Abstract
The αvβ3 integrin receptor plays an important role in human metastasis and tumor-induced angiogenesis. Cyclic Arg-Gly-Asp (cRGD) peptide represents a selective αvβ3 integrin ligand that has been extensively used for research, therapy, and diagnosis of neoangiogenesis. For developing photosensitizers with enhanced PDT efficacy, we here report the synthesis of a series of bifunctional agents in which the 3-(1′-hexyloxyethyl)-3-devinylpyropheophorbide-a (HPPH), a chlorophyll-based photosensitizer was conjugated to cRGD and the related analogs. The cell uptake, in vitro PDT efficacy of the conjugates were studied in αvβ3 integrin overexpressing U87 and 4T1 cell lines whereas the in vivo PDT efficacy and fluorescence-imaging potential of the conjugates were compared with the corresponding non-conjugated photosensitizer HPPH in 4T1 tumors. Compared to HPPH, the HPPH-cRGD conjugate in which the arginine and aspartic acid moieties were available for binding to two subunits of αvβ3 integrin showed faster clearance, enhanced tumor-imaging and PDT efficacy at 2–4 h post-injection. Molecular modeling studies also confirmed that the presence of HPPH moiety in HPPH-cRGD conjugate does not interfere with specific recognition of cRGD by αvβ3 integrin. Compared to U87 and 4T1 cells the HPPH-cRGD showed significantly low photosensitizing efficacy in A431 (αvβ3 negative) tumor cells, suggesting possible target-specificity of the conjugate.
Keywords: Photodynamic therapy, photosensitizer, HPPH, cRGD : Cyclic Arg-Gly-Asp
Introduction
Since the world-wide approval of Photofrin®, photodynamic therapy (PDT) has been accepted as an alternative clinical cancer treatment modality1–5. The utility of this approach is also being investigated in combination with surgery or chemotherapy. Similar to chemotherapy, PDT requires agents (photosensitizers) which exhibit selectivity for tumors and in common with radiotherapy, the mode of action involves the use of electromagnetic radiation in order to generate reactive oxygen species (ROS). However, PDT is much milder approach to cancer treatment than these two modalities and it exploits the biological consequences of localized oxidative damage inflicted by photodynamic processes.6 Three critical elements are required for the initial photodynamic processes to occur: a drug that can be activated by light (a photosensitizer), light and oxygen. Upon exposing the tumors with an appropriate wavelength of light, the photosensitizer produces an excited triplet-state that can interact with molecular oxygen to produce singlet oxygen responsible for inducing cell damage through direct and indirect cytotoxicity.7 In addition to photosensitizer, singlet oxygen formation and light dosimetry play important roles in PDT. The structure-activity relationship (SAR) and quantitative structure activity relationship (QSAR) studies in a series of alkyl ether analogs of pyropheophorbides have shown that overall lipophilicity and the position of various hydrophilic and hydrophobic groups in the molecules makes a remarkable difference in cell uptake, intracellular localization and long-term tumor cure.8,9 This approach has been quite successful in developing effective photosensitizers and a few of them are currently at various stages of clinical or preclinical trials.10–12
Conjugates between the photosensitizers and small molecules have also been designed to improve cell type target-specific agents and illustrate a new approach to optimize PDT.13 To overcome the difficulties in using large proteins and antibodies as targeting vehicles,14 there have been efforts to use smaller peptides as targeting vehicles.15 These peptides recognize fairly specific receptors that are over-expressed on certain tumor cells. One of the receptors, which has been of immense interest for targeting certain tumor imaging and/or therapeutic agents has been αvβ3 integrin known for its over-expression in both tumor cells and activated endothelial cells of the neovasculature during tumor regrowth, invasion, and metastasis.16,17 In recent years, a large number of cRGD (cyclic Arg-Gly-Asp) peptide have been labeled with a variety of radionuclides and the resulting products have shown significant target specificity for brain and breast cancers, known for over-expression of αvβ3 integrin.18 Conjugation of monovalent or multivalent cRGD peptides with certain cyanine dye-based fluorophores has also shown a significant enhanced tumor-specificity in 4T1 (breast) and U87 (brain) tumors. 19
For quite some time our laboratory has been exploring the utility of a variety of chlorophyll-a and bacteriochlorophyll-a based photosensitizers for the use in PDT.20,21 Among these compounds 2-(1′-hexyloxyetyhyl)-2-devinylpyropheophorbide a (HPPH) derived from chlorophyll-a and certain longer wavelength agents; purpurinimide (700 nm) and bacteriopurpurinimide (800 nm) showed excellent photosensitizing efficacy with limited skin phototoxicity.22–24
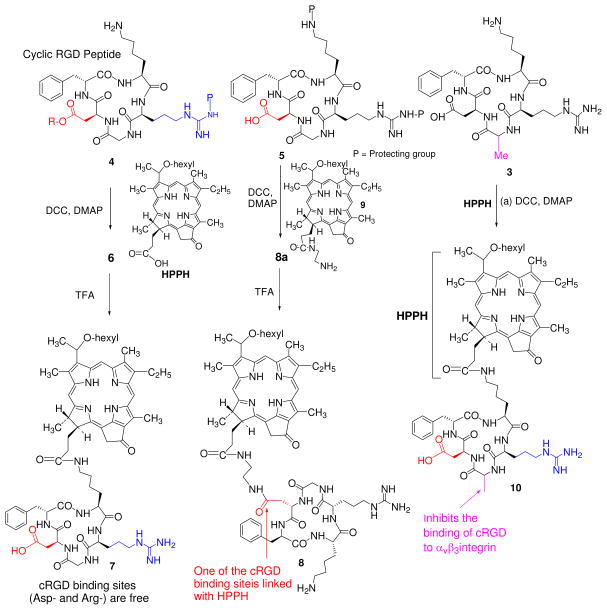
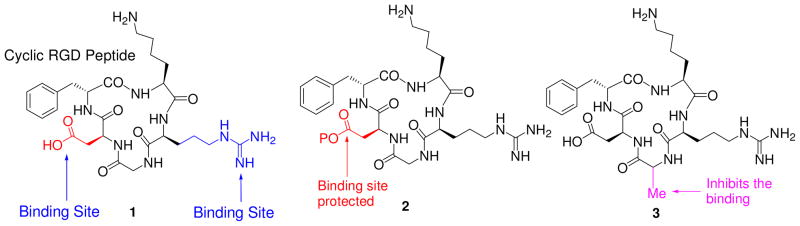
For a proof-of principle study we conjugated HPPH with cRGD peptide and to confirm the tumor-specificity of the conjugates, we selected three cRGD analogs for our synthetic strategy. In cRGD peptide 1, both the binding residues (Arg- and Asp-) were protected with an acid labile protecting group while the amino group in Lys- was left unprotected for conjugating to HPPH. In peptide 2 the amino- groups of Lys- and Arg- were protected while the carboxylic acid functionality of Asp- was left available for conjugating to HPPH derivative 9 and finally in peptide 3, the Gly- residue of peptide 1 was replaced with Alanine (Ala) because such a substitution in cRGD is known to prevent its binding to αvβ3 integrin25.
Results and Discussion
Chemistry
For the preparation of HPPH-cRGD conjugates 7 and 10. HPPH was reacted with cRGD peptides 3 and 4 following standard peptide chemistry and the resulting intermediates on treating with trifluoroacetic acid afforded the desired conjugates in 57% yield. The conjugate 8 in which HPPH was linked at the aspartic acid site was obtained by first converting the HPPH into derivative 9 containing an amino-functionality by previously reported methodology, which on subsequent reaction with cRGD analog 5 yielded the desired conjugate 8 in excellent yield. Our attempts to dissolve the compound 8 with common polar solvents were unsuccessful, however, it was determined that conjugate 8 was partly soluble in DMSO.
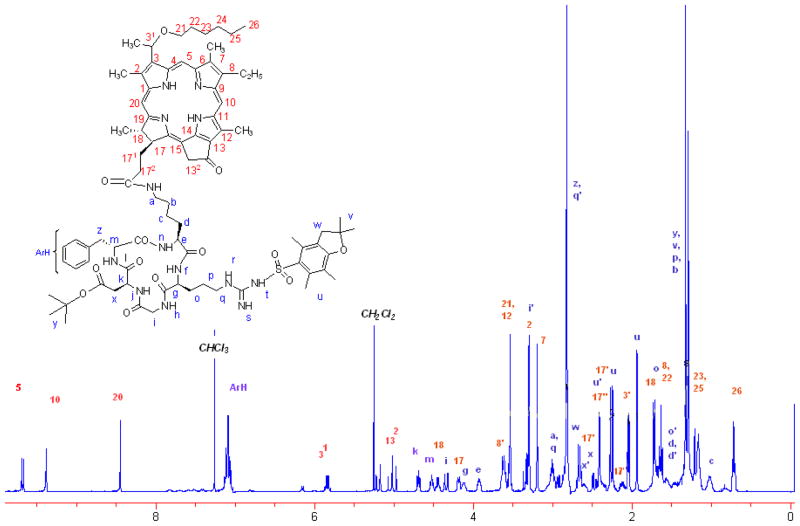
The reaction sequences for the synthesis of conjugates 7, 8 and 10 are depicted in Scheme 1 and their structures were confirmed by NMR and mass spectroscopic analysis. The 1H-NMR spectrum of the conjugate 6 showed an intricate pattern due to presence of RGD cyclic peptide. In particular, the splitting singlet (due to epimeric 31-H) at δ 9.73 indicated the presence of the 5H meso proton and NOE correlations between C-17 side chain of porphyrin moiety and side chain of the lysine unit of RGDfk peptide established the formation of the conjugate 6. From 2D NMR analysis, the structure of product 6 and the δ values of chemically equivalent protons are assigned and are listed in the table 1. The purity of the final products was ascertained by HPLC (supplemental data, Figure S6). Conjugation of the peptides to photosensitizer (HPPH) did not make any significant difference in their photophysical properties. Having the synthetic conjugates 7, 8 and 10 in hands, our next step was to compare the in vitro/in vivo photosensitizing efficacy, tumor uptake and intracellular localization characteristics between HPPH and the corresponding cRGD conjugates. The rationale of this study was to investigate the importance of the Arg- and Asp- amino acid residues in cRGD peptide in directing the target-specificity of the corresponding HPPH conjugates in αvβ3 over-expressed tumor models.
Scheme 1.
Synthesis of HPPH-cRGD analogs 7, 8 & 10
Table 1.
List of 1H-NMR values and their positions of conjugate 6
| Code | 1H-NMR Value (δ) | |
|---|---|---|
| 2 | 3.35 (s, 3H) | |
| 3 | 2.10 (dd, J=6.8, 1.2, 3H) | |
| 3′ | 5.90 (p, J=6.8,1H) | |
| 5 | 9.73 (splitted s, 1H) | |
| 7 | 3.25 (s, 3H) | |
| 8′ | 3.67 (m, 2H) | |
| 8” | 1.70 (t, J=7.6, 3H) | |
| 10 | 9.50 (s, 1H) | |
| 12 | 3.59 (s, 3H) | |
| 13” |
|
5.25 (d, J=19.6, 1H) 5.05 (d, J=19.6, 1H) |
| 17 | 4.24 (d, J=8.4, 1H) | |
| 17′ |
|
2.65 (m, 1H) 2.30 (m, 1H) |
| 17” |
|
2.45 (m, 1H) 2.18 (m, 1H) |
| 18 |
|
4.50 (dt, J=7.6, 6.0, 1H) 1.78 (d, J=7.2, 3H) |
| 20 | 8.50 (s, 1H) | |
| 21 | 3.62 (m, 2H) | |
| 22 | 1.74 (m, 2H) | |
| 23 | 1.40 (m, 2H) | |
| 24 | 1.27 (m, 2H) | |
| 25 | 1.24 (m, 2H) | |
| 26 | 0.78 (m, 3H) | |
| ArH | 7.15 (m, 5H) | |
| a | 3.05 (m, 2H) | |
| b | 1.40 (m, 2H) | |
| c | 1.10 (m, 2H) | |
| d |
|
1.65 (m, 1H) 1.52 (m, 1H) |
| e | 3.98 (m, 1H) | |
| g | 4.16 (m, 1H) | |
| i |
|
4.39 (dd, J=14.8, 1.6, 1H) 3.39 (m, 1H) |
| k | 4.74 (t, J=7.2, 1H) | |
| m | 4.56 (m, 1H) | |
| o |
|
1.76 (m, 1H) 1.63 (m, 1H) |
| p | 1.45 (m, 2H) | |
| q |
|
3.06 (m, 1H) 2.90 (m, 1H) |
| u |
|
2.00 (s, 3H) 2.31 (d, J=11.6, 3H) 2.47 (ss, J=2.8, 3H) |
| v | 1.38 (s, 6H) | |
| w | 2.74 (m, 2H) | |
| x |
|
2.70 (m, 1H) 2.52 (m, 1H) |
| y | 1.35 (s, 9H) | |
| z |
|
2.98 (m, 1H) 2.94 (m, 1H) |
Compared to HPPH, the cRGD-HPPH conjugate 7 showed significantly higher in vitro PDT efficacy and cell uptake
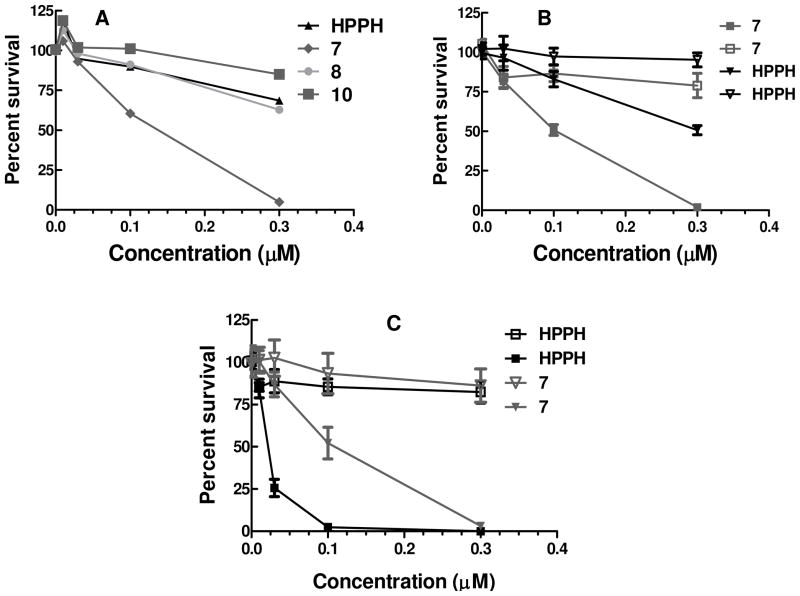
The initial in vitro photosensitizing efficacy of HPPH and the corresponding cRGD conjugates 7, 8 and 10 were performed in the U87 cell lines (αvβ3 positive). The cells were incubated with increasing concentrations of photosensitizers for 2 and 4h, and then exposed to 665 nm light and MTT assay was performed 48h later. None of the photosensitizers show any significant dark toxicity up to 1μM (Supplementary data, Figure S9) concentration. As can be seen from the results summarized in Fig. 3, under these experimental conditions compared to HPPH,, 7 and 8), the conjugate 7 containing the cRGD conjugate (cyclic Asp-, Gly-, Arg-) with both the -COOH and –NH2 functionalities in Asp- and Arg- available for binding to αvβ3 integrin was significantly more effective. No appreciable differences were observed between HPPH and other HPPH-cRGD conjugates in which either one of the binding residues (Asp-) was blocked or the Gly- was replaced with Ala, which is known to inhibit the integrin binding ability of cRGD. Further, the 2h or 4h (Data not shown) post-incubation of the photosensitizers did not show any significant difference in PDT efficacy. We further evaluated the photosensitizing efficacy of these photosensitizers in another αvβ3 positive cell line (4T1) at 2h incubation and as can be seen from Fig 3, the cRGD-HPPH conjugate proved to be the most effective. To identify the impact of the peptide moiety to target specificity to HPPH, we evaluated the efficacy of the photosensitizers in A431 cell line, reported as αvβ3 negative19. In vitro phototoxicity assays revealed that compared to HPPH the corresponding peptide conjugate 7 was less effective under similar experimental parameters. The lower activity of the other cRGD-HPPH conjugates in particular the conjugate 10 in which the Gly- amino acid residue is being replaced by Ala, is not attributable only due to overall lipophilicity of the PS alone and suggests a possible target-specificity of cRGD to integrin positive tumor cells.
Figure 3.
Comparative in vitro photosensitizing efficacy of the HPPH, and the corresponding peptide conjugates 7, 8 and 10 at variable photosensitizer concentrations in A: U87 (αvβ3 Concentration positive), B: 4T1 αvβ3 positive and C: A431 (αvβ3 negative) tumor cells respectively. The cells were incubated with photosensitizers for 2h before exposing to light. (665 nm, 0 and 2.0 J/cm2).The photosensitizer(s) alone, without exposing the cells to light did not show any cell kill. As an example, the dark toxicity results of the photosensitizer 7 are shown as hollow symbols.
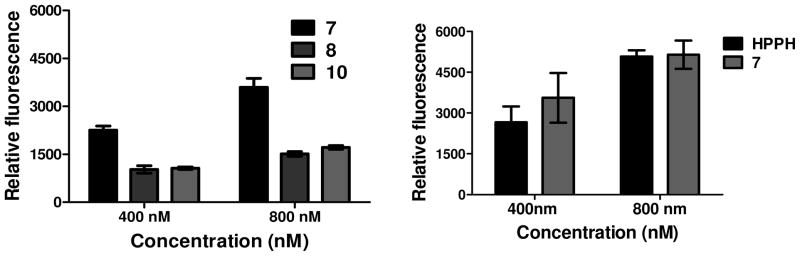
To assess whether the cRGD dependent changes in phototoxicity were due to altered photosensitizer levels in cells at the time of treatment, the cell uptake of the cRGD conjugates 7, 8 and 10 was determined in the U87 cell line at two concentrations (400 nM, 800 nM) at 2h and 4h post-incubation. From the data summarized in Figure 4, the highly effective HPPH-cRGD again showed the highest uptake, but it was significantly more at 4h (Supplementary data, Figure S10) than 2h incubation with similar PDT efficacy. The uptake of HPPH and its peptide conjugate 7 in 4T1 cells is comparable at 2h post incubation with the PS as determined by flow cytometry. We see similar uptake of HPPH and its peptide conjugate in U87 cells as well (Supplementary figure S10). We are currently investigating to understand the difference in activity of the HPPH as compared to its peptide conjugate 7 in both 4T1 as well as U87 cells in spite of both compounds showing similar uptake within these two αvβ3 overexpressing cell lines.
Figure 4.
In vitro uptake of (A) conjugates 7, 8 and 10 in two concentrations (400 nM and 800 nM) in U87 cells at 2h post-incubation, (B) Uptake of HPPH and conjugate 7 in two concentrations (400 and 800nm) in the same cell line and timepoint, measured by flow cytometry. HPPH and conjugate 7 produced similar up take in 4T1 cell lines (see Figure S10, Supplementary Information). However, there was a significant difference in both in vitro and in vivo PDT efficacy (see the text).
The presence of the cRGD moiety in HPPH alters its site of localization
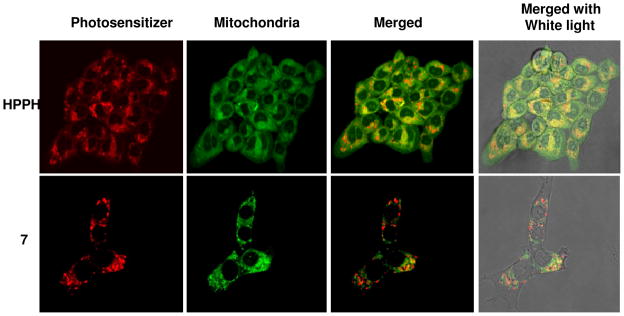
Previous studies with various porphyrin or reduced porphyrin (chlorins and bacteriochlorins) based compounds including the alkyl ether analogs of pyropheophorbide-a, showed that the most effective photosensitizers localize in mitochondria.26 It is also reported that the site-specificity alters by introducing certain small molecules such as steroids, vitamins and carbohydrate moieties to photosensitizers. In our previous study, fluorescence microscopy confirmed the predominantly mitochondrial location of HPPH and the altered site of localization to the lysosomes on introducing certain carbohydrate moieties (e.g. β-galactose) to HPPH27. In Figure 5 we show that the most effective conjugate 7 with cRGD introduced to HPPH altered the localization pattern to the cell membrane (Supplementary figure S11) as well as to punctuate cytoplasmic vesicles, a pattern observed with other RGD conjugates28. The altered cellular localization may trigger mechanisms that differ from those of predominantly mitochondrial localizing HPPH. The effects of the mechanisms may show cell type variability, which needs further study and is currently under investigation. Further localization studies of the conjugate 7 with ER, Golgi and lysosome organelle-specific probes are currently in progress.
Figure 5.
Comparative intracellular localization (false color images) of HPPH and the corresponding cRGD conjugate 7 with MitoTracker Green (mitochondrial probe) ipn 4T1 cells after incubating for 2h clearly indicates that introduction of cRGD moiety to HPPH changes its site of localization.
Compared to HPPH, the cRGD-HPPH conjugates 7 showed faster clearance in vivo and enhanced photodynamic efficacy
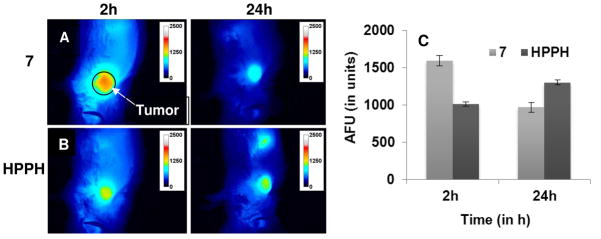
The encouraging in vitro results of HPPH-cRGD conjugate 7 prompted us to compare its in vivo uptake and photosensitizing efficacy. To look at its uptake in vivo we utilized fluorescence optical imaging as our compounds exhibit fluorescence upon excitation. We performed the fluorescence optical imaging experiment on BALB/c mice with s.c. 4T1 tumors (3 mice/group). Each set of mice was injected with HPPH or its peptide conjugate 7 (HPPH-cRGD) and animals were imaged at various time points from 2– 72 h. The tumor uptake of the peptide conjugate 7 (HPPH-cRGD) showed maximal uptake within the first 2h post injection as indicated by the fluorescence intensity and was visible in the tumor until 72h whereas HPPH showed maximum fluorescence intensity at 24h while visible until 72h. This indicates that conjugation of targeting peptides alters the maximal tumor uptake of HPPH.
Comparative PDT efficacy of HPPH and HPPH-cRGD Conjugates
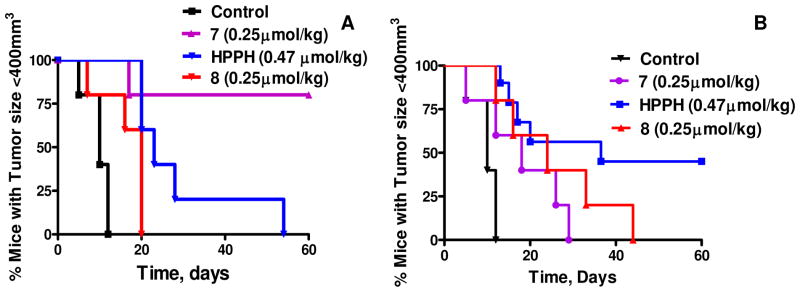
To determine the efficacy of the conjugates compared to HPPH in vivo we selected BALB/c mice bearing syngeneic 4T1 tumors. Groups of mice (10 mice/treatment group) bearing 4T1 tumors (average tumor size ~62.5 mm3) were injected with HPPH and the corresponding cRGD conjugate 7 (0.47 μmol/kg) in mice bearing αvβ3 integrin positive 4T1 tumors. Exposing the tumors to light (665 nm, 135J/cm2, 75mW/cm2) at 24h post injection gave 40% tumor cure at day 60 with HPPH. Reducing the dose to 0.25 μmol/kg did not produce any cures with either HPPH (data not shown) or HPPH-cRGD (7) when treated at 24h post injection, However, at the same dose of HPPH-cRGD (7) (0.25 μmol/kg) the mice treated at 4h post injection produced 80% tumor cure at day 60, whereas both HPPH (0.47 μmol/kg) and the HPPH-cRGD (Asp-blocked) (8) (0.25 μmol/kg) conjugate treated under similar treatment parameters gave minimal tumor response with no cures. In summary, at the 2h post-injection time point tumor response was in the order of HPPH-cRGD 7 > HPPH > HPPG-cRGD (Asp-blocked) 8. These data clearly indicates that cRGD remarkably alters the clearance time of HPPH from the tumor site.
Although the concept of using cRGD as a targeting moiety for the photosensitizer has been reported in the literature29, the results presented in this report happens to be first example to show the importance of the cRGD moiety in developing target-specific PDT agents. Further studies involving the mechanisms leading to increased cell kill as well as higher PDT efficacy with HPPH conjugated with single and multiple cRGD moieties are currently in progress.
Molecular modeling Studies
Molecular modeling was used to examine the molecular mechanisms responsible for the observed differences of in vitro/in vivo efficacies between the HPPH-cRGD conjugates, 7 and 8. Although molecular dynamics (MD) simulations and flexible docking are desirable for a study of ligand specific binding to target proteins, this approach would require huge computational resources for this system. In addition, it is well established that the ligand binding induces tertiary and quaternary structural change of the integrin αvβ3 system30 Thus we used an alternative approach, an anchored conformational analysis, where the effects of selective torsional angle variations in the linker and other parts of the ligand on the stability of the complex were examined systematically while the target protein and the target specific ligand moiety of the conjugates were fixed as found in the crystal structure. A similar approach was successfully applied to elucidate the difference in experimentally observed differences in in vitro activity of galectin targeted photosensitizers.
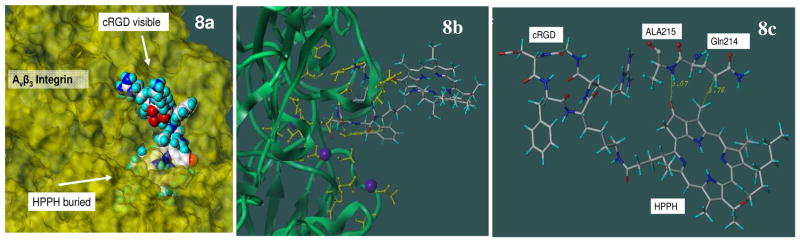
In the crystal structure of αvβ3 integrin and cRGD peptide, it was clearly shown that Asp (D) residue in the cRGD peptide plays an essential role in the specific recognition of αvβ3 integrin through interactions with various residues from the β3 subunit and Mn cations embedded in the β3 subunit. One of the Mn cations is directly coordinated with the Asp side chain group (COO-). This cationic Mn is also coordinated to Ser 121, Ser 123, and Glu 220. These residues in turn are coordinated to two other Mn cations that in turn form additional coordination bonds with other residues from the β3 subunit. In addition, the Asp side chain of cRGD peptide also makes a direct interaction with Asn 215. Thus this Asp residue is the key residue in this intricate network of interactions between cRDG, three Mn cations and integrin residues that are responsible for specific recognition of cRGD ligand by αvβ3 integrin. Creation of a linker through this Asp side chain in conjugate 8 makes it impossible to maintain this specific recognition scheme between the RGD peptide to αvβ3 integrin. This is demonstrated by the complex built by the superposition as described in the method. Figure 8a shows that the linker and HPPH atoms are now penetrated into integrin atoms, a clear indication that it is impossible to attach HPPH through the Asp side chain while maintaining the specific recognition of cRGD peptide found in the crystal. It is possible that the conjugate 8 still binds to αvβ3 integrin in a completely different manner, but this was not examined any further.
Figure 8.
Figure 8a: Structure of conjugate 7 in cRGD binding site of integrin using the crystal structure of cRGD-integrin complex. Conjugate 7 is shown in CPK representation with standard atom based colors. Integrin residues are shown in yellow surface representation. It is shown that the linker and HPPH atoms are clashed into integrin atoms as they are buried under the protein surface. Lys residue of cRGD is pointing away from the integrin. Figure 8b: Overview of the conjugate 7 interactions with integrin. Integrin backbones are represented by green ribbons. The conjugate 7 is represented by stick figure with standard atom based colors. The integrin residues involved in the interaction with cRGD residue and cationic Mn (blue balls) are shown in yellow ball and stick figure. Figure 8c: Specific additional interactions of energy optimized HPPH conjugate 7 with integrin residues.Conjugate 7 is shown in stick representation with standard atom based color coding. Integrin residues involved in the interactions are shown in ball-and-stick representation. The hydrogen bond between HPPH and Ala215 residues of integrin αv subunit is shown in green dotted lines. The distances involved in hydrogen bonding and the hydrophobic contact between HPPH and Gln214 are shown in angstrom (yellow line). For details see the “Materials and Methods”.
The same figure also shows that the Lys residue side chain of cRGD, which was used to create a linker to HPPH in the conjugate 7, points towards the solvents, away from the integrin. Molecular modeling of the conjugate 7 with the anchored conformational search described in the methods was performed to examine whether HPPH can be attached to the cRGD without interfering with the specific recognition of cRGD with integrin. In addition, it is also examined that whether the HPPH moiety will provide an additional stabilization for the cRGD-integrin complex. In brief, the selected torsional angles around the linker region were systematically altered to examine various possible conformations that were subjected to clustering of similar conformers. Once the stable conformers were identified from the previous step, the effects of torsional angles in the hexyloxyethyl moiety of HPPH on the stability of the complex were examined systematically. The two staged search used here was effective in finding the stable conformation of the ligand on the known cRGD-integrin complex by reducing the number of conformations that needed to be examined. Our anchored conformational analysis in the first stage indicated that in fact there are many linker conformations possible, which allows the conjugate 7 to interact more strongly with integrin than what are found in the crystal cRGD peptide or the initial extended conformer of conjugate 7, without interfering with the specific cRGD recognition by the integrin (the results not shown). One of such stable complexes is shown in figures 8b, 8c where 8.2 and 4.7 kcal/mol stabilizations over the crystal and initial conformer respectively were obtained through a hydrogen bonding between the keto group of the HPPH ring and an amino group of Ala 215 from the integrin αv subunit. In addition, there seems to be hydrophobic contracts between the neighboring methyl group of HPPH and the Gln214 side chain of integrin αv subunit. The second stage of the anchored conformational search on the hexyloxyethyl moiety of HPPH showed that no additional interaction with integrin was possible and thus no significant preference for various conformers of the hexyloxyethyl moiety of HPPH. Thus for the figures 8b and 8c, one of the conformers with low energy was selected for presentation. Another view of the same complex shown in figure 8c clearly demonstrates that the HPPH moiety can provide additional stabilization while maintaining specific RGD recognition by αvβ3 integrin.
Conclusions
The cyclic RGD peptide is one of the most extensively studied and used peptides for developing target-specific therapeutic and imaging agents. This approach has also been explored in developing improved PDT agents, but most of the examples are limited to synthesis and/or in vitro studies. The study presented here is the first example which illustrates the in vitro and in vivo characteristics of a series of HPPH-peptide conjugates and shows a remarkable impact of the cRGD moiety on the photosensitizing properties, tumor uptake, tumor-clearance and intracellular localization. Molecular modeling indicated that the presence of the HPPH moiety in the conjugate 8 would destroy specific recognition of RGD by αvβ3 integrin (because it was conjugated via the Asp residue, essential for binding to the αvβ3 integrin On the other hand, in conjugate 7, the HPPH moiety does not interfere with the specific recognition of cRGD byαvβ3 integrin and also provides an additional stabilization to the complex through hydrogen bonding and hydrophobic contacts to the protein making it a more effective candidate for specifically targeted PDT.
Materials and Methods
Chemistry
All chemicals were of reagent grade and used as such. Cyclic (RGDfK) peptides were purchased from Peptides International, Louisville, KY and were used as received. Solvents were dried using standard methods unless stated otherwise. Reactions were carried out under nitrogen atmosphere and were monitored by precoated (0.20 cm) silica TLC plastic sheet (20 cm × 20 cm) strips (POLYGRAM SIL N-HR) and or/UV-visible spectroscopy. UV-visible spectra were recorded on a Varian (Cary-50 Bio) spectrophotometer. 1H-NMR spectra were recorded on Brucker AMX 400 or Varian 400 spectrometers at 303 K in CDCl3 or ~10% of CD3OD or DMSO–d6 in CDCl3. All 2D 1H-NMR (COSY, TOCSY and NOESY) were run on Brucker AMX 400 MHz NMR spectrometer. Proton chemical shifts (δ) are reported in parts per million (ppm) relative to CDCl3 (7.26 ppm) or TMS (0.00 ppm). Coupling constants (J) are reported in Hertz (Hz) and s, d, t, q, p, m and br refer to singlet, doublet, triplet, quartet, pentet, multiplet and broad respectively. Mass spectral data (Electro Spray Ionization, ESI by fusion) were obtained from Biopolymer Facility, Roswell Park Cancer Institute, HRMS data were obtained from the Mass Spectrometry Facility, Michigan State University, East Lansing, MI.
HPLC Method
HPLC Analysis of conjugates was carried out using a Waters Delta 600 System consisting of the 600 Controller, 600 Fluid Handling Unit and 996 Photodiode Array Detector equipped with a Waters SunFire C18 column, 5 micron particle size, with dimensions 4.6 × 250mm. The mobile phase was isocratic: 100% Methanol at a flow of 1.0 ml/min. The component percentages are based on absorbance data from the 408 nm channel. (see supplemental data)
Synthesis of 3-devinyl-3-{1′-(hexyloxy)ethyl}pyropheophorbide-a-protected cyclo (RGDfK) conjugate (6)
To a solution of anhydrous DMF (2.0 mL), 3-devinyl-3-{1′-(hexyloxy) ethyl}pyropheo-phorbide-a (20 mg) (HPPH), protected cyclo(-RGDfK) (25 mg) (4), HOBt (10 mg), DMAP (5 mg), and EDCI (15 mg) were added and stirred under N2 at RT for 4 h. DMF was removed under high vacuum pump; residue treated with water and the solid crude was filtered. The purple color crude product was purified over silica column using 8%MeOH in CH2Cl2 as eluant to yield 30 mg (60%) of pure product (6); UV-vis(MeOH): 660(4.10×104), 603(7.68×103), 537(8.23×103), 506(7.87×103), 409(8.49×104). HRMS for C83H112N13O13S (MH+) (Calculated): 1530.8223; (Found): 1530.8221. 1H-NMR (10% CD3OD in CDCl3; 400 MHz): δ9.73 (splitted s, 1H, meso-H5); 9.50 (ss, J=1.6 1H, meso-H10); 8.50(s, 1H, meso-H20); 7.15(m, 5H, ArH, F); 5.90(p, J=6.8,1H, 31-H); 5.25(d, J=19.6, 1H, 132-CH2); 5.05(d, J=19.6, 1H, 132-CH2); 4.74(t, J=7.2, 1H, D-αCH); 4.56(m, 1H, F-αCH); 4.50(dt, J=7.6, 6.0, 1H, H-18); 4.39(dd, J=14.8, 1.6, 1H, G-αCH2); 4.24(d, J=8.4, 1H, H-17); 4.16(m, 1H, R-αCH); 3.98(m, 1H, K-αCH); 3.67(m, 2H, 8-CH2CH3); 3.62(m, 2H, OCH2CH2CH2CH2CH2CH3); 3.59(s, 3H, 12-CH3); 3.39(m, 1H, G-αCH2); 3.35(s, 3H, 2-CH3); 3.25(s, 3H, 7-CH3); 3.06(m, 1H, R-δCH2); 3.05(m, 2H, K-εCH2); 2.98(m, 1H, F-βCH2); 2.94(m, 1H, F-βCH2); 2.90(m, 1H, R-δCH2); 2.74(m, 2H, PbfCH2); 2.70 (m, 1H, D-βCH2); 2.65(m, 1H, 171-H); 2.52(m, 1H, D-βCH2); 2.47(ss, J=2.8, 3H, PbfArCH3); 2.45 (m, 1H, 172-H); 2.31(d, J=11.6, 3H, PbfArCH3); 2.30(m, 1H, 171-H); 2.18(m, 1H, 172-H); 2.10(dd, J=6.8, 1.2, 3H, 31-CH3); 2.00(s, 3H, PbfArCH3); 1.78(d, J=7.2, 3H, 18-CH3);1.76(m, 1H, R-βCH2); 1.74(m, 2H, OCH2CH2CH2CH2CH2CH3); 1.70(t, J=7.6, 3H, 8-CH2CH3); 1.65(m, 1H, K-βCH2); 1.63(m, 1H, R-βCH2); 1.52(m, 1H, K-βCH2); 1.45(m, 2H, R-γCH2); 1.40(m, 2H, K-δCH2); 1.40(m, 2H, OCH2CH2CH2CH2CH2CH3);1.38(s, 6H, PbfC(CH3)2); 1.35(s, 9H, tert-C(CH3)3); 1.27(m, 2H, OCH2CH2CH2CH2CH2CH3); 1.24(m, 2H, OCH2CH2CH2CH2CH2CH3); 1.10(m, 2H, K-γCH2); 0.78(m, 3H, OCH2CH2 CH2CH2CH2CH3). HPLC: 98.2 % of pure conjugate was obtained by following the method described above.
Synthesis of 3-devinyl-3-{1′-(hexyloxy)ethyl}pyropheophorbide-a–cyclo(RGDfK) conjugate (7)
TFA (1.5mL) was added to 3-devinyl-3-{1′-(hexyloxy)ethyl} pyropheophorbide-a- protected cyclo(RGDfK) conjugate (6) (15 mg) and reaction mixture was stirred for 4 hrs at room temperature. TFA was then removed under reduced pressure and compound was precipitated with cold ether, filtered to yield 12 mg (95%) product (7). UV-vis(MeOH): 660(4.00×104), 604(8.00×103), 536(8.66×103), 505(8.50×103), 408(8.03 × 104). HRMS for C66H88N13O10 (MH+) (Calculated): 1222.6777; (Found): 1222.6787 1H-NMR (10% CD3OD in CDCl3; 400 MHz): δ10.09 (splitted s, 1H, meso-H5); 9.77(ss, J=1.6 1H, meso-H10); 8.80(s, 1H, meso-H20); 7.00(m, 5H, ArH, F); 5.81(p, J=4.8,1H, 31-H); 5.28(d, J=19.8, 1H, 132-CH2); 5.04(d, J=19.8, 1H, 132-CH2); 4.63(m, 2H, D-αCH, F-αCH); 4.30(m, 4H, H-18;G-αCH2, H-17, R-αCH); 3.78(m, 3H, K-αCH, 8-CH2CH3); 3.60(m, 5H, OCH2CH2CH2CH2CH2CH3, 12-CH3); 3.48(m, 1H, G-αCH2); 3.31(s, 3H, 2-CH3); 3.24(s, 3H, 7-CH3); 3.03(m, 1H, R-δCH2); 2.93(m, 2H, K-εCH2); 2.68(m, 5H, F-βCH2, F-βCH2, R-δCH2, D-βCH2, 172-H); 2.50(m, 1H, D-βCH2); 2.35 (m, 2H, 171-H, 172-H); 2.26(m, 1H, 171-H); 2.00(dd, J=16.6, 4.4, 3H, 31-CH3); 1.72(d, J=7.2, 3H, 18-CH3); 1.66(m, 3H, R-βCH2, OCH2CH2CH2CH2CH2CH3); 1.57(t, J=6.8, 3H, 8-CH2CH3); 1.50(m, 3H, K-βCH2, R-βCH2); 1.44(m, 4H, R-γCH2, K-δCH2); 1.27(m, 4H, OCH2CH2CH2CH2CH2CH3); 1.14 (m, 2H, OCH2CH2CH2CH2CH2CH3); 0.79(m, 2H, K-γCH2); 0.63 (m, 3H, OCH2CH2 CH2CH2CH2CH3).
Synthesis of HPPH–cyclo(RGDfK) conjugate (8)
To a solution of anhydrous DMF (2.0 mL), HPPH derivative 931 (20 mg), protected cyclo(-RGDfK) (25 mg) (5), HOBt (10 mg), DMAP (5 mg), and EDCI (15 mg) were added and stirred under N2 at RT for 4 h. DMF was removed under high vacuum pump; residue treated with water and the crude solid was filtered. The purple color crude product was purified over silica column using 8%MeOH in CH2Cl2 as eluant to yield 25 mg (50%) of pure product. To this crude product, TFA (1.5mL) was added and reaction mixture was stirred for 4 hrs at room temperature. TFA was then removed under reduced pressure and compound was precipitated with cold ether, filtered to yield 11 mg (91%) product (8). UV-vis(MeOH): 661 (4.00×104); HRMS for C68H94N15O9 (MH+) (Calculated): 1264.7359; (Found): 1264.7351. 1H-NMR (DMSO–d6; 400 MHz): δ9.91 (splitted s, 1H, meso-H5); 9.84(s, 1H, meso-H10); 8.86(s, 1H, meso-H20); 7.20(m, 5H, ArH, F); 6.04(m,1H, 31-H); 5.32(d, J=19.8, 1H, 132-CH2); 5.12(d, J=19.8, 1H, 132-CH2); 4.72(m, 1H, D-αCH); 4.61(m, 2H, F-αCH, H-18); 4.38(dd, J=14.8, 1.6, 1H, G-αCH2); 4.20(m, 3H, H-17, R-αCH, K-αCH); 3.20–4.00(m, 13H, 8-CH2CH3, OCH2CH2CH2CH2CH2CH3, 12-CH3, G-αCH2, 2-CH3, -(NHCH2)2-, 3.04(s, 3H, 7-CH3); 2.90–3.08(m, 6H, R-δCH2, K-εCH2, F-βCH2, F-βCH2, R-δCH2); 2.63(m, 2H, D-βCH2, 171-H); 2.54(m, 1H, D-βCH2); 2.43 (m, 1H, 172-H); 2.27(m, 2H, 171-H, 172-H); 2.11(dd, J=6.0, 1.4, 3H, 31-CH3); 1.92(m, 2H, -(NHCH2)2-); 1.85(d, J=7.2, 3H, 18-CH3); 1.69(t, J=7.2, 3H, 8-CH2CH3); 1.26–1.84 (m,10H, K-βCH2, R-βCH2, R-γCH2, R-βCH2, OCH2CH2CH2CH2CH2CH3 K-δCH2); 1.40(m, 2H, OCH2CH2CH2CH2CH2CH3); 1.27(m, 4H, OCH2CH2CH2CH2CH2CH3); 0.89(m, 2H, K-γCH2); 0.73(m, 3H, OCH2CH2 CH2CH2CH2CH3). HPLC: 99.4 % of pure conjugate was obtained by following the method described above.
Synthesis of 3-devinyl-3-{1′-(hexyloxy)ethyl}pyropheophorbide-a–cyclo(RADfK) conjugate (10)
To a solution of anhydrous DMF (2.0 mL), 3-devinyl-3-{1′-(hexyloxy) ethyl}pyropheo-phorbide-a (20mg) (HPPH), cyclo(-RADfK) (26mg) (3), HOBt (10 mg), DMAP (5 mg), and EDCI (15 mg) were added and stirred under N2 at RT for 4 h. DMF was removed under high vacuum pump; residue treated with water and the solid crude was filtered. The purple color crude product was dried under reduced pressure and the residue obtained was washed with cold ether, dried to yield 11 mg (90%) product (10). UV-vis(MeOH): 661 (4.00×104), 604(8.00×103), 536(8.66×103), 505(8.50×103), 408(8.03×104). HRMS for C67H90N13O10 (MH+) (Calculated):1236.6933; (Found): 1236.6921. 1H-NMR (10% CD3OD in CDCl3; 400 MHz): δ9.42 (splitted s, 1H, meso-H5); 9.16 (s, 1H, meso-H10); 8.24 (s, 1H, meso-H20); 6.81 (m, 5H, ArH, F); 5.60 (p, J=6.8, 1H, 31-H); 4.95(d, J=19.6, 1H, 132-CH2); 4.78(d, J=19.6, 1H, 132-CH2); 4.25(m, 1H, D-αCH); 4.21(m, 3H, F-αCH, H-18, A-αCH2); 3.95(d, J=9.2, 1H, H-17); 3.73(m, 2H, R-αCH, K-αCH); 3.37(m, 7H, 8-CH2CH3, OCH2CH2CH2CH2CH2CH3, 12-CH3); 3.05(m, 3H, 7-CH3); 2.97(s, 3H, 2-CH3); 2.93(m, 1H, R-δCH2); 2.75(m, 3H, K-εCH2, F-βCH2); 2.50(m, 2H, F-βCH2, R-δCH2); 2.34 (m, 5H, A-αCH3, D-βCH2, 172-H); 2.18(m, 2H, D-βCH2, 171-H); 1.95(m, 1H, 172-H); 1.85(m, 1H, 171-H); 1.80(d, J=7.2, 3H, 31-CH3); 1.50(d, J=7.2, 3H, 18-CH3); 1.45(m, 3H, R-βCH2, OCH2CH2CH2CH2CH2CH3); 1.38(t, J=8.0, 3H, 8-CH2CH3); 0.75–1.10(m, 11H, K-βCH2, R-βCH2, R-γCH2, K-δCH2, OCH2CH2CH2CH2CH2CH3); 0.73(m, 2H, OCH2CH2CH2CH2CH2CH3); 0.57(m, 2H, K-γCH2); 0.43(t, J=6.8, 3H, OCH2CH2 CH2CH2CH2CH3). HPLC: 96.6 % of pure conjugate was obtained by following the method described above.
In vitro photosensitizing efficacy
The photosensitizing activity of the compound was determined as described before31. The tumor cell lines used are 4T1 (Mouse mammary tumor), U87 (Human Glioblastoma tumor) and A431 (Human epidermoid carcinoma) cell lines. The 4T1 tumor cells were grown in RPMI-1640 medium supplemented with 10% fetal bovine serum, L-glutamine, penicillin and streptomycin. U87 and A431 tumor cells were grown in DMEM (high glucose) with 10% fetal bovine serum, L-glutamine, penicillin, streptomycin, sodium pyruvate and non-essential amino acids. All types of tumor cells were maintained in an atmosphere of 5% CO2, 95% air and 100% humidity at 37ºC. For determining the PDT efficacy of the compounds, the cells were plated in 96 well plates at a cell density of 3000 cell/well in complete media. After 3h of incubation( to allow for attachment of cells to plate surface) at 37°C, the photosensitizers were added at variable concentrations and incubated at 37 °C for a further 2h without exposure to any light. Prior to light treatment, the cells were replaced with drug-free complete media. Cells were then illuminated with light from an argon-pumped dye laser set at 665 nm at a dose rate of 3.2 mW/cm2 for 0–2 J/cm2. After PDT the cells were incubated for a further 48h at 37°C in the dark. Following the 48h incubation, 10 μL of 5.0 mg/mL solution of 3-[4,5-dimethylthiazol-2-yl]-2–5-diphenyltetrazoliumbromide (MTT) in PBS (Sigma, St. Louis, MO) was added to each well. After 4 h incubation at 37 °C, the MTT and the media were removed, and 100 μL of DMSO was added to solubilize the formazan crystals. The 96- well plate was read on a microtiter plate reader (BioTek Instruments, Inc., ELx800 Absorbance Microplate Reader) at an absorbance of 570 nm. The results were plotted as a percent survival of the corresponding dark (drug, no light) control for each compound tested. Each data point represents the mean from three separate experiments, with 6 replicate wells and the error bars are the standard deviation.
In vivo photosensitizing efficacy
All studies were performed under approved protocols according to IACUC guidelines and described as before32. BALB/c mice were subcutaneously injected with 1 × 106 4T1 cells in 50 μL of PBS (on the right shoulder) and tumors were grown until they reached an average tumor volume of 62.5 mm3 (range 50–70 mm3). Before the laser irradiation, the mice were shaved, all the hair was removed from the inoculation site and the mice were injected intravenously with varying photosensitizer concentrations. At 2 and 24h post injection, mice were restrained without anesthesia (to avoid reduction in tumor reoxygenation) in plastic holders and then irradiated with laser light (665 nm) from an argon-pumped dye laser. The treatment parameters desired consisted of a light spot of 1 cm2 diameter and a total light dose of 135 J/cm2 delivered at a fluence rate of 75 mW/cm2. The mice were observed daily for signs of morbidity or tumor regrowth. If the tumor reappeared, the tumors were measured using two orthogonal measurements L and W (perpendicular to the L), and the volumes were calculated using the formula V= (L × W2)/2. Mice with a tumor volume which reached 400 mm3 (as defined by Institute’s IACUC standards) were euthanized according to IACUC guidelines. Mice were considered cures if the primary tumor did not show tumor volume ≥400mm3 at the end of 60 days of monitoring.
Intracellular localization of conjugates
To demonstrate the shift in sub cellular localization on conjugation of cRGD, HPPH or HPPH-cRGD 7 were co-incubated with the mitochondria specific fluorescent probe, Mitotracker Green (1μM for 1h). 4T1 cells were seeded at cell density of 0.5 × 105 cells on poly-L-Lysine coated glass bottom plates and cultured until attached. HPPH or HPPH-cRGDfK 7 were added to the cells at appropriate concentrations (typically 0.5 – 2.0 μM) were added to the cells, and the cells were incubated for 2h at 37°C, 5% CO2. The cells were rinsed briefly with phosphate buffered saline (PBS) and imaged on a spectral confocal microscope (TCS SP2, Leica Microsystems Semiconductor GmbH) with a HXC PL APO CS 63.0×1.40 oil immersion objective. The samples were excited by a pulsed diode laser at 405 nm (PDL800-D, PicoQuant GmbH). Filter combinations were as follows: for HPPH-containing compounds Ex 633nm and Em filter 640/50 nm; for MitoTracker Ex 543 nm, and Em BP 520/60. Images were analyzed by Image J (NIH) software. Cells were also previously imaged at 40× on an inverted fluorescence microscope (Zeiss Axiovert 200W, Germany) with a charged-coupled device camera (Dage Zeiss AxioCam MRm) using an AxioCam MRMRGrab Framegrabber and AxioVision LE 4.1 imaging software. Filter combinations were as follows: for HPPH-containing compounds Ex BP D410/40 nm, BeamSplitter FT 505dcxvu, and Em BP 675/50 nm; for MitoTracker Ex BP 565/30 nm BeamSplitter FT 585 nm and Em BP 520/60.
In vivo Fluorescence Optical Imaging
Fluorescence imaging of photosensitizer accumulation in the tumor was carried out on mice anesthetized with Ketamine/Xylazine (100/10) using a Nuance optical imaging camera system (Cambridge Research Inc., Woburn, MA). When the tumor reached 4–5 mm in diameter and prior to imaging, Nair was used to remove hair from the skin surrounding the 4T1 tumors. HPPH or its peptide conjugate HPPH-cRGD were injected i. v. at a dose of 0.3 μmol/kg. At 2h and 24h post injection, mice were imaged. Fluorescence excitation was achieved with laser light from an argon-pumped dye laser at 665 nm. Fluorescence emission images were acquired beyond 700 nm using a 695 long pass and 700 long pass filters in series. All data was subsequently analyzed using NIH’s Image J software.
Molecular Modeling
Construction of RGD-derivatives
The Semi-empirical MO, PM3, energy optimized structure of pyropheophorbide3 was used as a component of compounds 7 and 8. The exact crystal structure of the RGD segment found in the integrin – RGD complex (PDB:1L5G) was used to build the RGD – chlorin conjugates. N-methyl valine residue was replaced with lysine and appropriate segment was built to make the linker region between RGD and HPPH using SYBYL7.2 molecular modeling software (Tripos Inc., St. Louis, MO). To build compound 8, the Asp side chain of the RGD cyclic peptide was appropriately modified to create the linker region. All modification used standard bond length and bond angles of SYBYL7.2.
Construction of integrin -ligand complex
The crystal structure of αvβ3 extracellular segment complexed with RGD ligand (PDB:1L5G) was used as a template to model integrin αvβ3 with the RGD(Arg-Gly-Asp-Phe-Lys)- chlorin conjugate complexes. The modeled conjugates 7 and 8 were placed onto integrin αvβ3 using the RGD cyclic peptide (RGDfm-V) backbone atoms as the references for superposition.Since the RGD cyclic peptide in the conjugates 7 and 8 was not modified, this superposition operation resulted in a perfect fit.
Conformational Search of the RGD-chlorin conjugate
For the conjugate 7, we examined whether the pheophorbide moiety of HPPH provides additional stabilization for the RGD-conjugate integrin complex compared to the RGD cyclic peptide alone. The model complex structure just built from the above procedure is not considered to be the most stable ligand conformation in solution since the extended conformation of the linker was used to construct the conjugate. In addition, the hexyloxyethyl functionality may provide additional stabilization to the complex. Therefore, the systematic conformational search of RGD-HPPH conjugate at the RGD binding site of αvβ3 integrin was performed in two steps using Tripos SYBYL software version 7.2. The MMFF94 charges and Tripos Force Field were used for this systematic search with distance dependent dielectric function. First, nine torsional angles in the linker between the RGD ring and HPPH ring were systematically modified with the default set up except 60 degree interval and starting at current torsional angle. These 9 torsional angles in the linker region were selected first since these torsional angles will dictate the relative orientation of HPPH ring with respect to integrin residues.
From the systematic search result, the conformations were grouped into several subsets based on the relative orientation of HPPH ring with respect to integrin residues and the linker torsional angles. Several conformations from each set were subjected to further energy minimization. MMFF94 force field, MMFF94 atomic charges, distance dependent dielectric function and non bonding cutoff of 8 Å were used for the minimization with standard minimization parameters except a maximum iteration cycle of 300. During the minimization, all integrin atoms as well as key Mn2+ cations are fixed in space. The RGD cyclic peptide backbone atoms and the all the Arg, Gly, Asp side chain atoms, which are involved in specific recognition of αvβ3 integrin were also fixed during the optimization. The remaining atoms, Phe side chain atoms, the remnant of lysine side chain atoms, which forms a part of the linker region, all linker atoms, and HPPH atoms were optimized. Similar energy minimization was performed with the integrin-RGD complex crystal structure and the initial integrin-RGD-HPPH complex as the reference. The interaction energy between the integrin and the ligand was calculated as the difference between the complex energy and a sum of isolated protein and ligand energies. Once the most stable structure of HPPH-cRGD conjugate at αvβ3 integrin was obtained, then additional systematic conformational search and energy minimizations were performed for the all torsional angles within the hexyloxyethyl moiety of HPPH.
Supplementary Material
Figure 1.
Structures of the cRGD analogs selected for our proposed studies.
Figure 2.
1H-NMR assignment of HPPH-RGDfk-conjugate 6
Figure 6.
Whole body fluorescence images of representative BALB/c mice implanted with 4T1 tumors on the shoulder with HPPH or its peptide conjugate 7 at variable time points with a therapeutic dose (0.3 μmol kg−1, λex = 665 nm; λem = 710 nm). (A): 2h post injection (p.i.); (B): 24h p.i. (C). Average Fluorescent Intensity (AFU) of 3 mice+/− SD of a ROI (20 mm diameter) over the tumor in AU, arbitrary units
Figure 7.
In vivo photosensitizing efficacy of HPPH and its peptide conjugates 7 and 8 in BALB/c mice (10 mice/group) bearing 4T1 tumors and variable times post injection. The tumors were exposed to a laser light (665 nm, 135 J/cm2, 75mW/cm2) at dose shown per kg. (A) At 2h post injection, HPPH-cRGD was more effective than HPPH and showed 80% tumor response (8/10 mice were tumor-free on day 60). (B): At 24h post injection, HPPH was more effective than HPPH-cRGD conjugate and 4/10 mice were tumor-free on day 60.
Acknowledgments
The financial support from the NIH (CA127369), Roswell Park Alliance and the shared resources of the Roswell Park Cancer Center support grant CA16056 is highly appreciated. The authors are thankful to Carrie Batt for initial in vivo experiments. Mass spectrometry analyses were performed at Biopolymer Facility, Roswell Park Cancer Institute, Buffalo and Mass Spectrometry Facility at Michigan State University, East Lansing, MI, USA.
Abbreviations Used
- HPPH
3-(1′-hexyloxyethyl)3-devinylpyropheophorbide-a
- PDT
photodynamic therapy
- CD
cyanine dye
- PS
photosensitizer
- cRGD
cyclic aspartic acid (Asp), glycine (Gly) and arginine (Arg) peptides
- ROS
reactive oxygen species
- SAR
structure activity relationship
- QSAR
quantitative structure activity relationship
- FRET
fluorescence resonance energy transfer
- NMR
nuclear magnetic resonance
- HRMS
high resolution mass spectrometry
Footnotes
Supporting Information Available: 1H NMR spectra and HPLC chromatograms of the peptide-PS conjugates. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Dougherty TJ, Gomer CJ, Henderson BW, Jori G, Kessel D, Korbelik M, Moan J, Peng Q. Photodynamic Therapy. J Natl Cancer Inst. 1998;90:889–905. doi: 10.1093/jnci/90.12.889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dolmans DE, Fukumura D, Jain RK. Photodynamic therapy of cancer. Nat Rev Cancer. 2003;3:380–387. doi: 10.1038/nrc1071. [DOI] [PubMed] [Google Scholar]
- 3.Oleinick NL, Morris RL, Belichenko I. The role of apoptosis in response to photodynamic therapy: what, where, why and how? Photochem Photobiol Sci. 2002;1:1–21. doi: 10.1039/b108586g. [DOI] [PubMed] [Google Scholar]
- 4.Chen B, He C, Witte P, Hoopes PJ, Hasan T, Pogue BW. In: Vascular Targeting in photodynamic therapy in Advances in Photodynamic Therapy, Basic Translational and Clinical. Hamblin MR, Mroz P, editors. Artech House; Boston: 2008. [Google Scholar]
- 5.Gomer CJ. In: Photodynamic Therapy, Methods and Protocols. Gomer CJ, editor. Springer, Humana Press; New york: 2010. [Google Scholar]
- 6.Castano AP, Mroz P, Hamblin MR. Photodynamic therapy and anti-tumor immunity. Nature Reviews Cancer. 2006;6:535–545. doi: 10.1038/nrc1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.(a) Weishaupt KR, Gomer CJ, Dougherty TJ. Identification of singlet oxygen as the cytotoxic agent in photoinactivation of murine tumor. Cancer Res. 1976;90:889–899. [PubMed] [Google Scholar]; (b) Sherman WM, Allen CM, van Lier JE. Role of activated oxygen species in photodynamic therapy. Methods in Enzymol. 2000;319:376–386. doi: 10.1016/s0076-6879(00)19037-8. [DOI] [PubMed] [Google Scholar]
- 8.Pandey RK, Sumlin AB, Potter WR, Bellnier DA, Henderson BW, Constantine S, Aoudia M, Rodgers MA, Smith KM, Dougherty TJ. Structure and photodynamic efficacy among alkyl ether analogs of chlorophyll-a derivatives. Photochem Photobiol. 1996;63:194–205. doi: 10.1111/j.1751-1097.1996.tb02442.x. [DOI] [PubMed] [Google Scholar]
- 9.Henderson BW, Bellnier DA, Graco WR, Sharma A, Pandey RK, Vaughan L, Weishaupt KR, Rodgers MAJ, Smith KM, Dougherty TJ. A quantitative structure-activity relationship for a congeneric series of pyropheophorbide derivatives as photosensitizers for photodynamic therapy. Cancer Res. 1997;57:4000–4007. [PubMed] [Google Scholar]
- 10.Bellnier DA, Greco WR, Loewen GM, Nava V, Oseroff AO, Pandey RK, Tsuchida T, Dougherty TJ. Population pharmacokinetics of the photodynamic agent HPPH in cancer patients. Cancer Res. 2003;63:1806–1813. [PubMed] [Google Scholar]
- 11.Miller JD, Baron ED, Scull H, Hsia A, Berlin JC, McCormick T, Colussi V, Kenney ME, Cooper KD, Oleinick NL. Photodynamic therapy with the phthalocyanine photosensitizer Pc 4: the case experience with preclinical mechanistic and early clinical-translational studies. Toxicol Appl Pharmacol. 2007;224:290–9. doi: 10.1016/j.taap.2007.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Weersink RA, Bogaards A, Gertner M, Davidson SRH, Zhang K, Netchev G, Trachtenberg T, Wilson BC. J Photochem Photobiol, B. 2007;70:211–222. doi: 10.1016/j.jphotobiol.2005.01.008. [DOI] [PubMed] [Google Scholar]
- 13.Hamblin MR. In: Advances in Photodynamic Therapy, Basic Translational and Clinical. Hamblin MR, Mroz P, editors. Artech House; Boston: 2008. [Google Scholar]
- 14.(a) Solban N, Rizvi I, Hasan T. Targeted photodynamic therapy. Lasers in Surgery and Medicine. 2006;38(5):522–531. doi: 10.1002/lsm.20345. [DOI] [PubMed] [Google Scholar]; (b) Fabbrini M, Trachsel E, Soldano P, Bindi S, Alessi P, Bracci L, Kosmehl H, Zardi L, Neri D, Neri P. Selective occlusion of tumor blood vessels by targeted delivery of an antibody-photosensitizer conjugate. Int J Cancer. 2006;118:1805–1813. doi: 10.1002/ijc.21412. [DOI] [PubMed] [Google Scholar]
- 15.(a) Sibrian-Vazquez M, Jensen TJ, Vicente MGH. Synthesis, characterization, and metabolic stability of porphyrin-peptide conjugates bearing bifunctional signaling sequences. J Med Chem. 2008;51:2915–2923. doi: 10.1021/jm701050j. [DOI] [PubMed] [Google Scholar]; (b) Sibrian-Vazquez M, Jensen TJ, Hammer RP, Vicente MGH. Peptide-mediated Cell Transport of Water Soluble Porphyrin Conjugates. J Med Chem. 2006;49:1364–1372. doi: 10.1021/jm050893b. [DOI] [PubMed] [Google Scholar]
- 16.(a) Cheng Z, Wu Y, Xiong Z, Gambhir SS, Chen X. Near-infrared fluorescent RGD peptides for optical imaging of integrin αϖβ3 expression in living mice. Bioconjugate Chem. 2005;16:1433–1441. doi: 10.1021/bc0501698. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Mi Z, Guo H, Wai PY, Gao C, Kuo PC. Integrin-linked kniase regulates ostepontin-dependant MMP-2 and uPA expression to convey metastatic function in murine mammry epithelial cells. Carcinogenesis. 2006;27:1134–1145. doi: 10.1093/carcin/bgi352. [DOI] [PubMed] [Google Scholar]; (c) Sloan EK, Pouliot N, Stanley KL, Chia J, Moseley JM, Hards DK, Anderson RL. Tumor–specific expression of αvβ3 integrin promotes spontaneous metastasis of breast cancer to bone. Br Can Research. 2006;8 doi: 10.1186/bcr1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Toti US, Guru BR, Grill AE, Panyam J. Interfacial activity assisted surface functionalization: A novel approach to incorporate maleimide functional groups and cRGD peptide on polymeric nanoparticles for targeted drug delivery. Molecular Pharmaceutics. 2010;7:1108–1117. doi: 10.1021/mp900284c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Beer AJ, Haubner R, Goebel M, Luderschmidt S, Spiker ME, Wester HJ, Weber WA, Schwaiger M. Biodistribution and pharmacokinetics of the αvβ3-selective tracer 18F-galacto-RGD in cancer patients. J Nucl Med. 2005;46:1333–1341. [PubMed] [Google Scholar]
- 19.(a) Chen X, Conti PS, Moats RS. In vivo Near-Infrared Fluorescence Imaging of Integrin αvβ3 in Brain Tumor Xenografts. Cancer Research. 2004;64:8009–8014. doi: 10.1158/0008-5472.CAN-04-1956. [DOI] [PubMed] [Google Scholar]; (b) Hsu AR, Hou LC, Veeravagu A, Greve JM, Vogel H, Tse V, Chen X. In vivo near-infrared fluorescence imaging of αvβ3 in an orthotopic glioblastoma model. Molecular Imaging and Biology. 2006;8:315–323. doi: 10.1007/s11307-006-0059-y. [DOI] [PubMed] [Google Scholar]; (c) Haubner R, Weber WA, Beer AJ, Vabuliene E, Reim D, Sarbia M, Becker K-F, Goebel M, Wester H-J, Kesseler H, Schwiger M. Noninvasive visualization of the activated αvβ3 integrin in cancer patients by positron emission tomography and [18F]Galacto-RGD. PLOS Medicine. 2005;2:244–252. doi: 10.1371/journal.pmed.0020070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ethirajan M, Chen Y, Joshi P, Pandey RK. The role of porphyrin chemistry in tumor-imaging and photodynamic therapy. Chem Soc Rev, The Royal Society of Chemistry. 2011;40:340–362. doi: 10.1039/b915149b. [DOI] [PubMed] [Google Scholar]
- 21.(a) Chen Y, Li G, Pandey RK. Synthesis of bacteriochlorins and theit potential utility in PDT. Current Organic Chem. 2004;8:1105–1134. [Google Scholar]; (b) Gryshuk A, Chen Y, Goswami LN, Pandey S, Missert JR, Ohulchanskyy T, Potter W, Prasad PN, Oseroff AO, Pandey RK. Structure-activity relationship among purpurinimides and bacteriopurpurinimides: Trifluoromethyl substituent enhanced the photosensitizing efficacy. J Med Chem. 2007;50:1254–1267. doi: 10.1021/jm061036q. [DOI] [PubMed] [Google Scholar]
- 22.Pandey RK, Goswami LN, Chen Y, Gryshuk A, Missert JR, Oseroff AO, Dougherty TJ, Pandey RK, Dougherty TJ, Gomer CJ. Nature: A rich source for developing multifunctional agents. Tumor-imaging and photodynamic therapy. Lasers in Surgery and Medicine. 2006;38(Special Issue: Photodynamic Therapy):445–467. doi: 10.1002/lsm.20352. [DOI] [PubMed] [Google Scholar]
- 23.Chen Y, Graham A, Potter WR, Morgan J, Vaughan L, Bellnier DA, Henderson BW, Oseroff A, Dougherty TJ, Pandey RK. Bacteriopurpurinimides: Highly Stable and Potent Photosensitizers for Photodynamic Therapy. J Med Chem. 2002;45:255–258. doi: 10.1021/jm010400c. [DOI] [PubMed] [Google Scholar]
- 24.Dougherty TJ, Pandey RK, Nava HR, Smith JA, Douglass HO, Edge SB, Bellnier DA, OMalley L, Cooper M. Preliminary clinical data on a new photodynamic therapy photosensitizer, HPPH for treatment of obstructive esophageal cancer. PROC SPIE. 2000;3909:25–27. [Google Scholar]
- 25.Ruoslahti E. RGD and other recognition sequences for integrins. Annu Rev Cell Dev Biol. 1996;12:697–715. doi: 10.1146/annurev.cellbio.12.1.697. [DOI] [PubMed] [Google Scholar]
- 26.(a) Kessel D, Luo Y. Intracellular sites of photodamage as a factor in apoptotic cell death. J Porphyrins and Phthalocyanines. 2001;5:181–184. [Google Scholar]; (b) Kessel D, Luo Y, Dang Y, Chang CK. The role of subcellular localization in inhibition of apoptosis by photodynamic therapy. Photochem Photobiol. 1997;65:422–426. doi: 10.1111/j.1751-1097.1997.tb08581.x. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Morgan J, Potter WR, Oseroff A. Comparison of photodynamic targets in a carcinoma cell line and its mitochondrial DNA-deficient derivative. Photochem Photobiol. 2000;70:747–757. doi: 10.1562/0031-8655(2000)071<0747:coptia>2.0.co;2. [DOI] [PubMed] [Google Scholar]; (d) Kessel D, Oleinick NL. Initiation of autophagy by photodynamic therapy. Methods Enzymol. 2009;453:1–16. doi: 10.1016/S0076-6879(08)04001-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xiang Z, Morgan J, Pandey SK, Chen Y, Tracy E, Baumann H, Missert JR, Batt C, Jackson J, Bellnier DA, Henderson BW, Pandey RK. Conjugation of HPPH to carbohydrates changes its subcellular distribution and enhances photodynamic activity in vivo. J Med Chem. 2009;52:4306–4318. doi: 10.1021/jm9001617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Castel S, Pagan P, Mitjans F, Piulats J, Goodman S, Jonczyk A, Huber F, Vilaró S, Reina M. RGD peptides and monoclonal Antibodies, antagonists of αv-integrin, enter the cells by independent endocytic pathways. Laboratory Investigation. 2001;81:1615–26. doi: 10.1038/labinvest.3780375. [DOI] [PubMed] [Google Scholar]
- 29.Allen CM, Sharman WM, La Madeleine C, van Lier JE, Weber JM. Attenuation of photodynamically induced apoptosis by an RGD containing peptide. Photochem Photobiol Sci. 2002;4:246–54. doi: 10.1039/b109979e. [DOI] [PubMed] [Google Scholar]
- 30.Xiong JP, Stehle T, Zhang R, Joachimiak A, Frech M, Goodman SL, Arnaout MA. Crystal structure of the extracellular segment of integrin αvβ3 in complex with an Arg-Gly-Asp ligand. Science. 2002;296:151–5. doi: 10.1126/science.1069040. [DOI] [PubMed] [Google Scholar]
- 31.Pandey SK, Zheng X, Morgan J, Missert JR, Liu T-H, Shibata M, Bellnier DA, Oseroff AR, Henderson BW, Dougherty TJ, Pandey RK. Purpurinimide carbohydrate conjugates: effect of the position of the carbohydrate moiety in photosensitizing efficacy. Molecular Pharmaceutics. 2007;4:448–454. doi: 10.1021/mp060135x. [DOI] [PubMed] [Google Scholar]
- 32.Zheng X, Morgan J, Pandey SK, Chen Y, Tracy E, Baumann H, Missert JR, Batt C, Jackson J, Bellnier DA, Henderson BW, Pandey RK. Comparative positron-emission tomography (PET) imaging and phototherapeutic potential of I-labeled methyl-(3-1′-iodobenxyloxyethyl)pyropheophorbide-a vs. the corresponding glucose and galactose conjugates. J Med Chem. 2009;52:4306–4318. doi: 10.1021/jm8012213. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.