Abstract
Mammalian genomes contain billions of basepairs of DNA that must be highly compacted as chromatin to fit into the nano-scale of the nucleus, but yet be accessible to allow for transcription to occur. Binding to nucleosomal DNA is critical for ‘pioneer’ transcription factors such as the winged helix transcription factors Foxa1 and Foxa2 to regulate chromatin structure and gene activation. Here we report the genome-wide map of nucleosome positions in the mouse liver, with emphasis on transcriptional start sites, CpG islands, Foxa2 binding sites, and their correlation with gene expression. Despite the heterogeneity of liver tissue, we could clearly discern the nucleosome pattern of the predominant liver cell, the hepatocyte. By analyzing nucleosome occupancy and the distributions of heterochromatin protein 1 (Hp1), CBP (also known as Crebbp), and p300 (Ep300) in Foxa1/2-deficient livers we find, surprisingly, that the maintenance of nucleosome position and chromatin structure surrounding Foxa2 binding sites is independent of Foxa1/2.
Current ultra high-throughput sequencing technologies allow for high-resolution mapping of nucleosome positions throughout the genome1. Thus far, high-resolution maps of nucleosomes have been obtained for C. elegans, yeast, and human cell lines1–3, but not for mammalian tissues. Most transcription factors prefer to bind to double-stranded DNA at nucleosome-free regions1,4; however, previous in vitro studies have also shown that several ‘pioneer’ transcription factors such as the Foxa proteins, the glucocorticoid receptor (Nr3c1), and Sp1 are able to bind to nucleosomal DNA5–7. Gene ablation studies have shown that Foxa1 and Foxa2 redundantly regulate liver development and metabolism, whereas the role of Foxa3 in the liver is limited8–11. Foxa1 and Foxa2 have been suggested to act as ‘pioneer’ factors in liver development12. This model is supported by in vitro studies showing that Foxa proteins decompact chromatin and reposition nucleosomes by binding to nucleosome-occupied DNA at the Albumin (Alb1) enhancer5. However, the role of the Foxa factors with respect to nucleosome positioning in vivo remains to be determined.
RESULTS
Mapping nucleosomes with MNase-Seq or H3K4me1 ChIP-Seq
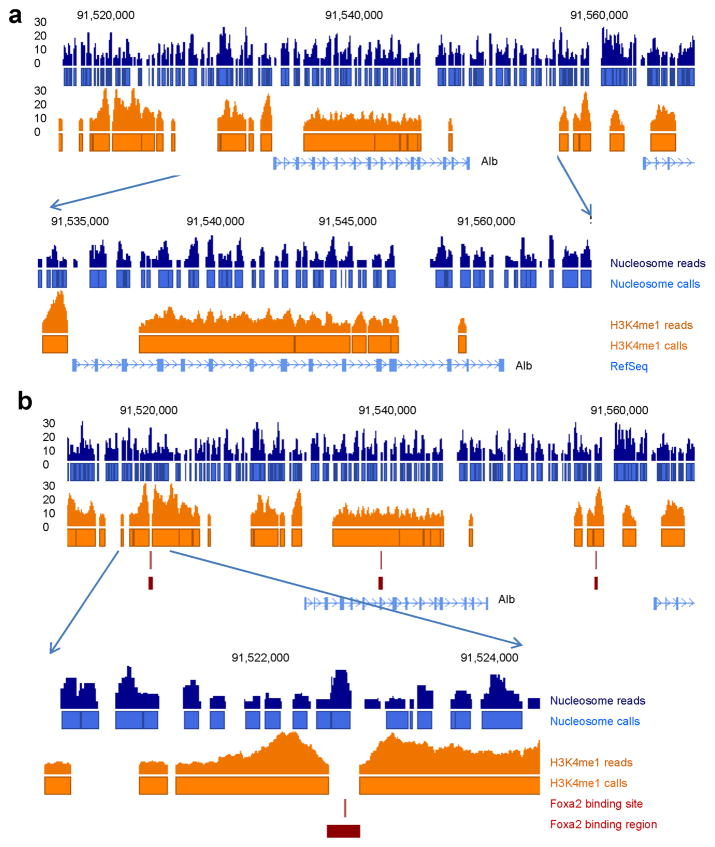
Presently, only very limited information is available regarding nucleosome positions in the mammalian liver. Recently, Hoffman and colleagues used limited reads from ChIP-Seq for H3K4me1 as the surrogate to approximate nucleosome positions in the mouse liver and concluded that H3K4me1 occupancy was sufficient to define most of the Foxa2 binding sites in the genome by forming ‘bi-modal’ nucleosomal boundaries surrounding Foxa2 binding sites13. This shortcut, while convenient and cost-saving, suffers from several short-comings. First, by definition, only histones with this specific modification, i.e. H3K3me1 were ‘counted’; thus, the vast majority of nucleosomes were missed. Secondly, ChIP-Seq for H3K4me1 is limited in resolution due to the sonication process; thus, the precise boundary of the nucleosome could not be determined. This issue is illustrated in Figure 1, where we compare nucleosome positions at the Albumin (Alb) locus obtained by Microccocal Nuclease digestion (this study, see below) to the nucleosome occupancy estimated by Hoffman and colleagues. Figure 1a demonstrates how, when relying only on H3K4me1 ChIP-Seq data, many adjacent nucleosomes were fused together into one nucleosomal area, while others were completely missed. In addition, the well-studied Foxa2 binding site in the Albumin enhancer had been reported as “bi-modal”, or flanked by nucleosomes, by ChIP-Seq for H3K4me113. However, we could not reproduce this finding, as our analysis clearly showed that there is a nucleosome positioned exactly over the Foxa2 binding site (Fig. 1b). Thus, the determination of nucleosome occupancy by Microccocal Nuclease digestion followed by ultra-high throughput sequencing is more powerful to resolve nucleosome position than relying solely on ChIP-Seq for H3K4me1.
Figure 1.
Mapping of nucleosome positions by Micrococcal nuclease is more informative than the use of H3K4me1 ChIP-Seq data. (a) Blue, nucleosome positions mapped by Micrococcal nuclease digestion followed by ultra-high throughput sequencing; orange, H3K4me1 ChIP-Seq data from mouse liver based on a cutoff of more than six reads per position13. Nucleosome positions estimated from H3K4me1 ChIP-Seq data are unable to resolve closely spaced nucleosomes and also miss multiple nucleosomes detected by mapping of nucleosomes with micrococcal nuclease. Near the 5′ end of the Alb1 gene, H3K4me1 ChIP-Seq data miss two important nucleosomes just downstream of the transcriptional start site, most likely because here H3 was methylated further to the me2 and me3 forms. (b) The well-known Foxa binding site in the albumin enhancer was called as “bi-modal”, i.e. nucleosome-free, by Hoffman and colleagues13, however, our data clearly show the site occupied by a nucleosome.
Nucleosome dynamics in the mouse liver
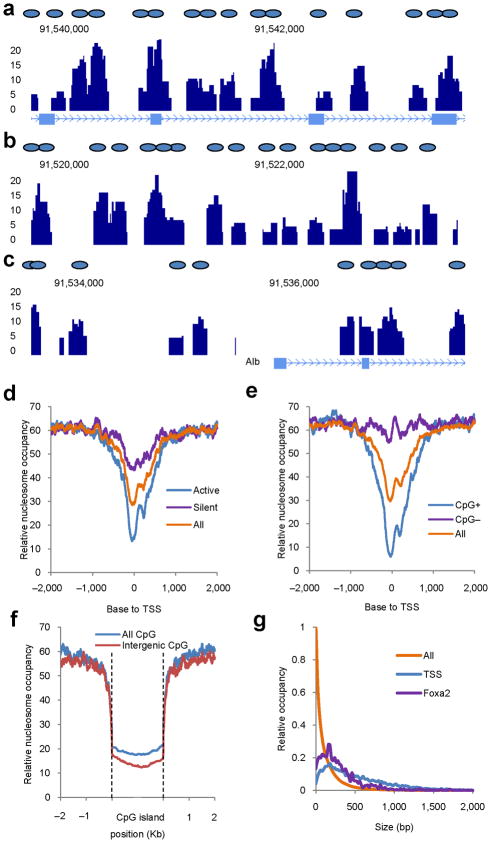
To investigate nucleosome occupancy in the mouse liver genome-wide, we isolated native chromatin and directly digested it with Microccocal Nuclease (MNase), which cannot digest DNA wrapped around the histone octamer (Supplementary Fig. 1). Using ultra high-throughput sequencing, we obtained ~124 million uniquely aligned sequence reads, resulting in a genome-wide map of nucleosome occupancy. Strikingly, despite the fact that the liver is not a homogenous tissue, the nucleosome profile exhibited a distinct pattern that reflected the status of chromatin in the predominant cell type, the hepatocyte. The nucleosome occupancy at the Alb1 locus is shown as an example in Figure 2a–c. Three major features of nucleosome occupancy were found at the Alb1 locus and other hepatocyte-expressed genes. First, nucleosome positions were similar in the majority of hepatocytes; second, nucleosomes were not equally spaced even in nucleosome-rich regions; third, a wide nucleosome-free region was found surrounding transcriptional start sites.
Figure 2.
Nucleosome dynamics in the mouse liver. (a–c) Nucleosome map of the mouse liver. Stack height profiles of nucleosome reads represent nucleosome occupancy in the gene body (a), enhancer (b) and surrounding the transcriptional start site (TSS) (c) of the Albumin gene. Blue ovals represent putative nucleosome positions. (d) Nucleosome distribution surrounding transcriptional start sites (‘All’, all annotated genes from the UCSC genome database; ‘Active’, 1,000 genes with the highest expression in the liver; ‘Silent’, 1,000 genes with no expression in the liver. (e) Nucleosome distribution surrounding TSS of genes associated with (CpG+) and without (CpG−) CpG islands. (f) Nucleosome distribution surrounding CpG islands. Either all CpG islands or those located more than 5 Kb distant to the nearest gene (Intergenic) were analyzed. (g) Sizes of nucleosome-free regions at Foxa2 binding sites, surrounding transcriptional start sites, or in the whole genome (All).
Genome-wide mapping of nucleosome positions provides the opportunity to study the link between nucleosome position and well-known genomic landmarks such as transcriptional start sites and CpG islands, the latter predominantly found near housekeeping genes14,15. The analysis of nucleosome distribution surrounding transcriptional start sites revealed that ~70% of them were nucleosome-free, with the nucleosome-free regions spanning approximately one to two nucleosome widths (Fig. 2d). Similar to what has been found in cultured cells1,16, transcriptional start sites of active genes showed lower nucleosome occupancy than those of silent genes (Fig. 2d). This is exemplified by the nucleosome occupancy of the Alb1 locus, which is active in the liver, and the Insulin 1 locus, which is not (Fig. 2c, Supplementary Fig. 2). Thus, the initiation of gene transcription in the liver depends, at least in part, on displacement of nucleosomes from transcriptional start sites. As expected, CpG island-associated genes showed significantly lower nucleosome occupancy surrounding transcriptional start sites than genes lacking CpG islands (Fig. 2e, Supplementary Fig. 3). CpG islands themselves were nucleosome-depleted even at intergenic locations (Fig. 2f, Supplementary Fig. 3). The genome-wide nucleosome coverage was about 60% (Fig. 2d–f). We also investigated the size of the nucleosome-free regions in the whole genome and near transcriptional start sites. The majority of nucleosome-free regions in the whole genome were less than 100 bp in size, while the nucleosome-free regions surrounding transcriptional start sites had a wider range (Fig. 2g), indicating uneven spacing between adjacent nucleosomes. In addition, we compared nucleosome spacing at gene body regions between active and silent genes; however, we found no significant differences (Supplementary Figure 4).
Genome-wide impact of Foxa2 on nucleosome positioning
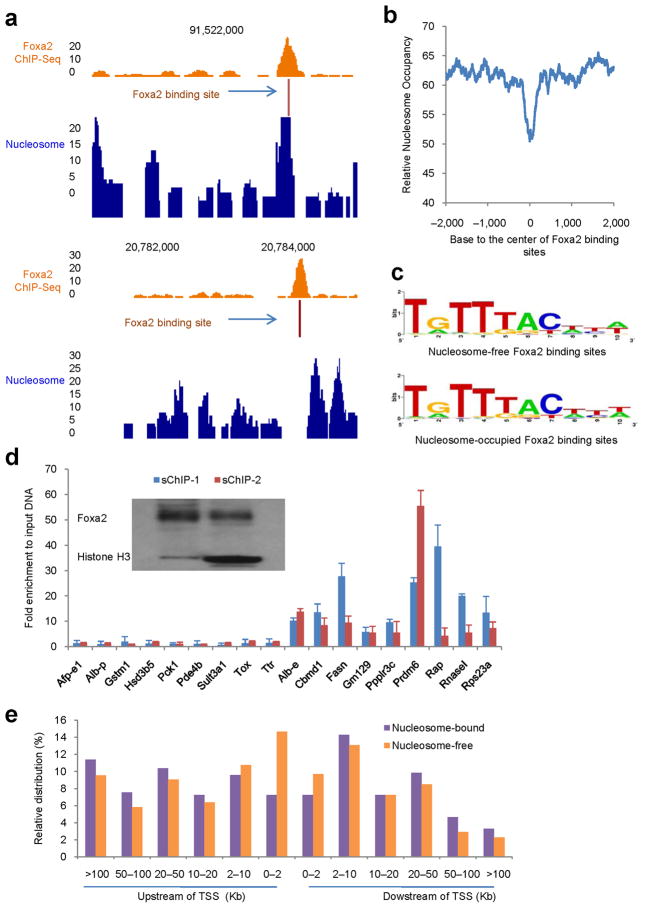
Foxa1 and Foxa2 have similar DNA binding domains (>95% amino acid identity) and share similar DNA target sequences17–19. We mapped all Foxa2 binding sites in the mouse liver by ChIP-Seq and identified 8,102 Foxa2 bound regions, corresponding to 4,435 genes. We found that Foxa2 bound to both nucleosome-free and –occupied DNA (two examples are shown in Fig. 3a). From the analysis of nucleosome occupancy at all Foxa2 binding sites, Foxa2 showed a limited preference for nucleosome-free regions (Fig. 3b). The majority of nucleosome-free regions surrounding Foxa2 binding sites were ~200 bp in width (Fig. 2g). Among all Foxa2 binding sites, at least half were nucleosome-free, while the rest showed evidence for co-occupancy with nucleosomes (Supplementary Fig. 5). The two sets of binding sites produced nearly identical positional weight matrices, indicating that no specific sequence directs Foxa2 to bind to nucleosomal DNA (Fig. 3c).
Figure 3.
Genome-wide distributions of Foxa2 and nucleosomes in the adult mouse liver. (a) Examples of Foxa2 occupancy at a nucleosome-bound (top) or nucleosome-free (bottom) binding site. The 10-bp sequence with the best match to the Foxa2 positional weight matrix is denoted as “Foxa2 binding site”. (b) Nucleosome occupancy surrounding Foxa2 binding sites. 0 indicates the center of 10-bp sequence with the best match to the Foxa2 positional weight matrix in each Foxa2 bound region. Note the relative nucleosome depletion at the center of Foxa2 bound sites. (c) Consensus binding sites of nucleosome-free and nucleosome-occupied Foxa2 do not differ significantly. (d) Direct interactions between Foxa2 and histones. Sequential ChIPs (sChIP) were performed with MNase-digested chromatin and analyzed by qPCR with 18 primers pairs. Nine of these corresponded to Foxa2 binding sites in nucleosome-free genomic regions (left side of graph), while nine corresponded to nucleosome-bound Foxa2 binding sites (right side of graph). The order of sChIP: sChIP-1 (anti-Foxa2 → anti-Histone H3) and sChIP-2 (anti-Histone H3 → anti-Foxa2). Regardless of ChIP order, all nucleosome-bound Foxa2 targets were confirmed as such by sChIP. Insert is the western blot using sChIP samples with anti-Foxa2 and anti-Histone H3 antibodies. (e) Distributions of Foxa2 binding sites at upstream and downstream of transcriptional start sites.
To confirm the interaction between Foxa2 and nucleosomes, we performed sequential ChIPs with anti-Foxa2 and anti-Histone H3 antibodies in both orders. The nucleosome-coupled Foxa2 targets were indeed occupied by nucleosomes, in contrast to the Foxa2 sites in nucleosome-free regions (Fig. 3d). In addition, we found Foxa2 binding sites even at significant distances from transcriptional start sites (Fig. 3e). There was no difference in the distribution of nucleosome-bound and –free Foxa2 binding sites, with the exception of the regions from transcriptional start sites to two kb upstream, which were enriched for nucleosome-free Foxa2 binding events (Fig. 3e). Finally, we investigated the correlation between nucleosome occupancy at Foxa2 binding sites and gene expression of the closest gene (Supplementary Fig. 6). 20% of the genes analyzed were associated with both nucleosome-free and –occupied Foxa2 binding sites. There was no difference in the level of gene expression between those genes that were only associated with nucleosome-free Foxa2 binding sites compared to those with only nucleosome-bound Foxa2 sites (Supplementary Fig. 6). These data indicate that nucleosome occupancy at Foxa2 binding sites does not correlate with gene expression levels in the liver.
Smale and colleagues reported recently that the methylation status of CpG sites near the Foxa binding site of the Alb1 enhancer plays an essential role in the differentiation of embryonic stem cells to endoderm in vitro20. Considering the recent finding that nucleosomal DNA is enriched with methylated CpGs21, we expected that the CpG sites in the Alb1 enhancer would be highly methylated and occupied by nucleosomes. Indeed, the classical Alb1 enhancer was bound by both Foxa2 and nucleosomes in the liver (Fig. 3a, upper panel).
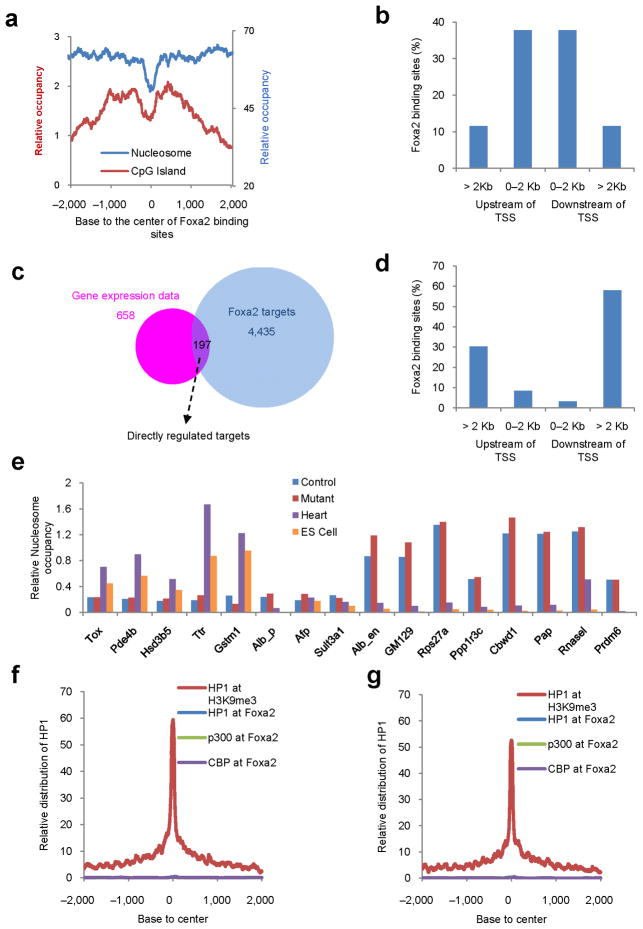
This finding seemed to be inconsistent with our data showing that intergenic CpG islands were nucleosome-depleted (Fig. 2f). Therefore, we investigated the relationship between CpG islands and Foxa2 binding sites on the genome-wide level. Surprisingly, the genome-wide distribution of CpG islands surrounding Foxa2 binding sites showed that CpG islands were depleted at Foxa2 binding sites but relatively enriched nearby, either upstream or downstream of the Foxa2 binding site (Fig. 4a, Supplementary Fig. 7). However, Foxa2 binding sites with nearby CpG islands only accounted for ~8% (720 sites) of all Foxa2 sites. Interestingly, this particular group of Foxa2 binding sites mostly resides within two kb of transcriptional start sites, in contrast to the much broader distribution of all Foxa2 binding sites (Fig. 4b, 3e). Thus, CpG islands form boundaries near a subset of Foxa2 binding sites.
Figure 4.
The maintenance of nucleosome position and chromatin structure is independent of Foxa1/2 in the adult liver. (a) Promoter-proximal Foxa2 binding sites are frequently surrounded by CpG islands. Distribution of CpG islands and nucleosomes surrounding Foxa2 binding sites. (b) Distribution of CpG bounded Foxa2 binding sites upstream and downstream of transcriptional start sites. (c) Intersection of differentially expressed genes in Foxa1/2 mutant liver and Foxa2 ChIP-Seq data. Gene expression data were differentially expressed genes from microarray analysis between Foxa1loxP/loxP;Foxa2loxP/loxP;AlfpCre (Mutant) and control mice; Foxa2 targets were Foxa2-targeted genes from ChIP-Seq analysis. (d) Distribution of Foxa2 binding sites upstream and downstream of transcriptional start sites for those sites associated with directly-regulated genes. A total of 369 Foxa2 binding sites were associated with the 197 differentially expressed genes. (e) Nucleosome occupancy surrounding Foxa2 binding sites associated with genes including thymocyte selection-associated high mobility group box (Tox), cAMP specific phosphodiesterase 4B (Pde4b), hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 5 (Hsd3b5), transthyretin (Ttr), glutathionine S-transferase m1 (Gstm1), albumin promoter (Alb_p), alpha fetoprotein enhancer (Afp_e), sulfotransferase family 3A, member 1 (Sult3a1), the albumin enhancer (Alb1_e), gene model 129 (GM129), ribosomal protein S27A (Rps27a), protein phosphatase 1, regulatory (inhibitor) subunit 3C (Ppp1r3c), COBW domain containing 1 (Cbwd1), poly A polymerase alpha (Pap), ribonuclease L (Rnasel) and PR domain containing 6 (Prdm6), in livers from Foxa1loxP/loxP;Foxa2loxP/loxP;AlfpCre (Mutant) and control mice, mouse heart and mouse embryonic stem cells (ES Cell). (f, g) Distribution of heterochromatin protein 1 (Hp1), CBP, and p300 surrounding Foxa2 binding sites, or Hp1 surrounding H3K9me3 sites in control (f) and Foxa1/2 mutant (g) livers. 0 corresponds to the center of Foxa2 binding sites or H3K9me3 sites, respectively.
Nucleosome position independent of Foxa1/2 in the adult liver
To investigate the impact of the Foxa proteins on nucleosome positioning in the adult liver, we performed loss-of-function analysis for both Foxa1 and Foxa2 using Cre-loxP technology11, in which Foxa1 and Foxa2 were specifically ablated in the liver at ~ day 14.5 of gestation. Gene expression profiling revealed that 658 genes were found to display at least a 2-fold change in mRNA expression level between mutant and control liver. By intersecting the set of differentially expressed genes with our Foxa2 target list from ChIP-Seq, we found 197 of the differentially expressed genes to be directly regulated by Foxa2 (Fig. 4c). Interestingly, the 369 Foxa2 binding sites regulating these 197 genes were localized mostly at enhancer regions at least two kb away from transcriptional start sites (Fig. 4d). In addition, there was no significant difference in the nucleosome occupancy pattern at Foxa2 binding of genes whose expression was altered in Foxa1/a2 deficient liver compared to those that were not (data not shown).
Next, we asked whether nucleosomes are able to re-occupy the nucleosome-free regions at Foxa binding sites when Foxa1 and Foxa2 are deleted in the liver. Strikingly, we found no change in nucleosome occupancy in Foxa1/2 mutant liver for sixteen randomly chosen genomic loci, regardless of whether they were nucleosome-free or nucleosome–occupied in normal liver (Fig. 4e). We could not analyze mice deficient for Foxa1, Foxa2 and Foxa3, since these mice are not viable. As alternative, we analyzed cells and tissues without any Foxa expression, including undifferentiated mouse embryonic stem (ES) cells and mouse heart. Several Foxa2 binding sites that were nucleosome-free in the liver were indeed occupied by nucleosomes in ES cells and heart (5 out of 8 regions tested), suggesting that Foxa proteins might be required to open chromatin at these regions (Fig. 4e). However, three of the nucleosome-free Foxa2 binding sites in the liver were still nucleosome-free even in heart and ES cells (Fig. 4e). Conversely, eight Foxa2 binding sites that were nucleosome-bound in the liver were completely nucleosome-free in ES cells and heart (Fig. 4e). None of the regions tested were occupied by nucleosomes in all four tissues or cell types. These data suggest that nucleosome positioning is independent of Foxa1/2 in the adult mouse liver, but do not preclude the possibility that Foxa factors reposition nucleosomes during embryonic development, or that Foxa3, Foxd3 and other Fox factors may bind to Foxa1/2 binding sites when Foxa1 and Foxa2 are absent. However, we previously showed that no liver forms when both Foxa1 and Foxa2 are ablated during gastrulation10, suggesting that Foxa3 is unable to compensate the loss of Foxa1/2. Foxd3 expression is high during early development but declines in adulthood, and is not detectable in the adult liver22.
Next, we investigated whether lack of Foxa1/2 leads to changes in chromatin structure. Hp1 and trimethylated histone H3 lysine 9 (H3K9me3) are genomic markers of heterochromatin23,24. Following ChIP-Seq, we plotted the Hp1 distribution surrounding all Foxa2 binding sites and H3K9me3 sites. While H3K9me3 sites were highly enriched for Hp1 binding, Foxa2 binding sites were Hp1-free, indicating that Foxa2 bound at euchromatic regions (Fig. 4f). Surprisingly, this was also true for these sites in Foxa1/2-deficient livers (Fig. 4g). CBP and p300 mark genomic regions under the transition from heterochromatin to euchromatin, and enhancer regions25,26. We also investigated genome-wide locations of CBP and p300 in both control and mutant livers by ChIP-Seq. Genome-wide distribution analysis showed that most (>90%) of CBP and p300 binding sites were localized at putative enhancer regions (> 5 kb from the transcription starting site) in both control and mutant livers (Supplementary Fig. 8). Just as for Hp1, CBP and p300 binding was also absent from most Foxa2 binding sites in both control and mutant livers (Fig. 4f, g). Thus, Foxa1/2 are dispensable for the maintenance of chromatin structure in the liver, at least at the regions surrounding Foxa2 binding sites.
Our study is the first to report genome-wide mapping of nucleosome positions in the mouse genome. Mapping of nucleosome positions in an organ rather than cultured cell lines is challenging because of tissue heterogeneity. Nevertheless, we were able to identify distinct nucleosome positions in the liver, because hepatocytes account for ~80% of liver nuclei. Our findings indicate further that nucleosome positions are nearly uniform in hepatocytes despite the zonation of the liver into periportal and pericentral regions. Our high-resolution maps of nucleosomes revealed nucleosome depletion at transcriptional start sites of house-keeping genes, highly expressed genes and some silent genes, and also at CpG-islands.
DISCUSSION
Although overall genome-wide Foxa2 binding did not correlate with reduced nucleosome occupancy and altered gene expression, we discovered that a subset of Foxa2 binding sites near promoters were bounded by CpG islands. Combined with genome-wide mapping of Foxa2 occupancy and gene ablation analysis, we found that Foxa1 and Foxa2 are dispensable for maintaining nucleosome positions and chromatin structure in the adult mouse liver. Our findings and prior in vitro studies would suggest that Foxa proteins act as ‘pioneer’ factors in the initial reprogramming of chromatin during endoderm development when other transcription factors are not abundant. While prior genetic data support the notion of the Foxa factors as pioneer factors10, other interpretations are also possible. For instance, the Foxa proteins could regulate a set of key regulatory molecules required for liver specification, and not act by globally modifying chromatin. Nevertheless, our current findings explain why hepatocyte differentiation was not affected dramatically when Foxa1/2 were ablated in the liver during late gestation11.
METHODS
Methods and any associated references are available in the online version of the paper at http://www.nature.com/nsmb/.
Supplementary Material
Acknowledgments
We thank Alan Fox, Olga Smirnova, Karrie Brondell, Andrew Chen, Amber Riblett and James LaRossa for excellent technical support. This study was supported by the NIDDK (P01-DK049210 to K.H.K.). Z. Li was supported by NSREC and JDRF postdoctoral fellowship awards. We would like to acknowledge support of the IDOM Functional Genomics Core by an NIDDK Research Center grant (P30DK19525).
Footnotes
AUTHOR CONTRIBUTIONS
Z.L. did the majority of experiments and computational analysis. J.S. and G.T. did a part of experiments and computational analysis. K.H.K. directed the whole study. Z.L. and K.H.K. designed the experiments and wrote the manuscript.
COMPETING INTERESTS STATEMENT
The authors declare no competing financial interests.
References
- 1.Schones DE, et al. Dynamic regulation of nucleosome positioning in the human genome. Cell. 2008;132:887–98. doi: 10.1016/j.cell.2008.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shivaswamy S, et al. Dynamic remodeling of individual nucleosomes across a eukaryotic genome in response to transcriptional perturbation. PLoS Biol. 2008;6:e65. doi: 10.1371/journal.pbio.0060065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Valouev A, et al. A high-resolution, nucleosome position map of C. elegans reveals a lack of universal sequence-dictated positioning. Genome Res. 2008;18:1051–63. doi: 10.1101/gr.076463.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beato M, Eisfeld K. Transcription factor access to chromatin. Nucleic Acids Res. 1997;25:3559–63. doi: 10.1093/nar/25.18.3559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McPherson CE, Shim EY, Friedman DS, Zaret KS. An active tissue-specific enhancer and bound transcription factors existing in a precisely positioned nucleosomal array. Cell. 1993;75:387–98. doi: 10.1016/0092-8674(93)80079-t. [DOI] [PubMed] [Google Scholar]
- 6.Chen H, Li B, Workman JL. A histone-binding protein, nucleoplasmin, stimulates transcription factor binding to nucleosomes and factor-induced nucleosome disassembly. EMBO J. 1994;13:380–90. doi: 10.1002/j.1460-2075.1994.tb06272.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li B, Adams CC, Workman JL. Nucleosome binding by the constitutive transcription factor Sp1. J Biol Chem. 1994;269:7756–63. [PubMed] [Google Scholar]
- 8.Kaestner KH, Hiemisch H, Schutz G. Targeted disruption of the gene encoding hepatocyte nuclear factor 3gamma results in reduced transcription of hepatocyte-specific genes. Mol Cell Biol. 1998;18:4245–51. doi: 10.1128/mcb.18.7.4245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shen W, Scearce LM, Brestelli JE, Sund NJ, Kaestner KH. Foxa3 (hepatocyte nuclear factor 3gamma) is required for the regulation of hepatic GLUT2 expression and the maintenance of glucose homeostasis during a prolonged fast. J Biol Chem. 2001;276:42812–7. doi: 10.1074/jbc.M106344200. [DOI] [PubMed] [Google Scholar]
- 10.Lee CS, Friedman JR, Fulmer JT, Kaestner KH. The initiation of liver development is dependent on Foxa transcription factors. Nature. 2005;435:944–7. doi: 10.1038/nature03649. [DOI] [PubMed] [Google Scholar]
- 11.Li Z, et al. Foxa1 and Foxa2 regulate bile duct development in mice. J Clin Invest. 2009 doi: 10.1172/JCI38201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zaret K. Developmental competence of the gut endoderm: genetic potentiation by GATA and HNF3/fork head proteins. Dev Biol. 1999;209:1–10. doi: 10.1006/dbio.1999.9228. [DOI] [PubMed] [Google Scholar]
- 13.Hoffman BG, et al. Locus co-occupancy, nucleosome positioning, and H3K4me1 regulate the functionality of FOXA2-, HNF4A-, and PDX1-bound loci in islets and liver. Genome Res. 20:1037–51. doi: 10.1101/gr.104356.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gardiner-Garden M, Frommer M. CpG islands in vertebrate genomes. J Mol Biol. 1987;196:261–82. doi: 10.1016/0022-2836(87)90689-9. [DOI] [PubMed] [Google Scholar]
- 15.Kundu TK, Rao MR. CpG islands in chromatin organization and gene expression. J Biochem. 1999;125:217–22. doi: 10.1093/oxfordjournals.jbchem.a022276. [DOI] [PubMed] [Google Scholar]
- 16.Yaragatti M, Basilico C, Dailey L. Identification of active transcriptional regulatory modules by the functional assay of DNA from nucleosome-free regions. Genome Res. 2008;18:930–8. doi: 10.1101/gr.073460.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lai E, Prezioso VR, Tao WF, Chen WS, Darnell JE., Jr Hepatocyte nuclear factor 3 alpha belongs to a gene family in mammals that is homologous to the Drosophila homeotic gene fork head. Genes Dev. 1991;5:416–27. doi: 10.1101/gad.5.3.416. [DOI] [PubMed] [Google Scholar]
- 18.Tuteja G, Jensen ST, White P, Kaestner KH. Cis-regulatory modules in the mammalian liver: composition depends on strength of Foxa2 consensus site. Nucleic Acids Res. 2008;36:4149–57. doi: 10.1093/nar/gkn366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lupien M, et al. FoxA1 translates epigenetic signatures into enhancer-driven lineage-specific transcription. Cell. 2008;132:958–70. doi: 10.1016/j.cell.2008.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xu J, et al. Transcriptional competence and the active marking of tissue-specific enhancers by defined transcription factors in embryonic and induced pluripotent stem cells. Genes Dev. 2009;23:2824–38. doi: 10.1101/gad.1861209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chodavarapu RK, et al. Relationship between nucleosome positioning and DNA methylation. Nature. 2010;466:388–92. doi: 10.1038/nature09147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sutton J, et al. Genesis, a winged helix transcriptional repressor with expression restricted to embryonic stem cells. J Biol Chem. 1996;271:23126–33. doi: 10.1074/jbc.271.38.23126. [DOI] [PubMed] [Google Scholar]
- 23.Fleischmann G, Filipski R, Elgin SC. Isolation and distribution of a Drosophila protein preferentially associated with inactive regions of the genome. Chromosoma. 1987;96:83–90. doi: 10.1007/BF00285889. [DOI] [PubMed] [Google Scholar]
- 24.Lachner M, O’Carroll D, Rea S, Mechtler K, Jenuwein T. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature. 2001;410:116–20. doi: 10.1038/35065132. [DOI] [PubMed] [Google Scholar]
- 25.Visel A, et al. ChIP-seq accurately predicts tissue-specific activity of enhancers. Nature. 2009;457:854–8. doi: 10.1038/nature07730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Heintzman ND, et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet. 2007;39:311–8. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.