Abstract
The chromatophore membrane of the photosynthetic diazotroph Rhodospirillum rubrum is of vital importance for a number of central processes, including nitrogen fixation. Using a novel amphiphile, we have identified protein complexes present under different nitrogen availability conditions by the use of two-dimensional Blue Native / SDS-PAGE and NSI-LC-LTQ-Orbitrap mass spectrometry. We have identified several membrane protein complexes, including components of the ATP synthase, reaction center, light harvesting and NADH dehydrogenase complexes. Additionally, we have identified differentially expressed proteins, such as subunits of the succinate dehydrogenase complex and other TCA cycle enzymes that are usually found in the cytosol, thus hinting at a possible association to the membrane in response to nitrogen deficiency. We propose a redox sensing mechanism that can influence the membrane subproteome in response to nitrogen availability.
Keywords: Rhodospirillum rubrum, nitrogen metabolism, Blue Native, chromatophore sub-proteome, amphiphile, Orbitrap
Introduction
The inner membrane of Gram-negative bacteria has a number of functions, e.g. permeability barrier, selective import of solutes into the cell, maintenance of transmembrane gradients and ATP generation, among others. In some species the inner membrane also forms invaginations, housing specialized functions such as storage of magnetic granules 1, 2 or photosynthesis 3. Rhodospirillum rubrum, a purple non-sulfur α-proteobacterium, is a nitrogen fixing photosynthetic bacterium in which the inner membrane forms invaginations, designated “chromatophores” 4, 5. The chromatophores of different phototrophic bacteria have been extensively studied in works on bioenergetics that allowed the understanding of many aspects of bacterial photosynthesis 6-8 and elucidation of the structure and mechanism of ATP synthase 9, 10. R. rubrum is a metabolically versatile organism, being able to grow in a variety of conditions, photoheterotrophically, photoautotrophically as well as chemoautotrophically. Phototrophic growth, however, coincides with a requirement for low oxygen concentrations 11. The nitrogen fixing capacity of R. rubrum was first described by Kamen and Gest in 1949 12. The reduction of atmospheric dinitrogen to ammonium is a property shared by a number of prokaryotes, in a reaction catalyzed by the oxygen-sensitive enzyme complex nitrogenase. This complex is composed of the dinitrogenase reductase or iron protein (encoded by nifH) and dinitrogenase or molybdenum iron protein (encoded by nifDK) and requires a large input of energy, in the form of ATP and reducing equivalents 13:
Due to the energy demand of nitrogen fixation and its oxygen sensitivity the nitrogenase complex is only expressed in nitrogen deficient and low intracellular oxygen conditions 14. The expression of nitrogenase and other nitrogen fixation-related proteins is under strict control, requiring the activation by the NifA protein. In R. rubrum and in related diazotrophs NifA requires the interaction with a PII protein, GlnB, to be active 15, 16. In Rhodobacter capsulatus and other purple non-sulfur bacteria the coordinated regulation of gene expression for nitrogen and carbon fixation, photosynthesis and electron transport were shown to be under the control of the two-component system Reg/Prr 17, 18. The integral membrane protein sensor RegB/PrrB perceives a signal that is transmitted by one of the cytochrome c oxidases. This signal, still unidentified but linked to the cellular redox status, induces the kinase activity of RegB/PrrB, which leads to phosphorylation of RegA/PrrA, regulating gene expression. In this way transcription of the nif (nitrogen fixation) and cbb (which includes genes for the enzymes of the Calvin cycle) operons is not only regulated in response to available nitrogen (through NifA) or carbon source (by the action of CbbR), but also to changes in the redox state of the cell 17. This coordinated regulation of nitrogen and carbon fixation pathways was also shown to occur in R. rubrum, with RuBisCO deletion mutant strains up regulating the expression of nif genes, even in photoheterotrophic conditions of nitrogen excess 19. Conversely, when nitrogenase expression is impaired by growth in nitrogen rich medium 20 or when activity is hindered by disrupting the electron transfer pathway to nitrogenase 21, wild-type R. rubrum was found to up regulate the expression of RuBisCO, presumably as an alternative electron sink. However, no Reg/Prr homologue has so far been described in R. rubrum and hence the identity of the regulating system coordinating nitrogen and carbon fixation pathways still remains unknown.
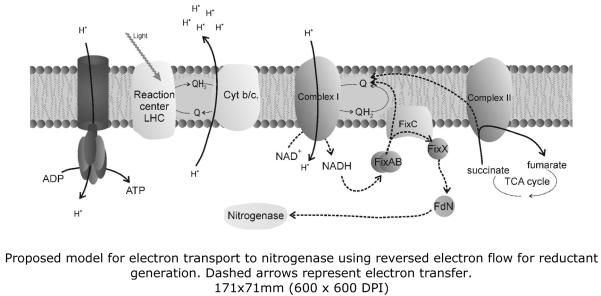
The chromatophore membrane, in addition to its role in photosynthesis and ATP generation, is also of vital importance for nitrogen fixation in R. rubrum. The major pathway for electron transport to nitrogenase under heterotrophic conditions was shown to require the formation of a membrane-associated complex – FixABCX 21. The proposed model for this electron transfer system suggests that the soluble FixAB proteins transfer electrons obtained from NAD(P)H to the membrane-associated FixC, which in turn reduces FixX. Reduced FixX is then able to reduce ferredoxin N, which is the main direct electron donor to nitrogenase 22, 23 (see Figure 5). Nitrogenase activity is also dependent on the existence of a proton-motive force (PMF). In addition to being required for the generation of ATP, PMF is also necessary for the efficient generation of reductant. Low oxygen concentrations stimulate nitrogenase activity in the dark whereas under anaerobic phototrophic conditions disruption of the electron transport chain by uncouplers will lead to inhibition of nitrogenase activity 22, 24. It was also suggested that FixC could interact with another membrane (or membrane-associated) protein, although so far no such partner has been identified. The addition of fluoroacetate to diazotrophically grown R. rubrum cultures inhibited nitrogenase activity, an effect that could be reversed by the addition of NAD(P)H 25. These results implicate that nitrogenase activity requires a working tricarboxylic acid (TCA) cycle and/or pyruvate dehydrogenase as fluoroacetate is a known potent inhibitor of aconitase 26. It was also shown that chromatophore suspensions could generate NADH from succinate only when illuminated and this interdependence between NADH generation and photosynthesis was proposed to be due to the reversal of the electron flow in the respiratory chain, using the PMF to overcome the thermodynamic barrier, with the net effect of generating more reductant 27.
Figure 5.
Proposed hypothetical model for electron transport to nitrogenase using reversed electron flow for reductant generation. Dashed arrows represent electron transfer.
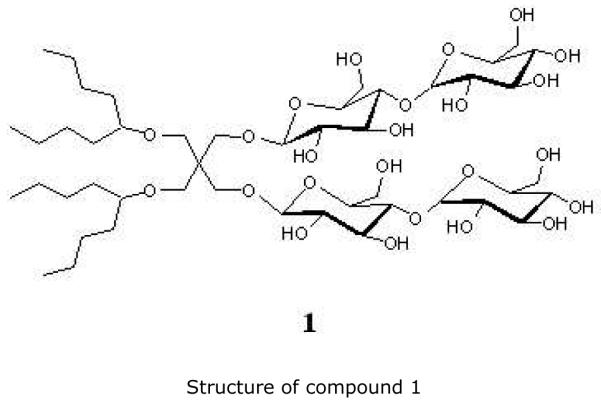
As the chromatophore membrane components play such an important role in the overall metabolism and they have such great influence on the process of nitrogen fixation, this study aimed to establish the identity of the membrane protein complexes whose expression/localization is regulated by nitrogen availability in R. rubrum. As no studies of the chromatophore membrane proteome of R. rubrum exist, we also identified some of the major complexes, in both cases using a combination of two-dimensional Blue Native (2D BN) / SDS-PAGE and mass spectrometry. We have used a novel amphiphile (figure 1), compound 1, which belongs to the recently developed class of “MNG amphiphiles”. Here the synthesis (see Supporting information) and compatibility with BN electrophoresis of amphiphile 1 are described for the first time. This amphiphile was designed for solubilization of membrane protein complexes from photosynthetic bacteria, maintaining protein-protein interactions and stabilizing components of the photosynthetic apparatus. Also, as the identification of membrane proteins using mass spectrometry is usually challenging, due to the highly hydrophobic and not easily ionizable nature of the peptides, we have used the high accuracy and high sensitivity of the nanospray ionization-liquid-chromatography-linear trap quadrupole-Orbitrap (NSI-LC-LTQ-Orbitrap) mass spectrometer 28, 29.
Figure 1.
Structure of compound 1
Materials and methods
1. Growth conditions and cell fractionation
R. rubrum S1 was grown photoheterotrophically as previously described 30, using 28 mM ammonium chloride (N+, nitrogen rich) or a mixture of 95% N2/5% CO2 (N−, nitrogen fixing) as nitrogen source. Three independent cultures for each growth condition were anaerobically harvested and lysed using a French Press cell as previously described 20. Lysates were centrifuged at 100 000g for 90 minutes, membrane pellets were collected and washed using 25 mM Tris (pH 7.5) with 1 mM phenylmethylsulfonyl fluoride (PMSF) and recentrifuged and further resuspended in ice-cold buffer – 25 mM Tris (pH 7.5), 10 % glycerol, 1 mM PMSF – flash-frozen in liquid nitrogen and stored at −80 °C until further use (corresponding to the “Whole membrane” fraction).
2. Chromatophore membrane enrichment
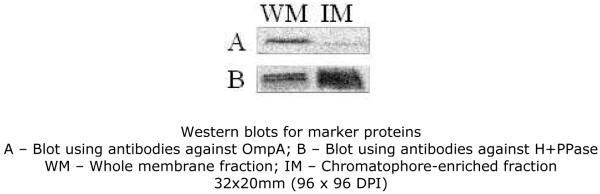
Whole membrane fractions were quantified using the Lowry protein quantification assay 31. The volume equivalent to 10 mg of protein was centrifuged at 220 000g for 15 minutes at 4 °C to remove glycerol and resuspended in 1 mL of 10 mM Hepes (pH 7.5) with 1 mM PMSF (buffer A). In order to separate the outer from the inner (chromatophore) membrane, the resuspended whole membrane fractions were loaded on top of sucrose gradients – 35% to 55% (w/v), in buffer A – and centrifuged in an SW32 rotor at 100 000g for 16 hours, at 4 °C 32. The colored bands were collected and diluted using buffer A, centrifuged at 100 000g for 90 minutes, 4 °C, to remove sucrose and the pellet was again resuspended in 1 mL of buffer A. A second 35-55% sucrose gradient centrifugation was performed for optimal separation. Two independent sucrose gradients were run for each culture. Coloured bands resulting from the second consecutive sucrose gradient were centrifuged as described, resuspended in 1 mL buffer A and again centrifuged at 220 000g for 15 minutes, 4 °C, to remove any trace of sucrose. Membrane pellets were resuspended in 750 μL of a buffer containing 10 mM Hepes (pH 7.5), 10 % glycerol and 1 mM PMSF, quantified using the Lowry assay, flash-frozen in liquid nitrogen and stored at −80 °C until further use – (“Chromatophore membrane fraction”). To test the purity of the preparation, 40 μg of whole membrane and of chromatophore membrane proteins were ran in 12.5% SDS-PAGE gels, transferred to PVDF membranes and probed with antibodies raised against Escherichia coli OmpA (an outer membrane protein 33) and R. rubrum proton-pumping pyrophosphatase (a chromatophore membrane protein 34). Blots were developed using SuperSignal West Pico Chemiluminescence kit (Thermo Scientific) and a DarkBox II CCD camera (Fujifilm).
3. Membrane protein sample preparation
The membrane proteins were solubilized using a novel amphiphile, compound 1, (belonging to the class of Maltose-neopentyl glycol (MNG) amphiphiles 35) developed for enhanced structural stability of membrane proteins (see Supporting information for synthesis details).
To test the optimal concentration of the detergent to use in the Blue Native system, a volume equivalent to 100 μg of purified chromatophore membrane proteins was centrifuged at 220 000g for 15 minutes, at 4 °C. The pellet was resuspended in 100 μL of membrane solubilization buffer, containing 750 mM 6-aminocaproic acid, 50 mM Bis-Tris/HCl (pH 7.0 at 4 °C) and an appropriate amount of detergent. Solubilization was allowed to proceed for 15 minutes on ice and unsolubilized material was removed by centrifugation at 220 000g for 15 minutes, at 4 °C. The supernatant was collected and Coomassie Brilliant Blue G250 (Serva) was added to a final concentration of 0.5% (w/v).
For membrane proteome analysis 400 μg of chromatophore membrane proteins were used, the detergent concentration was kept constant at 0.75% (w/v) and the pellet resulting from the first solubilization step was again resuspended in 100 μL of solubilization buffer, incubated for 15 minutes, re-centrifuged and the supernatant pooled with the previously obtained solubilized membrane protein fraction before addition of Coomassie Blue G250 to the pooled fractions.
4. Analysis of chromatophore membrane proteins by 2D BN/SDS-PAGE
For optimization of solubilization, 20 μL of different detergent concentration test samples prepared as described above were loaded onto 0.75 mm Blue Native PAGE gels, cast using the Hoefer SE250 system, and ran according to previously described protocols 36.
For membrane proteome analysis, 200 μL samples, prepared as described above, were loaded onto 1 mm Blue Native 7-15% first dimension gels (Hoefer SE600 system), and ran at 4 °C as described 36, until the Coomassie Blue G250 front was at the bottom of the gels. Ovalbumin (43 kDa), albumin (66 kDa), conalbumin (75 kDa), aldolase (158 kDa), ferritin (440 kDa) and thyroglobulin (669 kDa) (Gel Filtration Calibration Kit, GE Healthcare) were used as markers. Lanes from the first dimension were cut and frozen at −80 °C until further use. For the second dimension, 1.5 mm 10 % Tris-tricine SDS-PAGE gels 37 (BioRad Protean II system) were used. The lanes from the first dimension were incubated in equilibration buffer (50 mM Tris, pH 8.8, 2% SDS and 0.002% bromphenol blue), supplemented with 65 mM dithiothreitol (DTT) for 15 minutes followed by a second incubation in equilibration buffer supplemented with 135 mM iodoacetamide. The lanes were transferred to the top of the second dimension Tris-tricine gels and sealed in place with a 1% (w/v) low-melting agarose and 0.002% bromphenol blue solution, prepared using cathode buffer (0.2 M Tris, 0.2% SDS and 0.2 M tricine). Gels were run at 4 °C, using the above described cathode buffer and 0.2 M Tris (pH 8.9 at 4 °C) as anode buffer, at 30 V for 2 hours, followed by 130 V until the Coomassie Blue G250 front was 1 mm away from the bottom of the gel. Each independent chromatophore membrane sample was analyzed three times in the 2D BN/SDS-PAGE system.
5. Gel staining and analysis
Following gel run, gels were briefly rinsed with double distilled water and incubated in fixation solution (30% ethanol, 10% acetic acid) for 30 minutes, to remove excess SDS. The fixation step was repeated twice and the gels were rinsed with double distilled water for 2 × 15 minutes, before staining with Blue Silver stain 38, for a minimum of 20 hours. Gels were scanned with a GS-800 Calibrated Densitometer (BioRad) and analysed using PDQuest 7.3.0. (BioRad). For analysis purposes, the two gels with the best resolution (out of six) from each independent culture, one from each independent gradient, were selected, resulting in a matchset containing six gels per condition (twelve gels in total). The gels were normalized using the “Total density in gel image” method and spots were detected and matched using a Gaussian model. After manual treatment of spot assignments, to remove image artifacts and wrongly assigned or duplicated spots, a Student’s t-test, using 98% level of confidence, was used to detect statistically significant differences, including qualitative differences. Only two-fold or higher variations were considered to be significant. Furthermore, some of the most abundant spots or those in evident complexes (even though not changing in intensity between the two conditions) were also selected for identification.
6. In-gel digestion, peptide extraction and mass spectrometric analysis
57 spots were excised, washed and trypsin digested as previously described 20. Following digestion, 100 μl of 30% acetonitrile (ACN), 0.1% formic acid (FA) solution were added to each gel piece and sonication followed for 10 minutes. The liquid was collected to a fresh tube, while the gel piece was again sonicated in freshly added 100 μl of the same solution. The 200 μl from both washes was further acidified to 1% of FA, and then cleaned through a solid phase extraction cartridge Strata-X-C (strong cation exchange, Phenomenex, Torrance, California, USA). The eluate was dried in a Thermo Savant SPD111V Speedvac (Thermo Fischer Scientific, San Jose, CA, USA) and resuspended in 20 μl of 3% ACN, 0.1% FA prior to MS analysis.
MS was performed on a hybrid LTQ-Orbitrap Velos mass spectrometer (Thermo). An Agilent HPLC 1200 system (Agilent Technologies, Santa Clara, CA, USA) was used to inject 2μl of each sample and to provide the gradient for online reversed-phase nano-LC at a flow of 0.4 μl/min. Solvent A was 97% water (Milli-Q, Millipore Corporation, Billerica, Massachusetts, USA), 3% ACN, 0.1% FA; and solvent B was 5% water, 95% ACN, 0.1% FA. The non-linear gradient went from 2% B up to 40% B in 15 minutes, followed by a steep increase to 100% B in 5 minutes. A C18 guard desalting column (Agilent) was used prior to a 15 cm C18 picofrit column (100 μm internal diameter, 5 μm bead size, Nikkyo Technos Co., Tokyo, Japan) installed on to the NSI source of the LTQ-Orbitrap Velos instrument. We enabled “preview mode” for FTMS master scans, which proceeded at 30000 resolution (profile mode). Data-dependent MS/MS (centroid mode) followed in two stages: firstly, the top 5 ions from the master scan were selected for collision induced dissociation (CID, at 35% energy) with detection in the ion trap (ITMS); and after, the same 5 ions underwent higher energy collision dissociation (HCD, at 45% energy) with detection in the orbitrap (FTMS). The entire duty cycle lasted ~3.5s. HCD was used to complement the CID fragmentation method thereby yielding peptide identifications that would otherwise have been missed by a CID-only method (see supporting table 1). Precursors were isolated with a 2 m/z width and dynamic exclusion was used with 60 s duration.
All Orbitrap data was searched using Mascot 2.2 (Matrix Science Limited, London, UK) under the software platform Proteome Discoverer 1.1 (Thermo) against the fasta protein sequence database of the complete R. rubrum ATCC 11170 genome, as obtained from NCBI (ftp.ncbi.nih.gov/genomes). This was a target-decoy search with a 1% False Discovery Rate cut off limit, using a precursor mass tolerance of 10 ppm, and product mass tolerances of 0.02 Da for HCD-FTMS and 0.8 Da for CID-ITMS. The algorithm considered tryptic peptides with max. 2 missed cleavages, oxidation of methionine as variable modification, and carbamidomethylation of cysteine as fixed modification.
Results and Discussion
Membrane fractionation
In R. rubrum, as is common in Gram-negative bacteria, the outer membrane has a very abundant 39 set of integral membrane pore proteins, known as porins. One of these proteins, OmpA, is an integral outer-membrane protein involved, for example, in the process of amino acid import into the cell 40, 41. It was therefore deemed necessary to enrich our membrane sample in chromatophore (inner) membranes as much as possible. Western blots for OmpA show that, after two consecutive sucrose gradient runs, the amount of this protein in the enriched chromatophore fraction is negligible (Figure 2A), indicating an efficient separation of both membranes. It should however be noted that a very weak signal from OmpA is still visible in the chromatophore-enriched fraction blot, implying that a small portion of the outer membrane has co-purified with the chromatophore membrane. However, the increase in the western blot signal for the proton-pumping pyrophosphatase (H+–PPase) in the chromatophore membrane-enriched fraction demonstrated that chromatophore membrane proteins are greatly enriched after the two sucrose gradient runs (Figure 2B). For the subsequent discussion, we shall consider our membrane preparation as consisting mostly of chromatophore (inner) membrane protein complexes.
Figure 2.
Western blots for marker proteins
A – Blot using antibodies against OmpA; B – Blot using antibodies against H+PPase WM – Whole membrane fraction; IM – Chromatophore-enriched fraction
Solubilization and BN gel analysis
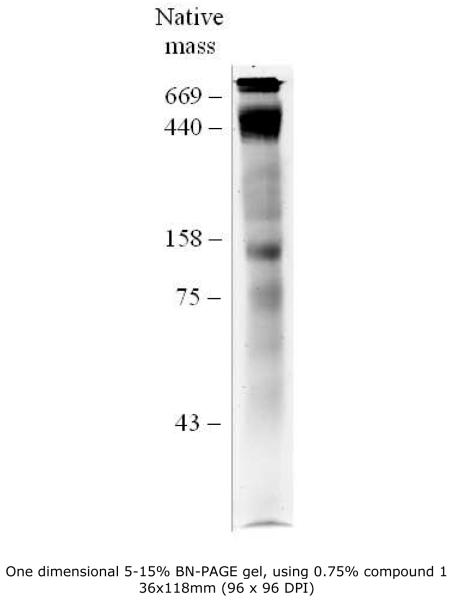
As compound 1 was a new and untested detergent for Blue Native (BN) electrophoresis, several tests were performed, in order to determine the optimal concentration for membrane protein complex solubilization. Compound 1 was fully compatible with the BN system used and already at 0.25% (w/v) a considerable amount of membrane protein complexes could be extracted. At concentrations of 1% (w/v) and above the solubilization seemed to be more efficient but at the cost of distortions in the band migration pattern and possible dissociation of some of the complexes. At 0.75% (w/v) however there are no visible distortions and the region between the 75 and 43 kDa markers is better resolved than with lower concentrations. The concentration for use in the membrane proteome study was fixed at 0.75% (w/v) (Figure 3). In comparison to dodecylmaltoside (DDM), at concentrations between 0.5% and 1% (w/v), first dimension gels using compound 1 showed more and better resolved bands in the lower molecular mass region referred above (data not shown).
Figure 3.
One dimensional 5-15% BN-PAGE gel, using 0.75% MNG-1
At this concentration several membrane complexes could be solubilized, including the FoF1 ATP synthase and the photosynthetic reaction center (RC) and light-harvesting (LH) complex and also the NADH dehydrogenase complex (Figure 4 and Table 1) which have been identified in other photosynthetic bacterial proteome studies 42, 43. The first, very high molecular mass band visible at the top of the first dimension gels does not resolve as well as the remaining complexes in the second dimension gel. This band contains, based on the high molecular mass and constituents (RC H and M proteins and the LH complex β subunit), a higher oligomeric state of the RC-LH complex, as recently described in a BN study of inner membrane complexes of Rhodobacter sphaeroides 44. Migrating at a lower molecular mass than this first band is a cluster of several closely migrating bands, composed of 2 different complexes, the RC-LH complex and the ATP synthase complex. The migration of the RC-LH complex as several different bands, above the 440 kDa marker, is most likely due to loss of different numbers of LH complex subunits during solubilization. The ATP synthase complex co-migrates in the first dimension with the RC-LH complex, due to a similarity in native complex mass. In recent studies of Rb. sphaeroides native inner membranes using atomic force microscopy and cryo-electron microscopy (CEM) the [RC-LH]2 complex was described as inducing the characteristic curvature of the inner membrane vesicles 45, 46. The ATP synthase complex could not, however, be seen in the vesicles studied and it was proposed to be located at the “neck” of the vesicles, not directly contacting with the [RC-LH]2 complex. It should be stressed at this point that, unlike Rb. sphaeroides, the photosynthetic apparatus of R. rubrum contains only one type of RC-LH complex 47 and that, even though the RC1-LH complexes from these two organisms are similar, some differences do exist 48.
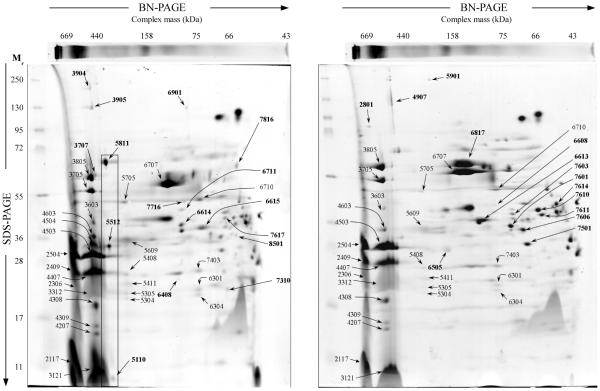
Figure 4.
Representative 2D-BN/SDS-PAGE gels of chromatophore-enriched membrane protein complexes from either nitrogen fixing (left) or nitrogen rich (right) conditions. Numbers represent spots analyzed by LTQ-Orbitrap mass spectrometry; numbers in bold represent spots with changes in intensity in the corresponding condition.
Table 1.
Proteins identified by LTQ-Orbitrap mass spectrometry
| A. Differentially expressed/localized proteins | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Normalized quantities a |
|||||||||||||
| Category | SSP | Gene Product/ Locus tag |
gi accession number |
Theoretical mass (kDa) |
Unique peptides |
PSM | Score (CID+ HCD) |
Coverage (%) |
N+ | N− | Ratio (N−/N+) |
Predicted localization |
TM helices |
| TCA cycle | 5110 | SDH subunit D | 83592539 | 13.5 | 2 | 7 | 269 | 15 | ND | 130.8 | NA | CM | 3 |
| 5512 | SDH Fe-S subunit beta | 83592541 | 29.5 | 22 | 85 | 2142 | 45 | 348.7 | 2196.3 | 6.3 | CMA | NA | |
| 5811 | SDH flavoprotein subunit alpha | 83592540 | 64.7 | 24 | 91 | 2911 | 47 | 1228.4 | 4352.8 | 3.6 | CMA | NA | |
| 6614 | Malate dehydrogenase | 83592546 | 33.5 | 20 | 121 | 4335 | 68 | ND | 1420.7 | NA | Cyt | NA | |
| 7716 | Succinyl-CoA synthetase beta subunit | 83592547 | 42.6 | 20 | 51 | 1477 | 51 | ND | 92.9 | NA | Cyt | NA | |
|
| |||||||||||||
| Amino acid metabolism |
6901 | O-acetylhomoserine/O-acetylserine sulfhydrylase |
83593171 | 46.0 | 12 | 32 | 965 | 40 | ND | 105.5 | NA | Cyt | NA |
| 7617 | Cysteine synthase | 83595123 | 35.4 | 17 | 49 | 1917 | 49 | 45.5 | 322.4 | 7.1 | Cyt | NA | |
|
| |||||||||||||
| Transport | 6615 | TRAP dicarboxylate transporter-DctP subunit | 83595007 | 36.1 | 23 | 96 | 2945 | 59 | ND | 1717.5 | NA | Per | NA |
|
| |||||||||||||
| Electron transport |
7816 | FixC like electron transfer flavoprotein dehydrogenase |
83591605 | 59.5 | 19 | 54 | 1799 | 36 | 49.8 | 223.9 | 4.5 | CMA | NA |
| 8501 | FixB like electron transfer flavoprotein alpha | 83594408 | 31.4 | 6 | 16 | 490 | 21 | ND | 66.2 | NA | Cyt | NA | |
|
| |||||||||||||
| Proteolysis | 6408 | Clp protease subunit ClpP | 83592885 | 23.9 | 9 | 22 | 800 | 34 | ND | 40.5 | NA | Cyt | NA |
|
| |||||||||||||
| Lipid synthesis | 6711 | Acetyl-CoA acetyltransferase | 83591614 | 40.2 | 16 | 60 | 1702 | 38 | ND | 268.2 | NA | Cyt | NA |
| 7310 | 50S ribosomal protein L6 | 83594005 | 19.4 | 6 | 8 | 371 | 27 | 38.1 | 124.1 | 3.2 | Cyt | NA | |
|
| |||||||||||||
| Photosynthesis | 3707 | Uroporphyrinogen III synthase HEM4 | 83594896 | 74.9 | 20 | 53 | 2516 | 41 | ND | 283.4 | NA | Cyt | NA |
|
| |||||||||||||
| Unknown | 3904 | Alpha-2-macroglobulin-like protein | 83591444 | 185.1 | 17 | 37 | 1543 | 10 | ND | 21.9 | NA | Unk | NA |
| 3905 | Alpha-2-macroglobulin-like protein | 83591444 | 185.1 | 26 | 57 | 2147 | 16 | ND | 21.9 | NA | Unk | NA | |
|
| |||||||||||||
| Transport | 6608 | Putative branched-chain amino acid periplasmic binding protein |
83594835 | 44.2 | 10 | 36 | 886 | 25 | 121.7 | ND | NA | Per | NA |
| 6613 | TRAP dicarboxylate transporter- DctP subunit | 83595007 | 36.1 | 33 | 151 | 4930 | 70 | 9255.9 | 59.38 | 0.01 | Per | NA | |
| 6817 | Extracellular solute-binding protein | 83593691 | 59.0 | 49 | 178 | 5720 | 64 | 25156.8 | 1173.6 | 0.05 | Per | NA | |
| 7601 | Periplasmic ribose binding protein | 83592701 | 33.2 | 30 | 144 | 5617 | 70 | 6207.6 | 273.3 | 0.04 | Per | NA | |
| 7603 | Periplasmic phosphate binding protein | 83591937 | 36.5 | 16 | 51 | 1637 | 50 | 926.7 | 14.9 | 0.02 | Per | NA | |
| 7606 | Basic membrane lipoprotein | 83593237 | 38.6 | 16 | 58 | 2167 | 45 | 4298.6 | 609.2 | 0.14 | Per | NA | |
| 7610 | Branched-chain ligand-binding receptor | 83593081 | 38.6 | 33 | 166 | 5161 | 61 | 4327.8 | 501.3 | 0.12 | Per | NA | |
| 7611 | Thiosulphate-binding protein | 83594729 | 38.5 | 16 | 53 | 1542 | 43 | 3215.9 | 396.2 | 0.12 | Per | NA | |
| 7614 | Extracellular solute-binding protein | 83592354 | 40.4 | 18 | 50 | 1685 | 38 | 3229.0 | 639.7 | 0.20 | Per | NA | |
|
| |||||||||||||
| Porins | 6505 | OmpA/MotB (porin associated protein) | 83594657 | 28.7 | 11 | 37 | 1509 | 40 | 219.5 | ND | NA | OM | NA |
| 7501 | OmpA family protein | 83591777 | 35.6 | 23 | 83 | 3121 | 39 | 5394.9 | 444.0 | 0.08 | OM | NA | |
|
| |||||||||||||
| Photosynthesis | 2801 | Uroporphyrinogen III synthase HEM4 | 83594896 | 74.9 | 30 | 82 | 2940 | 55 | 131.4 | ND | NA | Cyt | NA |
| Unknown | 4907 | Alpha-2-macroglobulin-like protein | 83591444 | 185.1 | 58 | 158 | 5686 | 34 | 1038.0 | 11.4 | 0.01 | Unk | NA |
| 5901 | Alpha-2-macroglobulin-like protein | 83591444 | 185.1 | 39 | 89 | 3100 | 24 | 104.1 | 14.6 | 0.14 | Unk | NA | |
| B. Constitutive proteins | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Category | SSP | Gene Product/ Locus tag |
gi accession number |
Theoretical mass (kDa) |
Unique peptides |
PSM | Score (CID + HCD) |
Coverage (%) |
Predicted localization |
TM helices |
| Energy metabolism |
3312 | ATP synthase delta subunit | 83592559 | 19.6 | 14 | 30 | 929 | 51 | CMA | NA |
| 3603 | ATP synthase gamma subunit | 83592561 | 32.4 | 7 | 20 | 553 | 23 | CMA | NA | |
| 3705 | ATP synthase beta subunit | 83592562 | 50.8 | 19 | 87 | 2973 | 52 | CMA | NA | |
| 3805 | ATP synthase alpha subunit | 83592560 | 55.2 | 30 | 109 | 3191 | 42 | CMA | NA | |
| 4207 | ATP synthase b’ subunit | 83594574 | 17.2 | 13 | 59 | 1889 | 66 | CM | 1 | |
| 4308 | ATP synthase b subunit | 83594573 | 19.6 | 15 | 60 | 2492 | 57 | CM | 1 | |
| 4309 | ATP synthase epsilon subunit | 83592563 | 14.3 | 7 | 60 | 1915 | 46 | CMA | NA | |
| 4603 | ATP synthase gamma subunit | 83592561 | 32.4 | 12 | 31 | 1194 | 39 | CMA | NA | |
| 5304 | NADH dehydrogenase subunit B | 83592891 | 20.2 | 5 | 15 | 594 | 29 | CMA | NA | |
| 5305 | NADH dehydrogenase subunit I | 83592898 | 18.6 | 5 | 10 | 197 | 20 | CMA | NA | |
| 5408 | NADH dehydrogenase subunit C | 83592892 | 24.8 | 18 | 56 | 1349 | 62 | CMA | NA | |
| 5411 | Ubiquinol-cytochrome c reductase, ironsulfur subunit |
83591704 | 19.6 | 4 | 14 | 184 | 18 | CM | 1 | |
| 5705 | NADH dehydrogenase subunit D | 83592893 | 44.3 | 10 | 23 | 666 | 28 | CMA | NA | |
| 6301 | NADH dehydrogenase subunit I | 83592898 | 18.6 | 8 | 18 | 390 | 22 | CMA | NA | |
| 6304 | NADH dehydrogenase subunit B | 83592891 | 20.2 | 6 | 21 | 765 | 32 | CMA | NA | |
| 6710 | NADH dehydrogenase subunit D | 83592893 | 44.3 | 14 | 38 | 715 | 32 | CMA | NA | |
| 7403 | NADH dehydrogenase subunit C | 83592892 | 24.8 | 28 | 100 | 2376 | 72 | CMA | NA | |
|
| ||||||||||
| Photosynthesis | 2117 | Light harvesting protein B870 (beta subunit) | 83594307 | 7.7 | 4 | 15 | 550 | 30 | CM | 1 |
| 2409 | Photosynthetic reaction center M-chain | 83594304 | 34.2 | 7 | 23 | 952 | 21 | CM | 5 | |
| 2504 | Photosynthetic reaction center H-chain | 83591956 | 27.9 | 21 | 67 | 2455 | 61 | CM | 1 | |
| 3121 | Light harvesting protein B870 (beta subunit) | 83594307 | 7.7 | 3 | 11 | 517 | 30 | CM | 1 | |
| 4407 | Photosynthetic reaction center M-chain | 83594304 | 34.2 | 8 | 37 | 861 | 21 | CM | 5 | |
| 4503 | Photosynthetic reaction center. H-chain | 83591956 | 27.9 | 17 | 49 | 1897 | 54 | CM | 1 | |
| 4504 | Photosynthetic reaction center. H-chain | 83591956 | 27.9 | 15 | 40 | 1439 | 45 | CM | 1 | |
|
| ||||||||||
| Transport | 5609 | Extracellular solute-binding protein | 83593422 | 32.4 | 8 | 16 | 449 | 23 | Per | NA |
| 6707 | Extracellular solute-binding protein | 83593691 | 59.0 | 41 | 223 | 7075 | 60 | Per | NA | |
|
| ||||||||||
| Unknown | 2306 | Putative membrane protein | 83593120 | 14.5 | 4 | 28 | 729 | 22 | CM | 3 |
FDR – false discovery rate; SSP – standard spot number assigned by PDQuest 7.3.0; TM– transmembrane; ND – not detected; NA – non applicable; CM – integral chromatophore membrane protein; CMA – chromatophore membrane associated protein; Cyt – cytosolic protein; Per – periplasmic protein; OM – outer membrane protein; Unk – unknown; PSM – peptide-spectrum matches; Score – cumulative Mascot ion score; Coverage – all accounted peptides are from PSMs (peptide-spectra match) of <1% FDR (false discovery rate), as determined by Mascot search against the target-decoy database
Quantification using the “Total density in gel image” normalization method from PDQuest 7.3.0.
Predicted using the PSORTb 3.0.2 server (http://www.psort.org/psortb/index.html)
Predicted using the TMHMM 2.0 server (http://www.cbs.dtu.dk/services/TMHMM/)
The Reaction Center – Light Harvesting core complex
The photosynthetic reaction center of R. rubrum is composed of three subunits (named M, L and H, organized with a stoichiometry of 1:1:1) and is surrounded by 16 pairs of helices, the light-harvesting alpha and beta subunits of the LH complex 47, 49. In our preparation, and as reported for Rb. sphaeroides 44 we were able to find the RC-LH core complex, with an apparent molecular mass of between 440 and 669 kDa, apart from the afore-mentioned higher oligomeric state complexes. We identified two of the three subunits pertaining to the RC complex, subunits M (spot 2409 and 4407) and H (spots 2504, 4503 and 4504). The higher order complexes may just be an artificial configuration as there is no indication for the formation of such complexes in vivo in R. rubrum membranes. The theoretical molecular mass for the RC-LH complex, as calculated from the available sequences, is of roughly 288 kDa. This complex has only been previously observed in a monomeric form in atomic force microscopy (AFM) and CEM studies of R. rubrum RC-LH complexes 49, 50. Unlike the case for other bacteria, such as Rb. sphaeroides or Rb. capsulatus 46, 51, 52, no PufX subunit has never found in association with the RC in R. rubrum and BlastP searches were unable to find proteins with sequences similar to those of PufX from either Rb. sphaeroides or Rb. capsulatus. This protein is thought to aid in the formation of the dimeric RC-LH complexes and pufX deletion strains of Rb. sphaeroides have monomeric closed RC-LH complexes, instead of the characteristic “S-shaped” dimeric forms 52, 53. These strains are, however, incapable of photosynthetic growth since diffusion of ubiquinol from the RC is delayed 53. The discrepancy between the theoretical value for the RC-LH complex (288 kDa) and the migration observed in our system (over 440 kDa) could very well be due to the binding of pigment and/or detergent molecules to the polypeptide chains in the complex. In fact, the light-harvesting β subunit, also known as the B870 protein (spots 2117 and 3121), which is not very well resolved during the second dimension run, migrates at a higher apparent molecular mass (~11 kDa) than would be predicted from the sequence (7.7 kDa), an effect also observed in the study of inner membrane complexes in Rb. sphaeroides 44, perhaps due to the strong nature of the interaction between these polypeptide chains and the photosynthetic pigments. Recently both the α and the β LH complex subunit structures from R. rubrum were determined using multidimensional NMR spectroscopy and were shown to be more similar to their counterparts in the LH2 rather than to those of the LH1 complex from Rb. sphaeroides 54. Taking into account the apparent migration of the LH complex subunits, instead of their theoretical mass, the calculated complex mass amounts to ca. 444 kDa, which is in better agreement with the migration observed in our system. It should be noted that a similar migration pattern (over 440 kDa, with multiple bands) was observed when the membranes were solubilized using DDM at concentrations between 0.5% and 1% (w/v) (data not shown).
The ATP synthase complex
The ATP synthase complex is among the most widely studied membrane complexes and many details of its constitution and activity have been fully established 55. In our study seven out of nine of the ATP synthase complex subunits could be extracted and resolved in the second dimension gel. The complex migrates in the first dimension between the 440 kDa and the 669 kDa markers, as the ATP synthase from Rb. sphaeroides, which migrates at roughly 580 kDa in a BN system 44. The predicted theoretical complex mass for the R. rubrum ATP synthase complex, using an α3β3γδε stoichiometry for the F1 part and abb’c12 for the Fo subcomplex (as in the case for the E. coli complex 55) is of approximately 540 kDa. As our system does not have enough resolution in the high mass range of the native first dimension, we are unable to calculate with accuracy the observed mass of the ATP synthase complex based on the migration pattern. We were able to identify five of the F1 subunits (α, β, γ, δ and ε, spots 3805, 3705, 3603 and 4603, 3312 and 4309, respectively) as well as the two subunits that constitute the “stator stalk” (b and b’, spots 4308 and 4207, respectively) 55. As in cyanobacteria and chloroplasts 56, 57, R. rubrum genome encodes both b and b’ subunits of the heterodimeric bb’ form of the stator stalk proteins 58, rather than the homodimeric b2 form observed in E. coli. Although the reason for this is not yet understood the heterodimeric form, rather than a homodimer, is present in photosynthetic bacteria but it was proposed that an asymmetric stator stalk dimer could increase efficiency or potential for regulation of the ATP synthase complex 56. We were unable to identify either of the Fo subunits a or c as also observed in other photosynthetic bacterial proteome studies on inner membrane protein complexes 42, 43. This may be due to their very hydrophobic nature and/or, in the case of the c subunits, to their small size and lack of positively charged amino acids, required for trypsin cleavage. This may exclude them both from being resolved in the second dimension and from being detected by mass spectrometry.
The NADH dehydrogenase complex (Complex I)
One of the largest known membrane complexes, the NADH dehydrogenase complex catalyzes the transfer of electrons from NADH to ubiquinone, with the concomitant transport of protons across the membrane 59. This complex,the first enzyme of the respiratory chain, has been successfully purified in a stable form from E. coli using DDM, lipids and divalent cations 60. We have identified a small number of the subunits of this 14 subunit complex in our study (subunits B, C, D and I). The complex migrates in the first dimension below the 440 kDa marker, which is much too low for the reported bacterial complex mass of about 550 kDa 61. In fact, the purified NADH dehydrogenase complex from E. coli was even shown to dimerize after purification, resulting in a complex eluting at roughly 1 MDa in gel filtration chromatography. However, these dimers are unstable and dissociate into monomers if stored at 4 °C for a few days 60. Several attempts have also been made to purify this complex from Rb. capsulatus but the complex was found to disaggregate upon detergent solubilization 62. This instability of the NADH dehydrogenase complex may be the cause of the migration pattern in our system, as the complex may have disaggregated during sample preparation and/or gel run. The subunits detected were found in two separate complexes, one migrating at a lower native mass, where only the B, C, D and I subunits were identified from it (spots 6301, 6304, 6710 and 7403) and another at a higher native mass (spots 5304, 5305, 5408 and 5705). The spot containing the iron-sulfur subunit of cytochrome c reductase (or complex III, spot 5411) migrates at roughly the same native complex mass as the second complex and this “heavier” complex may also contain other subunits that were neither visible in the second dimension gel nor selected for mass spectrometric analysis.
The NADH dehydrogenase complex has been proposed to play a role in reductant generation for nitrogenase, by using reversed electron flow and reversal of the proton pumping activity to regenerate NADH (Edgren, T., unpublished – see Figure 4). Such a reversed electron flow mechanism has also been proposed to occur in Rb. capsulatus grown under photoheterotrophic conditions, with succinate or lactate as substrates 62, as a mean to maintain the redox balance in the cell. In our statistical analysis it was not possible to detect any of the spots containing NADH dehydrogenase subunits as having a significant change in intensity throughout our dataset. However, these spots are on the limit of detection for our system and, should this variation be subtle, it may not be detected.
Differentially abundant membrane proteins in response to nitrogen source
So far we have only discussed those complexes whose expression is seemingly unaltered in response to nitrogen availability. However, some proteins appear to shift in abundance between nitrogen rich and nitrogen limited conditions, either due to differences in the expression level or in the relative localization (membrane association vs. soluble) in response to the nitrogen status. The role of the membrane proteome in R.rubrum concerning the regulation of nitrogen fixation activity has become evident in studies of the electron transfer to nitrogenase 21, 22, 63 and posttranslational regulation 64 where membrane protein complexes expressed and formed are vital for the reduction of molecular nitrogen to ammonia. This energy demanding process is dependent on NADH as a source of electrons through the Fix ABCX system and the activity of the nitrogenase protein is posttranslationally regulated by a reversible ADP – ribosylation that is partially signaled through a pathway in which reversible complex formation between a PII protein and AmtB1, a membrane protein, is central.
Electron transferring flavoproteins (ETF)
We were able to identify a FixC like protein (spot 7816), a membrane associated component involved in electron transfer, as being highly induced under nitrogen fixing conditions. From the migration in the BN dimension (being found just below the 66 kDa marker) it would seem that this protein is not in complex with any other protein, as it migrates in the first dimension close to its theoretical mass of 59.5 kDa. This electron transferring protein (belonging to the ETF-QO family) reveals a conserved FixC domain with FAD and NADH binding sites and a putative FixX domain (with ferredoxin like features) at the C terminal end of the protein. The FixC domain sequence though shows a higher degree of similarity than the FixX sequence.
We have also found an additional electron transfer flavoprotein (ETF alpha, spot 8501) to be up regulated in nitrogen fixing conditions, which contains a FixB like domain in accordance with the ETF alpha subunit in e.g. Azospirillum sp.B510. Interestingly the genes coding for these two ETF proteins are both adjacent to a gene encoding a 3-hydroxy-buturyl CoA dehydrogenase that is involved in the butyrate metabolic pathway for both these organisms thereby suggesting a possible function for the ETF protein in the butyrate metabolism.
As shown in earlier studies in our laboratory, the expression of the fixABCX operon involved in electron transfer to nitrogenase in R.rubrum is induced during diazotrophic growth 21 and additional regulated expression/localization of ETF gene products during nitrogen fixing conditions could possibly also contribute to the transfer of reducing equivalents to the energy demanding nitrogen fixation process, though this remains to be further proven by future biochemical studies.
TCA cycle related proteins
One of the most highly induced complexes in nitrogen fixing conditions is the succinate dehydrogenase (Sdh) complex. We identified three of the four subunits of this complex, subunit D (spot 5110), flavoprotein subunit alpha (5811) and the Fe-S subunit beta (5512), with all the corresponding spots increasing in intensity in nitrogen fixing conditions. In comparison to a proteome study performed in Rh. sphaeroides 44 we identified one more SDH subunit (SdhD) in addition to Sdh α and β. As previously mentioned, the RegA/RegB system found in Rb. capsulatus is able to sense changes in the cellular redox status and it could be that a similar and but as yet undescribed system is present in R. rubrum, thus affecting Sdh expression in response to changes in the redox state. It was also recently shown in R. rubrum that changes in the quinone pool affect the expression level of photosynthetic membrane components 65. Nitrogen fixation is a demanding process for the cell, requiring substantial reducing power and it may be that upon induction of nitrogen fixation the redox state of the quinone pool is also affected, changing the overall cellular redox balance. Sensing of redox changes, possibly at the level of the quinone pool, could explain the previously mentioned observations concerning the reciprocal regulation of nitrogenase and RuBisCO in response to different conditions, including nitrogen availability 20. It has previously been suggested that nitrogen and carbon fixation pathways are both regulated in R. rubrum as a way to keep the redox balance in the cell 19,20. We have also found two spots with increased intensity in nitrogen fixing conditions that correspond to two enzymes of the TCA cycle, malate dehydrogenase (spot 6614) and the succinyl-CoA synthetase beta subunit (spot 7716) whose genes are located next to each other in the R. rubrum genome. These enzymes are not predicted by the TMHMM server (http://www.cbs.dtu.dk/services/TMHMM/) to contain any transmembrane helices and are usually considered to be cytoplasmic. This higher intensity in nitrogen fixing conditions could be correlated with a higher level of expression, a differential localization in response to nitrogen status or simply an artifact of the preparation. It should be noted, however, that this effect could be consistently observed throughout our entire dataset. In accordance with this putative association to the chromatophore membrane, the two spots seem to migrate at the same native mass in the first dimension. The existence of a complex composed of TCA cycle enzymes has been previously proposed 66 and it is possible that in nitrogen fixing conditions there is a higher tendency for these proteins to associate to the chromatophore membrane. The unexpected membrane association of TCA cycle enzymes has also been observed in another proteome study of photosynthetic bacteria where a fraction of the malate dehydrogenase was found at the membrane 67. Early results from Anderson etal also supports a differential expression of succinate dehydrogenase and malate dehydrogenase showing higher in vitro activities in nitrogen fixing as compared to nitrogen rich R.rubrum cultures 68.
Taken together, both the ETF and TCA cycle proteins that exhibit an increase in nitrogen fixing conditions is in line with the increased demand for reductant in the reduction of dinitrogen catalyzed by nitrogenase. The generation of NADH through higher activities of the TCA cycle enzymes would support the electron transfer to nitrogenase through the reactions of the involved ETF proteins.
Transport systems
Several spots containing periplasmic binding proteins, belonging to different kinds of active transport systems, had higher intensity in nitrogen rich conditions, such as those containing proteins related to the transport of ribose (spot 7601), thiosulphate (spot 7611) or branched-chain amino acids (spot 6608). Interestingly, the spot for the periplasmic binding protein of the dicarboxylic acid tripartite ATP-independent periplasmic (TRAP) transporter seems to migrate at different native masses, depending on nitrogen availability (spot 6615 in nitrogen fixing and 6613 in nitrogen rich conditions). In nitrogen fixing conditions, the migration in the native dimension (close to the 75 kDa marker) could indicate that this protein is migrating by itself, as a dimer. In nitrogen rich conditions, however, this protein migrates at a higher native mass (between the 75 and the 158 kDa markers), which could indicate that it is interacting with another partner. The structure of a periplasmic binding protein from a related transporter system from Rb. sphaeroides was recently solved and this protein was shown to form a dimer, which may imply that dimerization of the receptor is necessary for efficient solute binding and transport across the membrane 69.
Conclusions
The chromatophore membrane of R. rubrum houses several protein complexes, among which the photosynthetic apparatus and the ATP synthase are dominant. We have studied the composition of the amphiphile-soluble membrane proteome under nitrogen fixing and nitrogen rich conditions, using a combination of 2D BN/SDS-PAGE and NSI-LC-LTQ-Orbitrap mass spectrometry. We have successfully tested a new amphiphile and showed its full compatibility with Blue Native electrophoresis in the conditions tested. In addition to the identification of most of the ATP synthase and photosynthetic RC-LH complex subunits, we have also identified several proteins whose spots vary in intensity between the two nitrogen availability conditions. The increased intensity in the spots containing Sdh complex subunits in nitrogen fixing conditions is one of the most striking differences as the expression of this complex had only been shown so far in other organisms to vary in response to carbon source and oxygen availability 70. Chromatophore membranes have been shown to catalyze light-dependent NAD+ reduction using succinate as a substrate and this was previously shown to be one of the steps in the reversed electron flow in the electron transport chain 71. Reversed electron flow is only observed to a relevant extent in anaerobic conditions, as in aerobic conditions electron flow would be diverted to oxygen reduction 72. Mathematical modeling of the electron transfer pathways in the chromatophore membrane of R. rubrum showed that under anaerobic phototrophic conditions electron flow in NADH dehydrogenase and Sdh is reversed, using the photosynthetically-generated proton gradient to overcome the thermodynamic barriers, with the overall outcome of NADH generation 73. This process allows the cell an operating TCA cycle and even if it seems wasteful to dissipate some of the proton gradient, this will easily be maintained by the photosynthetic process (see Figure 5).
Nitrogen fixing cells have a large requirement for reduction equivalents and it has been previously proposed that the changes in the redox status upon induction of nitrogen fixation lead to the down regulation of alternative electron sinks, such as carbon fixation through RuBisCO 20. Even though the present study also seems to point in the same direction, more detailed studies are required in order to identify the signal pathways involved in sensing and regulating the proteome changes in response to shifts in the cellular redox balance in R. rubrum.
Supplementary Material
Acknowledgements
This work was supported by grants from the Carl Tryggers Foundation to A. N., from the Swedish Research Council to S. N., from Fundação para a Ciência e a Tecnologia (Portugal), through the PhD fellowship SFRH/BD/23183/2005 to T. T. S., from the NIH grant P01 GM75913 to P. S. C. and S. H. G. and from the Lundbeck Foundation to S. R.. We thank Dr. Jan-Willem de Gier for supplying the OmpA antibodies and Timo Johannes Schmidt for excellent technical assistance with the sucrose density gradients and Western blots.
Footnotes
Supporting Information Supporting information is available: synthesis details and scheme for compound 1, with structures of the intermediates and final product; NMR and MS data for compound 1 and precursors and list of detailed MS peptide data is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Bazylinski DA, Frankel RB. Magnetosome formation in prokaryotes. Nat Rev Microbiol. 2004;2(3):217–30. doi: 10.1038/nrmicro842. [DOI] [PubMed] [Google Scholar]
- 2.Komeili A, Li Z, Newman DK, Jensen GJ. Magnetosomes are cell membrane invaginations organized by the actin-like protein MamK. Science. 2006;311(5758):242–5. doi: 10.1126/science.1123231. [DOI] [PubMed] [Google Scholar]
- 3.Tavano CL, Donohue TJ. Development of the bacterial photosynthetic apparatus. Curr Opin Microbiol. 2006;9(6):625–31. doi: 10.1016/j.mib.2006.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pardee AB, Schachman HK, Stanier RY. Chromatophores of Rhodospirillum rubrum. Nature. 1952;169(4294):282–3. doi: 10.1038/169282a0. [DOI] [PubMed] [Google Scholar]
- 5.Anderson IC, Fuller RC. Photophosphorylation by isolated chromatophores of the purple sulfur bacteria. Arch Biochem Biophys. 1958;76(1):168–79. doi: 10.1016/0003-9861(58)90131-0. [DOI] [PubMed] [Google Scholar]
- 6.Nishimura M. Studies on bacterial photophosphorylation. I. Kinetics of photophosphorylation in Rhodospirillum rubrum chromatophores by flashing light. Biochim Biophys Acta. 1962;57(1):88–95. doi: 10.1016/0006-3002(62)91082-x. [DOI] [PubMed] [Google Scholar]
- 7.Ginet N, Lavergne J. Equilibrium and kinetic parameters for the binding of inhibitors to the QB pocket in bacterial chromatophores: dependence on the state of QA. Biochemistry. 2001;40(6):1812–23. doi: 10.1021/bi001686a. [DOI] [PubMed] [Google Scholar]
- 8.Visser HM, Somsen OJ, van Mourik F, Lin S, van Stokkum IH, van Grondelle R. Direct observation of sub-picosecond equilibration of excitation energy in the light-harvesting antenna of Rhodospirillum rubrum. Biophys J. 1995;69(3):1083–99. doi: 10.1016/S0006-3495(95)79982-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Norling B, Strid A, Tourikas C, Nyren P. Amount and turnover rate of the F0F1-ATPase and the stoichiometry of its inhibition by oligomycin in Rhodospirillum rubrum chromatophores. Eur J Biochem. 1989;186(1-2):333–7. doi: 10.1111/j.1432-1033.1989.tb15213.x. [DOI] [PubMed] [Google Scholar]
- 10.Gromet-Elhanan Z, Khananshvili D, Weiss S, Kanazawa H, Futai M. ATP synthesis and hydrolysis by a hybrid system reconstituted from the beta-subunit of Escherichia coli F1-ATPase and beta-less chromatophores of Rhodospirillum rubrum. J Biol Chem. 1985;260(23):12635–40. [PubMed] [Google Scholar]
- 11.Gest H. Metabolic patterns in photosynthetic bacteria. Bacteriol Rev. 1951;15(4):183–210. doi: 10.1128/br.15.4.183-210.1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kamen MD, Gest H. Evidence for a nitrogenase system in the photosynthetic bacterium Rhodospirillum rubrum. Science. 1949;109(2840):560. doi: 10.1126/science.109.2840.560. [DOI] [PubMed] [Google Scholar]
- 13.Seefeldt LC, Hoffman BM, Dean DR. Mechanism of Mo-dependent nitrogenase. Annu Rev Biochem. 2009;78:701–22. doi: 10.1146/annurev.biochem.78.070907.103812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dixon R, Kahn D. Genetic regulation of biological nitrogen fixation. Nat Rev Microbiol. 2004;2(8):621–31. doi: 10.1038/nrmicro954. [DOI] [PubMed] [Google Scholar]
- 15.Zhang Y, Pohlmann EL, Roberts GP. Identification of critical residues in GlnB for its activation of NifA activity in the photosynthetic bacterium Rhodospirillum rubrum. Proc Natl Acad Sci U S A. 2004;101(9):2782–7. doi: 10.1073/pnas.0306763101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Araujo LM, Monteiro RA, Souza EM, Steffens MB, Rigo LU, Pedrosa FO, Chubatsu LS. GlnB is specifically required for Azospirillum brasilense NifA activity in Escherichia coli. Res Microbiol. 2004;155(6):491–5. doi: 10.1016/j.resmic.2004.03.002. [DOI] [PubMed] [Google Scholar]
- 17.Dubbs JM, Tabita FR. Regulators of nonsulfur purple phototrophic bacteria and the interactive control of CO2 assimilation, nitrogen fixation, hydrogen metabolism and energy generation. FEMS Microbiol Rev. 2004;28(3):353–76. doi: 10.1016/j.femsre.2004.01.002. [DOI] [PubMed] [Google Scholar]
- 18.Gibson JL, Dubbs JM, Tabita FR. Differential expression of the CO2 fixation operons of Rhodobacter sphaeroides by the Prr/Reg two-component system during chemoautotrophic growth. J Bacteriol. 2002;184(23):6654–64. doi: 10.1128/JB.184.23.6654-6664.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Joshi HM, Tabita FR. A global two component signal transduction system that integrates the control of photosynthesis, carbon dioxide assimilation, and nitrogen fixation. Proc Natl Acad Sci U S A. 1996;93(25):14515–20. doi: 10.1073/pnas.93.25.14515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Selao TT, Nordlund S, Noren A. Comparative proteomic studies in Rhodospirillum rubrum grown under different nitrogen conditions. J Proteome Res. 2008;7(8):3267–75. doi: 10.1021/pr700771u. [DOI] [PubMed] [Google Scholar]
- 21.Edgren T, Nordlund S. The fixABCX genes in Rhodospirillum rubrum encode a putative membrane complex participating in electron transfer to nitrogenase. J Bacteriol. 2004;186(7):2052–60. doi: 10.1128/JB.186.7.2052-2060.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Edgren T, Nordlund S. Two pathways of electron transport to nitrogenase in Rhodospirillum rubrum: the major pathway is dependent on the fix gene products. FEMS Microbiol Lett. 2006;260(1):30–5. doi: 10.1111/j.1574-6968.2006.00297.x. [DOI] [PubMed] [Google Scholar]
- 23.Edgren T, Nordlund S. Electron transport to nitrogenase in Rhodospirillum rubrum: identification of a new fdxN gene encoding the primary electron donor to nitrogenase. FEMS Microbiol Lett. 2005;245(2):345–51. doi: 10.1016/j.femsle.2005.03.024. [DOI] [PubMed] [Google Scholar]
- 24.Lindblad A, Nordlund S. Electron transport to nitrogenase in Rhodospirillum rubrum: Role of energization of the chromatophore membrane. Photosynth Res. 1997;53(1):23–8. [Google Scholar]
- 25.Brostedt E, Lindblad A, Jansson J, Nordlund S. Electron transport to nitrogenase in Rhodospirillum rubrum: the role of NAD(P)H as electron donor and the effect of fluoroacetate on nitrogenase activity. FEMS Microbiol Lett. 1997;150(2):263–7. [Google Scholar]
- 26.Gest H, Ormerod JG, Ormerod KS. Photometabolism of Rhodospirillum rubrum: light-dependent dissimilation of organic compounds to carbon dioxide and molecular hydrogen by an anaerobic citric acid cycle. Arch Biochem Biophys. 1962;97(1):21–33. doi: 10.1016/0003-9861(62)90039-5. [DOI] [PubMed] [Google Scholar]
- 27.Frenkel AW. Simultaneous reduction of diphosphopyridine nucleotide and oxidation of reduced flavin mononucleotide by illuminated bacterial chromatophores. J Am Chem Soc. 1958;80(13):3479–80. [Google Scholar]
- 28.Hu Q, Noll RJ, Li H, Makarov A, Hardman M, Cooks R. Graham. The Orbitrap: a new mass spectrometer. J Mass Spectrom. 2005;40(4):430–43. doi: 10.1002/jms.856. [DOI] [PubMed] [Google Scholar]
- 29.Yates JR, Cociorva D, Liao L, Zabrouskov V. Performance of a linear ion trap-Orbitrap hybrid for peptide analysis. Anal Chem. 2006;78(2):493–500. doi: 10.1021/ac0514624. [DOI] [PubMed] [Google Scholar]
- 30.Ormerod JG, Ormerod KS, Gest H. Light-dependent utilization of organic compounds and photoproduction of molecular hydrogen by photosynthetic bacteria; relationships with nitrogen metabolism. Arch Biochem Biophys. 1961;94(3):449–63. doi: 10.1016/0003-9861(61)90073-x. [DOI] [PubMed] [Google Scholar]
- 31.Lowry OH, Rosebrough NJ, Farr AL, Randall RJ. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951;193(1):265–75. [PubMed] [Google Scholar]
- 32.Collins ML, Niederman RA. Membranes of Rhodospirillum rubrum: physicochemical properties of chromatophore fractions isolated from osmotically and mechanically disrupted cells. J Bacteriol. 1976;126(3):1326–38. doi: 10.1128/jb.126.3.1326-1338.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chai TJ, Foulds J. Purification of protein A, an outer membrane component missing in Escherichia coli K-12 ompA mutants. Biochim Biophys Acta. 1977;493(1):210–5. doi: 10.1016/0005-2795(77)90274-4. [DOI] [PubMed] [Google Scholar]
- 34.Nyren P, Nore BF, Strid A. Proton-pumping N,N’-dicyclohexylcarbodiimide-sensitive inorganic pyrophosphate synthase from Rhodospirillum rubrum - purification, characterization and reconstitution. Biochemistry. 1991;30(11):2883–7. doi: 10.1021/bi00225a022. [DOI] [PubMed] [Google Scholar]
- 35.Chae Maltose-neopentyl glycol (MNG) amphiphiles for solubilization, stabilization and crystallization of membrane proteins. Nature Methods. 2010 doi: 10.1038/nmeth.1526. e. a. advance online publication, 31 October (doi: 10.1038/nmeth. 1526) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schagger H, von Jagow G. Blue native electrophoresis for isolation of membrane protein complexes in enzymatically active form. Anal Biochem. 1991;199(2):223–31. doi: 10.1016/0003-2697(91)90094-a. [DOI] [PubMed] [Google Scholar]
- 37.Schagger H, von Jagow G. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal Biochem. 1987;166(2):368–79. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- 38.Candiano G, Bruschi M, Musante L, Santucci L, Ghiggeri GM, Carnemolla B, Orecchia P, Zardi L, Righetti PG. Blue silver: a very sensitive colloidal Coomassie G-250 staining for proteome analysis. Electrophoresis. 2004;25(9):1327–33. doi: 10.1002/elps.200305844. [DOI] [PubMed] [Google Scholar]
- 39.Osborn MJ, Gander JE, Parisi E, Carson J. Mechanism of assembly of the outer membrane of Salmonella typhimurium. Isolation and characterization of cytoplasmic and outer membrane. J Biol Chem. 1972;247(12):3962–72. [PubMed] [Google Scholar]
- 40.Nikaido H, Vaara M. Molecular basis of bacterial outer membrane permeability. Microbiol Rev. 1985;49(1):1–32. doi: 10.1128/mr.49.1.1-32.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Manning PA, Pugsley AP, Reeves P. Defective growth functions in mutants of Escherichia coli K12 lacking a major outer membrane protein. J Mol Biol. 1977;116(2):285–300. doi: 10.1016/0022-2836(77)90217-0. [DOI] [PubMed] [Google Scholar]
- 42.Fejes AP, Beatty TJ, et al. Shotgun proteomic analysis of a chromatophore-enriched preparation from the purple phototrophic bacterium Rhodopseudomonas palustris. Photosynth Res. 2003;78:195–203. doi: 10.1023/B:PRES.0000006752.81486.74. [DOI] [PubMed] [Google Scholar]
- 43.Herranen M, A. E-M, et al. Towards Functional Proteomics of Membrane Protein Complexes in Synechocystis sp. 6803. Plant Physiol. 2004;134:470–481. doi: 10.1104/pp.103.032326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.D’Amici GM, Rinalducci S, Murgiano L, Italiano F, Zolla L. Oligomeric characterization of the photosynthetic apparatus of Rhodobacter sphaeroides R26.1 by nondenaturing electrophoresis methods. J Proteome Res. 2010;9(1):192–203. doi: 10.1021/pr9005052. [DOI] [PubMed] [Google Scholar]
- 45.Sturgis JN, Tucker JD, Olsen JD, Hunter CN, Niederman RA. Atomic force microscopy studies of native photosynthetic membranes. Biochemistry. 2009;48(17):3679–98. doi: 10.1021/bi900045x. [DOI] [PubMed] [Google Scholar]
- 46.Qian P, Bullough PA, Hunter CN. Three-dimensional reconstruction of a membrane-bending complex: the RC-LH1-PufX core dimer of Rhodobacter sphaeroides. J Biol Chem. 2008;283(20):14002–11. doi: 10.1074/jbc.M800625200. [DOI] [PubMed] [Google Scholar]
- 47.Ueda T, Ideguchi T, Kaino N, Kakuno T, Yamashita J, Horio T. Molecular organization of photochemical reaction complex in chromatophore membrane from Rhodospirillum rubrum as detected by immunochemical and proteolytic analyses. J Biochem. 1987;102(4):755–65. doi: 10.1093/oxfordjournals.jbchem.a122113. [DOI] [PubMed] [Google Scholar]
- 48.Davis CM, Parkes-Loach PS, Cook CK, Meadows KA, Bandilla M, Scheer H, Loach PA. Comparison of the structural requirements for bacteriochlorophyll binding in the core light-harvesting complexes of Rhodospirillum rubrum and Rhodobacter sphaeroides using reconstitution methodology with bacteriochlorophyll analogs. Biochemistry. 1996;35(9):3072–84. doi: 10.1021/bi951777l. [DOI] [PubMed] [Google Scholar]
- 49.Fotiadis D, Qian P, Philippsen A, Bullough PA, Engel A, Hunter CN. Structural analysis of the reaction center light-harvesting complex I photosynthetic core complex of Rhodospirillum rubrum using atomic force microscopy. J Biol Chem. 2004;279(3):2063–8. doi: 10.1074/jbc.M310382200. [DOI] [PubMed] [Google Scholar]
- 50.Karrasch S, Bullough PA, Ghosh R. The 8.5 A projection map of the light-harvesting complex I from Rhodospirillum rubrum reveals a ring composed of 16 subunits. EMBO J. 1995;14(4):631–8. doi: 10.1002/j.1460-2075.1995.tb07041.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Recchia PA, Davis CM, Lilburn TG, Beatty JT, Parkes-Loach PS, Hunter CN, Loach PA. Isolation of the PufX protein from Rhodobacter capsulatus and Rhodobacter sphaeroides: evidence for its interaction with the alpha-polypeptide of the core light-harvesting complex. Biochemistry. 1998;37(31):11055–63. doi: 10.1021/bi980657l. [DOI] [PubMed] [Google Scholar]
- 52.Barz WP, Vermeglio A, Francia F, Venturoli G, Melandri BA, Oesterhelt D. Role of the PufX protein in photosynthetic growth of Rhodobacter sphaeroides. 2. PufX is required for efficient ubiquinone/ubiquinol exchange between the reaction center QB site and the cytochrome bc1 complex. Biochemistry. 1995;34(46):15248–58. doi: 10.1021/bi00046a033. [DOI] [PubMed] [Google Scholar]
- 53.Comayras F, Jungas C, Lavergne J. Functional consequences of the organization of the photosynthetic apparatus in Rhodobacter sphaeroides: II. A study of PufX-membranes. J Biol Chem. 2005;280(12):11214–23. doi: 10.1074/jbc.M412089200. [DOI] [PubMed] [Google Scholar]
- 54.Wang ZY, Gokan K, Kobayashi M, Nozawa T. Solution structures of the core light-harvesting alpha and beta polypeptides from Rhodospirillum rubrum: implications for the pigment-protein and protein-protein interactions. J Mol Biol. 2005;347(2):465–77. doi: 10.1016/j.jmb.2005.01.017. [DOI] [PubMed] [Google Scholar]
- 55.Pedersen PL, Ko YH, Hong S. ATP synthases in the year 2000: evolving views about the structures of these remarkable enzyme complexes. J Bioenerg Biomembr. 2000;32(4):325–32. doi: 10.1023/a:1005594800983. [DOI] [PubMed] [Google Scholar]
- 56.Dunn SD, Kellner E, Lill H. Specific heterodimer formation by the cytoplasmic domains of the b and b’ subunits of cyanobacterial ATP synthase. Biochemistry. 2001;40(1):187–92. doi: 10.1021/bi001821j. [DOI] [PubMed] [Google Scholar]
- 57.Poetsch A, Berzborn RJ, Heberle J, Link TA, Dencher NA, Seelert H. Biophysics and bioinformatics reveal structural differences of the two peripheral stalk subunits in chloroplast ATP synthase. J Biochem. 2007;141(3):411–20. doi: 10.1093/jb/mvm045. [DOI] [PubMed] [Google Scholar]
- 58.Falk G, Walker JE. DNA sequence of a gene cluster coding for subunits of the F0 membrane sector of ATP synthase in Rhodospirillum rubrum. Support for modular evolution of the F1 and F0 sectors. Biochem J. 1988;254(1):109–22. doi: 10.1042/bj2540109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Walker JE. The NADH:ubiquinone oxidoreductase (complex I) of respiratory chains. Q Rev Biophys. 1992;25(3):253–324. doi: 10.1017/s003358350000425x. [DOI] [PubMed] [Google Scholar]
- 60.Sazanov LA, Carroll J, Holt P, Toime L, Fearnley IM. A role for native lipids in the stabilization and two-dimensional crystallization of the Escherichia coli NADH-ubiquinone oxidoreductase (complex I) J Biol Chem. 2003;278(21):19483–91. doi: 10.1074/jbc.M208959200. [DOI] [PubMed] [Google Scholar]
- 61.Yagi T, Yano T, Di Bernardo S, Matsuno-Yagi A. Procaryotic complex I (NDH-1), an overview. Biochim Biophys Acta. 1998;1364(2):125–33. doi: 10.1016/s0005-2728(98)00023-1. [DOI] [PubMed] [Google Scholar]
- 62.Dupuis A, Chevallet M, Darrouzet E, Duborjal H, Lunardi J, Issartel JP. The complex I from Rhodobacter capsulatus. Biochim Biophys Acta. 1998;1364(2):147–65. doi: 10.1016/s0005-2728(98)00025-5. [DOI] [PubMed] [Google Scholar]
- 63.Edgren T. Electron transport to nitrogenase in Rhodospirillum rubrum. Stockholm University; Stockholm: 2006. [Google Scholar]
- 64.Wang H, Franke CC, Nordlund S, Noren A. Reversible membrane association of dinitrogenase reductase activating glycohydrolase in the regulation of nitrogenase activity in Rhodospirillum rubrum; dependence on GlnJ and AmtB1. FEMS Microbiol Lett. 2005;253(2):273–9. doi: 10.1016/j.femsle.2005.09.049. [DOI] [PubMed] [Google Scholar]
- 65.Grammel H, Ghosh R. Redox-state dynamics of ubiquinone-10 imply cooperative regulation of photosynthetic membrane expression in Rhodospirillum rubrum. J Bacteriol. 2008;190(14):4912–21. doi: 10.1128/JB.00423-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lyubarev AE, Kurganov BI. Supramolecular organization of tricarboxylic acid cycle enzymes. Biosystems. 1989;22(2):91–102. doi: 10.1016/0303-2647(89)90038-5. [DOI] [PubMed] [Google Scholar]
- 67.Zeng X, Kaplan S, et al. Proteomic characterizion of the Rhodobacter Sphaeroides 2.4.1 Photosynthetic Membrane: Identification of New Proteins. J. Bacteriol. 2007;189(20):7464–7474. doi: 10.1128/JB.00946-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Anderson L, Fuller RC. Photosynthesis in Rhodospirillum rubrum. 3. Metabolic control of reductive pentose phosphate and tricarboxylic acid cycle enzymes. Plant Physiol. 1967;42(4):497–509. doi: 10.1104/pp.42.4.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gonin S, Arnoux P, Pierru B, Lavergne J, Alonso B, Sabaty M, Pignol D. Crystal structures of an extracytoplasmic solute receptor from a TRAP transporter in its open and closed forms reveal a helix-swapped dimer requiring a cation for alpha-keto acid binding. BMC Struct Biol. 2007;7:11. doi: 10.1186/1472-6807-7-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Park SJ, Tseng CP, Gunsalus RP. Regulation of succinate dehydrogenase (sdhCDAB) operon expression in Escherichia coli in response to carbon supply and anaerobiosis: role of ArcA and Fnr. Mol Microbiol. 1995;15(3):473–82. doi: 10.1111/j.1365-2958.1995.tb02261.x. [DOI] [PubMed] [Google Scholar]
- 71.Govindjee R, Sybesma C. The photoreduction of nicotinamide-adenine dinucleotide by chromatophore fractions from Rhodospirillum rubrum. Biophys J. 1972;12(7):897–908. doi: 10.1016/S0006-3495(72)86132-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Grammel H, Gilles ED, Ghosh R. Microaerophilic cooperation of reductive and oxidative pathways allows maximal photosynthetic membrane biosynthesis in Rhodospirillum rubrum. Appl Environ Microbiol. 2003;69(11):6577–86. doi: 10.1128/AEM.69.11.6577-6586.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Klamt S, Grammel H, Straube R, Ghosh R, Gilles ED. Modeling the electron transport chain of purple non-sulfur bacteria. Mol Syst Biol. 2008;4:156. doi: 10.1038/msb4100191. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.