Abstract
Transfer RNA (tRNA) translocates inside the ribosome during translation. We studied the interaction strengths between the ribosome and tRNA at various stages of translocation. We utilized an optical trap to measure the mechanical force to rupture tRNA from the ribosome. We measured the rupture forces of aminoacyl tRNA or peptidyl tRNA mimic from the ribosome in a prepeptidyl transfer state, the pretranslocational state, and the posttranslocational state. In addition, we measured the interaction strength between the ribosome and aminoacyl-tRNA in presence of viomycin. Based on the interaction strengths between the ribosome and tRNA under these conditions, 1), we concluded that tRNA interaction with the 30S subunit is far more important than the interaction with the 50S subunit in the mechanism of translocation; and 2), we propose a mechanism of translocation where the ribosomal ratchet motion, with the aid of EF-G, drives tRNA translocation.
Introduction
Translation, or protein synthesis by the ribosome, is a pivotal process to maintain cell viability (1–12). Aminoacyl tRNA (aa-tRNA) carries amino-acid residues required for translation. To synthesize a protein, the ribosome selects a series of aa-tRNAs dictated by the sequence of mRNA that is eventually translated into the sequence of amino-acid residues in the protein. The prokaryotic ribosome is composed of two subunits (30S and 50S) and has three transfer RNA (tRNA) accommodation sites (the E-, P-, and A-sites; see Fig. 1). Upon the selection of aa-tRNA according to the codon of mRNA in the 30S A-site, the nascent peptide chain is transferred to the aminoacyl group of the newly selected aa-tRNA. Upon the peptidyl transfer, the ribosome complex enters the pretranslocational (PRE) state where the two tRNAs form an equilibrium between classical and hybrid states. In classical states, tRNAs occupy either the P-site or the A-site. In hybrid states, each tRNA occupies two tRNA accommodation sites (one tRNA in 30S P- and 50S E-sites and the other in 30S A- and 50S P-sites as shown in Fig. 1). A GTPase elongation factor G (EF-G) binds to the ribosome in the PRE state followed by GTP hydrolysis and subsequent translocation (13–23). During translocation, the A-site tRNA (A-tRNA) moves to the P-site and the initial P-site tRNA (P-tRNA) moves to the E-site. Upon translocation, the ribosome complex enters the posttranslocational (POST) state. The E-site tRNA in a POST complex dissociates from the complex marking the end of a peptide-chain elongation cycle.
Figure 1.
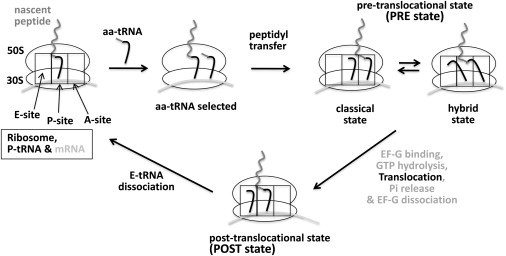
Aminoacyl-tRNA (aa-tRNA) selection and translocation in a peptide chain elongation cycle during translation. The prokaryotic ribosome is composed of two subunits, 30S and 50S (which have three tRNA accommodation sites, E, P, and A), shown as ellipses and rectangles, respectively. Upon the selection of aa-tRNA according to the codon of mRNA, the nascent peptidyl chain is transferred to the aminoacyl group of the newly selected aa-tRNA (the inverted L-shape). Upon peptidyl transfer, the two tRNAs form equilibrium between classical and hybrid states (the PRE state). EF-G binds to the ribosome in the PRE state followed by GTP hydrolysis and subsequent translocation. Upon translocation (the POST state), the aa-tRNA moves to the P-site and the initial P-tRNA moves to the E-site and eventually dissociates from the complex marking the end of a peptide chain elongation cycle.
Viomycin, an antibiotic drug, inhibits spontaneous and EF-G dependent translocation (15,16,24–26). Although the mechanism of translocation inhibition by viomycin remains unclear, it has been known that viomycin binds at the interface between the two ribosomal subunits, stabilizes some conformations of the ribosome, strengthens the subunit association, and suppresses the conformational fluctuations of PRE complexes (15,16,24–27). There are mixed results about whether the stalled PRE complexes in presence of viomycin are in the classical state or in the hybrid state (15,16,25–27).
The ribosome and aa-tRNA form multiple contacts according to biochemical assays and crystal structures, as contacts:
-
1.
Between the H69, h44 (A1492 and A1493), h18 (G530), and the tRNA anticodon stem loop.
-
2.
Between the H89, S12, and the tRNA CCA end and acceptor stem.
-
3.
Between the H38, L16, and the tRNA elbow regions (7,28–32).
It has been shown that changes in these ribosomal elements alter the fidelity of translation (33,34) and the efficiency of translocation (35). It has long been hypothesized that the ribosome is unlocked upon peptidyl transfer to weaken the interactions between the ribosome and tRNA/mRNA for subsequent translocation (36,37). Upon translocation, the ribosome is locked again and the interactions between the ribosome and tRNA/mRNA are restored. Experimental results supporting this hypothesis have been reported. In particular, the spontaneous ribosomal ratchet motion in the PRE state is suggested as a signature of the unlocked ribosome (38–40).
Major structural changes in the ribosome during unlocking are seen mostly in the 30S subunit. For instance, changes in the relative position of the head to the body domain in the unlocked state have been reported (19) and conformational changes in the head domain including h34 in the vicinity of the codon and some bridge elements in h44 upon unlocking of the ribosome in the PRE state were evidenced (32,41,42). A kinetics study has shown that the dissociation rate of peptidyl-tRNAPhe from the A-site in a PRE complex is considerably higher than the dissociation rate of Phe-tRNAPhe from the A-site in the prepeptidyl transfer state (43), although the absolute dissociation rates of tRNAs from the ribosome heavily depends on the identity of tRNAs and their aminoacyl groups (44–46). Although dissociation rates of aa-tRNAs in the A- and P-sites from the 70S ribosome and the 30S subunit have been reported previously (43–47), to our knowledge, there has not been any study reported to directly measure the interaction strengths between the ribosome and tRNA at various stages of translocation.
A tightly focused laser beam can trap a small object (48). The size of the object that can be easily trapped is typically in the range of a few hundred nanometers to a few micrometers, depending on the shape and the intensity of the beam and the refractive indices of the object and its surroundings, all of which determine the trapping force, or the stiffness of the trap (49). An optical trap is a potential well for the trapped object, and the shape of the well around the center of the trap is approximately a quadratic function of the displacement between the center of the trap and the center of the trapped object (49). Thus, the force exerted on the trapped object displaced from the beam center is linearly proportional to the displacement with a linearity constant defined as the trap stiffness. The stiffness of a trap can be determined by several different methods (49).
To study the interaction strength between two elements in a system, the rupture force of one element from the other measured with an optical trap can be used (50). Typically, in these measurements, one of the two elements is attached to a surface of the sample chamber and the other is attached to a small object (e.g., a plastic bead) that can be efficiently trapped in a focused laser beam. Then one of the two elements (i.e., the surface or the trapped bead) is moved to one direction at a constant speed typically in the range of tens of nanometers at approximately a few hundred nanometers per second. While the element attached to the trapped object is being pulled, the displacement of the trapped object from the trap center is monitored. The displacement at the moment of rupture is converted to the rupture force.
Here, we report interaction strengths between the ribosome and aa-tRNA at various stages of translocation measured as rupture forces. We used an optical trap to rupture aa-tRNA from the A- and P-sites of the ribosome under various conditions to study how the interaction strength between the ribosome and tRNA changes during the course of translocation.
Materials and Methods
Buffer conditions
For ribosome storage and rupture force measurements, we used a polymix buffer (51) that contains 5 mM phosphate (pH = 7.5), 90 mM KCl, 0.5 mM CaCl2, 4 mM (or 15 mM) MgCl2, 5 mM NH4(OAc), 1 mM DTT, 8 mM putrecine, and 1 mM spermidine. For factor storage, we used a buffer containing 10 mM Tris-HCl (pH = 7.2), 50 mM KCl, 5 mM 2-mercaptoethanol, and 50% glycerol. Viomycin sulfate was purchased from U.S. Pharmacopeia (Rockville, MD).
Ribosome, mRNA, and tRNA preparation
We followed a protocol published elsewhere to purify prokaryotic 70S tight-coupled ribosome from an Escherichia coli strain MRE600 (11,52). The 30S ribosomal subunit was prepared by a sucrose gradient run of the 70S particles dialyzed for 3 h at 4°C against the polymix buffer containing 1 mM Mg2+. The mRNA was purchased from Fisher Scientific (formerly Dharmacon, Lafayette, CO). The sequence of mRNA is derived from T4 gp32 and contained a biotin for surface immobilization at the 5′ end:
Purified tRNAfMET and tRNAPhe were purchased from Chemical Block (Limassol, Cyprus). We used purified tRNAfMET synthetase and tRNAPhe synthetase to aminoacylate tRNAs and used purified formyltransferase to formylate Met-tRNAfMET. N-acetyl-Phe-tRNAPhe (AcPhe-tRNAPhe) as a mimic of peptidyl tRNA was prepared by acetylation of Phe-tRNAPhe as described elsewhere (27,53). A succinimidyl ester functionalized polyethyleneglycol (Bio-PEG-SVA 5000 MW from Laysan Bio, Arab AL) was attached by conjugating the ester group to the acp3U47 of the tRNA. This PEG has a biotin at the other end to attach the molecule to a plastic bead coated with NeutrAvidin (ThermoScientific, Rockford, IL). The aminoacyl-tRNA used in the measurement has the structure of Bead-NeutrAvidin-biotin-PEG (5000 MW)-tRNA.
Factor purification
We used bacterial strains that overexpress tRNAfMET synthetase, tRNAPhe synthetase, formyltransferase, and EF-G to purify these enzymes after an IPTG-induced expression protocol. Briefly, LB media containing 50 μg/mL kanamycin was inoculated with an overexpressing strain, followed by 4∼6 h of incubation at 37°C to a midlog phase and subsequent IPTG induction for 2 h at 37°C. Grown cells were lysed and proteins were purified with a standard His-tag purification protocol using imidazole. Proteins were dialyzed against a 50% glycerol buffer and stored at −20°C.
Ribosome complex formation, aa-tRNA attachment to beads, and surface functionalization
A quantity of 500 nM 70S tight-coupled ribosome particles were incubated with 500 nM mRNA for 5 min at 37°C followed by the addition of fMet-tRNAfMet (final concentration 500 nM) and incubation at 30°C for 10 min. The mixture was filtered through a S300HR spin column (GE Healthcare, Piscataway NJ) to remove excess tRNA and mRNA. The quantity of 2 μM Phe-tRNAPhe was incubated with 0.5% plastic beads (1 μm diameter, coated with NeutrAvidin from ThermoScientific; 0.5% is equivalent to ∼0.02 nM, Catalog No. F-8777 from Invitrogen, Hercules, CA) overnight at room temperature. Free tRNAs were filtered out three times with a 100-kDa cutoff centrifugal filter device (Amicon filter from Millipore, Billerica, MA). A surface of a glass microscope slide was functionalized with an aminosilane reagent ((3-Aminopropyl)triethoxysilane from Sigma-Aldrich, St. Louis, MO) followed by PEGylation with a 1:100 mixture of SVA-PEG (MW 5000):Bio-SVA-PEG (MW 5000) (Laysan Bio).
Instrument and measurements
Streptavidin was incubated on the PEGylated microscope slide followed by a buffer wash and the preformed ribosome/mRNA/fMET-tRNAfMET complex was subsequently injected in the chamber. The concentration of the complex was at 30 pM to ensure an appropriate range of surface concentration to avoid tethering of a tRNA labeled bead to multiple ribosome complexes. Under this condition, we observe approximately one ribosome complex within 8.2–8.2 μm2 surface area. The typical surface concentration of the complex during the measurements can be verified by a fluorescence image of the ribosome complex under the measurement condition as shown in the Supporting Material. The sample chamber was rinsed again and the preformed Phe-tRNAPhe (2 μM) attached to the plastic beads was injected into the chamber followed by incubation for 2 h at room temperature. The sample slide was mounted on a modified model No. TE2000 inverted microscope (Nikon, Melville, NY).
Bead images were taken through a 100× 1.49 NA oil immersion objective lens (CFI Apochromat TIRF, Nikon, Japan) at 33 Hz with a charge-coupled device camera. Bright-field illumination was achieved with a tungsten lamp through a condenser mounted on the upper side of the sample slide (a schematic drawing of the setup is in the Supporting Material). The center position of a bead was determined in real-time by an algorithm that finds the centroid of the intensity pattern of the bead image on a charge-coupled device. The precision of the position determined with the method was typically within ±2 nm based on a series of position determinations of a bead stuck on a glass surface. The position of the trap beam was determined with a trapped free bead in a solution by measuring the position of the trapped bead. The stiffness of the optical trap was determined with the power spectrum method and confirmed with the equipartition method (49). We used a quadrant diode (diode and amplifier from Current Designs, Philadelphia, PA) as the high-frequency detection device for the stiffness determination. To record the positional fluctuations of a bead at a high frequency (5 kHz), we used the scattered trap beam by a trapped free bead imaged on a quadrant diode by the objective lens mounted on the upper side of the sample slide. The stiffness of the trap at 1.50 W 1064 nm trap beam (before entering the microscope) was 0.15 pN/nm.
After incubating the beads with the ribosome complex, fluctuating beads (i.e., beads attached to aa-tRNA accommodated into a ribosome/mRNA/fMet-tRNAfMET complex) were visually identified in the sample chamber. Once a fluctuating bead was identified, the bead was located at the center of the optical trap by moving a piezo-electrically driven microscope stage based on a P-733.3DD stage from Physik Instrumente (Karlsruhe, Germany). The piezo-stage was controlled by home-built software written in Visual C (Ver. 6.0; Microsoft, Redmond, WA). After locating the bead at the predetermined center of the optical trap precisely by controlling the piezo-stage with the software, the power of the trap laser beam (1064 nm 2.5 W max, COMPASS 1064–2500 mN; Coherent, Santa Clara, CA) was turned up to 1.50 W from 0.00 W (set to be at the minimum power and blocked with a shutter). The piezo-stage was moved to one direction at a rate of 100 nm/s until, shortly after a rupture event is identified, the motion of the piezo-stage reaches its maximum traveling distance to one direction (15 μm) or the bead displacement from the trap beam center reaches 300 nm (Fig. 2).
Figure 2.
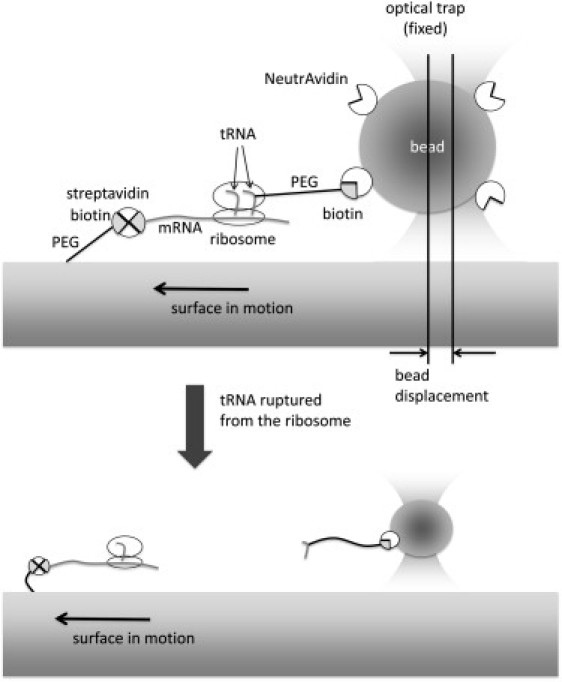
Experimental setup for the rupture force measurements. Please see the Materials and Methods for a detailed description. Briefly, the position of aa-tRNA attached to a polystyrene bead trapped in a focus of a laser beam is fixed, although the ribosome complex is pulled away from the tRNA by moving the microscope stage at a constant speed. An abrupt decrease in the displacement of the bead from the trap center is observed upon a rupture event as shown in Fig. 3.
Results
Fig. 3 shows typical rupture events of aa-tRNA (Phe-tRNAPhe) from a ribosome complex. The abrupt decreases in the displacement of the bead from the trap center at ∼4.5 s, 3.5 s, and 2.2 s, respectively, in the three examples indicate the rupture events of aa-tRNA from the complex. The maximum displacement of the bead from the trap center right before the rupture multiplied by the trap stiffness (= 0.15 pN/nm) yields the rupture force of tRNA. In the three examples, we found that 62 nm, 45 nm, and 30 nm of bead displacements from the trap center or 9.3 pN, 6.8 pN, and 4.5 pN of forces were required to rupture the tRNA form the ribosome complexes. We collected rupture forces of tRNA from these rupture events excluding rupture forces weaker than 2 pN to avoid any error due to too low rupture forces to accurately measure. Rupture forces from ∼70 to ∼250 rupture events were collected per case and rupture force histograms for six cases were constructed as shown in Fig. 4.
Figure 3.
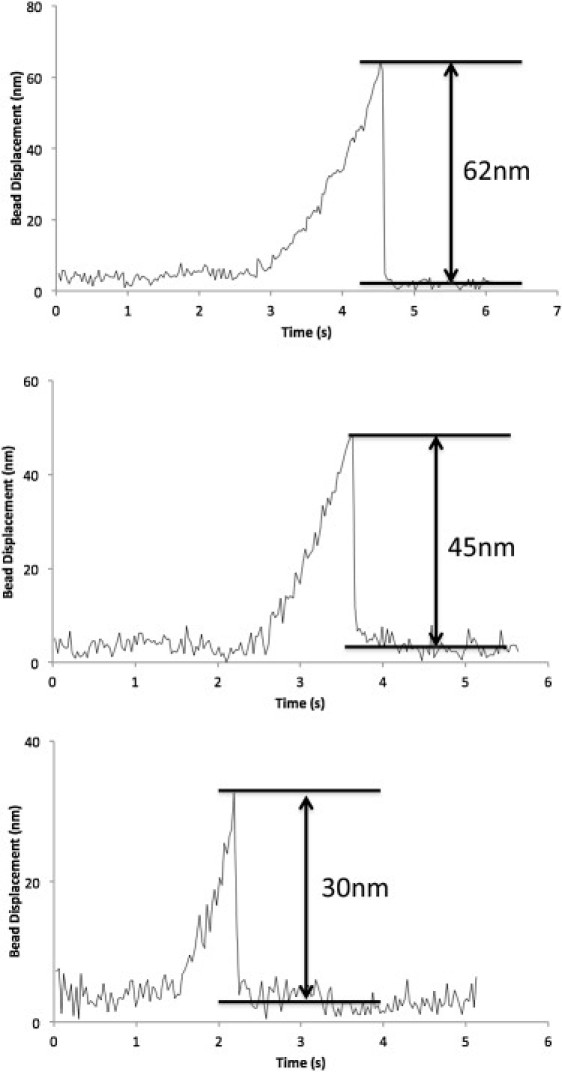
Three examples of trapped bead displacements from the trap center plotted against the elapsed time from the starting point of the constant speed ribosome pulling (100 nm/s) from the tRNA. The shown bead displacements of 62 nm, 45 nm, and 30 nm correspond to the rupture forces of 9.3 pN, 6.8 pN, and 4.5 pN, respectively.
Figure 4.
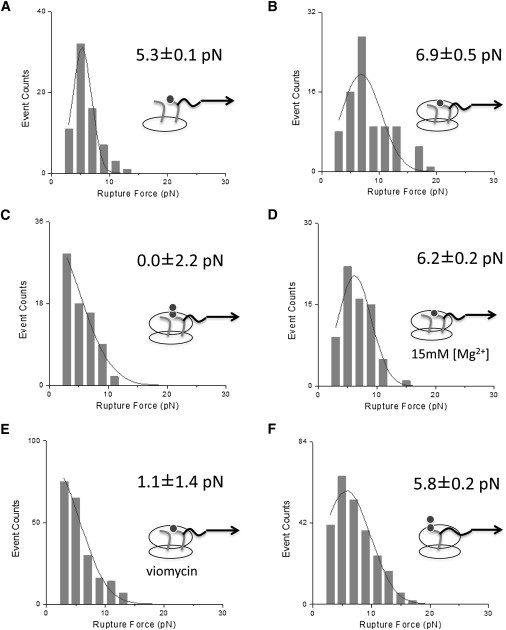
Histograms of tRNA rupture force from the ribosome. All the measurements were done at 4 mM [Mg2+] except for those in panel D. (A) Phe-tRNAPhe rupture from the A-site of the 30S subunit containing mRNA and tRNAfMet in the P-site. (B) Phe-tRNAPhe rupture from the A-site of the ribosome containing mRNA and tRNAfMet in the P-site. (C) AcPhe-tRNAPhe rupture from the A-site of the ribosome containing mRNA and tRNAfMet in the P-site. (D) Phe-tRNAPhe rupture from the A-site of the ribosome containing mRNA and tRNAfMet in the P-site at 15 mM [Mg2+]. (E) Phe-tRNAPhe rupture from the A-site of the ribosome containing mRNA and tRNAfMet in the P-site in presence of viomycin. (F) Phe-tRNAPhe rupture from the ribosome containing mRNA and fMet-tRNAfMet after incubating with EF-G/GTP.
Fig. 4 A shows the rupture force of Phe-tRNAPhe from the 30S subunit of the ribosome complexed with mRNA and deacylated tRNAfMet is 5.3 ± 0.1pN. The rupture force of Phe-tRNAPhe from the 70S ribosome complexed with mRNA and deacylated tRNAfMet is 6.9 ± 0.5 pN (Fig. 4 B), which is 1.6 ± 0.5 pN higher than the rupture force from the 30S only complex. It has been shown that the aa-tRNA accommodation efficiency by the ribosome is unaffected by the acylation state of P-tRNA (47,53,54). The tRNAs are in the classical states in a 70S complex with deacylated tRNA in the P-site and aa-tRNA in the A-site as previously shown (27). Thus, this ribosome complex likely resembles the conformational state right before peptidyl transfer. The rupture force of mRNA/tRNA from the ribosome has been previously measured (50). Based on the report which used a similar trap stiffness to ours (0.18 pN/nm vs. 0.15 pN/nm) and the same loading rate as ours (100 nm/s), tRNA rupture force out of a ribosome complex in these states can be indirectly estimated to be 5∼10 pN, which is in good agreement with our results.
After peptidyl transfer, the ribosome complex enters the PRE state where the ribosome is unlocked to allow the intersubunit ratchet motion (36–38,55) and the two tRNAs are in an equilibrium between classical states and hybrid states (56). At 4 mM [Mg2+], the equilibrium shift toward hybrid states and the dwell time of tRNAs in hybrid states is almost the same as the dwell time in classical states (27). Fig. 4 C shows the rupture force of AcPhe-tRNAPhe from a ribosome complex with deacylated P-tRNA (tRNAfMet). Because AcPhe-tRNAPhe is a mimic of peptidyl tRNA, combined with deacylated tRNA (tRNAfMet) in the P-site, this ribosome complex resembles a PRE complex.
The population of the beads tethered to these ribosome complexes is considerably lower than those tethered to the complexes in Fig. 4 B (i.e., the prepeptidyl transfer complexes). This result is in good agreement with a previous article reporting that the dissociation rate of AcPhe-tRNAPhe from a PRE complex is much higher than the dissociation rate of Phe-tRNAPhe from the A-site before peptidyl transfer (47). The rupture force of this peptidyl-tRNA mimic in a classical/hybrid state equilibrium (i.e., in the PRE state) is 0.0 ± 2.2 pN (Fig. 4 C, 0∼0.52 pN within a ∼98% confidence interval), which is 6.9 ± 2.3 pN lower than the force required to rupture Phe-tRNAPhe in the classical state before peptidyl transfer (Fig. 4 B).
In another set of rupture force measurements, we prepared a ribosome complex with mRNA, deacylated tRNAfMet, and Phe-tRNAPhe at 15 mM [Mg2+]. The rupture force of Phe-tRNAPhe from the A-site at 15 mM [Mg2+] was measured to be 6.2 ± 0.2 pN (Fig. 4 D), which is the same as the result at 4 mM [Mg2+] within error (6.9 ± 0.5 pN, Fig. 4 B). This result is in good agreement with the result from a previous single molecule study based on fluorescence resonance energy transfer (27). In the report, Phe-tRNAPhe in the A-site resides mostly in the classical state and its lifetime in the classical state is unaffected by changing [Mg2+] from 3.5 mM to 15 mM. Based on this report, it is expected that the rupture force of Phe-tRNAPhe in the A-site would not depend on Mg2+ concentration at >3.5 mM.
Next, to examine the effect of viomycin in the interaction between the ribosome and tRNA, we prepared a ribosome complex with mRNA, tRNAfMet, and Phe-tRNAphe and added 0.05 mM viomycin to the sample followed by 10-min incubation and subsequent rupture force measurements. Upon incubation, the population of the beads tethered to the ribosome complexes was considerably reduced to the degree observed with the PRE complexes (Fig. 4, C and E), which suggested that the rupture force of Phe-tRNAphe from the complex would be as low as that from the PRE complexes. Indeed, the measured rupture force of Phe-tRNAphe from the A-site in the presence of viomycin is 1.1 ± 1.4pN (Fig. 4 E), which is the same as the rupture force of AcPhe-tRNAPhe from the PRE complexes (0.0 ± 2.2 pN) within error, and is much smaller than the rupture force of Phe-tRNAPhe from the A-site in the classical state before peptidyl transfer (6.9 ± 0.5 pN). This result suggests that viomycin weakens the interactions between the ribosome and A-tRNA, possibly by stalling the complex in a PRE-like state.
Finally, we prepared a ribosome/mRNA/fMet-tRNAfMet/Phe-tRNAPhe complex and subsequently added EF-G/GTP to the complex to measure the rupture force of peptidyl tRNA (i.e., fMet-Phe-tRNAPhe) from the P-site. Previous studies on the tRNA dynamics in a ribosome/mRNA/fMet-tRNAfMet/Phe-tRNAPhe complex have shown that the two tRNAs undergo peptidyl transfer as soon as the two tRNAs are accommodated in the ribosome and subsequently form a PRE complex (47). Upon the addition of EF-G to this PRE complex, fMet-Phe-tRNAphe translocates to the P-site. The rupture force of this tRNA from the P-site of the ribosome is 5.8 ± 0.2 pN (Fig. 4 D), which is lower than the rupture force of Phe-tRNAPhe from the A-site by 1.1 ± 0.5 pN. A biochemical study (54) has reported that the dissociation rate of Phe-tRNAPhe from the P-site is slightly higher than that from the A-site, suggesting a slightly stronger interaction of tRNA with the A-site than with the P-site, which is in good agreement with our result.
Discussion
According to our measurements, the rupture force of Phe-tRNAPhe from the A-site of the 30S ribosomal complex is only 1.6 pN lower or 23% weaker than the rupture force of Phe-tRNAphe in the A-site of the 70S complex, indicating that the interaction between 50S A-site and aa-tRNA is much weaker than that between the 30S A-site and the tRNA. This may be because the tRNAs have strong codon-anticodon interactions with mRNA in the 30S subunit. In addition, the conformational rearrangements of the 30S subunit to a closed form upon the binding of a cognate aa-tRNA can also strengthen the interaction with aa-tRNA (57).
This kind of dynamic interaction between the 30S subunit and tRNAs during the course of tRNA selection may also be a key player in translocation. The stronger tRNA interaction with the 30S subunit than with the 50S subunit suggests that the interaction of tRNA with the 30S subunit is more important than with the 50S subunit in the mechanism of translocation for which the interactions between tRNAs/mRNA and the ribosome must be weakened under the locking/unlocking hypothesis. This suggestion is also supported by the fact that the most extensive conformational rearrangements in the ribosome upon the formation of the PRE state are in the 30S subunit (19).
The locking/unlocking hypothesis is in good agreement with our results, showing a dramatic decrease in the rupture force of tRNA from the A-site upon peptidyl transfer. The chemical change upon peptidyl transfer is only in the aminoacyl group of tRNAs, which interact only with the 50S subunit. Consequently, the rupture force from the PRE complex must be as high as at least the rupture force of tRNA from the 30S subunit (Fig. 4 A), unless there is any conformational change in the 30S subunit upon peptidyl transfer. This is contrary to what we observed—thereby verifying considerable conformational changes in the 30S subunit upon entering the PRE state.
These conformational changes in the 30S subunit resulted in significantly weakened interactions between the 30S and tRNA, which would facilitate the motions of tRNA during translocation. This facilitated motion of tRNA in a PRE complex due to the conformational rearrangements of the ribosome must persist whether tRNA and the ribosome is in the classical state or in the hybrid state. This is because our result shows almost negligible rupture force, despite tRNA and the ribosome spending half of the time in the classical state. Therefore, what characterizes the PRE state may be the conformational rearrangements of the ribosome that weaken the interaction between the ribosome and tRNA, regardless of the complex being in the classical or in the hybrid state.
According to our measurements, the rupture force of Phe-tRNAPhe from the A-site of the ribosome at 4 mM [Mg2+] is almost the same as the rupture force at 15 mM [Mg2+]. This result is in good agreement with a fluorescence-based study on the dwell times of aa-tRNA in the A-site at various [Mg2+] levels (27). These results support a hypothesis dictating that the interaction of tRNA with the ribosome in the classical state is kept constant over a wide range of [Mg2+]. The robust interaction between the ribosome and tRNA before peptidyl transfer would ensure proper alignment of the peptidyl group of P-tRNA with the aminoacyl group of the A-tRNA for efficient peptidyl transfer.
Viomycin binds at the subunit interface between H69 and h44 according to a crystal structure (26). Although it is now known that viomycin directly interacts with the ribosome to inhibit translocation, the mechanism still remains unclear. An early kinetics study has excluded the stabilization of tRNA binding to the ribosome from the mechanism of viomycin action (25). This study also reported that the unlocking rearrangement of the ribosome conformation preceding translocation is not affected by viomycin. An ensemble FRET study combined with chemical probing reported that the viomycin drives tRNA to hybrid states and stalls the ribosome in a ratcheted state that is identical to the hybrid state observed by a cryo-EM study (15). These results suggest that viomycin stalls the ribosome complex in the hybrid state where the interaction between the ribosome and tRNA is very weak according to our rupture force measurements.
On the other hand, a single molecule FRET study reported that tRNAs favor classical states in the presence of viomycin, but still show significant positioning in hybrid states (27). In line with this report, it was suggested that the drug stabilizes the peptidyl tRNA in the A-site and consequently inhibits translocation possibly by blocking structural fluctuations of the ribosome and tRNA based on a crystal structure (26). At the moment, based on our and previous results, we cannot conclude whether the ribosome complex bound with viomycin is stalled in the hybrid state or in the classical state. One possible explanation to embrace all of these results is that viomycin may create high-energy barriers surrounding the two conformations of the ribosome in the PRE state and the complexes can be trapped in one or the other conformation depending on the experimental conditions. In either conformation, the interaction between the ribosome and tRNA is very weak according to our measurements. Therefore, viomycin stalls the ribosome complex in a state where the interaction between tRNA and the ribosome is significantly weakened regardless of the conformation. Although weak tRNA interaction with the ribosome is a prerequisite for translocation under the locking/unlocking hypothesis, viomycin inhibits translocation.
Based on these results, we suggest that restoration of the weakened interaction between the ribosome and tRNA is critical in the mechanism of translocation. The restoration would likely coincide with conformational changes of the ribosome for relocking. To this end, we propose a hypothesis dictating that the structural changes of the ribosome accompanying locking may be what drives translocation. One possible scenario is that the back-ratcheting motion of the ribosome (38,39) in the PRE state (i.e., returning to the classical state from the hybrid state) drives translocation by forcing EF-G attached to the 50S subunit to push A-tRNA to the back-ratcheting direction of the ribosome.
This scenario is well supported by most of the existing results. For instance, EF-G can catalyze translocation only when there is tRNA in the A-site according to a biochemical study (58). As EF-G pushes A-tRNA for translocation by the aid of ribosome back-ratcheting in this model, A-tRNA is a critical element in the mechanism of translocation. More interestingly, early kinetics studies have shown that EF-G domains VI/V are required for efficient translocation (22,59). EF-G domains VI/V do not contain the GTP binding pocket nor directly interact with the GTPase-associated center in the 50S subunit. As the domains likely compete with A-tRNA anticodon stem loop for space in the 30S subunit, these domains may push A-tRNA during the back-ratcheting of the ribosome. Viomycin in this hypothesis inhibits translocation by stalling the ribosome complex in the classical or in the hybrid state and consequently by blocking the ratchet motion of the ribosome.
Conclusions
Based on the interaction strengths between the ribosome and tRNA at various stages of translocation, 1), we concluded that the tRNA interaction with the 30S subunit is far more important than the interaction with the 50S subunit in translocation; and 2), we suggest a mechanism of translocation where the ribosomal ratchet motion with the aid of EF-G drives tRNA translocation.
Acknowledgments
We thank Dr. Michael Ibba (Ohio State University) and Dr. Yves Mechulam (Laboratoire de Biochimie, Ecole Polytechnique) for sending us tRNA synthetase plasmids.
This work was supported by the National Institutes of Health (under grant No. GM0799601), a Searle Scholar Award, and a Camille and Henry Dreyfus New Faculty Award to T.-H.L.
Supporting Material
References
- 1.Green R., Noller H.F. Ribosomes and translation. Annu. Rev. Biochem. 1997;66:679–716. doi: 10.1146/annurev.biochem.66.1.679. [DOI] [PubMed] [Google Scholar]
- 2.Rodnina M.V., Wintermeyer W. Fidelity of aminoacyl-tRNA selection on the ribosome: kinetic and structural mechanisms. Annu. Rev. Biochem. 2001;70:415–435. doi: 10.1146/annurev.biochem.70.1.415. [DOI] [PubMed] [Google Scholar]
- 3.Hansen J.L., Schmeing T.M., Steitz T.A. Structural insights into peptide bond formation. Proc. Natl. Acad. Sci. USA. 2002;99:11670–11675. doi: 10.1073/pnas.172404099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ramakrishnan V. Ribosome structure and the mechanism of translation. Cell. 2002;108:557–572. doi: 10.1016/s0092-8674(02)00619-0. [DOI] [PubMed] [Google Scholar]
- 5.Dahlberg A.E. Ribosome structure. The ribosome in action. Science. 2001;292:868–869. doi: 10.1126/science.1061513. [DOI] [PubMed] [Google Scholar]
- 6.Thompson R.C., Dix D.B. Accuracy in protein synthesis. A kinetic study of the reaction of poly(U)-programmed ribosomes with a leucyl-tRNA2-elongation factor Tu-GTP complex. J. Biol. Chem. 1982;257:6677–6682. [PubMed] [Google Scholar]
- 7.Valle M., Zavialov A., Frank J. Incorporation of aminoacyl-tRNA into the ribosome as seen by cryo-electron microscopy. Nat. Struct. Biol. 2003;10:899–906. doi: 10.1038/nsb1003. [DOI] [PubMed] [Google Scholar]
- 8.Kurland C.G., Rigler R., Blomberg C. Allosteric mechanism for codon-dependent tRNA selection on ribosomes. Proc. Natl. Acad. Sci. USA. 1975;72:4248–4251. doi: 10.1073/pnas.72.11.4248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schrader J.M., Chapman S.J., Uhlenbeck O.C. Understanding the sequence specificity of tRNA binding to elongation factor Tu using tRNA mutagenesis. J. Mol. Biol. 2009;386:1255–1264. doi: 10.1016/j.jmb.2009.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vanzi F., Takagi Y., Goldman Y.E. Mechanical studies of single ribosome/mRNA complexes. Biophys. J. 2005;89:1909–1919. doi: 10.1529/biophysj.104.056283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee T.-H., Blanchard S.C., Chu S. The role of fluctuations in tRNA selection by the ribosome. Proc. Natl. Acad. Sci. USA. 2007;104:13661–13665. doi: 10.1073/pnas.0705988104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Berk V., Cate J.H. Insights into protein biosynthesis from structures of bacterial ribosomes. Curr. Opin. Struct. Biol. 2007;17:302–309. doi: 10.1016/j.sbi.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 13.Gao Y.G., Selmer M., Ramakrishnan V. The structure of the ribosome with elongation factor G trapped in the posttranslocational state. Science. 2009;326:694–699. doi: 10.1126/science.1179709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Spiegel P.C., Ermolenko D.N., Noller H.F. Elongation factor G stabilizes the hybrid-state conformation of the 70S ribosome. RNA. 2007;13:1473–1482. doi: 10.1261/rna.601507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ermolenko D.N., Spiegel P.C., Noller H.F. The antibiotic viomycin traps the ribosome in an intermediate state of translocation. Nat. Struct. Mol. Biol. 2007;14:493–497. doi: 10.1038/nsmb1243. [DOI] [PubMed] [Google Scholar]
- 16.Pan D., Kirillov S.V., Cooperman B.S. Kinetically competent intermediates in the translocation step of protein synthesis. Mol. Cell. 2007;25:519–529. doi: 10.1016/j.molcel.2007.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Seo H.-S., Abedin S., Cooperman B.S. EF-G-dependent GTPase on the ribosome. conformational change and fusidic acid inhibition. Biochemistry. 2006;45:2504–2514. doi: 10.1021/bi0516677. [DOI] [PubMed] [Google Scholar]
- 18.Savelsbergh A., Katunin V.I., Wintermeyer W. An elongation factor G-induced ribosome rearrangement precedes tRNA-mRNA translocation. Mol. Cell. 2003;11:1517–1523. doi: 10.1016/s1097-2765(03)00230-2. [DOI] [PubMed] [Google Scholar]
- 19.Stark H., Rodnina M.V., Wintermeyer W. Large-scale movement of elongation factor G and extensive conformational change of the ribosome during translocation. Cell. 2000;100:301–309. doi: 10.1016/s0092-8674(00)80666-2. [DOI] [PubMed] [Google Scholar]
- 20.Frank J., Agrawal R.K. A ratchet-like inter-subunit reorganization of the ribosome during translocation. Nature. 2000;406:318–322. doi: 10.1038/35018597. [DOI] [PubMed] [Google Scholar]
- 21.Agrawal R.K., Penczek P., Frank J. Visualization of elongation factor G on the Escherichia coli 70S ribosome: the mechanism of translocation. Proc. Natl. Acad. Sci. USA. 1998;95:6134–6138. doi: 10.1073/pnas.95.11.6134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Borowski C., Rodnina M.V., Wintermeyer W. Truncated elongation factor G lacking the G domain promotes translocation of the 3′ end but not of the anticodon domain of peptidyl-tRNA. Proc. Natl. Acad. Sci. USA. 1996;93:4202–4206. doi: 10.1073/pnas.93.9.4202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Inoue-Yokosawa N., Ishikawa C., Kaziro Y. The role of guanosine triphosphate in translocation reaction catalyzed by elongation factor G. J. Biol. Chem. 1974;249:4321–4323. [PubMed] [Google Scholar]
- 24.Modolell J., Vázquez D. The inhibition of ribosomal translocation by viomycin. Eur. J. Biochem. 1977;81:491–497. doi: 10.1111/j.1432-1033.1977.tb11974.x. [DOI] [PubMed] [Google Scholar]
- 25.Peske F., Savelsbergh A., Wintermeyer W. Conformational changes of the small ribosomal subunit during elongation factor G-dependent tRNA-mRNA translocation. J. Mol. Biol. 2004;343:1183–1194. doi: 10.1016/j.jmb.2004.08.097. [DOI] [PubMed] [Google Scholar]
- 26.Stanley R.E., Blaha G., Steitz T.A. The structures of the anti-tuberculosis antibiotics viomycin and capreomycin bound to the 70S ribosome. Nat. Struct. Mol. Biol. 2010;17:289–293. doi: 10.1038/nsmb.1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim H.D., Puglisi J.D., Chu S. Fluctuations of transfer RNAs between classical and hybrid states. Biophys. J. 2007;93:3575–3582. doi: 10.1529/biophysj.107.109884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stark H., Rodnina M.V., van Heel M. Ribosome interactions of aminoacyl-tRNA and elongation factor Tu in the codon-recognition complex. Nat. Struct. Biol. 2002;9:849–854. doi: 10.1038/nsb859. [DOI] [PubMed] [Google Scholar]
- 29.Moazed D., Robertson J.M., Noller H.F. Interaction of elongation factors EF-G and EF-Tu with a conserved loop in 23S RNA. Nature. 1988;334:362–364. doi: 10.1038/334362a0. [DOI] [PubMed] [Google Scholar]
- 30.Hausner T.P., Atmadja J., Nierhaus K.H. Evidence that the G2661 region of 23S rRNA is located at the ribosomal binding sites of both elongation factors. Biochimie. 1987;69:911–923. doi: 10.1016/0300-9084(87)90225-2. [DOI] [PubMed] [Google Scholar]
- 31.Vorstenbosch E., Pape T., Wintermeyer W. The G222D mutation in elongation factor Tu inhibits the codon-induced conformational changes leading to GTPase activation on the ribosome. EMBO J. 1996;15:6766–6774. [PMC free article] [PubMed] [Google Scholar]
- 32.Ogle J.M., Brodersen D.E., Ramakrishnan V. Recognition of cognate transfer RNA by the 30S ribosomal subunit. Science. 2001;292:897–902. doi: 10.1126/science.1060612. [DOI] [PubMed] [Google Scholar]
- 33.Powers T., Noller H.F. Selective perturbation of G530 of 16 S rRNA by translational miscoding agents and a streptomycin-dependence mutation in protein S12. J. Mol. Biol. 1994;235:156–172. doi: 10.1016/s0022-2836(05)80023-3. [DOI] [PubMed] [Google Scholar]
- 34.O'Connor M.O., Dahlberg A.E. The involvement of two distinct regions of 23 S ribosomal RNA in tRNA selection. J. Mol. Biol. 1995;254:838–847. doi: 10.1006/jmbi.1995.0659. [DOI] [PubMed] [Google Scholar]
- 35.Komoda T., Sato N.S., Suzuki T. The A-site finger in 23 S rRNA acts as a functional attenuator for translocation. J. Biol. Chem. 2006;281:32303–32309. doi: 10.1074/jbc.M607058200. [DOI] [PubMed] [Google Scholar]
- 36.Spirin A.S. A model of the functioning ribosome: locking and unlocking of the ribosome subparticles. Cold Spring Harb. Symp. Quant. Biol. 1969;34:197–207. doi: 10.1101/sqb.1969.034.01.026. [DOI] [PubMed] [Google Scholar]
- 37.Bretscher M.S. Translocation in protein synthesis: a hybrid structure model. Nature. 1968;218:675–677. doi: 10.1038/218675a0. [DOI] [PubMed] [Google Scholar]
- 38.Cornish P.V., Ermolenko D.N., Ha T. Spontaneous intersubunit rotation in single ribosomes. Mol. Cell. 2008;30:578–588. doi: 10.1016/j.molcel.2008.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ermolenko D.N., Majumdar Z.K., Noller H.F. Observation of intersubunit movement of the ribosome in solution using FRET. J. Mol. Biol. 2007;370:530–540. doi: 10.1016/j.jmb.2007.04.042. [DOI] [PubMed] [Google Scholar]
- 40.Zhang W., Dunkle J.A., Cate J.H. Structures of the ribosome in intermediate states of ratcheting. Science. 2009;325:1014–1017. doi: 10.1126/science.1175275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Matassova A.B., Rodnina M.V., Wintermeyer W. Elongation factor G-induced structural change in helix 34 of 16S rRNA related to translocation on the ribosome. RNA. 2001;7:1879–1885. [PMC free article] [PubMed] [Google Scholar]
- 42.Yusupova G.Z., Yusupov M.M., Noller H.F. The path of messenger RNA through the ribosome. Cell. 2001;106:233–241. doi: 10.1016/s0092-8674(01)00435-4. [DOI] [PubMed] [Google Scholar]
- 43.Lill R., Robertson J.M., Wintermeyer W. Affinities of tRNA binding sites of ribosomes from Escherichia coli. Biochemistry. 1986;25:3245–3255. doi: 10.1021/bi00359a025. [DOI] [PubMed] [Google Scholar]
- 44.Fahlman R.P., Dale T., Uhlenbeck O.C. Uniform binding of aminoacylated transfer RNAs to the ribosomal A and P sites. Mol. Cell. 2004;16:799–805. doi: 10.1016/j.molcel.2004.10.030. [DOI] [PubMed] [Google Scholar]
- 45.Shoji S., Abdi N.M., Fredrick K. Contribution of ribosomal residues to P-site tRNA binding. Nucleic Acids Res. 2009;37:4033–4042. doi: 10.1093/nar/gkp296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dale T., Fahlman R.P., Uhlenbeck O.C. Specificity of the ribosomal A site for aminoacyl-tRNAs. Nucleic Acids Res. 2009;37:1202–1210. doi: 10.1093/nar/gkn1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Semenkov Y.P., Rodnina M.V., Wintermeyer W. Energetic contribution of tRNA hybrid state formation to translocation catalysis on the ribosome. Nat. Struct. Biol. 2000;7:1027–1031. doi: 10.1038/80938. [DOI] [PubMed] [Google Scholar]
- 48.Ashkin A., Dziedzic J.M., Chu S. Observation of a single-beam gradient force optical trap for dielectric particles. Opt. Lett. 1986;11:288–290. doi: 10.1364/ol.11.000288. [DOI] [PubMed] [Google Scholar]
- 49.Neuman K.C., Block S.M. Optical trapping. Rev. Sci. Instrum. 2004;75:2787–2809. doi: 10.1063/1.1785844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Uemura S., Dorywalska M., Chu S. Peptide bond formation destabilizes Shine-Dalgarno interaction on the ribosome. Nature. 2007;446:454–457. doi: 10.1038/nature05625. [DOI] [PubMed] [Google Scholar]
- 51.Pettersson I., Kurland C.G. Ribosomal protein L7/L12 is required for optimal translation. Proc. Natl. Acad. Sci. USA. 1980;77:4007–4010. doi: 10.1073/pnas.77.7.4007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mishra P.P., Qureshi M.T., Lee T.H. Codon-dependent tRNA fluctuations monitored with fluorescence polarization. Biophys. J. 2010;99:3849–3858. doi: 10.1016/j.bpj.2010.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fredrick K., Noller H.F. Accurate translocation of mRNA by the ribosome requires a peptidyl group or its analog on the tRNA moving into the 30S P site. Mol. Cell. 2002;9:1125–1131. doi: 10.1016/s1097-2765(02)00523-3. [DOI] [PubMed] [Google Scholar]
- 54.Fahlman R.P., Uhlenbeck O.C. Contribution of the esterified amino acid to the binding of aminoacylated tRNAs to the ribosomal P- and A-sites. Biochemistry. 2004;43:7575–7583. doi: 10.1021/bi0495836. [DOI] [PubMed] [Google Scholar]
- 55.Valle M., Zavialov A., Frank J. Locking and unlocking of ribosomal motions. Cell. 2003;114:123–134. doi: 10.1016/s0092-8674(03)00476-8. [DOI] [PubMed] [Google Scholar]
- 56.Moazed D., Noller H.F. Intermediate states in the movement of transfer RNA in the ribosome. Nature. 1989;342:142–148. doi: 10.1038/342142a0. [DOI] [PubMed] [Google Scholar]
- 57.Ogle J.M., Murphy F.V., Ramakrishnan V. Selection of tRNA by the ribosome requires a transition from an open to a closed form. Cell. 2002;111:721–732. doi: 10.1016/s0092-8674(02)01086-3. [DOI] [PubMed] [Google Scholar]
- 58.Joseph S., Noller H.F. EF-G-catalyzed translocation of anticodon stem-loop analogs of transfer RNA in the ribosome. EMBO J. 1998;17:3478–3483. doi: 10.1093/emboj/17.12.3478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Savelsbergh A., Matassova N.B., Wintermeyer W. Role of domains 4 and 5 in elongation factor G functions on the ribosome. J. Mol. Biol. 2000;300:951–961. doi: 10.1006/jmbi.2000.3886. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


