Abstract
Viral hemorrhagic septicemia virus (VHSV) and infectious hematopoietic necrosis virus (IHNV) are members of the genus Novirhabdovirus within the Rhabdoviridae family, which can cause severe hemorrhagic disease in fresh- and saltwater fish worldwide. These viruses carry an additional nonvirion (NV) gene, which codes for the nonstructural NV protein that has been implicated to play a role in viral pathogenesis. To determine the precise biological function of this NV gene and its gene product, we generated NV-deficient and NV knockout recombinant VHSVs, using reverse genetics. Comparisons of the replication kinetics and markers for virus-induced apoptosis indicated that the NV-deficient and NV knockout mutant viruses induce apoptosis earlier in cell culture than the wild-type recombinant VHSV. These results suggest that the NV protein has an antiapoptotic function at the early stage of virus infection. Furthermore, we created a chimeric VHSV, in which the NV gene of VHSV was replaced by the IHNV NV gene, which was capable of suppressing apoptosis in cell culture. These results show that the NV protein of other members of Novirhabdovirus can restore the NV protein function. In this study, we also investigated the kinetics of VHSV replication during a single round of viral replication and examined the mechanism of VHSV-induced apoptosis. Our results show that VHSV infection induced caspases 3, 8 and 9 in cell culture.
INTRODUCTION
The Novirhabdovirus genus, belonging to the family Rhabdoviridae, was named after the presence of a unique gene, called the nonvirion (NV) gene, which is found between the glycoprotein (G) and polymerase (L) genes (38). Novirhabdovirus has four important type species: infectious hematopoietic necrosis virus (IHNV), viral hemorrhagic septicemia virus (VHSV), Hirame rhabdovirus, and snakehead rhabdovirus. To date, all members of the Novirhabdovirus genus are fish pathogens. The genome of Novirhabdovirus is composed of approximately 11 kb of single-stranded RNA, which contains the following five structural genes as other rhabdoviruses, the nucleocapsid protein (N), polymerase-associated phosphoprotein (P), matrix protein (M), surface glycoprotein (G), and virus polymerase (L) genes, as well as one nonstructural NV gene, which codes for NV protein that is expressed in infected cells but is not present in the virion particles (6, 33, 34, 38). The NV gene of Novirhabdovirus is approximately 500 nucleotides (nt) in length, which codes for the 12-kDa and 14-kDa NV proteins of IHNV and VHSV, respectively (25, 34).
Apoptosis, or programmed cell death, is an important physiological process for host defense against viral infection, and it occurs at the early stage of viral infection, thus limiting viral propagation (24). To overcome host resistance, many viruses carry antiapoptotic factors to inhibit apoptosis (20, 35). However, in some cases, viruses may trigger apoptosis at the late stage of viral replication to facilitate viral release and spread (18). The executioners in apoptosis are caspases, a family of cysteine-dependent, aspartate-directed proteinases. Caspases function as both effector caspases, such as caspases 3, 6, and 7, and initiator caspases, such as caspases 8 and 9 (36). Many members of the Rhabdoviridae family are known to induce apoptosis. For example, vesicular stomatitis virus (VSV) causes apoptosis through the initiator caspase 9 (21, 29). Rabies virus induces apoptosis through both caspase-dependent and -independent pathways (32). In the case of Novirhabdovirus, it was shown that VHSV infection results in massive apoptosis in renal lymphoid tissue and that carp leukocyte cells infected with turbot (Scophthalmus maximus L.) rhabdovirus induce the formation of discrete apoptotic bodies (13, 15). Also, it was observed that epithelioma papulosum cyprini (EPC) cells infected with wild-type IHNV produced lower levels of apoptosis than recombinant IHNV carrying the green fluorescence protein (GFP) gene rather than the NV gene (31). In earlier studies, it was shown that the matrix protein of Novirhabdovirus is involved in downregulation of host transcription and the induction of apoptosis (11). However, to date, the apoptotic kinetics of Novirhabdovirus and possible pathways of virus-induced apoptosis are unknown.
Complete nucleotide sequences of the NV genes of many IHNV and VHSV strains have been determined, which are highly variable between type species of Novirhabdovirus (3, 6, 26). Previous studies have demonstrated that the NV gene is not essential for viable virion production of IHNV and VHSV both in vitro and in vivo, but it is required for efficient viral replication and pathogenesis (1, 2, 7, 8, 37). The NV knockout recombinant IHNVs and VHSVs (rVHSVs) grow 1 to 2 logs lower in cell culture than the wild-type counterparts and are highly attenuated in the fish (1, 2, 7, 37). Besides its role in viral pathogenesis, the precise function of this NV protein is still unknown. Since the NV gene is present in all the IHNVs and VHSVs sequenced so far, we speculated that NV protein of Novirhabdovirus may have an antiapoptotic function. To date, none of the rhabdovirus proteins are shown to have antiapoptotic function.
In this study, we examined the apoptotic kinetics of VHSV and explored the possible pathways of virus-induced apoptosis. Furthermore, we generated the NV-deficient and NV knockout recombinant VHSVs, as well as a chimeric VHSV in which the NV gene of VHSV was replaced by the IHNV NV gene using reverse genetics, and investigated the role of the NV protein of Novirhabdovirus in apoptosis.
MATERIALS AND METHODS
Viruses, cells, and reagents.
The recombinant Great Lakes VHSV strain previously generated in our lab was propagated in EPC cells at 14°C (16). The cells were grown at 25°C in minimal essential medium (MEM) supplemented with 10% fetal bovine serum (FBS) and 2 mM l-glutamine. For preparation of recombinant virus stocks, confluent EPC cells grown at 25°C were infected at a multiplicity of infection (MOI) of 0.001 in MEM with 2% FBS. After 1 h of adsorption at 14°C, the inoculum was removed, and the cells were incubated at 14°C until extensive cytopathic effect (CPE) was observed. The supernatant was collected 3 or 4 days postinfection (p.i.), clarified, and stored at −80°C for further processing.
Construction of plasmids and rescue of mutant viruses.
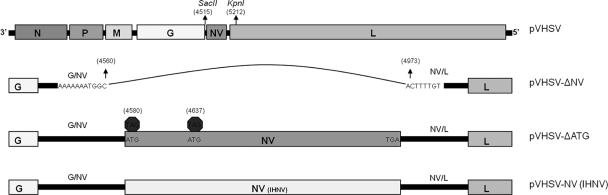
The full-length cDNA clone of VHSV utilized in this study was constructed, as described previously (1). The pVHSV-ΔNV clone was made by deleting the complete NV gene from nucleotides 4561 to 4982 (with respect to the antiviral sense of the complete genome). Construction of the engineered mutant clones are shown in Fig. 1. Gene manipulations were carried out by amplifying the C-terminal region of the G gene, the G/NV gene junction, and part of the NV/L gene border sequence of pVHSV full-length clone. The following primer pairs were used to amplify the above said region: primer pair SacIIF, TGAACACACCCGCGGAAACCACCATAG, and VHSV-NVkoR, CAAACAACAAAAGTGCCATTTTTTTCTATCTATCTAA, and primer pair VHSV-NVkoF, ATGGCACTTTTGTTGTTTGTAATCCTACA, and 5.3R, CTGGATTCGTGTAGAGTCAA. The resulting fragments were digested with respective restriction enzymes to replace the region between the SacII and KpnI restriction sites in the full-length pVHSV clone. This created an NV knockout cDNA clone of VHSV (pVHSV-ΔNV), as shown in Fig. 1.
Fig. 1.
Diagram of the recombinant VHSV genomes. The pVHSV-ΔNV plasmid was constructed by deleting the complete NV gene from nucleotide positions 4561 to 4982 (with respect to the antiviral sense of the complete genome). The pVHSV-ΔATG plasmid was constructed by mutating the first and second ATG (58th nucleotide position from the first ATG) codons of the NV ORF to TGA codons. The pVHSV-NV (IHNV) plasmid was made by replacing the ORF of VHSV NV with the ORF of the NV gene of the IHNV 220-90 strain. (Nucleotide positions relative to the VHSV complete genome are given in parentheses. G/NV, gene junction between G and NV; NV/L, gene junction between the NV and L genes.)
For the construction of the pVHSV-ΔATG clone, the first and second ATG (58th nucleotide position from the first ATG) codons of the NV open reading frame (ORF) were replaced with TGA to eliminate expression of the 12-kDa NV protein. The following primers were used to amplify the respective fragments from the pVHSV clone: (i) SacIIF, TGAACACACCCGCGGAAACCACCATAG, and VHSV-NVatg1R, CCGGCTTAATTACTACTTTCTGTCTCACAGGGGTGCCATTT; (ii) VHSV-NVatg1F, AAGTAGTAATTAAGCCGGCACACAGCACAACCA, and VHSV-NVatg2R, GTATGCGATCTACTCGTGGAGGACAAGT; and (iii) VHSV-NVatg2F, CCACGAGTAGATCGCATACAGACTAA, and 5.3R, CTGGATTCGTGTAGAGTCAA. The resulting fragments were fused by overlapping PCR, and the final fragment was digested with respective restriction enzymes to replace the region between the SacII and KpnI restriction sites in the full-length cDNA clone. This replacement created an NV-deficient full-length cDNA clone of VHSV (pVHSV-ΔATG), as shown in Fig. 1.
The rVHSV-NVIHNV chimeric virus was made by replacing the ORF of VHSV NV with the ORF of the NV gene of the IHNV 220-90 strain. The NV ORF of IHNV was amplified from the pIHNV (2) clone using the primer pairs that amplified (i) sequences between the stop codon of the G gene and the start codon of the NV gene (SacIIF, TGAACACACCCGCGGAAACCACCATAG, and VHS-NV2-IHN5R, GGTGGTCCATCTTTCTGTCTCACAGGGGTGCCATT), (ii) sequences between the stop codon of the NV gene and part of the coding region of the L gene (VHS-NV2-IHN3F, GCTTATCCCAGATAGACTCCTCCCCCTTCTCCCAGATA, and 5.3R, CCTGGATTCGTGTAGAGTCAA), and (iii) the IHNV NV ORF (VHS-NV2-IHN5F, CCTGTGAGACAGAAAGATGGACCACCGCGACATA, and VHS-NV2-IHN3R, GGAGGAGTCTATCTGGGATAAGCAAGAAA). These three fragments were fused to each other by two-step overlapping PCR and then digested with respective restriction enzymes to replace the region between the SacII and KpnI restriction sites in the full-length cDNA clone. This replacement created an NV-substituted full-length cDNA clone of VHSV (pVHSV-NVIHNV), as shown in Fig. 1. The NV knockout, NV-deficient mutant, and chimeric VHSVs were recovered by using reverse genetics, as described previously (1).
DNA fragmentation assay.
Mock-infected or virus-infected cells (∼3 × 106) grown in 25-cm2 tissue culture flasks were harvested at different time points, and low-molecular-weight DNAs were extracted. A DNA fragmentation assay was performed, as described previously (27). Briefly, the collected cells were washed in cold phosphate-buffered saline (PBS), resuspended in 500 μl of ice-cold lysis buffer (10 mM Tris [pH 7.5], 10 mM EDTA [pH 7.5], and 0.2% Triton X-100), and incubated at 4°C for 10 min. Lysates were centrifuged at 10,000 × g at 4°C for 1 min, and supernatants were subjected to phenol-chloroform-isoamyl alcohol (25:24:1 [vol/vol/vol]), by using 2 ml of phase lock gel (5 Prime GmbH, MD). DNAs were ethanol precipitated with 3 M sodium acetate. DNA samples were resuspended in 50 μl of sterile water and treated for 15 min at 37°C with RNase A (Fermentas, MD) at a final concentration of 1.0 μg/μl. All samples were run in a 1% agarose gel in 1× Tris-borate-EDTA buffer and stained with ethidium bromide.
Western blot analysis.
To detect viral protein expression levels, VHSV-infected EPC cells (25-cm2 tissue culture flasks) were harvested at the indicated time points, and the monolayer was washed twice with ice-cold PBS and incubated with 500 μl of RIPA buffer (25 mM Tris-HCl, [pH 7.6], 150 mM NaCl, 1% NP-40, 1% sodium deoxycholate, 0.1% SDS) at 4°C for 10 min. Cell lysate was centrifuged, and the supernatant was collected and stored at −20°C until used. For immunoblotting, the proteins were electrophoretically separated in an 11% sodium dodecyl sulfate-polyacrylamide gel. VHSV proteins were detected by Western blotting using polyclonal antibody against wild-type VHSV.
Plaque assay.
Infected cell cultures collected at different time intervals were stored at −80°C until used, and the titers of infectious progeny were determined by plaque assay. Briefly, confluent EPC cells were infected with recombinant virus stocks at the indicated multiplicity of infection (MOI) in individual T-25 flasks. Virus present in the infected cell culture supernatant was collected at different time intervals, clarified by centrifugation, and titrated on EPC cells by plaque assay, as described earlier (10). After 7 days of incubation at 14°C, the overlays were removed, and the cells were fixed and stained with a solution containing 25% formalin, 10% ethanol, 5% acetic acid, and 1% crystal violet for 5 min at room temperature. After rinsing of the cells with distilled water, the plaques were counted.
Caspase 3, 8, and 9 activity assays.
Caspase activity assays were performed using the procedures described in colorimetric assay kits obtained from BioVision (Mountain View, CA). EPC cells (3 × 106) grown in T-25 flask were mock infected or infected with recombinant viruses at an MOI of 1.0 and incubated for various intervals. The cells were trypsinized and washed with cold PBS. The cell pellets were resuspended in 50 μl of chilled cell lysis buffer and incubated on ice for 10 min. The cell lysates were harvested by centrifugation at 10,000 × g for 10 min. The supernatants were collected and frozen at −80°C until samples from all the time points were harvested. The colorimetric assay utilizes peptide substrates consisting of the consensus cleavage sequence for each caspase labeled with chromophore pNA (p-nitroanilide), such as DEVD-pNA (synthetic caspase 3 substrate), IETD-pNA (synthetic caspase 8 substrate), and LEHD- pNA (synthetic caspase 9 substrate), which were used in the study. Activated caspases in apoptotic cells cleave the synthetic substrates to release free pNA, which is then quantified using a microtiter plate reader. For each reaction, 50 μl of cell lysate and 50 μl of 2× reaction buffer containing 5 mM dithiothreitol (DTT) were mixed and then incubated with 50 mM of the caspase substrate at 37°C for 90 min. Finally, the reactions were analyzed in a microtiter plate reader (SpectraMax M5 microplate reader; Molecular Devices, Inc., CA) at 400 nm. Comparison of the absorbance of pNA from an apoptotic sample with an uninduced control allows determination of the fold increase in respective caspase activity. Results of all experiments are reported as means ± standard deviations (SD).
Apoptosis induction.
Protein synthesis inhibitor cycloheximide (CHI; 100 μg/ml) or the transcription inhibitor actinomycin D (ActD; 4 μg/ml) was used as a nonviral inducer of apoptosis. To determine the antiapoptotic nature of VHSV, the ActD was added to the infected cells after 2 h postinfection.
Statistical analysis.
A paired Student's t test was used to compare significances of individual time points. A P value of <0.05 was considered to be statistically significant.
RESULTS
Kinetics of VHSV replication and apoptosis.
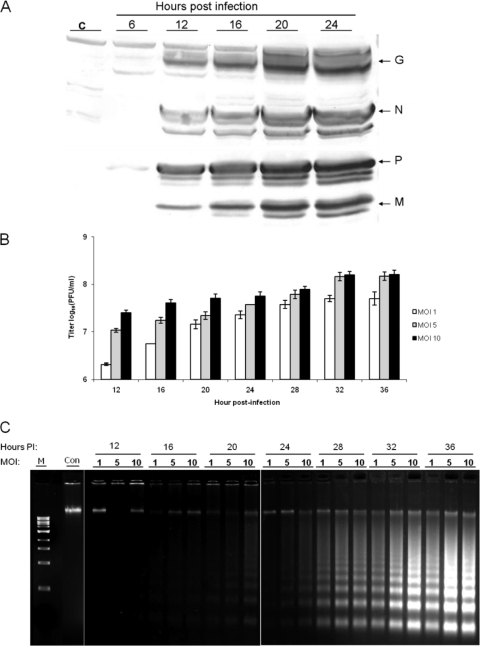
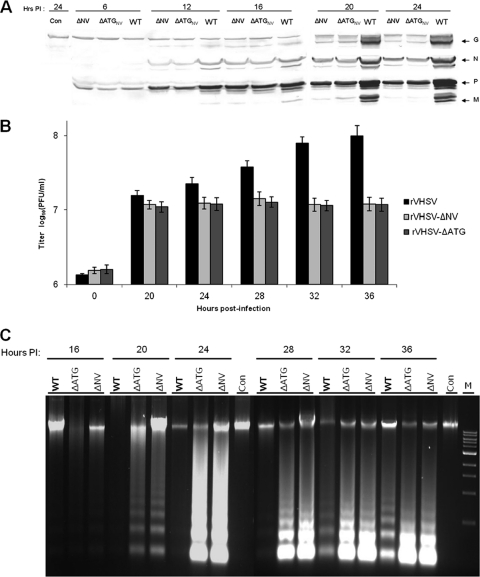
Initially, we investigated VHSV-induced apoptosis and possible pathways during the early stage of viral infection in order to provide valuable indicators for analysis of the NV protein function. First, the replication and apoptotic kinetics were studied by infecting EPC cells with different MOIs of rVHSV (MOIs of 1.0, 5.0, and 10.0). Viral protein expression was detected by immunostaining, and the production of infectious viral progeny was quantified by plaque assay at different time points. As shown in Fig. 2A, the increased viral protein expression in infected cells could be detected at 12 h postinfection (p.i.). Accordingly, there was very little viral protein detected prior to 12 h p.i., but the viral yields started increasing steadily after that (Fig. 2B). A DNA laddering assay was utilized to investigate VHSV-induced apoptosis, as shown in Fig. 2C. DNA fragmentation, which is a characteristic event in apoptotic cells, became detectable between 20 and 24 h and completed between 32 and 36 h p.i., suggesting that apoptosis occurred at the late stage of viral replication.
Fig. 2.
Kinetics of VHSV replication and induction of apoptosis in EPC cells during a single round of the viral life cycle. EPC cells were infected with rVHSV (MOI = 1.0, 5.0, and 10.0) and harvested at the indicated time intervals. (A) Kinetics of viral protein expression (MOI = 10.0). The virus-specific proteins (glycoprotein [G; 52 kDa], phosphoprotein [P; 26 kDa], nucleoprotein [N; 42 kDa], and matrix protein [M; 22 kDa]) in cell lysates were detected by Western blotting, as described in Materials and Methods. (B) Production of infectious viral progenies. Viral yields were quantified by plaque assay at the indicated times. Values of virus titers are shown as the means ± SD from three independent experiments. (C) DNA laddering in VHSV-infected EPC cells. At the indicated time points, infected and mock-infected cells were harvested for a DNA fragmentation assay.
VHSV infection induces caspases in cell culture.
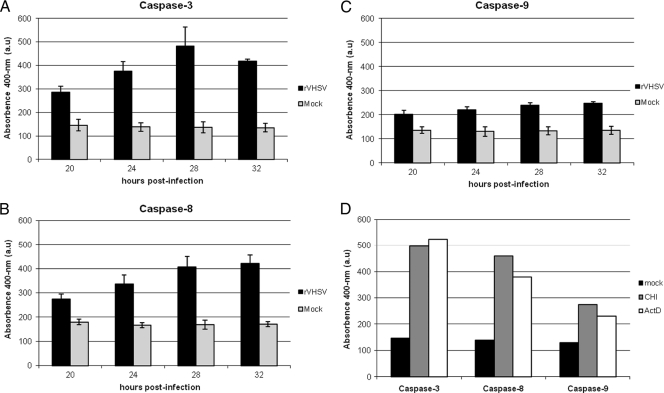
To investigate if VHSV induced any caspase pathways, caspase 3 activity in EPC cells during VHSV infection was examined at different time points. Caspase 3 activity was detected by a colorimetric method based on cleavage of the DEVD-pNA substrate. Figure 3A shows that caspase 3 was activated at approximately 20 h p.i. and that its activity increased over time, which correlates with the DNA laddering results. Both caspases 8 and 9 are upstream caspases in either extrinsic or intrinsic pathways. To determine which initiator caspase(s) is involved in VHSV-induced apoptosis, the activation of caspases 8 and 9 was examined. As shown in Fig. 3B and C, both caspases 8 and 9 were activated during VHSV infection. As a positive control, actinomycin D and cycloheximide were used, which induced both of the initiator caspases (Fig. 3D).
Fig. 3.
Caspase 3, 8, and 9 activities in VHSV-infected cells. EPC cells were mock infected or infected with rVHSV at an MOI of 5.0 PFU per cell, and cell lysates were prepared at the indicated times postinfection. The enzymatic activities of caspases 3, 8, and 9 were quantified as described in Materials and Methods. Caspase 3-like (A), caspase 8-like (B), and caspase 9-like (C) activities at the indicated times after infection (D). As a positive control for caspase 3, 8, and 9 activation, cycloheximide (CHI; 100 mg/ml) and actinomycin D (ActD, 4 μg/ml) were added to the cells. After 12 h of incubation at 4°C, cell lysates were prepared, and the enzymatic activities of caspases 3, 8, and 9 were determined. a.u, arbitrary units.
Antiapoptotic activity of VHSV.
Remarkably, induction of an antiapoptotic state by VHSV was evidenced by the resistance of the infected cells to a nonviral apoptotic inducer, actinomycin D. The treatment of uninfected cells with actinomycin D (4 μg/ml) for 20 h at 14°C resulted in the development of apoptosis in a significant proportion of cells (Fig. 4A). The yield of infectious VHSV titers was not significantly affected by actinomycin D treatment, and approximately 2.6 × 107 PFU/ml were produced by 20 h p.i. in the presence and absence of the drug (Fig. 4B). Moreover, actinomycin D-induced apoptosis was strongly suppressed in the VHSV-infected cells (Fig. 4C). All these data strongly suggest that VHSV infection orchestrates an antiapoptotic program in the infected cells. To exclude the possibility of viral proteins other than NV having antiapoptotic activity, the NV knockout VHSV was examined for its resistance to nonviral apoptotic inducer. Figure 4D shows that the NV knockout virus did not protect the EPC cells from actinomycin D-induced apoptosis, but instead, it exacerbated the apoptosis.
Fig. 4.
Inhibition of actinomycin D-induced apoptosis by VHSV. EPC cells were mock infected or VHSV infected and incubated at 14°C for 20 h in the absence and presence of 4 μg/ml actinomycin D (ActD). (A) Cytopathic effect induced by the recombinant VHSV in EPC cells. (B) Effect of ActD on viral titer at 20 h postinfection. (C) DNA fragmentation assay of the wild-type VHSV. (D) DNA fragmentation assay of rVHSV-ΔNV. Lanes M and C are a 1-kb DNA ladder marker and mock-infected control cells, respectively.
NV-deficient VHSV induces apoptosis earlier.
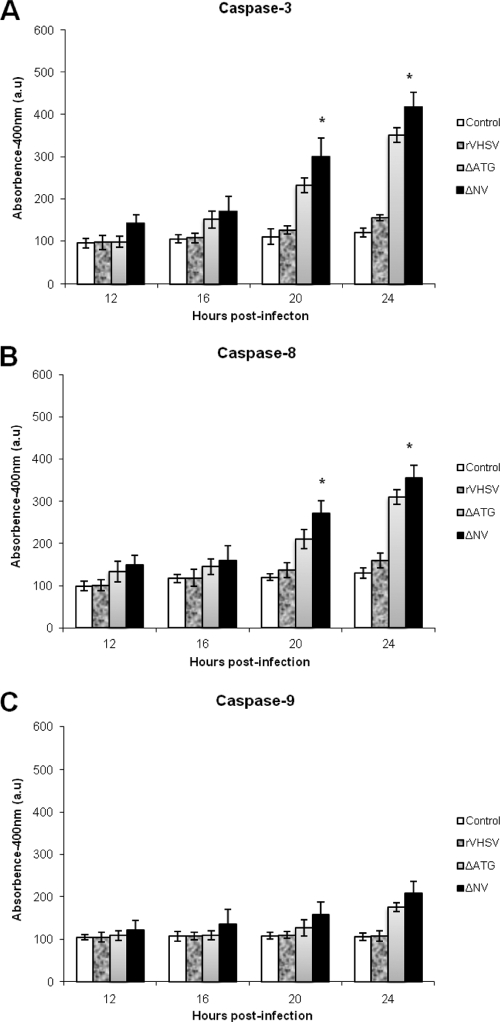
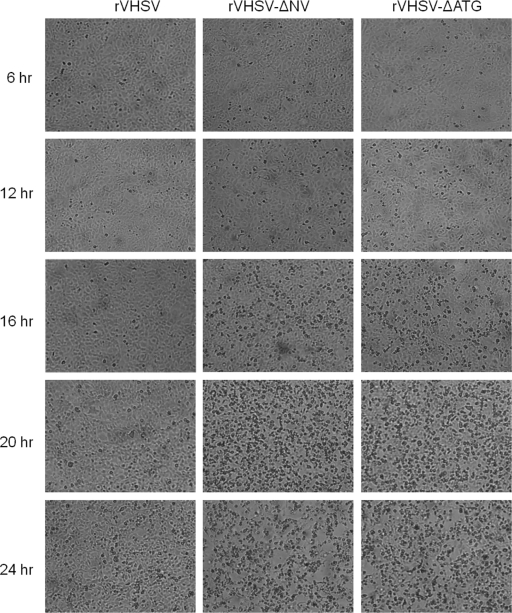
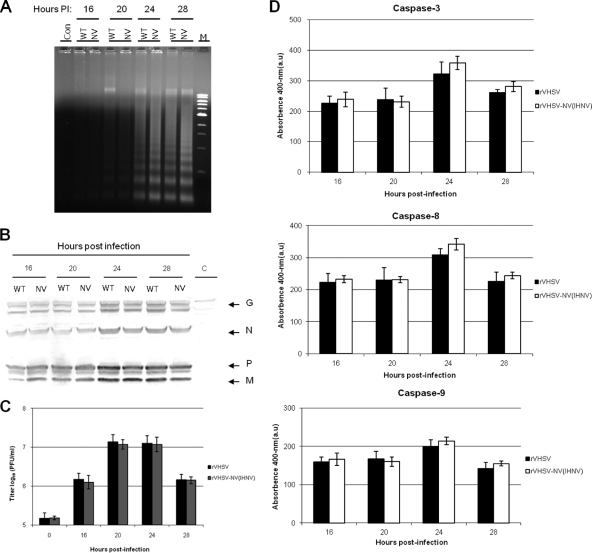
To ascertain the possible involvement of the VHSV NV protein in the control of apoptotic machinery, an NV knockout recombinant VHSV was generated by deleting the NV gene (rVHSV-ΔNV), and used in this study. To distinguish the role, if any, of the NV mRNA from the NV protein in the actual apoptotic machinery, we also constructed an NV-deficient recombinant VHSV (rVHSV-ΔATG) by mutating the start codon and an internal ATG of the NV ORF to a stop codon to ensure that no truncated NV protein will be produced from the mRNA. Having demonstrated that both caspase 8 and 9 activations could be used as markers for VHSV-induced apoptosis, we wanted to determine the function of the NV protein in apoptosis during a single round of viral replication. When EPC cells were infected with these three viruses, there was an appreciable difference in viral protein synthesis between the wild-type VHSV and its NV mutants (Fig. 5A). Before producing enough viral proteins, the cell machinery was completely shutdown by the NV mutant viruses, and cells had undergone complete lysis. However, rVHSV kept producing more viral proteins and thereby produced maximum progeny at 36 h p.i, whereas NV mutant viruses grew slower and produced 1 log lower progenies than its counterpart (Fig. 5B). As shown in Fig. 5C, rVHSV-ΔNV and rVHSV-ΔATG induced a stronger DNA laddering effect than the rVHSV at 20 h p.i., even though the latter was inoculated at an MOI of 5.0. Similarly, NV-deficient viruses induced higher caspase 3, 8, and 9 activities than rVHSV at 16, 20, and 24 h p.i (Fig. 6A to C). In addition, the CPE in rVHSV-ΔNV-infected cells occurred earlier and was greater than that in rVHSV-infected EPC cells during a single round of viral replication (Fig. 7). Taken together, our results indicate that rVHSV-ΔNV and rVHSV-ΔATG induce increased apoptosis compared to rVHSV, suggesting that the NV protein is antiapoptotic during the early stage of viral replication.
Fig. 5.
Replication and apoptotic kinetics of rVHSV, rVHSV-ΔNV, and rVHSV-ΔATG at the early stage of virus infection. EPC cells (∼5 × 106) were inoculated with equal amounts of rVHSV, rVHSV-ΔNV, and rVHSV-ΔATG at an MOI of 1.0 PFU per cell. (A) Cell lysates were harvested and analyzed for the virus-specific proteins by immunoblotting. (B) Infected cells were freeze-thawed, and viruses were quantified by plaque assay at the indicated time points. (C) The low-molecular-weight DNAs from cell lysates were extracted and analyzed by a DNA laddering assay. Con, control.
Fig. 6.
Caspase 3, 8, and 9 activities in rVHSV-, rVHSV-ΔNV-, and rVHSV-ΔATG-infected cells. EPC cells (∼3 × 106) were inoculated with equal amounts of rVHSV, rVHSV-ΔNV, and rVHSV-ΔATG at an MOI of 1.0 PFU per cell, and cell lysates were prepared at the indicated times postinfection. Caspase 3-like (A), caspase 8-like (B), and caspase 9-like (C) activities at the indicated times after infection. All assays were performed as described in Materials and Methods (*, P < 0.05 compared to that of rVHSV-ΔATG).
Fig. 7.
Cytopathic effect induced by the recombinant VHSVs in EPC cells. EPC cells were infected with recombinant VHSV (rVHSV panels), NV gene-deleted recombinant VHSV (rVHSV-ΔNV panels), and rVHSV with a mutated start codon of the NV gene (rVHSV-ΔATG panels). At different times postinfection, cell cultures were examined by light microscopy, and the cell morphology was photographed.
Apoptosis kinetics of chimeric VHSV expressing the NV gene of IHNV.
To demonstrate the antiapoptotic property of the NV protein regardless of the other Novirhabdovirus genome, we replaced the NV ORF of VHSV with the NV ORF of IHNV and compared the apoptosis kinetics of chimeric rVHSV-NVIHNV with that of the wild-type rVHSV. When EPC cells were infected with an equal amount of these viruses (MOI of 5.0), the electrophoretic pattern of DNA fragmentation produced by rVHSV-NVIHNV was similar to that of rVHSV (Fig. 8A), and there was no appreciable difference in viral protein synthesis between these two viruses (Fig. 8B). This result clearly demonstrates that the NV protein of IHNV does not alter the amount of VHSV protein synthesis in cell culture, which is further supported by the fact that the growth characteristic of rVHSV-NVIHNV was similar to that of rVHSV in EPC cells (Fig. 8C). The apoptotic kinetics of rVHSV-NVIHNV was further studied by measuring the caspase 3-, 8-, and 9-like activities. Our results indicate that there was a subtle difference in the levels of induced caspases between rVHSV and rVHSV-NVIHNV at different time points, and the latter induces slightly elevated but not statistically significant levels of all three caspase activities (Fig. 8D).
Fig. 8.
Comparison of replication and apoptotic kinetics of chimeric rVHSV-NVIHNV and rVHSV. EPC cells (∼1 × 106) were inoculated with equal amounts of rVHSV and rVHSV-NVIHNV at an MOI of 5.0 PFU per cell. (A) The low-molecular-weight DNAs from cell lysates were extracted and analyzed by a DNA laddering assay. (B) Cell lysates were harvested and analyzed for the virus-specific proteins by immunoblotting. C, control. (C) Infected cells were freeze-thawed, and viruses were quantified by plaque assay at the indicated time points. (D) Caspase 3-like, caspase 8-like, and caspase 9-like activities at the indicated times postinfection. Average titers and standard deviations (error bars) for the three replicates are shown. All assays were performed as described in Materials and Methods.
DISCUSSION
Viruses usually trigger apoptotic signals during infection of a cell that enables the host to limit virus replication and spread (19, 22). This apoptosis is more likely to be responsible for the pathogenesis of the infectious disease. At the same time, many viruses carry antiapoptotic genes to suppress apoptosis by which virus is able to produce more progenies by prolonging the survival of infected cells (4, 23).
In this study, the kinetics and signaling pathways of VHSV-induced apoptosis were studied in cell culture for the first time, based on a single round of replication. Our data show that VHSV-induced apoptosis occurs at the late stage during virus infection in EPC cells. An increase in viral protein expression was observed at 12 h p.i., whereas the viral yield was significantly higher after 16 h p.i. The appreciable DNA fragmentation and cytopathic effects starts appearing around 20 h p.i. Virus titer reaches the maximum between 32 and 36 h p.i and coincides with complete cell lysis, which occurs at a time after maximum progenies are released from infected cells.
We demonstrated that VHSV induced caspase 3, 8, and 9 activities in EPC cells, and the initiator caspases 8 and 9 and the effector caspase 3 were significantly induced, suggesting that VHSV-induced apoptosis is both receptor and mitochondrion mediated. In the case of VSV, it was shown that the VSV mutant of the matrix protein induces apoptosis through distinct pathways, and the investigators suggested that caspase 9 is activated by cross talk with caspase 8 (17). In our study, although we demonstrated a significant level of caspase 9 activity at the late stage of infection, the induction of caspase 9 was not as high as caspase 8, which could be due to cross talk between the caspase 8 and caspase 9 pathways.
The antiapoptotic state of VHSV was evidenced by the resistance of the infected cells to actinomycin D, a nonviral apoptotic inducer, which induces apoptosis by inhibiting the cellular transcription (30). When EPC cells were infected with VSHV and treated with actinomycin D, they showed greater resistance to actinomycin D treatment after 20 h p.i., whereas uninfected EPC cells treated with actinomycin D exhibited very significant apoptosis (Fig. 4). This result clearly indicates that VHSV elicits a strong antiapoptotic state by resisting apoptosis triggered not only by a viral inducer but also by a nonviral inducer at early stages of viral life cycle.
To study the function of the NV protein of VHSV, we generated an NV knockout mutant virus and compared the apoptosis and signaling pathways with those of the wild-type virus. We demonstrated that infection of EPC cells with the wild-type rVHSV leads to a limited extent of apoptotic cell death at early stages in infection, but the infection with an NV knockout VHSV results in extensive DNA fragmentation (due to apoptosis) and cytopathic effects at 20 h p.i. (Fig. 5C and 7). This result suggests that the NV protein is involved in antiapoptotic activity at the early stage of virus infection. By performing transfection experiments using an expression vector that codes for the NV protein, we observed that expression of NV in trans can completely restore the antiapoptotic activity of VHSV during infection (data not shown). By interfering with the apoptosis, NV functions as an antiapoptotic protein, but lack of any sequence homology with existing apoptotic proteins suggests that this may represent a new class of viral antiapoptotic proteins.
Many viruses carry genes encoding nonstructural proteins that effectively suppress or delay apoptosis, which enhance viral replication and consequently contribute to viral pathogenesis. In the case of respiratory syncytial virus (RSV), the two nonstructural (NS1 and NS2) proteins boost viral replication by delaying the apoptosis of the infected cell (9). The nonstructural protein (NS1) of influenza virus inhibits caspase 9 and glycogen synthase kinase 3β by activating the phosphatidylinositol 3-kinase (PI3K)/Akt pathway (14), thereby limiting the virus-induced cell death program. VP5, which is a nonstructural protein of infectious bursal disease virus (IBDV), has an antiapoptotic function at the early stage of virus infection (27). Similarly, a nonstructural protein, NS5A, encoded by hepatitis C virus, was shown to have an antiapoptotic function (12, 28). Here, we demonstrate that Novirhabdovirus carries a nonstructural protein, NV, which evades the cell defense and optimally produces viral progenies by delaying the programmed cell death triggered by the host cell.
By comparing VHSV-induced apoptotic characteristics of NV-deficient mutant viruses and wild-type VHSV, we provide important evidence that the Novirhabdovirus NV protein functions as an antiapoptotic protein during the early stage of VHSV replication, whose deficiency leads to the occurrence of apoptosis. First, the NV knockout and NV-deficient viruses cause greater apoptotic effects than rVHSV, as shown by DNA laddering and caspase 3, 8, and 9 activities. Second, early apoptosis is accompanied by decreased production of viral progeny, which suggests that the NV protein probably interacts with a cellular pathway to delay the apoptosis rather than virion release from the cell membrane. Therefore, the NV protein performs the function of inhibiting apoptosis initiated by viral replication and prevents infected cells from undergoing cell death before the virus finishes its life cycle. Thus, early apoptosis of infected cells results in reduced production of viral progeny. Consequently, the NV-deficient virus replicates slower and has a 1-log-lower titer than rVHSV (Fig. 5B).
To examine the role of the NV gene or its mRNA, rather than that of the NV protein, we also constructed an NV-deficient VHSV (rVHSV-ΔATG). We observed that rVHSV-ΔATG produced slighter effects than rVHSV-ΔNV in all the experiments, as follows: growth in cell culture, development and completion of CPE, and amount of caspases induced. At 20 h and 24 h p.i., rVHSV-ΔNV induced significantly higher caspase 3- and caspase 8-like activities than rVHSV-ΔATG (Fig. 6). It is not clear what the cause of these subtle effects is. However, it may be interpreted that NV protein is the one involved in apoptotic machinery and not the NV gene.
The antiapoptotic activity of the VHSV NV protein was further confirmed by investigating the NV protein of IHNV, which is a type species of Novirhabdovirus. For this purpose, we made a chimeric recombinant VHSV in which the VHSV NV ORF was replaced by the IHNV NV ORF. Chimeric rVHSV-NVIHNV exhibited growth properties similar to those of the wild-type rVHSV and no statistically significant differences in caspase induction, and DNA fragmentation and viral protein synthesis were observed between rVHSV and rVHSV-NVIHNV (Fig. 8).These results show that the NV protein of Novirhabdovirus is capable of suppressing apoptosis nonspecifically in EPC cells. Although the deduced amino acid sequence of the IHNV NV protein exhibits only 16% identity with that of the VHSV NV protein (3), it can restore the NV protein function and suppress apoptosis effectively regardless of the other Novirhabdovirus NV gene.
The antiapoptotic function of the NV protein can also correlate with attenuation of the virus in vivo. Recently, we reported that the NV-deficient VHSV caused only 16% cumulative percent mortality (CPM), compared to 100% CPM caused by rVHSV (1). Interestingly, fish which died of rVHSV-ΔNV showed symptoms similar to those caused by rVHSV. These findings suggested that the NV protein may not modulate the virulence of VHSV directly; instead, it promotes the viral replication by maintaining the cell viability, thereby causing high mortality. In other words, NV protein plays an indirect role in the pathogenesis of Novirhabdovirus by promoting the viral growth by delaying the cellular apoptosis program. To the best of our knowledge, this is the first report describing an antiapoptotic function of the NV protein in Novirhabdovirus.
It is very interesting that Novirhabdovirus expresses matrix protein, which induces apoptosis, and at the same time expresses NV protein to suppress apoptosis. Novirhabdovirus synthesizes RNA transcripts in a concentration gradient manner (5) in the following order, N > P > M> G> NV> L, which results in the production of a large amount of matrix (M) protein compared to production of a very small amount of NV protein. The virus survival is dependent upon effective exploitation of the existing cellular machinery. In this case, NV protein is produced only in catalytic amounts to prevent early apoptosis and is not produced enough to thwart late apoptosis, caused by abundant matrix protein, which would facilitate the viral release. Thus, Novirhabdovirus has evolved a strategy to adjust the subtle balance of early and late apoptosis to optimize viral replication efficiency. The involvement of the NV protein in the apoptosis pathway should be explored in detail with further studies.
Footnotes
Published ahead of print on 8 June 2011.
REFERENCES
- 1. Ammayappan A., Kurath G., Thompson T. M., Vakharia V. N. 2010. A reverse genetics system for the Great Lakes strain of viral hemorrhagic septicemia virus: the NV gene is required for pathogenicity. Mar. Biotechnol. doi:10.1007/s10126-010-9329-4 [DOI] [PubMed] [Google Scholar]
- 2. Ammayappan A., LaPatra S. E., Vakharia V. N. 2010. A vaccinia-virus-free reverse genetics system for infectious hematopoietic necrosis virus 220-90. J. Virol. Methods 167:132–139 [DOI] [PubMed] [Google Scholar]
- 3. Ammayappan A., Vakharia V. N. 2009. Molecular characterization of the Great Lakes viral hemorrhagic septicemia virus (VHSV) isolate from USA. Virol. J. 6:171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bagchi P., et al. 2010. Rotavirus nonstructural protein 1 suppresses virus-induced cellular apoptosis to facilitate viral growth by activating the cell survival pathways during early stages of infection. J. Virol. 84:6834–6845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Banerjee A. K. 1987. Transcription and replication of rhabdoviruses. Microbiol. Rev. 51:66–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Basurco B., Benmansour A. 1995. Distant strains of the fish rhabdovirus VHSV maintain a sixth functional cistron which codes for a nonstructural protein of unknown function. Virology 212:741–745 [DOI] [PubMed] [Google Scholar]
- 7. Biacchesi S., Lamoureux A., Merour F., Bernard J., Bremont M. 2010. Limited interference at the early stage of infection between two recombinant novirhabdoviruses: viral hemorrhagic septicemia virus and infectious hematopoietic necrosis virus. J. Virol. 84:10038–10050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Biacchesi S., Thoulouze M. I., Bearzotti M., Yu Y. X., Bremont M. 2000. Recovery of NV knockout infectious hematopoietic necrosis virus expressing foreign genes. J. Virol. 74:11247–11253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bitko V., et al. 2007. Nonstructural proteins of respiratory syncytial virus suppress premature apoptosis by an NF-kappaB-dependent, interferon-independent mechanism and facilitate virus growth. J. Virol. 81:1786–1795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Burke J. A., Mulcahy D. 1980. Plaquing procedure for infectious hematopoietic necrosis virus. Appl. Environ. Microbiol. 39:872–876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chiou P., Kim P., Carol H., Ormonde P., Leong J. C. 2000. Infectious hematopoietic necrosis virus matrix protein inhibits host-directed gene expression and induces morphological changes of apoptosis in cell cultures. J. Virol. 74:7619–7627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chung Y. L., Sheu M. L., Yen S. H. 2003. Hepatitis C virus NS5A as a potential viral Bcl-2 homologue interacts with Bax and inhibits apoptosis in hepatocellular carcinoma. Int. J. Cancer 107:65–73 [DOI] [PubMed] [Google Scholar]
- 13. Du C. S., Zhang Q. Y., Li C. L., Miao D. L., Gui J. F. 2004. Induction of apoptosis in a carp leucocyte cell line infected with turbot (Scophthalmus maximus L.) rhabdovirus. Virus Res. 101:119–126 [DOI] [PubMed] [Google Scholar]
- 14. Ehrhardt C., et al. 2007. Influenza A virus NS1 protein activates the PI3K/Akt pathway to mediate antiapoptotic signaling responses. J. Virol. 81:3058–3067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Eleouet J. F., et al. 2001. Comparative study of in-situ cell death induced by the viruses of viral haemorrhagic septicaemia (VHS) and infectious pancreatic necrosis (IPN) in rainbow trout. J. Comp. Path. 124:300–307 [DOI] [PubMed] [Google Scholar]
- 16. Fijan N., et al. 1983. Some properties of the Epitheliorna papulosum cyprini (EPC) cell line from carp Cyprinus carpio. Ann. Virol. (Inst. Pasteur). 134E:207–220 [Google Scholar]
- 17. Gaddy D. F., Lyles D. S. 2005. Vesicular stomatitis viruses expressing wild-type or mutant M proteins activate apoptosis through distinct pathways. J. Virol. 79:4170–4179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hay S., Kannourakis G. 2002. A time to kill: viral manipulation of the cell death program. J. Gen. Virol. 83:1547–1564 [DOI] [PubMed] [Google Scholar]
- 19. Hinshaw V. S., Olsen C. W., Dybdahl-Sissoko N., Evans D. 1994. Apoptosis: a mechanism of cell killing by influenza A and B viruses. J. Virol. 68:3667–3673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kelly G. L., et al. 2009. An Epstein-Barr virus anti-apoptotic protein constitutively expressed in transformed cells and implicated in burkitt lymphomagenesis: the Wp/BHRF1 link. PLoS Pathog. 5:e1000341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kopecky S. A., Lyles D. S. 2003. Contrasting effects of matrix protein on apoptosis in HeLa and BHK cells infected with vesicular stomatitis virus are due to inhibition of host gene expression. J. Virol. 77:4658–4669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Koyama A. H. 1995. Induction of apoptotic DNA fragmentation by the infection of vesicular stomatitis virus. Virus Res. 37:285–290 [DOI] [PubMed] [Google Scholar]
- 23. Koyama A. H., Irie H., Kato A., Nagai Y., Adachi A. 2003. Virus multiplication and induction of apoptosis by Sendai virus: role of the C proteins. Microbes Infect. 5:373–378 [DOI] [PubMed] [Google Scholar]
- 24. Koyama A. H., et al. 1998. Role of virus-induced apoptosis in a host defense mechanism against virus infection. J. Med. Invest. 45:37–45 [PubMed] [Google Scholar]
- 25. Kurath G., Leong J. C. 1985. Characterization of infectious hematopoietic necrosis virus mRNA species demonstrates a nonvirion rhabdovirus protein. J. Virol. 53:462–468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kurath G., Higman K. H., Bjorklund H. V. 1997. Distribution and variation of NV genes in fish rhabdoviruses. J. Gen. Virol. 78:113–117 [DOI] [PubMed] [Google Scholar]
- 27. Liu M., Vakharia V. N. 2006. Nonstructural protein of infectious bursal disease virus inhibits apoptosis at the early stage of virus infection. J. Virol. 80:3369–3377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Miyasaka Y., et al. 2003. Hepatitis C virus nonstructural protein 5a inhibits tumor necrosis factor-alpha mediated apoptosis in Huh7 cells. J. Infect. Dis. 188:1537–1544 [DOI] [PubMed] [Google Scholar]
- 29. Pearce A. F., Lyles D. S. 2009. Vesicular stomatitis virus induces apoptosis primarily through Bak rather than Bax by inactivating Mcl-1 and Bcl-XL. J. Virol. 83:9102–9112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Romanova L. I., et al. 2009. Antiapoptotic activity of the cardiovirus leader protein, a viral “security” protein. J. Virol. 83:7273–7284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Romero A., Figueras A., Thoulouze M. I., Bremont M., Novoa B. 2008. Recombinant infectious hematopoietic necrosis viruses induce protection for rainbow trout Oncorhynchus mykiss. Dis. Aquat. Organ. 80:123–135 [DOI] [PubMed] [Google Scholar]
- 32. Sarmento L., Tseggai T., Dhingra V., Fu Z. F. 2006. Rabies virus-induced apoptosis involves caspase-dependent and caspase-independent pathways. Virus Res. 121:144–151 [DOI] [PubMed] [Google Scholar]
- 33. Schütze H., Mundt E., Mettenleiter T. C. 1999. Complete genomic sequence of viral hemorrhagic septicemia virus, a fish rhabdovirus. Virus Genes 19:59–65 [DOI] [PubMed] [Google Scholar]
- 34. Schütze H., Enzmann P. J., Mundt E., Mettenleiter T. C. 1996. Identification of the nonvirion (NV) protein of fish rhabdoviruses viral haemorrhagic septicaemia virus and infectious haematopoietic necrosis virus. J. Gen. Virol. 77:1259–1263 [DOI] [PubMed] [Google Scholar]
- 35. Skaletskaya A., et al. 2001. A cytomegalovirus-encoded inhibitor of apoptosis that suppresses caspase-8 activation. Proc. Natl. Acad. Sci. U. S. A. 98:7829–7834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Thornberry N. A., Lazebnik Y. 1998. Caspases: enemies within. Science 281:1312–1316 [DOI] [PubMed] [Google Scholar]
- 37. Thoulouze M. L., Bouguyon E., Carpentier C., Bremont M. 2004. Essential role of the NV protein of Novirhabdovirus for pathogenicity in rainbow trout. J. Virol. 78:4098–4107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Tordo N., et al. 2004. Family Rhabdoviridae. In C. M. Fauquet, M. A. Mayo, J. Maniloff, U. Desselberger, and L. A. Ball (ed.), Virus taxonomy: eighth report of the international committee for taxonomy of viruses. Academic Press, San Diego, CA [Google Scholar]