Abstract
Custom-designed microarray analysis was utilized to evaluate expression levels of glutamate receptors (GluRs) and GluR-interacting protein genes within isolated dentate gyrus granule cells following axotomy of the principal input, the perforant path (PP). Dentate gyrus granule cells were evaluated by microdissection via laser capture microdissection, terminal continuation RNA amplification, and microarray analysis following unilateral PP transections at seven time points. Expression profiles garnered from granule cells on the side ipsilateral to PP transections were compared and contrasted with naive subjects and mice subjected to unilateral occipital cortex lesions. Selected microarray observations were validated by real-time quantitative PCR analysis. Postlesion time-dependent alterations in specific α-amino-3-hydroxy-5-methyl-4-isoxazole propionic acid receptors, kainate receptors, N-methyl-D-aspartate (NMDA) receptors, and GluR-interacting protein genes were found across the time course of the study, suggesting a neuroplasticity response associated with the transsynaptic granule cell alterations following axotomy of incoming PP terminals.
Keywords: AMPA receptor, Axotomy, Entorhinal cortex, Kainate receptor, Microarray, NMDA receptor, RNA amplification, Transsynaptic degeneration
Introduction
Understanding the molecular and cellular underpinnings of synaptic plasticity and neuroadaptive alterations within the central nervous system are of extreme importance for fundamental comprehension of how the brain responds to internal and external stimuli, including development and injury, among others. Although synaptic reorganization has been characterized in peripheral nervous system paradigms [1, 2], less is known about this phenomena within the brain. A relative exception is the study of reactive synaptogenesis within the dentate gyrus following perforant path (PP) transections. The PP supplies the main cortical input into the hippocampal formation and originates principally in stellate cells of layer II of the entorhinal cortex [3, 4]. Unilateral interruption of the PP (i.e., via electrolytic destruction, knife cut, or aspiration lesion) [5–7] leads to profound (>80%) synaptic loss within the ipsilateral (ipsi) outer molecular layer (OML), followed by sprouting, reactive synaptogenesis, and eventual normalization/partial recovery of physiological properties and behavior [8–10]. In rodent species, these plastic changes occur within the first few days to several weeks postlesion. Specifically, the majority of axon terminals from the severed PP fibers rapidly degenerate and die back within approximately 2–5 days postlesion (DPL) [11, 12], leaving a denervated zone in the ipsi OML. A sprouting of axon terminals then occurs to reinnervate the vacated synaptic sites on granule cell dendritic branches in the ipsi OML. Newly sprouted axon terminals arise from the contralateral (contra) entorhinal cortex (via the PP), associational/commissural pathways located in the inner molecular layer (IML), and septohippocampal cholinergic fibers [4, 13, 14]. Reactive synaptogenesis [5, 15] results from the formation of synapses of the newly sprouted axon terminals with the postsynaptic granule cell dendrites in the OML.
The PP is a glutamatergic pathway, and granule cells are enriched with glutamate receptors (GluRs), including ionotropic α-amino-3-hydroxy-5-methyl-4-isoxazole propionic acid (AMPA), kainate (KA), and N-methyl-D-aspartate (NMDA) receptor subunits and metabotropic GluRs [16–20]. The dentate gyrus also expresses other markers of glutamatergic neurotransmission including excitatory amino acid transporters [21, 22] and GluR-interacting proteins [23, 24]. Both long-term potentiation and long-term depression have been described within the circuit, underscoring the excitatory nature of this pathway that is important for learning and memory [25, 26].
PP transections cause alterations in the expression levels of many molecules within the dentate gyrus, including GluR subunits. For example, the GRIN1 (NMDA R1) subunit is upregulated at the mRNA and protein level following unilateral PP transections [6, 7, 27, 28]. Although many studies have focused on one (or a few) transcripts/proteins following PP transections at one (or a few) postlesion time points, there is a paucity of studies looking at coordinate changes in GluRs, within dentate gyrus granule cells following PP transections over an extended time course [7, 29]. We have demonstrated selected alterations in AMPA subunits GluR1 (GRIA1) and GluR2 (GRIA2) as well as several glial-associated markers including glial fibrillary acidic protein, interleukin-1, interleukin-6, and the receptor for advanced glycation end products following PP transections using a microarray-based approach employing regional dissections of the dentate gyrus [7]. Although many of these reported changes are likely to be from neuroplasticity associated with granule cells and dendrites, there is a profound glial activation following PP transections [30, 31], making the assignment of gene alterations to a specific cell type virtually impossible. Importantly, a regional study cannot discern expression level changes on individual populations of cells, necessitating an alternate experimental strategy to perform population cell gene expression profiling [7]. To this end, the present study employs laser capture microdissection (LCM) in concert with the terminal continuation (TC) RNA amplification method [32–35] and custom-designed microarray analysis [36, 37] to enable molecular fingerprinting of select GluRs and GluR-interacting protein genes within dentate gyrus granule cells.
A precise lesion was performed in adult mice to axotomize the PP without damaging the hippocampal formation and the postsynaptic cells of interest therein. A time course of cellular and molecular alterations in isolated dentate gyrus granule cells was performed on the ipsi side of unilateral PP transections in adult C57BL/6 mice at seven postlesion time points (1, 2, 5, 10, 14, 30, and 60 DPL) and compared to naive control subjects and mice subjected to an occipital cortex (OC) lesion without damage to the underlying angular bundle and PP [7, 38]. In this manner, a focused microarray approach was employed to evaluate the dynamic regulation of GluR subunits across the time course of the lesion.
Materials and methods
Lesion paradigm and tissue processing
Adult male C57BL/6 mice (2–4 months old; 20–30 g) were employed in the study. The experimental protocols were approved by the IACUC committee of the Nathan Kline Institute/NYU Langone Medical Center and were in full accordance with NIH guidelines. One group of mice received unilateral PP transection (n=6–10 per postlesion time point; 1, 2, 5, 10, 14, 30, and 60 DPL). Another group of mice received unilateral OC lesions (n=4–6 per postlesion time point; 1, 10, 14, 30, and 60 DPL) to serve as a surgical control for the PP transections [7, 38]. Naive mice (n=6; termed 0 DPL) served as experimental controls. Another group of mice (n=6 per time point) served as sham controls and were subjected to dural reflection without cortical damage and survived to 1 and 14 DPL for comparison with control, PP transections, and OC lesion subjects. For the surgical procedures, mice were anesthetized with an intramuscular injection of ketamine (80 mg/kg) followed by xylazine (13 mg/kg). Mice were placed prone into a stereotaxic apparatus (Kopf, Tujunga, CA, USA) and secured using a mouse skull adapter (Stoelting, Kiel, WI, USA), and temperature was maintained at 37 °C with a heating pad. A 1.0-mm craniotomy approximately 6.5 mm caudal to bregma and 2.0 mm lateral to the midline [39] was made using a high-speed dental drill with a trephine bit (Stoelting). The dura was incised with a 25-gauge needle, exposing the cortical mantle. The overlying occipital cortex and white matter was vacuum-aspirated with a blunt-tipped 22-gauge needle, exposing the angular bundle. The PP was then transected by aspiration of the angular bundle [7, 38, 40]. Similar to the PP transection, the OC lesion was performed at the same coordinates by aspiration of the overlying occipital cortex and white matter but without damaging the angular bundle or subcortical structures [7, 38, 40, 41]. Once surgical procedures were completed, the skull was sealed with bone wax, and the fascia and scalp were closed in a single layer using an autoclip. Animals resided in their home cage until the appropriate postlesion time point.
Mice were administered an overdose of ketamine/xylazine and perfused transcardially with ice-cold 4% paraformaldehyde in phosphate buffer (PB, 0.1 M; pH 7.4). Brains were paraffin-embedded, and 6-μm-thick tissue sections throughout the rostrocaudal extent of the dentate gyrus as well as the lesion site were processed for hematoxylin and eosin and cresyl violet to verify the extent of each surgical procedure [42]. Tissue sections were processed for immunocytochemistry using several antibodies including nonphosphorylated neurofilament proteins (RMdO20) [43] to delineate granule cells and hippocampal lamination patterns, and MAP2 (Sigma, St. Louis, MO) to identify granule cell dendrites in the IML and OML of the dentate gyrus as well as selected GluR subunits. Neurofilament immunostaining enables excellent visualization of individual granule cells in the densely packed granule cell layer and helps to avoid microaspiration of unwanted glial cells, blood vessels, and epithelial cells [44–46]. Neurofilament immunostaining demonstrates no difference in hybridization signal intensity in fixed hippocampal tissues compared to Nissl stains such as cresyl violet, thionin, and hematoxylin [42]. Deparaffinized tissue sections were blocked in a 0.1 M Tris (pH 7.6) solution containing 2% donor horse serum (DHS; Sigma) and 0.01% Triton X-100 for 1 h and then incubated with primary antibodies in a 0.1 M Tris/2% DHS solution overnight at 4 °C in a humidified chamber. Sections were processed with the ABC kit (Vector Labs, Burlingame, CA, USA) and developed with 0.05% diaminobenzidine (Sigma), 0.03% hydrogen peroxide, and 0.01 M imidazole in Tris buffer for 10 min [38, 42, 47]. Tissue sections were not coverslipped or counterstained and maintained in RNase-free 0.1 M Tris for subsequent downstream genetic applications.
LCM and TC RNA amplification
LCM was performed on granule cells obtained from the superior blade of the dentate gyrus for experimental consistency. Dentate gyrus granule cells were microaspirated via LCM (Arcturus PixCell IIe, Molecular Devices, Sunnyvale, CA, USA) as described previously on tissue sections dried in xylene (two incubations of fresh xylene for 15–30 min each) [42, 47] (Fig. 1). Approximately 50 dentate gyrus granule cells were microaspirated per reaction. This amount was determined experimentally to yield reproducible RNA for downstream genetic analyses without compromising the integrity of the dentate gyrus molecular fingerprint (i.e., glial and/or vascular cell contamination is minimal to nonexistent) [7, 38]. Although the possibility that glial elements are also accrued in the LCM procedure cannot be excluded, virtually no expression of markers typically associated with astrocytes and microglia have been demonstrated from microaspirated granule cells or CA1 hippocampal neurons [44, 45]. In contrast, when regional assessments of the hippocampus are performed by custom-designed microarray analysis, glial markers are abundant [7]. Each microdissection of 50 cells was subjected to TC RNA amplification [32–35] and used individually on custom-designed arrays synthesized in the laboratory (three to five arrays were assayed per time point per subject) [36, 37, 48] (Fig. 1 and Electronic supplementary material Fig. S1).
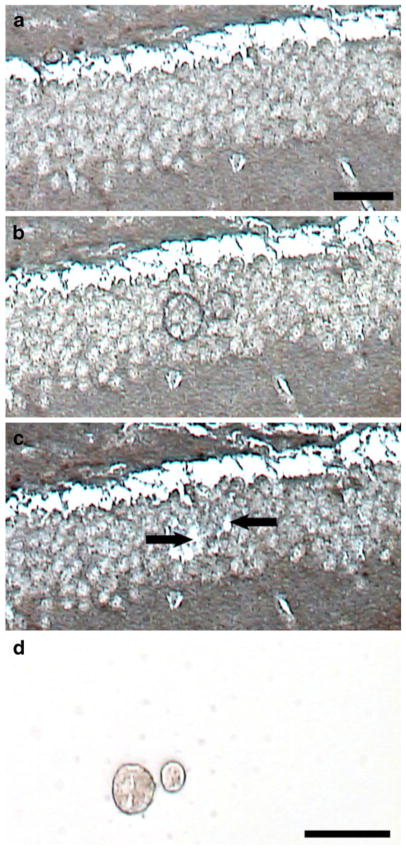
Fig. 1.
Microaspiration of neurofilament-immunoreactive granule cells using LCM. The tissue section is dehydrated in xylenes and not coverslipped (note the grainy appearance of the tissue). a Section prior to LCM. b Following one laser pulse, the thermoplastic film on the cap is adhered to individual granule cells. c The cap is removed from the LCM, leaving behind a space (arrows) where granule cells are microaspirated. d Representative captured cells are visualized by placing the cap on a clean slide for contrast. Scale bar a–c, 50 μm; d, 25 μm
RNA amplification of dentate gyrus granule cells (e.g., population cell analysis) was performed using the TC RNA amplification method. The TC RNA amplification protocol is available at http://cdr.rfmh.org/pages/ginsberglabpage.html. Briefly, dentate gyrus granule cells were homogenized in 500 μl of Trizol reagent (Invitrogen, Carlsbad, CA, USA), extracted with chloroform, and precipitated utilizing isopropanol. RNAs were reverse transcribed in the presence of poly d(T) primer (100 ng/μl) and TC primer (100 ng/μl) in 1× first strand buffer (Invitrogen), 2 μg of linear acrylamide (Applied Biosystems, Foster City, CA, USA), 10 mM dNTPs, 100 μM DTT, 20 U of SuperRNase Inhibitor (Applied Biosystems), and 200 U of reverse transcriptase (Superscript III, Invitrogen). Single stranded cDNAs were digested and then placed in a thermal cycler using a solution consisting of 10 mM Tris (pH 8.3), 50 mM KCl, 1.5 mM MgCl2, and 10 U RNase H (Invitrogen) in a final volume of 100 μl. The thermal cycler program was ran as follows: RNase H digestion step at 37 °C, 30 min; denaturation step 95 °C, 3 min; primer re-annealing step 60 °C, 5 min [34]. Samples were purified by column filtration (Montage, Millipore, Billerica, MA, USA). Hybridization probes were synthesized by in vitro transcription using 33P incorporation in 40 mM Tris (pH 7.5), 6 mM MgCl2, 10 mM NaCl, 2 mM spermidine, 10 mM DTT, 2.5 mM ATP, GTP, and CTP, 100 μM of cold UTP, 20 U of RNase inhibitor, 2 KU of T7 RNA polymerase (Epicentre, Madison, WI, USA), and 120 μCi of 33P-UTP (Perkin-Elmer, Boston, MA, USA) [32, 33]. The reaction was performed at 37 °C for 4 h. Radiolabeled TC RNA probes were hybridized to custom-designed cDNA arrays without further purification.
Custom-designed array platforms
Custom-designed arrays were synthesized in the laboratory as described previously [37, 48, 49]. Briefly, array platforms consisted of 1 μg of linearized cDNA purified from plasmid preparations adhered to high-density nitrocellulose (Hybond XL, GE Healthcare, Piscataway, NJ, USA). Each cDNA and/or expressed sequence-tagged cDNA (EST) was verified by sequence analysis and restriction digestion. cDNA clones and ESTs from mouse, rat, and human were employed. Approximately 220 cDNAs/ESTs were utilized on the current array platform. Abbreviations for individual genes were based upon NCBI/Unigene annotation. Arrays were prehybridized (2 h) and hybridized (16 h) in a solution consisting of 6× SSPE, 5× Denhardt’s solution, 50% formamide, 0.1% sodium dodecyl sulfate (SDS), and denatured salmon sperm DNA (Invitrogen) at 42 °C in a rotisserie oven [36, 37]. Following hybridization, arrays were washed sequentially in 2× SSC/0.1% SDS, 1× SSC/0.1% SDS, and 0.5× SSC/0.1% SDS for 20 min each at 42 °C. Arrays were stored in a phosphor screen overnight and developed on a phosphor imager (GE Healthcare).
Custom-designed microarray analysis
The statistical procedures for custom-designed microarray analysis have been described in detail [37, 45, 50]. Multiple (n=3–5) arrays were assayed per subject; however, statistical analysis was nested to each subject for increased statistical stringency [7, 36, 42]. Briefly, a global normalization approach was utilized whereby hybridization signal intensity for each feature on the custom-designed array was quantified by subtracting background using the empty vector pBluescript, and expression of each feature was expressed as a ratio of the total hybridization signal intensity of the individual array. A global normalization procedure effectively minimizes variation due to differences in specific activity and absolute quantity of individual probes [36, 37, 51, 52]. In this analysis scheme, an expression profile of relative changes in mRNA levels is enabled. Relative changes in total hybridization signal intensity for individual glutamatergic neurotransmission transcripts was analyzed by one-way analysis of variance (ANOVA) with post hoc analysis (Neumann–Keuls test). The level of significance was set at p<0.01 as the determinant for statistical significance, to reduce potential Type I errors [7, 42, 50]. p values (p<0.02 to <0.05) were considered as trends and reported accordingly. False discovery rates were also estimated as described previously [7, 53, 54]. Evaluation of additional classes of transcripts within dentate gyrus granule cells will comprise a separate report due to the extensive amount of data and validation requirements. Expression levels were tabulated and clustered using bioinformatics and graphics software (Gene-Linker Gold, Predictive Patterns, Kingston, ON, USA).
Real-time qPCR
Quantitative PCR (qPCR) on LCM captured cells from fixed tissues is currently under development in the laboratory and has been used for detection of abundant mRNAs such as the housekeeping gene glyceraldehyde-3-phosphate dehydrogenase (GAPDH) [35, 55]. To date, this approach is still being refined for use with moderately expressed genes including choline acetyltransferase or GluRs [55]. Therefore, qPCR was performed on microdissected frozen tissue samples as starting material. Briefly, the dentate gyrus region was micropunched from approximately 1-mm coronal tissue slabs using a dissecting stereomicroscope (Zeiss, Thornwood, NY, USA) [42] and stored at −80°C in microfuge tubes until used. TaqMan hydrolysis probes were employed for GluR1 (GRIA1), GluR2 (GRIA2), GluR3 (GRIA3), GluR4 (GRIA4), GRIN1, beta-actin (ACTB), and GAPDH. Samples were assayed on a real-time qPCR thermal cycler (7900HT, ABI). Standard curves and cycle threshold (Ct) were calculated using standards from total mouse brain RNA [32, 42]. Relative alterations in PCR product synthesis were analyzed by one-way ANOVA with post hoc analysis (Neumann–Keuls test). The level of statistical significance was set at (p<0.05). Amplicon specificity was evaluated by subcloning the amplicon products (Zero Blunt, Invitrogen) and performing sequence analysis [34].
Results
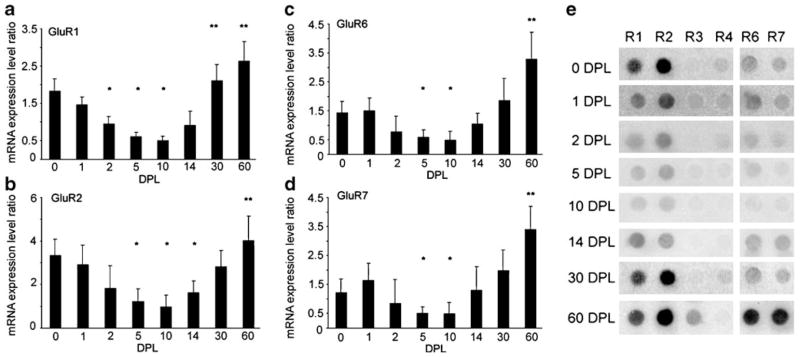
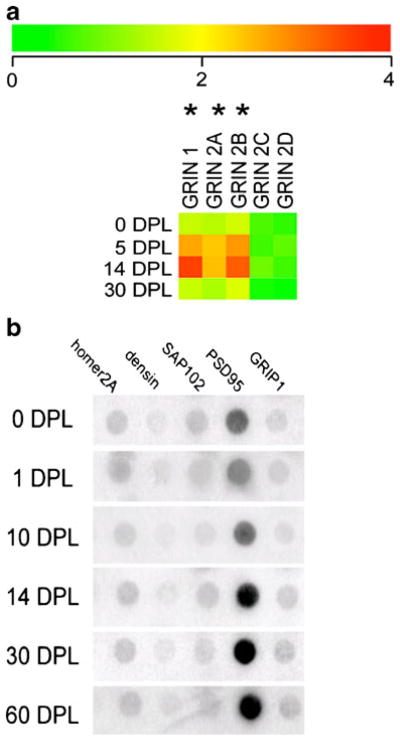
Single population expression analysis on the ipsi side of the PP transections at seven time points generated expression patterns of genes related to glutamatergic transmission. Other classes of transcripts were evaluated that will comprise a separate report due to the extensive amount of data. No differences in expression levels were found between the naive controls and the sham lesions. No significant alterations in ACTB or GAPDH were observed across the time course of the PP transections and OC lesions, demonstrating that expression levels of these housekeeping genes did not vary significantly in these injury paradigms. Moreover, no significant alterations in GluRs were observed following OC lesions or sham surgeries (Fig. 2). Baseline AMPA receptor GluR1 and GluR2 expression levels were high, whereas GluR3 and GluR4 expression levels were low. KA receptors GluR6 and GluR7 were moderately expressed. Transient downregulation of selective GluRs were observed following unilateral PP transections followed by long-term upregulation of specific AMPA and KA transcripts. Specifically, time course analysis of dentate gyrus granule cells demonstrated significant downregulation of GluR1 on the ipsi side of PP transections at 2, 5, and 10 DPL (p<0.01; Fig. 3a). Early downregulation of GluR1 was followed by a trend for long-term overexpression at 30 and 60 DPL (p<0.04; Fig. 3a). Downregulation of GluR2 was observed at 5, 10, and 14 DPL (p<0.01) followed by recovery at 30 DPL and a trend for overexpression at 60 DPL (p<0.03; Fig. 3b). No significant alterations were observed for AMPA receptors GluR3 or GluR4 across the lesion paradigm. KA receptors GluR6 (GRIK2) and GluR7 (GRIK3) displayed similar expression patterns with downregulation at 5 and 10 DPL (p<0.003) and recovery to unlesioned levels at 14 and 30 DPL, and displayed significant overexpression at 60 DPL (p<0.005; Fig. 3c, d). In contrast to the regulation of AMPA and KA receptors, expression profiling of NMDA receptors indicated a transient, significant upregulation of NMDA receptor subunits GRIN1 (NMDA R1), GRIN2A (NMDA R2A), and GRIN2B (NMDA R2B) at 5 DPL (p<0.01) and 14 DPL (p<0.005) that returned to baseline levels at 30 DPL (Fig. 4a). Low expression levels and no significant differences between time points were observed for GRIN2C (NMDA R2C) and GRIN2D (NMDA R2D). Regulation of select GluR-interacting protein genes were also found following PP transections. Specifically, differential regulation of PSD-95 was observed, as upregulation was found on the ipsi side to PP transections at 14, 30, and 60 DPL (p<0.01). In contrast, no changes in relative gene expression levels were found for other GluR-interacting protein genes interrogated including homer2A, densin-180, synapse-associated protein 102 (SAP102), or GluR-interacting protein 1 (GRIP1) (Fig. 4b).
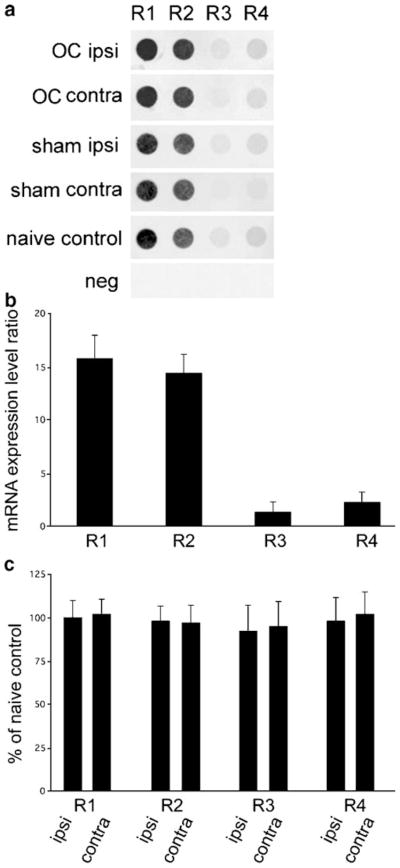
Fig. 2.
Lack of regulation of AMPA receptors in mouse dentate gyrus granule cells following unilateral OC lesions and sham surgeries. a Representative arrays illustrating no significant changes in GluR1, GluR2, GluR3, and GluR4 expression on ipsi and contra sides of the OC lesions and sham surgeries as compared to naive controls. b Histogram demonstrating the relative hybridization signal intensity levels for microarray analysis in a. c Histogram demonstrating no quantitative differences between the ipsi and contra sides of the OC lesions as compared to unlesioned controls (demarcated as 100%) for GluR1–GluR4
Fig. 3.
Time course analysis of select AMPA and KA receptors within dentate gyrus granule cells following unilateral PP transections. a Downregulation of GluR1 on the side ipsilateral to PP transections was observed at 2, 5, and 10 DPL (significant decrease, *p<0.01). Early downregulation was followed by a trend toward long-term overexpression at 30 and 60 DPL (a trend toward an increase, **p< 0.04). b. Downregulation of GluR2 was found at 5, 10, and 14 DPL (significant decrease, *p<0.01) followed by recovery at 30 DPL and a trend toward overexpression at 60 DPL (a trend toward an increase, **p<0.03). c Downregulation of GluR6 was found at 5 and 10 DPL, recovery to unlesioned levels at 14–30 DPL, and displayed abundant overexpression at 60 DPL (a significant increase, **p<0.003). d GluR7 expression levels effectively parallel GluR6 levels, with downregulation at 5 and 10 DPL, normalized expression at 14–30 DPL, and overexpression at 60 DPL (**p<0.005). e A representative microarray depicting select GluR subunits across the seven time points. The unlesioned control is demarcated as 0 DPL
Fig. 4.
Analysis of NMDA receptor subunits and GluR-interacting protein genes following unilateral PP transections. a Heatmap with color-coded bar for relative expression levels indicating transient upregulation (asterisk) of GRIN1, GRIN2A, and GRIN2B at 5 and 14 DPL on the ipsi side of PP transections as compared to untreated controls (denoted as 0 DPL). Expression levels returned to baseline control levels by 30 DPL. No differences were observed for GRIN2C and GRIN2D, although expression levels were near the limit of detection. b Custom-designed microarrays illustrating the expression of GluR-interacting protein genes at 1, 14, 30, and 60 DPL on the side ipsilateral to PP transections. Long-term upregulation of PSD-95 is found at 14, 30, and 60 DPL (p<0.01). In contrast, no changes in relative gene expression levels are found for homer2A, densin-180, SAP102, or GRIP1
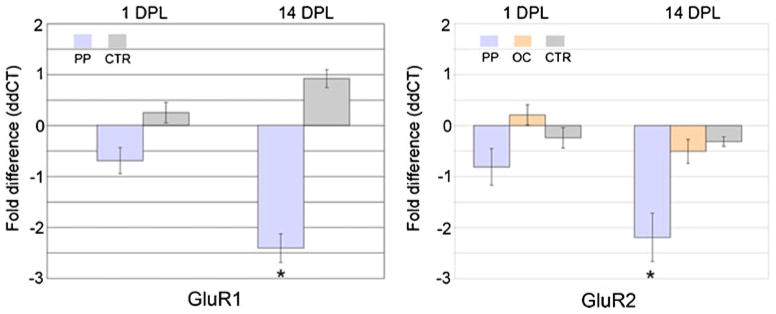
Validation of select microarray results was performed via real-time qPCR. Downregulation of GluR1 and GluR2 was observed in microdissected dentate gyrus from the ipsi side of PP transections at 14 DPL, consistent with the microarray findings (Fig. 5). No changes in GluR1 or GluR2 expression were found by qPCR following OC lesions, indicating the relative specificity of the PP lesion for alterations in select GluRs. Moreover, a lack of regulation of GluR3 and GluR4 was found, consistent with the microarray results (data not shown).
Fig. 5.
Histograms demonstrating relative expression levels of AMPA subunits GluR1 and GluR2 following unilateral PP transections and OC lesions at two time points (1 and 14 DPL) relative to untreated controls (CTR) within the ipsi dentate gyrus. The ddCT method is employed to determine relative gene level differences with GAPDH qPCR used as a control. No significant differences are found at 1 DPL. Consistent with the microarray results, significant downregulation (*p<0.02) of GluR1 (~2.4-fold) and GluR2 (~2.2-fold) is observed via qPCR on the ipsi side of PP transections relative to CTR and OC lesions
Discussion
This study was designed to evaluate AMPA, KA, and NMDA subunit expression levels following PP transections, a central nervous system axotomy paradigm. RNA amplification and custom-designed microarray analysis of dentate granule cells was performed at several postlesion time points to generate an expression profile of granule cells following transection of the principal glutamatergic afferent. Expression profile analysis of injured granule cells indicates alterations in select ionotropic GluR expression, suggesting an excitotoxic mechanism as part of the neurodegenerative program, although other programs of degeneration cannot be excluded at this point. Expression profiling following unilateral PP transections reveals a dynamic regulation of several AMPA, KA, and NMDA subunits as well as GluR-interacting proteins across the time course of the lesion. Initial downregulation of AMPA and KA receptors is followed by normalization of expression levels and ultimately, overexpression of several genes relevant to glutamatergic neurotransmission, including PSD-95, which may be related to the reorganization of pre- and postsynaptic machinery following axotomy. These observations are consistent with downregulation of GluR1 and GluR2/3, but not GRIP1 protein levels in regional hippocampal homogenates following unilateral PP transections [7]. In contrast, select NMDA receptor upregulation was observed 5–14 DPL and then returned to baseline levels. These changes in receptor expression parallel structural and synaptic alterations that occur over the course of the lesion paradigm. Specifically, transneuronal loss of select AMPA and KA GluRs as well as upregulation of select NMDA receptor subunit expression on postsynaptic granule cells on the ipsi side of PP transections coincides with loss of presynaptic entorhinal cortex PP terminals (approximately 5–14 DPL), possibly as a result of sublethal excitotoxic glutamate release from the axotomized terminals [6, 27, 56, 57]. Transneuronal recovery of GluR subunit expression (approximately 14–30 DPL) and overexpression (approximately 30–60 DPL) of postsynaptic GluRs on granule cells takes place as reactive synaptogenesis occurs adjacent to postsynaptic sites. Transneuronal alterations of GluRs and glutamate transporters on granule cells appear to have profound long-term effects on the physiology of the hippocampal formation following PP transections [8, 58], and the current study has begun to identify relevant transcripts for further observation. Alterations in GluR subunits following PP transections are also relevant toward understanding the pathophysiology of epilepsy, as seizures that are caused by PP stimulation can be blocked by the uncompetitive NMDA receptor channel blocker dizocilpine maleate (MK-801) [59, 60]. Future studies combining the PP transection paradigm with blockade of GluRs through pharmacological delivery of excitatory amino acid antagonists and subsequent microarray analysis of dentate gyrus granule cells will be employed to assess a potential mechanism of sublethal excitotoxicity. If our hypothesis is correct, then various combinations of non-NMDA and NMDA antagonists will attenuate alterations in gene expression observed in dentate gyrus granule cells following PP transections.
One effect of the PP transections may be traumatic depolarization and subsequent release of presynaptic glutamate [61–63] into extracellular interstitium of the dentate gyrus that, in turn, causes sublethal and/or delayed excitotoxicity within postsynaptic, somatodendritic targets. Glutamatergic neurotransmission is essential for normative brain function; however, excessive concentrations of glutamate, blockade of glutamate uptake, and/or hyperactivation of GluRs on neurons will cause excitotoxicity [64–66]. Excitotoxicity can occur as an acute event or act through a delayed neurodegenerative mechanism in vivo, causing both apoptotic and necrotic forms of cell death as well as chronic, sublethal neurodegeneration [67–70]. Long-term transneuronal changes have been found within the dentate gyrus in animal models following excitotoxic insult [71]. Neurodegenerative changes following PP transections resemble aspects of cellular alterations that occur following excitotoxicity. For example, parvalbumin-immunoreactive interneurons in the hilus display short- and long-term changes in response to PP transections [57, 72–74], consistent with excitotoxicity. These morphological changes are blocked by MK-801 [57], strongly implicating excitotoxicity following PP transections through an NMDA receptor mediated mechanism. Additional evidence that axotomy of glutamatergic pathways causes transneuronal excitotoxic damage comes from the fimbria–fornix transection paradigm [67, 68, 75]. Similar to PP transections [57], pretreatment with MK-801 prior to fimbria–fornix transections attenuated cellular damage [67].
In summary, expression profile analysis of dentate gyrus granule cells following unilateral PP transections and OC lesions demonstrated selective regulation of specific genes related to glutamatergic neurotransmission during the time course of the injury paradigm. These observations have implications toward understanding the mechanisms that underlie dynamic GluR reorganization and are relevant toward understanding the pathophysiology of neurodegenerative disorders that display selective vulnerability within these discrete circuits. Furthermore, alterations in GluR subunits are relevant to understanding the cellular and molecular underpinnings of activity-dependent responses within hippocampal and forebrain circuits and are also directly relevant to uncovering the mechanism(s) underlying synaptic and neurodegenerative changes in the brains of humans with a variety of neurodegenerative disorders.
Supplementary Material
Acknowledgments
I thank Dr. Shaoli Che, Irina Elarova, John T. Le, and Marc D. Ruben for expert technical assistance. Support for this project comes from the NINDS (NS43939), NIA (AG09466 and AG17617), and the Alzheimer’s Association.
Footnotes
Electronic supplementary material The online version of this article (doi:10.1007/s00216-010-3826-1) contains supplementary material, which is available to authorized users.
References
- 1.Aldskogius H, Arvidsson J, Grant G. The reaction of primary sensory neurons to peripheral nerve injury with particular emphasis on transganglionic changes. Brain Res. 1985;357:27–46. doi: 10.1016/0165-0173(85)90006-2. [DOI] [PubMed] [Google Scholar]
- 2.Santos AF, Caroni P. Assembly, plasticity and selective vulnerability to disease of mouse neuromuscular junctions. J Neurocytol. 2003;32:849–862. doi: 10.1023/B:NEUR.0000020628.36013.88. [DOI] [PubMed] [Google Scholar]
- 3.Squire LR, Stark CE, Clark RE. The medial temporal lobe. Annu Rev Neurosci. 2004;27:279–306. doi: 10.1146/annurev.neuro.27.070203.144130. [DOI] [PubMed] [Google Scholar]
- 4.Steward O, Scoville SA. Cells of origin of entorhinal cortical afferents to the hippocampus and fascia dentata of the rat. J Comp Neurol. 1976;169:347–370. doi: 10.1002/cne.901690306. [DOI] [PubMed] [Google Scholar]
- 5.Steward O, Vinsant SL. The process of reinnervation in the dentate gyrus of adult rats: a quantitative electron microscopic analysis of terminal proliferation and reactive synaptogenesis. J Comp Neurol. 1983;214:370–386. [Google Scholar]
- 6.Gazzaley AH, Benson DL, Huntley GW, Morrison JH. Differential subcellular regulation of NMDAR1 protein and mRNA in dendrites of dentate gyrus granule cells after perforant path transection. J Neurosci. 1997;17:2006–2017. doi: 10.1523/JNEUROSCI.17-06-02006.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ginsberg SD. Glutamatergic neurotransmission expression profiling in the mouse hippocampus after perforant-path transection. Am J Geriatr Psychiatry. 2005;13:1052–1061. doi: 10.1176/appi.ajgp.13.12.1052. [DOI] [PubMed] [Google Scholar]
- 8.Hardman R, Evans DJ, Fellows L, Hayes B, Rupniak HT, Barnes JC, Higgins GA. Evidence for recovery of spatial learning following entorhinal cortex lesions in mice. Brain Res. 1997;758:187–200. doi: 10.1016/s0006-8993(97)00223-0. [DOI] [PubMed] [Google Scholar]
- 9.Steward O, Loesche J, Horton WC. Behavioral correlates of denervation and reinnervation of the hippocampal formation of the rat: open field activity and cue utilization following bilateral entorhinal cortex lesions. Brain Res Bull. 1977;2:41–48. doi: 10.1016/0361-9230(77)90023-5. [DOI] [PubMed] [Google Scholar]
- 10.Reeves TM, Smith DC. Reinnervation of the dentate gyrus and recovery of alternation behavior following entorhinal cortex lesions. Behav Neurosci. 1987;101:179–186. doi: 10.1037//0735-7044.101.2.179. [DOI] [PubMed] [Google Scholar]
- 11.Matthews DA, Cotman C, Lynch G. An electron microscopic study of lesion-induced synaptogenesis in the dentate gyrus of the adult rat. I. Magnitude and time course of degeneration. Brain Res. 1976;115:1–21. doi: 10.1016/0006-8993(76)90819-2. [DOI] [PubMed] [Google Scholar]
- 12.Matthews DA, Cotman C, Lynch G. An electron microscopic study of lesion-induced synaptogenesis in the dentate gyrus of the adult rat. II. Reappearance of morphologically normal synaptic contacts. Brain Res. 1976;115:23–41. doi: 10.1016/0006-8993(76)90820-9. [DOI] [PubMed] [Google Scholar]
- 13.Deller T, Nitsch R, Frotscher M. Phaseolus vulgaris-leucoagglutinin tracing of commissural fibers to the rat dentate gyrus: evidence for a previously unknown commissural projection to the outer molecular layer. J Comp Neurol. 1995;352:55–68. doi: 10.1002/cne.903520105. [DOI] [PubMed] [Google Scholar]
- 14.Nadler JV, Cotman CW, Lynch G. Histochemical evidence of altered development of cholinergic fibers in the rat dentate gyrus following lesions. I. Time course after complete unilateral entorhinal lesion at various ages. J Comp Neurol. 1977;171:561–587. doi: 10.1002/cne.901710409. [DOI] [PubMed] [Google Scholar]
- 15.Marrone DF, Petit TL. The role of synaptic morphology in neural plasticity: structural interactions underlying synaptic power. Brain Res Brain Res Rev. 2002;38:291–308. doi: 10.1016/s0165-0173(01)00147-3. [DOI] [PubMed] [Google Scholar]
- 16.Martin LJ, Blackstone CD, Levey AI, Huganir RL, Price DL. Cellular localizations of AMPA glutamate receptors within the basal forebrain magnocellular complex of rat and monkey. J Neurosci. 1993;13:2249–2263. doi: 10.1523/JNEUROSCI.13-05-02249.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Martin LJ, Blackstone CD, Huganir RL, Price DL. Cellular localization of a metabotropic glutamate receptor in rat brain. Neuron. 1992;9:259–270. doi: 10.1016/0896-6273(92)90165-a. [DOI] [PubMed] [Google Scholar]
- 18.Petralia RS, Wenthold RJ. Light and electron immunocytochemical localization of AMPA-selective glutamate receptors in the rat brain. J Comp Neurol. 1992;318:329–354. doi: 10.1002/cne.903180309. [DOI] [PubMed] [Google Scholar]
- 19.Petralia RS, Yokotani N, Wenthold RJ. Light and electron microscope distribution of the NMDA receptor subunit NMDAR1 in the rat nervous system using a selective anti-peptide antibody. J Neurosci. 1994;14:667–696. doi: 10.1523/JNEUROSCI.14-02-00667.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Petralia RS, Wang Y-X, Wenthold RJ. Histological and ultrastructural localization of the kainate receptor subunits, KA2 and GluR6/7, in the rat nervous system using selective antipeptide antibodies. J Comp Neurol. 1994;349:85–110. doi: 10.1002/cne.903490107. [DOI] [PubMed] [Google Scholar]
- 21.Rothstein JD, Martin LJ, Levey AI, Dykes-Hoberg M, Jin L, Wu D, Nash N, Kuncl RW. Localization of neuronal and glial glutamate transporters. Neuron. 1994;13:713–725. doi: 10.1016/0896-6273(94)90038-8. [DOI] [PubMed] [Google Scholar]
- 22.Hein C, Horvath E, Kugler P. Glutamate transporter expression in astrocytes of the rat dentate gyrus following lesion of the entorhinal cortex. Eur J Neurosci. 2001;13:1839–1848. doi: 10.1046/j.0953-816x.2001.01559.x. [DOI] [PubMed] [Google Scholar]
- 23.Dong H, Zhang P, Song I, Petralia RS, Liao D, Huganir RL. Characterization of the glutamate receptor-interacting proteins GRIP1 and GRIP2. J Neurosci. 1999;19:6930–6941. doi: 10.1523/JNEUROSCI.19-16-06930.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hirbec H, Francis JC, Lauri SE, Braithwaite SP, Coussen F, Mulle C, Dev KK, Coutinho V, Meyer G, Isaac JT, Collingridge GL, Henley JM. Rapid and differential regulation of AMPA and kainate receptors at hippocampal mossy fibre synapses by PICK1 and GRIP. Neuron. 2003;37:625–638. doi: 10.1016/s0896-6273(02)01191-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rosenzweig ES, Barnes CA. Impact of aging on hippocampal function: plasticity, network dynamics, and cognition. Prog Neurobiol. 2003;69:143–179. doi: 10.1016/s0301-0082(02)00126-0. [DOI] [PubMed] [Google Scholar]
- 26.Nakao K, Ikegaya Y, Yamada MK, Nishiyama N, Matsuki N. Fimbrial control of bidirectional synaptic plasticity of medial perforant path-dentate transmission. Synapse. 2003;47:163–168. doi: 10.1002/syn.10168. [DOI] [PubMed] [Google Scholar]
- 27.Adams MM, Gazzaley AH, Morrison JH. Attenuated lesion-induced N-methyl-D-aspartate receptor (NMDAR) plasticity in the dentate gyrus of aged rats following perforant path lesions. Exp Neurol. 2001;172:244–249. doi: 10.1006/exnr.2001.7794. [DOI] [PubMed] [Google Scholar]
- 28.Iwakiri M, Mizukami K, Ishikawa M, Hidaka S, Asada T. Alterations of NMDAR1 and NMDAR2a/B immunoreactivity in the hippocampus after perforant pathway lesion. Neuropathology. 2002;22:154–160. doi: 10.1046/j.1440-1789.2002.00434.x. [DOI] [PubMed] [Google Scholar]
- 29.Ying G, Huang C, Jing N, Zhou C. Identification of differentially expressed genes in the denervated rat hippocampus by cDNA arrays. Neurosci Lett. 2001;306:121–125. doi: 10.1016/s0304-3940(01)01882-1. [DOI] [PubMed] [Google Scholar]
- 30.Jensen MB, Gonzalez B, Castellano B, Zimmer J. Microglial and astroglial reactions to anterograde axonal degeneration: a histochemical and immunocytochemical study of the adult rat fascia dentata after entorhinal perforant path lesions. Exp Brain Res. 1994;98:245–260. doi: 10.1007/BF00228413. [DOI] [PubMed] [Google Scholar]
- 31.Savaskan NE, Nitsch R. Molecules involved in reactive sprouting in the hippocampus. Rev Neurosci. 2001;12:195–215. doi: 10.1515/revneuro.2001.12.3.195. [DOI] [PubMed] [Google Scholar]
- 32.Alldred MJ, Che S, Ginsberg SD. Terminal continuation (TC) RNA amplification enables expression profiling using minute RNA input obtained from mouse brain. Int J Mol Sci. 2008;9:2091–2104. doi: 10.3390/ijms9112091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Alldred MJ, Che S, Ginsberg SD. Terminal continuation (TC) RNA amplification without second strand synthesis. J Neurosci Methods. 2009;177:381–385. doi: 10.1016/j.jneumeth.2008.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Che S, Ginsberg SD. Amplification of transcripts using terminal continuation. Lab Invest. 2004;84:131–137. doi: 10.1038/labinvest.3700005. [DOI] [PubMed] [Google Scholar]
- 35.Che S, Ginsberg SD. RNA amplification methodologies. In: McNamara PA, editor. Trends in RNA research. Nova Science; Hauppauge: 2006. pp. 277–301. [Google Scholar]
- 36.Ginsberg SD. RNA amplification strategies for small sample populations. Methods. 2005;37:229–237. doi: 10.1016/j.ymeth.2005.09.003. [DOI] [PubMed] [Google Scholar]
- 37.Ginsberg SD. Transcriptional profiling of small samples in the central nervous system. Meth Mol Biol. 2008;439:147–158. doi: 10.1007/978-1-59745-188-8_10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ginsberg SD, Che S. RNA amplification in brain tissues. Neurochem Res. 2002;27:981–992. doi: 10.1023/a:1020944502581. [DOI] [PubMed] [Google Scholar]
- 39.Paxinos G, Franklin KBJ. The mouse brain in stereotaxic coordinates. 2. Academic; San Diego: 2001. [Google Scholar]
- 40.Ginsberg SD, Martin LJ, Rothstein JD. Regional deafferentation down-regulates subtypes of glutamate transporter proteins. J Neurochem. 1995;65:2800–2803. doi: 10.1046/j.1471-4159.1995.65062800.x. [DOI] [PubMed] [Google Scholar]
- 41.Ginsberg SD, Rothstein JD, Price DL, Martin LJ. Fimbria–fornix transections selectively down-regulate subtypes of glutamate transporter and glutamate receptor proteins in septum and hippocampus. J Neurochem. 1996;67:1208–1216. doi: 10.1046/j.1471-4159.1996.67031208.x. [DOI] [PubMed] [Google Scholar]
- 42.Ginsberg SD, Che S. Combined histochemical staining, RNA amplification, regional, and single cell analysis within the hippocampus. Lab Invest. 2004;84:952–962. doi: 10.1038/labinvest.3700110. [DOI] [PubMed] [Google Scholar]
- 43.Lee VM-Y, Carden MJ, Schlaepfer WW, Trojanowski JQ. Monoclonal antibodies distinguish several differentially phosphorylated states of the two largest rat neurofilament subunits (NF-H and NF-M) and demonstrate their existence in the normal nervous system of adult rats. J Neurosci. 1987;7:3474–3488. doi: 10.1523/JNEUROSCI.07-11-03474.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ginsberg SD, Hemby SE, Lee VM-Y, Eberwine JH, Trojanowski JQ. Expression profile of transcripts in Alzheimer’s disease tangle-bearing CA1 neurons. Ann Neurol. 2000;48:77–87. [PubMed] [Google Scholar]
- 45.Ginsberg SD, Hemby SE, Mufson EJ, Martin LJ. Cell and tissue microdissection in combination with genomic and proteomic applications. In: Zaborszky L, Wouterlood FG, Lanciego JL, editors. Neuroanatomical tract tracing 3: molecules, neurons, and systems. Springer; New York: 2006. pp. 109–141. [Google Scholar]
- 46.Ginsberg SD, Elarova I, Ruben M, Tan F, Counts SE, Eberwine JH, Trojanowski JQ, Hemby SE, Mufson EJ, Che S. Single cell gene expression analysis: implications for neurodegenerative and neuropsychiatric disorders. Neurochem Res. 2004;29:1054–1065. doi: 10.1023/b:nere.0000023593.77052.f7. [DOI] [PubMed] [Google Scholar]
- 47.Ginsberg SD, Che S. Expression profile analysis within the human hippocampus: comparison of CA1 and CA3 pyramidal neurons. J Comp Neurol. 2005;487:107–118. doi: 10.1002/cne.20535. [DOI] [PubMed] [Google Scholar]
- 48.Ginsberg SD. Microarray use for the analysis of the CNS. In: Squire LR, editor. Encyclopedia of neuroscience. Vol. 5. Academic; Oxford: 2009. pp. 835–841. [Google Scholar]
- 49.Ginsberg SD, Che S, Counts SE, Mufson EJ. Single cell gene expression profiling in Alzheimer’s disease. NeuroRx. 2006;3:302–318. doi: 10.1016/j.nurx.2006.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ginsberg SD, Che S, Wuu J, Counts SE, Mufson EJ. Down regulation of trk but not p75 gene expression in single cholinergic basal forebrain neurons mark the progression of Alzheimer’s disease. J Neurochem. 2006;97:475–487. doi: 10.1111/j.1471-4159.2006.03764.x. [DOI] [PubMed] [Google Scholar]
- 51.Kacharmina JE, Crino PB, Eberwine J. Preparation of cDNA from single cells and subcellular regions. Meth Enzymol. 1999;303:3–18. doi: 10.1016/s0076-6879(99)03003-7. [DOI] [PubMed] [Google Scholar]
- 52.Hemby SE, Trojanowski JQ, Ginsberg SD. Neuron-specific age-related decreases in dopamine receptor subtype mRNAs. J Comp Neurol. 2003;456:176–183. doi: 10.1002/cne.10525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Broberg P. A comparative review of estimates of the proportion unchanged genes and the false discovery rate. BMC Bioinform. 2005;6:199. doi: 10.1186/1471-2105-6-199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Reiner A, Yekutieli D, Benjamini Y. Identifying differentially expressed genes using false discovery rate controlling procedures. Bioinformatics. 2003;19:368–375. doi: 10.1093/bioinformatics/btf877. [DOI] [PubMed] [Google Scholar]
- 55.Counts SE, He B, Che S, Ginsberg SD, Mufson EJ. Galanin fiber hyperinnervation preserves neuroprotective gene expression in cholinergic basal forebrain neurons in Alzheimer’s disease. J Alzheimers Dis. 2009;18:885–896. doi: 10.3233/JAD-2009-1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kovac AD, Kwidzinski E, Heimrich B, Bittigau P, Deller T, Nitsch R, Bechmann I. Entorhinal cortex lesion in the mouse induces transsynaptic death of perforant path target neurons. Brain Pathol. 2004;14:249–257. doi: 10.1111/j.1750-3639.2004.tb00061.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nitsch R, Frotscher M. Reduction of posttraumatic transneuronal “early gene” activation and dendritic atrophy by the N-methyl-D-aspartate receptor antagonist MK-801. Proc Natl Acad Sci USA. 1992;89:5197–5200. doi: 10.1073/pnas.89.11.5197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bartesaghi R. Hippocampal-entorhinal relationships: electrophysiological analysis of the ventral hippocampal projections to the ventral entorhinal cortex. Neuroscience. 1994;61:457–466. doi: 10.1016/0306-4522(94)90426-x. [DOI] [PubMed] [Google Scholar]
- 59.Kelsey JE, Sanderson KL, Frye CA. Perforant path stimulation in rats produces seizures, loss of hippocampal neurons, and a deficit in spatial mapping which are reduced by prior MK-801. Behav Brain Res. 2000;107:59–69. doi: 10.1016/s0166-4328(99)00107-2. [DOI] [PubMed] [Google Scholar]
- 60.Kienzler F, Norwood BA, Sloviter RS. Hippocampal injury, atrophy, synaptic reorganization, and epileptogenesis after perforant pathway stimulation-induced status epilepticus in the mouse. J Comp Neurol. 2009;515:181–196. doi: 10.1002/cne.22059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Takahashi M, Liou S-Y, Kunihara M. Ca2+- and Cl-dependent, NMDA receptor-mediated neuronal death induced by depolarization in rat hippocampal organotypic cultures. Brain Res. 1995;675:249–256. doi: 10.1016/0006-8993(95)00078-5. [DOI] [PubMed] [Google Scholar]
- 62.Storm-Mathisen J, Opsahl MW. Aspartate and/or glutamate may be transmitters in hippocampal efferents to septum and hypothalamus. Neurosci Lett. 1978;9:65–70. doi: 10.1016/0304-3940(78)90049-6. [DOI] [PubMed] [Google Scholar]
- 63.Schramm M, Eimerl S, Costa E. Serum and depolarizing agents cause acute neurotoxicity in cultured cerebellar granule cells: role of the glutamate receptor responsive to N-methyl-D-aspartate. Proc Natl Acad Sci USA. 1990;87:1193–1197. doi: 10.1073/pnas.87.3.1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Martin LJ, Al-Abdulla NA, Brambrink AM, Kirsch JR, Sieber FE, Portera-Cailliau C. Neurodegeneration in excitotoxicity, global cerebral ischemia, and target deprivation: a perspective on the contributions of apoptosis and necrosis. Brain Res Bull. 1998;46:281–309. doi: 10.1016/s0361-9230(98)00024-0. [DOI] [PubMed] [Google Scholar]
- 65.Olney JW. Excitotoxicity, apoptosis and neuropsychiatric disorders. Curr Opin Pharmacol. 2003;3:101–109. [PubMed] [Google Scholar]
- 66.Sheldon AL, Robinson MB. The role of glutamate transporters in neurodegenerative diseases and potential opportunities for intervention. Neurochem Int. 2007;51:333–355. doi: 10.1016/j.neuint.2007.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ginsberg SD, Portera-Cailliau C, Martin LJ. Fimbria–fornix transection and excitotoxicity produce similar neurodegeneration in septum. Neuroscience. 1999;88:1059–1071. doi: 10.1016/s0306-4522(98)00288-7. [DOI] [PubMed] [Google Scholar]
- 68.Ginsberg SD, Martin LJ. Axonal transection in adult rat brain induces transsynaptic apoptosis and persistent atrophy of target neurons. J Neurotrauma. 2002;19:99–109. doi: 10.1089/089771502753460277. [DOI] [PubMed] [Google Scholar]
- 69.Martin LJ, Sieber FE, Traystman RJ. Apoptosis and necrosis occur in separate neuronal populations in hippocampus and cerebellum after ischemia and are associated with differential alterations in metabotropic glutamate receptor signaling pathways. J Cereb Blood Flow Metab. 2000;20:153–167. doi: 10.1097/00004647-200001000-00020. [DOI] [PubMed] [Google Scholar]
- 70.Portera-Cailliau C, Price DL, Martin LJ. Non-NMDA and NMDA receptor-mediated excitotoxic neuronal deaths in adult brain are morphologically distinct: further evidence for an apoptosis-necrosis continuum. J Comp Neurol. 1997;378:88–104. [PubMed] [Google Scholar]
- 71.Miettinen R, Kotti T, Tuunanen J, Toppinen A, Riekkinen P, Sr, Halonen T. Hippocampal damage after injection of kainic acid into the rat entorhinal cortex. Brain Res. 1998;813:9–17. doi: 10.1016/s0006-8993(98)00915-9. [DOI] [PubMed] [Google Scholar]
- 72.Diekmann S, Ohm TG, Nitsch R. Long-lasting trans-neuronal changes in rat dentate granule cell dendrites after entorhinal cortex lesion. A combined intracellular injection and electron microscopy study. Brain Pathol. 1996;6:205–214. doi: 10.1111/j.1750-3639.1996.tb00846.x. [DOI] [PubMed] [Google Scholar]
- 73.Nitsch R, Bader S, Frotscher M. Reorganization of input synapses of parvalbumin-containing neurons in the rat fascia dentata following entorhinal lesion. Neurosci Lett. 1992;135:33–36. doi: 10.1016/0304-3940(92)90129-u. [DOI] [PubMed] [Google Scholar]
- 74.Nitsch R, Frotscher M. Transneuronal changes in dendrites of GABAergic parvalbumin-containing neurons of the rat fascia dentata following entorhinal lesion. Hippocampus. 1993;3:481–490. doi: 10.1002/hipo.450030409. [DOI] [PubMed] [Google Scholar]
- 75.Ginsberg SD, Martin LJ. Ultrastructural analysis of the progression of neurodegeneration in the septum following fimbria–fornix transection. Neuroscience. 1998;86:1259–1272. doi: 10.1016/s0306-4522(98)00136-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.