Abstract
Protein modification is critical for the regulation of protein functions. Cross-talks among different types of protein modifications should yield concerted and coordinated regulatory networks for physiological functions. Here we have employed system-wide and quantitative phosphoproteomics analyses to reveal a global cross-talk for SUMOylation-modulated phosphorylation. Furthermore, as specific examples, we have shown that the α subunit of casein kinase II is SUMOylated and that this affects the phosphorylation of its substrates. SUMO-regulated phosphorylation is involved in cell cycle control. Our data demonstrate an interplay between protein SUMOylation and phosphorylation and imply a regulatory role for this SUMOylation-modulated phosphorylation.
Keywords: Cell Cycle, Phosphotyrosine, Protein Phosphorylation, Proteomics, SUMOylation
Introduction
The small ubiquitin-like modifiers (SUMOs)3 belong to a subfamily of an ubiquitin-like protein superfamily. The covalent protein modification by SUMO affects a broad range of cellular activities. There are four human SUMO genes: SUMO1, SUMO2, SUMO3, and SUMO4. SUMO2 and SUMO3 share 97% amino acid sequence identity, whereas∼50% identity is shared by SUMO2/3 and SUMO1 (1). The SUMO2/3 proteins have the ability to form poly-SUMO chains through their internal K11 linkage, but SUMO1 does not. SUMO1 can only be conjugated to poly-SUMO2/3 chains for the termination of poly-SUMO chains (2). SUMO4 shows a similarity to SUMO2/3, but it is unclear whether it is a pseudogene.
Although many advances in understanding the biochemistry of SUMOylation have been made during the past decades, progresses in large-scale identification of SUMO substrates were only made in recent years. In contrast to the traditional approach for the identification of SUMOylated proteins, which was mostly based on immunoblotting of a known protein (a hypothesis-driven process), an affinity enrichment/purification strategy coupled with mass spectrometry-based protein analysis techniques (an unbiased screening process) is currently the most widely used approach (3–5). So far, ∼800 putative SUMO substrates have been identified (3–9). These large-scale analyses have revealed important functions of the SUMO system in many biological processes, including RNA transcription, cell cycle, mRNA processing and splicing, as well as DNA metabolism and repair. However, the exact molecular mechanism by which the SUMO system plays its role in these cellular processes is poorly understood. There is much evidence from studies of individual proteins suggesting that SUMOylation and phosphorylation processes may be connected and cross-controlled (10–13), but whether this is a general regulatory mechanism remains to be studied.
Phosphorylation is the most extensively studied reversible posttranslational protein modification. Over 77,880 non-redundant phosphorylation sites on ∼12,000 non-redundant proteins have been identified so far, according to PhosphoSitePlus, a database compiled by Cell Signaling Technology, Inc. More than 80% of the phosphorylation sites were identified by high throughput methods, primarily the mass spectrometry-based techniques. More recently, with the advantage of stable isotope labeling by amino acid in cell culture (SILAC) labeling, phosphoproteomics analysis has gone quantitative, and a few important signaling events have been studied using quantitative phosphoproteomics techniques from a dynamic and unbiased perspective.
Cross-talks between various protein posttranslational modifications may represent an important regulatory circuit for protein functions. Phosphorylation-dependent SUMOylation has been investigated, and an evolutionarily conserved phosphorylation-dependent SUMO-modification motif was identified. A number of transcription factors, such as heat shock factors, MEF2A, GATA-1, and ERRγ, have been identified as SUMO-modified proteins in which the SUMOylations are dependent on their phosphorylation-dependent SUMO modification motif (12). Meanwhile, evidence for phosphorylation-obstructed SUMO modification was also found. For example, phosphorylation of the AIB1 protein by the MAPK pathway was found to be inhibitory to its SUMO modification (14). These studies highlight the important regulatory mechanism for phosphorylation-modulated SUMOylation in regulating cellular events.
Although extensive studies have been carried out on phosphorylation-regulated SUMOylation, large-scale studies on SUMO-modulated phosphorylations have not been performed. In this study, we have analyzed the changes of the phosphoproteome landscape in response to alterations of SUMOylation at a global level. We discovered that, in response to a decreased SUMO level, the phosphorylation of some proteins was altered significantly (we defined them as up- or down-regulation by at least 2-fold). A significant portion of these phosphoproteins was previously known to be SUMO-modified. Our data indicate that extensive cross-talks exist between protein SUMOylation and phosphorylation. Through additional biochemical studies on the basis of information extracted from our phosphoproteomics data, we identified casein kinase II α as a new SUMO-modified protein, and investigated the influence of decreased SUMOylation on cell cycle progression.
EXPERIMENTAL PROCEDURES
Plasmids, Antibodies, and Reagents
HA-SUMO1 and HA-SUMO2 plasmids were obtained from Dr. C. Lima at the Sloan-Kettering Cancer Institute. HA-CKII α was subcloned into pcDNA3.0 with the XhoI and EcoRI restriction sites. Antibodies against SUMO1 and SUMO2/3 were produced by NewEast Biosciences. HA antibody was from Roche. A pan-phospho-tyrosine antibody, P-Tyr-100, was purchased from Cell Signaling Technology, Inc. Cdc2 and pTyr15 Cdc2 antibodies were from Signalway Antibody, and the FAK and pTyr397 FAK antibodies were from Santa Cruz Biotechnology, Inc. Ginkgolic acid was purchased from Sigma. Propidium iodide was purchased from Beyotime.
RNA Knockdown
SUMO1 and SUMO2/3 shRNA plasmids were constructed by inserting oligonucleotides containing SUMO1 or SUMO2/3 siRNA sequences (SUMO1-siRNA, 5′-CACATCTCAAGAAACTCAA-3′; SUMO2/3-siRNA, 5′-GTCAATGAGGCAGATCAGA-3′) into the pSuper vector (OligoEngine). A pSuper plasmid encoding siRNA against the prokaryotic protein LacZ was used as a control. Cells were collected and analyzed 48 h after transfection.
Western Blot Analysis and Immunoprecipitation
For Western blot analysis, 50 μg of total cellular proteins were separated by SDS-PAGE, transferred electrophoretically onto a PVDF membrane (Millipore), and blocked for 1 h with TBST (Tris-Buffered Saline Tween-20) containing either 5% nonfat milk or 5% BSA, incubated with a primary antibody overnight at 4 °C, and followed by washing three times with TBST. After adding a horseradish peroxidase-conjugated secondary antibody and incubating for 1 h at room temperature in TBST containing 5% nonfat milk, chemiluminescent substrates (Millipore) were added and incubated for 5 min at room temperature.
For immunoprecipitation, cells were washed three times with ice-cold PBS. Cell lysates were prepared by resuspending cells in Nonidet P-40-containing radioimmune precipitation assay buffer (50 mm Tris-HCl (pH 7.4), 1% Nonidet P-40, 0.25% Na-deoxycholate, 150 mm NaCl). Protein G-agarose (GE Healthcare) and antibodies were added to cell lysates and rotated overnight at 4 °C. To collect immunoprecipitates, samples were centrifuged at 400 × g for 5 min. The pellet was washed with ice-cold PBS three times and then boiled in 2.5 × SDS loading buffer at 95 °C for 5 min. The supernatant was collected and loaded for Western blot analysis.
Cell Culture and SILAC Labeling
For cell culture, HEK 293T cells were cultured in DMEM with 10% FBS and penicillin/streptomycin. Ginkgolic acid and the DMSO control were added directly into the cells for the indicated time points. For SILAC labeling, cells were washed twice with PBS and cultured in DMEM medium containing either 12C6, 14N4 Arg, 12C6, 14N2 Lys (Sigma), or 13C6, 15N4 Arg, 13C6, 15N2 Lys (Sigma). Cells were cultured in a humidified incubator (5% CO2) at 37 °C for at least six doubling times. The concentrations of the amino acids Arg and Lys used in SILAC labeling of HEK293T cells were 0.398 mm and 0.798 mm, respectively.
For cell extract preparation, cells were treated with or without 100 μm ginkgolic acid for 6 h and harvested by centrifugation at 400 × g. Cell pellets were washed three times with cold PBS, then lysed in radioimmune precipitation assay buffer (25 mm Tris-HCl (pH 7.6), 150 mm NaCl, 1% Nonidet P-40, 1% sodium deoxycholate, 0.1% SDS) containing phosphatase inhibitor mixture (PhosSTOP, Roche) and protease inhibitor mixture (COMPLETE, Roche) for 0.5 h on ice. The supernatant was collected after centrifugation at 16,000 × g for 0.5 h.
Trypsin Digestion, C18 Desalt, and Phosphopeptide Enrichment by IMAC
For trypsin digestion, protein extract was precipitated with 3 volumes of 50% acetone/50% ethanol/0.1% acetic acid on ice for 2 h and then centrifuged at 10,000 × g for 15 min. Pellets were resuspended in 8 m urea, 0.2 m Tris (pH 8), 4 mm CaCl2, reduced in 10 mm DTT for 1 h at 56 °C, and then alkylated with 40 mm iodoacetamide for 30 min in the dark at room temperature. Trypsin (Worthington) was added in a 1:50 (trypsin/protein) w/w ratio after diluting the pellets seven times and rotated overnight at 37 °C.
For C18 desalt and IMAC, tryptic peptides were loaded onto a 2 g Sep-Pak C18 column (Waters), washed twice with 10 ml 1% acetic acid, eluted with 7 ml 80% acetonitrile containing 0.1% acetic acid, dried using a SpeedVac (Labconco), dissolved in 400 μl 1% acetic acid, and loaded onto a minicolumn of 40 μl of IMAC resin that was prepared as described previously (15). The IMAC column was washed twice with 40 μl of wash buffer containing 25% acetonitrile, 100 mm NaCl, and 0.1% acetic acid, washed once each with 40 μl of 1% acetic acid and 20 μl of deionized water, eluted with 120 μl 6% NH4OH, and SpeedVac-dried.
HILIC and LC-MS/MS
Phosphopeptides fractionation by HILIC was performed on an Agilent 1200 HPLC system (Agilent Technologies) equipped with a TSKgel Amide-80 column (2.0 × 150 mm, 5-μm particle size, 200-Å pore size) (TOSOH Bioscience). A 60-min elution gradient was used with 90% acetonitrile, 0.005% trifluoroacetic acid as buffer A, and 0.005% trifluoroacetic acid as buffer B. The gradient elution profile was composed of 0–12% B for 5 min, 12–30% B for 25 min, and 30–90% B for 5 min and then maintained at 90% B for 5 min, followed by 10–100% A for 5 min, and ending at 100% A for 15 min. The flow rate was 0.15 ml/min. UV absorbance was monitored at 215 nm. Fractions were collected every 2 min and dried by SpeedVac.
For LC-MS/MS analyses, a QSTAR ELITE mass spectrometer (Applied Biosystems) coupled with an online Eksigent nano multidimensional liquid chromatography system utilizing a nanospray ionization source was used. Peptides were first concentrated onto a CapTrap column (0.5 × 2 mm, MICHROM Bioresources, Inc.) followed by elution into an analytical column (MAGIC C18AQ, 100 μm × 150 mm, 3-μm particle size, 200-Å pore size, MICHROM Bioresources, Inc.). Mobile phase A (2% acetonitrile, 0.1% formic acid) and mobile phase B (98% acetonitrile, 0.1% formic acid) were used to establish a 130-min gradient comprised of 5 min 5% B, then 25 min 5–15% B, followed by 55 min 15–40% B, then 15 min 40–80% B, maintained at 80% B for 10 min, then 5 min 80–5% B, and finally maintained at 5% B for 15 min. The flow rate was ∼300 nL/min. For MS/MS analysis, each scan cycle consisted of one full-scan mass spectrum (with m/z ranging from 400 to 1800 and charge states from 2 to 5) followed by five MS/MS events. The threshold count was set to 30, and the exclusion window was 90 s. Mass tolerance was 50 mDa. Automatic collision energy and automatic MS/MS accumulation were selected (16).
Data Identification and Quantification, and Bioinformatics
For identification and quantification, raw data from QSTAR ELITE were loaded by Mascot Daemon (version 2.2.2) (Matrix Science, London, UK) to an in-house Mascot server (version 2.2) (Matrix Science, London, UK) and Distiller (version 2.2.1.2) (Matrix Science, London, UK). Peak lists were generated by Distiller and searched against a target/decoy SwissProt human protein database (version 57.7; 20,405 sequences) by the Mascot server. Spectra match criteria were set as follows: Fixed modification was carbamidomethyl at the Cys residue and variable modifications were oxidation at the Met residue and phosphorylation at the Ser, Thr, or Tyr residues. Additionally, Arg-10 and Lys-8 were set as exclusive modifications, and taxonomy was set to “human.” Peptide and MS/MS tolerances were set as 50 ppm and 0.2 Da, respectively. The peptide charges were 2+, 3+, 4+, or 5+, with two missed cleavages allowed. The significance threshold was p < 0.05. For quantitation analysis, raw data from database searching were processed by Mascot Distiller. We set the fraction, correlation, and standard error as 0.5, 0.9, and 0.2, respectively. The median of all quantitation data from non-phosphopeptides was used to normalize the peptide ratios.
For bioinformatics, gene ontology analysis was done by David Bioinformatics. SUMOylated protein in the protein list was manually checked according to published data. SUMOylation site prediction was performed by SUMOplot and SUMOsp 2.0 (17) with high stringency. Kinase analysis was done by presenting sequences of proteins identified in the list to the software PTMs Peptide Scanner and manually checking the kinase for the peptides identified by MS.
Cell Cycle Analysis
For cell cycle analysis, cells treated with DMSO and ginkgolic acid were washed two times with ice-cold PBS, fixed in 70% ethanol at −20 °C, collected by centrifuge, and stained with propidium iodide. Stained cells were then analyzed by flow cytometer (Beckman Coulter Epics XL).
RESULTS
System-wide Cross-talk between Protein SUMOylation and Phosphorylation
To establish that there are system-wide cross-talks between protein SUMOylation and phosphorylation, we examined the changes of the phosphoproteome landscape in response to alterations of SUMOylation. Ginkgolic acid, a newly discovered specific inhibitor of SUMOylation (18), was used to reduce the global SUMOylation. Protein SUMOylation and phosphorylation were evaluated by immunoblotting with antibodies against SUMOs or phospho-tyrosine. Treatment of HEK293 cells with ginkgolic acid at 100 μm for 6 h led to a significant decrease in SUMO1 and SUMO2/3 conjugation, as well as in global protein tyrosine phosphorylation (Fig. 1, A and B). These results indicate that tyrosine phosphorylation could be influenced by SUMO modification. Moreover, when compared with cells treated with pervanadate, a strong pan-tyrosine phosphatase inhibitor, ginkgolic acid-treated cells showed different patterns of changes in protein tyrosine phosphorylation (Fig. 1B). This implies that SUMO regulation of protein tyrosine phosphorylation is different from the phosphatase-dependent alteration.
FIGURE 1.
Influence of SUMOylation on protein phosphorylation. A, inhibition of SUMO modification by ginkgolic acid. HEK293T cells were treated with 100 μm ginkgolic acid or DMSO as a control for 6 h before harvest, then lysed and probed with anti-SUMO1 and SUMO2/3 antibody. Actin was used as a loading control. IB, immunoblotting. B, the same samples from A and pervanadate-treated samples were blotted with pan-anti-phospho-tyrosine antibody. Actin was used as a loading control. C, HEK293T cells were transfected with pcDNA3.0 control, HA-SUMO1, or HA-SUMO2 plasmids and were starved for 18 h after 24 h of transfection. Cells were then harvested, lysed, and blotted with anti-SUMO1, SUMO2/3, and HA antibodies. D, the same samples from C were blotted with pan-anti-phospho-tyrosine antibody. Actin was used as a loading control. E, SUMO1 or SUMO2/3 shRNA plasmids, as well as LacZ plasmid control, were transfected into HEK293T cells for 48 h. Cells were harvested, lysed, and blotted with anti-SUMO1 and SUMO2 antibody. Actin was used as a loading control. F, the same samples from A and E were blotted with anti-FAK and anti-pTyr-FAK antibody. Actin was used as a loading control. Pv, pervanadate.
To further demonstrate cross-talks between SUMOylation and tyrosine phosphorylation, we used a gain of function approach. We transfected HEK293 cells with HA-tagged SUMO1 or SUMO2 plasmids and analyzed protein tyrosine phosphorylation. SUMO1 and SUMO2/3 immunoblotting confirmed the increase of cellular SUMO modification after SUMO transfection (Fig. 1C). Importantly, increased protein tyrosine phosphorylation was observed in SUMO-transfected cells (Fig. 1D). This provides further evidence supporting that there is a global cross-talk between SUMOylation and phosphorylation. Because of the lack of high quality and specific pan-antibodies against phospho-serine and phospho-threonine, potential cross-talks between SUMOylation and serine/threonine phosphorylation were not tested here. The extensive cross-talk observed between SUMOylation and tyrosine phosphorylation can serve as a foretaste for much larger-scale cross-talks between SUMOylation and phosphorylation.
To confirm the conclusion on the basis of data from ginkgolic acid treatment, we used an alternative method to decrease SUMO modification in HEK293T cells. We designed siRNAs against SUMO1 or SUMO2/3 to decrease their expression. Immunoblotting results showed that decreased expression of SUMO1 or SUMO2/3 led to decreased global tyrosine phosphorylation (Fig. 1E), supporting the extensive cross-talks between SUMOylation and phosphorylation.
To further investigate the mechanism by which SUMOylation could affect global protein tyrosine phosphorylation, we tested a potential effect of SUMOylation on the activity of protein tyrosine kinases. Using FAK, a non-receptor tyrosine kinase, as a specific example of tyrosine kinases, we demonstrated that SUMOylation reduction decreased the activity of FAK. The kinase activity of FAK is dependent on its autophosphorylation on Tyr-397, which, in turn, is positively modulated by SUMOylation (13). We examined whether GA treatment could alter FAK autophosphorylation at Tyr-397 (as a readout of the activation of FAK). As shown in Fig. 1F, GA treatment showed a decrease in tyrosine phosphorylation of Tyr-397 in the SUMOylated form of FAK compared with the DMSO-treated group. This result was further supported by experiments using siRNAs against SUMO1 and SUMO2/3 (Fig. 1F). Hence, SUMOylation could affect the activity of tyrosine kinase, leading to changes in global protein tyrosine phosphorylation.
Quantitative Phosphoproteomics Analysis of Global Cross-talks between SUMOylation and Phosphorylation
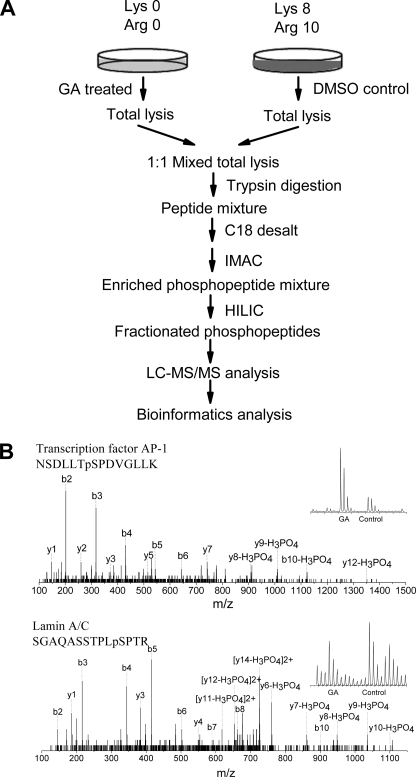
Although the immunoblotting-based approach provided evidence for system-wide cross-talks between SUMOylation and phosphorylation, little details at the molecular level could be extracted from the Western blot data. Hence, we applied SILAC-based quantitative phosphoproteomics strategy to systematically investigate the cross-talk between SUMOylation and phosphorylation (Fig. 2A). HEK293 cells were cultured in SILAC media for more than five generations to ensure >95% labeling. Cells were then treated with ginkgolic acid for 6 h. Total cellular proteins were digested with trypsin, and phosphopeptides were enriched by an IMAC method (15). After fractionating the enriched phosphopeptides by HILIC, phosphopeptides were analyzed by LC-MS/MS techniques. Examples of mass spectra used for the identification of phosphopeptides and for the quantification of changes in phosphorylation are shown in Fig. 2B.
FIGURE 2.
Quantitative proteomics approach used to reveal global interplay between phosphorylation and SUMOylation. A, flow chart of SILAC-based quantitative phosphoproteomics procedure. B, selected examples of mass spectra used in detection and quantitation of phosphopeptides. One phosphopeptide is originated from transcription factor AP-1, and the other one is from Lamin-A/C.
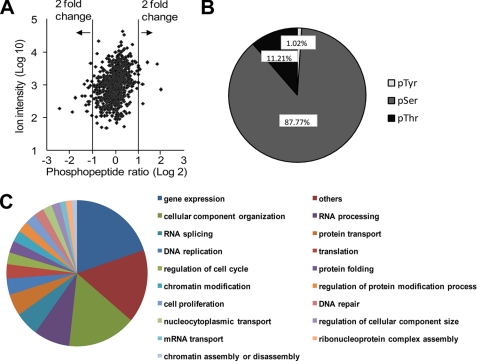
Peptide identification and quantification were based on the Mascot program with a confidence setting of >95%. The false discovery rate for the level of peptides was 3.72% (see the detailed list of peptides identified by LC-MS/MS in supplemental Table S1). A total of 739 phosphopeptides, originated from 508 unique proteins, were identified and quantified following the workflow outlined in Fig. 2A. Among these phosphorylation sites, only 3.65% was significantly altered when a 2-fold up- or down- regulation was set as the cutoff (Fig. 3A). A detailed list is provided in supplemental Table S2, in which all the phosphopeptides were manually inspected. The identified phosphopeptides consist of 689 serine phosphorylation (pSer), 88 threonine phosphorylation (pThr), and eight tyrosine phosphorylation (pTyr) (Fig. 3B). These phosphoproteins are from a variety of different biological functional groups (Fig. 3C).
FIGURE 3.
Summary of data from large-scale phosphopeptide identification and quantitation. A, scatter plot. The x axis represents the log 2 ratio (L/H) value, the y axis represents the log10 ion intensity value. A log 2 value of +1 or −1 represents a 2-fold alteration in phosphorylation. B, distribution of phospho-serine, threonine, and tyrosine sites among detected phosphopeptides. C, biological functional groups of identified phosphoproteins.
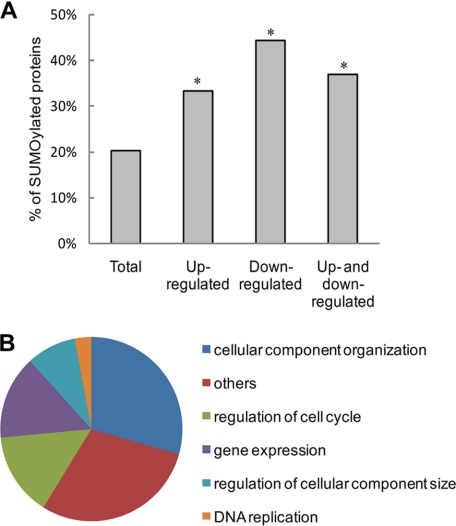
When phosphoproteins from our data (supplemental Table S2) were compared with known SUMOylated proteins according to the published large-scale SUMOylation studies (3, 6), we noticed that the SUMOylated proteins were specifically enriched in the significantly up- or down- regulated groups (we defined this as at least 2-fold changes) (Fig. 4A) from 20.3% in the overall phosphorylated proteins to 37.0% in the significantly altered population. Our data strongly suggest that there is a generalized regulation of phosphorylation by SUMOylation. Furthermore, in contrast to the distribution of all phosphorylated proteins identified in this study (Fig. 3C), we found that the up- or down- regulated phosphoproteins are mainly concentrated in a few biological processes, such as regulation of cell cycle, gene expression, and DNA replication (Fig. 4B). These biological processes are known to be regulated by protein SUMOylation.
FIGURE 4.
Bioinformatics analysis of identified proteins. A, bar graph showing the percentage of known published SUMOylated proteins in our identified protein list. *, p < 0.01 relative to the “total” group. B, gene ontology analysis by DAVID. The pie chart shows the biological process distribution of phosphoproteins with more than 2-fold changes in protein phosphorylation.
Regulation of Casein Kinase II α by SUMOylation
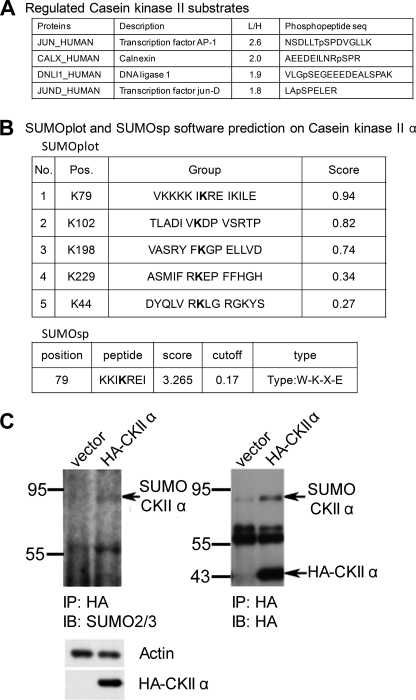
From our phosphoprotein list, we observed that several substrates for casein kinase II have their phosphorylation rates increased (Fig. 5A). Because not all casein kinase II substrates in our list are SUMOylated, we suspect that casein kinase II itself might be regulated by SUMOylation.
FIGURE 5.
SUMO modification of casein kinase II α. A, substrates of casein kinase II. B, SUMOplot and SUMOsp 2.0 prediction of the casein kinase II α subunit. The stringency of SUMOsp 2.0 was set to be at high stringency. C, SUMO modification of casein kinase II α. The HA-tagged casein kinase II α subunit was expressed in 293T cells. After 24-h transfection, cells were lysed and immunoprecipitated (IP) with anti-HA antibody and then probed with anti-SUMO2/3 antibody. Total lysates of transfected cells were blotted with anti-HA antibody and anti-actin antibody. IB, immunoblotting.
Casein kinase II is a tetramer that consists of one α subunit, one α' subunit, and two β subunits. While the homologous α and α' subunits are the catalytic subunits, the β subunits are the regulatory subunits. We submitted sequences of the three subunits to SUMOylation prediction software SUMOplot and SUMOsp 2.0, and found that the α and α' subunits are very likely to be modified by SUMO, whereas the β subunit is very unlikely to be modified (Fig. 5B). We therefore constructed an HA-tagged α subunit of casein kinase II and analyzed whether it is SUMO-modified (because of the high homology in sequences, we did not exam the α' subunit). HA-tagged casein kinase II α proteins from transfected HEK293T lysates were immunoprecipitated, followed by immunoblotting with antibodies against SUMO2/3. As shown in Fig. 5C, casein kinase II α was indeed modified by SUMO2. To our knowledge, this was the first report proving that casein kinase II α subunit is a SUMOylated protein.
Cell Cycle Regulation by SUMO-regulated Phosphorylation
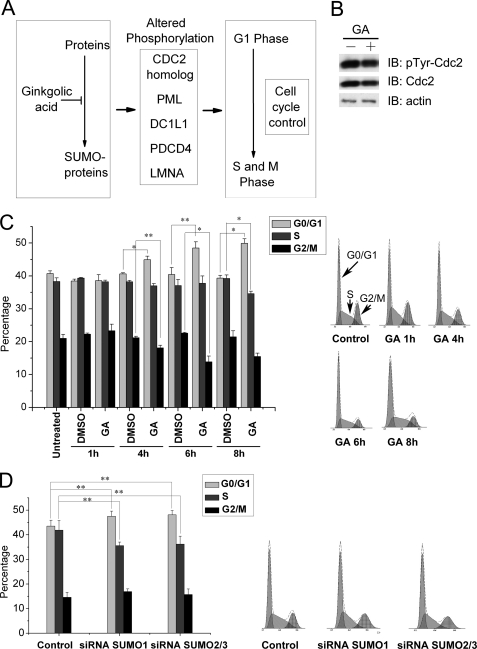
From our quantitative phosphoproteomics data, we also found that phosphorylation of a number of proteins involved in cell cycle control was up- or down-regulated upon ginkgolic acid treatment (Fig. 6A). Some of the phosphorylation sites identified were previously linked to the involvement of the protein in cell cycle control (19, 20). For example, the Cdc2 homolog is involved in cell cycle regulation. Our data clearly showed that the tyrosine phosphorylation reduction of the Cdc2 homolog correlated with the SUMOylation reduction (Fig. 6B). It needs to be noted that for some proteins, the overall protein expression levels could also be altered following SUMOylation reduction. The Cdc2 homolog could be such a protein. We observed the reduction in both protein levels as well as protein phosphorylation upon inhibition of SUMOylation.
FIGURE 6.
Effect on cell cycle by SUMO-regulated phosphorylation. A, proteins involved in cell cycle control were enriched in up- and down-regulated groups. B, HEK293T cells were treated with DMSO and 100 μm ginkgolic acid for 6 h before harvest, then lysed, and blotted (IB) with anti-Cdc2 and anti-pTyr Cdc2 antibody. Actin was used as a loading control. C, flow cytometry analysis of the cell cycle upon ginkgolic acid treatment. Representative original flow cytometry data are shown on the right. HEK 293T cells were treated with DMSO or 100 μm ginkgolic acid for 1 h, 4 h, 6 h, and 8 h. Cells were harvested, stained with propidium iodide, and analyzed by flow cytometry. n = 3; *, p < 0.05; **, p < 0.01. D, flow cytometry analysis of the cell cycle after knockdown of SUMO expression. Representative original flow cytometry data are shown on the right. HEK 293T cells were transfected with shRNA plasmids against LacZ control, SUMO1, or SUMO2/3 for 48 h. Cells were harvested, stained with propidium iodide, and analyzed by flow cytometry for cell cycle changes. n = 6; **, p < 0.01. All the original flow cytometry analysis of cell cycle were included in Supplemental Fig. S3.
We then explored whether SUMOylation reduction can induce an alteration in cell cycle. As shown in Fig. 6C, treating cells with ginkgolic acid caused an increase of cells at the G0/G1 phase. The most significant increase of cells in the G0/G1 phase and the concomitantly decrease of cells in G2/M phase occurred after 6 h of ginkgolic acid treatment. Hence, it is possible that one mechanism for SUMO-regulation in cell cycle control is through cross-talks between SUMOylation and phosphorylation of proteins involved in cell cycle control.
Furthermore, we investigated the influence on cell cycle control when SUMOylation is reduced by either siRNAs against SUMO1 and SUMO2/3 or by overexpression of SENPs (Sentrin/SUMO-specific proteases). Our results showed that siRNA treatment also caused cell accumulation at the G0/G1 phase, which is similar to the GA treatment data but with a slight increase of cells in G2/M phase (Fig. 6D). In addition, we found that overexpression of SENPs, especially SENP2, led to increased cell arrest in G2/M phase, as reported previously (data not shown) (21). Therefore, our results indicated that alteration in SUMO modification could regulate the cell cycle. Moreover, our data showed that different means of SUMOylation reduction might lead to different cell distribution patterns.
DISCUSSION
Using posttranslational modifications to regulate or fine-tune the functions of a protein is one of the most important features of proteome. As two of the most common and reversible posttranslational modifications, phosphorylation and SUMOylation are actively involved in various cellular processes. Hence, it makes sense for these two modifications to actively interplay with each other to coordinate cellular events.
In this study, we have systematically investigated the cellular phosphoproteome changes under SUMO inhibition. Using immunoblotting techniques, we have demonstrated that protein tyrosine phosphorylation was positively correlated with SUMOylations. The biological implication of this discovery may be quite significant because tyrosine phosphorylation plays key roles in cellular signaling, especially in cancers. Of the 518 putative protein kinases in the human genome, ∼100 are predicted to be tyrosine kinases, and more than 50% of these tyrosine kinases are related to cancer development (22). Many growth factor receptors are tyrosine kinases. From our phosphoproteomic analyses, the limited sequencing information obtained on tyrosine phosphorylation prohibited us to extract any sequencing level signatures about the cross-talks between SUMOylation and tyrosine phosphorylation.
Large-scale analyses for SUMOylated proteins implicate that SUMOs are involved in many important biological processes, such as transcriptional regulation, cell cycle control, mRNA metabolism, and DNA repair (3–5, 9). However, the underlying molecular mechanisms are not well understood. Because the reversible protein phosphorylation serves as a switch to control many protein functions, SUMO-regulated protein phosphorylation could well be one of the important mechanisms for SUMO regulation. In this study, we discovered that SUMOylated proteins were specifically enriched in the ginkgolic acid-regulated phosphoprotein group, making a direct link between SUMOylation and phosphorylation. Gene ontology analysis of the up- and down-regulated group showed that these phosphoproteins are mainly involved in transcriptional regulation, cell cycle regulation, and DNA replication, which are consistent with the known roles of SUMO in these cellular processes (3, 5, 23, 24).
Protein phosphorylation is controlled by protein kinases and phosphatases. SUMO regulation of kinases and phosphatases might be the simplest mechanism for SUMO regulation of protein phosphorylation. For example, tyrosine phosphatase 1B is modified by SUMO, and its enzyme activity is down-regulated upon SUMO modification (10). This may partially explain our observation that tyrosine phosphorylation is positively coordinated with SUMOylation.
Casein kinase II is a ubiquitously expressed and constitutively active kinase with more than 300 substrates participating in various cellular events, including cell cycle regulation, transcriptional control, and viral infection (25, 26). Given its constitutively active nature, its regulation is of vital importance in coordinating various cellular events. The discovery that the casein kinase II α subunit could be modified and regulated by SUMO reveals a key regulatory mechanism in modulating casein kinase II and might explain the different activities shown by casein kinase II. In addition to casein kinase II α, we have also shown the regulation of FAK kinase activity by SUMOylation. These two examples with protein kinases demonstrate that one of the molecular mechanisms by which SUMOylation could regulate protein phosphorylation is through direct modulation of the activity of protein kinases.
The SUMO system has been implicated to have an important role in regulating the cell cycle (5). Many important cell cycle-related proteins are reported to be SUMOylated (3). However, the consequences of SUMO modification and how SUMOylation participates in cell cycle control are far from clear. On the basis of our phosphoproteome data, here we speculate that one possible mechanism for SUMO regulation in cell cycle control is through cross-talks between SUMOylation and phosphorylation of proteins involved in cell cycle control. Our data indicated that alteration in SUMO modification could modulate the cell cycle. Furthermore, different means of SUMOylation reduction might lead to different cell distribution patterns. Multiple factors could contribute to these different effects. First, different SUMO reduction methods are with different treatment time schemes. GA treatment was between 4–8 h. For siRNA and SENP overexpression treatments, samples were analyzed 48 h after transfection. These time differences in sample treatment might lead to cell population differences. Second, neither siRNA nor SENP approaches would yield a complete SUMOylation elimination. These treatments only affect a subset of SUMO substrates. Hence, the overall SUMOylated proteins in siRNA- or SENP-treated samples are not expected to be the same as those treated with GA. Thus, the GA, siRNA, and SENP approaches may not have the same functional consequences. Nevertheless, all these data showed an interconnection between SUMOylation and cell cycle control. The detailed molecular mechanisms need to be studied further. Our data could provide a new direction for studying cell cycle control, which is SUMO-regulated protein phosphorylation.
Supplementary Material
This work was supported by National Basic Research Program of China Grant 2007CB914200, National Science Foundation of China Grants 31028015 and 30921001, PCSIRT Grant IRT0745, and 111 Project of China Grant B06018.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S3 and Tables S1 and S2.
- SUMO
- small ubiquitin-like modifier
- SILAC
- stable isotope labeling by amino acid in cell culture
- FAK
- focal adhesion kinase
- IMAC
- immobilized metal affinity chromatography
- HILIC
- hydrophilic interaction chromatography
- GA
- ginkgolic acid
- SENP
- sentrin-specific protease.
REFERENCES
- 1. Tatham M. H., Jaffray E., Vaughan O. A., Desterro J. M., Botting C. H., Naismith J. H., Hay R. T. (2001) J. Biol. Chem. 276, 35368–35374 [DOI] [PubMed] [Google Scholar]
- 2. Mertins P., Eberl H. C., Renkawitz J., Olsen J. V., Tremblay M. L., Mann M., Ullrich A., Daub H. (2008) Mol. Cell Proteomics 7, 1763–1777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Golebiowski F., Matic I., Tatham M. H., Cole C., Yin Y., Nakamura A., Cox J., Barton G. J., Mann M., Hay R. T. (2009) Sci. Signal. 2, ra24. [DOI] [PubMed] [Google Scholar]
- 4. Denison C., Rudner A. D., Gerber S. A., Bakalarski C. E., Moazed D., Gygi S. P. (2005) Mol. Cell. Proteomics 4, 246–254 [DOI] [PubMed] [Google Scholar]
- 5. Nie M., Xie Y., Loo J. A., Courey A. J. (2009) PLoS ONE 4, e5905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Vertegaal A. C., Andersen J. S., Ogg S. C., Hay R. T., Mann M., Lamond A. I. (2006) Mol. Cell. Proteomics 5, 2298–2310 [DOI] [PubMed] [Google Scholar]
- 7. Wykoff D. D., O'Shea E. K. (2005) Mol. Cell. Proteomics 4, 73–83 [DOI] [PubMed] [Google Scholar]
- 8. Matic I., van Hagen M., Schimmel J., Macek B., Ogg S. C., Tatham M. H., Hay R. T., Lamond A. I., Mann M., Vertegaal A. C. (2008) Mol. Cell. Proteomics 7, 132–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Panse V. G., Hardeland U., Werner T., Kuster B., Hurt E. (2004) J. Biol. Chem. 279, 41346–41351 [DOI] [PubMed] [Google Scholar]
- 10. Dadke S., Cotteret S., Yip S. C., Jaffer Z. M., Haj F., Ivanov A., Rauscher F., 3rd, Shuai K., Ng T., Neel B. G., Chernoff J. (2007) Nat. Cell Biol. 9, 80–85 [DOI] [PubMed] [Google Scholar]
- 11. Hietakangas V., Ahlskog J. K., Jakobsson A. M., Hellesuo M., Sahlberg N. M., Holmberg C. I., Mikhailov A., Palvimo J. J., Pirkkala L., Sistonen L. (2003) Mol. Cell. Biol. 23, 2953–2968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hietakangas V., Anckar J., Blomster H. A., Fujimoto M., Palvimo J. J., Nakai A., Sistonen L. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 45–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kadaré G., Toutant M., Formstecher E., Corvol J. C., Carnaud M., Boutterin M. C., Girault J. A. (2003) J. Biol. Chem. 278, 47434–47440 [DOI] [PubMed] [Google Scholar]
- 14. Wu H., Sun L., Zhang Y., Chen Y., Shi B., Li R., Wang Y., Liang J., Fan D., Wu G., Wang D., Li S., Shang Y. (2006) J. Biol. Chem. 281, 21848–21856 [DOI] [PubMed] [Google Scholar]
- 15. Albuquerque C. P., Smolka M. B., Payne S. H., Bafna V., Eng J., Zhou H. (2008) Mol. Cell. Proteomics 7, 1389–1396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chen X., Wu D., Zhao Y., Wong B. H., Guo L. (2010) J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 879, 25–34 [DOI] [PubMed] [Google Scholar]
- 17. Ren J., Gao X., Jin C., Zhu M., Wang X., Shaw A., Wen L., Yao X., Xue Y. (2009) Proteomics 9, 3409–3412 [DOI] [PubMed] [Google Scholar]
- 18. Fukuda I., Ito A., Hirai G., Nishimura S., Kawasaki H., Saitoh H., Kimura K., Sodeoka M., Yoshida M. (2009) Chem. Biol. 16, 133–140 [DOI] [PubMed] [Google Scholar]
- 19. Palamarchuk A., Efanov A., Maximov V., Aqeilan R. I., Croce C. M., Pekarsky Y. (2005) Cancer Res. 65, 11282–11286 [DOI] [PubMed] [Google Scholar]
- 20. Ward G. E., Kirschner M. W. (1990) Cell 61, 561–577 [DOI] [PubMed] [Google Scholar]
- 21. Zhang X. D., Goeres J., Zhang H., Yen T. J., Porter A. C., Matunis M. J. (2008) Mol. Cell 29, 729–741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Manning G., Whyte D. B., Martinez R., Hunter T., Sudarsanam S. (2002) Science 298, 1912–1934 [DOI] [PubMed] [Google Scholar]
- 23. Makhnevych T., Sydorskyy Y., Xin X., Srikumar T., Vizeacoumar F. J., Jeram S. M., Li Z., Bahr S., Andrews B. J., Boone C., Raught B. (2009) Mol. Cell 33, 124–135 [DOI] [PubMed] [Google Scholar]
- 24. Yang X. J., Grégoire S. (2006) Mol. Cell 23, 779–786 [DOI] [PubMed] [Google Scholar]
- 25. Meggio F., Pinna L. A. (2003) FASEB J. 17, 349–368 [DOI] [PubMed] [Google Scholar]
- 26. Pinna L. A. (2002) J. Cell Sci. 115, 3873–3878 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.