Abstract
Cellular proteostasis (or protein homeostasis) depends on the timely folding and disposal of conformationally damaged polypeptides during their life span at all subcellular locations. This process is particularly important for membrane proteins confined to the cell surface with critical regulatory role in cellular homoeostasis and intercellular communication. Accumulating evidences indicate that membrane proteins exported from the endoplasmic reticulum (ER) are subjected to peripheral quality control (QC) along the late secretory and endocytic pathways, as well as at the plasma membrane (PM). Recently identified components of the PM QC recognition and effector mechanisms responsible for ubiquitination and lysosomal degradation of conformationally damaged PM proteins uncovered striking similarities to and differences from that of the ER QC machinery. Possible implications of the peripheral protein QC activity in phenotypic modulation of conformational diseases are also outlined.
Introduction
Preventing the accumulation of misfolded, aggregation prone and potentially cytotoxic polypeptides that are generated by mutations, transcriptional and translational errors or cellular and environmental stresses are essential to preserve protein homeostasis [1–3]. The global proteostasis network encompasses regulatory mechanism of transcription, translation and protein folding, vesicular transport as well as degradation pathways [3]. The balance of protein folding and degradation, at least in part, depends on the folding energetics of the client protein, influenced by posttranslational modifications, oligomerization and the lipid environment, as well as the activity of the relevant folding and degradation machinery [3]. Molecular chaperones and co-chaperones can shield exposed hydrophobic residues and stabilize folding intermediates to suppress aggregation and promote folding of newly synthesized membrane proteins at the ER [4]. Chaperones and co-chaperones also participate in triage decision by targeting nonnative polypeptides for degradation via ubiquitin (Ub) proteasome system (UPS) [4,5]. This requires retrotranslocation of misfolded membrane proteins from the ER into the cytoplasm and processive cleavage by the 26S proteasome [1,5].
Incompletely understood conformational surveillance mechanisms determine the fate of non-native membrane proteins in post-ER compartments. Membrane proteins with limited or delayed conformational defects can escape the ER and be retrieved from the cis-Golgi compartment back to the ER or targeted from the trans-Golgi network by vesicular transport carriers into vacuoles/lysosomes [2]. The Endosomal Sorting Complex Required for Transport (ESCRT)-dependent concentration and inward budding of ubiquitinated native cargoes from the limiting membrane of endosome provide a solution for the topological problem of polytopic membrane protein degradation [6]. Here we review recent progresses in identifying some of the constituents of the PM proteostasis mechanism that participate in the timely removal and degradation of damaged membrane proteins.
The substrate specificity of the plasma membrane QC
The selective recognition and elimination of conformationally defective membrane proteins from post-ER compartments has been postulated more than a decade ago [2]. A Golgi QC mechanism was proposed for the rapid vacuolar/lysosomal disposal of several substrates in both yeast and mammalian cells [2]. The mutant form of the PM H+-ATPase (Pma1–10), the α factor receptor (Ste2–3p), the arginine permease (Can1ts) and the destabilized general amino acid permease (Gap1) are rapidly degraded from the yeast PM [7–9]. In mammalian cells unliganded MHC I, mutant variants of CFTR, α2-receptors, transferrin receptor, bile salt export pump (BSEP), megalencephalic leukoencephalopathy with subcortical cyst 1 (MLC1), influenza hemagglutinin, vasopressin V-2 receptor (V2R), dopamine D4.4 receptor (DRD4) and Na+-H+ exchanger 6 (NHE6) with perceived or documented structural defects are also rapidly eliminated from cell surface [10–17] (see Table 1). Destabilizing point mutations are primarily localized in the cytoplasmic and transmembrane segments in these polypeptides. Accelerated PM disposal of H+/K+-ATPase, κ and δ opiod receptor, Kv1.4 potassium channel, glucose transporter 1 (GLUT1) and CFTR, however, could be also triggered by impaired N-linked glycosylation at the exofacial surface [18–22]. The global conformational defect of these PM proteins may be attributed to impaired targeting to the calnexin-calreticulin chaperone cycle at the ER [23] and/or direct structural destabilization of the native fold in a chaperone-independent manner [22,24]. The rapid degradation of the glycosylation-deficient CFTR from the PM was induced by the combination of these mechanisms [22].
Table 1.
Peripheral protein QC substrates
| membrane protein | mutation/condition | degron | PM stability related disease | reference | |
|---|---|---|---|---|---|
| Mammalian | |||||
| bile salt export pump (BSEP) | E297G (Cy) | 2–3 Ub | ↓ | progressive familial | [36] |
| D482G (Cy) | intrahepatic cholestasis type 2 (PFIC2) | ||||
| CFTR | rΔF508 (Cy) | poly/multimono Ub | ↓ | cystic fibrosis (CF) | [**44], [*27] |
| Δ70 (Cy truncation) | poly/multimono Ub | ↓ | [*27] | ||
| N894D, N900D (Ex) | poly/multimono Ub | ↓ | [22] | ||
| Na/H exchanger (NHE6) | Δ255–256 (TM) | poly/multimono Ub | ↓ | Angelman syndrome | [35] |
| MLC1 | multiple (TM or Cy) | ND | ↓ | megalencephalic leukoencephalopathy with subcortical cysts (MLC) |
[66] |
| HERG | low K+ | Ub | ↓ | type 2 long QT syndrome | [37] |
| LDL receptor | high salt or low pH (Ex) | ND | ↓ | hypercholesterolemia | [16] |
| Dopamine D4.4 receptor | M345T (TM) | poly/multimono Ub | ↓ | attention deficit hyperactivity disorder | [**28] |
| Vasopressin V2 receptor | W164S (TM) | poly/multimono Ub | ↓ | nephrogenic diabetes insipidus | [**28] |
| alpha-2A adrenergic receptor | D79N (TM) | ND | ↓ | cardiovascular diseases | [14] |
| N422D (TM) | ND | ↓ | [14] | ||
| Δi3loop (Cy) | ND | ↓ | [69] | ||
| CD4tl-lambdaC | L57C (Cy) | poly/multimono Ub | ↓ | model protein | [**28] |
| H+/K+-ATPase β subunit | N99Q, N130Q, N161Q, N222Q (Ex) | ND | ↓ | gastric, autoimmune diseases | [18] |
| κ opioid receptor | N25/39Q (Ex) | ND | ↓ | pain control, neuronal phenotypes | [19] |
| δ opioid receptor | N18Q/N33Q (Ex) | ND | ↓ | pain control, neuronal phenotypes | [20] |
| GLUT1 | N45Y, Q or D (ex) | ND | ↓ | GLUT1 deficiency syndrome | [70] |
| EGFR | L858R (Cy), exon 19 deletion (Cy) | poly/multimono Ub | ↓ | cancer suspectibility | [71] |
| ErbB2 | Hsp90 inhibition | poly/multimono Ub | ↓ | breast cancer | [72] |
| TGFBR2 | Hsp90 inhibition | poly/multimono Ub | ↓ | tumor suspectibility | [73] |
| Yeast | |||||
| Pma1 | Icb1-100 | poly/multimono Ub | ↓ | NA | [29] |
| Pma1-7 | poly/multimono Ub | ↓ | NA | [7] | |
| Pma1-10 | poly/multimono Ub | ↓ | NA | [52] | |
| Gap1 | absence of sphingolipids | poly/multimono Ub | ↓ | NA | [9] |
Abbreviations: Cy, cytosolic; Ex, extracellular; TM, transmembrane; Ub, ubiquitin; ND, not determined; ↓, decreasing stability; CFTR, cystic fibrosis transmembrane conductance regulator; HERG, human ether-à-go-go related gene; LDL, low-density lipoprotein; GLUT, glucose transporter; EGFR, epidermal growth factor receptor; ErbbB2, v-erb-b2 erythroblastic leukemia viral oncogene homolog 2; TGFBR2, transforming growth factor (TGF)-beta type II Pma1, H(+)-ATPase; Gap1, general amino acid permease
As opposed to signaling induced posttranslational modification, structural perturbation that is necessary and sufficient to target a PM proteins for degradation [25,26] remains to be determined. The increased protease susceptibility of the Pma1–10, the low temperature rescued (r)ΔF508-CFTR, as well as the glycosylation-deficient and C-terminally truncated CFTR is consistent with a causal relationship between PM protein unfolding and accelerated lysosomal degradation [10,13,22,27]. A direct correlation was recently established between unfolding, ubiquitination and PM disposal of a transmembrane model protein, composed of the C-terminally truncated CD4 molecule fused to the temperature-sensitive N-terminal domain of bacteriophage λ(CD4T-λm) [28]. Thermodynamic destabilization of the cytosolic λm domain was sufficient to increase the CD4-λm PM turnover, internalization and lysosomal delivery [28]. Direct perturbations of transmembrane domains by insertion of charge residues or depletion of lipid rafts enriched in sphingolipid can also sensitize PM proteins for conformational destabilization and subsequent recognition by the peripheral QC [9,11,15,29]. Structural destabilization of the Pma1 and Gap1 transmembrane domains in strains defective of sphingoid base synthesis could be mechanistically similar to the farnesol-induced conformational change of the HMG-CoA reductase at the ER [30].
We propose that a subset of tyrosine kinase receptors (TKR, e.g. ErbB2, Ron, EGF, Met and EphA2 receptors) represents conditional substrates for the peripheral QC machinery [31,32]. Several kinase domains of TKRs are maintained in the their native fold by dynamic interaction with the molecular chaperone Hsp90 that recognizes poorly defined conformational flexibility of client proteins [33]. Inhibiting the Hsp90 ATP binding with benzoquinon ansamycins leads to the forced dissociation of the Hsp90-TKR complex and subsequent unfolding of the kinase domain. This culminates in the Ub-dependent disposal of the TKR by accelerated internalization and a combination of proteasomal and lysosomal proteolysis, a pharmacological intervention applied to down-regulate oncogenic TKR for cancer treatment [33,34].
Polyubiquitination signals the degradation of nonnative membrane proteins from the PM
Since poly- and multiple-mono-Ub can serve as efficient endocytic and lysosomal targeting signals, these posttranslational modifications are exploited for the regulated disposal of both native and nonnative PM proteins in mammalian cells [25,31] (Table 1). Increased ubiquitination of mutant variants of BSEP, Pma1, DRD4, V2R, NHE6 and CFTR was documented at either the PM or post-Golgi compartments [7,27,35,36]. Conversely, down-regulation of the E1 Ub-activating enzyme delayed the peripheral turnover of mutant CFTRs, DRD4 and V2R, as well as CD4T-λm in ts20 cells [22,27,28]. Extracellular K+-depletion and high salt or acidity also triggered Ub-dependent degradation of the wild-type HERG channel and LDL receptor, respectively, presumably by conformational destabilization [16,37]. Thermal unfolding of CD4T-λm was indeed coincided with its ubiquitination at the PM, monitored by bioluminescence resonance energy transfer in real-time [28].
Recent evidences suggest that structural and functional promiscuity of the poly-Ub chain is greater than originally proposed. Besides K48, most other Ub-linkages are involved in proteasomal degradation of misfolded polypeptides [38]. In addition to K63, K11-, K29- and K48-linked poly-Ub chains can be recognized as internalization and lysosomal sorting signals [39]. Although we lack systematic analysis of poly-Ub configuration in nonnative PM proteins, the K63-linked Ub-chain was more abundant in unfolded CD4T-λm at the PM, while the ER-entrapped CD4T-λm and cytosolic EGFP-λm contained preferentially K48-linked Ub-chains [28], consistent with the emerging model that an overlapping set of Ub-chains can participate in proteasome- and lysosomal-dependent protein degradation with variable efficiency [38,39].
The ubiquitination machinery of the peripheral QC system
Components of the ubiquitination machinery involved in conformationally-defective PM protein degradation have been recently identified by two different approaches in higher eukaryotes. A proteomic analysis was utilized to isolate the ubiquitination machinery in complex with the thermally unfolded CD4T-λm chimera confined to the PM. The unfolded CD4T-λm chimera was immunoisolated under non-denaturing conditions from HEK293 cells and the complex composition was analysed by liquid chromatography and mass spectrometry. The proteomic analysis revealed that CHIP (C-terminal Hsp70 interacting protein), a cytosolic E3 Ub-ligase, as well as Hsc70 and Hsp90 were selectively associated with unfolded, but not the native CD4T-λC at the PM [28]. CHIP consists of an N-terminal tetratricopeptide (TPR) domain that binds Hsc70, Hsp70 and Hsp90 molecular chaperones, a central helical domain mediating CHIP dimerization and a C-terminal U-box domain responsible for the binding of E2 Ub-conjugating enzymes (e.g. UbcH5 and Ubc13) and the Ub-ligase activity [40–42]. CHIP function as a QC E3 Ub ligase that selectively ubiquitinates conformationally defective cytosolic and ER polypeptides has been established [41]. CHIP TPR domain binds to Hsc70 and Hsp90, enabling complex formation with unfolded CD4T-λC, suggesting that CHIP function is not restricted to damaged ER and cytosolic polypeptides disposal [28,41,42]. Although CD4T-λC unfolding recruited Hsc70/Hsp70/Hsp90 to the PM [28], chaperone-independent substrate recognition by CHIP cannot be ruled out [43].
As a parallel approach, phenotypic small interfering RNA (siRNA) screens were performed in HeLa cells to isolate the E3 Ub ligase(s) responsible for unfolded rΔF508-CFTR elimination from the PM. This assay also isolated CHIP as the E3 Ub ligase responsible for the ubiquitination and degradation of unfolded rΔF508-CFTR in two cell models [44]. Biochemical assays proved that Hsc70 and Hsp90 in concert with a subset of co-chaperones (see below) were required for the rΔF508-CFTR ubiquitination and disposal from the PM [44]. In accord with their cellular abundance, the Hsc70-CHIP complex appears to play a more important role in unfolded CFTR recognition than the Hsp90-CHIP machinery [42,44–46] (Fig. 2). The conformation-sensitive ubiquitination of the rΔF508, but not the wild-type CFTR by Hsc70-CHIP complex was confirmed using an in vitro reconstitution assay [44]. The contribution of CHIP-dependent ubiquitination to the accelerated disposal of conformationally defective V2R and DRD4 from the PM was also confirmed [28,44], underlying the multiple substrate recognition capacity of the peripheral QC system.
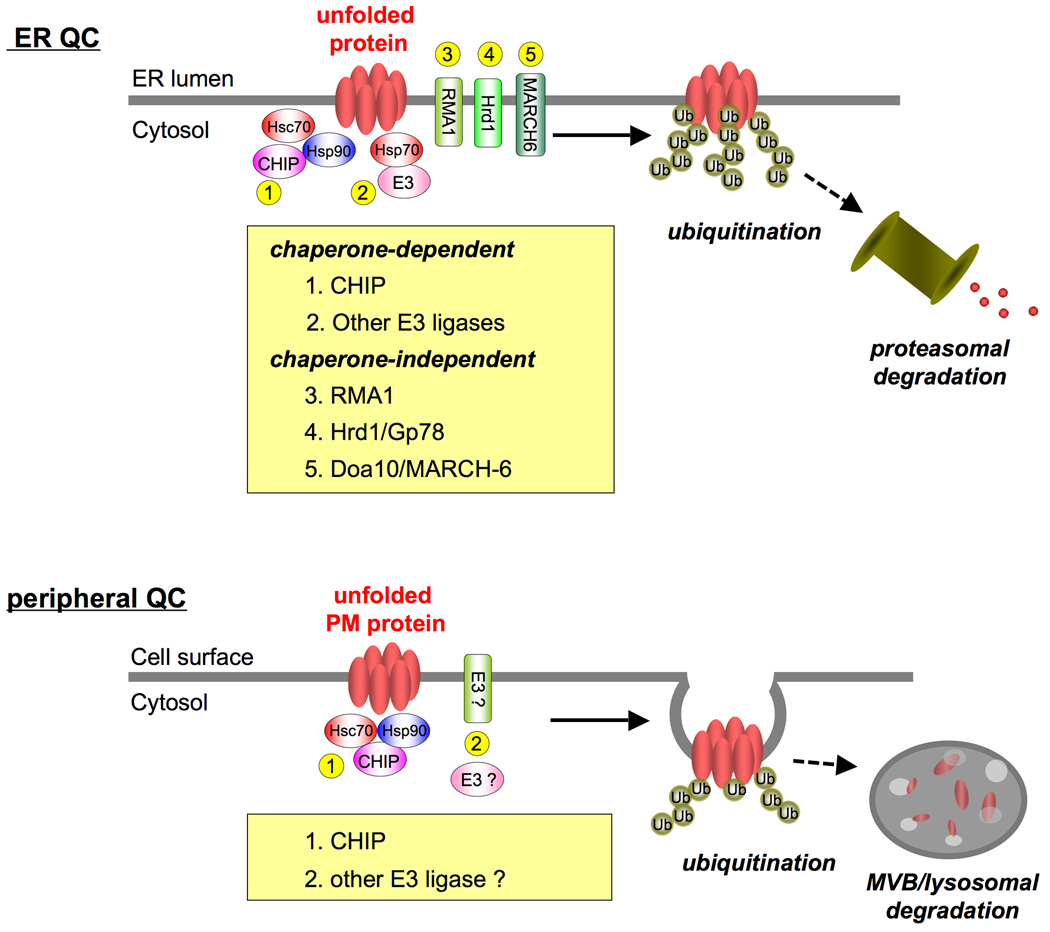
Figure 2. Ubiquitination machinery of the ER and peripheral QC systems.
Both chaperone-dependent (e.g. Hsc70/Hsp90-CHIP) and -independent (e.g. RMA1, Hrd1/Gp78 and Doa10/MARCH-6) ubiquitination pathways can contribute to the ER QC system [5,57]. This redundancy likely explains the limited phenotype of the Hsc70 or CHIP ablation on the ERAD efficiency of a subset of misfolded substrates [44]. Chaperone-binding cytoplasmic Ub-ligases (e.g. Ubr1/2 [42], UBE3A [54] and Cul5 [32]) may be also involved in the ER QC of membrane proteins with exposing cytoplasmic misfolding. In contrast, the redundancy of ubiquitination machinery appears to be limited for the peripheral QC, explaining the pronounced phenotypic consequences of Hsc70 or CHIP ablation on the peripheral degradation of nonnative PM proteins [44]. Although the contribution of additional Ub-ligase(s) remains to be uncovered, the PM-localized Ub-ligase, such as Gp78/AMFR [68] and other cytoplasmic Ub-ligase may play a role in the peripheral protein QC [44].
The phenotypic siRNA screens also revealed that co-chaperones DNAJA1 (Hdj2), DNAJB2 (HSJ1), Aha1 and HOP together with an E2 Ub-conjugating enzyme UbcH5 are essential constituents of the rΔF508-CFTR ubiquitination machinery in post-Golgi compartments [44]. Ablation of the J-domain protein DNAJA1 profoundly attenuated the rΔF508-CFTR ubiquitination, implying that DNAJA1 has a role in the Hsc70-dependent recognition of non-native PM proteins similar to its involvement in the ER QC [4,47]. The Hsc70-Hsp90 coupling factor HOP and the Hsp90 co-chaperone Aha1 probably enhance Hsp90 interaction with the PM client protein [44,48]. Supporting their degradative role in CFTR processing, ablation of HOP or Aha1 facilitates the non-native ΔF508-CFTR biosynthetic maturation [49,50]. DNAJB2, an Ub-interacting motif (UIM) containing J-domain protein, appears to regulate the rΔF508-CFTR ubiquitination only at the post-endocytic stage. This molecular mechanism remains to be established, but it may either enhance the ubiquitination or attenuate deubiquitination of the rΔF508-CFTR [51]. Likewise, it is plausible that deubiquitinating enzymes along the endocytic pathway regulate the degradation efficiency of non-native PM proteins as demonstrated for the mutant Pma1 and the wild-type CFTR [52,53].
E3 Ub-ligases can recognize non-native client proteins either via chaperone interactions (e.g. Ubr1/2 [42], UBE3A [54] and Cul5 [32]) or directly. The latter mechanism may prevail for a subset of client proteins of CHIP [43], Hrd1 [55] and San1 [56] in the cytoplasm, ER and nucleus, respectively. Intriguingly, direct substrate recognition by San1 is mediated by intrinsically disordered N- and C-terminal domains with embedded conserved recognition motifs [56]. In light of biological importance, it is conceivable that multiple ubiquitination mechanisms are involved in the peripheral protein QC, a possibility supported by the finding that CHIP ablation was unable to completely block the elimination of unfolded CFTR, DRD4 and V2R from the PM, while other Ub-ligase (e.g. Hrd1 and Gp78) knock-down could partially block the rΔF508-CFTR removal in HeLa cells [44].
Based on the redundancy of QC machineries in general [1,5,57], we envision that multiple Ub-dependent and, perhaps, Ub-independent degradation pathways as well, could be involved in the peripheral protein QC. For instance, BAG-1 stimulates lysosomal degradation without affecting the unfolded rΔF508-CFTR ubiquitination at the post-Golgi compartments [44] probably by facilitating interaction of the QC complex with endocytic and ESCRT adaptors (see below) [44], as well as providing physical links to proteasome-mediated degradation [58] and autophagy [59]. While Hsc70 strongly regulates the CHIP-mediated ubiquitination for lysosomal degradation of unfolded rΔF508-CFTR at the cell surface [44], Hsc70 is also involved in the chaperone-mediated autophagy of aberrant proteins [60]. Further studies will be required to clarify the contribution of these alternative degradation mechanisms in the context of the peripheral QC.
Comparison of the peripheral and ER/cytoplasmic QC
Intriguingly, constituents of the peripheral QC machinery are also involved in the ER and cytosolic QC [5,41,47,49,51], suggesting that similar principles may govern the recognition of structurally defective proteins at different cellular locations. This notion is in line with the capacity of CHIP-UbcH5 to synthesize Ub-chains with all possible linkages [40], conferring recognition signals for proteasomal degradation and Ub-binding endocytic adaptors for endo-lysosomal sorting [61]. One of the unique features of the ER/cytoplasmic proteostasis is that parallel and complementary pathways, including chaperone-dependent and -independent E3 ligases contribute to triage decision of misfolded polypeptides [55,62,63]. This redundancy appears to enhance the recognition flexibility and fidelity of the QC system to triage a significant fraction of newly synthesized proteins at the ER and cytoplasm (Fig. 2). In accord, CHIP or chaperone/co-chaperone ablation had modest effect on the ΔF508-CFTR ERAD, while significantly delayed the PM rΔF508-CFTR degradation [44].
Endocytic adaptors for the lysosomal targeting of non-native PM protein
Rapid endocytosis of aberrant PM proteins is probably mediated by Ub-binding clathrin adaptors (e.g. epsin1 and eps15/eps15R) similar to that of signaling-induced downregulation of native polypeptides [25,31,52]. These clathrin adaptors can recognize both K63- and K48-linked poly-Ub chain [31,64]. In addition, BAG-1, an Ub-like domain containing Hsc70 co-chaperone, may link the chaperone-PM protein complex to Ub-binding adaptors to the internalization and lysosomal sorting machinery through its Ub-like domain [44]. The severe recycling defect in concert with lysosomal rerouting of mutant CFTRs, V2R, DRD4 and CD4T-λC chimera implies that non-native PM proteins are subjected to conformation-dependent post-endocytic sorting. ESCRT components Hrs (Hepatocyte growth factor-regulated tyrosine kinase substrate), Stam1 (signal-transducing adaptor molecule), and TSG101 (tumor susceptibility gene 101) are essential for ubiquitinated native cargo delivery into MVB/lysosomes [6]. Down-regulation of Hrs, Stam1 or TSG101 also delayed the degradation of unfolded PM proteins and retained them in early endosome, revealing the anticipated function of ESCRT in peripheral QC of PM proteins [22,28,44].
The possible role of peripheral QC as modifier of the loss-of-function cellular phenotype
Promiscuous substrate specificity of Hsc70/Hsp90 towards non-native polypeptides implies that the peripheral QC probably contributes to triage decision of numerous PM proteins and the pathogenesis of certain conformational diseases. The phenotype of mutant BSEP and CFTR, associated with progressive familial intrahepatic cholestasis type 2 disease and cystic fibrosis (CF), respectively, suggest that the metabolic destabilization of these transporters correlates with their ubiquitination at the PM [27,36]. The severity of cholestasis and CF is inversely proportional with the PM density of the BSEP and CFTR, respectively [10,65]. In light of the limited fidelity of the ER QC and the possibility of delayed unfolding of mutants at the PM, it is tempting to speculate that the peripheral QC may exacerbate the phenotype of conformational diseases by prematurely disposing partially functional mutants from the PM. Indeed, selected mutants of CFTR, MLC1, V2R and BSEP could escape the ER QC and targeted for endo-lysosomal degradation from the PM in both primary cells and heterologous expression systems [10,27,36,66,67].
Conclusions and perspectives
Recently cellular and biochemical processes recognizing and disposing non-native PM proteins as part of the peripheral QC mechanism have begun to be elucidated. The unexpected complexity of the peripheral proteostatic mechanism is exemplified by the coordinated function of chaperones, co-chaperones, Ub-conjugating and -ligating enzymes, as well as Ub-binding PM and endosomal adaptors and the ESCRT machinery that complements the function of proteostasis networks of the ER, mitochondria, cytosol and nucleus. Despite this progress, a number of questions remain to be addressed. Do alternative degradation pathways (e.g. autophagocytosis and proteasome) contribute to peripheral proteostasis? What is the conformational sensitivity of the peripheral QC machinery in relation to the ER QC? Are other chaperone-dependent and -independent E3 ligases involved in the peripheral QC in analogy to the proteostasis networks of the ER and cytoplasm? Finally, can chemical or biological modulation of the peripheral proteostasis help alleviating the loss-of-function/expression phenotype of PM proteins in conformational disease and be exploited in therapeutic applications? Answers to these questions will help establish the molecular basis and significance of the peripheral QC systems in the complex cellular proteostasis networks in health and diseases.
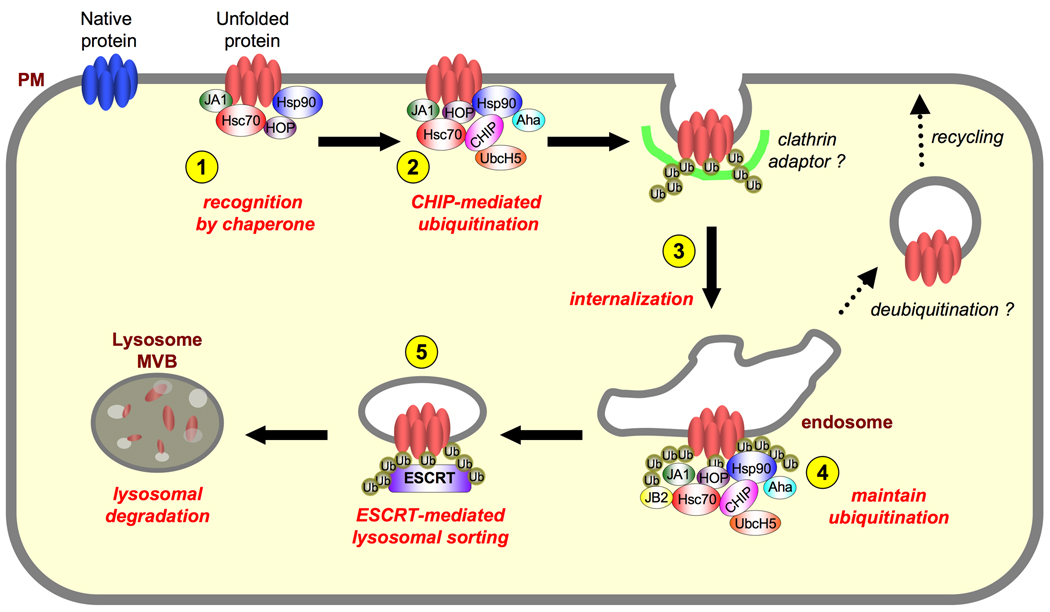
Figure 1. Working model for the peripheral protein QC network.
1. The cytoplasmic region of conformationally defective PM proteins is selectively recognized by Hsc70 in concert with DNAJA1 (JA1) and, possibly by the Hsp90/HOP/Aha1 machinery. 2. Prolonged interaction with the chaperone/co-chaperone complex recruits CHIP-UbcH5, leading to ubiquitination of conformationally damaged PM proteins. 3. Ubiquitinated nonnative PM proteins are rapidly endocytosed, possibly by clathrin-mediated internalization upon recruitment of Ub-binding endocytic adaptors. 4–5. Depending on the folding propensity of the cargo molecule and the proteostasis network state, dynamic interaction with chaperones and co-chaperones may favor the client protein refolding, deubiquitination (e.g. [53]) and subsequent recycling to the PM. Alternatively, irreversible unfolding of the PM protein would lead to persistent ubiquitination by CHIP-UbcH5 and/or by other E3 ligase(s), providing efficient sorting signals for ESCRT-dependent cargo concentration, intraluminal budding and multivesicular endosome formation for delivery into the degradative lysosomal compartment.
Acknowledgements
The experimental work performed in the laboratory of GL was supported by the Canadian Institute for Health Research, Canadian Cystic Fibrosis Foundation (CCFF), Cystic Fibrosis Foundation, NIH, NIDDK and Canadian Foundation for Innovation. TO was a recipient of a Postdoctoral Fellowship from the CCFF. GL is the recipient of a Canada Research Chair.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and recommended reading
Papers of particular interest, published within the annual period of review, have been highlighted as:
* of special interest
** of outstanding interest
- 1.Ellgaard L, Helenius A. Quality control in the endoplasmic reticulum. Nat Rev Mol Cell Biol. 2003;4(3):181–191. doi: 10.1038/nrm1052. [DOI] [PubMed] [Google Scholar]
- 2.Arvan P, Zhao X, Ramos-Castaneda J, Chang A. Secretory pathway quality control operating in Golgi, plasmalemmal, and endosomal systems. Traffic. 2002;3(11):771–780. doi: 10.1034/j.1600-0854.2002.31102.x. [DOI] [PubMed] [Google Scholar]
- 3. Powers ET, Morimoto RI, Dillin A, Kelly JW, Balch WE. Biological and chemical approaches to diseases of proteostasis deficiency. Annu Rev Biochem. 2009;78:959–991. doi: 10.1146/annurev.biochem.052308.114844.. An excellent review on the major determinants of proteostasis networks and their contribution to conformational diseases, applicable to the peripheral protein QC.
- 4.Young JC, Agashe VR, Siegers K, Hartl FU. Pathways of chaperone-mediated protein folding in the cytosol. Nat Rev Mol Cell Biol. 2004;5(10):781–791. doi: 10.1038/nrm1492. [DOI] [PubMed] [Google Scholar]
- 5.Vembar SS, Brodsky JL. One step at a time: endoplasmic reticulum-associated degradation. Nat Rev Mol Cell Biol. 2008;9(12):944–957. doi: 10.1038/nrm2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Raiborg C, Stenmark H. The ESCRT machinery in endosomal sorting of ubiquitylated membrane proteins. Nature. 2009;458(7237):445–452. doi: 10.1038/nature07961. [DOI] [PubMed] [Google Scholar]
- 7.Gong X, Chang A. A mutant plasma membrane ATPase, Pma1-10, is defective in stability at the yeast cell surface. Proc Natl Acad Sci U S A. 2001;98(16):9104–9109. doi: 10.1073/pnas.161282998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li Y, Kane T, Tipper C, Spatrick P, Jenness DD. Yeast mutants affecting possible quality control of plasma membrane proteins. Mol Cell Biol. 1999;19(5):3588–3599. doi: 10.1128/mcb.19.5.3588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lauwers E, Grossmann G, Andre B. Evidence for coupled biogenesis of yeast Gap1 permease and sphingolipids: essential role in transport activity and normal control by ubiquitination. Mol Biol Cell. 2007;18(8):3068–3080. doi: 10.1091/mbc.E07-03-0196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Benharouga M, Haardt M, Kartner N, Lukacs GL. COOH-terminal truncations promote proteasome-dependent degradation of mature cystic fibrosis transmembrane conductance regulator from post-Golgi compartments. J Cell Biol. 2001;153(5):957–970. doi: 10.1083/jcb.153.5.957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fayadat L, Kopito RR. Recognition of a single transmembrane degron by sequential quality control checkpoints. Mol Biol Cell. 2003;14(3):1268–1278. doi: 10.1091/mbc.E02-06-0363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ljunggren HG, Stam NJ, Ohlen C, Neefjes JJ, Hoglund P, Heemels MT, Bastin J, Schumacher TN, Townsend A, Karre K, et al. Empty MHC class I molecules come out in the cold. Nature. 1990;346(6283):476–480. doi: 10.1038/346476a0. [DOI] [PubMed] [Google Scholar]
- 13.Sharma M, Benharouga M, Hu W, Lukacs GL. Conformational and temperature-sensitive stability defects of the delta F508 cystic fibrosis transmembrane conductance regulator in post-endoplasmic reticulum compartments. J Biol Chem. 2001;276(12):8942–8950. doi: 10.1074/jbc.M009172200. [DOI] [PubMed] [Google Scholar]
- 14.Wilson MH, Highfield HA, Limbird LE. The role of a conserved inter-transmembrane domain interface in regulating alpha(2a)-adrenergic receptor conformational stability and cell-surface turnover. Mol Pharmacol. 2001;59(4):929–938. doi: 10.1124/mol.59.4.929. [DOI] [PubMed] [Google Scholar]
- 15.Zaliauskiene L, Kang S, Brouillette CG, Lebowitz J, Arani RB, Collawn JF. Down-regulation of cell surface receptors is modulated by polar residues within the transmembrane domain. Mol Biol Cell. 2000;11(8):2643–2655. doi: 10.1091/mbc.11.8.2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schaheen B, Dang H, Fares H. Derlin-dependent accumulation of integral membrane proteins at cell surfaces. J Cell Sci. 2009;122(Pt 13):2228–2239. doi: 10.1242/jcs.048892. [DOI] [PubMed] [Google Scholar]
- 17.Lukacs GL, Chang XB, Bear C, Kartner N, Mohamed A, Riordan JR, Grinstein S. The delta F508 mutation decreases the stability of cystic fibrosis transmembrane conductance regulator in the plasma membrane. Determination of functional half-lives on transfected cells. J Biol Chem. 1993;268(29):21592–21598. [PubMed] [Google Scholar]
- 18.Vagin O, Turdikulova S, Sachs G. The H,K-ATPase beta subunit as a model to study the role of N-glycosylation in membrane trafficking and apical sorting. J Biol Chem. 2004;279(37):39026–39034. doi: 10.1074/jbc.M405453200. [DOI] [PubMed] [Google Scholar]
- 19.Li J-G, Chen C, Liu-Chen L-Y. N-Glycosylation of the Human κ Opioid Receptor Enhances Its Stability but Slows Its Trafficking along the Biosynthesis Pathway†. Biochemistry. 2007;46(38):10960–10970. doi: 10.1021/bi700443j. [DOI] [PubMed] [Google Scholar]
- 20.Markkanen PMH, Petäjä-Repo UE. N-Glycan-mediated Quality Control in the Endoplasmic Reticulum Is Required for the Expression of Correctly Folded δ -Opioid Receptors at the Cell Surface. J Biol Chem. 2008;283(43):29086–29098. doi: 10.1074/jbc.M801880200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Watanabe I, Zhu J, Recio-Pinto E, Thornhill WB. Glycosylation Affects the Protein Stability and Cell Surface Expression of Kv1.4 but Not Kv1.1 Potassium Channels. J Biol Chem. 2004;279(10):8879–8885. doi: 10.1074/jbc.M309802200. [DOI] [PubMed] [Google Scholar]
- 22.Glozman R, Okiyoneda T, Mulvihill CM, Rini JM, Barriere H, Lukacs GL. N-glycans are direct determinants of CFTR folding and stability in secretory and endocytic membrane traffic. J Cell Biol. 2009;184(6):847–862. doi: 10.1083/jcb.200808124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lederkremer GZ. Glycoprotein folding, quality control and ER-associated degradation. Curr Opin Struct Biol. 2009;19(5):515–523. doi: 10.1016/j.sbi.2009.06.004. [DOI] [PubMed] [Google Scholar]
- 24.Hanson SR, Culyba EK, Hsu TL, Wong CH, Kelly JW, Powers ET. The core trisaccharide of an N-linked glycoprotein intrinsically accelerates folding and enhances stability. Proc Natl Acad Sci U S A. 2009;106(9):3131–3136. doi: 10.1073/pnas.0810318105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Rotin D, Staub O. Role of the ubiquitin system in regulating ion transport. Pflugers Arch. 2010;461(1):1–21. doi: 10.1007/s00424-010-0893-2.. A comprehensive review summarizing the cellular and biochemical regulatory mechanisms of native ion channel and transporter at the cell surface.
- 26. Lin CH, MacGurn JA, Chu T, Stefan CJ, Emr SD. Arrestin-related ubiquitin-ligase adaptors regulate endocytosis and protein turnover at the cell surface. Cell. 2008;135(4):714–725. doi: 10.1016/j.cell.2008.09.025.. Using genetic approaches, this paper identifies arrestin-related adaptor (ART) family members that account for the Rsp5-dependent down-regulation of yeast amino acid transporters from the PM.
- 27. Sharma M, Pampinella F, Nemes C, Benharouga M, So J, Du K, Bache KG, Papsin B, Zerangue N, Stenmark H, Lukacs GL. Misfolding diverts CFTR from recycling to degradation: quality control at early endosomes. J Cell Biol. 2004;164(6):923–933. doi: 10.1083/jcb.200312018.. The authors provide evidences for the role of ubiquitination in the recycling defect and preferential lysosomal sorting of conformationally defective CFTR channels as a part of the peripheral QC in mammalian cells.
- 28. Apaja PM, Xu H, Lukacs GL. Quality control for unfolded proteins at the plasma membrane. J Cell Biol. 2010;191(3):553–570. doi: 10.1083/jcb.201006012.. The authors demontrate that a conditionally unfolded PM model protein and mutant G protein coupled receptors are removed from the PM by a CHIP- and chaperone-dependent ubquitination process.
- 29.Wang Q, Chang A. Sphingoid base synthesis is required for oligomerization and cell surface stability of the yeast plasma membrane ATPase, Pma1. Proc Natl Acad Sci U S A. 2002;99(20):12853–12858. doi: 10.1073/pnas.202115499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shearer AG, Hampton RY. Lipid-mediated, reversible misfolding of a sterol-sensing domain protein. The EMBO journal. 2005;24(1):149–159. doi: 10.1038/sj.emboj.7600498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Traub LM, Lukacs GL. Decoding ubiquitin sorting signals for clathrin-dependent endocytosis by CLASPs. J Cell Sci. 2007;120(Pt 4):543–553. doi: 10.1242/jcs.03385. [DOI] [PubMed] [Google Scholar]
- 32.Ehrlich ES, Wang T, Luo K, Xiao Z, Niewiadomska AM, Martinez T, Xu W, Neckers L, Yu XF. Regulation of Hsp90 client proteins by a Cullin5-RING E3 ubiquitin ligase. Proc Natl Acad Sci U S A. 2009;106(48):20330–20335. doi: 10.1073/pnas.0810571106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Taipale M, Jarosz DF, Lindquist S. HSP90 at the hub of protein homeostasis: emerging mechanistic insights. Nat Rev Mol Cell Biol. 2010;11(7):515–528. doi: 10.1038/nrm2918. [DOI] [PubMed] [Google Scholar]
- 34.Trepel J, Mollapour M, Giaccone G, Neckers L. Targeting the dynamic HSP90 complex in cancer. Nat Rev Cancer. 2010;10(8):537–549. doi: 10.1038/nrc2887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Roxrud I, Raiborg C, Gilfillan GD, Stromme P, Stenmark H. Dual degradation mechanisms ensure disposal of NHE6 mutant protein associated with neurological disease. Exp Cell Res. 2009;315(17):3014–3027. doi: 10.1016/j.yexcr.2009.07.012. [DOI] [PubMed] [Google Scholar]
- 36.Hayashi H, Sugiyama Y. Short-chain ubiquitination is associated with the degradation rate of a cell-surface-resident bile salt export pump (BSEP/ABCB11) Mol Pharmacol. 2009;75(1):143–150. doi: 10.1124/mol.108.049288. [DOI] [PubMed] [Google Scholar]
- 37.Guo J, Massaeli H, Xu J, Jia Z, Wigle JT, Mesaeli N, Zhang S. Extracellular K+ concentration controls cell surface density of IKr in rabbit hearts and of the HERG channel in human cell lines. J Clin Invest. 2009;119(9):2745–2757. doi: 10.1172/JCI39027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Xu P, Duong DM, Seyfried NT, Cheng D, Xie Y, Robert J, Rush J, Hochstrasser M, Finley D, Peng J. Quantitative proteomics reveals the function of unconventional ubiquitin chains in proteasomal degradation. Cell. 2009;137(1):133–145. doi: 10.1016/j.cell.2009.01.041.. Using quantitative mass spectrometry, the authors demonstrate that most Ub-chain configuration, including the K11-linked chain, can be recognized and degraded by proteasome in yeast.
- 39. Boname JM, Thomas M, Stagg HR, Xu P, Peng J, Lehner PJ. Efficient Internalization of MHC I Requires Lysine-11 and Lysine-63 Mixed Linkage Polyubiquitin Chains. Traffic. 2009 doi: 10.1111/j.1600-0854.2009.01011.x.. This paper unequivocally shows that in addition to K63-, K11-linked poly-Ub chain serves as an efficient endocytic signal in mammalian cells.
- 40.Zhang M, Windheim M, Roe SM, Peggie M, Cohen P, Prodromou C, Pearl LH. Chaperoned ubiquitylation--crystal structures of the CHIP U box E3 ubiquitin ligase and a CHIP-Ubc13-Uev1a complex. Mol Cell. 2005;20(4):525–538. doi: 10.1016/j.molcel.2005.09.023. [DOI] [PubMed] [Google Scholar]
- 41.Meacham GC, Patterson C, Zhang W, Younger JM, Cyr DM. The Hsc70 co-chaperone CHIP targets immature CFTR for proteasomal degradation. Nat Cell Biol. 2001;3(1):100–105. doi: 10.1038/35050509. [DOI] [PubMed] [Google Scholar]
- 42. Heck JW, Cheung SK, Hampton RY. Cytoplasmic protein quality control degradation mediated by parallel actions of the E3 ubiquitin ligases Ubr1 and San1. Proc Natl Acad Sci U S A. 2010;107(3):1106–1111. doi: 10.1073/pnas.0910591107.. See ref *54
- 43.Rosser MF, Washburn E, Muchowski PJ, Patterson C, Cyr DM. Chaperone functions of the E3 ubiquitin ligase CHIP. J Biol Chem. 2007;282(31):22267–22277. doi: 10.1074/jbc.M700513200. [DOI] [PubMed] [Google Scholar]
- 44. Okiyoneda T, Barriere H, Bagdany M, Rabeh WM, Du K, Hohfeld J, Young JC, Lukacs GL. Peripheral protein quality control removes unfolded CFTR from the plasma membrane. Science. 2010;329(5993):805–810. doi: 10.1126/science.1191542.. Using genetic approaches, this paper identified chaperone/co-chaperone components of the peripheral protein QC machinery and their role in the ubiquitination dependent and independent PM removal and the ESCRT-dependent lysosomal sorting of misfolded CFTR.
- 45. Kundrat L, Regan L. Balance between folding and degradation for Hsp90-dependent client proteins: a key role for CHIP. Biochemistry. 2010;49(35):7428–7438. doi: 10.1021/bi100386w.. See ref *46
- 46. Stankiewicz M, Nikolay R, Rybin V, Mayer MP. CHIP participates in protein triage decisions by preferentially ubiquitinating Hsp70-bound substrates. The FEBS journal. 2010;277(16):3353–3367. doi: 10.1111/j.1742-4658.2010.07737.x.. Refs. *45 and *46 established the relative contribution and biochemical basis of Hsc70 and Hsp90-dependent CHIP ubiquitination of misfolded soluble model proteins.
- 47.Younger JM, Ren HY, Chen L, Fan CY, Fields A, Patterson C, Cyr DM. A foldable CFTR{Delta}F508 biogenic intermediate accumulates upon inhibition of the Hsc70-CHIP E3 ubiquitin ligase. J Cell Biol. 2004;167(6):1075–1085. doi: 10.1083/jcb.200410065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Koulov AV, Lapointe P, Lu B, Razvi A, Coppinger J, Dong MQ, Matteson J, Laister R, Arrowsmith C, Yates JR, 3rd, Balch WE. Biological and structural basis for Aha1 regulation of Hsp90 ATPase activity in maintaining proteostasis in the human disease cystic fibrosis. Mol Biol Cell. 2010;21(6):871–884. doi: 10.1091/mbc.E09-12-1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang X, Venable J, LaPointe P, Hutt DM, Koulov AV, Coppinger J, Gurkan C, Kellner W, Matteson J, Plutner H, Riordan JR, et al. Hsp90 cochaperone Aha1 downregulation rescues misfolding of CFTR in cystic fibrosis. Cell. 2006;127(4):803–815. doi: 10.1016/j.cell.2006.09.043. [DOI] [PubMed] [Google Scholar]
- 50.Marozkina NV, Yemen S, Borowitz M, Liu L, Plapp M, Sun F, Islam R, Erdmann-Gilmore P, Townsend RR, Lichti CF, Mantri S, et al. Hsp 70/Hsp 90 organizing protein as a nitrosylation target in cystic fibrosis therapy. Proc Natl Acad Sci U S A. 2010;107(25):11393–11398. doi: 10.1073/pnas.0909128107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Westhoff B, Chapple JP, van der Spuy J, Hohfeld J, Cheetham ME. HSJ1 is a neuronal shuttling factor for the sorting of chaperone clients to the proteasome. Curr Biol. 2005;15(11):1058–1064. doi: 10.1016/j.cub.2005.04.058. [DOI] [PubMed] [Google Scholar]
- 52.Liu Y, Chang A. Quality control of a mutant plasma membrane ATPase: ubiquitylation prevents cell-surface stability. J Cell Sci. 2006;119(Pt 2):360–369. doi: 10.1242/jcs.02749. [DOI] [PubMed] [Google Scholar]
- 53. Bomberger JM, Barnaby RL, Stanton BA. The deubiquitinating enzyme USP10 regulates the post-endocytic sorting of cystic fibrosis transmembrane conductance regulator in airway epithelial cells. J Biol Chem. 2009;284(28):18778–18789. doi: 10.1074/jbc.M109.001685.. This paper proposes that deubiquitination regulates the post-endocytic sorting of CFTR and promotes the channel recycling by preventing lysosomal delivery.
- 54. Mishra A, Godavarthi SK, Maheshwari M, Goswami A, Jana NR. The ubiquitin ligase E6-AP is induced and recruited to aggresomes in response to proteasome inhibition and may be involved in the ubiquitination of Hsp70-bound misfolded proteins. J Biol Chem. 2009;284(16):10537–10545. doi: 10.1074/jbc.M806804200.. Refs. *42 and *54 demonstrate the functional signficance of additional chaperone-dependent E3 Ub-ligase in soluble and integral membraen protein QC.
- 55. Sato BK, Schulz D, Do PH, Hampton RY. Misfolded membrane proteins are specifically recognized by the transmembrane domain of the Hrd1p ubiquitin ligase. Mol Cell. 2009;34(2):212–222. doi: 10.1016/j.molcel.2009.03.010.. The authors demonstrate the direct recognition of misfolded membrane proteins mediated by the transmembrane domains of the Hrd1 Ub-ligase in the ER.
- 56.Rosenbaum JC, Fredrickson EK, Oeser ML, Garrett-Engele CM, Locke MN, Richardson LA, Nelson ZW, Hetrick ED, Milac TI, Gottschling DE, Gardner RG. Disorder targets misorder in nuclear quality control degradation: a disordered ubiquitin ligase directly recognizes its misfolded substrates. Mol Cell. 2011;41(1):93–106. doi: 10.1016/j.molcel.2010.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Hirsch C, Gauss R, Horn SC, Neuber O, Sommer T. The ubiquitylation machinery of the endoplasmic reticulum. Nature. 2009;458(7237):453–460. doi: 10.1038/nature07962.. An excellent review on the ubiquitination mechanism by the ER QC.
- 58.Luders J, Demand J, Hohfeld J. The ubiquitin-related BAG-1 provides a link between the molecular chaperones Hsc70/Hsp70 and the proteasome. J Biol Chem. 2000;275(7):4613–4617. doi: 10.1074/jbc.275.7.4613. [DOI] [PubMed] [Google Scholar]
- 59.Gurusamy N, Lekli I, Gherghiceanu M, Popescu LM, Das DK. BAG-1 induces autophagy for cardiac cell survival. Autophagy. 2009;5(1):120–121. doi: 10.4161/auto.5.1.7303. [DOI] [PubMed] [Google Scholar]
- 60.Orenstein SJ, Cuervo AM. Chaperone-mediated autophagy: molecular mechanisms and physiological relevance. Seminars in cell & developmental biology. 2010;21(7):719–726. doi: 10.1016/j.semcdb.2010.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Finley D. Recognition and processing of ubiquitin-protein conjugates by the proteasome. Annu Rev Biochem. 2009;78:477–513. doi: 10.1146/annurev.biochem.78.081507.101607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Younger JM, Chen L, Ren HY, Rosser MF, Turnbull EL, Fan CY, Patterson C, Cyr DM. Sequential quality-control checkpoints triage misfolded cystic fibrosis transmembrane conductance regulator. Cell. 2006;126(3):571–582. doi: 10.1016/j.cell.2006.06.041. [DOI] [PubMed] [Google Scholar]
- 63.Morito D, Hirao K, Oda Y, Hosokawa N, Tokunaga F, Cyr DM, Tanaka K, Iwai K, Nagata AK. Gp78 cooperates with RMA1 in endoplasmic reticulum-associated degradation of CFTRDeltaF508. Mol Biol Cell. 2008;19(4):1328–1336. doi: 10.1091/mbc.E07-06-0601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Barriere H, Nemes C, Lechardeur D, Khan-Mohammad M, Fruh K, Lukacs GL. Molecular basis of oligoubiquitin-dependent internalization of membrane proteins in Mammalian cells. Traffic. 2006;7(3):282–297. doi: 10.1111/j.1600-0854.2006.00384.x. [DOI] [PubMed] [Google Scholar]
- 65.Lam P, Pearson CL, Soroka CJ, Xu S, Mennone A, Boyer JL. Levels of plasma membrane expression in progressive and benign mutations of the bile salt export pump (Bsep/Abcb11) correlate with severity of cholestatic diseases. Am J Physiol Cell Physiol. 2007;293(5):C1709–C1716. doi: 10.1152/ajpcell.00327.2007. [DOI] [PubMed] [Google Scholar]
- 66.Duarri A, Teijido O, Lopez-Hernandez T, Scheper GC, Barriere H, Boor I, Aguado F, Zorzano A, Palacin M, Martinez A, Lukacs GL, et al. Molecular pathogenesis of megalencephalic leukoencephalopathy with subcortical cysts: mutations in MLC1 cause folding defects. Hum Mol Genet. 2008;17(23):3728–3739. doi: 10.1093/hmg/ddn269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Robben JH, Knoers NV, Deen PM. Characterization of vasopressin V2 receptor mutants in nephrogenic diabetes insipidus in a polarized cell model. Am J Physiol Renal Physiol. 2005;289(2):F265–F272. doi: 10.1152/ajprenal.00404.2004. [DOI] [PubMed] [Google Scholar]
- 68.Fairbank M, St-Pierre P, Nabi IR. The complex biology of autocrine motility factor/phosphoglucose isomerase (AMF/PGI) and its receptor, the gp78/AMFR E3 ubiquitin ligase. Molecular bioSystems. 2009;5(8):793–801. doi: 10.1039/b820820b. [DOI] [PubMed] [Google Scholar]
- 69.Edwards SW, Limbird LE. Role for the Third Intracellular Loop in Cell Surface Stabilization of the α2A–Adrenergic Receptor. J Biol Chem. 1999;274(23):16331–16336. doi: 10.1074/jbc.274.23.16331. [DOI] [PubMed] [Google Scholar]
- 70.Asano T, Takata K, Katagiri H, Ishihara H, Inukai K, Anai M, Hirano H, Yazaki Y, Oka Y. The role of N-glycosylation in the targeting and stability of GLUT1 glucose transporter. FEBS Lett. 1993;324(3):258–261. doi: 10.1016/0014-5793(93)80129-i. [DOI] [PubMed] [Google Scholar]
- 71.Shimamura T, Lowell AM, Engelman JA, Shapiro GI. Epidermal growth factor receptors harboring kinase domain mutations associate with the heat shock protein 90 chaperone and are destabilized following exposure to geldanamycins. Cancer Res. 2005;65(14):6401–6408. doi: 10.1158/0008-5472.CAN-05-0933. [DOI] [PubMed] [Google Scholar]
- 72.Marx C, Held JM, Gibson BW, Benz CC. ErbB2 trafficking and degradation associated with K48 and K63 polyubiquitination. Cancer Res. 2010;70(9):3709–3717. doi: 10.1158/0008-5472.CAN-09-3768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wrighton KH, Lin X, Feng XH. Critical regulation of TGFbeta signaling by Hsp90. Proc Natl Acad Sci U S A. 2008;105(27):9244–9249. doi: 10.1073/pnas.0800163105. [DOI] [PMC free article] [PubMed] [Google Scholar]