speculation that FluB PB2 recognizes the cap structure in the flexible pocket, we made substitution mutations at position 325 in the presence of an Asp363 mutation (Gln → Asp), which should extend too much into the pocket where Gln325 is present. Interestingly, the transcription activity and the cap-binding activity of the Asp363 mutant were restored to the wild-type levels by the Arg325 mutation (Fig. 4E and 5) without changing the replication activity. The transcription activity of the Asp363 single mutant was decreased, possibly because the longer distance between Asp363 and the guanine residue may make hydrogen bonds weak (see Fig. S4B in the supplemental material). These results suggest that the hydrogen bond made by Arg325 with the ribose of the guanosine could support the recognition of the cap structure (Fig. 4E, 5A, 5C, and 5D; see Fig. S4C in the supplemental material). Crystal structure analyses of wild-type FluB PB2 and the mutant containing Arg325 and Asp363 are needed to support our hypothesis.
In summary, our results indicate that the substrate specificity and the residues essential in the cap recognition are different between FluA and FluB polymerases. In the case of the FluA polymerase, m7G-capped RNA is cleaved specifically, and the stacking interactions of His357 and Phe404 with the methylated guanine base and of Phe323 with the ribose of m7GTP and the hydrogen bonds made by Glu361 and Lys376 on the methylated guanine are essential for cap recognition as observed in other cap-binding proteins. In contrast, in the case of the FluB polymerase, unmethylated capped RNA is cleaved as well as m7G-capped RNA, and the stacking interaction which is made only by Trp359 with the guanine base and the hydrogen bonds which are made by Glu363 on the guanine base and by Gln325 with the ribose of m7GTP are enough for cap recognition.
Supplementary Material
ACKNOWLEDGMENTS
We thank Y. Suzuki and T. Gotanda (Kitasato Institute, Research Center for Biologicals, Saitama, Japan) for providing the purified influenza virus A/Panama/2007/99 and B/Shanghai/361/02 virions. We also thank T. Ogino (Lemer Research Institute, Cleveland Clinic, Cleveland, OH) and A. Kawaguchi (Kitasato Institute for Life Sciences, Kitasato University, Tokyo, Japan) for critical discussion.
This research was supported in part by a grant-in-aid from the Ministry of Education, Culture, Sports, Science, and Technology of Japan (to K.N.).
Footnotes
Supplemental material for this article may be found at http://jvi.asm.org/.
Published ahead of print on 18 May 2011.
REFERENCES
- 1. Argos P. 1988. A sequence motif in many polymerases. Nucleic Acids Res. 16:9909–9916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Biswas S. K., Boutz P. L., Nayak D. P. 1998. Influenza virus nucleoprotein interacts with influenza virus polymerase proteins. J. Virol. 72:5493–5501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bouloy M., Morgan M. A., Shatkin A. J., Krug R. M. 1979. Cap and internal nucleotides of reovirus mRNA primers are incorporated into influenza viral complementary RNA during transcription in vitro. J. Virol. 32:895–904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bouloy M., Plotch S. J., Krug R. M. 1980. Both the 7-methyl and the 2′-O-methyl groups in the cap of mRNA strongly influence its ability to act as primer for influenza virus RNA transcription. Proc. Natl. Acad. Sci. U. S. A. 77:3952–3956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bouloy M., Plotch S. J., Krug R. M. 1978. Globin mRNAs are primers for the transcription of influenza viral RNA in vitro. Proc. Natl. Acad. Sci. U. S. A. 75:4886–4890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Braam J., Ulmanen I., Krug R. M. 1983. Molecular model of a eucaryotic transcription complex: functions and movements of influenza P proteins during capped RNA-primed transcription. Cell 34:609–618 [DOI] [PubMed] [Google Scholar]
- 7. Chen Z., et al. 1998. Comparison of the ability of viral protein-expressing plasmid DNAs to protect against influenza. Vaccine 16:1544–1549 [DOI] [PubMed] [Google Scholar]
- 8. Dias A., et al. 2009. The cap-snatching endonuclease of influenza virus polymerase resides in the PA subunit. Nature 458:914–918 [DOI] [PubMed] [Google Scholar]
- 9. Engelhardt O. G., Fodor E. 2006. Functional association between viral and cellular transcription during influenza virus infection. Rev. Med. Virol. 16:329–345 [DOI] [PubMed] [Google Scholar]
- 10. Fodor E., et al. 2002. A single amino acid mutation in the PA subunit of the influenza virus RNA polymerase inhibits endonucleolytic cleavage of capped RNAs. J. Virol. 76:8989–9001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Fraenkel-Conrat H., Steinschneider A. 1967. Stepwise degradation of RNA: periodate followed by aniline cleavage. Methods Enzymol. 12B:243–246 [Google Scholar]
- 12. Guilligay D., et al. 2008. The structural basis for cap binding by influenza virus polymerase subunit PB2. Nat. Struct. Mol. Biol. 15:500–506 [DOI] [PubMed] [Google Scholar]
- 13. Hara K., Schmidt F. I., Crow M., Brownlee G. G. 2006. Amino acid residues in the N-terminal region of the PA subunit of influenza A virus RNA polymerase play a critical role in protein stability, endonuclease activity, cap binding, and virion RNA promoter binding. J. Virol. 80:7789–7798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. He X., et al. 2008. Crystal structure of the polymerase PA(C)-PB1(N) complex from an avian influenza H5N1 virus. Nature 454:1123–1126 [DOI] [PubMed] [Google Scholar]
- 15. Hodel A. E., Gershon P. D., Shi X., Wang S. M., Quiocho F. A. 1997. Specific protein recognition of an mRNA cap through its alkylated base. Nat. Struct. Biol. 4:350–354 [DOI] [PubMed] [Google Scholar]
- 16. Hu G., Gershon P. D., Hodel A. E., Quiocho F. A. 1999. mRNA cap recognition: dominant role of enhanced stacking interactions between methylated bases and protein aromatic side chains. Proc. Natl. Acad. Sci. U. S. A. 96:7149–7154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Iwatsuki-Horimoto K., et al. 2008. Limited compatibility between the RNA polymerase components of influenza virus type A and B. Virus Res. 135:161–165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Izaurralde E., Stepinski J., Darzynkiewicz E., Mattaj I. W. 1992. A cap binding protein that may mediate nuclear export of RNA polymerase II-transcribed RNAs. J. Cell Biol. 118:1287–1295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kawaguchi A., Naito T., Nagata K. 2005. Involvement of influenza virus PA subunit in assembly of functional RNA polymerase complexes. J. Virol. 79:732–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Labadie K., Dos Santos Afonso E., Rameix-Welti M. A., van der Werf S., Naffakh N. 2007. Host-range determinants on the PB2 protein of influenza A viruses control the interaction between the viral polymerase and nucleoprotein in human cells. Virology 362:271–282 [DOI] [PubMed] [Google Scholar]
- 21. Lamb R. A., Choppin P. W. 1977. Synthesis of influenza virus polypeptides in cells resistant to alpha-amanitin: evidence for the involvement of cellular RNA polymerase II in virus replication. J. Virol. 23:816–819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Li C., Hatta M., Watanabe S., Neumann G., Kawaoka Y. 2008. Compatibility among polymerase subunit proteins is a restricting factor in reassortment between equine H7N7 and human H3N2 influenza viruses. J. Virol. 82:11880–11888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mazza C., Segref A., Mattaj I. W., Cusack S. 2002. Large-scale induced fit recognition of an m(7)GpppG cap analogue by the human nuclear cap-binding complex. EMBO J. 21:5548–5557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Neumann G., et al. 1999. Generation of influenza A viruses entirely from cloned cDNAs. Proc. Natl. Acad. Sci. U. S. A. 96:9345–9350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Neumann G., Zobel A., Hobom G. 1994. RNA polymerase I-mediated expression of influenza viral RNA molecules. Virology 202:477–479 [DOI] [PubMed] [Google Scholar]
- 26. Niedzwiecka A., et al. 2002. Biophysical studies of eIF4E cap-binding protein: recognition of mRNA 5′ cap structure and synthetic fragments of eIF4G and 4E-BP1 proteins. J. Mol. Biol. 319:615–635 [DOI] [PubMed] [Google Scholar]
- 27. Obayashi E., et al. 2008. The structural basis for an essential subunit interaction in influenza virus RNA polymerase. Nature 454:1127–1131 [DOI] [PubMed] [Google Scholar]
- 28. Ogino T., Kobayashi M., Iwama M., Mizumoto K. 2005. Sendai virus RNA-dependent RNA polymerase L protein catalyzes cap methylation of virus-specific mRNA. J. Biol. Chem. 280:4429–4435 [DOI] [PubMed] [Google Scholar]
- 29. Plotch S. J., Tomasz J., Krug R. M. 1978. Absence of detectable capping and methylating enzymes in influenza virions. J. Virol. 28:75–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Shimizu K., Handa H., Nakada S., Nagata K. 1994. Regulation of influenza virus RNA polymerase activity by cellular and viral factors. Nucleic Acids Res. 22:5047–5053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sugiyama K., et al. 2009. Structural insight into the essential PB1-PB2 subunit contact of the influenza virus RNA polymerase. EMBO J. 28:1803–1811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Tomassini J. E. 1996. Expression, purification, and characterization of orthomyxovirus: influenza transcriptase. Methods Enzymol. 275:90–99 [DOI] [PubMed] [Google Scholar]
- 33. Tomoo K., et al. 2002. Crystal structures of 7-methylguanosine 5′-triphosphate (m(7)GTP)- and P(1)-7-methylguanosine-P(3)-adenosine-5′,5′-triphosphate (m(7)GpppA)-bound human full-length eukaryotic initiation factor 4E: biological importance of the C-terminal flexible region. Biochem. J. 362:539–544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Tomoo K., et al. 2003. Structural features of human initiation factor 4E, studied by X-ray crystal analyses and molecular dynamics simulations. J. Mol. Biol. 328:365–383 [DOI] [PubMed] [Google Scholar]
- 35. Turan K., et al. 2004. Nuclear MxA proteins form a complex with influenza virus NP and inhibit the transcription of the engineered influenza virus genome. Nucleic Acids Res. 32:643–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Yuan P., et al. 2009. Crystal structure of an avian influenza polymerase PA(N) reveals an endonuclease active site. Nature 458:909–913 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.