Abstract
To analyze the relationship between antisense transcription and alternative splicing, we developed a computational approach for the detection of antisense-correlated exon splicing events using Affymetrix exon array data. Our analysis of expression data from 176 lymphoblastoid cell lines revealed that the majority of expressed sense–antisense genes exhibited alternative splicing events that were correlated to the expression of the antisense gene. Most of these events occurred in areas of sense–antisense (SAS) gene overlap, which were significantly enriched in both exons and nucleosome occupancy levels relative to nonoverlapping regions of the same genes. Nucleosome occupancy was highly correlated with Pol II abundance across overlapping regions and with concomitant increases in local alternative exon usage. These results are consistent with an antisense transcription-mediated mechanism of splicing regulation in normal human cells. A comparison of the prevalence of antisense-correlated splicing events between individuals of Mormon versus African descent revealed population-specific events that may indicate the continued evolution of new SAS loci. Furthermore, the presence of antisense transcription was correlated to alternative splicing across multiple metazoan species, suggesting that it may be a conserved mechanism contributing to splicing regulation.
Much of the complexity of mammalian biology can be attributed to the regulation of gene expression via changes in the level, splicing, and localization of RNA (Wang et al. 2008; Licatalosi and Darnell 2010). One type of regulation occurs between genes that are encoded in an overlapping and opposite orientation. Such sense–antisense (SAS) gene pairs encode proteins and noncoding RNAs that play key roles in development, and have been implicated in diseases such as cancer (Vanhee-Brossollet and Vaquero 1998; Tufarelli et al. 2003; Reis et al. 2004; Chen et al. 2005; Engstrom et al. 2006). Antisense transcripts have been identified at 50%–70% of mammalian loci (Carninci et al. 2005; RIKEN Genome Exploration Research Group et al. 2005), yet despite their prevalence, regulatory roles have only been elucidated for a small subset of SAS genes (for review, see Vanhee-Brossollet and Vaquero 1998; Lavorgna et al. 2004). Since a large proportion (40%) of antisense transcripts are noncoding RNAs, they may act predominantly as regulators of sense gene expression (Mattick 2004).
In a limited number of cases, antisense transcription has been correlated to sense gene splicing (Mihalich et al. 2003; Louro et al. 2007; Annilo et al. 2009) or shown to regulate sense gene splicing (Krystal et al. 1990; Kuersten and Goodwin 2003; Yan et al. 2005; Beltran et al. 2008; Allo et al. 2009). One well-characterized example is antisense-mediated splicing regulation of the thyroid hormone receptor THRA by the antisense transcript NR1D1 (Hastings et al. 1997). At this locus, coexpressed sense and antisense transcripts can form double-stranded RNA (dsRNA) over the region of SAS overlap, leading to splice site masking and a consequent shift in mRNA isoform production. Similar changes in splicing can be achieved by the addition of synthetic antisense oligonucleotides (Garcia-Blanco et al. 2004) or can be triggered by siRNAs produced from endogenous antisense transcripts (Allo et al. 2009). In vitro and in vivo, synthetic antisense oligonucleotides can modulate splicing reactions in favor of specific isoforms of disease-related genes, suggesting the possibility of therapeutic strategies that influence disease outcomes (Garcia-Blanco et al. 2004).
To date, there are no genome-wide studies that have investigated the relationship between alternative splicing and antisense transcription in the human genome. We therefore set out to investigate this relationship, with the objectives of assessing both the correlation between antisense transcription and splicing events in normal human cells (i.e., antisense-correlated splicing), and investigating possible mechanisms for antisense-mediated splicing regulation.
Results
Exon splicing is strongly correlated to antisense gene expression
Our goal was to examine the relationship between alternative splicing and antisense transcription. To establish the parameters of this relationship in normal human tissues, we analyzed expression data from 176 lymphoblastoid cell lines (LCLs) (Huang et al. 2007). These data were generated using Affymetrix Human Exon 1.0 ST arrays, which measure expression using 1.4 million probesets representing known and predicted exons on both strands over the genome. Eighty-seven Center d'Etude Polymorphisme Humain individuals from Utah (CEU) and 89 Yoruba individuals from Iabadan, Nigeria (YRI) were included in the analysis (Huang et al. 2007).
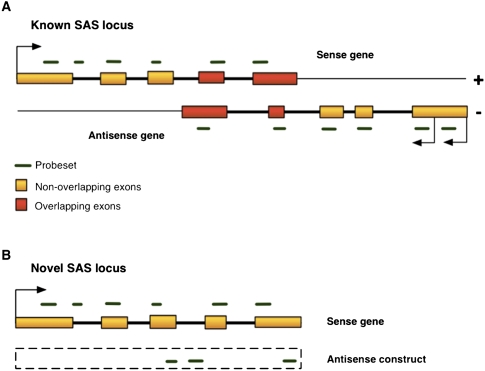
Probesets mapping to the sense strand of Ensembl exons were used to measure sense gene expression (see Methods) (Fig. 1A). We focused our analysis on a total of 3530 genes found in 1765 SAS loci, each with two gene members (Supplemental Table 1). When analyzing the expression of these genes (denoted “known SAS”), we used probesets mapping within the coordinates of the annotated boundaries of each of the two genes in a pair.
Figure 1.
Probesets mapping to known and novel SAS genes. (A) Schematic diagram of a hypothetical known SAS gene pair shows the structural arrangement of overlapping exons (red rectangles) and nonoverlapping exons (orange rectangles). In our analysis, each partner gene is, in turn, treated as the antisense gene. Probesets (horizontal green dashes) map to the sense strand of the gene in either exons or introns. (B) A novel SAS sense gene. Since the structure of the transcribed antisense RNA is unknown, an antisense construct (dashed-line box) spans the genomic coordinates of the sense gene and approximates antisense expression. All antisense probesets encompassed by that region are used to infer the expression of the antisense construct. If the actual antisense transcript extends beyond the sense gene boundaries, the antisense construct expression under-represents the actual level of antisense transcription at that locus.
To analyze antisense transcription at genes without an annotated antisense gene partner, we measured expression using probesets on the opposite strands of such genes (Fig. 1B). We found probesets, mapping to the opposite strand of 8313 genes, which had been designed based on previous evidence of transcription, such as ESTs (expressed sequence tags) (Liu et al. 2003). The category of genes without annotated antisense gene partners is hereafter referred to as “novel SAS,” and the expression values of the probesets mapping antisense to these genes were summed into an “antisense construct” (Fig. 1B; Supplemental Table 2). Probesets mapping to introns were also analyzed since these may represent alternative splice variants of annotated genes, including those with novel exons, or exons with alternative 3′- and 5′-splice sites. Retained introns may cause frameshifts in some isoforms and trigger nonsense-mediated decay (NMD), while novel exons may impart altered functionality to the encoded protein. We could not discern between these two possibilities using the exon array data.
The alternative splicing of each probeset was measured by normalizing its expression to the expression of the gene in each of the 176 samples (as described in Methods). The resulting value (the “splice index,” SI) represented an exon's relative inclusion (high SI) or exclusion (low SI) in the expressed mRNA isoforms. SI values were calculated for a total of 2995 probesets in 258 known SAS genes expressed above background in the 176 LCL samples, as well as 4187 probesets in 215 novel SAS genes. Next, the relationship between the splicing of sense gene exons and antisense gene expression was inferred using Spearman correlations (as described in Methods). Briefly, we measured correlations between (1) the SI values of each probeset across the 176 samples, and (2) the expression of the antisense gene (or construct) in the same samples. Spearman correlation P-values were corrected for multiple testing using the stringent Bonferroni method, yielding a conservative set of results.
This analysis revealed a widespread relationship between splicing and antisense transcription in human LCLs. Of the 258 known SAS genes, the vast majority (191 genes, 74.1%) had probesets whose relative inclusion in the expressed mRNAs (i.e., splicing) was significantly correlated to antisense gene expression (i.e., antisense-correlated splicing events; Bonferroni-corrected P < 0.05). Overall, 24% of the 2995 expressed probesets had inclusion levels that were significantly correlated to antisense gene expression (Fig. 2B; Supplemental Table 1, Bonferroni-corrected P < 0.05; Supplemental Table 3). Of these 191 known SAS genes, 75.4% had antisense-correlated splicing events in both gene partners, as would be expected from a reciprocal relationship (see example in Supplemental Fig. 3). On average, 32.3% of known SAS gene exons had antisense-correlated splicing, suggesting that expressed alternative isoforms differed significantly from each other (Supplemental Fig. 2).
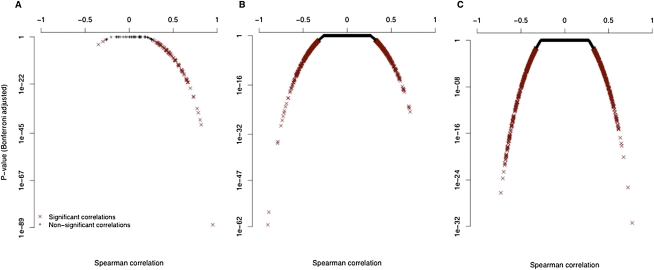
Figure 2.
SAS genes tend to have concordant expression and antisense-correlated splicing events. (A) The majority of SAS gene pairs have positively correlated gene expression values across the 176 LCL samples. (B,C) Individual probesets in known (B) and novel (C) SAS genes, have either positive or negative correlations (x-axis) between their SI values and the expression of the antisense gene or construct (the y-axis is the Bonferroni-corrected P-values of Spearman correlations).
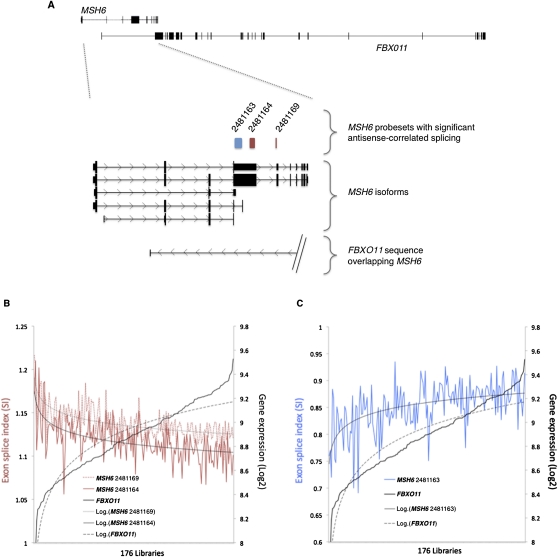
An example of antisense-correlated splicing is shown for the MSH6 gene (mutS homolog 6), which is involved in DNA-mismatch repair (Fig. 3A). At this locus, the splicing of three MSH6 probesets was significantly correlated to the expression of the antisense gene, FBXO11 (F-box protein 11). These three probesets mapped to two MSH6 exons; two probesets had SI values that were negatively correlated to FBXO11 expression, indicating that the corresponding exons were excluded from MSH6 mRNA isoforms present during high FBXO11 expression (Fig. 3A). The third probeset had SI values that were positively correlated to FBXO11 expression, indicating that the corresponding exon was preferentially included during higher FBXO11 expression. Interestingly, the MSH6 exon that encodes the core DNA mismatch repair motif was profiled by two probesets (Fig. 3A). The splicing of the 5′-most probeset was positively correlated to FBXO11 expression (id = 2481163; r = 0.56) (Fig 3C), while the inclusion of the 3′-most probeset was negatively correlated to FBXO11 expression (id = 2481164; r = −0.59) (Fig 3B). The 3′-most probeset maps within the coding region of the DNA mismatch repair motif and thus distinguishes those MSH6 isoforms that contain the motif from those isoforms that do not. Since the SI values of this probeset and of that of another downstream probeset (id = 2481160; r = −0.63) (Fig 3B) are negatively correlated to antisense gene expression, our analysis is compatible with the notion that FBXO11 expression is positively correlated to short MSH6 isoforms.
Figure 3.
Antisense-correlated splicing events at the MSH6 gene. (A) The MSH6 (+ strand) and FBXO11 (− strand) known SAS locus is represented at the top of the panel, with dotted lines indicating the area of interest displayed in the bottom panel. MSH6 isoforms are depicted as thick (coding) or thin (noncoding) black bars (exons) connected by lines (introns) with directional arrows (“>” indicates + strand). The span of the longest known FBXO11 isoform is depicted on the − strand (“<”). Three MSH6 probesets have either positive (r = 0.56; blue rectangle) or negative (r = −0.59; r = −0.63; red rectangles) correlations between their respective SI values (i.e., probeset inclusion in the mRNA) and the antisense gene expression (i.e., FBXO11 expression). Probeset locations correspond to their genomic location in the MSH6 locus. In B and C the probeset SI values (left-hand y-axis) are shown along with the antisense gene expression (right-hand y-axis) for all 176 LCL samples (x-axis); samples are ordered by increasing FBXO11 expression. Trendlines (log2) for each data series are superimposed on the graphs.
Similarly to the known SAS gene category, 78.1% (168) of the 215 novel SAS genes had significant antisense-correlated splicing events. A total of 19.8% of the 4167 expressed exons in these genes had antisense-correlated SI values (Fig. 3C; Supplemental Table 4). Genes contained (1) exons with positive antisense-correlated splicing, indicating their inclusion in isoforms coexpressed with the antisense gene; (2) exons with negative antisense-correlated splicing, indicating their exclusion from expressed isoforms; and (3) exons whose splicing was uncorrelated with antisense transcription, indicating either constitutive expression or splicing regulation mediated by independent factors. On average, 23.1% of novel SAS gene exons had antisense-correlated splicing, suggesting that expressed alternative isoforms differed significantly from each other (Supplemental Fig. 2). More than a third of exons with antisense-correlated splicing events encoded protein domains (Supplemental Results), suggesting that these events have the potential to modify primary amino acid sequences.
We reanalyzed the CEU and YRI data separately (see Supplemental Results), since previous studies have observed population-specific differences in gene expression patterns (Spielman et al. 2007; Storey et al. 2007; Zhang et al. 2008). Such differences were also evident in our data, as a higher proportion of novel SAS genes showed antisense-correlated splicing events unique to the YRI (35.6%) versus CEU (24.1%) individuals (Supplemental Fig. 4). Although these novel SAS genes were not differentially expressed, they had a significantly greater variability in SI values (P-value = 8.3 × 10−8; considering all expressed probesets, as described in Supplemental Results) than the known SAS genes. This indicates that novel SAS genes have greater levels of alternative splicing relative to known SAS genes in the YRI individuals.
Antisense expression affects both splicing and expression of sense genes
Although previous studies identified correlations between antisense transcription and sense gene expression (Chen et al. 2005; RIKEN Genome Exploration Research Group et al. 2005), the correspondence between antisense-correlated changes in splicing and gene expression was undetermined. Using the 258 known SAS genes expressed in LCLs, we calculated correlations between gene expression values of partner genes across the 176 samples. Significant gene-level correlations were found for 68.2% of pairs (176 genes, Bonferroni corrected P < 0.05) (Fig. 2A; Supplemental Table 5). As shown in the previous section, antisense-correlated splicing events occurred at 74.0% of the 258 known SAS genes. Thus, our results are compatible with a model in which antisense transcription affects splicing and expression of the partner gene to a similar extent. For 170 genes, antisense transcription was significantly correlated to both sense gene expression and splicing. A few genes had antisense-correlated changes only in splicing (21 genes) or only in expression (six genes). As observed in previous studies (Chen et al. 2005; RIKEN Genome Exploration Research Group et al. 2005), the expression of most SAS gene pairs was positively correlated, indicating concordant expression (Fig. 2A).
Regions of SAS overlap are enriched in exons with antisense-correlated splicing events
RNA-masking of splice sites via dsRNA formation underlies antisense-mediated splicing regulation of genes such as THRA (also known as TRα) (Hastings et al. 1997), highlighting a functional consequence of SAS sequence overlap. To determine the relative importance of SAS sequence overlap in our data, we ascertained whether probesets that overlapped an antisense gene (“overlapping probesets”) were more likely to exhibit antisense-correlated splicing events than probesets outside of the annotated overlap (“nonoverlapping probesets”). We defined regions of overlap as exonic or intronic gene regions that mapped on the opposite strand of an annotated gene, and within its boundaries (depicted in Fig. 1A). This was done to enable detection of interactions between intronic regions of pre-mRNA molecules.
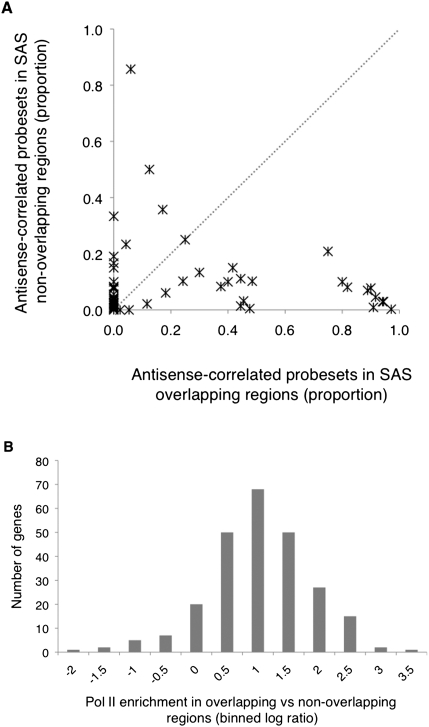
Of the 191 known SAS genes with antisense-correlated splicing events, 75 had at least two overlapping and two nonoverlapping probesets expressed above background. For each of these genes, we compared the proportion of probesets with antisense-correlated splicing in overlapping versus nonoverlapping regions (proportions were corrected for the total number of expressed probesets) (see Methods). We reasoned that if sequence overlap was not an important factor, the proportions of these two groups would be equal. Instead, we observed a 2.5-fold increase in the frequency of antisense-correlated probesets within SAS overlaps (t-test, P-value = 4.6 × 10−3) (Fig. 4A). Physical overlap therefore seems to be an important aspect of the observed antisense-correlated splicing events, likely indicating that sequence overlap is a key feature of the mechanism of splicing control acting at these loci.
Figure 4.
Antisense-correlated splicing events and Pol II occupancy levels are enriched in SAS overlaps. (A) The x-axis coordinate of each gene shows the corrected fraction of all expressed overlapping exons with antisense-correlated splicing. The y-axis shows the corrected fraction of all expressed nonoverlapping exons with antisense-correlated splicing. The fraction of probesets inside and outside the regions of overlap were corrected for overall number of probesets (as in Methods). The gray (dotted) line represents equal proportions of antisense-correlated overlapping and nonoverlapping probesets (B). The log10 ratios of overlapping versus nonoverlapping Pol II peaks/kilobase for 248 known SAS genes reveals Pol II enrichment in SAS overlaps (log ratios >0) at the majority of genes.
Regions of SAS overlap are enriched in exons and nucleosomes
Recent analyses (Nahkuri et al. 2009; Schwartz et al. 2009; Spies et al. 2009; Tilgner et al. 2009) of publicly available ChIP-seq data from human T-cells (Schones et al. 2008) found that nucleosome occupancy is elevated in exons relative to introns and indicated that this enrichment decreases the rate of RNA polymerase II (Pol II) elongation (Schwartz et al. 2009). Indeed, nucleosomes constitute chromatin “roadblocks” that act to slow Pol II elongation rate (Kulaeva et al. 2009), and slower Pol II elongation rates have, in turn, been shown to increase the rate of alternative splicing (de la Mata et al. 2003). Thus, one hypothesis for the observed increase in the rate of antisense-correlated alternative splicing events in SAS overlaps may involve a decreased polymerase speed in those regions, indirectly caused by increased exon frequency. An increased exon frequency in SAS overlaps can reasonably be expected since such regions contain sequences encoded by both the sense and the antisense genes.
To investigate this hypothesis, we asked whether areas of SAS overlap were enriched in exons relative to flanking (nonoverlapping) regions in the same genes. Calculating the frequency of Ensembl-annotated exons in SAS genes (per kilobase; see Methods), revealed a 7.2-fold increased exon/kilobase frequency in overlapping (3.1 exons/kb) versus flanking nonoverlapping regions (0.43 exons/kb; Welch's t-test, P < 2.2 × 10−16). This finding suggests that regions of overlap have a greater frequency of nucleosomes, as expected from previous studies (Schwartz et al. 2009; Spies et al. 2009; Tilgner et al. 2009), and as confirmed by us (Supplemental Fig. S5; Supplemental Results).
We expected the increased frequency of Pol II “roadblocks” (i.e., nucleosomes) in SAS overlaps to cause attenuated Pol II elongation speed in these regions. Given the documented effects of decreased Pol II speed on alternative splicing (de la Mata et al. 2003), we also expected an increased local frequency of alternatively spliced exons. To test these predictions, we ascertained the levels of both Pol II occupancy, and of alternative splicing in regions of SAS overlap relative to nonoverlapping regions, as described next.
Increased Pol II occupancy in regions of SAS overlap
Pol II occupancy levels were analyzed using publicly available ChIP-seq data, from one of the 176 lymphoblastoid cell lines (GM12878), that were generated as part of the ENCODE project (ENCODE Project Consortium 2004). We sought to determine whether Pol II peaks were enriched in regions of SAS sequence overlap relative to flanking nonoverlapping regions in individual sense or antisense genes. Pol II occupancy was used as a surrogate measure of Pol II speed, since areas with stalled or slowly moving complexes were more likely to be observed as bound by Pol II in a ChIP-seq experiment than areas with fast moving Pol II complexes. Thus, Pol II peaks were expected to represent regions of DNA through which Pol II exhibits slow elongation speeds. To assess Pol II occupancy, areas with significant enrichment of signal over background (i.e., “peaks”) were enumerated independently in overlapping and nonoverlapping regions of known SAS genes (see Supplemental Results). We found that a total of 248 expressed known SAS genes harbored 488 Pol II peaks in distinct regions: 85 peaks (17.4%) occurred in known promoters, 212 peaks (43.4%) were in nonoverlapping regions, and 191 peaks (39.1%) were in overlapping regions. Regions of overlap spanned an average of 11.1% of the total gene lengths. By calculating the log ratio of overlapping versus nonoverlapping Pol II occupancy levels (peaks/kb), a 5.5-fold enrichment was observed in areas of overlap for 85.9% of the 248 known SAS genes (Fig. 4B, Mann-Whitney Test, P = 2.4 × 10−19). This enrichment corresponded with the anticipated effect of increased nucleosome frequency on Pol II speed in areas of SAS overlaps, which led us to expect local changes in splicing outcomes.
Higher rate of alternative splicing in areas of SAS overlap
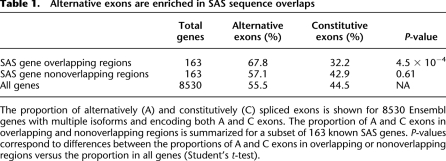
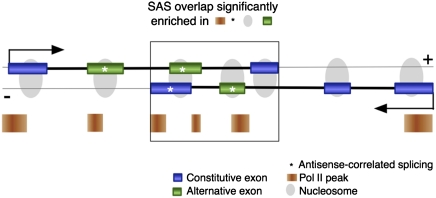
To investigate changes in alternative splicing in overlapping versus nonoverlapping regions, we identified constitutive and alternative exons for 8530 Ensembl genes with multiple isoforms. The 149,032 exons encoded by these genes were categorized as “constitutive” if present in all annotated gene isoforms (45.5% of exons), and “alternative” if found in only a subset of isoforms (55.5% of exons). Next, all exons encoded in the 2668 known SAS genes were subdivided into those found in overlapping and nonoverlapping regions, as previously described. Of these, we analyzed 163 genes that expressed both alternative and constitutive exons, had at least two exons in overlapping regions, and had at least two exons in nonoverlapping regions. A total of 57.1% of exons in nonoverlapping regions were alternatively spliced, similar to the proportion of alternative exons in all 8530 genes with multiple isoforms (Table 1) (Student's t-test, P = 0.6). However, when considering exons in overlapping regions, 67.8% of exons were alternatively spliced, a significant increase from the overall proportion (Table 1) (Student's t-test, P = 4.5 × 10−4). Elevated levels of alternative splicing thus correlate with the local decrease in Pol II transcriptional speed, and this, in turn, is compatible with the notion that antisense transcription ultimately increases the diversity of alternative isoforms expressed from SAS loci (Fig. 5).
Table 1.
Alternative exons are enriched in SAS sequence overlaps
Figure 5.
Model of distinct features enriched in SAS overlapping regions. Features over-represented in the SAS overlap (large rectangle) include exon frequency (blue or green rectangles connected by a thick black line), the proportion of alternative (green) versus constitutive (blue) exons, Pol II peak frequency (orange rectangles), and the proportion of exons with antisense-correlated splicing patterns (*). Nucleosomes (gray ovals) are localized to exons and are therefore enriched in the area of SAS overlap. (Black arrows) Transcriptional direction.
Known and novel SAS genes have more annotated isoforms
The relationship between antisense transcription and alternative splicing could indicate that genes with antisense transcripts may encode a greater diversity of transcript isoforms compared to those lacking antisense transcripts. We tested this hypothesis by analyzing 5169 known SAS genes, 7823 novel SAS genes, and 7929 non-SAS genes, and found that known and novel SAS genes were, indeed, associated with a larger number of distinct isoforms compared to non-SAS genes (average of 2.3 and 2.3, vs. 1.8, respectively; Welch t-test, P = 3.2 × 10−84 [Supplemental Fig. S1]; Welch t-test, P-values in Supplemental Table 2). However, we also found that, on average, known and novel SAS genes were in general significantly longer (83.7 kb and 70.6 kb, vs. 22.9 kb), and had more introns (9.4 and 9.9 vs. 6.6 [Supplemental Fig. S1]; P-values in Supplemental Table 2). This led us to consider the possibility that the multiple alternative isoforms found in known and novel SAS genes may simply be due to the increased chance of observing alternative transcription in longer genes.
To investigate this possibility, we segregated the non-SAS and known SAS genes into bins of increasing gene length, and asked whether known SAS genes within each bin had a significantly different number of isoforms relative to the non-SAS genes in the same bin (Supplemental Results). We found that known SAS genes of length >11.4 kb (the 50th percentile of non-SAS gene lengths and the 14th percentile of the known SAS gene lengths) had a significantly greater number of transcript isoforms compared to non-SAS genes in the same length bin. Together with previous observations of antisense-regulated splicing events (Krystal et al. 1990; Hastings et al. 1997; Kuersten and Goodwin 2003; Yan et al. 2005; Beltran et al. 2008), these results are consistent with a putative role for antisense transcription in splicing regulation.
Antisense transcription coincides with alternative splicing throughout metazoan evolution
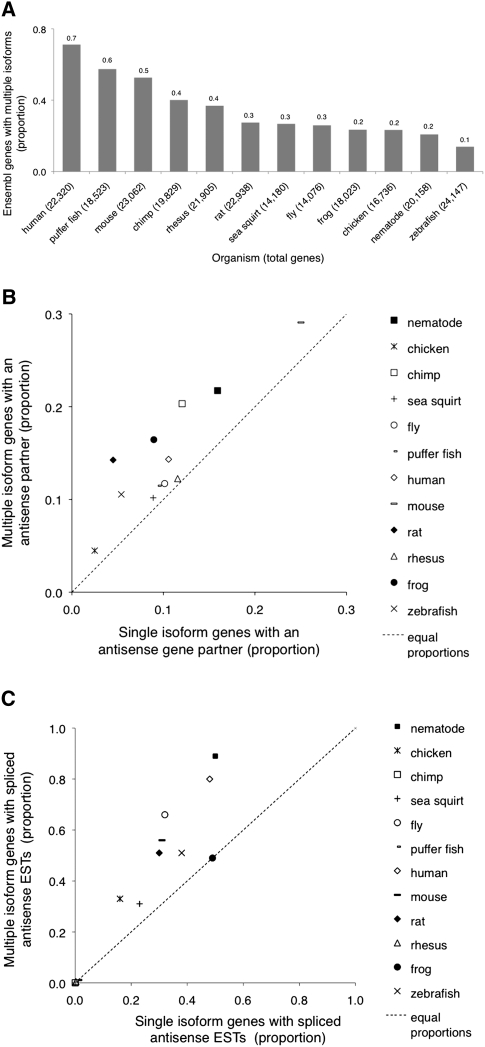
Since antisense transcription has been observed in numerous organisms (Dahary et al. 2005; Zhang et al. 2006), we hypothesized that the relationship between splicing and antisense transcription has been conserved throughout evolution. To address this possibility, we measured the concordance between alternative splicing and antisense transcription in 12 species, including human, mouse, rat, chimp, rhesus monkey, Drosophila, chicken, Xenopus, sea sponge, Fugu, worm, and zebrafish. We first divided genes into two categories: those with multiple annotated isoforms, and those with a single known isoform (Fig. 6A). In each species, we then compared the proportion of known SAS genes in each category and found that a significantly higher proportion of multiple-isoform genes had known antisense gene partners in nearly all species (11 of 12) (Fig. 6B; corresponding P-values in Supplemental Table 4).
Figure 6.
High concordance between alternative splicing and antisense transcription in multiple species. (A) The proportion of all genes with multiple isoforms in twelve species. (B) Genes with multiple isoforms are enriched in known SAS pairs and (C) in EST evidence for novel antisense transcription (novel SAS genes). The dotted lines represent equal proportions of SAS genes or antisense ESTs among genes with multiple or single isoforms. Note that some organisms have very few ESTs (see Supplemental Table 4B).
We next measured novel antisense transcription by using species-specific ESTs that mapped to the antisense strand of annotated genes (Methods). Antisense ESTs were found in a significantly larger proportion of genes with multiple rather than single isoforms (Fig. 6C; Supplemental Table 4), and this relationship remained significant for the subset of genes with highly expressed ESTs (see Supplemental Results). Antisense ESTs were also more highly expressed at loci with multiple isoforms, indicating that antisense transcription is stronger at these loci (data not shown). Together, these findings indicate that antisense transcription is a general feature of genes with multiple transcripts throughout evolution.
Discussion
Others have reported on the abundance of antisense transcription in mammalian transcriptomes (Chen et al. 2004; Kapranov et al. 2005; RIKEN Genome Exploration Research Group et al. 2005; Engstrom et al. 2006) and on the frequent coexpression of SAS gene partners (Reis et al. 2004; Chen et al. 2005; Kiyosawa et al. 2005). However, the general functional implications of antisense transcription remain to be elucidated. In this study, we show that both known and novel instances of antisense transcription are strongly correlated to sense gene splicing, affecting 20%–24% of exons at 74%–79% of expressed genes with annotated (known SAS) or unannotated (novel SAS) antisense transcripts, respectively. We refer to this phenomenon as antisense-correlated splicing. Although a few examples of antisense-correlated splicing events have been reported in the literature (Hastings et al. 1997; Mihalich et al. 2003; Yan et al. 2005; Annilo et al. 2009), we provide for the first time evidence linking antisense transcription to alternative splicing across the majority of SAS loci expressed in human lymphoblastoid cell lines.
Altering the complement of proteins associated with the Pol II C-terminal domain (CTD) can affect splicing either by altering the elongation speed of the polymerase or by making specific splicing factors available cotranscriptionally (Listerman et al. 2006), thus affecting the alternative expression of many genes. In contrast to such trans-acting effects of classical splicing regulatory mechanisms (Wang and Burge 2008), a distinguishing aspect of antisense-mediated splicing regulation is its effect on individual cis-encoded genes. This effect is particularly notable in areas of SAS sequence overlap, since overlapping regions were enriched in antisense-correlated alternative splicing events. Our findings indicate that these regions are distinguished from flanking nonoverlapping regions by a greater frequency of exons and elevated nucleosome occupancy. The increased frequency of nucleosomes in regions of SAS overlaps was associated with decreased Pol II speed, and then further associated with a significant increase in alternative exon usage in areas of SAS overlap (Fig. 5). A similar increase in nucleosome occupancy has previously been linked to another type of alternative processing: actively used polyadenylation signals (PAS) in T-cells (Spies et al. 2009). In conjunction, these observations underscore the potential role that sequence-based determinants of nucleosome positioning (such as nucleosome binding affinity of exonic and PAS-associated sequences) may play in alternative polyadenylation and splicing.
Previous work on the fibronectin 1 (FN1) locus showed that siRNAs, produced from endogenous antisense transcripts, can trigger local heterochromatinization and cause inclusion of an alternatively spliced exon via the transcriptional gene silencing pathway (Allo et al. 2009). As we found in our more general observations, the Allo et al. (2009) study suggested that exon inclusion was also dependent on decreased Pol II speed at the FN1 locus. Although we did not find evidence for siRNA-mediated heterochromatinization using publicly available data (see Supplemental Results), we cannot exclude the possibility that this may be one mechanism through which antisense-correlated alternative splicing events are generally regulated. In fact, our results would notionally support this possibility, since the increased frequency of nucleosomes in areas of SAS overlaps could act as a target for the deposition of heterochromatin marks. Furthermore, our results show that Pol II speed is decreased over regions of overlap, which is consistent with the findings at the FN1 locus. An alternative mechanism of decreasing Pol II speed at SAS loci could involve transcriptional interference from polymerases transcribing an antisense gene (Shearwin et al. 2005; Galburt et al. 2007). Our results also indicated that a higher frequency of exons may be sufficient to result in a decrease in Pol II elongation speed. The relative contributions of these mechanisms to the regulation of alternative splicing events at SAS loci will be an interesting focus of future studies.
In contrast to known SAS genes, the antisense transcripts at novel SAS loci do not correspond to annotated genes with identifiable exons, and at least some of these may thus correspond to noncoding RNAs. The prevalence of novel SAS genes was higher than that of known SAS genes, indicating the importance of noncoding RNAs to the regulation of alternative splicing at SAS loci. The novel SAS class of genes was also the most functionally diverse, relative to known SAS genes and to genes without any detectable antisense transcription (Supplemental Results). In line with our previous findings (Morrissy et al. 2009), novel SAS genes were enriched in known cancer genes, and the Gene Ontology terms were consistent with this observation (Supplemental Results).
We considered genes of comparable length and found that genes with antisense transcription have an increased number of annotated isoforms compared to genes without antisense transcription, as expected from the positive relationship between splicing and antisense expression. In general, however, antisense transcription was positively associated with longer genes, at both known and novel SAS loci. This association is not surprising, since longer genomic regions are more likely, simply by chance, to accrue functional promoter sequences, for instance, from transposable element (TE) insertions that can drive both coding and noncoding RNA transcription in the antisense orientation (Faulkner et al. 2009; Romanish et al. 2009). We speculate that antisense transcription arising by chance can be consequently selected for as a means of increasing the variety of isoforms expressed from novel SAS genes. In line with this hypothesis, the novel SAS genes (but not known SAS genes) had a greater variability of splicing in the Yoruban individuals, which are drawn from a more genetically diverse population than the CEU individuals (Mormon individuals from Utah). One plausible explanation for this observation is that the noncoding antisense RNA transcripts expressed in the YRI individuals differ in terms of structure (i.e., extent of SAS overlap) from the corresponding transcripts in the Mormon population. This suggests that the evolution of new gene isoforms could still be an active process in the YRI population. Corroborating evidence for the negative selection on the separation of SAS overlaps has previously been documented between the human, mouse, and Fugu genomes (Dahary et al. 2005).
The use of exon arrays in this work reflects the availability of numerous samples of data that provide both strand-specific and exon-level expression. These requirements have precluded the use of RNA-seq data (Bentley et al. 2008; Valouev et al. 2008), unless such data are generated using strand-specific libraries. A recent in-depth comparison of RNA-seq library construction methods showed that it is both simple and cost-effective to create strand-specific libraries that can be sequenced on the Illumina platform (Parkhomchuk et al. 2009; Levin et al. 2010); hence, future studies may well benefit from access to such data. The prevalent correlations between antisense transcription and alternative splicing of sense genes described here provide another strong argument for the continued and widespread adoption of strand-specific expression-profiling protocols. Compared to microarrays, strand-specific RNA-seq data would not only increase the detectable dynamic range of alternatively expressed exons, but it would also provide relative measures of a subset of differentially expressed sense gene isoforms (i.e., those with distinct splice sites). Alternatively, single-molecule sequencing methods (Bowers et al. 2009) would allow full discrimination of specific gene isoforms expressed in a correlated manner to antisense transcription.
We found a strong concordance between known antisense transcription and genes with multiple isoforms in amphibians, fishes, insects, birds, nematodes, and mammals. In conjunction with detectable alterations in chromatin-state and Pol II processivity at human known SAS loci, these observations advocate for a conserved role of antisense transcription in the regulation of alternative splicing. In support of this speculation, we found similar rates of antisense-correlated splicing events in a diverse series of human tissues, including normal tissues (neuronal, mesenchymal, and epithelial tissues) as well as cancerous samples (AS Morrissy and MA Marra, in prep.). Subsets of these events were specific to individual normal tissues or to cancer, indicating the potential relevance of these events to cancer biology.
Methods
Ensembl genes
Ensembl (Hubbard et al. 2002) gene annotations (including gene, cDNA, and exon coordinates; release 49) were downloaded via the Ensembl Perl API. Genes whose genomic coordinates overlapped by at least one base and that were encoded on opposite strands were categorized as known SAS genes. Gene regions that mapped within the genomic boundaries of another gene on the opposing strand were defined as “overlapping.” Flanking regions in the same genes were defined as “nonoverlapping.”
Exons were classified as alternative (A) or constitutive (C) if they were found in a subset or in all of the annotated isoforms of a gene, respectively. Only known SAS genes with both A and C exons and at least two expressed exons in the overlapping and two expressed exons in the nonoverlapping SAS region were considered.
Public data sets
Lymphoblastoid cell lines
Publicly available CEU and YRI Affymetrix Human Exon 1.0 ST Array data (http://media.affymetrix.com:80/support/technical/technotes/exon_array_design_technote.pdf) were downloaded from the Gene Expression Omnibus (GEO, GSE7792) (Barrett et al. 2009). A total of 18,041 genes had probesets mapping to both the positive and negative strands of the genome, and 10,636 genes had probesets mapping only to the sense strand. An additional 366 genes had probesets mapping only to the antisense strand and likely reflect changes in gene annotations since probeset design. Array data were background-corrected and normalized using the PLIER algorithm (Expression Console; www.affymetrix.com/support/technical/software_downloads.affx). The log2 of the resulting expression values was used in further analyses. Probesets were filtered for expression above background (Griffith et al. 2008) in at least 20% of samples. Gene-level expression values were calculated for genes that had a minimum of 20% of probesets expressed in at least 20% of samples and a minimum of two expressed probesets. For novel SAS genes, an “antisense construct” was generated to represent the unknown antisense transcript. The boundaries of the antisense construct were set to the genomic boundaries of the sense gene, but only probesets mapping to the opposite strands were considered (Fig. 1B). Probesets mapping in this region were used to calculate the antisense construct expression in an analogous way to annotated genes.
Multiple species data
Current gene annotations and EST data were downloaded from the UCSC Genome Browser (Rosenbloom et al. 2010) for human (Homo sapiens), Fugu (Takifugu rubripes), mouse (Mus musculus), chimp (Pan troglodytes), rhesus (Macaca mulatta), rat (Rattus norvegicus), sea squirt (Ciona intestinalis), Drosophila (Drosophila melanogaster), Xenopus (Xenopus tropicalis), chicken (Gallus gallus), nematode (Caenorhabditis elegans), and zebrafish (Danio rerio). Only ESTs with known orientation were considered (intronEST table).
Pol II data
ChIP-seq data were downloaded from the UCSC Genome Browser (Primary Table: wgEncodeYaleChIPseqRel2SignalGm12878Pol2). For the Pol II analysis, known SAS genes were required to have at least one Pol II peak in the gene body (i.e., not including the first exon of the gene).
Splice index calculations
Gene expression was calculated as the mean of all probesets mapping to the sense strand of that gene or antisense construct. Probesets that mapped to introns as well as exons were included, since they may represent alternatively spliced exons, intron retention events, or other unannotated splicing variations, such as alternative 5′- or 3′-splice-site usage. Each probeset is therefore referred to as an exon. The splice index was the expression of the exon normalized to the expression of the whole gene:
The Spearman's rank correlation coefficient of each sense exon splicing index and the antisense gene (or construct) expression was calculated for all SAS genes, using the cor.test function in R (R Development Core Team 2008). Associated correlation P-values (Best and Roberts 1975) were multiple-test-corrected using the Bonferroni method (Wright 1992). In known SAS gene pairs, each gene partner was, in turn, analyzed as the sense gene and as the antisense gene. Correlations (and associated P-values) between gene expression values were calculated using the same methods.
Relative to probesets that were not antisense-correlated, correlated probesets did not have biases in any of the following features: number of independent probes, cross-hybridization type, or probe count (Chi-square test, respective P-values = 0.98, 0.80, 1.00).
Probesets with antisense-correlated splicing did not differ in melting temperature (Tm) relative to probesets without antisense-correlated splicing, in the same genes (t-test, p > 0.5). We used the nearest-neighbor method to predict melting temperatures of nucleic acid duplexes (SantaLucia 1998). GC content is a strong determinant of the hybridization energy of double-stranded DNA; however, interactions between neighboring bases along the helix mean that stacking energies are significant. The nearest-neighbor model accounts for this by considering pairs of adjacent bases along the backbone at a time. Each of these has enthalpic and entropic parameters, the sum of which determine melting temperature (SantaLucia 1998).
The majority of probesets did not overlap probesets on the opposing strand, indicating that intensity signals from sense and antisense genes are independent of each other (e.g., 31,004 [∼10%] and 12,542 [∼4%] of 321,393 probesets mapping sense to genes overlap a probeset on the opposing strand by at most 1 bp or 100 bp, respectively). In the SAS genes analyzed in this study, no sense probesets overlap antisense probesets, suggesting that any bias that might be introduced by such overlap is not a factor.
Exon frequency calculations
The frequency of exons per kilobase (exons/kb) was calculated for 1765 known SAS gene pairs. For each gene pair, the number of exons per kilobase in the overlapping region (including exons from both strands) was compared to the number of exons per kilobase in nonoverlapping regions of both genes. For this analysis, overlapping alternative exons (i.e., sharing the same genomic location, but differing in 5′ or 3′ ends) were only counted once.
Enrichment of antisense-correlated splicing events in overlapping versus nonoverlapping regions
The proportion of antisense-correlated splicing events in SAS overlaps was analyzed at 75 genes that had at least two overlapping and two nonoverlapping probesets expressed above background, and at least one probeset whose splicing was correlated to expression of the antisense gene. Overlapping probesets were those that mapped to regions of the genome shared by both the sense and the antisense gene, while nonoverlapping probesets were those mapping to flanking regions of the genome (i.e., spanned by the sequence of only the sense or the antisense gene) (see Fig. 1). The proportion of antisense-correlated probesets (i.e., those whose SI values were significantly correlated to antisense gene expression across 176 samples) was calculated relative to the number of total expressed probesets in the region of interest (overlapping or nonoverlapping). To account for the (generally) larger number of expressed probesets in nonoverlapping regions, the calculated proportion was normalized according to the following equation, where Covp is the number of probesets in the region of interest (in this case overlapping), and where Eovp and Etotal are, respectively, the number of expressed probesets in the overlapping region or the whole gene:
Antisense transcription in multiple species
For each species, we (1) identified genes that overlapped by at least 1 bp and were encoded on opposing strands (known SAS), and (2) enumerated the number of annotated isoforms in Ensembl. Genes were divided into those with single or multiple isoforms, and the proportion of known SAS genes in each category was computed for individual species. To analyze the concordance between novel antisense transcription and number of annotated isoforms, ESTs with orientation information (i.e., spliced ESTs) were downloaded from the UCSC Browser. The proportion of genes with at least one EST mapping to the opposing strand was calculated for genes with single or multiple annotated isoforms, in each species. For a more stringent analysis, the median antisense EST count at annotated genes was calculated for all species, and only genes with antisense EST counts above the median were further considered.
Acknowledgments
We are grateful for funding provided by the University of British Columbia, the Michael Smith Foundation for Health Research (MSFHR), the Natural Sciences and Engineering Research Council (NSERC), Genome British Columbia, the Terry Fox Foundation (TFF), the Canadian Institutes of Health Research (CIHR), the National Cancer Institute of Canada (NCIC), and the BC Cancer Foundation (BCCF). A.S.M. was supported by CIHR and MSFHR. M.G. was supported by NSERC, TFF, and NCIC and was a Senior Graduate Trainee of the MSFHR and Genome BC. M.A.M. is an MSFHR scholar and Terry Fox Young Investigator.
Authors’ contributions: A.S.M. and M.A.M. conceived the analyses. A.S.M. designed and performed all computational analyses and created the figures and tables. M.G. conducted microarray data preprocessing and contributed analysis concepts. A.S.M. and M.A.M. prepared the manuscript, aided by M.G.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.113431.110.
References
- Allo M, Buggiano V, Fededa JP, Petrillo E, Schor I, de la Mata M, Agirre E, Plass M, Eyras E, Elela SA, et al. 2009. Control of alternative splicing through siRNA-mediated transcriptional gene silencing. Nat Struct Mol Biol 16: 717–724 [DOI] [PubMed] [Google Scholar]
- Annilo T, Kepp K, Laan M 2009. Natural antisense transcript of natriuretic peptide precursor A (NPPA): structural organization and modulation of NPPA expression. BMC Mol Biol 10: 81 doi: 10.1186/1471-2199-10-81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett T, Troup DB, Wilhite SE, Ledoux P, Rudnev D, Evangelista C, Kim IF, Soboleva A, Tomashevsky M, Marshall KA, et al. 2009. NCBI GEO: archive for high-throughput functional genomic data. Nucleic Acids Res 37: D885–D890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beltran M, Puig I, Pena C, Garcia JM, Alvarez AB, Pena R, Bonilla F, de Herreros AG 2008. A natural antisense transcript regulates Zeb2/Sip1 gene expression during Snail1-induced epithelial–ìmesenchymal transition. Genes Dev 22: 756–769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentley DR, Balasubramanian S, Swerdlow HP, Smith GP, Milton J, Brown CG, Hall KP, Evers DJ, Barnes CL, Bignell HR, et al. 2008. Accurate whole human genome sequencing using reversible terminator chemistry. Nature 456: 53–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Best DJ, Roberts DE 1975. Algorithm AS 89: The upper tail probabilities of Spearman's Rho. J R Stat Soc Ser C Appl Stat 24: 377–379 [Google Scholar]
- Bowers J, Mitchell J, Beer E, Buzby PR, Causey M, Efcavitch JW, Jarosz M, Krzymanska-Olejnik E, Kung L, Lipson D, et al. 2009. Virtual terminator nucleotides for next-generation DNA sequencing. Nat Methods 6: 593–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carninci P, Kasukawa T, Katayama S, Gough J, Frith MC, Maeda N, Oyama R, Ravasi T, Lenhard B, Wells C, et al. 2005. The transcriptional landscape of the mammalian genome. Science 309: 1559–1563 [DOI] [PubMed] [Google Scholar]
- Chen J, Sun M, Kent W, Huang X, Xie H, Wang W, Zhou G, Shi R, Rowley J 2004. Over 20% of human transcripts might form sense–antisense pairs. Nucleic Acids Res 32: 4812–4820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Sun M, Hurst LD, Carmichael GG, Rowley JD 2005. Genome-wide analysis of coordinate expression and evolution of human cis-encoded sense-antisense transcripts. Trends Genet 21: 326–329 [DOI] [PubMed] [Google Scholar]
- Dahary D, Elroy-Stein O, Sorek R 2005. Naturally occurring antisense: Transcriptional leakage or real overlap? Genome Res 15: 364–368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Mata M, Alonso CR, Kadener S, Fededa JP, Blaustein M, Pelisch F, Cramer P, Bentley D, Kornblihtt AR 2003. A Slow RNA polymerase II affects alternative splicing in vivo. Mol Cell 12: 525–532 [DOI] [PubMed] [Google Scholar]
- ENCODE Project Consortium 2004. The ENCODE (ENCyclopedia Of DNA Elements) Project. Science 306: 636–640 [DOI] [PubMed] [Google Scholar]
- Engstrom PG, Suzuki H, Ninomiya N, Akalin A, Sessa L, Lavorgna G, Brozzi A, Luzi L, Tan SL, Yang L, et al. 2006. Complex loci in human and mouse genomes. PLoS Genet 2: e47 doi: 10.1371/journal.pgen.0020047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faulkner GJ, Kimura Y, Daub CO, Wani S, Plessy C, Irvine KM, Schroder K, Cloonan N, Steptoe AL, Lassmann T, et al. 2009. The regulated retrotransposon transcriptome of mammalian cells. Nat Genet 41: 563–571 [DOI] [PubMed] [Google Scholar]
- Galburt EA, Grill SW, Wiedmann A, Lubkowska L, Choy J, Nogales E, Kashlev M, Bustamante C 2007. Backtracking determines the force sensitivity of RNAP II in a factor-dependent manner. Nature 446: 820–823 [DOI] [PubMed] [Google Scholar]
- Garcia-Blanco M, Baraniak A, Lasda E 2004. Alternative splicing in disease and therapy. Nat Biotechnol 22: 535–546 [DOI] [PubMed] [Google Scholar]
- Griffith M, Tang MJ, Griffith OL, Morin RD, Chan SY, Asano JK, Zeng T, Flibotte S, Ally A, Baross A, et al. 2008. ALEXA: a microarray design platform for alternative expression analysis. Nat Methods 5: 118 doi: 10.1038/nmeth0208-118 [DOI] [PubMed] [Google Scholar]
- Hastings M, Milcarek C, Martincic K, Peterson M, Munroe S 1997. Expression of the thyroid hormone receptor gene, erbAα, in B lymphocytes: Alternative mRNA processing is independent of differentiation but correlates with antisense RNA levels. Nucleic Acids Res 25: 4296–4300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang RS, Duan S, Bleibel WK, Kistner EO, Zhang W, Clark TA, Chen TX, Schweitzer AC, Blume JE, Cox NJ, et al. 2007. A genome-wide approach to identify genetic variants that contribute to etoposide-induced cytotoxicity. Proc Natl Acad Sci 104: 9758–9763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard T, Barker D, Birney E, Cameron G, Chen Y, Clark L, Cox T, Cuff J, Curwen V, Down T, et al. 2002. The Ensembl genome database project. Nucleic Acids Res 30: 38–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapranov P, Drenkow J, Cheng J, Long J, Helt G, Dike S, Gingeras TR 2005. Examples of the complex architecture of the human transcriptome revealed by RACE and high-density tiling arrays. Genome Res 15: 987–997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiyosawa H, Mise N, Iwase S, Hayashizaki Y, Abe K 2005. Disclosing hidden transcripts: Mouse natural sense–antisense transcripts tend to be poly(A) negative and nuclear localized. Genome Res 15: 463–474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krystal GW, Armstrong BC, Battey JF 1990. N-myc mRNA forms an RNA–RNA duplex with endogenous antisense transcripts. Mol Cell Biol 10: 4180–4191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuersten S, Goodwin EB 2003. The power of the 3′ UTR: translational control and development. Nat Rev Genet 4: 626–637 [DOI] [PubMed] [Google Scholar]
- Kulaeva OI, Gaykalova DA, Pestov NA, Golovastov VV, Vassylyev DG, Artsimovitch I, Studitsky VM 2009. Mechanism of chromatin remodeling and recovery during passage of RNA polymerase II. Nat Struct Mol Biol 16: 1272–1278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavorgna G, Dahary D, Lehner B, Sorek R, Sanderson CM, Casari G 2004. In search of antisense. Trends Biochem Sci 29: 88–94 [DOI] [PubMed] [Google Scholar]
- Levin JZ, Yassour M, Adiconis X, Nusbaum C, Thompson DA, Friedman N, Gnirke A, Regev A 2010. Comprehensive comparative analysis of strand-specific RNA sequencing methods. Nat Methods 7: 709–715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Licatalosi DD, Darnell RB 2010. RNA processing and its regulation: global insights into biological networks. Nat Rev Genet 11: 75–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Listerman I, Sapra AK, Neugebauer KM 2006. Cotranscriptional coupling of splicing factor recruitment and precursor messenger RNA splicing in mammalian cells. Nat Struct Mol Biol 13: 815–822 [DOI] [PubMed] [Google Scholar]
- Liu G, Loraine A, Shigeta R, Cline M, Cheng J, Valmeekam V, Sun S, Kulp D, Siani-Rose M 2003. NetAffx: Affymetrix probesets and annotations. Nucleic Acids Res 31: 82–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louro R, Nakaya H, Amaral P, Festa F, Sogayar M, da Silva A, Verjovski-Almeida S, Reis E 2007. Androgen responsive intronic non-coding RNAs. BMC Biol 5: 4 doi: 10.1186/1741-7007-5-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattick J 2004. RNA regulation: a new genetics? Nat Rev Genet 5: 316–323 [DOI] [PubMed] [Google Scholar]
- Mihalich A, Reina M, Mangioni S, Ponti E, Alberti L, Vigano P, Vignali M, Di Blasio AM 2003. Different basic fibroblast growth factor and fibroblast growth factor-antisense expression in eutopic endometrial stromal cells derived from women with and without endometriosis. J Clin Endocrinol Metab 88: 2853–2859 [DOI] [PubMed] [Google Scholar]
- Morrissy AS, Morin RD, Delaney A, Zeng T, McDonald H, Jones S, Zhao Y, Hirst M, Marra MA 2009. Next-generation tag sequencing for cancer gene expression profiling. Genome Res 19: 1825–1835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nahkuri S, Taft RJ, Mattick JS 2009. Nucleosomes are preferentially positioned at exons in somatic and sperm cells. Cell Cycle 8: 3420–3424 [DOI] [PubMed] [Google Scholar]
- Parkhomchuk D, Borodina T, Amstislavskiy V, Banaru M, Hallen L, Krobitsch S, Lehrach H, Soldatov A 2009. Transcriptome analysis by strand-specific sequencing of complementary DNA. Nucleic Acids Res 37: e123 doi: 10.1093/nar/gkp596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team 2008. R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna: http://www.R-project.org [Google Scholar]
- Reis E, Nakaya H, Louro R, Canavez F, Flatschart A, Almeida G, Egidio C, Paquola A, Machado A, Festa F, et al. 2004. Antisense intronic non-coding RNA levels correlate to the degree of tumor differentiation in prostate cancer. Oncogene 23: 6684–6692 [DOI] [PubMed] [Google Scholar]
- RIKEN Genome Exploration Research Group and Genome Science Group (Genome Network Project Core Group) and the FANTOM Consortium, Katayama S, Tomaru Y, Kasukawa T, Waki K, Nakanishi M, Nakamura M, Nishida H, Yap CC, Suzuki M, Kawai J, et al. 2005. Antisense transcription in the mammalian transcriptome. Science 309: 1564–1566 [DOI] [PubMed] [Google Scholar]
- Romanish MT, Nakamura H, Lai CB, Wang Y, Mager DL 2009. A novel protein isoform of the multicopy human NAIP gene derives from intragenic Alu SINE promoters. PLoS ONE 4: e5761 doi: 10.1371/journal.pone.0005761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenbloom KR, Dreszer TR, Pheasant M, Barber GP, Meyer LR, Pohl A, Raney BJ, Wang T, Hinrichs AS, Zweig AS, et al. 2010. ENCODE whole-genome data in the UCSC Genome Browser. Nucleic Acids Res 38: D620–D625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- SantaLucia J Jr 1998. A unified view of polymer, dumbbell, and oligonucleotide DNA nearest-neighbor thermodynamics. Proc Natl Acad Sci 95: 1460–1465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schones DE, Cui K, Cuddapah S, Roh TY, Barski A, Wang Z, Wei G, Zhao K 2008. Dynamic regulation of nucleosome positioning in the human genome. Cell 132: 887–898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz S, Meshorer E, Ast G 2009. Chromatin organization marks exon–intron structure. Nat Struct Mol Biol 16: 990–995 [DOI] [PubMed] [Google Scholar]
- Shearwin KE, Callen BP, Egan JB 2005. Transcriptional interference—a crash course. Trends Genet 21: 339–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spielman RS, Bastone LA, Burdick JT, Morley M, Ewens WJ, Cheung VG 2007. Common genetic variants account for differences in gene expression among ethnic groups. Nat Genet 39: 226–231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spies N, Nielsen CB, Padgett RA, Burge CB 2009. Biased chromatin signatures around polyadenylation sites and exons. Mol Cell 36: 245–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storey JD, Madeoy J, Strout JL, Wurfel M, Ronald J, Akey JM 2007. Gene-expression variation within and among human populations. Am J Hum Genet 80: 502–509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilgner H, Nikolaou C, Althammer S, Sammeth M, Beato M, Valcarcel J, Guigo R 2009. Nucleosome positioning as a determinant of exon recognition. Nat Struct Mol Biol 16: 996–1001 [DOI] [PubMed] [Google Scholar]
- Tufarelli C, Sloane Stanley JA, Garrick D, Sharpe JA, Ayyub H, Wood WG, Higgs DR 2003. Transcription of antisense RNA leading to gene silencing and methylation as a novel cause of human genetic disease. Nat Genet 34: 157–165 [DOI] [PubMed] [Google Scholar]
- Valouev A, Ichikawa J, Tonthat T, Stuart J, Ranade S, Peckham H, Zeng K, Malek JA, Costa G, McKernan K, et al. 2008. A high-resolution, nucleosome position map of C. elegans reveals a lack of universal sequence-dictated positioning. Genome Res 18: 1051–1063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanhee-Brossollet C, Vaquero C 1998. Do natural antisense transcripts make sense in eukaryotes? Gene 211: 1–9 [DOI] [PubMed] [Google Scholar]
- Wang Z, Burge CB 2008. Splicing regulation: From a parts list of regulatory elements to an integrated splicing code. RNA 14: 802–813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, Burge CB 2008. Alternative isoform regulation in human tissue transcriptomes. Nature 456: 470–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright SP 1992. Adjusted P-values for simultaneous inference. Biometrics 48: 1005–1013 [Google Scholar]
- Yan M-D, Hong C-C, Lai G-M, Cheng A-L, Lin Y-W, Chuang S-E 2005. Identification and characterization of a novel gene Saf transcribed from the opposite strand of Fas. Hum Mol Genet 14: 1465–1474 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Liu XS, Liu QR, Wei L 2006. Genome-wide in silico identification and analysis of cis natural antisense transcripts (cis-NATs) in ten species. Nucleic Acids Res 34: 3465–3475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Duan S, Kistner EO, Bleibel WK, Huang RS, Clark TA, Chen TX, Schweitzer AC, Blume JE, Cox NJ, et al. 2008. Evaluation of genetic variation contributing to differences in gene expression between populations. Am J Hum Genet 82: 631–640 [DOI] [PMC free article] [PubMed] [Google Scholar]