Abstract
The large post-reproductive life span reported for the free-living hermaphroditic nematode, Caenorhabditis elegans, which lives for about 10 days after its 5-day period of self-reproduction, seems at odds with evolutionary theory. Species with long post-reproductive life spans such as mammals are sometimes explained by a need for parental care or transfer of information. This does not seem a suitable explanation for C elegans. Previous reports have shown that C elegans can regain fertility when mated after the self-fertile period but did not report the functional limits. Here, we report the functional life span of the C elegans germ line when mating with males. We show that C elegans can regain fertility late in life (significantly later than in previous reports) and that the end of this period corresponds quite well to its 3-week total life span. Genetic analysis reveals that late-life fertility is controlled by conserved pathways involved with aging and dietary restriction.
Keywords: Reproduction, Aging, Evolution, Dietary restriction, Genetics
MULTICELLULAR organisms must respond to changes in the environment by altering their pattern of allocation of resources between growth (soma) and reproduction (germ line) in order to maximize lifetime reproductive fitness. These responses are mediated by a complex set of still largely undefined interacting genetic networks that integrate energy acquisition and utilization, developmental and reproductive timing, and behavioral and sexual interactions. The tilt in the balance of these interactions toward early reproduction and the ultimate precedence of the germ line over the soma are hypothesized to be the ultimate causes of aging and senescence—bodies simply cannot be maintained indefinitely against inevitable environmental damage when that maintenance comes at the cost of a reproductive disadvantage. In its most extreme form, there will be no natural selection to maintain the soma once reproduction has ceased, unless there is some secondary contribution that the old can make to young relatives. Indeed, evolutionary theory predicts that one should see a “wave of death” late in life in most organisms (1–3). Thus, it is perhaps not surprising that mortality dramatically increases after female (hermaphrodite for Caenorhabditis elegans, which are essentially females that produce their own sperm; the hermaphrodite gonad is explained in detail in the Results) reproductive senescence in species that range from flies to humans (1,3–5). Caenorhabditis elegans has been an unexplained outlier in this trend, spending approximately 70% of its life in a reproductively senescent state—much more, for instance, than that of primates and other mammals (4). Is this simply an artifact of laboratory rearing or are there important portions of the biology of this nematode that have been overlooked in this important model of aging and senescence?
Caenorhabditis elegans, which exist mostly as hermaphrodites, typically finish self-reproduction by about the fifth day of adult life (6,7). Surprisingly, these animals can live for another 2 weeks (6,8). Many mutations that extend organismal life span (Age) have been identified in the nematode. The first longevity mutant, age-1, shows as much as a 10-fold extension of somatic life (8,9); as in the wild type, all of this life extension is post-reproductive (10). In a few Age mutants, a modest extension of the period of self-fertility is seen (a relatively low number of progeny for a few days beyond wild type), but it comes at a cost of total progeny production (11–14). In addition to these results, two studies have recently demonstrated that C elegans can in fact display delayed reproductive senescence when hermaphrodite sperm is supplemented with sperm from males via mating (11,15); importantly, neither study determined the age limits of cross-fertility. It also now appears that there are multiple developmental checkpoints in which development and reproduction can be halted and then restarted in response to starvation (16–23). We would expect such factors to be important in longevity and reproduction because allocation to somatic maintenance versus reproductive output should vary in response to the availability and uncertainty of resources. Thus, there is reason to believe that standard picture of “normal” reproduction and longevity in C elegans, especially in the face of environmental mediators, may be incomplete.
One gap in our understanding of reproductive life span is that no prior study has reported the functional life span of the germ line with respect to mating late in life. The latest age at which cross-fertility was examined in prior studies was Day 10 of adulthood (11). The maximum age at which hermaphrodites could become impregnated by males was not determined. Thus, the functional life span of the hermaphrodite germ line is not known when male mating is taken into account; life span (or functional life span) cannot be determined without observing function until cessation. Here, we measure germ line longevity by examining the capacity for C elegans hermaphrodites to respond to cross-fertilization after self-reproductive senescence—right up to the longevity barrier generated by somatic senescence. We find that a set of known longevity-extension mutants, representative of most classes of life-span extension, have little effect on germ line maintenance, whereas dietary input is critical. Thus, the structure of the genetic systems governing the interplay between reproduction and longevity in C elegans may be more complex than previously believed.
EXPERIMENTAL PROCEDURES
Strains and Growth Conditions
All strains were grown and assayed at 20°C on standard NGM under standard laboratory conditions except for SM190 animals, and their wild-type controls, which were grown according to Panowski and colleagues (24) followed by assay at 20°C as in (24). Strains used in these experiments were as follows: N2 CGCb (wild type), SM190 (pha-4(zu225)), NP717 (coelomocytes-less mutant), JK574 (fog-2(q71)), CB1370 (daf-2(e1370)), CF1038 (daf-16(mu86)), DA465 (eat-2(ad465)), TJ1052 (age-1(hx546)), MQ887(isp-1(qm150)), RB950 (cup-4(ok837)), NS3099 (nhr-49(nr2041)), TM2984 (nlp-7(tm2984)), EU1 (skn-1(zu67)), and PS3551 (hsf-1(sy441)). We note that the use of single alleles does not eliminate the possibility that some of the effects may be to linked point mutations that we were not able to outcross. However, we feel that the use of single alleles was warranted by an unavailability of other viable alleles, the genetic breadth of the study, and/or the fact that RNA interference by feeding could not be employed in a bacterial deprivation scenario and soaking or injecting RNA interference methods in older animals may be highly variable and would likely add confounding variables to mating and starvation scenarios.
Late-Life Mating Protocols for 20°C
All experiments were conducted by maintaining 150–175 hypochlorite-synchronized (25) animals on male-free, OP50-seeded 10-cm NGM Petri dishes until mating was initiated. When mating was initiated, individual old hermaphrodites were removed from mass culture and placed on an OP50-seeded 6-cm NGM Petri dish; progeny production was recorded each day via the number of hatched larvae detected on the plate. For experiments imposing dietary restriction (DR) through starvation, adults were moved to fresh plates each day; on the fifth day of adulthood, all animals were washed into a 50-mL conical vial and rinsed three times with S-Basal containing 200 μg/mL of ampicillin. All animals resided in the last wash for a total of 20 minutes at 20°C, giving them at least 25 defecation cycles (at 1 per 45 seconds) in the antibiotic-laden S-basal. This is important to get rid of bacteria in the gut. Late-life cross-progeny production without an antibiotic wash occurs indistinguishably, but the incidence of contamination and experimental failure is high (>50%). After washing, half of animals were transferred to large OP50-seeded NGM; the other half was transferred to large unseeded NGM plates. Aging animals were transferred to fresh plates daily or every other day; AL and DR groups were transferred equally. Subsequent matings took place as noted in the text. Actual numbers of old hermaphrodites assayed varied from 10 to 25 per condition per trial; see raw data in Supplementary Excel File 1 for details of each experiment.
Microscopy
Animals were mounted on 2% agarose pads with tricaine/levamisole anesthetic according to McCarter and colleagues (26). Nomarski DIC optics were used to examine animals through a 40× objective to view gonad morphology, gonad sheath contractions, ovulation, and fertilization. Animals were mounted for no more than 2 hours in all experiments longitudinally examining gonad morphology on Days 11 and 12 of adulthood and for no more than 30 minutes for studies examining fertilization kinetics.
Experimental Design and Statistical Analysis
Initial experiments detected positive effects in cross-progeny production for fog-2 mutants and starved wild-type animals with sample sizes of as few as 10 old hermaphrodites (see raw data in Supplementary Excel File 1, first three trials). Thus, to examine more genotypes simultaneously, we utilized a subsequent experimental design wherein we examined 10 old hermaphrodites per condition. When we observed no or significantly less than wild-type, ad libitum (AL) cross-progeny production in a mutant at Day 14, we performed assays at Day 11, where we could observe at least some AL cross-progeny production. This baseline of AL cross-progeny production allowed us to determine if cross-progeny production could be enhanced by bacterial deprivation treatment. Numbers of animals per trial and in total are similar to prior reports examining cross-progeny production after expiration of self-fertility (see raw data in Supplementary Excel File 1 and Table 1). Previous studies examining cross-fertility and reproductive life span used as few as six individuals per trial (15) and total numbers of animals for late-life cross-fertility ranged from 7 to 99 (11). Nevertheless, we do suggest and exercise caution when interpreting our negative results. All data sets were analyzed for normal distribution and then compared with the appropriate control group using a t test or a Mann–Whitney U test, in accordance with the results of the Kolmogrov–Smirnov normality test. No data sets for experiments in Figure 4 were determined to be normally distributed.
Table 1.
Late-Life Cross-Progeny Production
| Meta Analysis | Progeny Production |
p Values* |
#Trials | #AL | #BD | |||
| Genotype | AL AVG ± SD | BD AVG ± SD | BD vs AL | AL vs wt | BD vs wt | |||
| wild-type D11 mating | 2.4 ± 5.1 | 20.1 ± 26.2 | <.001 | N/A | N/A | 4 | 55 | 45 |
| wild-type D14 mating | 0.5 ± 1.4 | 14.3 ± 20.5 | <.001 | N/A | N/A | 5 | 60 | 60 |
| wild-type ts D11 mating | 4.1 ± 1.4 | 10.3 ± 20.8 | .85 | N/A | N/A | 2 | 20 | 20 |
| wild-type ts D14 mating | 2.6 ± 3.4 | 3.8 ± 6.0 | .96 | N/A | N/A | 2 | 20 | 20 |
| pha-4 ts D11 mating | 3.1 ± 5.3 | 9.8 ± 12.6 | .06 | .16 | .42 | 2 | 20 | 20 |
| pha-4 ts D14 mating | 0.6 ± 1.1 | 4.5 ± 6.2 | .01 | .02 | .71 | 2 | 20 | 20 |
| fog-2 D11 mating | 9.4 ± 11.1 | 14.1 ± 21.7 | .78 | <.001 | .46 | 2AL/1 BD | 23 | 10 |
| fog-2 D14 mating | 2.8 ± 5.0 | 10.0 ± 14.2 | .009 | .002 | .45 | 3 | 28 | 30 |
| daf-16 D11 mating | 0.5 ± 1.4 | 0.6 ± 1.1 | .33 | .10 | <.001 | 3 | 30 | 30 |
| daf-16 D14 mating | 0.0 ± 0.0 | 0.1 ± 0.6 | .16 | <.001 | <.001 | 3 | 30 | 30 |
| hsf-1 D11 mating | 0.1 ± 0.4 | 0.6 ± 1.0 | .02 | <.001 | <.001 | 3 | 30 | 30 |
| hsf-1 D14 mating | 0.1 ± 0.2 | 0.1 ± 0.5 | 1.00 | <.001 | <.001 | 2 | 20 | 20 |
| skn-1 D14 mating | 0.1 ± 0.5 | 0.8± 3.9 | .69 | .11 | <.001 | 3 | 28 | 30 |
| nhr-49 D11 mating | 0.3 ± 1.8 | 0.03 ± 0.2 | 1.00 | .001 | <.001 | 3 | 30 | 30 |
| nhr-49 D14 mating | 0.0 ± 0.0 | 0.0 ± 0.0 | 1.00 | <.001 | <.001 | 2 | 20 | 20 |
| cup-4 D11 mating | 7.0 ± 7.3 | 16.2 ± 22.4 | .13 | .001 | .55 | 3 | 30 | 30 |
| nlp-7 D11 mating | 0.2 ± 0.8 | 4.5 ± 7.0 | .007 | .02 | <.001 | 3 | 30 | 30 |
| NP717 D14 mating | 0.4 ± 0.8 | 0.6 ± 2.1 | .81 | .35 | <.001 | 2 | 20 | 20 |
Notes: AL = ad libitum; AVG = average; BD = bacterially deprived; wt = wild type.
All data sets analyzed failed tests for normal distribution.
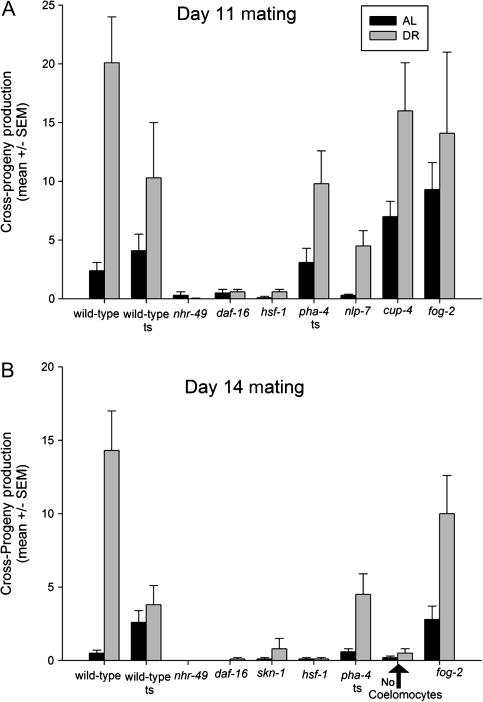
Figure 4.
Late-life progeny production after ad libitum feeding or dietary restriction by bacterial deprivation (BD). (A) Progeny production (mean ± SEM) for wild-type and mutant animals mated on Day 11 of adulthood after being fed ad libitum or after a period of BD (see Experimental Procedures and Table 1). “Wild-type ts” denotes wild-type animals that were raised at 25°C and then shifted to 20°C, as controls for the temperature sensitive pha-4(zu225) allele. (B) Progeny production after mating on Day 14 of adulthood. Data represent all trials combined (see also Table 1 and Supplementary Excel File 1 for individual trial data).
RESULTS
The Functional Life Span of the Caenorhabditis elegans Germ Line
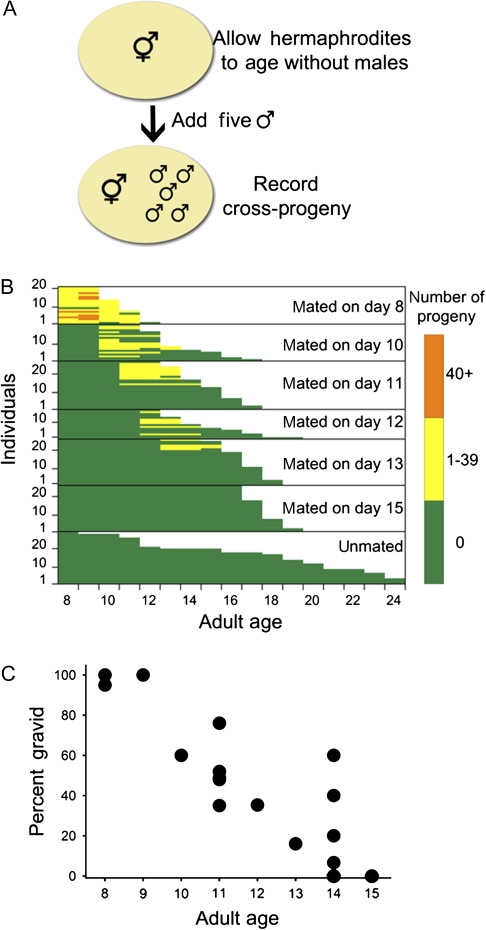
We first asked if it is possible to obtain cross-progeny later in life, after the exhaustion of self-sperm and the consequent loss of self-reproduction. Prior publications indicated that male sperm could be utilized by a hermaphrodite gonad that had been depleted of self-sperm (27,28), and a recent publication reported that self-sperm–depleted hermaphrodites can produce cross-progeny via mating with males, as late as Day 10 of adulthood (11). To determine the latest age on which cross-progeny could still be produced, we mated individual wild-type (N2 and CGCb) hermaphrodites at various times during their life span with five young wild-type males to determine the last day on which cross-progeny were produced (Figure 1). About 300 self-progeny are produced in the first 5 days of life and then self-progeny production drop to zero (6,7,29). However, all N2 hermaphrodites retained the ability to reproduce, when these hermaphrodites were mated with five wild-type males on the eighth day of adulthood (Figure 1B). Cross-fertility declined more or less linearly throughout life (Figure 1C). At Day 10 of adulthood, 60% of the hermaphrodites could still regain fertility, falling to 16% at 13 days of adult life and to 0 by Day 15 (Figure 1B); no hermaphrodites produced any cross-progeny when mated on Day 15 of adulthood or later in three independent trials (n = 60). This last day of potential for cross-fertility is similar to reported mean wild-type life spans that ranged from 12 to 18 days at 20°C (6,8). The intersection of organismic life span with the end of the period of potential cross-fertility provides a feasible explanation for the long post-self-reproductive life typically observed in C elegans. In addition and consistent with at least one prior report (30), we observe a trade-off between life span and cross-progeny production at all ages (Figure 1B, p < .001, Mann–Whitney U test comparing median life span for each mated age vs age-matched unmated controls).
Figure 1.
Methods and results of measuring late-life fertility of Caenorhabditis elegans hermaphrodites (N2 CGCb), depleted for self-sperm. (A) Experimental design showing method for assessment of late-life fertility in individual hermaphrodites (see Experimental Procedures for additional description). (B) Fertility and survival of wild-type hermaphrodites that were either mated on various days of adulthood or left unmated. Each horizontal bar shows daily fertility colorimetrically and survival of an individual hermaphrodite; matings and counts started at Day 8 of adulthood. (C) Summary data from several different experiments. Percent of hermaphrodites at various adult ages that were able to regain fertility and produce any cross-progeny (n = 10–25); for Day 15 no hermaphrodites produced any cross-progeny in three independent experiments (n = 60).
Reactivation of Old Gonads in Response to Mating
The hermaphrodite gonad of C elegans consists of two U-shaped tubular ovotestes joined at one arm of each U by the uterus. The anterior and posterior gonads each have a spermatheca but share a uterus and a vulva. At the distal end of each gonad is a distal tip cell controlling proliferation of a population of proximal germ line stem cells, which divide mitotically, producing a syncytium containing many nuclei that migrate down each gonad arm (from the distal tip cell toward the uterus), gaining an intact cell membrane, undergoing meiosis, and becoming mature oocytes by the time they arrive at the spermatheca (Figure 2A). Mature oocytes are ovulated into the spermatheca, fertilized with sperm, and pass into the uterus for a few cell divisions until they are laid (26,27). About 160 sperm are produced in each gonad arm during the fourth larval stage, before the first oocytes are generated (26,28). These “self” sperm are stored in the spermatheca for use throughout the first days of adulthood. However, if mating with a male occurs, male sperm are preferentially used for fertilization.
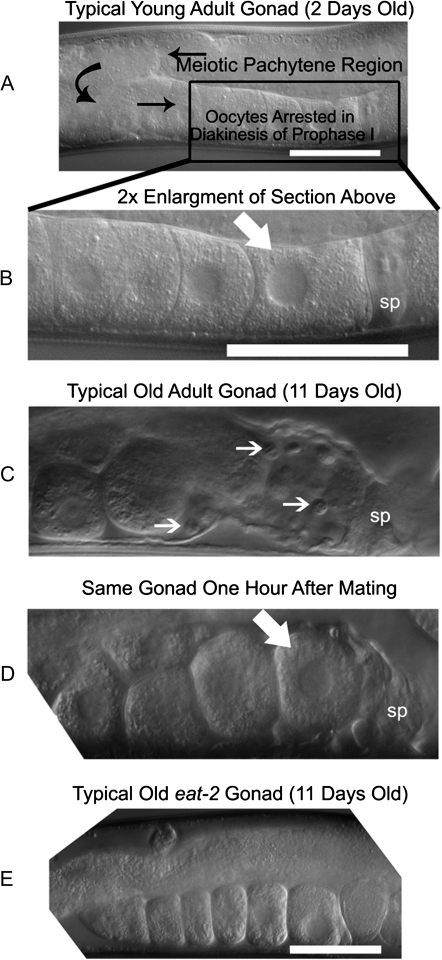
Figure 2.
DIC micrographs of young and old hermaphrodite gonads (N2 CGCb). (A) Typical self-fertile hermaphrodite on the second day of adulthood (A, B, and E) white scale bar is 50 μM. Black arrows indicate the flow of cells, starting with the germ cells in the germ line and ending with oocytes as labeled. (B) Shows 2× enlargement of the region of the gonad containing the oocytes and spermatheca. The white arrow points to the proximal oocyte, nearest the spermatheca (labeled “sp”) and the next oocyte to be fertilized. (C) Typical sperm-depleted gonad on the 11th day of adulthood. Small white arrows point to unengulfed cells remaining in the flaccid gonad sheath. (D) The same gonad as in (C) after 1 hour of mating. The flaccid space, previously occupied by unengulfed cells is now occupied by a proximal oocyte (white arrow) awaiting ovulation and fertilization. We observe this rapid “rejuvenation” under the microscope in 30%–60% of animals that are able to produce progeny (82 mated wild-type gonads were examined from 41 individuals). (E) Typical gonad of an unmated 11-day-old adult eat-2(ad465) mutant (30 unmated gonads, from 15 individuals; see also Supplementary Figure 1).
To determine how the gonad changes morphologically with age and with mating, we examined gonads on Day 11 of adulthood and followed them longitudinally using Nomarski DIC optics. In young animals, the gonad is well organized and morphologically intact (Figure 2A and B). During gamete maturation, germ line cells mature as they move through the gonad, becoming oocytes arrested in diakinesis by the time they arrive at the proximal gonad next to the spermatheca (Figure 2A). In young adults, a proximal oocyte is always present next to the spermatheca, awaiting fertilization (Figure 2A and B). As animals age, the gonad becomes physically disorganized (31); no proximal oocyte is found next to the spermatheca in old hermaphrodites (Figure 2C). Instead, there are “unengulfed” cells, not present in young animals, which may be dead germ line nurse cells or cell bodies left over from oocyte differentiation (32) (Figure 2C). Unfertilized oocytes are sometimes observed in the spermatheca of some older unmated hermaphrodites (15%–30% of gonads in three experiments).
We found that 11-day-old adult hermaphrodites produced embryos within 3 hours of mating with males. Somatic gonad sheath contractions could be seen after 1 hour of mating, indicating that male sperm are able to induce oocyte maturation and ovulation in a time-frame similar to young adults (26,27). We analyzed old gonads longitudinally before and after mating. Within an hour and a half after mating, unengulfed cells can no longer be found in the gonad and a proximal oocyte lies next to the spermatheca; the proximal oocyte in Figure 2D is probably the same oocyte visible in Figure 2C, about 50 μM from the spermatheca. The fate of the apparently unengulfed cells remains unknown, but it seems likely that they were pushed into the uterus. Thus, old senescent gonads are rapidly reactivated in response to mating late in life.
Long-Lived Mutants and Reproductive Life Span
More than 700 gerontogene manipulations are now known to modulate the life of the C elegans soma (33); we asked whether representatives of different classes of these genes also modulate the functional life span of the hermaphrodite gonad. Various reports have suggested that some prolongation of germ line life span does occur in long-lived (Age) mutants; however, systematic studies of other Age mutants found negative or no effects. For example, some daf-2 mutant alleles do cause prolonged self-reproduction, albeit often with reduced overall fertility (7,12,14). At least one study has examined cross-progeny production of Age mutants, mated early in life (11), but we wanted to know if cross-progeny production is extended in Age mutants when mating is initiated late.
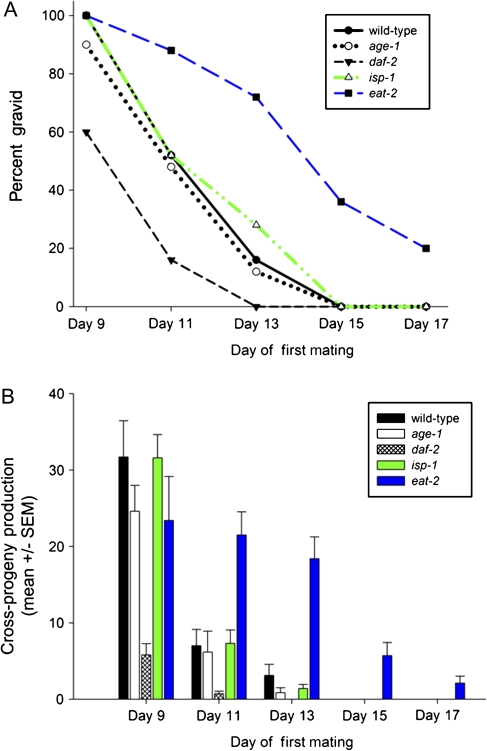
We examined late-life cross-progeny production in long-lived mutants representative of three major pathways known to extend life span in C elegans: insulin-like signaling (age-1, daf-2) (8,34), mitochondrial dysfunction (isp-1) (35), and DR (eat-2) (36); the effects of these mutations on somatic life span are shown in Table 2. We found that the age-1(hx546) and isp-1(qm150) mutations had no effect on the period of cross-progeny production or the number of progeny that were produced in late life (Figure 3A and B), despite their dramatic effect on hermaphrodite life span. The daf-2(e1370) mutation significantly decreased the age at which late-life cross-progeny could be produced as well as the number of progeny produced (p < .001; Figure 3A and B). In dramatic contrast to these results, animals with the eat-2(ad465) mutation showed both an increase in the fraction of hermaphrodites that could become fertile late in life and a significant increase in the number of progeny as compared with wild-type animals mated on Days 11, 13, 15, and 17 of adulthood (p < .001 for all comparisons with wild type; Figure 3A and B). Therefore, mutations that extend the life of the soma do not generally confer extension of late-life cross-fertility.
Table 2.
Measured and Reported Somatic Life Spans
| Study | wt Life Span (d) | Mutation/Condition | Mutant Life Span | %Change |
| Present study* | 17, 17.5 | BD(D5 Adults) | 19, 25 | +12, +43 |
| Lee and colleagues (37) † | 14.6 | BD(D4 Adults) | 19.4 | +24.5 |
| Kaberlein and colleagues (38) | 19 | BD(D2 Adults) | 30 | +50 |
| Friedman and Johnson (8) | 15 | age-1(hx546) | 25.5 | +65 |
| Kenyon and colleagues (39) | 20 | daf-2(e1370) | 42 | +110 |
| Lakowski and Hekimi (36) | 19.5 | eat-2(ad465) | 25.1 | +29 |
| Feng and colleagues (40) | 20.3 | isp-1(qm150) | 33 | +65 |
| Van Gilst and colleagues (41) | 17 | nhr-49(nr2041) | 7 | −59 |
| Wilson and colleagues (42) † | 12.7 | skn-1(zu67) | 9.9 | −28 |
| Lin and colleagues (43) ‡ | 20 | daf-16(mu86) | 16 | −20 |
| Panowski and colleagues (24) ‡ | 22 | pha-4(zu225) | 20 | −9.8 |
| Hajdu-Cronin and colleagues (44) | 19 | hsf-1(sy441) | 12.4 | −35 |
| Arantes-Olivera and colleagues (45) | 19.4 | fog-2(q71) | 20.7(not significant) | +6.7 |
| Park and colleagues (46) | 18.6 | nlp-7(tm2984) | 16.2 | −14.8 |
| Park and colleagues (46) | 18.6 | cup-4(ok837) | 17.4 | −6.9 |
Notes: AL = ad libitum; AVG = average; BD = bacterially deprived; wt = wild type.
Median life spans reported for two different cohorts of unmated animals randomly selected from the raw data. The “(D5 Adults)” designates that bacterial deprivation was initiated at Day 5 of adulthood. Subsequent notations follow the same pattern.
These studies used the fem-1(hc17) background as a surrogate for wild type.
There were no numbers in the text of these manuscripts except percent difference; data extrapolated from figures.
Figure 3.
Frequency and magnitude of late-life fertility after cessation of self-reproduction. (A) Percent of wild-type and Age (long-lived) mutant hermaphrodites that are able to produce cross-progeny after self-sperm depletion. (B) Mean (± SEM) number of cross-progeny produced for wild-type and Age mutants after mating on different days of adulthood. Progeny production was measured for at least 15 individuals for each data point.
To determine if there was any morphological basis for the performance difference between wild-type and Age mutants (especially eat-2 mutants), we analyzed the structural integrity of the gonad in mated and unmated individuals on Day 11 of adulthood. Animals were binned into six categories using a qualitative three-factor binary analysis, examining (a) the presence of the gonad, (b) physical orientation of the gonad superstructure, and (c) the presence of unengulfed cells or inappropriate location of oocytes. Both mated and unmated 11- and 12-day-old eat-2 mutants exhibited significantly better gonad morphology than wild-type animals (Figure 2E; p < .05, Mann–Whitney U test in Supplementary Figure 1 compared with wild-type animals on either day). Old daf-2(e1370) mutants also had significantly better gonad morphology than wild-type animals, but this conferred no benefit for cross-progeny production to those animals, suggesting that morphology alone does not explain differential late-life fertility.
Other Forms of DR Also Increase Late-Life Cross-Fertility
The differential effects observed in eat-2 results in this study, and others (11,47) prompted us to ask whether other modes of imposing DR might also extend the life span of the germ line. Previous murine studies have shown that animals on a DR diet, returned to an AL diet later in life, exhibit enhanced oocyte quality or fertility relative to animals that have been fed AL their entire lives (48–51). In C elegans, there are multiple ways of imposing DR (52); we used complete starvation later in life (38). We imposed complete starvation at Day 5 of adulthood [preventing “bagging” from starvation (37)] until the day when mating was initiated when animals were transferred to individual plates with fresh food. Animals that underwent starvation produced more cross-progeny than AL controls (Figure 4A and B; for detailed statistics, see Table 1). After mating at Day 11 of adulthood, animals on an AL diet produced an average of 2.4 ± 5.1 cross-progeny, whereas animals that had been on DR produced significantly more: 20.1 ± 26.2 cross-progeny (p < .001). When animals were mated on Day 14 of adulthood, AL controls generated 0.5 ± 1.4 cross-progeny, significantly less than the DR production of 14.3 ± 20.5 (p < .001). Thus, at least two distinct methods of imposing DR resulted in similar extension of late-life reproduction.
Transcription Factors Regulating Germ Line Reactivation
We asked whether enhanced reproductive capacity by starvation–DR is dependent upon the same downstream transcription factors that are required for somatic enhancement of life span by DR (52). We therefore examined mutants that were deficient in daf-16, skn-1, pha-4, and hsf-1 to see if they interfered with the enhanced reproductive potential after DR. We also examined nhr-49, which encodes a nuclear hormone receptor that is required for the fasting response and maintenance of the germ line during starvation-induced reproductive diapause in young adults (22,41,53). The effects of these mutations on the soma are shown in Table 2.
The transcription factors NHR-49/HNF-4 and DAF-16/FOXO are absolute requirements for late-life cross-fertility. Null nhr-49(nr2041) mutants mated on Day 11 or 14 produced almost no progeny under any condition (p ≤ .001 for all comparisons; Figure 4A and B; Table 1). Furthermore, nhr-49(nr2041) animals could not increase cross-progeny production in response to DR at either day tested. Animals lacking DAF-16 produced reduced numbers of progeny under AL conditions (Day 11) but failed to reach statistical significance. The daf-16 mutants also failed to increase cross-progeny production in response to DR (p < .001 for daf-16 DR vs wild-type DR; Figure 4A; Table 1). By Day 14, daf-16(mu86) mutants did not produce any cross-progeny under AL conditions and did not significantly increase progeny production in response to DR (Figure 4B; Table 1).
We tested animals, containing the skn-1(zu67) mutation, which do not express intestinal SKN-1 but do express functional SKN-1 in sensory neurons. These animals were able to produce wild-type levels of cross-progeny when mated on Day 14 of adulthood under AL conditions; skn-1 mutants were not examined at Day 11 because of apparent wild-type germ line perdurance to Day 14 of adulthood. The skn-1(zu67) animals fail to increase cross-progeny production either relative to AL skn-1(zu67) animals or matched with wild-type DR controls (Figure 4B; Table 1).
The transcription factors HSF-1 and PHA-4/FOXA are required for wild-type germ line life span but not for increasing late-life cross-progeny production in response to DR. The hsf-1(sy441) mutants fail to produce wild-type levels of cross-progeny when mated at Day 11 (0.1 ± 0.4, p < .001 vs AL wild type) but do significantly increase progeny production in response to DR (0.1 ± 0.4 AL vs 0.6 ± 1.0 DR, p = .02), but levels are very much reduced in comparison with the wild-type control. Animals mated on Day 14 showed no response to DR, producing significantly fewer progeny than wild type, possibly due to the decrepit state of 14-day-old hsf-1(sy441) mutants (Figures 4A and B; Table 1). The pha-4(zu225) mutation significantly reduces cross-progeny production by Day 14 of adulthood under AL conditions, producing only 0.6 ± 1.1 compared with paired wild-type temperature-shifted controls, which average 2.6 ± 3.4 (p = .02, Figure 4B; Table 1). Yet, animals that were starved and do not produce wild-type levels of PHA-4 can significantly increase cross-progeny production to an average of 4.5 ± 6.2, which is not significantly different than their wild-type temperature-shifted DR controls (p = .01 vs AL pha-4, Figure 4B; Table 1).
Genes and Cells Involved With Somatic and Germ Line Response to DR
We recently reported that downstream effectors of SKN-1, nlp-7, and cup-4 were required for DR enhancement of somatic life span (46,54). Furthermore, cup-4 acts through the coelomocyte cells (55). Thus, we tested the requirements of nlp-7, cup-4, and the coelomocytes in late-life fertility under AL and DR conditions.
NLP-7 is not required for germ line response to DR but is required for normal AL germ line function (Figure 4A; Table 1). The nlp-7(tm2984) null mutants produce significantly fewer cross-progeny than wild-type controls under AL conditions, with an average of only 0.2 ± 0.8 compared with the wild-type average of 2.4 ± 5.1 (p = .02). However, the nlp-7 mutants were able to significantly increase cross-progeny production in response to DR to an average of 4.5 ± 7.0 (p = .007). The nlp-7 mutants did not produce wild-type levels of cross-progeny in response to starvation, but this was probably due to the initially lower AL fertility.
We had previously uncovered a role for cup-4, a coelomocyte-specific gene, in mediating DR (46,54), and we asked whether cup-4 may also have a role in preserving the germ line in response to DR. CUP-4 is not required for wild-type fertility under either condition, and mutants may actually increase fertility. Under AL conditions, animals that do not express CUP-4 produce an average of 7.0 ± 7.3, which is significantly more cross-progeny than wild-type AL controls (p = .001; Figure 4A; Table 1). After DR, cup-4(ok837) animals increase cross-progeny production to an average of 16.2 ± 22.4, which is not significantly different from the DR controls.
The Greenwald lab has constructed a strain of nematode in which the six coelomocyte cells are ablated through the expression of Diphtheria toxin A, permitting analyses of animals with no functional coelomocytes (56). The coelomocyte cells were not detected as a requirement for AL late-life cross-fertility (Figure 4B; Table 1), although fertility is very low. However, the coelomocyte-ablated animals are unable to increase cross-progeny production in response to DR.
The functional effects of our genetic analysis are summarized in Figure 5; a summary of the effects of genotypes and treatments on somatic life span is shown in Table 2. For additional descriptions of the phenotypes for each genotype used as well as molecular functions of the gene products, see Supplementary Table 1. Raw mating and life span data are available in Supplementary Excel File 1.
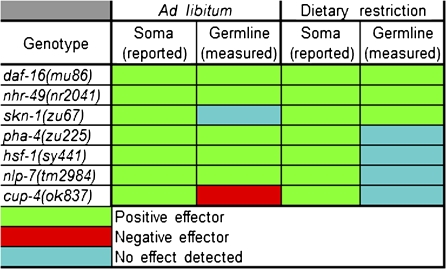
Figure 5.
Graphical representation of genetic effects on life span and cross-fertility under AL and DR conditions. A colorimetric table summarizing the effects of genotype on life span and late-life cross-fertility is shown. The references for the reported life span effects are presented in Table 2.
The Period of Self-Fertility Alters Late-Life Fertility
We were not able to recover rates of egg production in old adults that approached those seen in young hermaphrodites. There may be some wear and tear on the gonad or depletion of gamete generation capacity during the self-fertility period. To test the hypothesis that the self-fertility period alters the functional life span of the germ line, we examined fog-2 mutants. The fog-2(q71) mutant does not produce sperm and is essentially female, producing no progeny unless mated (26). The fog-2(q71) animals produce significantly more cross-progeny than wild-type animals, when mated at Days 11 and 14 (Figure 4A and B; Table 1): 9.4 ± 11.1 on Day 11 and 2.8 ± 5.0 on Day 14 compared with the wild-type average of 2.4 ± 5.1 and 0.5 ± 1.4 (p < .001, p = .002), respectively. Thus, the production of progeny early in life influences the ability of animals to produce cross-progeny late in life.
Discussion
Genetic Mechanisms for Late-Life Fertility and DR-Induced Enhancement
We have found that some genes required for life-span extension in response to DR are not required for DR-based extension of fertility, indicating that there may be independent genetic circuits regulating soma and germ line responses to DR. Of particular interest is the observation that while the SKN-1 transcription factor is required for DR extension of both life span (57) and fertility, NLP-7 and CUP-4, downstream effectors of SKN-1 in DR life-span enhancement, are not required for starvation-induced enhancement of late-life fertility. This observation suggests that there may be other currently unidentified SKN-1 effectors that act specifically to modulate the germ line response to DR.
Unexpectedly, loss of CUP-4, an ion channel that functions in coelomocyte endocytosis, leads to increased AL fertility. Given that this effect is not duplicated by loss of coelomocytes themselves, we speculate that CUP-4 plays a specific role in either reducing an intercellular signaling factor that promotes germ line function or producing a negative germ line regulator.
PHA-4 and HSF-1, two transcription factors required for normal fertility under AL conditions, are also not essential for DR-induced fertility increases. We interpret these findings to indicate that while these transcription factors may be important for maintaining optimal germ line function, they may not be involved in the initiation of the extended fertility induced by DR. Interestingly, PHA-4 localizes to the somatic gonad, including the distal tip cell, where PHA-4 may be exerting influence on germ line proliferation (58,59); alternatively, the influence of PHA-4 on the germ line could be due to activity in other tissues, such as the intestine. Finally, two transcription factors, NHR-49 and DAF-16, appear to be required for all aspects of somatic life (41,60) and fertility, including enhancement of late-life fertility induced by starvation. These results are summarized in Figure 5.
Evolutionary Aspects of Late-Life Cross-Fertility
We have shown that in C elegans hermaphrodites, reproduction can be restored via mating with males after the expiration of the self-fertile period several days later than previously reported. These sexual interactions later in life serve to rejuvenate the germ line, expanding reproductive capacity and reproductive life span (expiration of cross-fertility is Day 14 of adulthood or Day 17 of total life span for wild-type animals) to the point of near intersection with median life span of C elegans [12–20 days for wild-type at 20°C (6,8); Table 2]. Thus, the apparent post-reproductive life span of C elegans is significantly diminished when cross-fertility is taken into account, making the post-reproductive life span of C elegans more comparable with that of fruit flies rather than mammals. These results also suggest that there may be strong selection for outcrossing with males in the wild and suggest that there may be a relationship with the frequency of males in wild populations and their ability to be responsive to male mating late in life.
How likely is it that these mating-induced effects have shaped the life history evolution of these nematodes? Studies of genetic variation in natural populations of C elegans suggest that cross-fertilization may be fairly rare (61–64). However, a number of natural isolates maintain high frequencies of males in the laboratory (61), and several studies have detected high levels of polymorphism within some natural populations that must be the result of mating with males (64,65). The most likely explanation for these patterns is that cross-fertilization does in fact occur in nature at a low but significant level but that the offspring from these matings may be at a fitness disadvantage, perhaps because of segregating incompatibility factors (61,64,66). Thus, despite the fact that C elegans does self-fertilize frequently, the mating-induced reproductive effects that we observe here—especially since they significantly increase total reproduction—are likely to be a significant part of the natural ecology of this species.
Age Mutations’ Diverse Effects on Germ Line Life Span
When we asked if Age mutants extended reproductive longevity, as they do the longevity of the soma, we found unaltered or earlier termination of ability to produce late-life cross-progeny for the daf-2, age-1, and isp-1 mutants. Thus, in these mutants, the life-extension and reduced-mortality effects on the soma are not shared by the germ line, consistent with the Disposable Soma Theory of Aging, which suggests that there must be trade-offs between the soma and germ line (5). In contrast, a mutation in eat-2 does extend the life span of both the soma and the germ line. However, one “cost” of the eat-2(ad465) mutation is an increased generation time; thus, there may still be a trade-off when one considers the amount of extra time required for development, consistent with at least one prior report (67) and current mathematical models (68). Yet, despite the trade-off for an eat-2 mutant, aging wild-type animals seem to experience only benefits to the soma and germ line from a period of starvation after the self-fertile period.
DR-Based Preservation of Germ Line Function Is Conserved in Mammals
We have discovered that two forms of DR, one genetically mediated by slower rates of food uptake in an eat-2 mutant and the other purely environmentally mediated via starvation, prolong the late-life cross-fertile period that occurs after the expiration of self-fertility. Mammalian studies in which mice were under DR and then returned to AL feeding conditions also demonstrated increased fertility or oocyte quality relative to AL controls (48–51). This conservation of germ line response to DR between worms and mice indicates that the phenomenon may be broadly conserved across phyla. Studies in Drosophila have not reported this phenomenon of germ line life-span extension after DR, but this may be due to the fact that flies were continually deprived of calories and not returned to AL conditions; a period of caloric restriction followed by AL feeding in flies may produce a similar result to that seen in worms and mice.
Orchestration of Germ Line DR Response by Distinct Transcription Factors
Major transcription factors that coordinate somatic life-span enhancements in response to DR (52) are also used in maintaining and enhancing the life span of germ line. Note that we are still cautious about interpreting lower fertility as a specific response indicating direct molecular interactions, rather than as an organism-wide sickness as is obvious when one examines genetic alterations in important proteins, such as HSF-1 (31). The transcription factor mutants are all short-lived relative to wild-type animals and thus physiologically older than wild-type animals (Figure 5; Table 2). However, a strict interpretation of our data suggests that utilization of a particular transcription factor during AL or DR conditions by the soma or germ line can be distinct. This study indicates that post-reproductive life span within C elegans is actually the result of a balance between the self-fertile capacity, the availability of mates, and access to food; all of which are orchestrated by the differential action of transcription factors in the soma and germ line in response to the environment.
Novel Functions for Genes in Germ Line Maintenance and Repression
Our study revealed that at least three genes involved in somatic life-span extension in response to DR have novel roles in the germ line. Loss of pha-4 results in significantly reduced cross-progeny production (by Day 14) under AL conditions but not under starvation–DR conditions (bacterial dilution was not tested), indicating that pha-4 is required for maintaining the germ line during normal aging but not required for enhancing the germ line capacity for reproduction in response to bacterial deprivation. Thus, in contrast to the essential role of PHA-4 in DR-based somatic life span enhancement (24), PHA-4 plays a minor role in the normal aging of the germ line as it does in the soma but is not involved in DR-based (starvation) enhancement of the germ line life span. PHA-4 is not required for somatic life-span extension in response to intermittent fasting (69); thus, different forms of DR can have distinct genetic requirements in distinct tissues (52).
Similar to pha-4, two other genes (cup-4 and nlp-7) that are required for life extension in response to DR are not required for extension of the germ line life span in response to bacterial deprivation. Although nlp-7 is required for AL fertility, cup-4 is unique in that it is the only gene that appears to actively restrict AL fertility. Thus, cup-4 effectively limits the productivity of the germ line during AL conditions. This supports a model wherein CUP-4 represses germ line function to alter resource allocation between soma and germ line. This role for CUP-4 in resource allocation is congruent with the disposable soma theory (5) because it demonstrates that there are gene products that are actively repressing germ line function; this leaves the resources that would be used for germ line function free to be used elsewhere in the soma.
Many Pathways May Result in Different Forms of Germ Line Preservation
The preservation of the germ line after reproductive senescence is probably distinct from adult reproductive diapause (ARD) that occurs when C elegans are starved as young adults (22). First, animals can maintain germ line integrity to reproduce at Day 14 of adulthood with or without initiating starvation; the frequency and magnitude of the phenotype increase when starvation is initiated. Second, unlike ARD, the reactivation of senescent, sperm-depleted gonads happens after the self-fertility period and requires food and mating, not just food. Finally, another difference between late-life fertility and ARD is that we found oocytes to be arrested and ready for fertilization within 1 hour, unlike the shriveled mass of approximately 35 germ cells in ARD. However, the requirement of nhr-49 is absolute for late-life fertility as well as for ARD, indicating at least some functional pathway overlap.
Previous studies in which animals were mated earlier in life (11,15) produced results distinct from those found in this study, where animals are mated after a period of reproductive senescence at later ages. For example, both Hughes and colleagues and Luo and colleagues found that the daf-2(e1370) mutation (also examined in the present study) showed a great increase in reproductive life span as measured by percent of the population producing any cross-progeny. In contrast, we found that daf-2(e1370) animals displayed the largest reduction in the fraction of animals producing cross-progeny and the lowest number of cross-progeny per individual, among all Age mutants tested. There are two distinct differences between these studies and ours. First, in prior studies where daf-2 animals displayed a greater fraction of animals producing cross-progeny, said animals were mated as young adults on the first day of adulthood and not after a period of reproductive senescence as in this study. Second, animals tested for cross-fertility with age in previous studies were younger, with Day 10 of adulthood being the latest age at which matings were initiated. We find that wild-type animals equal (eat-2) or out-perform all Age mutants, with respect to late-life cross-progeny production (Figure 3 vs Figure 4), provided they experience a period of starvation after reproductive senescence. Importantly, the sole measure of percent fertile does not reflect the magnitude of cross-progeny production and thus does not reflect fitness. What matters for Darwinian fitness is progeny per genotype and generation time (70–72).
Notably, we do not find our results to be in conflict with prior reports (11,15,47); instead, we find our results to be reflective of changing physiological states in different epochs of life. The notion that animals at a later phase of life have different genetic influences on fertility because they are in distinct physiological states is supported prior reports showing distinct transcriptional and physiological profiles with age (73–75). The fact that distinct genotypes respond differently to the same treatment at different ages is itself evidence that physiology and thus influences on physiology change with age.
The Cost of Self-Fertility and Rejuvenating Effect of DR
The fact that worms with virgin uteruses (fog-2 animals) produce more cross-progeny than animals undergoing the self-fertile period (during which they produce about 300 progeny) is consistent with a scenario in which self-fertility negatively affects late-life cross-fertility. This could result from either wear and tear on the physical gonad (due to mating pathology or egg throughput) or depletion of some other unknown factor affecting oocyte production. Mating pathology is certainly a driving force in the death of mated young adults (30) or older adults examined in this study. Oocyte depletion due to a diminished germ line stem cell division capacity in the wild type is also possible. However, the germ line of these animals is not exhausted: old animals (with or without a self-fertile period) produced nowhere near the over 1,000 cross-progeny that can be produced over the life span of an individual worm that is heavily mated with males (26). It is difficult to be certain of the major force(s) that causes animals to stop producing cross-progeny late in life. The large variance in cross-progeny production indicates that there may be a strong stochastic component in how the hermaphrodite gonad decays and, therefore, many different physiological states with age.
In a prior report (11), fog-2 animals were mated at Day 10 of adulthood and 28 individuals were found to produce 7 ± 4 cross-progeny, whereas 99 wild-type animals were found to produce 3.6 ± 0.6 cross-progeny. There was a trend for fog-2 animals to produce more progeny, but it was not significant at the ages tested. However, at a later epoch of life than has previously been examined, animals that do not experience ovulation produce more cross-progeny, indicating that the self-fertility period can influence late-life fertility when examined at later ages in paired trials. Importantly, we again do not find our results to be in conflict with the prior report examining fog-2(q71) animals’ cross-fertile capacity after the expiration of self-fertility; instead, we interpret these results to be reflective of an increasing influence of gonad wear and tear with age. It might not matter if the somatic gonad was used or not for mid-life reactivation via fertilization with male sperm [mating at Day 10 of adulthood, reported in Hughes and colleagues (11)], but clearly, there is a difference in late-life cross-progeny production between old (mated at Day 11 or 14 of adulthood, Figure 4A and B; Table 1) wild-type animals that experience well over 300 ovulation events and fog-2(q71) hermaphrodites, which experience a greatly decreased number of ovulation events, and hence less use of the somatic gonad machinery.
Starvation and Preservation of Somatic and Germ Line Life Span
We have shown that DR via complete starvation extends germ line life span of wild-type C elegans. In addition to starvation-induced preservation of late-life fertility after self-fertility, starvation initiated in early adulthood preserves the self-fertile capacity until food is replenished (22). We also noted that DR-based preservation of the germ line was conserved in mammals as well. Why is this so? One explanation may be that starvation is a universal stress that animals have had to endure since the beginning of life on earth; resources become depleted periodically. Organisms have had to find ways to endure until resources become available or become extinct. If an organism only preserved the life of its soma and not its germ line, then that organism would never be able to produce the next generation; if species encountered starvation as a whole and no individual could endure starvation and subsequently reproduce, then that species would become extinct. Thus, preservation of the soma without preservation of the germ line negates Darwinian fitness. Hence, starvation and other forms of DR may initiate a coordinated program to preserve both somatic and germ line function until resources become available.
SUPPLEMENTARY MATERIAL
Supplementary material can be found at: http://biomedgerontology.oxfordjournals.org/.
FUNDING
National Institutes of Health, National Institute on Aging, and the Glenn Foundation
Supplementary Material
Acknowledgments
The authors would like to thank National Institutes of Health, National Institute on Aging, and the Glenn Foundation for funding their research. These authors declare no competing financial interests.
Author contributions: D.W., A.R.M., C.D.L, and T.E.J. designed all experiments. A.R.M., D.W., S.-K.P., J.R.C., P.M.T., and T.E.J. conducted all experiments. A.R.M. and D.W. performed statistical analysis on data. T.E.J., P.C.P., C.D.L., and A.R.M. generated the manuscript. All authors concurred on the final version of the manuscript.
References
- 1.Kirkwood TB, Shanley DP. The connections between general and reproductive senescence and the evolutionary basis of menopause. Ann N Y Acad Sci. 2010;1204:21–29. doi: 10.1111/j.1749-6632.2010.05520.x. [DOI] [PubMed] [Google Scholar]
- 2.Hamilton WD. The moulding of senescence by natural selection. J Theor Biol. 1966;12:12–45. doi: 10.1016/0022-5193(66)90184-6. [DOI] [PubMed] [Google Scholar]
- 3.Sgro CM, Partridge L. A delayed wave of death from reproduction in Drosophila. Science. 1999;286:2521–2524. doi: 10.1126/science.286.5449.2521. [DOI] [PubMed] [Google Scholar]
- 4.Cohen AA. Female post-reproductive lifespan: a general mammalian trait. Biol Rev Camb Philos Soc. 2004;79:733–750. doi: 10.1017/s1464793103006432. [DOI] [PubMed] [Google Scholar]
- 5.Kirkwood TB. Evolution of ageing. Nature. 1977;270:301–304. doi: 10.1038/270301a0. [DOI] [PubMed] [Google Scholar]
- 6.Klass MR. Aging in the nematode Caenorhabditis elegans: major biological and environmental factors influencing life span. Mech Ageing Dev. 1977;6:413–429. doi: 10.1016/0047-6374(77)90043-4. [DOI] [PubMed] [Google Scholar]
- 7.Mendenhall AR, LeBlanc MG, Mohan DP, Padilla PA. Reduction in ovulation or male sex phenotype increases long-term anoxia survival in a daf-16-independent manner in Caenorhabditis elegans. Physiol Genomics. 2009;36:167–178. doi: 10.1152/physiolgenomics.90278.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Friedman DB, Johnson TE. A mutation in the age-1 gene in Caenorhabditis elegans lengthens life and reduces hermaphrodite fertility. Genetics. 1988;118:75–86. doi: 10.1093/genetics/118.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ayyadevara S, Alla R, Thaden JJ, Shmookler Reis RJ. Remarkable longevity and stress resistance of nematode PI3K-null mutants. Aging Cell. 2008;7:13–22. doi: 10.1111/j.1474-9726.2007.00348.x. [DOI] [PubMed] [Google Scholar]
- 10.Tissenbaum HA, Johnson TE. Aging Processes in Caenorhabditis elegans. Cold Spring Harbor, NY: Cold Spring Harbor Monograph Series; 2008. pp. 153–184. [Google Scholar]
- 11.Hughes SE, Evason K, Xiong C, Kornfeld K. Genetic and pharmacological factors that influence reproductive aging in nematodes. PLoS Genet. 2007;3:e25. doi: 10.1371/journal.pgen.0030025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gems D, Sutton AJ, Sundermeyer ML, et al. Two pleiotropic classes of daf-2 mutation affect larval arrest, adult behavior, reproduction and longevity in Caenorhabditis elegans. Genetics. 1998;150:129–155. doi: 10.1093/genetics/150.1.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Larsen PL, Albert PS, Riddle DL. Genes that regulate both development and longevity in Caenorhabditis elegans. Genetics. 1995;139:1567–1583. doi: 10.1093/genetics/139.4.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lithgow GJ, White TM, Hinerfeld DA, Johnson TE. Thermotolerance of a long-lived mutant of Caenorhabditis elegans. J Gerontol. 1994;49:270–276. doi: 10.1093/geronj/49.6.b270. [DOI] [PubMed] [Google Scholar]
- 15.Luo S, Shaw WM, Ashraf J, Murphy CT. TGF-beta Sma/Mab signaling mutations uncouple reproductive aging from somatic aging. PLoS Genet. 2009;5:e1000789. doi: 10.1371/journal.pgen.1000789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gerisch B, Weitzel C, Kober-Eisermann C, et al. A hormonal signaling pathway influencing C. elegans metabolism, reproductive development, and life span. Dev Cell. 2001;1:841–851. doi: 10.1016/s1534-5807(01)00085-5. [DOI] [PubMed] [Google Scholar]
- 17.Baugh LR, Sternberg PW. DAF-16/FOXO regulates transcription of cki-1/Cip/Kip and repression of lin-4 during C. elegans L1 arrest. Curr Biol. 2006;16:780–785. doi: 10.1016/j.cub.2006.03.021. [DOI] [PubMed] [Google Scholar]
- 18.Johnson TE, Mitchell DH, Kline S, et al. Arresting development arrests aging in the nematode Caenorhabditis elegans. Mech Ageing Dev. 1984;28:23–40. doi: 10.1016/0047-6374(84)90150-7. [DOI] [PubMed] [Google Scholar]
- 19.Apfeld J, Kenyon C. Cell nonautonomy of C. elegans daf-2 function in the regulation of diapause and life span. Cell. 1998;95:199–210. doi: 10.1016/s0092-8674(00)81751-1. [DOI] [PubMed] [Google Scholar]
- 20.McElwee JJ, Schuster E, Blanc E, et al. Diapause-associated metabolic traits reiterated in long-lived daf-2 mutants in the nematode Caenorhabditis elegans. Mech Ageing Dev. 2006;127:458–472. doi: 10.1016/j.mad.2006.01.006. [DOI] [PubMed] [Google Scholar]
- 21.Weinkove D, Halstead JR, Gems D, Divecha N. Long-term starvation and ageing induce AGE-1/PI 3-kinase-dependent translocation of DAF-16/FOXO to the cytoplasm. BMC Biol. 2006;4:1. doi: 10.1186/1741-7007-4-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Angelo G, Van Gilst MR. Starvation protects germline stem cells and extends reproductive longevity in C. elegans. Science. 2009;326:954–958. doi: 10.1126/science.1178343. [DOI] [PubMed] [Google Scholar]
- 23.Lee RY, Hench J, Ruvkun G. Regulation of C. elegans DAF-16 and its human ortholog FKHRL1 by the daf-2 insulin-like signaling pathway. Curr Biol. 2001;11:1950–1957. doi: 10.1016/s0960-9822(01)00595-4. [DOI] [PubMed] [Google Scholar]
- 24.Panowski SH, Wolff S, Aguilaniu H, et al. PHA-4/Foxa mediates diet-restriction-induced longevity of C. elegans. Nature. 2007;447:550–555. doi: 10.1038/nature05837. [DOI] [PubMed] [Google Scholar]
- 25.Fabian TJ, Johnson TE. Production of age-synchronous mass cultures of Caenorhabditis elegans. J Gerontol. 1994;49:B145–B56. doi: 10.1093/geronj/49.4.b145. [DOI] [PubMed] [Google Scholar]
- 26.McCarter J, Bartlett B, Dang T, Schedl T. On the control of oocyte meiotic maturation and ovulation in Caenorhabditis elegans. Dev Biol. 1999;205:111–128. doi: 10.1006/dbio.1998.9109. [DOI] [PubMed] [Google Scholar]
- 27.Ward S, Carrel JS. Fertilization and sperm competition in the nematode Caenorhabditis elegans. Dev Biol. 1979;73:304–321. doi: 10.1016/0012-1606(79)90069-1. [DOI] [PubMed] [Google Scholar]
- 28.Kimble J, Ward S. Germ-line development and fertilization. In: Wood WB, editor. The Nematode Caenorhabditis elegans. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1988. pp. 191–213. [Google Scholar]
- 29.Johnson TE. Aging can be genetically dissected into component processes using long-lived lines of Caenorhabditis elegans. Proc Natl Acad Sci U S A. 1987;84:3777–3781. doi: 10.1073/pnas.84.11.3777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gems D, Riddle DL. Longevity in Caenorhabditis elegans reduced by mating but not gamete production. Nature. 1996;379:723–725. doi: 10.1038/379723a0. [DOI] [PubMed] [Google Scholar]
- 31.Garigan D, Hsu AL, Fraser AG, et al. Genetic analysis of tissue aging in Caenorhabditis elegans: a role for heat-shock factor and bacterial proliferation. Genetics. 2002;161:1101–1112. doi: 10.1093/genetics/161.3.1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gumienny TL, Lambie E, Hartwieg E, et al. Genetic control of programmed cell death in the Caenorhabditis elegans hermaphrodite germline. Development. 1999;126:1011–1022. doi: 10.1242/dev.126.5.1011. [DOI] [PubMed] [Google Scholar]
- 33.Magalhães JPd. Human Ageing Genomic Resources. October 31, 2009. http://genomics.senescence.info/genes/index.htm. Accessed December 14, 2009. [Google Scholar]
- 34.Dorman JB, Albinder B, Shroyer T, Kenyon C. The age-1 and daf-2 genes function in a common pathway to control the lifespan of Caenorhabditis elegans. Genetics. 1995;141:1399–1406. doi: 10.1093/genetics/141.4.1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rea SL, Ventura N, Johnson TE. Relationship between mitochondrial electron transport chain dysfunction, development, and life extension in Caenorhabditis elegans. PLoS Biol. 2007;5 doi: 10.1371/journal.pbio.0050259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lakowski B, Hekimi S. The genetics of caloric restriction in Caenorhabditis elegans. Proc Natl Acad Sci U S A. 1998;95:13091–13096. doi: 10.1073/pnas.95.22.13091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee GD, Wilson MA, Zhu M, et al. Dietary deprivation extends lifespan in Caenorhabditis elegans. Aging Cell. 2006;5:515–524. doi: 10.1111/j.1474-9726.2006.00241.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kaeberlein TL, Smith ED, Tsuchiya M, et al. Lifespan extension in Caenorhabditis elegans by complete removal of food. Aging Cell. 2006;5:487–494. doi: 10.1111/j.1474-9726.2006.00238.x. [DOI] [PubMed] [Google Scholar]
- 39.Kenyon C, Chang J, Gensch E, et al. A C. elegans mutant that lives twice as long as wild type. Nature. 1993;366:461–464. doi: 10.1038/366461a0. [DOI] [PubMed] [Google Scholar]
- 40.Feng J, Bussiere F, Hekimi S. Mitochondrial electron transport is a key determinant of life span in Caenorhabditis elegans. Dev Cell. 2001;1:633–644. doi: 10.1016/s1534-5807(01)00071-5. [DOI] [PubMed] [Google Scholar]
- 41.Van Gilst MR, Hadjivassiliou H, Jolly A, Yamamoto KR. Nuclear hormone receptor NHR-49 controls fat consumption and fatty acid composition in C. elegans. PLoS Biol. 2005;3:e53. doi: 10.1371/journal.pbio.0030053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wilson MA, Shukitt-Hale B, Kalt W, et al. Blueberry polyphenols increase lifespan and thermotolerance in Caenorhabditis elegans. Aging Cell. 2006;5:59–68. doi: 10.1111/j.1474-9726.2006.00192.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lin K, Hsin H, Libina N, Kenyon C. Regulation of the Caenorhabditis elegans longevity protein DAF-16 by insulin/IGF-1 and germline signaling. Nat Genet. 2001;28:139–145. doi: 10.1038/88850. [DOI] [PubMed] [Google Scholar]
- 44.Hajdu-Cronin YM, Chen WJ, Sternberg PW. The L-type cyclin CYL-1 and the heat-shock-factor HSF-1 are required for heat-shock-induced protein expression in Caenorhabditis elegans. Genetics. 2004;168:1937–1949. doi: 10.1534/genetics.104.028423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Arantes-Oliveira N, Apfeld J, Dillin A, Kenyon C. Regulation of life-span by germ-line stem cells in Caenorhabditis elegans. Science. 2002;295:502–505. doi: 10.1126/science.1065768. [DOI] [PubMed] [Google Scholar]
- 46.Park SK, Link CD, Johnson TE. Life-span extension by dietary restriction is mediated by NLP-7 signaling and coelomocyte endocytosis in C. elegans. FASEB J. 2009;24:383–392. doi: 10.1096/fj.09-142984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Huang C, Xiong C, Kornfeld K. Measurements of age-related changes of physiological processes that predict lifespan of Caenorhabditis elegans. Proc Natl Acad Sci U S A. 2004;101:8084–8089. doi: 10.1073/pnas.0400848101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rikke BA, Liao CY, McQueen MB, et al. Genetic dissection of dietary restriction in mice supports the metabolic efficiency model of life extension. Exp Gerontol. 2010;45:691–701. doi: 10.1016/j.exger.2010.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nelson JF, Karelus K, Bergman MD, Felicio LS. Neuroendocrine involvement in aging: evidence from studies of reproductive aging and caloric restriction. Neurobiol Aging. 1995;16:837–843. doi: 10.1016/0197-4580(95)00072-m. ; discussion. 855–856. [DOI] [PubMed] [Google Scholar]
- 50.Johnston SL, Grune T, Bell LM, et al. Having it all: historical energy intakes do not generate the anticipated trade-offs in fecundity. Proc Biol Sci. 2006;273:1369–1374. doi: 10.1098/rspb.2005.3456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Selesniemi K, Lee HJ, Tilly JL. Moderate caloric restriction initiated in rodents during adulthood sustains function of the female reproductive axis into advanced chronological age. Aging Cell. 2008;7:622–629. doi: 10.1111/j.1474-9726.2008.00409.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Greer EL, Brunet A. Different dietary restriction regimens extend lifespan by both independent and overlapping genetic pathways in C. elegans. Aging Cell. 2009;8:113–127. doi: 10.1111/j.1474-9726.2009.00459.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Van Gilst MR, Hadjivassiliou H, Yamamoto KR. Caenorhabditis elegans nutrient response system partially dependent on nuclear receptor NHR-49. Proc Natl Acad Sci U S A. 2005;102:13496–13501. doi: 10.1073/pnas.0506234102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Park SK, Tedesco PM, Johnson TE. Oxidative stress and longevity in Caenorhabditis elegans as mediated by SKN-1. Aging Cell. 2009;8:258–269. doi: 10.1111/j.1474-9726.2009.00473.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Patton A, Knuth S, Schaheen B, et al. Endocytosis function of a ligand-gated ion channel homolog in Caenorhabditis elegans. Curr Biol. 2005;15:1045–1050. doi: 10.1016/j.cub.2005.04.057. [DOI] [PubMed] [Google Scholar]
- 56.Fares H, Greenwald I. Genetic analysis of endocytosis in Caenorhabditis elegans: coelomocyte uptake defective mutants. Genetics. 2001;159:133–145. doi: 10.1093/genetics/159.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.An JH, Blackwell TK. SKN-1 links C. elegans mesendodermal specification to a conserved oxidative stress response. Genes Dev. 2003;17:1882–1893. doi: 10.1101/gad.1107803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Azzaria M, Goszczynski B, Chung MA, et al. A fork head/HNF-3 homolog expressed in the pharynx and intestine of the Caenorhabditis elegans embryo. Dev Biol. 1996;178:289–303. doi: 10.1006/dbio.1996.0219. [DOI] [PubMed] [Google Scholar]
- 59.Kalb JM, Lau KK, Goszczynski B, et al. pha-4 is Ce-fkh-1, a fork head/HNF-3alpha, beta, gamma homolog that functions in organogenesis of the C. elegans pharynx. Development. 1998;125:2171–2180. doi: 10.1242/dev.125.12.2171. [DOI] [PubMed] [Google Scholar]
- 60.Henderson ST, Bonafe M, Johnson TE. daf-16 protects the nematode Caenorhabditis elegans during food deprivation. J Gerontol A Biol Sci Med Sci. 2006;61:444–460. doi: 10.1093/gerona/61.5.444. [DOI] [PubMed] [Google Scholar]
- 61.Anderson JL, Morran LT, Phillips PC. Outcrossing and the maintenance of males within C. elegans populations. J Hered. 2010;101(Suppl. 1):S62–S74. doi: 10.1093/jhered/esq003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rockman MV, Kruglyak L. Recombinational landscape and population genomics of Caenorhabditis elegans. PLoS Genet. 2009;5:e1000419. doi: 10.1371/journal.pgen.1000419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cutter AD, Felix MA, Barriere A, Charlesworth D. Patterns of nucleotide polymorphism distinguish temperate and tropical wild isolates of Caenorhabditis briggsae. Genetics. 2006;173:2021–2031. doi: 10.1534/genetics.106.058651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Barriere A, Felix MA. Natural variation and population genetics of Caenorhabditis elegans. WormBook. 2005:1–19. doi: 10.1895/wormbook.1.43.1. doi:10.1895/wormbook.1.43.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sivasundar A, Hey J. Population genetics of Caenorhabditis elegans: the paradox of low polymorphism in a widespread species. Genetics. 2003;163:147–157. doi: 10.1093/genetics/163.1.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Seidel HS, Rockman MV, Kruglyak L. Widespread genetic incompatibility in C. elegans maintained by balancing selection. Science. 2008;319:589–594. doi: 10.1126/science.1151107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Saul N, Pietsch K, Menzel R, et al. The longevity effect of tannic acid in Caenorhabditis elegans: disposable Soma meets hormesis. J Gerontol A Biol Sci Med Sci. 2010;65:626–635. doi: 10.1093/gerona/glq051. [DOI] [PubMed] [Google Scholar]
- 68.Hou C, Bolt KM, Bergman A. Energetic basis of correlation between catch-up growth, health maintenance, and aging. J Gerontol A Biol Sci Med Sci. 2011;66:627–638. doi: 10.1093/gerona/glr027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Honjoh S, Yamamoto T, Uno M, Nishida E. Signalling through RHEB-1 mediates intermittent fasting-induced longevity in C. elegans. Nature. 2009;457:726–730. doi: 10.1038/nature07583. [DOI] [PubMed] [Google Scholar]
- 70.Chen J, Senturk D, Wang JL, et al. A demographic analysis of the fitness cost of extended longevity in Caenorhabditis elegans. J Gerontol A Biol Sci Med Sci. 2007;62:126–135. doi: 10.1093/gerona/62.2.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Jenkins NL, McColl G, Lithgow GJ. Fitness cost of extended lifespan in Caenorhabditis elegans. Proc Biol Sci. 2004;271:2523–2526. doi: 10.1098/rspb.2004.2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Walker DW, McColl G, Jenkins NL, et al. Evolution of lifespan in C. elegans. Nature. 2000;405:296–297. doi: 10.1038/35012693. [DOI] [PubMed] [Google Scholar]
- 73.Herndon LA, Schmeissner PJ, Dudaronek JM, et al. Stochastic and genetic factors influence tissue-specific decline in ageing C. elegans. Nature. 2002;419:808–814. doi: 10.1038/nature01135. [DOI] [PubMed] [Google Scholar]
- 74.Lund J, Tedesco P, Duke K, et al. Transcriptional profile of aging in C. elegans. Curr Biol. 2002;12:1566–1573. doi: 10.1016/s0960-9822(02)01146-6. [DOI] [PubMed] [Google Scholar]
- 75.Budovskaya YV, Wu K, Southworth LK, et al. An elt-3/elt-5/elt-6 GATA transcription circuit guides aging in C. elegans. Cell. 2008;134:291–303. doi: 10.1016/j.cell.2008.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.