Abstract
Sequences in the 5′ untranslated region (5′UTR) of hepatitis C virus (HCV) RNA is important for modulating both translation and RNA replication. The translation of the HCV genome depends on an internal ribosome entry site (IRES) located within the 341-nucleotide 5′UTR, while RNA replication requires a smaller region. A question arises whether the replication and translation functions require different regions of the 5′UTR and different sets of RNA-binding proteins. Here, we showed that the 5′-most 157 nucleotides of HCV RNA is the minimum 5′UTR for RNA replication, and it partially overlaps with the IRES. Stem-loops 1 and 2 of the 5′UTR are essential for RNA replication, whereas stem-loop 1 is not required for translation. We also found that poly(C)-binding protein 2 (PCBP2) bound to the replication region of the 5′UTR and associated with detergent-resistant membrane fractions, which are the sites of the HCV replication complex. The knockdown of PCBP2 by short hairpin RNA decreased the amounts of HCV RNA and nonstructural proteins. Antibody-mediated blocking of PCBP2 reduced HCV RNA replication in vitro, indicating that PCBP2 is directly involved in HCV RNA replication. Furthermore, PCBP2 knockdown reduced IRES-dependent translation preferentially from a dual reporter plasmid, suggesting that PCBP2 also regulated IRES activity. These findings indicate that PCBP2 participates in both HCV RNA replication and translation. Moreover, PCBP2 interacts with HCV 5′- and 3′UTR RNA fragments to form an RNA-protein complex and induces the circularization of HCV RNA, as revealed by electron microscopy. This study thus demonstrates the mechanism of the participation of PCBP2 in HCV translation and replication and provides physical evidence for HCV RNA circularization through 5′- and 3′UTR interaction.
INTRODUCTION
In positive-strand RNA viruses, such as hepatitis C virus (HCV), the viral genome serves multiple roles in the virus life cycle: (i) as an mRNA for the translation of the viral proteins; (ii) as a template for the synthesis of negative-strand RNA, which, in turn, serves as the template for producing more positive-strand RNA; and (iii) as a nascent vRNA genome packaged into new virus particles. HCV starts to assemble its viral particles around lipid droplets and exits the cell through the secretory pathway (34). Since the same positive-strand RNA can participate in different steps of the viral life cycle, the regulation between these processes is very important and may be mediated by various host factors interacting with the viral RNA or viral proteins. Signals required for the replication and translation of positive-strand RNA viruses usually are located in the 5′- and 3′-terminal regions of the viral RNA. The sequences required for RNA replication and translation often overlap, and the regulatory mechanism can be separated or shared.
The HCV genome is a 9.6-kb uncapped linear single-stranded RNA (ssRNA) molecule with positive polarity. It contains 5′ and 3′ untranslated regions (UTRs), including control elements required for translation and replication (47). The HCV UTRs flank an uninterrupted open reading frame encoding a single polyprotein of 3,011 amino acids, which is processed into structural (C, E1, E2, and p7) and nonstructural (NS2, NS3, NS4A, NS4B, NS5A, and NS5B) proteins by host and viral proteases (42). The translation of the HCV genome, which lacks a 5′ cap, depends on an internal ribosome entry site (IRES) within the 341-nucleotide 5′UTR (28, 50). Computer analysis and structure probing predict four distinct RNA domains in the HCV 5′UTR (45). The short stem-loop 1 is formed by residues 5 to 20 and is not required for the IRES activity, while stem-loops 2, 3, and 4 are identified as the IRES region.
Current data indicate that the 5′UTR of HCV functions as a platform to recruit viral and cellular proteins and not only directs IRES-dependent protein synthesis but also plays roles in viral RNA replication (15, 58). Several cellular RNA-binding proteins, including eukaryotic initiation factor 3 (eIF3), 40S ribosomal subunit (29), La autoantigen (4), NS1-associated protein 1 (NSAP1; also known as synaptotagmin-binding cytoplasmic RNA-interacting protein [SYNCRIP]) (9, 31), polypyrimidine-tract-binding protein (PTB) (3, 26), heterogeneous nuclear ribonucleoprotein L (hnRNP L), and poly(C)-binding protein (PCBP) (20), have been shown to bind to the 5′ end and/or 3′ end of HCV RNA and may regulate translation and/or replication. Some of the host factors directly regulate HCV RNA replication either by participating in the formation of the RNA replication complex (e.g., VAP-33) (19) or by binding to the viral RNA (e.g., PTB and La). Many host proteins interacting with the HCV 5′UTR have dual functions. La autoantigen, PTB, and SYNCRIP were found not only to regulate RNA translation but also to modulate its replication (7, 11, 35). Questions arise about whether the regulatory sequences for protein synthesis and RNA replication overlap or are separable and whether the regulation of HCV replication and translation is coupled.
The poliovirus 5′UTR has been shown to interact with multiple cellular RNA-binding proteins. PCBP2 (also named hnRNP E2 or αCP-2), a host protein known to bind to the 5′UTR of the poliovirus genome, regulates the switch between poliovirus translation and replication (43). PCPB2 is a member of the cellular heterogeneous nuclear ribonucleoprotein (hnRNP) family, which is expressed in both the nucleus and cytoplasm. hnRNPs are well known for their abilities to bind to cellular proteins and RNAs to facilitate many biological processes, such as mRNA stabilization, transcriptional regulation, translational control, and apoptotic program activation (37, 40). PCBP2 contains three highly conserved hnRNP K homology (KH) domains, the first and third of which mediate ribohomopoly(C) [poly(rC)] binding, and the second domain may enhance binding affinity and/or specificity (10, 12). The protein also has been shown to form homodimers (18) and to interact with other hnRNPs (30). PCBP2 also binds to the HCV 5′UTR (16, 51), but whether it binds to the IRES region or the replication region is still unclear. The role of PCBP2 in regulating HCV translation has been investigated but remains inconclusive (14, 16). Also, its role in HCV RNA replication is not clear. It is possible that PCBP2, similarly to PTB and La, regulates both the translation and replication of HCV RNA.
In this study, we showed that the first 157 nucleotides of the HCV 5′UTR are necessary for RNA replication. The screening of the cellular proteins binding to the HCV 5′UTR identified PCBP2 as a protein binding to the replication region of the 5′UTR. Further investigation showed that PCBP2 plays roles in both HCV RNA replication and translation. We showed that PCBP2 was able to bind both the 5′- and 3′UTR to form an RNA-protein complex. Moreover, electron microscopy (EM) showed that PCBP2 induced changes of RNA conformation from linear to circular, suggesting that PCBP2 is involved in the circularization of the HCV genome. This circularization may be important for HCV RNA replication and is common among RNA viruses.
MATERIALS AND METHODS
Cells.
Huh7 cells were grown at 37°C in Dulbecco's modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and nonessential amino acids. Huh7N1b replicon cells (32) harboring an HCV subgenomic replicon RNA were grown in the same medium containing 0.7 mg/ml G418 (CalBioChem).
In vitro transcription, electroporation, and selection of G418-resistant cells.
Plasmid DNA was linearized with XbaI, purified by phenol extraction and ethanol precipitation, and dissolved in 0.1× Tris-EDTA (TE) buffer. Five μg of restricted plasmid DNA was in vitro transcribed into RNA by a commercial kit (Promega). After incubation for 2 h at 37°C, transcription was terminated by adding RNase-free DNase. RNA was purified with acidic phenol and chloroform, precipitated with isopropanol, and dissolved in RNase-free water. The RNA concentration was determined by the measurement of the optical density at 260 nm, and the integrity of transcribed RNA was checked by denaturing agarose gel electrophoresis. For electroporation, 0.1 to 10 μg of in vitro transcript was adjusted using total cellular RNA to a final amount of about 40 μg and mixed with 400 μl of a suspension of 107 Huh7 cells per ml. After one pulse at 960 μF and 220 V in an Electro Cell Manipulator (BTX ECM630), cells were left on ice for 15 min and then transferred to 10 ml of complete DMEM containing 125 μl dimethylsulfoxide (DMSO) and seeded in a 10-cm-diameter culture dish. After 24 h, medium was replaced by complete DMEM containing G418 (800 μg/ml). Cells were refreshed with medium containing G418 every 2 days. Two to 4 weeks later, plates were stained with crystal violet.
Preparation of cell extracts.
Huh7 cells were harvested and washed twice in cold phosphate-buffered saline. A range of 5 ′ 108 to 1 ′ 109 cells were resuspended in five packed-cell volumes of buffer A (10 mM HEPES [pH 7.9], 1.5 mM MgCl2, 10 mM KCl, 0.5 mM dithiothreitol [DTT]) and incubated for 10 min on ice. After centrifugation for 10 min at 2,000 rpm in a 224 swing bucket rotor, the pellet was used for nuclear extract preparation and the supernatant was used for cytoplasmic extract preparation. For the latter, the supernatant was mixed with a 0.11 volume of buffer B (0.3 M HEPES [pH 7.9], 1.4 M KCl, 0.03 M MgCl2) and centrifuged at 40,000 rpm (100,000 × g) in a Beckman 50 Ti ultracentrifuge rotor for 60 min at 4°C. The supernatant was collected and dialyzed against buffer D (5 mM HEPES [pH 7.8], 25 mM KCl, 2 mM MgCl2, 3.8% glycerol, 0.1 mM EDTA, 2 mM DTT). For the preparation of nuclear extract, the pellet was resuspended with a 0.67 volume of buffer C (20 mM HEPES [pH 7.9], 25% glycerol, 0.42 M NaCl, 1.5 mM MgCl2, 0.2 mM EDTA, 0.5 mM phenylmethylsulfonyl fluoride [PMSF], 0.5 mM DTT). After incubation for 30 min on ice, the sample was centrifuged at 13,000 rpm for 30 min. The supernatant was collected and dialyzed against buffer D. After centrifugation at 13,000 rpm for 20 min, aliquots of supernatants were stored at −80°C.
RNA affinity purification.
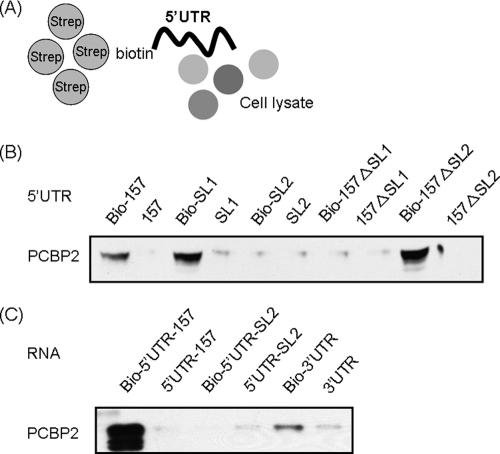
For the biotinylation of RNA, different RNA fragments were in vitro transcribed by T7 RNA polymerase with biotin-UTP (Ambion) using the MEGAshortscript kit (Ambion). Fifty microliters of streptavidin beads (Invitrogen) was washed with bead-washing buffer (5 mM Tris-HCl, 0.5 mM EDTA, 1 M NaCl) twice and incubated with 2 μg of biotinylated RNA for 1 h at room temperature. The beads were washed with bead-washing buffer five times and incubated with Huh7 cytoplasmic extract (0.5 mg), 400 U of RNasin (Promega), and 40 μg of yeast tRNA (Ambion) in binding buffer (25 mM KCl, 20 mM HEPES [pH 7.6], 2.5 mM MgCl2, 0.1 mM EDTA, 10% glycerol, 0.5 mM DTT, 0.1 mM PMSF) for 30 min at room temperature and then incubated for 2 h at 4°C with rotation. The RNA-protein mixture was washed with protein-washing buffer (50 mM KCl, 20 mM HEPES [pH 7.6], 2.5 mM MgCl2, 0.1 mM EDTA, 10% glycerol, 0.5 mM DTT, 0.1 mM PMSF) five times. The eluates were boiled in protein loading buffer and separated by SDS-PAGE. The gel was stained with Coomassie brilliant blue, and individual bands were excised from the gel and analyzed by matrix-assisted laser desorption ionization (MALDI) mass spectrometry. Proteins binding to the biotinylated RNA were investigated by immunoblotting (see Fig. 3A).
Fig. 3.
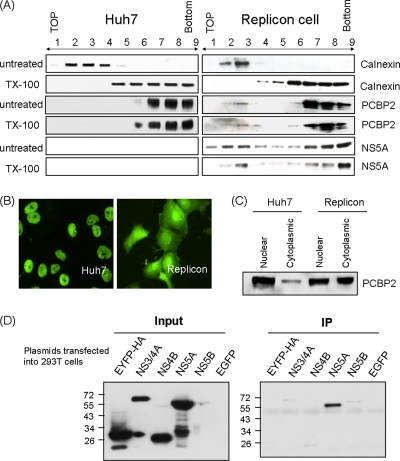
Identification of PCBP2 binding to HCV 5′UTR and 3′UTR by streptavidin-biotin RNA-protein binding assay. (A) Huh7 cytoplasmic extract was incubated with various biotin-labeled RNA and selected with streptavidin beads. Proteins binding to the RNA were blotted with mouse anti-PCBP2 antibody. (B) PCBP2 bound to the fragments containing nt 1 to 157 of the HCV 5′UTR, SL1, and the 157-nt fragment deleting SL2, but it did not bind to SL2 or the 157-nt fragment deleting SL1. (C) PCBP2 also bound to the 3′ end of HCV RNA, but it did so much more weakly than to the 5′ end.
Antibodies.
The primary antibodies used in this study include rabbit anti-hnRNP-E1/E2 (Sigma), mouse anti-PCBP2 monoclonal antibody (Abnova), anti-HCV NS3 monoclonal antibody (Leica), anti-HCV NS5A monoclonal antibody (BioDesign), anti-calnexin monoclonal antibody, and anti-VAPA monoclonal antibody (Chemicon).
PCBP2 knockdown in HCV replicon cells and HCV-infected cells.
The production of lentivirus expressing PCBP2-short hairpin RNA (shRNA) followed the protocol from the National RNAi Core Facility, Academia Sinica, Taiwan (http://rnai.genmed.sinica.edu.tw/file/protocol/2_LentivirusProductionV4.pdf). Four shPCBP2 clones (TRCN0000074683 to TRCN0000074686) were used. The viral titer was determined in Huh7 cells by a cell viability assay (relative infectous unit [RIU] method) according to the instructions of the National RNAi Core (http://rnai.genmed.sinica.edu.tw/file/protocol/4_1_EstimationLentivirusTiterRIUV1.pdf). HCV replicon Rep1.1 cells (32) and Huh7.5 cells were infected with shPCBP2 lentivirus at a multiplicity of infection (MOI) of 3 in the presence of Polybrene (Sigma) (8 μg/ml). Infected cells were incubated for 24 h, and then media were replaced with selective media containing puromycin (3 μg/ml). At 3 days after lentivirus infection, Huh7.5 cells were infected with JC1 virus at an MOI of 1. The JC1 viruses were produced according to a previously described method (8). Cells were harvested 2 to 3 days after JC1 virus infection.
Membrane flotation and detergent solubilization assays.
The membrane flotation assay was performed as previously described (38). Cells were seeded in two 100-mm-diameter plates, lysed in 1 ml of hypotonic buffer (10 mM Tris-HCl [pH 7.5], 10 mM KCl, 5 mM MgCl2), and passed through a 25-guage needle 20 times. Nuclei and unbroken cells were removed by centrifugation at 1,000 × g for 5 min at 4°C. The supernatant was left untreated or was treated with 1% Triton X-100 for 1 h at 4°C. Cell lysates were mixed with 3 ml of 72% sucrose in low-salt buffer (LSB; containing 50 mM Tris-HCl [pH 7.5], 25 mM KCl, and 5 mM MgCl2) and overlaid with 4 ml of 55% sucrose, followed by 1.5 ml of 10% sucrose in LSB. The sucrose gradient was centrifuged at 38,000 rpm in a Beckman SW41 Ti rotor for 14 h at 4°C. After centrifugation, 1-ml fractions were taken from the top of the gradient. To each fraction was added 1.7 ml of LSB to dilute sucrose, and then the fractions were concentrated by being passed through an Amicon Ultra 100k filter (Millipore). Each fraction was separated by SDS-PAGE and transferred to a nitrocellulose membrane. After being blocked, the membrane was incubated with the primary antibody, followed by the appropriate species-specific horseradish peroxidase conjugate. Bound antibody was detected by the ECL-plus system (Amersham).
Cell-free RNA replication assay and antibody-mediated blocking experiment.
Cell lysates of 5 × 107 replicon or control Huh7 cells were prepared by optimizing previously described conditions (5). The cells grown in 100-mm-diameter dishes were washed with ice-cold phosphate-buffered saline (PBS), followed by treatment with ice-cold 1-ml/dish hypotonic buffer for 20 min. Cells then were scraped and centrifuged at 2,000 rpm for 5 min. The cell pellet was resuspended in 1 ml incomplete replication buffer (100 mM HEPES [pH 7.4]; 50 mM NH4Cl; 10 mM KCl; 1 mM spermidine; 1 mM [each] ATP, GTP, and CTP; 10 μM UTP; 10 mM MgCl2; 0.3 mM MnCl2; 10 mM DTT), and lysed with a Dounce homogenizer for 40 strokes of the pestle. The cell suspension was centrifuged at 1,600 rpm for 5 min at 4°C, and then the supernatant was centrifuged at 16,000 × g for 20 min at 4°C. The supernatant was stored at −80°C.
For antibody-mediated blocking experiments, 40 μl of cytoplasmic fraction was treated with 1% Triton X-100 at 4°C for 1 h and incubated with 0.1 to 10 μg of the indicated antibody at 4°C for 4 h with rotation. Each sample then was incubated with [32P]UTP (30 μCi; 800 Ci/mmol), 10 μg of actinomycin D per ml, and 800 U of RNase inhibitor per ml (Promega) for 3 h at 30°C. RNA from the total mixture was extracted with TRIzol reagent (Invitrogen) and purified. The RNA products were analyzed by gel electrophoresis on 1% formaldehyde agarose gels.
Luciferase reporter assay.
After Huh7 cells were infected with shLacZ or shPCBP2, the cells were transfected with a bicistronic plasmid (psiCHECK2) carrying firefly luciferase (FLuc) and renilla luciferase (RLuc) genes mediated by the HCV IRES and cap, respectively. At 20 h posttransfection, luciferase activities were determined using the dual-luciferase reporter assay system (Promega).
5′-3′UTR coprecipitation assay.
For coprecipitation assays, 2 μg of biotinylated RNA, prepared by in vitro transcription, were bound to 50 μl streptavidin beads for 30 min at RT. The 32P-labeled RNA probe was incubated with bovine serum albumin (BSA) or recombinant PCBP2 (Abnova) in the presence of 10 μg of tRNA. The RNA-protein complex then was incubated with the RNA-coated streptavidin beads at 4°C for 2 h. The beads were collected and washed, and the precipitated RNA was analyzed on a denaturing 5% Tris-borate polyacrylamide gel and autoradiographed.
Preparation of RNA template for EM.
For the visualization of RNA, we designed an RNA template which consists of single-stranded 5′- and 3′UTRs of HCV flanking ∼2,000 bp of double-stranded RNA to avoid an RNA secondary structure. The ∼2,000-bp fragment from the full-length HCV-1b replicon (32) sequence was amplified, digested with AscI and ClaI, and ligated to generate a new plasmid, pUC-HCV-2000, which consists of the viral 5′- and 3′UTRs flanking the neomycin-coding sequence, part of the encephalomyocarditis virus (EMCV) IRES, and part of the HCV NS5b coding sequence. The PCR fragments carrying T7 promoter were amplified from sense and antisense portions of pUC-HCV-2000 to produce the partially double-stranded RNA. RNA transcripts, including HCV-5′-3′, HCV-5′-Δ3′, HCV-Δ5′-Δ3′, and their complementary sequences, were obtained by in vitro transcription using T7 RNA polymerase and treated with RNase-free DNase I to remove DNA templates. The integrity of the RNA was verified by electrophoresis on agarose gels. Partially double-stranded RNA molecules were prepared by annealing equal amounts of positive- and negative-strand RNA in annealing buffer (10 mM HEPES, pH 7.4, 100 mM KCl, 0.2 mM EDTA) at 95°C and slowly cooling down to 4°C.
EM.
Protein-RNA complexes were formed by incubating 200 ng of BSA or PCBP2 (Abnova) with 100 ng of RNA in 20 μl of RNA-protein binding buffer (20 mM HEPES and 50 mM KCl) for 20 min at room temperature and stabilized by cross-linking with 0.6% glutaraldehyde at room temperature for 10 min. Fifty-microliter droplets, consisting of RNA 0.5 ng/μl, ammonium acetate 0.25 M, and cytochrome c (7 ng/μl), were incubated for 5 to 20 min at room temperature. RNA diffused to the drop surface and was picked up on Formvar-coated grids (300 mesh). The grids were stained with 10 μl of 8% uranyl acetate in 5 ml of 75% ethanol for 15 s, washed with 5 ml of 90% ethanol for 30 s, and air dried. The RNA conformation was observed with a transmission electron microscope (Tecnai G2 Spirit Twin; FEI Company).
RESULTS
Experimental approach.
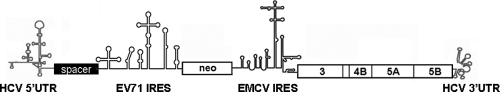
To distinguish the sequences required for translation and for replication, we constructed an HCV replicon system. In the pUC-HCV1b-neo45 replicon (32), neo gene expression is driven by the HCV IRES and allows colony formation upon G418 selection. We inserted an EV71 IRES between the HCV 5′UTR and neo gene and an 86-nt spacer between two IRES elements to avoid potential cross-influence between them. The IRES of the EMCV directs the translation of the NS3-5B region. To facilitate the modification of the HCV 5′UTR, a unique restriction site for BglII, right behind the HCV 5′ UTR, was generated. Thus, this design allows the analysis of sequence requirements of the HCV 5′UTR for RNA replication without affecting translation (Fig. 1). To determine whether this modified HCV replicon, pUC-HCV1b-sp-EVI-neo45, was able to self amplify, we transfected its transcripts into Huh7 cells. After 2 weeks of G418 selection many colonies were detected, indicating that EV71 IRES was able to drive neo gene expression and that the replicon RNA was able to replicate.
Fig. 1.
Schematic presentation of the genome organization of HCV subgenomic replicon pUC-HCV1b-sp-EVI-neo45. The EV71 IRES directs the translation of the neomycin phosphotransferase gene, while the EMCV IRES directs the translation of the NS3 to NS5B region, which is flanked at the 3′ end by the 3′UTR. The 86-nucleotide-long spacer element (black box) is inserted between the HCV 5′UTR and EV71 IRES to avoid possible interference between the two IRES. The RNA sequence of the 86-nucleotide-long spacer is AUGAGCACGAAUCCUAAACCUCAAAGAAAAACCAAAGGGCGCGCCAGCUCGGAUCCACUAGUCCAGUGUGGUGGAAUUGCCCUUGA.
Construction of HCV replicons with various deletions of HCV 5′UTR to determine the replication region.
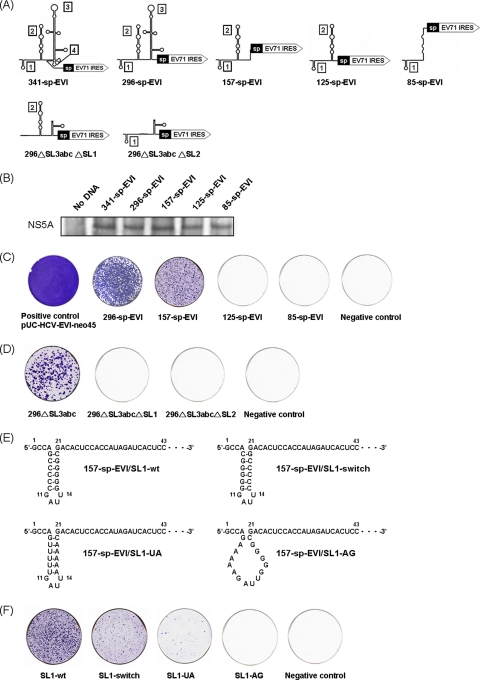
To map which stem-loops of the 5′UTR are required for RNA replication, we constructed a series of mutant replicons in which combinations of the stem-loops of the HCV 5′UTR were deleted (Fig. 2A). To ensure that the mutations in the 5′UTR did not affect the translation driven by EV71 and EMCV IRES, the mutant RNAs were in vitro translated by a coupled transcription/translation system (Promega), and the protein products (NS5A) were detected by immunoblotting. There were no differences in the amounts of NS5A translated (Fig. 2B). In the previous research, subgenomic replicons carrying three IRES elements were used to study HCV RNA replication and proved not to affect translation activity after mutating the HCV 5′UTR (15). Therefore, the effects of 5′UTR mutations on HCV replication and translation can be independently assessed. Mutant RNAs were transfected into Huh7 cells, and G418 selection and colony formation was evaluated. Colonies were formed with the 296-sp-EVI and 157-sp-EVI replicons, but the efficiency of colony formation was lower than that of RNA carrying the full-length HCV 5′UTR. In comparison, 85-sp-EVI and 125-sp-EVI did not form any colonies. These data showed that the first 157 nucleotides in the 5′ end comprising stem-loops 1 and 2 of the 5′UTR are sufficient for HCV RNA replication, but the replication efficiency is significantly enhanced by the presence of stem-loops 3 and 4 (Fig. 2C).
Fig. 2.
Colony formation assay of HCV replicons with different 5′UTR deletions. (A) Predicted secondary RNA structures of the full-length and mutated HCV 5′UTR. (B) In vitro translation of the mutant replicons. The protein products were determined by immunoblotting. (C) One μg in vitro-transcribed RNA of each mutant construct was transfected into 1 × 107 naïve Huh7 cells by electroporation. Two weeks after transfection and G418 selection, cells were fixed and stained with crystal violet. (D) Replicon deleting stem-loop (SL) 4 and SL3abc still replicated in Huh7 cells and formed colonies, but deleting SL1 or SL2 from the 296▵SL3abc replicon inhibited colony formation. (E) Predicted sequences and structures of wild-type and mutant 5′UTR SL1 RNA elements. The stem of the stem-loop 1 structure was disrupted by the nucleotide replacement of cytidines at positions 6 to 10 with adenosines (SL1-AG). The stem-loop structure was restored by either switching the side of the nucleotides formed in the stem (SL1-switch) or replacement with A/U base pairs (SL1-UA). (F) Switching or substituting the nucleotides while maintaining the SL1 structure allowed colony formation (SL1-switch and SL1-UA), whereas the disruption of the SL1 structure inhibited it (SL1-AG).
To investigate the requirement of stem-loops 1 and 2 of the HCV 5′UTR in RNA replication, we deleted each part from the pUCnt296▵SL3abc backbone, which is a replication-competent 5′UTR mutant replicon. Deleting either stem-loop 1 or 2 from this replicon resulted in the complete loss of colony formation, indicating that both stem-loops 1 and 2 of the HCV 5′UTR are required for RNA replication (Fig. 2D). Previously it has been shown that SL2, but not SL1, is part of the IRES (24). Thus, SL1 is involved in RNA replication exclusively, whereas SL2 is involved in both replication and translation.
To determine whether the sequence and/or structure of the 5′UTR stem-loop 1 are required for HCV RNA replication, we mutated SL1 RNA elements of the subgenomic replicon 157-sp-EVI by nucleotide substitution. The stem was disrupted by the replacement of cytidines (C) at nucleotides 6 to 10 with adenosines (A) (157-sp-EVI/SL1-AG). To maintain the stem of SL1 with different sequences, the nucleotides of the stem were switched by compensatory mutations (157-sp-EVI/SL1-switch) or by replacing C-G base pairs with U-A base pairs (157-sp-EVI/SL1-UA) (Fig. 2E). The disruption of the SL1 stem by nucleotide substitutions inhibited colony formation (SL1-AG). The maintenance of the stem structure by compensatory mutations with different nucleotides (SL1 switch) or by the replacement of the C/G base-paired stem with A/U base pairs (SL1-UA) allowed HCV RNA replication to occur at decreased efficiency. The results indicated that the maintenance of the 5′UTR SL1 structure was sufficient for HCV RNA replication, but the sequence of the stem still is important for efficient replication (Fig. 2F).
Cellular protein PCBP2 is identified as a protein binding to the replication region of HCV 5′- and 3′UTR.
To understand the mechanism of the regulation of RNA replication by the 5′UTR sequence, we searched for the cellular proteins binding to this region. We first performed RNA-protein binding experiments using Huh7 cellular lysates and the biotin-labeled 157-nt replication region of HCV 5′UTR RNA. RNA-binding proteins were pulled down together with biotin-labeled RNA by streptavidin beads (Fig. 3A). The proteins were separated by SDS-PAGE and identified by MALDI-mass spectrometry (Table 1). Among the proteins identified, PCBP2 had been shown to be involved in HCV RNA translation (14), but its effects on RNA replication have not been studied. Since PCBP2 also regulates poliovirus RNA replication, we focused on this protein. We first confirmed the binding of PCBP2 to the HCV 5′UTR by immunoblotting; the results indicated that PCBP2 bound to the HCV 5′UTR replication region, especially to the stem-loop 1 RNA fragment (Fig. 3B). It did not bind to SL2. PCBP2 was unable to bind to the replication region RNA without SL1, but it bound to the same RNA fragment without SL2. Therefore, SL1 of the HCV 5′UTR is important for PCBP2 binding. (Fig. 3B).
Table 1.
Cellular proteins binding to the replication region of the HCV 5′UTR
| Protein | GenBank accession no. |
|---|---|
| RNA helicase A | L13848.1 |
| Heat shock protein 90 | P40292 |
| Interleukin enhancer-binding factor 3 | Q12906 |
| Heat shock 70-kDa protein 1A | P08107 |
| Insulin-like growth factor 2 mRNA-binding protein 1a | Q9NZI8 |
| Heterogeneous nuclear ribonucleoprotein M | P52272 |
| Polypyrimidine tract-binding protein 1b | P26599 |
| Poly(rC)-binding protein 2 | Q15366 |
| Elongation factor 1A | P68104 |
| Glyceraldehyde-3-phosphate dehydrogenase | P04797 |
| β-Actin | P60709 |
In HCV, 3′ ends of the RNA genome enhance translation initiation (27) and also RNA replication (56). Therefore, we studied whether PCBP2 also bound to the 3′UTR. The results showed that PCBP2 also bound to the 3′ end of HCV RNA, but it did so much more weakly than it did to the 5′ end (Fig. 3C).
In vivo knockdown of PCBP2 suppresses HCV replication.
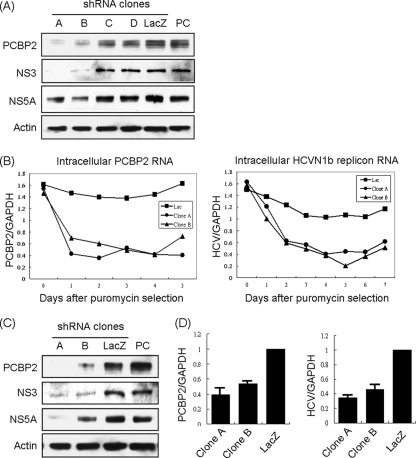
To determine the biological role of PCBP2 in HCV RNA replication, we monitored the effects of PCBP2 knockdown on HCV RNA replication and translation. Several shRNAs targeting different regions of PCBP2 were used. The replicon cells expressing PCBP2 shRNA clones A and B showed a substantial reduction of the endogenous PCBP2, while clones C and D did not have significant effects. Correspondingly, the protein expression levels of HCV NS3 and NS5A also were decreased in replicon cells expressing clones A and B but not C and D (Fig. 4A). In the time course assay of the RNA expression level, the knockdown of PCBP2 in HCV replicon cells resulted in a rapid reduction of PCBP2 RNA and HCV RNA with almost the same kinetics (Fig. 4B). In HCV-infected Huh7.5 cells, the expression of shPCBP2 clones A and B also resulted in a reduction in the amounts of PCBP2 and a corresponding decrease of HCV NS3 and NS5A proteins (Fig. 4C) as well as of HCV RNA (Fig. 4D). The results indicated that the expression level of PCBP2 correlated with HCV RNA and nonstructural protein levels, suggesting that PCBP2 is involved in HCV RNA replication and/or translation.
Fig. 4.
Effects of PCBP2 knockdown in HCV replicon cells and HCV-infected cells. (A) The HCV replicon Rep1.1 cells were infected with lentiviruses containing four different shPCBP2 clones. After puromycin selection for 3 days, cell lysates were used for immunoblotting with different antibodies to detect the various proteins. PC (positive control) stands for the replicon cell lysates without the lentivirus infection. (B) The HCV replicon cells were infected with shPCBP2 clones A and B and unrelated shLacZ and then incubated in the presence of puromycin for different days. The RNAs harvested at different time points were quantified by quantitative reverse transcription-PCR (qRT-PCR). Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) served as an internal control. (C) Huh7.5 cells were infected with lentiviruses containing shPCBP2 (clones A and B) or shLacZ. After puromycin selection for 2 days, the cells were infected with JC1 virus. At 3 days after JC1 virus infection, intracellular proteins and RNAs were analyzed by immunoblotting (C) and qRT-PCR (D), respectively.
Some PCBP2 is located in detergent-resistant membrane fraction and interacts with HCV nonstructural proteins in HCV replicon cells.
HCV RNA replication occurs in the detergent-resistant membrane (DRM) fraction (5, 13, 49), suggesting that the viral replication complex is associated with the lipid raft (2, 19). Since the nonstructural proteins and RNA of HCV are associated with the DRM structure, we examined whether PCBP2 also was associated with the DRM structure. The membrane flotation analysis showed that the majority of PCBP2 was located in the cytosolic fractions (fractions 5 to 9) (Fig. 5A) in both Huh7 and HCV replicon Rep1.1 cells; however, a small fraction of it was associated with the membrane fractions of the replicon cells but not of Huh7 cells (fractions 2 to 4) (Fig. 5A). These membrane-bound PCBP2 proteins could not be removed by Triton X-100 treatment at 4°C. HCV NS5A also was located in the DRM fraction of the replicon cells (fractions 2 and 3) (Fig. 5A). As a control, calnexin, a marker protein of the endoplasmic reticulum (ER) membrane, was detected exclusively in the detergent-soluble membrane fraction (fractions 2 and 3) in both Huh7 and replicon cells (Fig. 5A), which is characteristic of unmodified ER. The data suggested that some PCBP2 proteins are located together with NS5A in the DRM fraction in the replicon cells but not in the cells without the HCV replicon, indicating that PCBP2 is specifically recruited to the HCV replication complex.
Fig. 5.
(A) Membrane flotation analysis of the location of PCBP2. Cell lysates from Huh7 and replicon cells left untreated or treated with Triton X-100 were separated by sucrose gradient sedimentation as described in Materials and Methods. Each fraction was blotted with various antibodies. Fractions 2 to 4 represent the membrane fraction. Fractions 5 to 9 represent the cytosolic fraction. (B) The immunofluorescence of PCBP2 in the naïve Huh7 cells and replicon cells. (C) The nuclear and cytoplasmic extracts of Huh7 and replicon cells were separated and blotted with PCBP2 antibody. (D) Immunoprecipitation assay of the HCV viral proteins interacting with PCBP2. Plasmids expressing NS3/4A (pCI-NS3/4A-HA), NS4B (pUI-NS4B-HA), NS5A (pUI-NS5A-HA), NS5B (pCAG2-NS5B-HA), and EYFP (pCAG-EYFP-HA), all carrying an HA tag, were transfected into 293T cells individually. The cell lysates were immunoprecipitated with rabbit anti-PCBP2 antibody, and each NS protein was detected with mouse anti-HA antibody by immunoblotting.
The immunostaining results further showed that PCBP2 was located mostly in the nucleus of Huh7 cells, while it was expressed in both the nucleus and cytoplasm of the replicon cells (Fig. 5B). This result was confirmed by the biochemical separation of nuclear and cytoplasmic fractions (Fig. 5C). These data together suggested that PCBP2 is associated with the replication complex in the cytoplasm and plays an important role in HCV replication.
Since PCBP2 and viral NS proteins are located in the DRM, PCBP2 may interact with the HCV nonstructural proteins of the replication complex. To identify whether and which of the HCV NS proteins interact(s) with PCBP2, plasmids expressing NS3/4A, NS4B, NS5A, NS5B, and EYFP, all carrying a hemagglutinin (HA) tag (8, 32), were transfected into 293T cells individually. The cell lysates were immunoprecipitated with rabbit anti-PCBP2 antibody, and the coprecipitated NS protein was detected with mouse anti-HA antibody by immunoblotting. Among all of the HCV nonstructural proteins, only NS5A was found to interact with PCBP2 (Fig. 5D). The results indicated that PCBP2 is a part of the HCV RNA replication complex in the DRM, and it interacts with both HCV RNA and a nonstructural protein.
PCBP2 is required for HCV RNA replication in vitro.
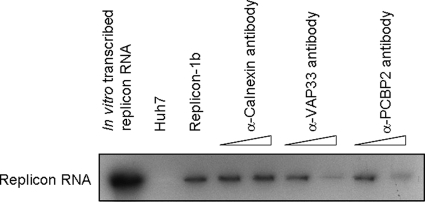
The data so far showed that the level of PCBP2 protein correlated with the HCV RNA titer, suggesting that the reduction of PCBP2 resulted in the inhibition of HCV replication. This effect may be due to the direct inhibition of RNA replication or indirectly results from the inhibition of translation, i.e., the viral NS protein synthesis was decreased, causing the inhibition of viral RNA synthesis. To determine whether PCBP2 directly affects HCV RNA replication, we performed an in vitro replication assay using crude membrane fractions of the HCV replicon cells and assessed the effect of the antibody-mediated blocking of proteins (5, 19). The replicon cell lysates were incubated with different amounts of mouse anti-PCBP2 monoclonal antibody to block the endogenous PCBP2. After incubation, samples were used for the cell-free synthesis of viral RNA. The results showed that the blocking of PCBP2 repressed the RNA replication activity in an antibody concentration-dependent manner. As a control, antibody against VAP-33, which is a critical factor in HCV RNA replication (22), also inhibited HCV RNA replication. The anti-calnexin antibody did not block HCV RNA replication (Fig. 6). The result suggested that PCBP2 is directly involved in HCV RNA replication.
Fig. 6.
Antibody-mediated blocking of PCBP2 repressed HCV replication in in vitro replication assay. HCV replicon cell lysates were incubated with increasing amounts of anti-calnexin, anti-VAP-33, or anti-PCPB2 antibodies to block the indicated proteins. The samples then were incubated with [α-32P]UTP in a cell-free RNA-dependent RNA polymerase assay. The HCV RNA product was detected by autoradiography after separation by formaldehyde agarose gel electrophoresis. In vitro-transcribed replicon RNA served as a marker to indicate the size of HCV replicon RNA.
PCBP2 also is involved in HCV IRES-mediated translation.
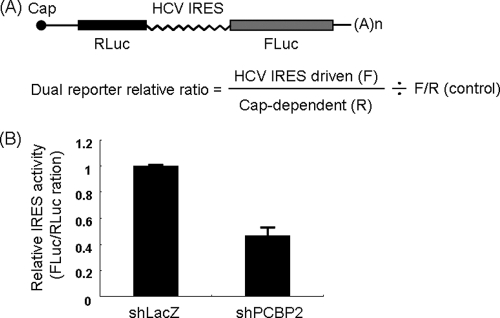
To determine whether PCBP2 is involved in translation, a bicistronic plasmid was transfected into Huh7 cells expressing either shLacZ or shPCBP2. The renilla luciferase gene was driven by cap-dependent translation, while the firefly luciferase gene was under the control of IRES-dependent translation (Fig. 7A). The results showed that, in the presence of shPCBP2, the IRES activity was significantly lower than that of cap-dependent translation, indicating that PCBP2 is involved in IRES-mediated translation (Fig. 7B).
Fig. 7.
PCBP2 is involved in HCV IRES-mediated translation. (A) The structure of the bicistronic plasmid psiCHECK containing renilla and firefly luciferase genes mediated by cap or IRES-dependent translation, respectively. (B) Huh7 cells were infected with shLacZ or shPCBP2, and 3 days later they were transfected with psiCHECK plasmid. Cell lysates were collected at 20 h after transfection, and a luciferase assay was performed. HCV IRES activity relative to the cap-dependent translation was calculated based on the two luciferase activities.
These results indicate that PCBP2 is a dual-function protein that mediates both HCV RNA replication and translation.
Linkage of HCV 5′UTR to 3′UTR through PCBP2.
Previous data showed that PCBP2 binds to both the 5′- and 3′UTR of HCV RNA (Fig. 3); furthermore, PCBP2 oligomerizes by interacting with itself (16), suggesting that PCBP2 is involved in mediating interactions between both ends of the viral genome. To test this notion, we applied streptavidin beads loaded with the biotinylated HCV 5′UTR to test for its possible interaction/coprecipitation with a 32P-labeled 3′UTR in the presence or absence of PCBP2 (Fig. 8A). We observed a significant level of 5′ to 3′ RNA interaction in the presence of PCBP2. In contrast, unrelated protein BSA did not cause coprecipitation between both ends of HCV RNA. Increasing amounts of the unlabeled 5′- or 3′UTR RNA competed away the binding between the 5′- and 3′UTR, suggesting that PCBP2 linkage to both ends of HCV RNA is a specific interaction (Fig. 8B). Furthermore, SL1 alone, but not SL2, of the 5′UTR could mediate the interaction through PCBP2 linkage (Fig. 8C). Hence, these data supported the conclusion that PCBP2 promotes specific interactions between the HCV 5′- and 3′UTR.
Fig. 8.
5′-3′UTR coprecipitation assay. (A) Schematic sketch of the experimental protocol (see Materials and Methods for details). (B) Autoradiographs of the RNAs bound to the streptavidin beads using different combinations of proteins and RNA fragments as indicated. BSA was used as an unrelated protein. Increasing amounts of unlabeled 5′UTR and 3′UTR, but not the unrelated RNA, competed away the binding of 32P-3′UTR to bio-5′UTR. (C) SL1, but not SL2, of the HCV 5′UTR binds to the 3′UTR in the presence of PCBP2.
PCPB2 induces HCV RNA circularization.
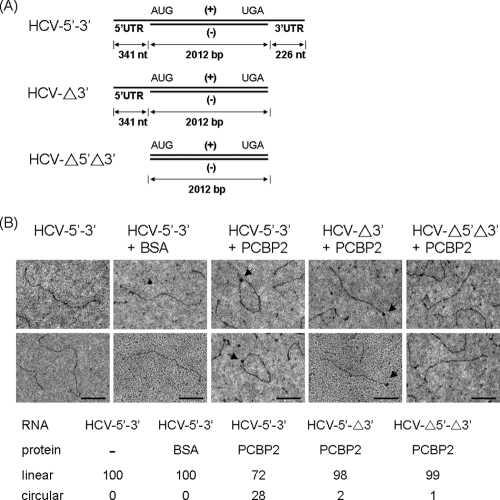
The PCBP2-mediated interaction between the HCV 5′UTR and 3′UTR suggests that PCBP2 forms a protein bridge to induce HCV RNA circularization. To substantiate this concept, we observed protein-mediated changes of the HCV RNA conformation by electron microscopy (EM). To avoid the secondary structures of single-stranded RNAs, we designed RNA molecules that consisted of the single-stranded HCV 5′- and 3′UTRs flanking ∼2,000-bp double-stranded RNA of HCV internal sequences (see Materials and Methods) (Fig. 9A). After interacting with PCBP2 or unrelated protein BSA, the RNA molecules were observed by EM. In the presence of BSA, the RNA molecules appeared in linear conformation. In the presence of PCBP2, a large portion of RNA was circular. Significantly, there appeared to be a protein dot in the middle of the circular RNA. The RNAs deleting one or both ends of HCV UTRs failed to undergo circularization, even though one end of the RNA molecules appeared to be coated with proteins (Fig. 9B). These data indicated that PCBP2 interacts with both ends of the viral UTRs and induces the circularization of HCV RNA molecules.
Fig. 9.
HCV RNA circularizes in the presence of PCBP2. (A) Scheme of the structure of RNA molecules used (see Materials and Methods). (B) RNA electron micrographs. RNA molecules with 5′- and/or 3′UTRs were incubated with PCBP2 or BSA and observed under EM. One hundred molecules were counted for each set, and the ratio of circular to linear RNA molecules is indicated. Arrows indicate protein molecules binding at the end of RNA or on the circularized RNA. Bar, 200 nm.
DISCUSSION
Positive-strand RNA viruses, such as HCV, poliovirus, and coronavirus, amplify their genomes to thousands of copies in only a few hours; thus, efficiency and speed are important characteristics of the viral replication machinery. The genomic RNA initially directs the synthesis of viral proteins. Once the viral proteins are synthesized, the viral RNA is copied, starting from the 3′ end, to generate a complementary negative-stranded RNA that, in turn, is transcribed into a nascent positive-strand genomic RNA. Thus, the genomic RNA must be recognized by both the translation machinery and the viral replication apparatus, and the regulation between these processes is very important and may be affected by various host factors interacting with the viral RNA or viral proteins. Recent evidence indicates that, besides the virus-encoded factors, host cell proteins play an essential role in the replication of positive-strand RNA viruses.
Signals required for the replication and translation of positive-strand RNA viruses usually are located in the 5′- and 3′-terminal regions of the viral RNA. The sequences required for RNA replication and translation often overlap, and the regulatory mechanisms can be separated or shared. In this study, we used a replicon system which allows the high-level self replication of HCV RNA in Huh7 cells to identify the regulatory sequences (36). In this transient replication system, we chose the IRES of EV71 to drive neo gene expression for several reasons: (i) the translational activity of EV71 IRES is higher (33), and (ii) both in structure and in functional terms, the HCV IRES is different from the EV71 IRES, and therefore it should not contain sequences or structures that could compensate for deleted sequences of the HCV 5′UTR. The HCV 5′UTR is well known to mediate IRES-dependent translation (50). Previous data showed that the HCV 5′UTR also participates in RNA replication, but the functional region and how it regulates both mechanisms remained unclear (15). In this study, we showed that the 5′-terminal 157 nucleotides, including stem-loops 1 and 2, of the HCV genome are necessary for RNA replication. Stem-loop 2 of this replication region is also a part of the IRES-mediated translation region, indicating that this region participates in both replication and translation. Whether the stem-loop 2 sequence of the HCV 5′UTR mediates the switching between these two functions remains unclear.
Previous studies have indicated that the HCV RNA replication complexes consist of viral and host protein complexes located on the DRM (13, 55). We have identified several potential factors that bind to the replication region of the HCV 5′UTR, one of which is PCBP2. PCBP2 previously was known to interact with the 5′UTR of the HCV genome, but its binding site and biological function have not been clarified (16). Some research indicates that the role of PCBP2 in HCV is to mediate IRES-dependent translation (14, 41), yet it is unclear how PCBP2 does so. Some researchers report that PCBP2-depleted rabbit reticulocyte lysate still supports translation from HCV IRES-dependent mRNA (16). Other researchers conclude that ribohomopoly(C) competitor RNA, via PCBP sequestration, inhibits the translation of poliovirus mRNAs without affecting the translation of an HCV IRES-dependent mRNA (39), and HCV IRES-dependent translation in yeast is not dependent on PCBP2 (46). Also, a study showed that the HCV RNA level was reduced by PCBP2 short interfering RNA (siRNA) knockdown (44). Thus, the precise role of PCBP2 in HCV translation was not clear. Our studies here showed that PCBP2 regulates both the replication and translation of HCV RNA. Current results show that several other cellular proteins, including PTB (26), Lsm1-7 (48), and IGF2BP1 (54), also bind to the HCV 5′UTR and regulate IRES-dependent translation. Whether PCBP2 directly regulates translation or indirectly functions as a complex with other cellular factors needs further investigation.
PCBP2 also functions in both the translation (6) and replication of poliovirus RNA (52, 53). Furthermore, PCBP2 can mediate the switch from viral translation to RNA replication of poliovirus (17). The initiation of translation is facilitated by the binding of PCBP2 to the poliovirus IRES in the 5′UTR and the bridging of RNA to the ribosomes. The presence of ribosomes on viral RNA prevents the viral RNA polymerase from initiating negative-strand RNA elongation. After PCBP2 is cleaved by the accumulated viral proteinase 3CD, the binding of ribosomes to the IRES is inhibited. The translation process is stopped, but the cleaved form of PCBP2 still can facilitate the interaction of the 5′ RNP complex with the 3′ poly(A) tract via a bridging interaction with poly(A)-binding protein (PABP). Therefore, the translating ribosomes on the RNA template are cleared, and the latter interaction would lead to the initiation of negative-strand RNA synthesis (43). Whether the mechanism of PCBP2 involvement in HCV replication and translation is similar to, or different from, that of poliovirus requires further investigation.
Our data show that PCBP2 interacts with HCV RNA and proteins. The localization of PCBP2 to the DRM fractions in HCV replicon cells indicates that PCBP2 is associated with the RNA replication complex. Furthermore, the finding that antibody-mediated blocking of PCBP2 led to the inhibition of HCV RNA replication in vitro strongly suggests that PCBP2 is directly involved in HCV RNA replication. It is possible that PCBP2 recruits other factors to facilitate the assembly of the replication complex. It is particularly interesting that it interacts with NS5A, an essential protein in HCV RNA replication. Our studies also found that PCBP2 affects the HCV IRES-dependent translation activity in the shRNA knockdown experiment. Thus, we established that PCBP2 is a dual-function protein, being involved in both RNA replication and translation of HCV. So far, several host proteins, including La autoantigen, polypyrimidine-tract-binding protein (PTB) (1, 7, 11), and SYNCRIP (9, 35), have been identified to bind to the HCV 5′UTR and regulate both translation and replication. PTB binds to three separate sites on the HCV 5′UTR, two of which are located at the region of IRES (3, 26). The dual functions of the host RNA-binding proteins may be a common characteristic in viral life cycles. We found that PCBP2 binds predominantly to SL1, which is required exclusively for RNA replication, but not SL2, a region involved in both replication and translation. Perhaps its role in HCV replication and translation is through the interaction between the 5′ and 3′UTR, since it also binds to the 3′UTR of HCV RNA.
Genome circularization may be a common feature of positive-strand RNA viruses, but the specific details of circularization vary from virus family to family, such as RNA-RNA interaction for flaviviruses (21), RNA-protein-protein-RNA interaction for picornaviruses (23), and cRNA-RNA sequences for dengue virus (57). Genome circularization may provide several advantages for viral replication, including the coordination of translation and RNA synthesis, the localization of the viral polymerase at the appropriate start site (allowing the viral and cellular proteins to interact with the terminal ends of the viral genome and, in turn, initiating translation or replication efficiently), and a control mechanism for the integrity of the viral genome. In our study, PCBP2 was found to bind to both the 5′ and 3′ ends of the HCV genome and also formed an RNA-protein complex. Furthermore, it recruits both ends of the genome to change the RNA conformation from linear to circular. Such RNA circularization explains how 3′UTR sequences can regulate translation (25) and possibly also RNA replication. Several other proteins, including PTB, NF/NFAR proteins, LSm1-7, and IGF2BP1, also were found to bind to both ends of the HCV genome and be involved in replication and/or translation (25, 26, 48, 54), suggesting that there is a set of protein complexes which are recruited to the ends of the HCV genome, forming a bridge between 5′ and 3′ ends to regulate and switch the machineries of translation and replication in turn. Our study here provided physical evidence for the possibility of such RNA circularization.
ACKNOWLEDGMENTS
We especially thank the National RNAi Core Facility for providing the shRNA clones and the technical support from the EM facilities, Mass Spectrometer Facility, and Microarray Core Facility of the Institute of Molecular Biology and Core Facilities for Proteomics and Glycomics of the Institute of Biology Chemistry of Academia Sinica, Taiwan. We also thank Jing-Ying Huang for constructing the dual reporter plasmid psiCHECK2 and the technical and material support of our laboratory members, including Wen-chi Su and Vikas Saxena.
Footnotes
Published ahead of print on 1 June 2011.
REFERENCES
- 1. Aizaki H., Choi K. S., Liu M., Li Y. J., Lai M. M. 2006. Polypyrimidine-tract-binding protein is a component of the HCV RNA replication complex and necessary for RNA synthesis. J. Biomed. Sci. 13:469–480 [DOI] [PubMed] [Google Scholar]
- 2. Aizaki H., Lee K. J., Sung V. M., Ishiko H., Lai M. M. 2004. Characterization of the hepatitis C virus RNA replication complex associated with lipid rafts. Virology 324:450–461 [DOI] [PubMed] [Google Scholar]
- 3. Ali N., Siddiqui A. 1995. Interaction of polypyrimidine tract-binding protein with the 5′ noncoding region of the hepatitis C virus RNA genome and its functional requirement in internal initiation of translation. J. Virol. 69:6367–6375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ali N., Siddiqui A. 1997. The La antigen binds 5′ noncoding region of the hepatitis C virus RNA in the context of the initiator AUG codon and stimulates internal ribosome entry site-mediated translation. Proc. Natl. Acad. Sci. U. S. A. 94:2249–2254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ali N., Tardif K. D., Siddiqui A. 2002. Cell-free replication of the hepatitis C virus subgenomic replicon. J. Virol. 76:12001–12007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bedard K. M., Walter B. L., Semler B. L. 2004. Multimerization of poly(rC) binding protein 2 is required for translation initiation mediated by a viral IRES. RNA 10:1266–1276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chang K. S., Luo G. 2006. The polypyrimidine tract-binding protein (PTB) is required for efficient replication of hepatitis C virus (HCV) RNA. Virus Res. 115:1–8 [DOI] [PubMed] [Google Scholar]
- 8. Chen Y. C., et al. 2010. Polo-like kinase 1 is involved in hepatitis C virus replication by hyperphosphorylating NS5A. J. Virol. 84:7983–7993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Choi K. S., Mizutani A., Lai M. M. 2004. SYNCRIP, a member of the heterogeneous nuclear ribonucleoprotein family, is involved in mouse hepatitis virus RNA synthesis. J. Virol. 78:13153–13162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Dejgaard K., Leffers H. 1996. Characterisation of the nucleic-acid-binding activity of KH domains. Different properties of different domains. Eur. J. Biochem. 241:425–431 [DOI] [PubMed] [Google Scholar]
- 11. Domitrovich A. M., Diebel K. W., Ali N., Sarker S., Siddiqui A. 2005. Role of La autoantigen and polypyrimidine tract-binding protein in HCV replication. Virology 335:72–86 [DOI] [PubMed] [Google Scholar]
- 12. Du Z., et al. 2007. X-ray crystallographic and NMR studies of protein-protein and protein-nucleic acid interactions involving the KH domains from human poly(C)-binding protein-2. RNA 13:1043–1051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. El-Hage N., Luo G. 2003. Replication of hepatitis C virus RNA occurs in a membrane-bound replication complex containing nonstructural viral proteins and RNA. J. Gen. Virol. 84:2761–2769 [DOI] [PubMed] [Google Scholar]
- 14. Fontanes V., Raychaudhuri S., Dasgupta A. 2009. A cell-permeable peptide inhibits hepatitis C virus replication by sequestering IRES transacting factors. Virology 394:82–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Friebe P., Lohmann V., Krieger N., Bartenschlager R. 2001. Sequences in the 5′ nontranslated region of hepatitis C virus required for RNA replication. J. Virol. 75:12047–12057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Fukushi S., et al. 2001. Interaction of poly(rC)-binding protein 2 with the 5′-terminal stem loop of the hepatitis C-virus genome. Virus Res. 73:67–79 [DOI] [PubMed] [Google Scholar]
- 17. Gamarnik A. V., Andino R. 1998. Switch from translation to RNA replication in a positive-stranded RNA virus. Genes Dev. 12:2293–2304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gamarnik A. V., Andino R. 1997. Two functional complexes formed by KH domain containing proteins with the 5′ noncoding region of poliovirus RNA. RNA 3:882–892 [PMC free article] [PubMed] [Google Scholar]
- 19. Gao L., Aizaki H., He J. W., Lai M. M. 2004. Interactions between viral nonstructural proteins and host protein hVAP-33 mediate the formation of hepatitis C virus RNA replication complex on lipid raft. J. Virol. 78:3480–3488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gontarek R. R., et al. 1999. hnRNP C and polypyrimidine tract-binding protein specifically interact with the pyrimidine-rich region within the 3′NTR of the HCV RNA genome. Nucleic Acids Res. 27:1457–1463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hahn C. S., et al. 1987. Conserved elements in the 3′ untranslated region of flavivirus RNAs and potential cyclization sequences. J. Mol. Biol. 198:33–41 [DOI] [PubMed] [Google Scholar]
- 22. Hamamoto I., et al. 2005. Human VAP-B is involved in hepatitis C virus replication through interaction with NS5A and NS5B. J. Virol. 79:13473–13482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Herold J., Andino R. 2001. Poliovirus RNA replication requires genome circularization through a protein-protein bridge. Mol. Cell 7:581–591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Honda M., et al. 1996. Structural requirements for initiation of translation by internal ribosome entry within genome-length hepatitis C virus RNA. Virology 222:31–42 [DOI] [PubMed] [Google Scholar]
- 25. Isken O., et al. 2007. Nuclear factors are involved in hepatitis C virus RNA replication. RNA 13:1675–1692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ito T., Lai M. M. 1999. An internal polypyrimidine-tract-binding protein-binding site in the hepatitis C virus RNA attenuates translation, which is relieved by the 3′-untranslated sequence. Virology 254:288–296 [DOI] [PubMed] [Google Scholar]
- 27. Ito T., Tahara S. M., Lai M. M. 1998. The 3′-untranslated region of hepatitis C virus RNA enhances translation from an internal ribosomal entry site. J. Virol. 72:8789–8796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ji H., Fraser C. S., Yu Y., Leary J., Doudna J. A. 2004. Coordinated assembly of human translation initiation complexes by the hepatitis C virus internal ribosome entry site RNA. Proc. Natl. Acad. Sci. U. S. A. 101:16990–16995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kieft J. S., Zhou K., Grech A., Jubin R., Doudna J. A. 2002. Crystal structure of an RNA tertiary domain essential to HCV IRES-mediated translation initiation. Nat. Struct. Biol. 9:370–374 [DOI] [PubMed] [Google Scholar]
- 30. Kim J. H., Hahm B., Kim Y. K., Choi M., Jang S. K. 2000. Protein-protein interaction among hnRNPs shuttling between nucleus and cytoplasm. J. Mol. Biol. 298:395–405 [DOI] [PubMed] [Google Scholar]
- 31. Kim J. H., et al. 2004. A cellular RNA-binding protein enhances internal ribosomal entry site-dependent translation through an interaction downstream of the hepatitis C virus polyprotein initiation codon. Mol. Cell. Biol. 24:7878–7890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lai C. K., Jeng K. S., Machida K., Lai M. M. 2008. Association of hepatitis C virus replication complexes with microtubules and actin filaments is dependent on the interaction of NS3 and NS5A. J. Virol. 82:8838–8848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lee J. C., et al. 2005. High-efficiency protein expression mediated by enterovirus 71 internal ribosome entry site. Biotechnol. Bioeng. 90:656–662 [DOI] [PubMed] [Google Scholar]
- 34. Lindenbach B. D., Rice C. M. 2005. Unravelling hepatitis C virus replication from genome to function. Nature 436:933–938 [DOI] [PubMed] [Google Scholar]
- 35. Liu H. M., et al. 2009. SYNCRIP (synaptotagmin-binding, cytoplasmic RNA-interacting protein) is a host factor involved in hepatitis C virus RNA replication. Virology 386:249–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lohmann V., et al. 1999. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science 285:110–113 [DOI] [PubMed] [Google Scholar]
- 37. Makeyev A. V., Liebhaber S. A. 2002. The poly(C)-binding proteins: a multiplicity of functions and a search for mechanisms. RNA 8:265–278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Mizutani A., Fukuda M., Ibata K., Shiraishi Y., Mikoshiba K. 2000. SYNCRIP, a cytoplasmic counterpart of heterogeneous nuclear ribonucleoprotein R, interacts with ubiquitous synaptotagmin isoforms. J. Biol. Chem. 275:9823–9831 [DOI] [PubMed] [Google Scholar]
- 39. Murray K. E., Roberts A. W., Barton D. J. 2001. Poly(rC) binding proteins mediate poliovirus mRNA stability. RNA 7:1126–1141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ostareck-Lederer A., Ostareck D. H., Hentze M. W. 1998. Cytoplasmic regulatory functions of the KH-domain proteins hnRNPs K and E1/E2. Trends Biochem. Sci. 23:409–411 [DOI] [PubMed] [Google Scholar]
- 41. Pawlotsky J. M., Chevaliez S., McHutchison J. G. 2007. The hepatitis C virus life cycle as a target for new antiviral therapies. Gastroenterology 132:1979–1998 [DOI] [PubMed] [Google Scholar]
- 42. Penin F., Dubuisson J., Rey F. A., Moradpour D., Pawlotsky J. M. 2004. Structural biology of hepatitis C virus. Hepatology 39:5–19 [DOI] [PubMed] [Google Scholar]
- 43. Perera R., Daijogo S., Walter B. L., Nguyen J. H., Semler B. L. 2007. Cellular protein modification by poliovirus: the two faces of poly(rC)-binding protein. J. Virol. 81:8919–8932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Randall G., et al. 2007. Cellular cofactors affecting hepatitis C virus infection and replication. Proc. Natl. Acad. Sci. U. S. A. 104:12884–12889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Rijnbrand R. C., Lemon S. M. 2000. Internal ribosome entry site-mediated translation in hepatitis C virus replication. Curr. Top. Microbiol. Immunol. 242:85–116 [DOI] [PubMed] [Google Scholar]
- 46. Rosenfeld A. B., Racaniello V. R. 2005. Hepatitis C virus internal ribosome entry site-dependent translation in Saccharomyces cerevisiae is independent of polypyrimidine tract-binding protein, poly(rC)-binding protein 2, and La protein. J. Virol. 79:10126–10137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Sarnow P. 2003. Viral internal ribosome entry site elements: novel ribosome-RNA complexes and roles in viral pathogenesis. J. Virol. 77:2801–2806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Scheller N., et al. 2009. Translation and replication of hepatitis C virus genomic RNA depends on ancient cellular proteins that control mRNA fates. Proc. Natl. Acad. Sci. U. S. A. 106:13517–13522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Shi S. T., Lee K. J., Aizaki H., Hwang S. B., Lai M. M. 2003. Hepatitis C virus RNA replication occurs on a detergent-resistant membrane that cofractionates with caveolin-2. J. Virol. 77:4160–4168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Spahn C. M., et al. 2001. Hepatitis C virus IRES RNA-induced changes in the conformation of the 40s ribosomal subunit. Science 291:1959–1962 [DOI] [PubMed] [Google Scholar]
- 51. Spångberg K., Schwartz S. 1999. Poly(C)-binding protein interacts with the hepatitis C virus 5′ untranslated region. J. Gen. Virol. 80:1371–1376 [DOI] [PubMed] [Google Scholar]
- 52. Spear A., Sharma N., Flanegan J. B. 2008. Protein-RNA tethering: the role of poly(C) binding protein 2 in poliovirus RNA replication. Virology 374:280–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Toyoda H., Franco D., Fujita K., Paul A. V., Wimmer E. 2007. Replication of poliovirus requires binding of the poly(rC) binding protein to the cloverleaf as well as to the adjacent C-rich spacer sequence between the cloverleaf and the internal ribosomal entry site. J. Virol. 81:10017–10028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Weinlich S., et al. 2009. IGF2BP1 enhances HCV IRES-mediated translation initiation via the 3′UTR. RNA 15:1528–1542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Yang G., et al. 2004. Newly synthesized hepatitis C virus replicon RNA is protected from nuclease activity by a protease-sensitive factor(s). J. Virol. 78:10202–10205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Yi M., Lemon S. M. 2003. 3′ Nontranslated RNA signals required for replication of hepatitis C virus RNA. J. Virol. 77:3557–3568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. You S., Padmanabhan R. 1999. A novel in vitro replication system for Dengue virus. Initiation of RNA synthesis at the 3′-end of exogenous viral RNA templates requires 5′- and 3′-terminal complementary sequence motifs of the viral RNA. J. Biol. Chem. 274:33714–33722 [DOI] [PubMed] [Google Scholar]
- 58. Zhao W. D., Wimmer E. 2001. Genetic analysis of a poliovirus/hepatitis C virus chimera: new structure for domain II of the internal ribosomal entry site of hepatitis C virus. J. Virol. 75:3719–3730 [DOI] [PMC free article] [PubMed] [Google Scholar]