Abstract
Estimation of influenza infection rates is important for determination of the extent of epidemic spread and for calculation of severity indicators. The authors compared estimated infection rates from paired and cross-sectional serologic surveys, rates of influenza like illness (ILI) obtained from sentinel general practitioners (GPs), and ILI samples that tested positive for influenza using data from similar periods collected during the 2009 H1N1 epidemic in Singapore. The authors performed sensitivity analyses to assess the robustness of estimates to input parameter uncertainties, and they determined sample sizes required for differing levels of precision. Estimates from paired seroconversion were 17% (95% Bayesian credible interval (BCI): 14, 20), higher than those from cross-sectional serology (12%, 95% BCI: 9, 17). Adjusted ILI estimates were 15% (95% BCI: 10, 25), and estimates computed from ILI and laboratory data were 12% (95% BCI: 8, 18). Serologic estimates were least sensitive to the risk of input parameter misspecification. ILI-based estimates were more sensitive to parameter misspecification, though this was lessened by incorporation of laboratory data. Obtaining a 5-percentage-point spread for the 95% confidence interval in infection rates would require more than 1,000 participants per serologic study, a sentinel network of 90 GPs, or 50 GPs when combined with laboratory samples. The various types of estimates will provide comparable findings if accurate input parameters can be obtained.
Keywords: epidemics; estimation; infection; influenza, human; population surveillance; serologic tests; statistics as topic
Assessing the spread and severity of influenza epidemics is necessary to calibrate response and mitigation strategies (1). The World Health Organization and many individual countries have made substantial investments to measure epidemic indicators. One important indicator is the epidemic's infection rate, which is crucial to quantify overall morbidity and to obtain an accurate denominator for calculating complication and mortality rates used to classify severity; the latter, in turn, guides prioritization of interventions for mitigating epidemic severity.
The 2009 influenza pandemic showed the urgency of such assessments for activation of appropriate responses, especially early in a pandemic. Because complete case counts are not feasible (2), during the 2009 epidemic public health officials in many countries attempted to estimate infection rates using whatever data were available. This included estimating clinical attack rates from influenza like illness (ILI) surveillance, determining infection rates through serologic surveys, and even using nontraditional methods such as Internet searches (3, 4). However, existing data collection plans are vital, since extrapolating from ILI surveillance necessitates estimating rates of primary-care consultation among influenza cases, which are influenced by population health-care-seeking behaviors (5), while serologic surveys require substantial planning and laboratory support (6).
With myriad estimation methods in use, it is important to determine their comparability and stability to misspecification of input parameters, to allow better interpretation of estimates over different countries and successive influenza epidemics. In this study, we answered these questions by comparing results of different methods in a single setting.
MATERIALS AND METHODS
To illustrate the different methods used worldwide to estimate infection rates during the 2009 H1N1 influenza pandemic, we performed a literature search with the PubMed search engine (US National Library of Medicine), spanning May 1, 2009, to August 1, 2010, using the search terms “influenza attack rate” and “influenza infection rate.” The inclusion criterion was all English-language articles that provided infection rate estimates and explicitly described the methods used to derive the estimates.
Different methods for estimating infection rates
From the common methods used globally (5, 7–20) (Table 1), we selected 4 representative generic methods (Table 2), together with generic equations and minimum data requirements, to determine their comparability. The 4 methods were serologic cohort and cross-sectional studies, sentinel general practitioner (GP) ILI surveillance, and laboratory surveillance to supplement GP data.
Table 1.
Results From Studies That Estimated Infection Rates for H1N1 Influenza A, 2009
| First Author, Year (Reference No.) | Study Location | Study Period | Estimated Infection Rate | Method of Estimation | Details |
| Lipsitch, 2009 (7) | Mexico | April 2009 | 0.11%–0.35% during the month of April 2009 (population of 106,682,518) | Surveillance data from travelers | International public health records surveyed to estimate infection rates among travelers to Mexico |
| Cases among Mexican residents = cases in travelers × (Mexican population × 30 days)/(traveler population × duration of travel) | |||||
| D'Ortenzio, 2010 (8) | Réunion Island, France | May 2009–September 2009 | 12.85% (104,067/810,000) | Sentinel physician network, cross-sectional ARI prevalence survey | Incidence of ARI consultations gathered from social insurance data, adjusted by the proportion of sentinel physician consultations |
| Health-care-seeking behavior in persons with ARI from a cross-sectional survey | |||||
| Calculated by extrapolating the proportion of randomly selected ARI patients testing H1N1-positive in the total estimated no. of ARI cases | |||||
| Dawood, 2010 (9) | Hunter New England, Australia | June 1, 2009–August 30, 2009 | 6.2% (range, 4.4%–8.2%) | Syndromic surveillance and laboratory data | Incidence of ILI from an online self-reporting ILI surveillance system |
| 53,383 (range, 37,828–70,597) out a population of 866,565 | Proportion of ILI samples that tested H1N1-positive from national laboratories | ||||
| Using these data, the proportion of ILI cases due to H1N1 was estimated and extrapolated to the general population. | |||||
| Gordon, 2010 (10) | Nicaragua | June 1, 2009–November 15, 2009 | 20.1% among children aged 2–14 years | Syndromic surveillance, laboratory testing | Cohort of children selected from an existing dengue study |
| Testing criteria were fever with cough, sore throat, or rhinorrhea | |||||
| Samples were tested by RT-PCR to determine the H1N1 clinical attack rate. | |||||
| No extrapolation to the general population was done. | |||||
| Flahault, 2009 (5) | France | September 2009–December 2009 | 10.6% among pregnant women | Cross-sectional seroprevalence | Cross-sectional seroprevalence study from serum obtained from pregnant women in weeks 48–49 of 2009 |
| 1,712,000 cases (95% CI: 1,112,700, 2,311,300) in persons aged 20–39 years | Cumulative seroprevalence was then estimated for the population aged 20–39 years. | ||||
| Moghadami, 2010 (11) | Iran | December 2009 | 58.9% (1,504/2,553) | Cross-sectional seroprevalence | Single-sample cross-sectional seroprevalence study |
| Serum samples from randomly selected participants in the community | |||||
| Miller, 2010 (12) | England, United Kingdom | August 2009–September 2009 | Age group, years | Cross-sectional seroprevalence | Cross-sectional seroprevalence study involving pre- and postpandemic samples from blood collected for other purposes |
| <5: 21.3% (95% CI: 8.8, 40.3) | |||||
| 5–14: 42.0% (95% CI: 26.3, 58.2) | Infection rates were estimated by subtracting prepandemic seroprevalence from postpandemic seroprevalence. | ||||
| 15–24: 20.6% (95% CI: 1.6, 42.4) | |||||
| 25–44: 6.2% (95% CI: −2.8, 18.7) | |||||
| 45–64: −2.7% (95% CI: −10.3, 7.1) | |||||
| ≥65: 0.9% (95% CI: −8.8, 13.3) | |||||
| Chan, 2010 (13) | Taiwan, Republic of China | October 2009–November 2009 | 30.8% among health-care workers | Cross-sectional seroprevalence | Single-sample cross-sectional seroprevalence study |
| 12.6% among controls | Serum samples taken from hospital staff and controls | ||||
| Ross, 2010 (14) | Pittsburg, Pennsylvania, United States | Mid-November–early December 2009 | 21% (unadjusted) | Cross-sectional seroprevalence | Cross-sectional seroprevalence study with pre- and postpandemic samples |
| Range from 5% for persons aged 70–79 years to 45% for persons aged 10–19 years | Prepandemic samples only from young adults aged 18–24 years | ||||
| Baseline 6% among young adults aged 18–24 years | Postpandemic samples from laboratory specimens collected for other purposes over a wide age range | ||||
| Allwinn, 2010 (15) | Germany | November 2009 | 12% (27/225) with titer of ≥1:40 (unadjusted) | Cross-sectional seroprevalence | First sample from blood donors previously recruited for a serum survey of the spread of enterovirus 71 infection |
| Baseline 13.1% (19/145) with titers of 1:>32 | Second sample from randomly selected patients at a local university hospital | ||||
| Grills, 2010 (16) | Australia | August 2009–October 2009 | 10% in adults aged 18–65 years | Cross-sectional seroprevalence | Participants in a health monitoring program were tested opportunistically. |
| Baseline prepandemic seropositive rate from another study was subtracted from the result. | |||||
| Chen, 2010 (17) | Singapore | June 22, 2009–October 15, 2009 | 13.5% in community-dwelling adults | Serologic cohort study | Multisample seroepidemiologic cohort study |
| 6.5% in hospital staff | Serial serum samples from individuals | ||||
| 29.4% in military personnel | Seroconversion was determined by a 4-fold rise in titers. | ||||
| 1.2% in long-term-care patients | |||||
| Crum-Cianflone, 2009 (18) | San Diego, California, United States | April 21, 2009–May 8, 2009 | 0.53% (101 per 100,000) from April 21, 2009, to May 8, 2009 | Complete testing of ILI cases | Complete RT-PCR testing of all ILI cases from a captive population of local US military beneficiaries |
| Colizza, 2009 (19) | Mexico | April 2009 | 0.11%–1.31% (121,000–1,394,000 cases as of April 30, 2009) | Mathematical modeling | Model with a geographically structured metapopulation approach |
| Use of a population-level census, human mobility flows, and disease dynamics to model disease evolution and infections | |||||
| Presanis, 2009 (20) | Milwaukee, Wisconsin, and New York, New York, United States | April 2009–July 2009 | Not shown; used as a denominator to determine hospitalization and case-fatality rates | Mathematical modeling | Estimation using mathematical model and probabilities of ILI with consultations, consultations that were tested, and proportion positive. |
| Data from physician consultations, laboratory, and telephone survey | For New York, a telephone survey was conducted to determine self-reported ILI status. |
Abbreviations: ARI, acute respiratory illness; CI, confidence interval; ILI, influenzalike illness; RT-PCR, reverse-transcriptase polymerase chain reaction.
Table 2.
Methods Used for Estimating Rates of Influenza Infection During the 2009 H1N1 Outbreak in Singapore
| Method and Data Requirements | Advantages (+) and Disadvantages (−) |
| Method 1: paired serologic surveysa | |
| Seroconversion data from cohort study | + Detects subclinical cases |
| Sensitivity of the serologic test to detect true infection | − Difficulties in timely data collection during an evolving pandemic |
| Total population size (to determine confidence interval for the estimate) | − No estimate of clinical infection rate |
| − Availability of results is dependent on sampling intervals | |
| Method 2: cross-sectional serologic surveysb | |
| Proportion of persons with high pre- and postpandemic titers | + Relative ease of data collection in comparison with paired serologic surveys |
| Sensitivity to detect change in titers (proportion of true infections that have high postpandemic and low prepandemic titers using the cutoff titer) | − Risk of underestimation because of persons with high baseline titers |
| Total population size (to determine confidence interval for the estimate) | − Difficult to generalize to population when using banked samples |
| Method 3: syndromic surveillance for ILIc | |
| Data on all ILI consultations from sentinel GPs | + Allows for “real-time” estimation of infection rate |
| Proportion of influenza cases involving consultation for ILI | + Data collection is possible with minimal resources |
| Proportion of ILI consultations due to influenza | − Unable to capture subclinical infections |
| Market share of GPs surveyed among the total population | − Dependent on clinician reporting |
| Total population size | − Difficulties in estimating input parameters |
| − Large margin of error if given inaccurate data | |
| Method 4: syndromic surveillance for ILI with virologic datad | |
| Data on all ILI consultations from sentinel GPs | + Margin of error is reduced in comparison with method 3 |
| Market share of GPs surveyed among the total population | + Allows for “real-time” estimation of infection rate |
| Proportion of influenza cases involving consultation for ILI | − Additional resources required for laboratory testing |
| Laboratory proportion of ILI samples that test positive for influenza | − Dependent on sensitivity of laboratory test |
| Sensitivity of the laboratory test | |
| Total population size |
Abbreviations: GP, general practitioner; ILI, influenzalike illness.
Method 1 infection rate = (no. of persons who seroconverted)/[(total no. followed up) × (sensitivity of the serologic test)].
Method 2 infection rate = [(proportion with high postpandemic titers) − (proportion with high prepandemic titers)]/(sensitivity to detect true change in titers).
Method 3 infection rate = (no. of ILI cases)/[(market share of GPs surveyed) × population × (proportion of influenza cases that involved consultation for ILI) × (proportion of ILI consultations due to influenza)].
Method 4 infection rate = (no. of ILI cases)/[(market share of GPs surveyed) × population × (proportion of influenza cases that involved consultation for ILI) × (proportion of ILI samples that tested positive/sensitivity of the laboratory test)].
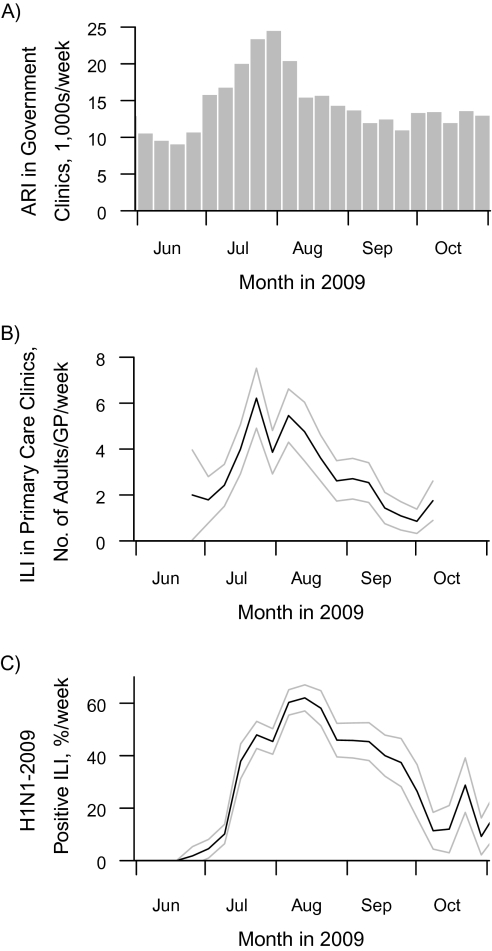
Using data from the first wave of the 2009 H1N1 epidemic in Singapore, a tropical city-state, we compared infection rates estimated using these methods. Singapore was ideal for this study, because the first epidemic wave's temporal progression was well-defined—beginning in late June, peaking in early August, and ending by September (Figure 1)—and several surveillance programs and studies were performed simultaneously in the adult population, facilitating comparison of different methods.
Figure 1.
Sources of available data on influenza infection in Singapore from June to October 2009. A) Numbers of cases of acute respiratory illness (ARI) diagnosed in government clinics, in thousands per week; B) numbers of adult cases of influenza like illness (ILI) reported by primary-care general practitioner (GP) sentinel clinics per GP per week; C) percentage of ILI cases that tested positive for H1N1-2009 influenza per week. Lighter lines, 95% confidence interval.
Data sources
We used data sources available from June 2009, at the first suggestion of community transmission in Singapore, to October 2009, 1 month after numbers of respiratory illness cases returned to baseline levels (Figure 1). ILI cases were defined as cases involving new-onset respiratory symptoms with body temperature greater than 38.0°C (100.4°F), following World Health Organization definitions (21, 22). We performed aggregated and age-stratified analyses among 5 age groups: 20–24, 25–34, 35–44, 45–54, and ≥55 years. Data sources included:
1) A paired seroincidence adult cohort study (17). Multiple blood samples were obtained from each participant, including a baseline sample taken up to June 27, 2009 (before the local epidemic); a second sample taken between August 20, 2009, and August 29, 2009 (4 weeks after the epidemic's peak); and a postepidemic sample taken between October 6, 2009, and October 11, 2009 (4 weeks after the epidemic subsided). Fortnightly telephone surveys were used to collect data on clinical symptoms and health-care consultations. Data from 727 participants with paired serum samples were used.
2) A sentinel GP network of 23 GPs nationwide reporting ILI cases, initiated in June 2009 (23). Individual patient consultations involving ILI were recorded using a standardized template and submitted daily, together with basic demographic details.
3) Laboratory-based national surveillance by the Ministry of Health using samples from ILI patients visiting sentinel primary health-care clinics. Samples were tested for 2009 influenza A virus (H1N1) by means of reverse-transcriptase polymerase chain reaction (RT-PCR) (24), producing weekly age-stratified data on the proportion of ILI samples positive for H1N1-2009. When combined with ILI surveillance data, this negates the need to estimate the proportion of ILI consultations due to influenza (ILI consultations include conditions not due to influenza), leaving only the proportion of influenza cases who seek medical consultation for ILI to be determined.
Data from the serologic and GP studies were collected under the approval of the National University of Singapore Institutional Review Board. Laboratory data were part of the Ministry of Health's ongoing influenza epidemiology surveillance program, and no ethics review was required.
Statistical methods and computation of infection rates
In addition to the main data, each method required supplementary data ranging from simple test sensitivity for paired serologic data to consultation rates given infection and infection rates given ILI consultation. While serologic surveys intrinsically account for asymptomatic infections, ILI-based estimates need to be complemented with prior information to allow for nonreporting of symptomatic cases and asymptomatic infections. Because the latter information was available from the serologic surveys, we used parameters derived from the serologic surveys for the ILI-based estimates.
To allow full propagation of parametric uncertainty, we used an objective Bayesian approach, taking flat prior distributions in the absence of data and informative priors only when suitable external data were available. We used as many data as were available from these studies, including some which would not be available in other settings using only 1 source of data. Full details on the statistical methods used and the distributions of key parameters can be found in the Web Appendix (http://aje.oxfordjournals.org/).
Method 1: paired serologic surveys.
To estimate infection rates from paired serologic surveys, we defined overall seroconversion as a 4-fold or greater rise in titer on hemagglutination inhibition (HAI) testing between baseline titers and subsequent samples for the same individual. Since not all influenza infections may be detected by HAI (because of sample timing, insufficient titer increases, or measurement error), we adjusted the seroconversion rates by HAI sensitivity using data from our study and another study (17, 25). Because there was no clear evidence on HAI false-positive rates, we did not adjust for this possibility.
Method 2: cross-sectional serologic surveys.
To estimate infection rates from cross-sectional sampling similarly to other studies, we defined the cross-sectional seroprevalence at each sampling point as the proportion with HAI titers of ≥40 (12, 14). We then subtracted baseline seroprevalence from final seroprevalence and adjusted the results by the sensitivity of a single postinfection sample to detect HAI titers of ≥40 in patients confirmed to have infection.
Method 3: ILI data from sentinel GPs.
When using ILI data from sentinel GP sites, we computed the number of ILI consultations per sentinel GP day and scaled this to the population using the relative proportion of ILI seen by the average GP, using data on primary-care consultations from a national survey (26). In addition, we estimated the ratio of all ILI consultations to influenza infections through data on symptoms and health-care-seeking behavior available from our serologic cohort study (adjusting for HAI sensitivity), assuming that the serologic study was representative of the general population in terms of symptom presentation and health-care-seeking behavior. We then estimated the number of community influenza infections given the ILI observed. In the absence of such data, other approaches must be taken to scale the estimates from the sampled data to the general population appropriately (see Discussion).
Method 4: laboratory surveillance and ILI data from sentinel GPs.
We also used laboratory data to supplement sentinel GP ILI data, replacing the proportion of ILI consultations due to influenza with the proportion of ILI samples that tested positive for H1N1-2009 by RT-PCR, while adjusting for the imperfect sensitivity of the RT-PCR assay in detecting influenza cases (25). Ideally, validation should be performed in the same laboratory using the same virus strain and correlated with epidemiologic data; because this was not possible, we performed sensitivity analysis to account for it. We then incorporated the fraction of infections without a primary care consultation for ILI from our cohort study as above.
Because of the poor specificity of acute respiratory illness in estimating influenza (27), we did not include analysis relying on acute respiratory illness only.
Sensitivity analyses
Because not all countries have access to relevant supporting data, especially on ILI consultation rates, some methods require extrapolation from other settings. Therefore, we performed Bayesian sensitivity analyses to determine the robustness of these methods to misspecification of key input parameters and the resulting impact on inferred infection rates. We set Dirac delta priors on one parameter at a time, keeping all other priors as above and varying the single parameter in question over a plausible range, as might be done operationally when no accurate data are available. The parameters examined were the sensitivity of the tests, the ratio of all ILI consultations to influenza cases, and the market share of sentinel GPs—factors that may vary by strain, location, and time.
Analysis of sample-size effects
Finally, to appreciate the effect of sample size on the spread of estimates for future surveys, we performed bootstrap analysis on our existing data. For methods 1 and 2, we simulated, using a binomial distribution, the proportion of infections which might be observed to seroconvert with different numbers of paired sera or to have antibodies at titers ≥40 in different numbers of baseline and follow-up samples, respectively, assuming that the true infection rate corresponded to our estimate. For methods 3 and 4, we simulated the observations for a situation in which the sentinel GP ILI data had been derived from different numbers of GPs, by resampling with replacement from the available GPs (we restricted resampling to GPs who submitted data for at least 50% of all days). To estimate the effect of laboratory samples on method 4, we used the binomial distribution to simulate positive proportions which might be observed in each week, assuming that laboratory samples were distributed uniformly each week across the epidemic. The corresponding formulae were applied to the estimates derived from the bootstrap with 100,000 resamples for each method and sample size. Since the availability of external data in future outbreaks is unpredictable, we did not attempt to incorporate parametric uncertainty from external data in these analyses. We used a 5- to 10-percentage-point spread in the 95% confidence interval of the infection rate estimate as reasonable for classifying epidemic severity or for evaluating the success of interventions.
RESULTS
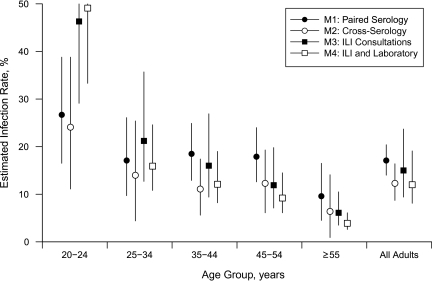
Figure 2 shows the estimated infection rates calculated from the various estimation methods based on the Singapore studies. The overall infection rate estimated using paired seroconversion samples was 17% (95% Bayesian credible interval (BCI): 14, 20). Using estimates derived from paired seroconversion data as the comparison group, the overall estimate derived from cross-sectional serologic sampling (obtained with baseline and final titers from the serologic cohort study as independent samples) was lower at 12% (95% BCI: 9, 17), also observed across all age groups. Estimates from ILI rates (15%, 95% BCI: 10, 25) and estimates from the combination of ILI and laboratory data (12%, 95% BCI: 8, 18) provided overall estimates close to the serologic estimates, although there were variations among various age groups.
Figure 2.
Rates of H1N1-2009 influenza infection estimated from various methods, aggregated and by age group, Singapore, 2009. For details on methods 1–4 (M1–M4), see Table 2. ILI, influenza like illness. Whiskers, 95% Bayesian credible interval.
The substantial overlap in 95% Bayesian credible intervals for all 4 methods, along with fairly close point estimates, suggests that accurate determination of input variables can produce similar results regardless of the estimation method. The actual and effective sample sizes available to us led to estimates from ILI alone being the most uncertain, while seroconversion data gave the most precise estimates, although this may have been different if resources had allowed for different relative sample sizes. Estimates using the combination of ILI and laboratory data were less sensitive than ILI alone but more sensitive than the seroconversion estimates.
From the sensitivity analyses (Web Figure 1), serologic cohort estimates were very robust to misspecification of the external input parameter (test sensitivity), as were cross-sectional serologic estimates. The latter, however, were strongly influenced by misspecification of the level of baseline prepandemic titers. Because substantial proportions of persons had baseline antibodies to H1N1-2009 (12, 17, 28), accurately determining baseline rates is important, and cross-sectional estimates that assumed no baseline titers (similar to the 0% baseline value in Web Figure 1C) would bias infection rate estimates upwards.
ILI estimates were very sensitive and changed substantially with key parameters of market share per GP, proportion of influenza cases who seek medical consultation for ILI, and proportion of ILI consultations due to influenza. Estimates derived from combining ILI data with laboratory data only required determining the proportion of influenza cases that sought medical consultation for ILI, which we obtained from our serologic study questionnaire. Infection rate estimates were very sensitive to misspecification of this parameter; however, were it determined with greater accuracy, this method would provide extremely accurate estimates, as shown by the very narrow Bayesian credible intervals in Web Figure 1H.
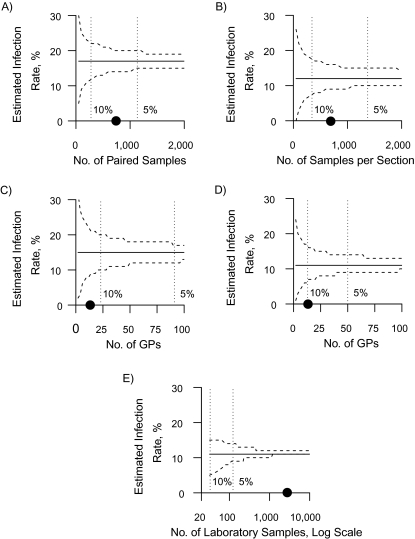
Figure 3 shows the sample sizes required to obtain 5- and 10-percentage-point spreads in mean infection rates for the 95% confidence intervals. For paired serologic estimates, 300 participants were required in order to achieve a 10-percentage-point spread, and 1,150 were required for a 5-percentage-point spread. For cross-sectional serologic estimates, more persons are needed per survey to achieve a similar spread. For ILI estimates alone, the required number of GPs that report daily had to be 20 and 90 to achieve spreads of 10- and 5-percentage points, respectively (Singapore had an estimated 2,138 GPs in 2009) (26). Adding laboratory data reduces the number of GPs needed by almost half, while the total number of laboratory samples required over the entire study period was less than 200 for a 5-percentage-point spread.
Figure 3.
Change in the 95% confidence interval (dashed lines) for the mean estimated H1N1 influenza infection rate (solid line) with different sample sizes using 4 different estimation methods, Singapore, 2009. A) Method 1; B) method 2; C) method 3; D) method 4; E) method 4 (see Table 2). Sample sizes which resulted in a 5- and 10-percentage-point spreads in the confidence interval for the mean estimates are shown with dotted vertical lines; the actual sample size used in the Singapore studies is shown with a circle on the x-axis. The total number of general practitioners (GPs) in Singapore in 2009 was approximately 2,138.
Table 1 summarizes data on the 15 papers selected from our literature search, out of 295 identified. These studies did not all use the same methods, and it is difficult to compare their results because most investigators did not completely adjust for key parameters such as test sensitivity, asymptomatic cases, or baseline titers. Three studies used surveillance data—from travelers to Mexico early in the epidemic (7), from a sentinel physician network (8), and from online surveillance tools together with laboratory data (9); and all used different methods of scaling data to the population level. Of the serologic surveys, only 1 study used paired samples (17); 7 studies were cross-sectional, with different sample origins (5, 11–16); and only 2 adjusted for prepandemic seroprevalence (12, 16). Two studies performed laboratory testing of all ILI cases but in unique small-scale military (18) and pediatric cohort (10) settings, while 2 used mathematical modeling of primary data (19, 20).
DISCUSSION
Estimation of epidemic infection rates is important in order to evaluate disease morbidity and to obtain accurate denominators for severity indicators, such as hospitalizations or case fatality. Attempts have been made to determine infection rates through different methods during different time periods (Table 1). However, none describe the relative comparability and robustness of these estimation methods in a single setting. Public health professionals and policy-makers should understand the advantages and disadvantages of these methods to incorporate data collection into preparedness plans and to account for possible errors.
Serologic surveys provide reliable estimates of infection rates, since they determine antibodies even for asymptomatic cases (17, 29). Serial sampling from individuals in the context of H1N1-2009 is important because baseline antibodies were present from cross-reactivity to different strains (28). Serial sampling requires preplanning and good timing to establish cohorts with baseline blood samples before the epidemic's onset. Therefore, few countries have been able to perform serologic cohort studies (6). Serologic surveys are only available after each sampling interval, depending on laboratory capacity; will usually not provide real-time estimates; and are unable to detect temporary rises in titers that may arise from mild infections, which may be important for subsequent immunity. A further weakness of serologic surveys is that they do not estimate clinical infection rates unless clinical surveys are conducted simultaneously.
Cross-sectional serologic surveys have disadvantages similar to those of cohort studies but are easier to conduct without individual follow-up, and samples can be obtained from other collection sources (e.g., blood banks). However, upon subtraction of baseline prepandemic levels, they may produce lower estimates than cohort studies because of overcompensation for baseline titers (12, 17, 28). This may result in estimates with negative infection rates, which are difficult to interpret (12). This may also be a problem when producing age-stratified estimates if baseline antibody levels differ by age (12, 14, 28). Other surveys used only a single postpandemic sample without baseline adjustment (5, 11, 13–15), which may have resulted in overestimation; estimates were as high as 58.9% in one study (11) and were 21% in another study, which also had a 6% baseline prevalence of antibodies (14). Unless accurate baseline estimates are available, cross-sectional surveys will be less accurate than paired surveys. The sample source may also make it difficult to generalize results—some studies obtained blood collected for other purposes, including blood donations and health monitoring programs, which may not represent the general population (12, 14–16).
It is clear from our sensitivity analyses that serologic survey estimates result in narrower ranges and are less sensitive to misspecification of input parameters. However, how serologic titers decrease over time is unknown, especially if samples are taken at long intervals. This can be averted by conducting serologic cohort studies with multiple samplings at shorter intervals.
ILI-derived estimates are easily obtained from sentinel GPs, and in Singapore they were similar to serologic and laboratory estimates. However, ILI estimates are very sensitive to changes in input parameters, and these must be determined accurately. Adjustment for nonreporting and asymptomatic infection can be achieved via a “scaling-up factor” or by using prior information on ILI consultations for all influenza infections. The latter is intrinsically difficult to obtain, since it is ideally based on data from confirmed cases, which were available for our study (17). In other settings, strategies might involve extrapolating from past epidemics or other regions. For example, because different adjustment factors were used in the studies by D'Ortenzio et al. (8) and Dawood et al. (9), these estimates are unlikely to be comparable. Adding accurate laboratory testing data to ILI addresses the otherwise substantial difficulty in estimating ILI consultations due to influenza, and results in estimates that are less sensitive to parameter misspecification. This does not obviate the need to estimate the proportion of influenza cases who seek medical consultation for ILI, which we did via our serologic cohort (17), although this proportion can also be estimated through local surveys carried out among ILI cases (since consultation is influenced by local health-care-seeking behaviors) (2), adjusted by the proportion of ILI among influenza cases, which is a biologic variable that presumably can be extrapolated from other regions. The need for reliable extraneous data is the main weakness of consultation data, especially in heterogeneous environments.
Infection rates differ across age groups, with the highest infection rates being seen in younger adults, confirming that young adults (and perhaps children) had higher infection rates during the 2009 H1N1 pandemic. Estimates from different estimation methods also differ across age groups: Greater differences exist between estimates in the younger age groups, with ILI-derived estimates being biased upwards relative to serology (although the 95% Bayesian credible intervals overlap). This shows the difficulties in estimating attack rates for different age categories through surveillance, without having accurate scaling factors and concomitantly larger sample sizes to accurately determine age-specific infection rates.
Data from primary health-care surveillance and laboratories are more suited than serologic studies to providing real-time data with which to map an epidemic's development and develop predictive models (23). ILI-derived estimates with laboratory data can also be continually used to monitor seasonal influenza infection rates and are already part of many routine surveillance systems. The Mexican studies carried out at the epidemic's start to determine early extent of spread (7, 19) and the localized San Diego, California, outbreak (18) provided real-time estimates for early planning. However, additional laboratory data may not be readily available in low-resource settings and may be difficult to obtain in a heterogeneous setting with different sociodemographic profiles within a country.
Another potential obstacle to accurate estimation of infection rates is the sample size required for sufficient accuracy. Serologic studies required to achieve a 5-percentage-point spread (≥1,000 participants per survey) may be difficult to perform in settings with fewer resources. ILI estimates may be easier to collect if GPs are able to routinely report ILI cases, since 4% of all GPs can achieve a 5-percentage-point spread. Including laboratory samples further reduces the number of GPs required, while only requiring a small number of samples over the epidemic period because of good correlation between influenza-positive laboratory samples and the epidemic curve. With a consistent sentinel GP network and laboratory testing program, method 4 can be routinely used to estimate infection rates for regular influenza seasons and the relative burden of disease from different strains.
Two limitations of our study were the lack of pediatric data for comparison (these data were collected differently from data on adults) and small sample sizes when stratifying by age for some analyses. Researchers who aim to estimate age-group-specific infection rates will need to increase the sample size proportionally, which could result in very large studies. In this paper, we have clearly displayed the differences between the methods in a single population, and these concepts are applicable to other populations and settings. Although this study was based on Singapore's H1N1-2009 epidemic, the methods proposed are applicable globally to other infectious diseases.
Estimates of infection rates from serologic data and ILI data with or without laboratory data can provide comparable results if input parameters are accurately determined. Each method has advantages and disadvantages which should be considered when comparing estimates. The epidemic timing, objectives of data collection, and availability of resources will also determine the method used. Countries with sufficient resources may consider using multiple estimation methods to cover the disadvantages of some while benefiting from the advantages of others.
Supplementary Material
Acknowledgments
Author affiliations: Department of Epidemiology and Public Health, Yong Loo Lin School of Medicine, National University of Singapore, Singapore, Singapore (Vernon J. Lee, Mark I. Chen, Wei-Yen Lim); National Centre for Epidemiology and Population Health, Australian National University, Canberra, Australia (Vernon J. Lee, Paul M. Kelly); Biodefence Centre, Ministry of Defence, Singapore, Singapore (Vernon J. Lee, Jonathan Yap, Jocelyn Ong); Department of Clinical Epidemiology, Tan Tock Seng Hospital, Singapore, Singapore (Mark I. Chen, Jimmy B. S. Ong); Emerging Infectious Diseases Program, Duke-NUS Graduate Medical School, Singapore, Singapore (Mark I. Chen); National Public Health Laboratory, Ministry of Health, Singapore, Singapore (Raymond T. P. Lin, Tze Minn Mak); WHO Collaborating Center for Reference and Research for Influenza, Victorian Infectious Diseases Reference Laboratory, Melbourne, Australia (Ian Barr); Department of Medicine, National University Hospital, Singapore, Singapore (Lee Gan Goh); College of Family Physicians, Singapore, Singapore (Lee Gan Goh); Department of Infectious Diseases, Tan Tock Seng Hospital, Singapore, Singapore (Yee Sin Leo); Australian Capital Territories Health, Canberra, Australia (Paul M. Kelly); and Department of Statistics and Applied Probability, Faculty of Science, National University of Singapore, Singapore, Singapore (Alex R. Cook).
This project was funded by the National Medical Research Council of Singapore (grants NMRC/H1N1O/002/2009 and NMRC/H1N1R/005/2009).
The authors thank the National University of Singapore and the Singapore Ministry of Health for providing the data for this study; the Melbourne WHO Collaborating Centre for Reference and Research on Influenza (which is supported by the Australian Government Department of Health and Ageing) for laboratory and technical assistance; and Teo Guo Ci for useful comments on the statistical approach.
The funders played no role in the study design, data collection and analysis, the decision to publish, or preparation of the manuscript.
Conflict of interest: V. J. L. has received unrelated research grants from GlaxoSmithKline. A. R. C. has received research funding from the National University of Singapore. No other conflicts of interest, financial or otherwise, are present.
Glossary
Abbreviations
- BCI
Bayesian credible interval
- GP
general practitioner
- HAI
hemagglutination inhibition
- ILI
influenzalike illness
- RT-PCR
reverse-transcriptase polymerase chain reaction
References
- 1.World Health Organization. Assessing the Severity of an Influenza Pandemic. Geneva, Switzerland: World Health Organization; 2009. ( http://www.who.int/csr/disease/swineflu/assess/disease_swineflu_assess_20090511/en/index.html). (Accessed June 15, 2010) [Google Scholar]
- 2.Lipsitch M, Hayden FG, Cowling BJ, et al. How to maintain surveillance for novel influenza A H1N1 when there are too many cases to count. Lancet. 2009;374(9696):1209–1211. doi: 10.1016/S0140-6736(09)61377-5. [DOI] [PubMed] [Google Scholar]
- 3.Ginsberg J, Mohebbi MH, Patel RS, et al. Detecting influenza epidemics using search engine query data. Nature. 2009;457(7232):1012–1014. doi: 10.1038/nature07634. [DOI] [PubMed] [Google Scholar]
- 4.Cook AR, Chen MIC, Pin Lin RT. Internet search limitations and pandemic influenza, Singapore [letter] Emerg Inf Dis. 2010;16(10):1647–1649. doi: 10.3201/eid1610.100840. (doi: 10.3201/eid1610.100840) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Flahault A, de Lamballerie X, Hanslik T, et al. Symptomatic infections less frequent with H1N1pdm than with seasonal strains. PLoS Curr. 2009;1 doi: 10.1371/currents.RRN1140. RRN1140. (doi: 10.1371/currents.RRN1140) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.World Health Organization. Seroepidemiological studies of pandemic influenza A (H1N1) 2009 virus. Wkly Epidemiol Rec. 2010;85(24):229–235. [PubMed] [Google Scholar]
- 7.Lipsitch M, Lajous M, O'Hagan JJ, et al. Use of cumulative incidence of novel influenza A/H1N1 in foreign travelers to estimate lower bounds on cumulative incidence in Mexico. PLoS One. 2009;4(9):e6895. doi: 10.1371/journal.pone.0006895. (doi: 10.1371/journal.pone.0006895) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.D'Ortenzio E, Renault P, Jaffar-Bandjee MC, et al. A review of the dynamics and severity of the pandemic A(H1N1) influenza virus on Réunion Island, 2009. Clin Microbiol Infect. 2010;16(4):309–316. doi: 10.1111/j.1469-0691.2010.03171.x. [DOI] [PubMed] [Google Scholar]
- 9.Dawood FS, Hope KG, Durrheim DN, et al. Estimating the disease burden of pandemic (H1N1) 2009 virus infection in Hunter New England, Northern New South Wales, Australia, 2009. PLoS One. 2010;5(3):e9880. doi: 10.1371/journal.pone.0009880. (doi: 10.1371/journal.pone.0009880) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gordon A, Saborío S, Videa E, et al. Clinical attack rate and presentation of pandemic H1N1 influenza versus seasonal influenza A and B in a pediatric cohort in Nicaragua. Clin Infect Dis. 2010;50(11):1462–1467. doi: 10.1086/652647. [DOI] [PubMed] [Google Scholar]
- 11.Moghadami M, Moattari A, Tabatabaee HR, et al. High titers of hemagglutination inhibition antibodies against 2009 H1N1 influenza virus in southern Iran. Iran J Immunol. 2010;7(1):39–48. [PubMed] [Google Scholar]
- 12.Miller E, Hoschler K, Hardelid P, et al. Incidence of 2009 pandemic influenza A H1N1 infection in England: a cross-sectional serological study. Lancet. 2010;375(9720):1100–1108. doi: 10.1016/S0140-6736(09)62126-7. [DOI] [PubMed] [Google Scholar]
- 13.Chan YJ, Lee CL, Hwang SJ, et al. Seroprevalence of antibodies to pandemic (H1N1) 2009 influenza virus among hospital staff in a medical center in Taiwan. J Chin Med Assoc. 2010;73(2):62–66. doi: 10.1016/S1726-4901(10)70003-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ross T, Zimmer S, Burke D, et al. Seroprevalence following the second wave of pandemic 2009 H1N1 influenza. PLoS Curr. 2010;2 doi: 10.1371/currents.RRN1148. RRN1148. (doi: 10.1371/currents.RRN1148) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Allwinn R, Geiler J, Berger A, et al. Determination of serum antibodies against swine-origin influenza A virus H1N1/09 by immunofluorescence, haemagglutination inhibition, and by neutralization tests: how is the prevalence rate of protecting antibodies in humans? Med Microbiol Immunol. 2010;199(2):117–121. doi: 10.1007/s00430-010-0143-4. [DOI] [PubMed] [Google Scholar]
- 16.Grills N, Piers LS, Barr I, et al. A lower than expected adult Victorian community attack rate for pandemic (H1N1) 2009. Aust N Z J Public Health. 2010;34(3):228–231. doi: 10.1111/j.1753-6405.2010.00518.x. [DOI] [PubMed] [Google Scholar]
- 17.Chen MI, Lee VJ, Lim WY, et al. 2009 influenza A(H1N1) seroconversion rates and risk factors among distinct adult cohorts in Singapore. JAMA. 2010;303(14):1383–1391. doi: 10.1001/jama.2010.404. [DOI] [PubMed] [Google Scholar]
- 18.Crum-Cianflone NF, Blair PJ, Faix D, et al. Clinical and epidemiologic characteristics of an outbreak of novel H1N1 (swine origin) influenza A virus among United States military beneficiaries. Clin Infect Dis. 2009;49(12):1801–1810. doi: 10.1086/648508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Colizza V, Vespignani A, Perra N, et al. Estimate of novel influenza A/H1N1 cases in Mexico at the early stage of the pandemic with a spatially structured epidemic model. PLoS Curr. 2009;1 doi: 10.1371/currents.RRN1129. RRN1129. (doi: 10.1371/currents.RRN1129) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Presanis AM, De Angelis D New York City Swine Flu Investigation Team, et al. The severity of pandemic H1N1 influenza in the United States, from April to July 2009: a Bayesian analysis. PLoS Med. 2009;6(12):e1000207. doi: 10.1371/journal.pmed.1000207. (doi: 10.1371/journal.pmed.1000207) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.World Health Organization. WHO Recommended Surveillance Standards, Second Edition. Geneva, Switzerland: World Health Organization; 2009. ( http://www.who.int/csr/resources/publications/surveillance/WHO_CDS_CSR_ISR_99_2_EN/en). (Accessed June 15, 2010) [Google Scholar]
- 22.Hayden FG, Belshe R, Villanueva C, et al. Management of influenza in households: a prospective, randomized comparison of oseltamivir treatment with or without postexposure prophylaxis. J Infect Dis. 2004;189(3):440–449. doi: 10.1086/381128. [DOI] [PubMed] [Google Scholar]
- 23.Ong JBS, Chen MIC, Cook AR, et al. Real-time epidemic monitoring and forecasting of H1N1-2009 using influenza-like illness from general practice and family doctor clinics in Singapore. PLoS One. 2010;5(4):e10036. doi: 10.1371/journal.pone.0010036. (doi: 10.1371/journal.pone.0010036) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.World Health Organization. CDC Protocol of Realtime RTPCR for Influenza A(H1N1) Geneva, Switzerland: World Health Organization; 2009. ( http://www.who.int/csr/resources/publications/swineflu/CDCRealtimeRTPCR_SwineH1Assay-2009_20090430.pdf). (Accessed June 15, 2010) [Google Scholar]
- 25.Zambon M, Hays J, Webster A, et al. Diagnosis of influenza in the community: relationship of clinical diagnosis to confirmed virological, serologic, or molecular detection of influenza. Arch Intern Med. 2001;161(17):2116–2122. doi: 10.1001/archinte.161.17.2116. [DOI] [PubMed] [Google Scholar]
- 26.Integrated Service Division, Ministry of Health, Singapore. Ministry of Health Primary Care Survey 2005. Singapore, Singapore: Ministry of Health; 2005. [Google Scholar]
- 27.Cook AR, Lee HC, Ong JBS, et al. Predicting the influenza A (H1N1-2009) epidemic in Singapore using influenza-like-illness monitoring. Epidemiol News Bull. 2010;36(1):1–6. [Google Scholar]
- 28.Hancock K, Veguilla V, Lu X, et al. Cross-reactive antibody responses to the 2009 pandemic H1N1 influenza virus. N Engl J Med. 2009;361(20):1945–1952. doi: 10.1056/NEJMoa0906453. [DOI] [PubMed] [Google Scholar]
- 29.Reed C, Katz JM. Serological surveys for 2009 pandemic influenza A H1N1. Lancet. 2010;375(9720):1062–1063. doi: 10.1016/S0140-6736(09)62194-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.