Abstract
Purpose
In our previous studies we showed that upregulating claudin-6 (CLDN6) expression may contribute to preventing breast cancer, and that 17β-estradiol induces a concentration- and time-related effect on CLDN6 mRNA and protein expression in MCF-7 cells. However, the mechanisms of 17β-estradiol regulation of CLDN6 are still unclear. We determined the role of estrogen receptors in the regulation of CLDN6 expression in human breast cancer tissues and a cell line.
Methods
CLDN6, estrogen receptor alpha (ERα) and estrogen receptor beta (ERβ) expression in breast cancer tissues were examined using immunohistochemistry. The human breast cancer cell line, MCF-7, which expresses ERα but not ERβ was used. CLDN6 and ERα expression were measured by reverse transcriptase-PCR, Western blotting and immunofluorescent staining. Treatments with propyl pyrazole triol (PPT) and ICI 182, 780 (ICI) were performed.
Results
The results revealed that CLDN6 expression was related to ERα in breast cancer tissues (p=0.033). PPT, an ERα-selective ligand, upregulated CLDN6 expression at 10-5 mol/L after 24 hours. The effect of PPT on regulating CLDN6 expression in MCF-7 cells was blocked by ICI.
Conclusion
These findings suggest that Erα reulates CLDN6 expression in breast cancer tissues and that 17β-estradiol induces CLDN6 expression through an ERα pathway in MCF-7 cells.
Keywords: Breast carcinoma, Claudins, Estrogen, Estrogen receptor alpha, Tight junctions
INTRODUCTION
Tight junctions are located at the extreme apical region of junction-associated complexes in epithelial and endothelial cells, where they play important roles maintaining cell polarity, cell adherence and regulating cell proliferation and differentiation [1,2]. Tight junctions are composed of the junctional adhesion molecules, claudins (CLDNs) and occludins [3-5]. CLDNs are major components of tight junctions, forming the backbone of the tight junction strands [2]. CLDNs expression has been reportedly altered in several cancers [6,7]. Whether upregulated or downregulated, the structure and functions of tight junctions are often abnormal in a number of cancers [8]. Tight junction dysfunction has been presumed as a mechanism for the loss of cell adhesion, and an important step for the progression of cancer to metastasis. The expression and functions of CLDNs may be highly tissue- and cell-specific, as CLDNs may be useful molecular markers for many different cancers because of their highly specific expression patterns [7,9]. CLDN proteins may also be promising targets for antibody-based therapy because they are transmembrane proteins with two relatively large extracellular loops [10].
CLDN6 is a member of the CLDN family and plays important roles maintaining the permeability barrier and transepithelial resistance of epidermal cells [11]. In our previous work, we found that CLDN6 expression was downregulated in human and rat mammary cancer cell lines [12]. Furthermore, MCF-7 cells transfected with CLDN6 grew slowly and had a higher death rate than control cells. Anchorage-independent growth, invasive and migratory traits also decreases substantially in CLDN6-expressing cells, and transepithelial electrical resistance increases in the CLDN6-transfected cells [13]. Osanai et al. [14] found that decreased expression of CLDN6 may increase breast tumor formation suggesting that CLDN6 may act as a cancer suppressor, and its downregulation may contribute to the malignant progression of certain types of breast cancers. Conversely, increased CLDN6 expression may decrease breast tumor formation and contribute to the prevention of breast cancer.
In our previous studies, we found that 17β-estradiol upregulates CLDN6 expression in MCF-7 cells [15], and according to current reports, CLDN1, 3, and 4 expression are strongly related to the expression of estrogen receptors (ERs) in breast cancer tissues [16]. However, it is presently unclear how CLDN6 expression is related to ERs. This study sought to determine the role of ERs in the regulation of CLDN6 expression. We examined the CLDN6, ERα and ERβ expression in breast cancer tissues to determine if there might be a correlation between CLDN6 and ERs expression.
METHODS
Immunohistochemical analysis of CLDN6 and ERs
All invasive breast cancers used in our study were obtained from Jilin University. The permission for this study was granted by the Ethical Committee of the School of Basic Medical Sciences, Jilin University. Eighty patients with surgically resected invasive breast carcinoma were investigated. All patients were women (age, 23-65 years) not undergoing any treatments. All samples were designated as infiltrating ductal carcinomas by the specialist committee of Jilin University. All tissues were fixed in 10% paraformaldehyde, and then paraffin-embedded tissues were cut into 4 µm slices, which were stained by immunohistochemistry using the UltraSensitive TMS-P kit (Maxim, Fuzhou, China), according to the manufacturer's instructions. All specimens were stained with 1:1,000 diluted anti-CLDN6 antibody (Santa Cruz Biotechnologies, Santa Cruz, USA), anti-ERα antibody (Bios, Beijing, China) and anti-ERβ (Bios), respectively. DAB color reagent (Bios) was used. Immunostaining was observed under light microscopy with 200x magnification, and five different visual fields in each section were examined. If there was a discrepancy in staining among the five visual fields from the same patient, the overall staining was assessed. Scoring was performed as follows: negative (-), <10% positive tumor cells; positive (+), ≥10% positive tumor cells.
Cell culture and treatment
Human breast adenocarcinoma cell lines (MCF-7 and MDA-MB-231) were cultured in Dulbecco's modified Eagle's medium (DMEM) (GIBCO, Grand Island, USA) containing 10% fetal bovine serum (FBS) (BD, Tokyo, Japan), at 37℃ in an atmosphere containing 5% CO2.
MCF-7 cells were harvested, washed twice by centrifugation in PBS, and then cultured in 6-well plates in DMEM containing 10% FBS for 12 hours. The culture medium was replaced with DMEM without FBS for 12 hours. Cells were treated for 1 hour to 24 hours with different concentrations of propyl pyrazole triol (PPT) (Sigma, St. Louis, USA) (ranging from 10-8 mol/L to 10-5 mol/L), 5×10-9 mol/L 17β-estradiol (Cayman, Denver, USA) and 10-7 mol/L 182780 ICI (ICI) for 24 hours. Control cells were cultured in DMEM only. Vacant vehicle control cells were cultured with DMEM including DMSO at a final concentration of 0.007%.
Reverse transcriptase-PCR (RT-PCR)
Total RNA was extracted from tissue using TRIzol reagent (Invitrogen, Carlsbad, USA). The RNeasy kit (TaKaRa, Dalian, China) was used according to the manufacturer's instructions. RNA concentration was measured at an absorbance at 260 nm. Total RNA (0.5 µg) was used for reverse transcription reactions using M-MLV reverse transcriptase (TaKaRa) and random primers (TaKaRa) at 42℃ for 60 minutes. One µL of the final mixture was used for subsequent PCR reactions using Taq DNA polymerase (TaKaRa). Amplification conditions were as follows: PCR reactions were denatured at 94℃ for 2 minutes, followed by 28 cycles of 94℃ for 30 seconds, 50-60℃ for 30 seconds, and 72℃ for 1 minute. This sequence was followed by a final extension step of 10 minutes at 72℃. β-actin was used as an endogenous control.
The PCR primers were used as follows: CLDN6, 5'-cagtcagctccttcaacct-3' (sense) and 5'-CCATCCAGAAGTGGCAGTG-3' (antisense). β-actin, 5'-CCACTGCGTCGCGGGG-3' (sense) and 5'-GGCAGCCAGCTCAGCCATG-3' (antisense). ERα, 5'-CCTACTACCTGGAGAACGAG-3' (sense) and 5'-CTCT-TCGGTCTTTTCGTATG-3' (antisense), and ERβ, 5'-AAAA-GAATCATTCAATGACA-3' (sense) and 5'-ATTAACACCT-CCATCCAACA-3' (antisense) [17].
The reaction products were resolved on 1.5% agarose gels and visualized by ethidium bromide staining. The images were observed and photographed under an ultraviolet lamp using the Gel Imaging System (Bio-Rad Laboratories Inc., Hercules, USA). The results were analyzed by Quantity One 4.4.1 software (Bio-Rad Laboratories Inc.).
Western blotting
Total protein was extracted using 360 µL of protein extraction fluid (Hua Te Sheng, Beijing, China) with 40 µL (10 mM) phenylmethanesulfonyl fluoride according to the supplier's instructions. Total protein concentration was determined using a BCA Protein Assay Kit (Pierce Chemical Co., Rockford, USA).
For each sample, 60 µg of denatured total protein per lane was resolved by SDS/PAGE (15% acrylamide), and then transferred to a nitrocellulose membrane (Millipore, Billerica, USA) using a Semi-DRY Transfer cell (Bio-Rad Laboratories Inc.). The membrane was blocked with 5% defatted milk (in 25 mM Tris, pH 8.0, 125 mM NaCl, 0.1% Tween 20) for 1 hour at room temperature and then incubated with 1:1,000 diluted anti-CLDN6 (Santa Cruz Biotechnologies) or anti-ERα (Santa Cruz Biotechnologies) antibody at 4℃ overnight. After washing three times with PBS, the membrane was incubated with a horseradish peroxidase conjugated anti-goat IgG or anti-rabbit IgG (1:1,000; Santa Cruz Biotechnologies) secondary antibody at room temperature for 1 hour. After washing, the immunoreactive bands were visualized using an ECL Western blotting system (GE, Buckinghamshire, UK) and exposed to X-ray film (Eastman Kodak, Xiamen, China). Anti-β-actin (Santa Cruz Biotechnologies) antibody was used as the endogenous control.
Immunofluorescence staining
MCF-7 cells were seeded onto small coverslips and treated with 5×10-9 mol/L 17β-estradiol (Cayman, Denver, USA), 10-5 mol/LPPT and 10-7 mol/L ICI for 24 hours. After washing three times with PBS, the cells were fixed with 4% paraformaldehyde for 30 minutes at room temperature. The cells were incubated with 0.3% H2O2 and 30 µL goat serum for 30 minutes, respectively. The cells were stained with 1:500 diluted anti-CLDN6 antibodies (Santa Cruz Biotechnologies) at 4℃ overnight, and 1:50 diluted rhodamine (red)-conjugated anti-goat IgG was used as a secondary antibody. The cells were photographed under a confocal microscope (Olympus, Tokyo, Japan).
Statistical analysis
All data were analyzed with SPSS statistics software (version 13.0; SPSS Inc., Chicago, USA). Relationships between CLDN6 and ERs were assessed by the Pearson's chi-square test. The relative amount of the target mRNA or protein expression was calculated by the ratio of the integrated optical density (IOD) of the target mRNA or target protein to the IOD of the housekeeping gene or protein β-actin. The data are presented as means±standard errors (SE) and evaluated using independent t-tests. A p-value <0.05 was considered statistically significant.
RESULTS
Correlation between expression of CLDN6 and ERs
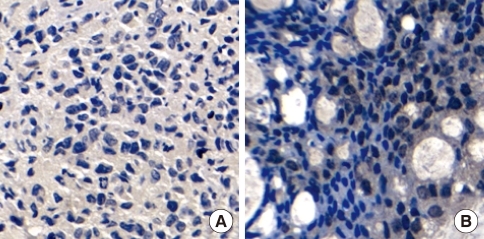
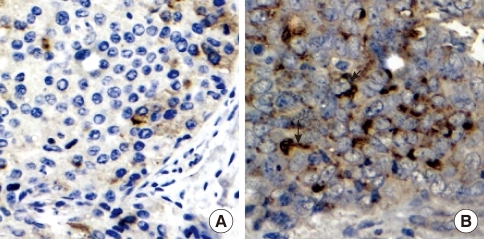
CLDN6 and ERs expression were examined in 80 breast cancer tissues by immunohistochemical analysis to determine whether CLDN6 expression was regulated by estrogen in human breast cancers. All breast cancer tissue sections were stained with specific antibodies to CLDN6, ERα and ERβ, respectively. ERα was located at cell nuclei (Figure 1), and CLDN6 was located on cell membranes (Figure 2). ERβ was located at cell nuclei and cytoplasm (data not shown).
Figure 1.
Estrogen receptor-α (ERα) expression in breast carcinoma, as detected by immunohistochemistry. Staining was specific for tumor tissue, and nuclear staining was observed. Bar=20 µm. (A) Negative expression of ERα in a breast carcinoma. (B) Positive expression of ERα in a breast carcinoma.
Figure 2.
Claudin-6 (CLDN6) expression in a breast carcinoma, as detected by immunohistochemistry. Staining was specific for tumor tissue, and membrane staining was observed. Bar=20 µm. (A) Negative CLDN6 expression in a breast carcinoma. (B) Positive CLDN6 expression in a breast carcinoma.
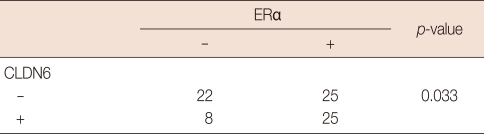
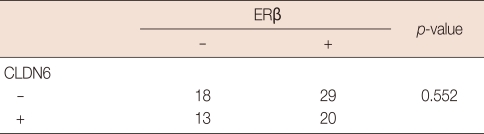
Fifty tumors were stained positive for ERα, and 30 tumors were negative (Table 1). Half (25/50) of the 50 ERα-positive tumors were stained positively for CLDN6, but only 27% (8/30) of the ERα-negative tumors stained positively for CLDN6. The correlation was found between CLDN6 and ERα (p=0.033). However, 40.8% (20/49) of the ERβ-positive tumors stained positively for CLDN6, and 41.9% (13/31) of the ERβ-negative tumors stained positively for CLDN6. No correlation was found between CLDN6 and ERβ (p=0.552) (Table 2).
Table 1.
Correlation between CLDN6 protein and ERα protein in breast cancers
The expression of claudin-6 (CLDN6) and estrogen receptor-α (ERα) was determined by immunohistochemistry, and the correlation between CLDN6 and ERα was determined by the Pearson's chi-square test.
Table 2.
Correlation between CLDN6 protein and ERβ protein in breast cancers
The expression of claudin-6 (CLDN6) and estrogen receptor-β (ERβ) was determined by immunohistochemistry, and the correlation between CLDN6 and ERβ was determined by the Pearson's chi-square test.
Expression of ERα and ERβ in MCF-7 cells
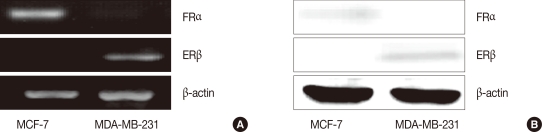
In our previous works, we found that 17β-estradiol induces CLDN6 mRNA and protein expression in MCF-7 cells and that the effects of 17β-estradiol could be blocked by a selective estrogen receptor antagonist, ICI [15]. ERα and ERβ expression was examined in MCF-7 cells to determine with ER subset was involved in the process of 17β-estradiol regulated CLND6 expression. As s result, MCF-7 cells expressed ERα, but not ERβ (Figure 3). MDA-MB-231 cells were used as an ERβ positive control.
Figure 3.
Estrogen receptor-α (ERα) and estrogen receptor-β (ERβ) expression in MCF-7 cells, determined by RT-PCR (A) and Western blot assay (B). MCF-cells, different with MDA MB 231 cells, only expressed ERα, but not ERβ.
PPT induced CLDN6 expression in MCF-7 cells with concentration and time dependence
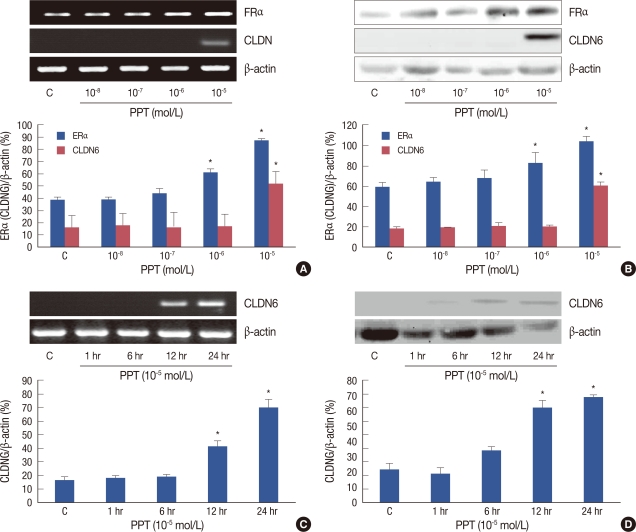
To elucidate the contribution of ERα in the regulation of CLDN6 expression, MCF-7 cells were treated with different concentrations of PPT, a selective ERα agonist (from 10-8 mol/L to 10-5 mol/L), for 1 to 24 hours. ERα and CLDN6 expression were examined using RT-PCR and Western blotting. We found that 10-5 mol/L and 10-6 mol/L PPT could profoundly upregulates ERα expression, but only 10-5 mol/L PPT induced the expression of CLDN6 mRNA and protein levels after 24 hours (Figure 4).
Figure 4.
MCF-7 cells were treated with different concentrations of propyl pyrazole triol (PPT) for 1 to 24 hr. PPT induced estrogen receptor-α (ERα) and claudin-6 (CLDN6) expression in a concentration- and time-dependent manner. (A) RT-PCR was used to determine ERα and CLDN6 expression in MCF-7 cells treated with different concentrations of PPT for 24 hr. The results showed that 10-6 mol/L and 10-5 mol/L PPT had the greatest effect on inducing ERα expression but only 10-5 mol/L PPT induced CLDN6 expression. (B) The Western blot assay was performed to detect ERα and CLDN6 protein expression in MCF-7 cells treated with different concentrations of PPT for 24 hr. The changes in protein levels were coincident with mRNA levels. (C) CLDN6 expression in MCF-7 cells treated with 10-5 mol/L PPT for 1 to 24 hr. The results showed that 10-5 mol/L PPT for 24 hr had the greatest effect on inducing ERα and CLDN6 mRNA expression levels. (D) The Western blot assay confirmed that protein levels were associated with mRNA levels.
*p<0.05, vs. MCF-7 cells (statistically significant).
Effect of ER antagonist on PPT-induced CLDN6 expression
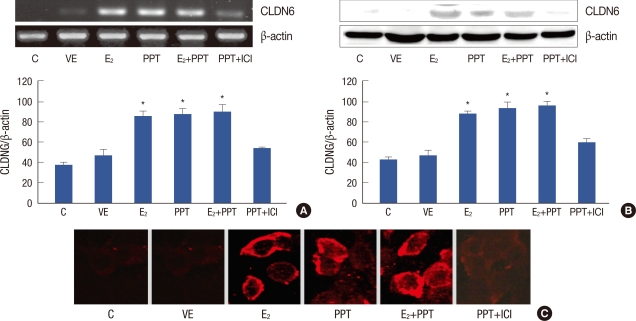
To confirm that CLDN6 expression was regulated by PPT through ERα pathway, MCF-7 cells were treated with ICI, a selective estrogen receptors antagonist, coincident with PPT treatment. CLDN6 mRNA expression was detected using RT-PCR. ICI significantly blocked PPT-induced CLDN6 mRNA expression (Figure 5A). CLDN6 protein expression was detected using Western blotting. The results confirmed that 17β-estradiol and PPT induced CLDN6 protein expression, and that ICI could block the effect (Figure 5B). Immunofluorescent staining showed that 17β-estradiol and PPT induced CLDN6 expression which was located at cell membranes (Figure 5C).
Figure 5.
Effect of ICI182780 (ICI) on propyl pyrazole triol (PPT)-induced claudin-6 (CLDN6) expression. (A) The effect of ICI on PPT-induced CLDN6 mRNA expression was determined by RT-PCR. Lane 1 and Lane 2 showed the expression of CLDN6 in control cells (C) and vacant vehicle control cells (VE) respectively. Lanes 3-5 showed that 5×10-9 mol/L 17β-estradiol (E2) and 10-5 mol/L PPT for 24 hr induced CLDN6 expression respectively or collectively. Lane 6 shows that ICI significantly blocked the effect of PPT. (B) The Western blot assay revealed CLDN6 protein expression in the six groups. Lanes 3-5 showed that CLDN6 protein expression was significantly upregulated in MCF-7 cells treated with 17-β-estradiol or (and) PPT for 24 hr. Lane 6 shows that the effect of PPT could be blocked by ICI. (C) Immunofluoresce staining showed CLDN6 expression on the membranes of MCF-7 cells treated with 17β-estradiol or (and) PPT for 24 hr.
*p<0.05, vs. MCF-7 cells (statistically significant).
DISCUSSION
Infiltrating ductal carcinomas of the breast are believed to arise from the terminal mammary duct. During the process of tumorigenesis and development, mammary epithelial cells acquire mesenchymal characteristics (epithelial/mesenchyme transition, EMT), with altered cell polarity and adhesion, which contributes to the dissemination of tumor cells through blood vessels and lymphatics from the original site [18]. Current reports suggest that tight junction protein expression or locations of specific tight junction-associated expression are altered before the initiation of EMT [19].
Tight junctions are located at the extreme apical region of the junction complex in epithelial and endothelial cells, where they can seal intracellular gaps, maintain cell polarity, adhesion, and permeability as well as regulate cell proliferation and differentiation [1,2].
Tight junctions mainly consist of CLDNs, occludin, junctional adhesion molecules. However, CLDNs are major constituents and determine the barrier properties of tight junctions [18]. CLDN6, one of 24 members of the CLDN family, was first identified by searching for sequences in genomic databases [5]. CLDN6 is similar to CLDN1, 7, 8, 10, 14, 15, 17, and 19 [20], as it four transmembrane domains, and two extracellular loops [21]. CLDN6 is located on chromosome 16 p13.3 and encodes a 23 kDa membrane protein of 219 amino acids [5]. Recent reports show that CLDN6 plays important roles in cells under both physiological and pathological conditions. For example, CLDN6 can regulate adipogenesis and fat deposition [22], it is required for normal blastocyst formation [23], and its expression occurs in early development, with particular attention to definitive endoderm derivatives [24]. CLDN6 is expressed very early in epidermal morphogenesis [1], and it is important for maintaining permeability barriers and transepithelial resistance in epidermal cells [11].
The current study herein has shown that CLDN6 was silenced in esophageal squamous cell carcinoma [25]. In our previous work, we have identified CLDN6 as a potential mammary cancer suppressor gene, which may contribute to the mammary cancer resistant phenotype observed in Copenhagen rats, and CLDN6 expression was undetectable or at low levels in human and rat mammary cancer cell lines [12].
In addition, we demonstrated that MCF-7 cells transfected with CLDN6 grow slower, show an increased rate of cell death, have reduced 2-D and 3-D colony-forming ability, and decreased invasiveness and cell migration [13]. These results suggest that CLDN6 could have beneficial effects for inhibiting carcinogenesis and the malignant progression of certain types of breast cancers. Our further studies found that 17β-estradiol upregulates CLDN6 mRNA and protein expression in MCF-7 cells [15]. However, the regulatory mechanisms involved in the process of 17β-estradiol induced CLDN6 expression are not fully understood.
A significant association exists between CLDN1, 3, and 4, and ERs in breast cancers [16]. However, to our knowledge, no information is available about CLDN6. So, we examined CLDN6 and ERs expression in the tissues of 80 patients with breast cancers using immunohistochemistry, where the results showed a significant association between CLDN6 and ERα expression. In 50 ERα-positive tumors, 25 (50%) stained positive for CLDN6, but only eight of 30 ERα-negative tumors (27%) were positive for CLDN6 (Table 1). But no significant association was observed between CLDN6 and ERβ expression (Table 2). Previous reports have shown that ERα-negative tumors represent a more invasive phenotype and a poorer prognosis compared with ERα-positive breast cancers [26]. The decreased expression of CLDN6 protein in the ERα-negative breast cancers was coincident with our hypothesis that CLDN6 acts as an anti-oncogene, and its expression might be regulated by estrogen via ERα.
A recent report shows that 17β-estradiol has biphasic effects on occludin expression [17]. In our previous study, we found that 17β-estradiol significantly upregulated CLDN6 expression with a maximal effect at 5×10-9 mol/L for 24 hours and that the effect of 17β-estradiol could be blocked by the ERs antagonist ICI [15]. To investigate which ER subset was involved in the process of 17β-estradiol induced CLDN6 expression, we examined the expression of ERs in MCF-7 cells. Similar to an earlier report, MCF-7 cells expressed ERα, but not ERβ (Figure 3) [17]. The ERα agonist PPT and the ERs antagonist ICI were employed in the present study, and the results revealed that CLDN6 mRNA and protein expression could be induced by PPT (Figure 4), and that the induction could be blocked by ICI (Figure 5). We detected the location of CLDN6 protein in MCF-7 cells using immunofluorescent staining, but only the MCF-7 cells treated with 17β-estradiol or (and) PPT showed CLDN6 expression located at the plasma membranes (Figure 5C).
Estrogen has a wide array of cell and tissue-specific effects. The biological effects of estrogen are mediated by ERα and ERβ [27], which are members of a large superfamily of nuclear receptors regulating the expression of target genes via two pathways. The first is a genomic pathway, and the second is non-genomic pathway The latter is mediated by membrane-associated ERs and occurs so rapidly that the effects of estrogen do not depend on RNA and protein synthesis activation [28]. In the classical genomic pathway, estrogen binds to nuclear receptors, the receptors dimerize and bind to specific response elements known as estrogen response elements (EREs) located in target gene promoter [28]. However, a number of studies have shown that ERs can regulate the expression of several genes without binding directly to DNA, but through modulating the function of other transcription factors by protein-protein interactions in the nucleus [29]. The interaction of ERs with the activator protein 1 (AP-1) transcription factor complex is an example of such an ERE-independent genomic actions [30]. This mode of activation regulates the expression of specific downstream genes and may require hours to days in a single cell type [29]. According to our previous works, 17β-estradiol could upregulate CLDN6 mRNA and protein expression in MCF-7 cells after 24 hours [15]. In this study, PPT induced CLDN6 mRNA and protein expression at 24 hours (Figure 4), indicating that estrogen could modify the expression of CLDN6 mRNA and protein through a genomic pathway mediated by ERα.
Our study provides the first direct evidence that CLDN6 expression is associated with ERα in breast cancer tissues, and that PPT could induces CLDN6 mRNA and protein expression in MCF-7 cells. A further study showed that this effect could be blocked by the ER antagonist, ICI. We concluded that estrogen could induce CLDN6 expression via an ERα pathway in breast cancer tissues and MCF-7 cells.
References
- 1.Tsukita S, Furuse M, Itoh M. Multifunctional strands in tight junctions. Nat Rev Mol Cell Biol. 2001;2:285–293. doi: 10.1038/35067088. [DOI] [PubMed] [Google Scholar]
- 2.Matter K, Balda MS. Signalling to and from tight junctions. Nat Rev Mol Cell Biol. 2003;4:225–236. doi: 10.1038/nrm1055. [DOI] [PubMed] [Google Scholar]
- 3.McCarthy KM, Skare IB, Stankewich MC, Furuse M, Tsukita S, Rogers RA, et al. Occludin is a functional component of the tight junction. J Cell Sci. 1996;109(Pt 9):2287–2298. doi: 10.1242/jcs.109.9.2287. [DOI] [PubMed] [Google Scholar]
- 4.Lui WY, Lee WM, Cheng CY. Transforming growth factor-beta3 perturbs the inter-Sertoli tight junction permeability barrier in vitro possibly mediated via its effects on occludin, zonula occludens-1, and claudin-1. Endocrinology. 2001;142:1865–1877. doi: 10.1210/endo.142.5.8116. [DOI] [PubMed] [Google Scholar]
- 5.Morita K, Furuse M, Fujimoto K, Tsukita S. Claudin multigene family encoding four-transmembrane domain protein components of tight junction strands. Proc Natl Acad Sci U S A. 1999;96:511–516. doi: 10.1073/pnas.96.2.511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kramer F, White K, Kubbies M, Swisshelm K, Weber BH. Genomic organization of claudin-1 and its assessment in hereditary and sporadic breast cancer. Hum Genet. 2000;107:249–256. doi: 10.1007/s004390000375. [DOI] [PubMed] [Google Scholar]
- 7.Resnick MB, Konkin T, Routhier J, Sabo E, Pricolo VE. Claudin-1 is a strong prognostic indicator in stage II colonic cancer: a tissue microarray study. Mod Pathol. 2005;18:511–518. doi: 10.1038/modpathol.3800301. [DOI] [PubMed] [Google Scholar]
- 8.Oliveira SS, Morgado-Diaz JA. Claudins: multifunctional players in epithelial tight junctions and their role in cancer. Cell Mol Life Sci. 2007;64:17–28. doi: 10.1007/s00018-006-6314-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cheung ST, Leung KL, Ip YC, Chen X, Fong DY, Ng IO, et al. Claudin-10 expression level is associated with recurrence of primary hepatocellular carcinoma. Clin Cancer Res. 2005;11(2 Pt 1):551–556. [PubMed] [Google Scholar]
- 10.Offner S, Hekele A, Teichmann U, Weinberger S, Gross S, Kufer P, et al. Epithelial tight junction proteins as potential antibody targets for pancarcinoma therapy. Cancer Immunol Immunother. 2005;54:431–445. doi: 10.1007/s00262-004-0613-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Troy TC, Rahbar R, Arabzadeh A, Cheung RM, Turksen K. Delayed epidermal permeability barrier formation and hair follicle aberrations in Inv-Cldn6 mice. Mech Dev. 2005;122:805–819. doi: 10.1016/j.mod.2005.03.001. [DOI] [PubMed] [Google Scholar]
- 12.Quan C, Lu SJ. Identification of genes preferentially expressed in mammary epithelial cells of Copenhagen rat using subtractive hybridization and microarrays. Carcinogenesis. 2003;24:1593–1599. doi: 10.1093/carcin/bgg129. [DOI] [PubMed] [Google Scholar]
- 13.Wu Q, Liu Y, Ren Y, Xu X, Yu L, Li Y, et al. Tight junction protein, claudin-6, downregulates the malignant phenotype of breast carcinoma. Eur J Cancer Prev. 2010;19:186–194. doi: 10.1097/CEJ.0b013e328337210e. [DOI] [PubMed] [Google Scholar]
- 14.Osanai M, Murata M, Chiba H, Kojima T, Sawada N. Epigenetic silencing of claudin-6 promotes anchorage-independent growth of breast carcinoma cells. Cancer Sci. 2007;98:1557–1562. doi: 10.1111/j.1349-7006.2007.00569.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu YF, Wu Q, Xu XM, Ren Y, Yu LN, Quan CS, et al. Effects of 17 beta-estradiol on proliferation and migration of MCF-7 cell by regulating expression of claudin-6. Zhonghua Bing Li Xue Za Zhi. 2010;39:44–47. [PubMed] [Google Scholar]
- 16.Blanchard AA, Skliris GP, Watson PH, Murphy LC, Penner C, Tomes L, et al. Claudins 1, 3, and 4 protein expression in ER negative breast cancer correlates with markers of the basal phenotype. Virchows Arch. 2009;454:647–656. doi: 10.1007/s00428-009-0770-6. [DOI] [PubMed] [Google Scholar]
- 17.Ye L, Martin TA, Parr C, Harrison GM, Mansel RE, Jiang WG. Biphasic effects of 17-beta-estradiol on expression of occludin and transendothelial resistance and paracellular permeability in human vascular endothelial cells. J Cell Physiol. 2003;196:362–369. doi: 10.1002/jcp.10315. [DOI] [PubMed] [Google Scholar]
- 18.Angelow S, Ahlstrom R, Yu AS. Biology of claudins. Am J Physiol Renal Physiol. 2008;295:F867–F876. doi: 10.1152/ajprenal.90264.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Blanco D, Vicent S, Elizegi E, Pino I, Fraga MF, Esteller M, et al. Altered expression of adhesion molecules and epithelial-mesenchymal transition in silica-induced rat lung carcinogenesis. Lab Invest. 2004;84:999–1012. doi: 10.1038/labinvest.3700129. [DOI] [PubMed] [Google Scholar]
- 20.Krause G, Winkler L, Mueller SL, Haseloff RF, Piontek J, Blasig IE. Structure and function of claudins. Biochim Biophys Acta. 2008;1778:631–645. doi: 10.1016/j.bbamem.2007.10.018. [DOI] [PubMed] [Google Scholar]
- 21.Colegio OR, VanItallie C, Rahner C, Anderson JM. Claudin extracellular domains determine paracellular charge selectivity and resistance but not tight junction fibril architecture. Am J Physiol Cell Physiol. 2003;284:C1346–C1354. doi: 10.1152/ajpcell.00547.2002. [DOI] [PubMed] [Google Scholar]
- 22.Hong YH, Hishikawa D, Miyahara H, Nishimura Y, Tsuzuki H, Gotoh C, et al. Up-regulation of the claudin-6 gene in adipogenesis. Biosci Biotechnol Biochem. 2005;69:2117–2121. doi: 10.1271/bbb.69.2117. [DOI] [PubMed] [Google Scholar]
- 23.Moriwaki K, Tsukita S, Furuse M. Tight junctions containing claudin 4 and 6 are essential for blastocyst formation in preimplantation mouse embryos. Dev Biol. 2007;312:509–522. doi: 10.1016/j.ydbio.2007.09.049. [DOI] [PubMed] [Google Scholar]
- 24.Anderson WJ, Zhou Q, Alcalde V, Kaneko OF, Blank LJ, Sherwood RI, et al. Genetic targeting of the endoderm with claudin-6CreER. Dev Dyn. 2008;237:504–512. doi: 10.1002/dvdy.21437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tsunoda S, Smith E, De Young NJ, Wang X, Tian ZQ, Liu JF, et al. Methylation of CLDN6, FBN2, RBP1, RBP4, TFPI2, and TMEFF2 in esophageal squamous cell carcinoma. Oncol Rep. 2009;21:1067–1073. doi: 10.3892/or_00000325. [DOI] [PubMed] [Google Scholar]
- 26.Putti TC, El-Rehim DM, Rakha EA, Paish CE, Lee AH, Pinder SE, et al. Estrogen receptor-negative breast carcinomas: a review of morphology and immunophenotypical analysis. Mod Pathol. 2005;18:26–35. doi: 10.1038/modpathol.3800255. [DOI] [PubMed] [Google Scholar]
- 27.Nilsson S, Makela S, Treuter E, Tujague M, Thomsen J, Andersson G, et al. Mechanisms of estrogen action. Physiol Rev. 2001;81:1535–1565. doi: 10.1152/physrev.2001.81.4.1535. [DOI] [PubMed] [Google Scholar]
- 28.Losel R, Wehling M. Nongenomic actions of steroid hormones. Nat Rev Mol Cell Biol. 2003;4:46–56. doi: 10.1038/nrm1009. [DOI] [PubMed] [Google Scholar]
- 29.Gottlicher M, Heck S, Herrlich P. Transcriptional cross-talk, the second mode of steroid hormone receptor action. J Mol Med. 1998;76:480–489. doi: 10.1007/s001090050242. [DOI] [PubMed] [Google Scholar]
- 30.O'Lone R, Frith MC, Karlsson EK, Hansen U. Genomic targets of nuclear estrogen receptors. Mol Endocrinol. 2004;18:1859–1875. doi: 10.1210/me.2003-0044. [DOI] [PubMed] [Google Scholar]