Abstract
Membrane contact sites (MCSs) are formed by the close apposition of membranes of two organelles. They are zones where signals and small molecules, such as lipids and calcium, are exchanged between intracellular compartments. The past few years have seen considerable progress in our understanding of how MCSs form and facilitate the exchange of lipids and signals. Here we summarize what has been learned about MCSs between the endoplamic reticulum (ER) and the plasma membrane, the ER and mitochondria, and the ER and endosomes or lysosomes. These findings suggest that we are just beginning to understand how MCSs form and function.
Introduction
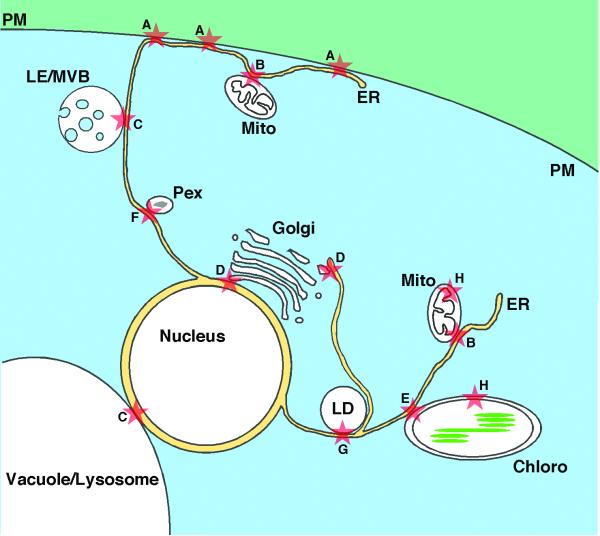
Compartmentalization in eukaryotic cells induces the need for inter-organelle trafficking of information and metabolites such as lipids. Mechanisms of intracellular lipid exchange are summarized in Box 1. One location where lipids, other small molecules, and signals are exchanged between organelles is membrane contact sites (MCSs), regions where the membranes of two organelles are closely apposed [1, 2]. At these sites the membranes of two organelles come within 10 to 30 nm of one another. MCSs are ubiquitous in all cells types. Many are observed between the endoplasmic reticulum (ER) and a second organelle (Figure 1). MCSs have also been observed within organelles that contain internal membranes such as mitochondria, chloroplasts, and multivesicular bodies. In the past few years there has been considerable progress in our understanding of how small molecules, such as lipids and calcium, and signals are exchanged at MCS as well as how some MCSs are formed. Here we focus on recent advances in our knowledge of lipid trafficking and signaling at MCSs between the ER and other organelles, and also focus on how MCSs are formed. For reviews of calcium trafficking at these sites see [3, 4].
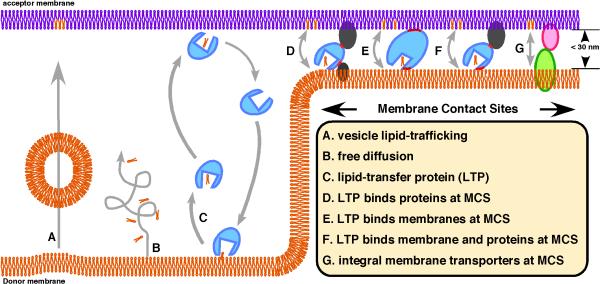
Box 1. Mechanisms of intracellular lipid exchange.
Lipid exchange between cellular compartments occurs by both vesicular and non-vesicular mechanisms, [52, 53]. One mechanism of non-vesicular lipid transfer between membranes is the spontaneous transfer of lipid monomers (B). This process is too slow to be physiologically relevant for most classes of lipid, with the notable exceptions of sterols and some lysolipids. Some non-vesicular lipid exchange requires lipid-transfer proteins (LTPs) (C). There are five large families of LTPs and most cell types express many of them [54]. LTPs bind a single lipid molecule in a hydrophobic tunnel or cleft and can shuttle the bound lipid between membranes. Rather than diffusing long distances through the cytosol many LTPs may function at MCSs, where they could rapidly shuttle between membranes (D-F). Some lipid exchange at MCS does not seem to require LTPs but instead may be mediated by integral membrane transporters (G).
Figure 1. Location of membrane contact sites.
A. Endoplasmic reticulum (ER) - plasma membrane (PM) B. ER - Mitochondria (Mito) C. ER - Late endosome (LE)/ multivesicular bodies (MVB) and ER - lysosome. In yeast there are contacts between the nucleus and vacuole. D. ER – Golgi complex. E. ER and chloroplast (chloro). F. ER – peroxisome (Pex) G. ER and lipid droplets (LD). H. Contact sites between the inner and outer membranes of mitochondria and chloroplasts. Similar contacts are also found in Gram negative bacteria (not shown).
MCSs between the ER and the plasma membrane (PM)
Close contacts between the ER and PM have been observed in all cells and can be quite extensive; such as in muscle cells where these contacts are needed for calcium exchange between these organelles.
How close contacts between the ER and PM are established is not well understood. Several proteins that regulate ER-PM contact site formation have been identified. In mammalian cells the ER calcium sensor STIM1 and the PM calcium channel ORAI1 interact when cells are depleted of calcium and drive the formation of ER-PM contacts [3, 5]. In muscle cells, junctophilins are thought to maintain close contacts and perhaps a specific spacing between the ER and PM. These integral membrane proteins reside in the ER and probably simultaneously bind the PM [6, 7]. Another mechanism for induction of ER-PM contacts has recently been suggested [8]. This work showed that dimerization of a coatomer-binding peptide anchored to ER membranes induced ER-PM contacts in a microtubule dependent fashion [8]. Although the mechanism is not yet understood, this work suggests that vesicular trafficking may be required to bring the ER close to the PM. It is not clear if simply bringing the ER near the PM is enough to cause contacts to form or if other steps are necessary. In yeast the ER proteins Ice2 and Scs2 are thought to be important for ER-PM contacts since cells missing both proteins have a significant decrease in the amount of ER found closely apposed to the PM [9].
There is growing evidence that sterols and signals are exchanged between the ER and PM at close contacts between these organelles. It has been known for some time that sterols can be transferred between the ER and PM by non-vesicular pathways [10-13]. In yeast, this sterol exchange between the ER and PM may be mediated by oxysterol-binding protein (OSBP)-related proteins (ORPs), lipid-binding proteins conserved from yeast to humans [14, 15]. Yeast has seven ORPs, which are called Osh proteins, and it has been found that all of them can bind sterols and exchange them between liposomes in vitro [16••]. The structure of one of these proteins, Osh4/Kes1, has been solved and found to form a nearly complete β-barrel that binds a single sterol inside [17]. In a strain missing all the Osh proteins sterol exchange between the ER and PM has been shown to slow significantly [18]. However, whether Osh proteins mediate most of the non-vesicular sterol exchange between the ER and PM or other compartments has been questioned [19]. In mammalian cells, recent work implicates some mammalian ORPs in non-vesicular sterol trafficking from the PM to the ER and lipid droplets (LDs) [20•] and other ORPs have been shown to bind and transfer cholesterol in vitro [21].
Four of the seven Osh protein are enriched on regions of the ER that are close to the PM, presumably at ER-PM contact sites [16••]. Two of these Osh proteins, Osh2 and Osh3, have domains that are required for targeting them to ER-PM contact sites [22, 23]. They contain a pleckstrin homology (PH) domain, which binds phosphoinositides (PIPs) in the PM and a two phenylalanines in an acidic tract (FFAT) motif, which is bound by the ER membrane proteins Scs2 and Scs22. The mammalian homologues of Scs2 and Scs22 are known as VAMP-associated proteins (VAPs). The PH and FFAT domains of Osh proteins are required for targeting them to ER-PM contact sites. However, two of the Osh protein that localize to ER-PM contacts lack PH and FFAT motifs and it is not known how they are targeted to these sites.
The core lipid-binding domain found in all Osh proteins has the ability to bind two membranes simultaneously [16••]. Membrane binding by this domain was studied in Osh4/Kes1 and was found to have at least two membrane-binding surfaces, one near the mouth of the sterol-binding pocket and a second at the distal site. The distal binding surface is required for Osh4/Kes1 to function in cells and is close to residues needed for PIP binding on the surface of the protein [24] that is not part of the sterol-binding pocket. PIP-binding at this site also regulates sterol binding. Thus, Osh proteins and perhaps all ORPs may be able to bind PIPs (or other lipids) in one membrane while simultaneously extracting or delivering sterols to a second membrane. This makes them well suited to transfer lipids or signals at MCSs.
A new model of how Osh proteins mediate signaling at ER-PM MCSs has been suggested by recent work from Stephan et al [25••]. The authors propose that Osh proteins function as PIP sensors that modulate PIP levels in cells by regulating the PIP phosphatase Sac1. Sac1 is an integral membrane protein in the ER and its active site is in the cytosol. Osh3 interacts with Sac1 at ER-PM contacts sites where it regulates Sac1 activity. Remarkably, this regulation can be reconstituted in vitro and Sac1 in one membrane can dephosphorylate PIPs in a second membrane. Osh proteins and the VAP homologues Scs2 and Scs22 regulate this process. Thus, Sac1 in the ER can hydrolyse PIPs on the PM or at any other MCS between the ER and a second membrane. This conclusion is consistent with a recent structural study of Sac1 [26] and earlier studies that suggested Sac1 can regulate PIP levels in the PM even though it is anchored in the ER [27, 28]. A role for Osh proteins in regulating PIP metabolism has also been found in other studies [24, 29]. How Osh proteins regulate Sac1, what role sterol-binding plays, and what signaling pathways are involved remain to be discovered.
Another method of signaling between the ER and PM at contact sites has been suggested by studies on the tyrosine phosphatase PTP1B. This enzyme is anchored to the cytosolic surface of the ER. A number of studies have reported that PTP1B directly binds signaling substrates on the PM including PKC∂, insulin receptor, EGFR and EphA3 [30-32]. Thus PTP1B probably functions at ER-PM contact sites.
MCSs between ER and mitochondria
Close contacts between the ER and mitochondria have been noted in a number of studies. For example, it has been found that in HeLa cells 5 to 20% of the surface of the mitochondrial network is in contact with the ER [33]. Similarly, a study in yeast found about 100 close contacts (< 30 nm) between the ER and mitochondria per cell [34]. These contacts are thought to be important for the exchange of calcium [4] and lipids [35] between the ER and mitochondria.
In the last few years there has been substantial progress in our understanding of how MCSs between the ER and mitochondria are formed. In mammalian cells the dynamin-like GTPase Mitofusin 2 (Mfn2), which is involved in mitochondrial fusion, has been suggested by the Scorrano group to tether the ER and mitochondria [36•]. Most Mfn2 is localized to the outer mitochondrial membrane (OMM) but the authors found that a fraction was found in the ER. This ER-localized fraction of Mfn2 forms dimers with Mfn2 in the OMM and with Mitofusin 1, which is also in the OMM, tethering ER and mitochondria. Importantly, in Mfn2-/- cells calcium trafficking between the ER and mitochondria is reduced, consistent with a role for Mfn2 in tethering. Recent work has suggested that tethering is also regulated by trichloplein/mitostatin, a keratin-binding protein [37].
A different complex involved in maintaining contacts between the ER and mitochondria in yeast has been identified. Kornmann and colleagues devised a cleaver screen to isolate mutants with a defect in ER-mitochondrial tethering. They discovered a complex that was named ERMES (ER-mitochondria encounter structure) [38••]. This complex contains the ER protein Mmm1, two proteins in the OMM, Mdm10 and Mdm34, and the cytosolic protein Mdm12. Deletion of any one of these proteins causes the complex to dissociate. Consistent with a role for the ERMES complex in tethering, lipid exchange between the ER and mitochondria was found to decrease in ERMES mutants. Whether the ERMES complex is only a tether or participates more directly in lipid exchange between the ER and mitochondria remains an open question. Interestingly, three of the four ERMES complex proteins contain a domain predicted to form a tubular structure that may allow lipid exchange between membranes [39, 40].
MCSs between ER and lysosomes or endosomes
In yeast, formation of the MCS between the nucleus and the vacuole, called the nucleus-vacuole junction (NVJ), requires the interaction of the ER membrane protein Nvj1 with Vac8, which is anchored on the vacuole [41]. NVJ formation is necessary for microautophagy of the nucleus (PMN), which can be induced by stress conditions such as nitrogen starvation or the presence of rapamycin [42]. Whether the NVJ is also a site where lipids or signals are exchanged between the ER and vacuole is not known. The lipid-binding proteins Osh1/Swh1, which is an ORP, and the fatty acid elongase Tsc13 localize to the NVJ and both are involved in PMN formation [43, 44]. Recent findings from the Mayer group have revealed that sterol and sphingolipid synthesis mutants have a defect in PMN formation [45•]. They suggest that a lipid microdomain is present in the membranes of the NVJ and that this microdomain is necessary to recruit proteins to the NVJ for the PMN formation [45•].
The NVJ is not found in higher eukaryotes. However, a number of recent studies in mammalian cells suggest the importance of MCSs between the ER and organelles in the endocytic trafficking pathway such as lysosomes, endosomes, and multivesicular bodies (MVBs). The ER tyrosine phosphatase PTP1B was shown to interact with one of its substrates, EGFR, on the surface of MVBs and localize to close contacts between the ER and MVBs [46]. A second example of signaling between the ER and the endomembrane system is provided by elegant studies on ORP1L, which demonstrate how this protein regulates MCSs between the ER and endosomes [47, 48]. ORP1L binds the small GTPase Rab7 and Rab7-interacting lysosomal protein on endosomes [47]. This association recruits the p150Glued subunit of the dynein-dynactin motor to endosomes [48]. Under low cholesterol conditions, ORP1L undergoes a conformational change that allows its FFAT domain to interact with VAP proteins on the ER, leading to the release of p150Glued and to the formation of close contacts between the ER and endosomes [49••]. Therefore, ORP1L acts as a cholesterol sensor that regulates contacts between the ER and endosomes. Whether cholesterol exchange occurs at these junctions and what role, if any, ORP1L plays in this transfer remains to be determined.
Another ORP, ORP5, has been recently implicated in cholesterol exchange between late endosomes/lysomes (LE/LY) and the ER, perhaps at MCSs between these organelles. Efficient trafficking of cholesterol derived from low-density lipoproteins (LDLs) out of LE/LYs requires Neimann Pick Type C protein 1 (NPC1), an integral membrane protein in LE/LYs [50]. Yang and colleagues found that ORP5 interacts with NPC1 and is needed for trafficking of LDL-derived cholesterol out of LE/LYs [51•]. ORP5 is an integral membrane protein in the ER and may be enriched at MCSs between the ER and LE/LYs. ORP5 is able to transfer sterol between liposomes in vitro and could mediate cholesterol transfer between the ER and LE/LYs [51•]. Alternatively, its primary function may be to regulate MCSs formation between the ER and LE/LYs.
Conclusion
The past few years have seen a dramatic increase in our knowledge of how MCSs are formed and how lipids and signals can be exchanged between organelles at these sites, yet much remains to be elucidated. MCSs almost certainly play critical roles in organelle function. For example, contacts between lipid droplets (LDs) and ER-LD MCS are thought to be important sites of lipid exchange and signaling. How MCSs form and are regulated remain important questions. Some MCSs, such as those between the ER and mitochondria, may be maintained by a number of low affinity interactions rather than by a single complex. Better techniques for isolating and characterizing proteins in MCSs will help resolve this issue. In many cases, the mechanisms of lipid exchange and signaling at MCSs remain poorly understood as does the energetics and regulation of these processes. This minireview has focused on only a few of the known MCSs in cells. Given the large number and diversity of these contacts it is likely that what we know now is only the tip of the iceberg. The next few years are likely to see a revolution in our understanding of how MCSs mediate intracellular trafficking of lipids, calcium, and signals between intracellular compartments.
Acknowledgments
We thank Chris Stefan for sharing unpublished findings, Ashley Slack for reading the manuscript and apologize to colleagues whose work we could not included because of space limitations.
This work was supported by the Intramural Research Fund of the National Institute of Diabetes and Digestive and Kidney diseases.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and recommended reading
Papers of particular interest, published within the annual period of review, have been highlighted as:
• of special interest
•• of outstanding interest
- 1.Lebiedzinska M, Szabadkai G, Jones AW, Duszynski J, Wieckowski MR. Interactions between the endoplasmic reticulum, mitochondria, plasma membrane and other subcellular organelles. Int J Biochem Cell Biol. 2009;41:1805–1816. doi: 10.1016/j.biocel.2009.02.017. [DOI] [PubMed] [Google Scholar]
- 2.Levine T, Loewen C. Inter-organelle membrane contact sites: through a glass, darkly. Curr Opin Cell Biol. 2006;18:371–378. doi: 10.1016/j.ceb.2006.06.011. [DOI] [PubMed] [Google Scholar]
- 3.Deng X, Wang Y, Zhou Y, Soboloff J, Gill DL. STIM and Orai: dynamic intermembrane coupling to control cellular calcium signals. J Biol Chem. 2009;284:22501–22505. doi: 10.1074/jbc.R109.018655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hayashi T, Rizzuto R, Hajnoczky G, Su TP. MAM: more than just a housekeeper. Trends Cell Biol. 2009;19:81–88. doi: 10.1016/j.tcb.2008.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lewis RS. The molecular choreography of a store-operated calcium channel. Nature. 2007;446:284–287. doi: 10.1038/nature05637. [DOI] [PubMed] [Google Scholar]
- 6.Garbino A, van Oort RJ, Dixit SS, Landstrom AP, Ackerman MJ, Wehrens XH. Molecular evolution of the junctophilin gene family. Physiol Genomics. 2009;37:175–186. doi: 10.1152/physiolgenomics.00017.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Takeshima H, Komazaki S, Nishi M, Iino M, Kangawa K. Junctophilins: a novel family of junctional membrane complex proteins. Mol Cell. 2000;6:11–22. doi: 10.1016/s1097-2765(00)00003-4. [DOI] [PubMed] [Google Scholar]
- 8.Lavieu G, Orci L, Shi L, Geiling M, Ravazzola M, Wieland F, Cosson P, Rothman JE. Induction of cortical endoplasmic reticulum by dimerization of a coatomer-binding peptide anchored to endoplasmic reticulum membranes. Proc Natl Acad Sci U S A. 2010;107:6876–6881. doi: 10.1073/pnas.1002536107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Loewen CJ, Young BP, Tavassoli S, Levine TP. Inheritance of cortical ER in yeast is required for normal septin organization. J Cell Biol. 2007;179:467–483. doi: 10.1083/jcb.200708205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Baumann NA, Sullivan DP, Ohvo-Rekila H, Simonot C, Pottekat A, Klaassen Z, Beh CT, Menon AK. Transport of newly synthesized sterol to the sterol-enriched plasma membrane occurs via nonvesicular equilibration. Biochemistry. 2005;44:5816–5826. doi: 10.1021/bi048296z. [DOI] [PubMed] [Google Scholar]
- 11.Heino S, Lusa S, Somerharju P, Ehnholm C, Olkkonen VM, Ikonen E. Dissecting the role of the golgi complex and lipid rafts in biosynthetic transport of cholesterol to the cell surface. Proc Natl Acad Sci U S A. 2000;97:8375–8380. doi: 10.1073/pnas.140218797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li Y, Prinz WA. ATP-binding cassette (ABC) transporters mediate nonvesicular, raft-modulated sterol movement from the plasma membrane to the endoplasmic reticulum. J Biol Chem. 2004;279:45226–45234. doi: 10.1074/jbc.M407600200. [DOI] [PubMed] [Google Scholar]
- 13.Urbani L, Simoni RD. Cholesterol and vesicular stomatitis virus G protein take separate routes from the endoplasmic reticulum to the plasma membrane. J Biol Chem. 1990;265:1919–1923. [PubMed] [Google Scholar]
- 14.Raychaudhuri S, Prinz WA. The diverse functions of oxysterol-binding proteins. Annu Rev Cell Dev Biol. 2010;26:157–177. doi: 10.1146/annurev.cellbio.042308.113334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ngo MH, Colbourne TR, Ridgway ND. Functional implications of sterol transport by the oxysterol-binding protein gene family. Biochem J. 2010;429:13–24. doi: 10.1042/BJ20100263. [DOI] [PubMed] [Google Scholar]
- 16 ••.Schulz TA, Choi MG, Raychaudhuri S, Mears JA, Ghirlando R, Hinshaw JE, Prinz WA. Lipid-regulated sterol transfer between closely apposed membranes by oxysterol-binding protein homologues. J Cell Biol. 2009;187:889–903. doi: 10.1083/jcb.200905007. The core lipid-binding domain found in all ORPs is shown to be able to bind two membranes simultaneously. ORP binding to one membrane at an MCS may regulate its ability to extract or delivery sterols to a second membrane, suggesting how these proteins could facilitate lipid exchange or signaling at MCSs.
- 17.Im YJ, Raychaudhuri S, Prinz WA, Hurley JH. Structural mechanism for sterol sensing and transport by OSBP-related proteins. Nature. 2005;437:154–158. doi: 10.1038/nature03923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Beh CT, Cool L, Phillips J, Rine J. Overlapping functions of the yeast oxysterol-binding protein homologues. Genetics. 2001;157:1117–1140. doi: 10.1093/genetics/157.3.1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Beh CT, Alfaro G, Duamel G, Sullivan DP, Kersting MC, Dighe S, Kozminski KG, Menon AK. Yeast oxysterol-binding proteins: sterol transporters or regulators of cell polarization? Mol Cell Biochem. 2009;326:9–13. doi: 10.1007/s11010-008-9999-7. [DOI] [PubMed] [Google Scholar]
- 20 •.Jansen M, Ohsaki Y, Rega L Rita, Bittman R, Olkkonen VM, Ikonen E. Role of ORPs in Sterol Transport from Plasma Membrane to ER and Lipid Droplets in Mammalian Cells. Traffic. 2011;12:218–231. doi: 10.1111/j.1600-0854.2010.01142.x. This study shows that ORP1S and ORP2 are involved in sterol transport from the PM to LD via the ER.
- 21.Ngo M, Ridgway ND. Oxysterol binding protein-related Protein 9 (ORP9) is a cholesterol transfer protein that regulates Golgi structure and function. Mol Biol Cell. 2009;20:1388–1399. doi: 10.1091/mbc.E08-09-0905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Loewen CJ, Roy A, Levine TP. A conserved ER targeting motif in three families of lipid binding proteins and in Opi1p binds VAP. EMBO J. 2003;22:2025–2035. doi: 10.1093/emboj/cdg201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Levine TP, Munro S. Dual targeting of Osh1p, a yeast homologue of oxysterol-binding protein, to both the Golgi and the nucleus-vacuole junction. Mol Biol Cell. 2001;12:1633–1644. doi: 10.1091/mbc.12.6.1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li X, Rivas MP, Fang M, Marchena J, Mehrotra B, Chaudhary A, Feng L, Prestwich GD, Bankaitis VA. Analysis of oxysterol binding protein homologue Kes1p function in regulation of Sec14p-dependent protein transport from the yeast Golgi complex. J Cell Biol. 2002;157:63–77. doi: 10.1083/jcb.200201037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25 ••.Stefan CJ, Manford AG, Baird D, Yamada-Hanff J, Mao Y, Emr SD. Osh proteins regulate phosphoinositide metabolism at ER-plasma membrane contact sites. Cell. 2011;144:389–401. doi: 10.1016/j.cell.2010.12.034. This study finds that yeast ORPs, called Osh proteins, regulate the phosphatase Sac1 at ER-PM contact sites and may function as lipid sensors. VAP homologs, ER-resident proteins that bind some Osh proteins, are also required.
- 26.Manford A, Xia T, Saxena AK, Stefan C, Hu F, Emr SD, Mao Y. Crystal structure of the yeast Sac1: implications for its phosphoinositide phosphatase function. EMBO J. 2010;29:1489–1498. doi: 10.1038/emboj.2010.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Baird D, Stefan C, Audhya A, Weys S, Emr SD. Assembly of the PtdIns 4-kinase Stt4 complex at the plasma membrane requires Ypp1 and Efr3. J Cell Biol. 2008;183:1061–1074. doi: 10.1083/jcb.200804003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Foti M, Audhya A, Emr SD. Sac1 lipid phosphatase and Stt4 phosphatidylinositol 4-kinase regulate a pool of phosphatidylinositol 4-phosphate that functions in the control of the actin cytoskeleton and vacuole morphology. Mol Biol Cell. 2001;12:2396–2411. doi: 10.1091/mbc.12.8.2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fairn GD, McMaster CR. The roles of the human lipid-binding proteins ORP9S and ORP10S in vesicular transport. Biochem Cell Biol. 2005;83:631–636. doi: 10.1139/o05-064. [DOI] [PubMed] [Google Scholar]
- 30.Anderie I, Schulz I, Schmid A. Direct interaction between ER membrane-bound PTP1B and its plasma membrane-anchored targets. Cell Signal. 2007;19:582–592. doi: 10.1016/j.cellsig.2006.08.007. [DOI] [PubMed] [Google Scholar]
- 31.Flint AJ, Tiganis T, Barford D, Tonks NK. Development of “substrate-trapping” mutants to identify physiological substrates of protein tyrosine phosphatases. Proc Natl Acad Sci U S A. 1997;94:1680–1685. doi: 10.1073/pnas.94.5.1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nievergall E, Janes PW, Stegmayer C, Vail ME, Haj FG, Teng SW, Neel BG, Bastiaens PI, Lackmann M. PTP1B regulates Eph receptor function and trafficking. J Cell Biol. 2010;191:1189–1203. doi: 10.1083/jcb.201005035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rizzuto R, Pinton P, Carrington W, Fay FS, Fogarty KE, Lifshitz LM, Tuft RA, Pozzan T. Close contacts with the endoplasmic reticulum as determinants of mitochondrial Ca2+ responses. Science. 1998;280:1763–1766. doi: 10.1126/science.280.5370.1763. [DOI] [PubMed] [Google Scholar]
- 34.Achleitner G, Gaigg B, Krasser A, Kainersdorfer E, Kohlwein SD, Perktold A, Zellnig G, Daum G. Association between the endoplasmic reticulum and mitochondria of yeast facilitates interorganelle transport of phospholipids through membrane contact. Eur J Biochem. 1999;264:545–553. doi: 10.1046/j.1432-1327.1999.00658.x. [DOI] [PubMed] [Google Scholar]
- 35.Osman C, Voelker DR, Langer T. Making heads or tails of phospholipids in mitochondria. J Cell Biol. 2011;192:7–16. doi: 10.1083/jcb.201006159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36 •.de Brito OM, Scorrano L. Mitofusin 2 tethers endoplasmic reticulum to mitochondria. Nature. 2008;456:605–610. doi: 10.1038/nature07534. This study finds that interactions between mitofusins in the ER and in the outer membrane of mitochondria tether these organelles. Calcium signaling between the ER and mitochondria is reduced in cells lacking mitofusins.
- 37.Cerqua C, Anesti V, Pyakurel A, Liu D, Naon D, Wiche G, Baffa R, Dimmer KS, Scorrano L. Trichoplein/mitostatin regulates endoplasmic reticulum-mitochondria juxtaposition. EMBO Rep. 2010;11:854–860. doi: 10.1038/embor.2010.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38 ••.Kornmann B, Currie E, Collins SR, Schuldiner M, Nunnari J, Weissman JS, Walter P. An ER-mitochondria tethering complex revealed by a synthetic biology screen. Science. 2009;325:477–481. doi: 10.1126/science.1175088. A clever genetic screen was used to identify a complex of four proteins that tethers the ER and mitochondria in yeast. Lipid exchange between the ER and mitochondria slowed in mutants lacking this complex.
- 39.Kopec KO, Alva V, Lupas AN. Homology of SMP domains to the TULIP superfamily of lipid-binding proteins provides a structural basis for lipid exchange between ER and mitochondria. Bioinformatics. 2010;26:1927–1931. doi: 10.1093/bioinformatics/btq326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee I, Hong W. Diverse membrane-associated proteins contain a novel SMP domain. FASEB J. 2006;20:202–206. doi: 10.1096/fj.05-4581hyp. [DOI] [PubMed] [Google Scholar]
- 41.Pan X, Roberts P, Chen Y, Kvam E, Shulga N, Huang K, Lemmon S, Goldfarb DS. Nucleus-vacuole junctions in Saccharomyces cerevisiae are formed through the direct interaction of Vac8p with Nvj1p. Mol Biol Cell. 2000;11:2445–2457. doi: 10.1091/mbc.11.7.2445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Roberts P, Moshitch-Moshkovitz S, Kvam E, O’Toole E, Winey M, Goldfarb DS. Piecemeal microautophagy of nucleus in Saccharomyces cerevisiae. Mol Biol Cell. 2003;14:129–141. doi: 10.1091/mbc.E02-08-0483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kvam E, Goldfarb DS. Nvj1p is the outer-nuclear-membrane receptor for oxysterol-binding protein homolog Osh1p in Saccharomyces cerevisiae. J Cell Sci. 2004;117:4959–4968. doi: 10.1242/jcs.01372. [DOI] [PubMed] [Google Scholar]
- 44.Kvam E, Gable K, Dunn TM, Goldfarb DS. Targeting of Tsc13p to nucleus-vacuole junctions: a role for very-long-chain fatty acids in the biogenesis of microautophagic vesicles. Mol Biol Cell. 2005;16:3987–3998. doi: 10.1091/mbc.E05-04-0290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45 •.Dawaliby R, Mayer A. Microautophagy of the Nucleus Coincides with a Vacuolar Diffusion Barrier at Nuclear-Vacuolar Junctions. Mol Biol Cell. 2010 doi: 10.1091/mbc.E09-09-0782. Mutants with defects in PMN formation were isolated. These include strains with defects in sterol and sphingolipid biosynthesis. The authors postulate that sterol and sphingolipid- enriched raft-like domains form in the membranes at NVJs.
- 46.Eden ER, White IJ, Tsapara A, Futter CE. Membrane contacts between endosomes and ER provide sites for PTP1B-epidermal growth factor receptor interaction. Nat Cell Biol. 2010;12:267–272. doi: 10.1038/ncb2026. [DOI] [PubMed] [Google Scholar]
- 47.Johansson M, Lehto M, Tanhuanpaa K, Cover TL, Olkkonen VM. The oxysterol-binding protein homologue ORP1L interacts with Rab7 and alters functional properties of late endocytic compartments. Mol Biol Cell. 2005;16:5480–5492. doi: 10.1091/mbc.E05-03-0189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Johansson M, Rocha N, Zwart W, Jordens I, Janssen L, Kuijl C, Olkkonen VM, Neefjes J. Activation of endosomal dynein motors by stepwise assembly of Rab7-RILP-p150Glued, ORP1L, and the receptor betalll spectrin. J Cell Biol. 2007;176:459–471. doi: 10.1083/jcb.200606077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49 ••.Rocha N, Kuijl C, van der Kant R, Janssen L, Houben D, Janssen H, Zwart W, Neefjes J. Cholesterol sensor ORP1L contacts the ER protein VAP to control Rab7-RILP-p150 Glued and late endosome positioning. J Cell Biol. 2009;185:1209–1225. doi: 10.1083/jcb.200811005. Neefjes and co-workers show that ORP1L regulates close contacts between the ER and endosomes. ORP1L binds proteins on endosomes and, in response to low cholesterol conditions, undergoes a conformational change that allows it to also bind VAPs on the ER, promoting the association between the ER and endosomes.
- 50.Watari H, Blanchette-Mackie EJ, Dwyer NK, Glick JM, Patel S, Neufeld EB, Brady RO, Pentchev PG, Strauss JFr. Niemann-Pick C1 protein: obligatory roles for N-terminal domains and lysosomal targeting in cholesterol mobilization. Proc Natl Acad Sci U S A. 1999;96:805–810. doi: 10.1073/pnas.96.3.805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51 •.Du X, Kumar J, Ferguson C, Schulz TA, Ong YS, Hong W, Prinz WA, Parton RG, Brown AJ, Yang H. A role for oxysterol-binding protein-related protein 5 in endosomal cholesterol trafficking. J Cell Biol. 2011;192:121–135. doi: 10.1083/jcb.201004142. ORP5, an integral membrane protein in the ER, is shown to mediate trafficking of LDL-derived cholesterol from LE/LYs, perhaps at MCSs between the ER and LE/LYs. ORP5 may directly facilitate cholesterol exchange or indirectly regulate cholesterol transport between the ER and LE/LYs.
- 52.Lev S. Non-vesicular lipid transport by lipid-transfer proteins and beyond. Nat Rev Mol Cell Biol. 2010;11:739–750. doi: 10.1038/nrm2971. [DOI] [PubMed] [Google Scholar]
- 53.Voelker DR. Genetic and biochemical analysis of non-vesicular lipid traffic. Annu Rev Biochem. 2009;78:827–856. doi: 10.1146/annurev.biochem.78.081307.112144. [DOI] [PubMed] [Google Scholar]
- 54.D’Angelo G, Vicinanza M, De Matteis MA. Lipid-transfer proteins in biosynthetic pathways. Curr Opin Cell Biol. 2008;20:360–370. doi: 10.1016/j.ceb.2008.03.013. [DOI] [PubMed] [Google Scholar]