Abstract
DNA is a remarkable macromolecule that functions primarily as the carrier of the genetic information of organisms ranging from viruses to bacteria to eukaryotes. The ability of DNA polymerases to efficiently and accurately replicate genetic material represents one of the most fundamental yet complex biological processes found in nature. The central dogma of DNA polymerization is that the efficiency and fidelity of this biological process is dependent upon proper hydrogen-bonding interactions between an incoming nucleotide and its templating partner. However, the foundation of this dogma has been recently challenged by the demonstration that DNA polymerases can effectively and, in some cases, selectively incorporate non-natural nucleotides lacking classic hydrogen-bonding capabilities into DNA. In this review, we describe the results of several laboratories that have employed a variety of non-natural nucleotide analogs to decipher the molecular mechanism of DNA polymerization. The use of various non-natural nucleotides has lead to the development of several different models that can explain how efficient DNA synthesis can occur in the absence of hydrogen-bonding interactions. These models include the influence of steric fit and shape complementarity, hydrophobicity and solvation energies, base-stacking capabilities, and negative selection as alternatives to rules invoking simple recognition of hydrogen bonding patterns. Discussions are also provided regarding how the kinetics of primer extension and exonuclease proofreading activities associated with high-fidelity DNA polymerases are influenced by the absence of hydrogen-bonding functional groups exhibited by non-natural nucleotides.
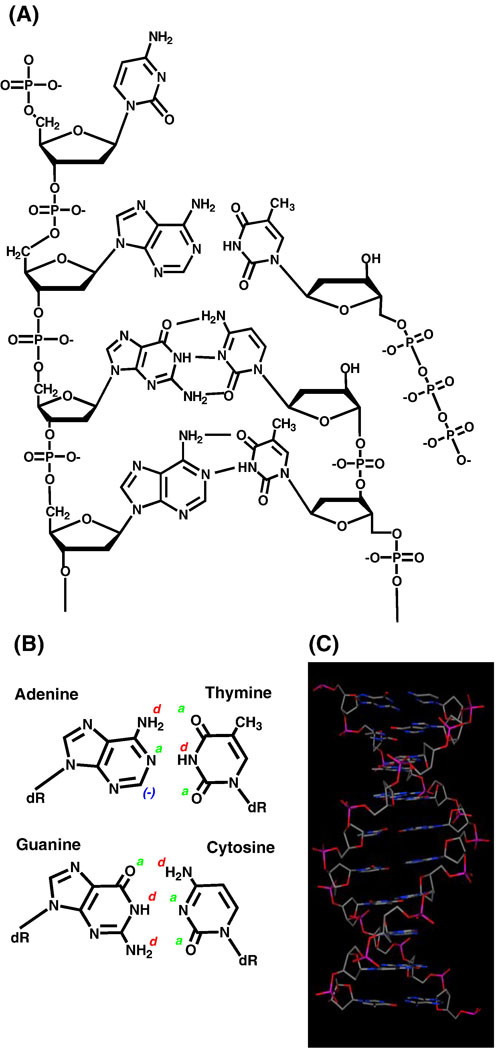
DNA polymerases are responsible for chromosome replication, and many play essential roles in DNA repair and recombination. These enzymes add mononucleotides onto the 3′-end of a primer strand using the complementary strand as a template (Figure 1A). Viewing DNA in this simple two-dimensional projection gives the impression that hydrogen-bonding interactions between the template base and the base of the incoming nucleotides are the most powerful physical forces that stabilize the conformation and structure of nucleic acid. By inference, these hydrogen-bonding interactions are thought to be the primary determinants in base pair recognition during the polymerization reaction. In this case, the mutual recognition of adenine (A)1 by thymine (T) and of guanine (G) by cytosine (C) involves hydrogen-bonding interactions between each partner (Figure 1B). At the atomic level, the non-sp2 hybridized amino groups are good hydrogen bond donors (denoted as d) while the oxo and the sp2 hybridized amino groups within the heterocyclic rings are hydrogen bond acceptors (denoted as a). For these preferred tautomers, the pattern for an A:T base pair uses complementarity d*a*(−) to a*d*a hydrogen bonding interactions while the G:C base pair uses complementarity a*d*d tod*a*a interactions. These base pairing patterns are commonly referred to as Watson-Crick base pairs and are the predominant pattern used to stabilize DNA.2
Figure 1.
(A) DNA as presented in linear, two-dimensional projections. (B) Hydrogen bonding interactions between natural nucleobases. (C) Three-dimensional representation of typical B-form DNA highlighting the influence of hydrogen-bonding interactions, steric constraints, π-π stacking interactions, and hydrophobicity on its structure.
Although hydrogen bonding interactions are a prominent feature that influences the conformation and tertiary stucture of DNA, other physical features such as π–π stacking interactions, desolvation/hydrophobic effects, and geometrical constraints contribute extensively to the stability of nucleic acid (reviewed in [1]). In solution, DNA exists as a double helix that resembles an intertwining spiral staircase where the nucleobases are stacked above and below one another (Figure 1C). In this native form, each base is rotated ~36° around the helical axis relative to the next base pair such that roughly 10 base pairs make a complete turn of 360°. While hydrogen-bonding, π–π stacking interactions, solvation and hydrophobic effects, and geometrical constraints play important roles in defining the structure of DNA (reviewed in [2]), their roles during DNA polymerization remain remarkably elusive. One example is with respect to the roles of hydrophobicity and desolvation energies. Hydrophobicity defines the tendency of a molecule to repel water whereas desolvation energy defines the quantity of energy required to remove water from a molecule. Although these terms are sometimes used interchangeably, each provides a unique biophysical consequence toward stabilizing nucleobase interactions during DNA polymerization. For example, it is evident from the structures of duplex DNA that the interior of the helix is hydrophobic since it is devoid of water. This hydrophobic environment is essential for the formation of the correct hydrogen-bonding network between each base pair. However, creating a hydrophobic environment during DNA polymerization is challenging since desolvation must occur on the templating and incoming nucleobase.
DNA polymerases are fascinating enzymes as they maintain remarkable specificity despite the fact that the heteropolymeric nature of the genomic message dictates that the substrate requirement changes during each cycle of nucleotide incorporation. As such, it is remarkable that most polymerases are strict in their ability to selectivity incorporate only one of four potential deoxynucleoside 5′-monophosphates (dNMPs) opposite a template base while being flexible enough to recognize four distinct pairing partners (A:T, G:C, T:A, and C:G). In fact, replicative DNA polymerases display incredible fidelity as they have error frequencies of only 1 mistake every 106 opportunities [3–5]. Even more impressive is the fact that these enzymes perform the repetitive cycle of nucleotide binding, base-pairing, phosphodiester bond formation, product release, and movement to the next templating position at rates greater than 100 bp/sec[6]. The underlying molecular events describing the remarkable speed and accuracy of DNA polymerases are generally defined by the rate and equilibrium constants for all the individual reactions involved in polymerization cycle. These include the binding of the substrates DNA and dNTP, conformational changes, phosphoryl transfer, and kinetic steps associated with product release. However, the details for how polymerases cope with the intricate biophysical features of both DNA and dNTP substrates are often ignored. In this review, we describe the work of several laboratories that have used various non-natural nucleotides to define the contribution of the aforementioned biophysical forces on the kinetics and selectivity of nucleotide incorporation. Each distinct section will describe the application of a unique set of non-natural nucleotides toward understanding DNA polymerization. This is provided as a historical perspective describing the rationale for designing the nucleotide analog of interest, a description of pertinent results, and a discussion of mechanistic implications. In addition, we describe the mechanistic information gained from applying these non-natural nucleotides on the behavior of DNA polymerases during primer elongation and exonuclease proofreading.
Lessons Learned from Replicating Alternative Hydrogen-Bonding Patterns
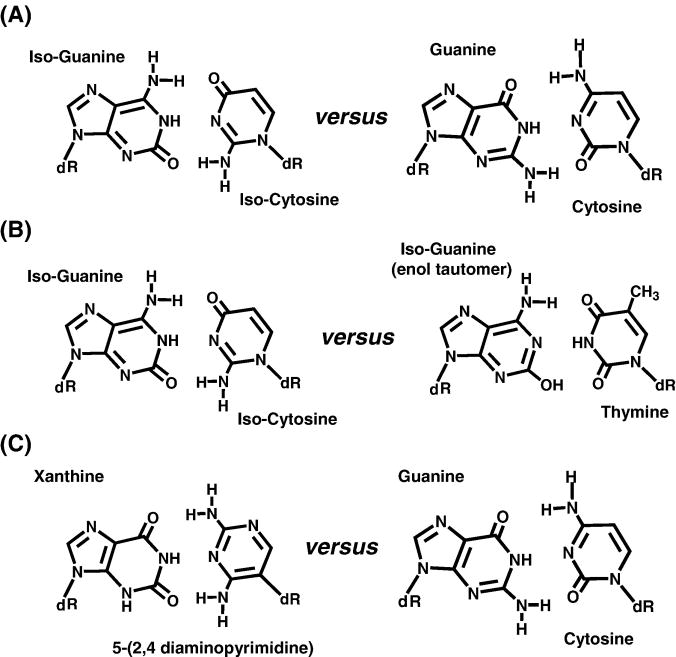
One of the first published efforts of rationally designing an alternative base-pair that could be efficiently replicated was reported by Steve Benner’s group [7]. The strategy was remarkably simple and straightforward: construct a base pair that is geometrically identical to an existing Watson-Crick base pair by manipulating the spatial arrangements of hydrogen-bonding acceptor-donor pairs. An intriguing base pairing combination consisting of iso-cytosine paired with iso-guanine was initially tested (Figure 2A). While this novel base pair has differing arrangements of hydrogen-bond acceptor-donor pairs compared to natural A:T and C:G base pairs, this combination is predicted to maintain the proper interglycosyl bond distance and angles relative to a Watson-Crick base pair. The E. coli Klenow fragment was used to test whether each non-natural base pair could be formed enzymatically. Although the iso-cytosine: iso-guanine base pair can be formed [8], the overall fidelity of this new base pair is low as iso-guanine can be easily incorporated opposite a templating thymine and vice versa [7]. The ease for forming an iso-guanine: thymine base pair is attributed to the ability of the non-natural nucleotide to undergo tautomerization from the desired keto form to the thermodynamically preferred enol form (Figure 2B) [7].
Figure 2.
(A) Structural comparison of non-natural base pairs of iso-guanine and iso-cytosine with the natural base pair guanine: cytosine. (B) Tautomerization of (C) Structural comparison of non-natural base pair of xanthine and 5-(2,4 diaminopyrimidine) with the natural base pair guanine: cytosine.
To improve fidelity, attempts were made using 5-(2,4-diaminopyrimidine) and xanthine to form a new base pair. As illustrated in Figure 2C, this new base pair combination resembles G:C but is different with respect to hydrogen bonding arrangements in the minor groove. It was shown that the E. coli Klenow fragment incorporated deoxyxanthosine monophosphate opposite 5-(2,4-diaminopyrimidine) with relatively high efficiency [9]. In addition, this base pair exhibits high fidelity as deoxyxanthosine monophosphate is poorly incorporated opposite the natural pyrimidines, T and C. Unfortunately, the Klenow fragment could not incorporate the triphosphate form of deoxyribose 5-(2,4-diaminopyrimidine) opposite a templating xanthine. Thus, these base pairs represent one of the first examples of asymmetric DNA polymerization in which the polymerase displays an unprecedented preference for forming one base pair (xanthine opposite 5-(2,4-diaminopyrimidine)) compared to its complement (5-(2,4-diaminopyrimidine) opposite xanthine.
Hydrogen Bonding coupled with Steric Guidance
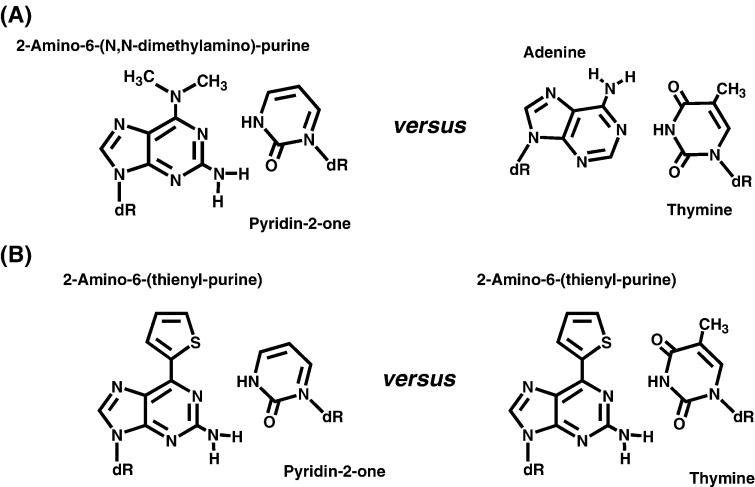
An alternative strategy developed by Yokoyama’s group was to use positive/negative reinforcement to generate novel base pair combinations [10]. Positive reinforcement is achieved through Watson-Crick hydrogen-bonding interactions between two nucleobases and are used to optimize enthalpic contributions for their association. Negative reinforcement is gained through the contributions of steric guidance in which the introduction of bulky side groups lacking hydrogen-bonding potential are included to sterically hinder mispair formation. An example of this design is the formation of the pyridin-2-one:2-amino-6-(N,N-dimethylamino)purine base pair (Figure 3A) [11]. As illustrated, enthalpic interactions are gained through a Watson-Crick type of hydrogen-bonding interactions of the heterocyclic nitrogen and the oxo group in pyridin-2-one with the complementary heterocyclic nitrogen and exocyclic amino group in 2-amino-6-(N,N-dimethylamino)purine. In addition, the 6-dimethyl-amino group acts as a steric block to prevent interactions with either thymine or cytosine. Steric hindrance is avoided since pyridin-2-one does not possess a functional group that can interact with the 6-dimethyl-amino group. Indeed, this design works as the Klenow fragment efficiently incorporates pyridin-2-one nucleotide opposite 2-amino-6-(N,N-dimethylamino)purine [11]. However, selectivity is again a major complication as dTMP is incorporated opposite 2-amino-6-(N,N-dimethylamino)purine with an efficiency comparable to that of pyridin-2-one [11].
Figure 3.
(A) Structures of the 2-amino-6-(N,N-dimethylamino)purine: pyridin-2-one base pair. (B) Base pairing interactions of 2-amino-(6-thienyl)purine with pyridin-2-one versus cytosine.
To improve selectivity, the 6-dimethylamino group was replaced with the bulkier 6-(2-thienyl) group that was predicted to prevent interactions with either natural pyrimidine (Figure 3B) [12]. As predicted, this modification enhances selectivity as the pyridin-2-one analog is incorporated opposite 2-amino-6-(2-thienyl)purine more efficiently than the misincorporation of any natural nucleotide [12]. This work clearly demonstrates that polymerases can function by using positive/negative reinforcement as a mechanism for nucleotide discrimination. However, the complex composition of the nucleobase makes it difficult to distinguish which molecular force, hydrogen bonding or steric exclusion, is more important for polymerization.
DNA Polymerization in the Absence of Hydrogen Bonds
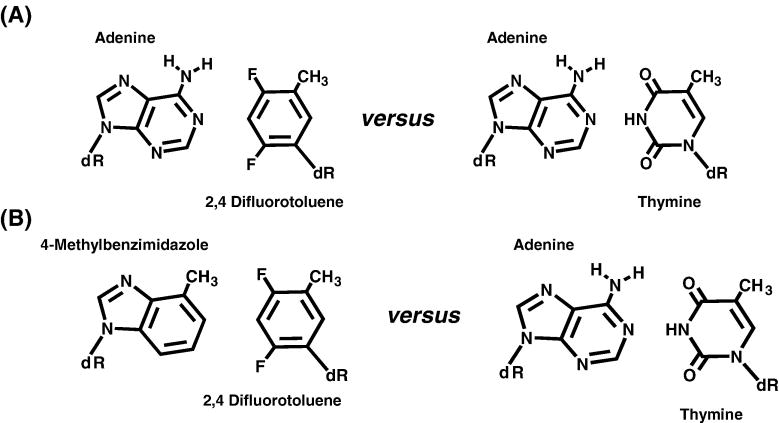
Work by Yokoyama’s group highlights that enzymatic formation of a base pair can be influenced through steric hindrance to dictate the fidelity of DNA synthesis. The next question is if solely the shape of the base pair influences DNA polymerization. This can only be addressed by measuring the incorporation of nucleotides that are completely devoid of classical hydrogen-bonding functional groups. Perhaps the most cited example of efficient replication in the complete absence of classical hydrogen-bonding interactions comes from the Kool laboratory demonstrating that 2,4-difluorotoluene monophosphate (dFMP) (Figure 4A), the non-hydrogen-bonding isostere of dTTP, is effectively incorporated opposite adenine [13]. Using the E. coli Klenow fragment, Kool and co-workers showed that the catalytic efficiency for incorporating dFMP opposite adenine is only 100-fold lower than that for incorporating dTMP opposite adenine. The Vmax for inserting dFMP opposite adenine is only ~3-fold slower than that using dTMP, the natural pairing partner [13]. However, the Km value for dFTP is 95 μM, which is ~40-fold higher than the Km for dTTP [13]. The significant perturbation in Km values for the non-natural nucleotide suggests that hydrogen-bonding functional groups play a significant role in binding affinity. However, an accurate comparison between the Kd values for the natural and non-natural nucleotide must be performed before this conclusion can be unambiguously assigned. Regardless, these remarkable pieces of information illustrate that the shape of the formed base pair plays an important role during DNA polymerization.
Figure 4.
Structural comparison of non-natural base pair of adenine:2,4-difluorotoluene with the natural base pair adenine: thymine.
Although the catalytic efficiency for using dFTP in polymerization reactions with a template A is low, incorporation of dFMP opposite template G, C, and T is not observed at all [13]. Thus, dFTP is used by the E. coli Klenow fragment and several other DNA polymerases with high fidelity but with reduced catalytic efficiencies compared with natural nucleotides. Collectively, these results indicate that hydrogen-bonding interactions between the incoming nucleotide and the templating nucleobase are not needed for polymerization but are required for optimal efficiency. However, the more provocative implication is that DNA polymerases do not use hydrogen-bonding interactions to achieve fidelity during nucleotide selection. Instead, complementary base pairs are proposed to adopt a shape that is viewed as correct by a polymerase which then allows for rapid polymerization. On the other hand, incorrect base pairs are proposed to adopt non-optimal shapes that prevent rapid polymerization. This mechanism has been coined the “shape complementarity” model to account the efficiency and fidelity of most DNA polymerases [14–16].
The shape complementarity model has been thoroughly examined by the Kool laboratory by measuring the kinetics for enzymatically forming new base pairs that are completely devoid of hydrogen bonding interactions [17–20]. One example is the design of 4-methylbenzimidazole as a complementary pairing partner for 2,4-difluorotoluene (Figure 4B). The pairing combination of 4-methylbenzimidazole and 2,4-difluorotoluene is geometrically similar, if not identical, to an adenine: thymine base pair [17]. In fact, the methyl group of 4-methylbenzimidazole moiety is approximately the same size as the amino group of adenine while the fluoro groups of 2,4-difluorotoluene are similar in size to the oxo group of thymine. However, both non-natural groups are incapable of forming classical hydrogen-bonding interactions with each other. In spite of this deficiency, kinetic studies reveal that the catalytic efficiency for incorporating 2,4-difluortoluene triphosphate opposite a templating 4-methylbenzimidazole is only 200-fold lower than for incorporating dTMP opposite adenine [17]. In addition, the efficiency for incorporating dTMP opposite a templating 4-methylbenzimidazole is low due to perturbations in both Km and Vmax values. This marks an important milestone in understanding the mechanism of DNA polymerases as it demonstrates the potential to achieve nucleotide selectivity without relying on hydrogen-bonding interactions for nucleotide incorporation. However, it is important to note that this achievement is limited since the non-hydrogen bonded primer terminus cannot be easily extended. Thus, there are significant difficulties in extending beyond nucleic acid containing non-natural nucleotides in the primer strand, the template strand, or both strands.
The importance of steric complementarity on nucleotide selection has been further analyzed by systematically evaluating the effects of base-pair size on the kinetics of nucleotide incorporation [21–23]. In these studies, the total size of a base pair was increased in small increments of 0.25 Å using a series of nonpolar thymidine shape mimics that contained different halides at positions corresponding to 2- and 4- of thymine (Figure 5). Steady-state rates of nucleotide incorporation reported with the high-fidelity bacteriophage T7 DNA polymerase revealed that nucleotide incorporation efficiencies decreased by 280-fold for non-natural base pairs that are 0.4 Å larger than the optimum size of a natural pair (10.6 Å). Base pairs that are 0.3 Å smaller than the optimum size also had lower catalytic efficiencies [21]. These data were collectively interpreted with respect to the active site tightness of the polymerase being used as the primary determinant for nucleotide selection during polymerization [21].
Figure 5.
Structures of nonpolar thymidine mimics that contain various halides at positions corresponding to 2- and 4- of thymine.
More recently, the influence of steric constraints and shape complementarity on nucleotide selectivity was evaluated by quantifying the effects of functional group size at specific positions of the non-natural nucleotide [23]. In these studies, the shape of the nucleobase was systematically altered by introducing different combinations of H, F, Cl, and Br atoms at variable positions of toluene as a nucleobase scaffold. Surprisingly, subtle changes in the size and location of the substituent groups produced large kinetic effects on the catalytic efficiency for their incorporation [23]. Although these analogs are all closely related in size, the efficiency in enzymatic incorporation varied amongst each other by 3,500-fold [23]. These results imply that base pair shape is just as important as overall size [23]. Unfortunately, a clear correlation between the shape of the nucleobase and its Km and/or Vmax value is not observed. Thus, while the E. coli Klenow fragment is highly sensitive to small changes in the shape and size of the incoming nucleotide, it is not possible to derive a defined structure-activity relationship explaining how each factor influences binding affinity and/or phosphoryl transfer.
Using Hydrophobic Non-Natural Nucleotides to Lubricate the Wheels of Replication
The helical nature of duplex DNA makes it intuitively obvious as to the roles that desolvation plays during replication. It is clear that water molecules surrounding the functional groups of the incoming dNTP must be removed prior to the formation of hydrogen bonds within the interior of the DNA helix. However, demonstrating how and when desolvation occurs along the DNA polymerization pathway is a daunting challenge.
Goodman and Petruska were amongst the first to critically evaluate the role of nucleobase hydrophobicity and desolvation as the primary determinant for maintaining fidelity and enhancing the efficiency of incorporation [24]. However, some very innovative studies evaluating desolvation during DNA polymerization have come from the collective studies of Romesberg and Schultz [25–28]. Their efforts represent rational attempts to generate new base pair combinations without the use of hydrogen-bonding interactions or shape-complementarity. Their studies probed the use of hydrophobic packing interactions as the primary molecular force used to enhance nucleotide incorporation and fidelity. This work was guided by other studies on the effects of hydrophobicity on protein structure and stability (reviewed in [29] and [30]) and lead to the hypothesis that interactions between two hydrophobic nucleobases should be strong and selective during DNA polymerization. The basis for selectivity arises from the argument that forming a base pair between a hydrophobic, non-natural base and a hydrophilic, natural base will be highly disfavored due to the energetic penalty associated with their formation.
Of several pairing combinations examined, the base pair formed between 7-azaindole and isocarbostyril (Figure 6) is the most noteworthy due to its unique thermodynamic and kinetic properties. Despite the lack of hydrogen bonding interactions, the 7-azaindole: isocarbostyril pair is only slightly less stable than an A:T base pair [25]. The higher than expected stability likely reflects entropic stabilization of the bases within the DNA helix. More importantly, the hydrophobic base pair can be enzymatically formed as isocarbostyril monophosphate is incorporated opposite a template 7-azaindole with an efficient kcat/Km value of 2.7*105 M−1min−1 [25]. This value is only 100-fold lower than that measured for the incorporation of dTMP opposite adenine. In addition, formation of the hydrophobic, non-natural base pair is kinetically symmetrical as the kcat/Km value for incorporating 7-azaindole monophosphate opposite a template templating isocarbostyril is identical to the value measured for incorporating isocarbostyril opposite 7-azaindole (compare 1.8*105 M−1min−1 versus 2.7*105 M−1min−1, respectively [25].
Figure 6.
Structures of 7-azaindole and isocarbostyril nucleosides
Selectivity is also maintained as incorporating a natural dNTP opposite either of these hydrophobic bases is highly unfavorable. For example, the catalytic efficiencies for the incorporation of a natural dNMP opposite 7-azaindole are ~100-fold lower than for inserting isocarbostyril nucleotide opposite 7-azaindole. The increase in replication fidelity is consistent with the rationale for using hydrophobicity as the driving force for polymerization. Unfortunately, this increase in fidelity does not extend to the enzymatic formation of hydrophobic base pairs. In fact, fidelity for replicating two hydrophobic bases is lost as it is exceedingly easy to enzymatically form self-pairs that consist of 7-azaindole:7-azaindole and isocarbostyril: isocarbostyril [25]. In these examples, the formation of either self pair is kinetically identical to forming the mixed pair of 7-azaindole: isocarbostyril [25]. Despite the lack of shape complementarity, formation of the 7-azaindole:7-azaindole self-pair occurs with reasonable efficiency as it is only 200-fold slower than forming natural base pairs [25]. As a result, three combinations of non-natural base pairs are possible as opposed to a single, unique base pair as originally intended. Thus, non-natural nucleotides that can be easily desolvated are thermodynamically more stable in the interior of DNA. This feature correlates with their facile incorporation opposite other hydrophobic nucleobases present in the templating strand. Indeed, an often overlooked result reported by Moran et al. [13] is that while 2,4-difluorotoluene is considered to be a selective partner for adenine, the catalytic efficiency for forming a self-pair between two 2,4-difluorotoluenes is actually higher than that measured for incorporation opposite adenine. In fact, the Km value of 53 μM for 2,4-difluorotoluene opposite itself is 2-fold lower than the value of 95 μM measured for incorporation opposite adenine [13]. Collectively, these data suggest that reductions in desolvation energies lower entropic penalties and favor nucleotide incorporation. However, reductions in desolvation energies also produce negative effects on selectivity as a hydrophobic base can be efficiently incorporated opposite any other hydrophobic base regardless of its shape and/or size [13].
Negative Selection as a Mechanism for Maintaining Polymerase Fidelity
Most models of replication fidelity invoke some form of “positive” selection in which the polymerase accepts the correct incoming nucleotide due to energetically favorable interactions with the templating base. Historically, these positive interactions have been attributed to the formation of hydrogen-bonds between the incoming dNTP and the templating base. It is clear from the previous sections that polymerization can occur even when the incoming nucleobase is devoid of hydrogen-bonding functional groups. However, these studies also reveal the negative consequences of replication in the absence of hydrogen-bonding potential that include a precarious decrease in the fidelity of DNA synthesis.
The Kuchta laboratory has developed a unique model invoking both positive and negative selection as a way to achieve fidelity [31–36]. This model postulates that during dNTP binding, the polymerase allows the incoming dNTP and the template base to adopt the lowest free energy conformation during their interaction. If binding is optimal, then phosphoryl transfer can occur. Phosphoryl transfer is prevented as a consequence of incorrect pairing which hinders nucleotide binding. Through this mechanism, the polymerase can sample a wide variety of nucleotides. However, only the dNTP that adopts the lowest free energy conformation will be accepted due to the formation of favorable interactions. This model differs from positive selection models which suggest that only a limited number of possibly correct partners can be sampled by a DNA polymerase.
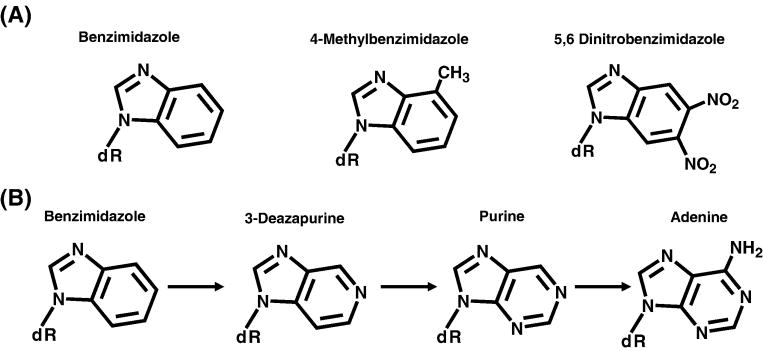
Substantial evidence for this model of polymerase fidelity comes from comprehensive studies examining the incorporation of purine analogs by a variety of DNA polymerases [31–36]. For brevity, we review only the data presented for the eukaryotic polymerase, pol alpha (pol α) as these provide compelling evidence for a model invoking both positive and negative selection. Despite being a high-fidelity enzyme, pol α incorporates non-natural dNMP analogs such as benzimidazole triphosphate, 4-methoxy benzimidazole triphosphate, and 5,6 dinitro-benzimidazole triphosphate (Figure 7A) opposite any four natural templating bases with remarkable efficiencies [35]. Rates for their incorporation are significantly faster than those measured during incorrect incorporation and surprisingly approach those measured for the enzymatic incorporation of a natural dNMP opposite it correct partner. To further understand how pol α discriminates between a correct versus an incorrect dNTP, the Kuchta lab used an innovative approach to “evolve” a low fidelity non-natural nucleotide such as benzamidazole triphosphate into a high fidelity nucleotide by judiciously adding hydrogen-bonding functional groups at various positions of the benzamidazole moiety (Figure 7B). In the converse approach, high-fidelity natural nucleotides were “de-evolved” into low fidelity substrates by removing functional groups capable of hydrogen-bonding interactions. For example, pol α displays low fidelity during the incorporation of benzimidazole opposite any of the four natural nucleobases. However, the addition of a heterocylic nitrogen corresponding to the N-3 of a purine (1-deazapurine-dNTP) increases the efficiency of incorporation opposite pyrimidines and provides discrimination against misincorporation. Likewise, removal of N-3 from adenine or guanine leads to a several-fold reduction in fidelity for the incorporation of the corresponding 3-deazaadenine and 3-deazaguanine triphosphates opposite cytidine and thymine, respectively [35]. These data provide compelling evidence that key functional groups behave as negative selectors involved in nucleotide binding. It is important to emphasize that the shape of the incoming nucleotide is not used as a positive or negative effector by pol α [35]. This is evident as pol α misincorporates dNMPs such as benzimidazole nucleotide and 4-methylbenzimidazole nucleotide which resemble dATP as effectively as non-natural nucleotides such as 5,6-dinitrobenzimidazole triphosphate that differ from dATP in both shape and size [35].
Figure 7.
(A) Structures for benzimidazole, 4-methylbenzimidazole, 5,6-dinitrobenzimidazole. (B) Example of the “evolution” of the low fidelity non-natural nucleotide, benzimidazole, into a high-fidelity natural nucleotide. Intermediates such as 3-deazapurine and purine display incremental enhancements in fidelity for selective pairing partners.
What it Takes to Make a Mistake: Using Non-Natural Nucleotides to Understand Translesion DNA Synthesis
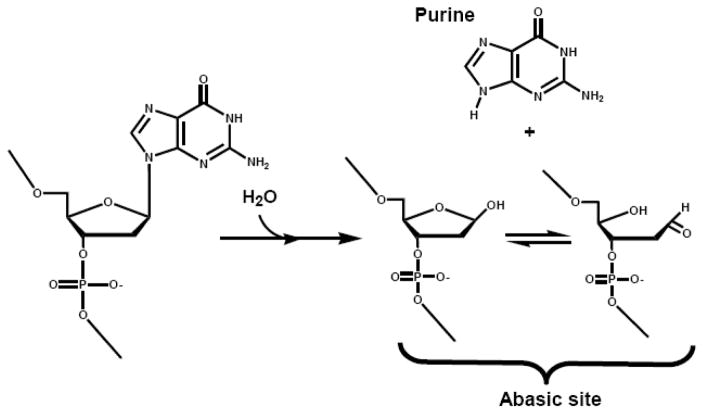
The previous sections have described the utility of non-natural nucleotides as mechanistic probes for correct DNA synthesis. However, DNA polymerases can also replicate damaged DNA templates in which the coding information present on the DNA template is altered. The ability of DNA polymerases to inappropriately replicate damaged DNA is a leading cause of mutagenesis (reviewed in [37]) and plays important roles in the development of diseases such as cancer (reviewed in [38]). A wide variety of DNA lesions exist and have been evaluated for mutagenic potential both in vitro (reviewed in [39]) and in vivo (reviewed in [40]). Perhaps the most interesting DNA lesion is the abasic site as this is the most prevalent and arguably the most pro-mutagenic lesion found in nature. The abasic site is a prototypical non-coding DNA lesion that is formed by the hydrolysis of the bond between the C1′ of the sugar moiety and the N9 of a purine or the N1 of a pyrimidine (Figure 8). The biological significance and consequence for misincorporating nucleotides opposite this DNA lesion have been extensively reviewed elsewhere [41]. Briefly, the lack of coding information present at an abasic site predicts that all four dNMPs should be incorporated with equal efficiencies. Surprisingly, the vast majority of high fidelity polymerases preferentially incorporate dAMP opposite this lesion [42–44]. This phenomenon has been coined the “A-rule” [45] and provides an interesting conundrum to investigate which biophysical property of adenine accounts for the unprecedented selectivity for insertion opposite this non-instructional lesion. Our discussions here focus on the mechanistic work performed using the abasic site as a model to evaluate how DNA synthesis occurs in the absence of hydrogen-bonding information and steric influence.
Figure 8.
Mechanism for the non-enzymatic formation of an abasic site, a non-instructional DNA lesion.
Several laboratories have used non-natural nucleotides to study the mechanism of translesion DNA synthesis. One example reported by the Kool laboratory was the investigation of shape complementarity during replication opposite an abasic site [46]. They demonstrated that the E. coli Klenow fragment incorporates pyrene nucleotide (dPMP) (Figure 9) opposite an abasic site ~100-fold more efficiently than any natural dNMP, including dAMP [46]. The Km of 3 μM for dPTP opposite the lesion is 27-fold lower than the Km of 85 μM measured for dATP [46]. Likewise, the kcat of 9 sec−1 for pyrene triphosphate is 10-fold faster than that measured for the incorporation of dAMP [46]. Theoretical modeling studies showed that the overall shape and size of the pyrene: abasic site mispair is nearly identical to that of natural adenine: thymine pair [46]. Thus, the kinetic data for the facile insertion of pyrene monophosphate were interpreted with respect to the shape complementary model such that the “void” present at an abasic site can be easily filled by the bulky aromatic nucleobase. However, other physical factors present on pyrene including π-electron density and hydrophobicity could also contribute to its incorporation opposite this non-instructional DNA lesion.
Figure 9.
Structure of pyrene triphosphate that is incorporated opposite an abasic site.
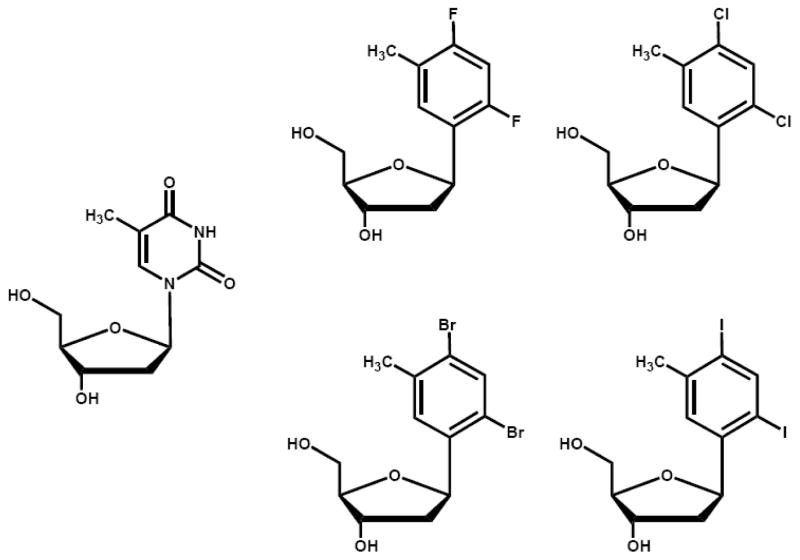
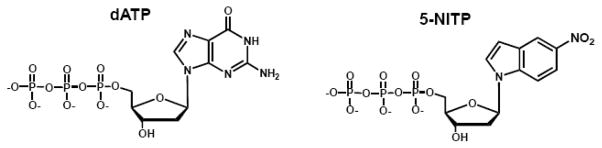
To further evaluate the contributions of shape complementary, base-stacking, and solvation energies toward nucleotide incorporation, the Berdis and Lee laboratories have measured the insertion of a series of modified nucleotides opposite an abasic site. 5-nitro-indolyl-2′-deoxyriboside triphosphate (5-NI nucleotide) (Figure 10) is an example of a unique non-natural nucleotide that is incorporated opposite an abasic site ~1,000-fold more efficiently than dAMP, the preferred natural nucleotide [47]. This enhancement for the high fidelity bacteriophage T4 DNA polymerase is not due to an increase in binding affinity as the Km of 18 μM for 5-NI nucleotide is only 2-fold lower that the Km of 36 μM for dATP [44, 47]. Instead, 5-NI nucleotide is incorporated opposite an abasic site with a fast polymerization rate constant of 126 sec−1 that is ~800 times faster than the value of 0.15 sec−1 measured with dATP. It is remarkable that the kinetic parameters measured for 5-NI nucleotide incorporation opposite an abasic site are nearly identical to those for the enzymatic formation of a natural Watson-Crick base pair, i.e., dAMP opposite thymine [44].
Figure 10.
Structural comparisons of the non-natural nucleotide, 5-NITP, with natural purine nucleotide, dATP.
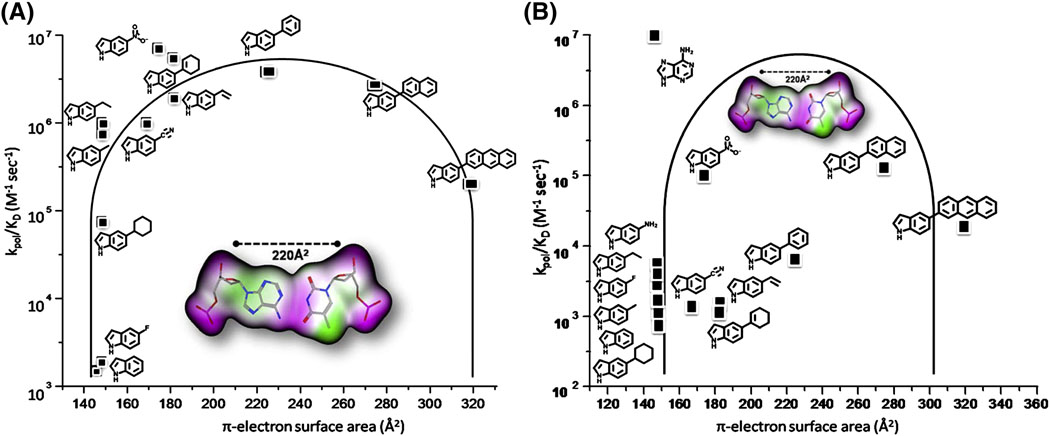
These data clearly emphasize previous observations that DNA polymerization can occur in the complete absence of hydrogen bonding interactions between nucleobases [13, 17, 21]. However, they also argue against shape complementarity since 5-NI nucleotide is substantially smaller than pyrene triphosphate and cannot completely occupy the void present at the DNA lesion. Unfortunately, it is difficult to unambiguously determine which biophysical feature of 5-NI nucleotide is responsible for its enhanced kinetic behavior during translesion DNA synthesis. Indeed, the nitro group is unique as it possesses several distinct physical parameters including electron withdrawing potential, hydrophobic nature, π-electron density, and zwitterionic character. The importance of these physical features was investigated using a medicinal chemistry approach in which the nitro moiety was systematically replaced with functional groups that differ by electron withdrawing potential, hydrophobic nature, π-electron density, and zwitterionic character. The structures for the resulting library of nucleotide analogs are provided in Figure 11. The kinetic parameters for the incorporation of these analogs opposite an abasic site were then used to generate a structure-activity relationship for incorporation opposite the non-instructional abasic site [48–51]. The data from the studies is summarized in Figure 12A, which provides a plot of the catalytic efficiency of each 5-substituted indolyl deoxynucleotide as a function of its π-electron surface area. The parabolic shape of the plot reveals that the efficiency of nucleotide incorporation opposite an abasic site depends upon the optimal π-electron surface area that most closely mimics that for a natural Watson-Crick base pair.
Figure 11.
Structures of various 5-substituted indolyl-2′-deoxyribose-5′-triphosphates used to probe the influence of π-electron stacking interactions on translesion DNA synthesis.
Figure 12.
(A) Structure-activity relationship for the incorporation of various 5-substituted indolyl-deoxynucleotides opposite an abasic site. (B) Structure-activity relationship for the incorporation of various 5-substituted indolyl-deoxynucleotides opposite a templating thymine.
This model differs from the steric fit/shape complementary model in two important aspects. First, the steric fit model predicts that analogs that are similar in shape and size should be incorporated with nearly identical efficiencies opposite the lesion. This prediction is not observed when comparing the kinetics of incorporation for three nucleotide analogs, 5-cyclohexyl, 5-cyclohexene, and 5-phenyl-indole-2′deoxyriboside triphosphates, which are similar in shape and size but that differ in π-electron surface area. Specifically, the 5-phenyl derivative is incorporated opposite an abasic site with an extremely high catalytic efficiency of 3.8*106 M−1sec−1 [49]. In fact, the kpol of 53 sec−1 and Kd of 14 μM are very similar to those measured for the 5-nitro analog [47]. Visual inspection reveals that these analogs clearly differ in shape and size but are similar with respect to the presence of π-electron surface density. Data obtained for the 5-cyclohexyl and 5-cyclohexene-containing nucleotides support the conclusion that π-electron density plays an important role, as the catalytic efficiency for the 5-cyclohexene-indole analog is 75-fold greater than that for the 5-cyclohexyl-indole derivative. The higher efficiency is reflected in a substantial increase in the kpol value (compare 25 versus 0.7 sec−1, respectively) rather than an influence on nucleotide binding and indicates that π-electron density enhances the rate of incorporation opposite the abasic site [48].
An additional argument against the T4 DNA polymerase relying exclusively on steric fit comes from a thorough examination of the kinetics for incorporating non-natural nucleotides opposite a template thymine. Figure 12B reports the catalytic efficiency for their incorporation as a function of the overall size of the formed base pair.3 It is clear that a parabolic trend in catalytic efficiency does not exist when only the size of the 5-substituted indole nucleobase. Specifically, analogs such as the 5-amino-, 5-fluoro-, and 5-methyl-indole derivative are poorly incorporated despite being predicted pairing partners of thymine. In fact, analogs such as 5-napthyl- and 5-anthracene-indolyl deoxynucleotides which lack any shape complementary for thymine are incorporated with catalytic efficiencies that are higher than expected based solely upon steric and/or shape constraints.
Who Needs Fidelity? Examples and Applications for the Promiscuous Incorporation of Universal Nucleotides
The lack of fidelity exhibited by various non-natural nucleotides led to the concept of developing a “universal” nucleotide, i.e., one that is incorporated opposite multiple templating nucleobases with identical efficiencies [52]. The ability of a nucleobase to pair with multiple partners would allow for the development of various technologies to overcome ambiguities arising from the degeneracy of the genetic code. Such nucleotides would find applications as PCR primers, DNA sequencing reagents, interference RNA, and oligonucleotide probes for identifying mutations and polymorphisms in genomic material.
The earliest “universal bases” are the azole carboxamide derivatives (Figure 13A) developed by Bergstrom and Davisson [53–55]. These analogs are ambiguous since rotation about the amide bond provides two alternative and mutually exclusive hydrogen-bonding patterns. Secondly, while each non-natural base is similar in shape and size, they differ with respect to chemical composition which provides each with a unique electronic signature [55]. The ability of these analogs to allow degenerate replication was first examined by measuring the ability of Taq DNA polymerase to perform PCR on a DNA template containing each azole derivative [53]. Since each pyrimidine-like analog presents two alternative hydrogen-bonding patterns, it was predicted that dAMP and dGMP would be incorporated with nearly equal efficiencies. Surprisingly, Taq DNA polymerase showed an unexpectedly strong bias toward incorporating dAMP opposite several of the non-natural nucleotides [53]. In fact, it is remarkable that some analogs are actually replicated as purines. One interesting example is 1,2-pyrazole-3-carboxamide which is replicated as purine since dCTP and dTTP are utilized with equal frequencies [53].
Figure 13.
(A) Chemical structures of various pyrole and pyrazole nucleotide analogs. (B) Structures of various triazole carboxamide nucleotide analogs.
Subsequent attempts to optimize the ambiguous nature of these analogs have focused on manipulating their electronic features through permutations in the number and location of heterocyclic nitrogens. In this design, the pyrazole-3-carboxamide was changed to a class of triazoles that include (1H)-1,2,3-triazole-4-carboxamide, (2H)-1,2,3-triazole-4-carboxamide, and 1,2,4-triazole-4-carboxamide (Figure 13B) [56]. All three analogs are rather promiscuous as each allows for the incorporation of multiple dNMPs. Of these three analogs, (1H)-1,2,3-triazole-4-carboxamide displays the highest degree of selectivity as the catalytic efficiency for incorporating dGMP is 10-fold higher than dAMP and greater than 50-fold more efficient than the incorporation of the pyrimidines dCMP or dTMP [56]. In contrast, 1,2,4-triazole-3-carboxamide and (2H)-1,2,3-triazole-4-carboxamide behave as universal nucleotides since nearly identical catalytic efficiencies are reported for the incorporation of various dNMPs.
Another interesting class of promiscuous non-natural nucleotides is 5-NI nucleotide. As mentioned earlier, this non-natural nucleotide is incorporated opposite an abasic site with an unexpectedly high catalytic efficiency [47]. However, 5-NI nucleotide is unique as it can act as a universal pairing partner. For example, the introduction of a single 5-nitroindole residue into a DNA primer allows for ambiguous pairing and subsequent elongation of the modified primer [57]. However, the efficiency significantly worsens as the number of 5-nitroindole residues in the primer increases [57] and reflects the ability of the oligonucleotide to form secondary structures caused by the “self-pairing” capabilities of 5-nitroindole.
In addition, 5-NI nucleotide is incorporated opposite any templating nucleobase by various DNA polymerases [47, 58, 59]. Initial qualitative studies using the E. coli Klenow fragment showed that the efficiency for incorporation was remarkably similar for all four templating nucleobases [47]. More quantitative studies with the high fidelity bacteriophage T4 DNA polymerase confirm that 5-NI nucleotide is a very promiscuous nucleotide as it can be incorporated opposite any of the four natural bases with unexpectedly high catalytic efficiencies of ~104 M−1sec−1 [47]. It is surprising that 5-NI nucleotide is incorporated opposite adenine and thymine with identical efficiencies since the 5-nitroindole: adenine pair is predicted to be less thermodynamically favored compared to the 5-nitroindole: thymine base pair [59]. The inability to correlate catalytic efficiency with base pair size again argues against a model invoking steric fit as the sole determinant in defining polymerization efficiency.
Probing the Mechanisms of Primer Extension and Proofreading using Non-Natural Nucleotides
DNA replication is a multi-faceted process encompassing several discrete activities that include nucleotide binding and phosphoryl transfer followed by elongation after incorporation of a correct nucleotide or exonuclease editing of a mispair formed after insertion opposite a non-complementary nucleotide. In the sections that follow, we will explore how primer extension and exonuclease proofreading activities of high-fidelity DNA polymerases are influenced by non-natural nucleotides that lack hydrogen-bonding functional groups.
Lessons Learned from Monitoring the Kinetics of Elongating Non-Natural Base Pairs
Most reports examining the extension of non-natural base pairs have been driven by application-based technologies such as developing novel PCR and cloning techniques [60] as well as designing novel biopolymers as diagnostic and/or therapeutic agents [61]. However, the most widely cited application for the creation of a new functional base-pair is for expansion of the genetic code [62–64]. This involves creating nucleic acid containing uniquely coding nucleobases distinct from naturally occurring bases. Developing a new genetic code is the first step in creating an “artificial life-form” that could be used as a biotechnological platform for protein engineering [65] in addition to producing novel bioactive molecules such as antibiotics and anti-cancer agents [66]. Despite these lofty goals, detailed studies on the kinetics of extending beyond non-natural base pairs have lagged behind studies examining the kinetics of their formation. Much of this deficiency lies in the fact that non-natural nucleotides are poorly elongated. However, the Kool and Romesberg laboratories have independently taken the lead in developing a set of rules that loosely explain (or predict) the ability of a non-natural base pair to be efficiently elongated.
Is Elongation Dependent upon Proper Hydrogen-Bonding Interactions?
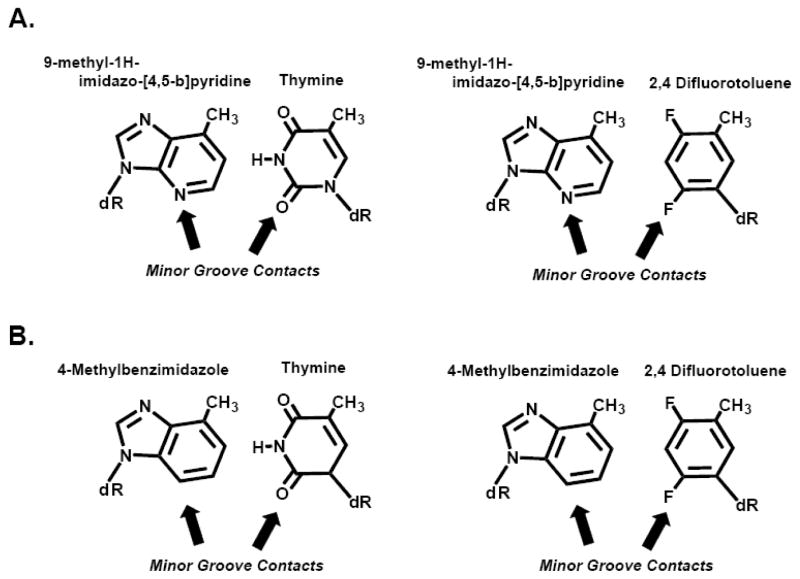
Kool’s group was amongst the first to quantitatively evaluate the ability of DNA polymerases to extend beyond mixed combinations of natural: non-natural and non-natural: non-natural base pairs. An obvious prediction from their earlier work was that the A:dF base pair should be efficiently extended since it accurately mimics the shape and size of a natural A:T pair.(Note: The first base listed in any basepair combination indicates its location in the primer strand while the second base is in the templating strand.) It was somewhat suprising that this prediction is inaccurate since the exonuclease-deficient form of the E. coli Klenow fragment extends beyond the A:dF pair ~100-fold less efficiently than a natural A:T base pair [18]. This reduction is caused by a ~40-fold higher Km for the next correct nucleotide rather than through a negative effect on the Vmax for incorporation. Additional experiments demonstrate that the extension kinetics for this non-natural base pair is asymmetrical as the efficiency of ~6*103 % min−1μM−1 for extending beyond dF:A is 100-fold lower than the value of ~2 *105 % min−1μM−1 measured using A:dF [18]). The predominant effect is on the Vmax for nucleotide incorporation as extending the dF:A base pair is ~50-fold slower than the A:dF combination [18]. The difference in extension rates for identical yet asymmetrical base pairs argues that shape complementary plays a minor role in regulating the kinetics of elongation. As a result, it was proposed that extension beyond certain mispairs is governed by hydrogen-bonding groups present in the minor groove of nucleic acid that make appropriate contacts with the DNA polymerase.
This hypothesis was investigated by replacing A with the isosteric analog, dQ (9-methyl-1H-imidazo-[4,5-b]pyridine) (Figure 14A). This non-natural nucleotide lacks the N1 of adenine, but retains the N3 that could participate in minor groove interactions. As a result, the dQ:dF non-natural base pair is extended as efficiently as the mixed A:dF pairing combination since minor groove interactions are maintained as predicted [18]. However, it is quite surprising that the E. coli Klenow fragment extends the dQ:dF base pair ~10-fold more efficiently than the dQ:T base pair since the later contains a hydrogen-bonding functional group while the former does not [18].
Figure 14.
(A) Comparison of the base pair formed between 9-methyl-1H-imidazo-[4,5-b]pyridine (dQ) and thymine (T) versus the base pair formed between 9-methyl-1H-imidazo-[4,5-b]pyridine (dQ) and 2,4-difluorotoluene (dF). (B) Comparison of the base pair formed between 4-methylimidazole (dZ) and thymine (T) versus the base pair formed between 4-methylimidazole (dZ) and 2,4-difluorotoluene (dF). As described in the text, all four base pairs are similar with respect to shape and size. However, differences in the minor groove contacts with respect to hydrogen bonding functional groups influence the kinetics of elongation.
More dramatic effects were observed when dQ is replaced with the related dZ analog (4-methylbenzimidazole) that differs only by the removal of the N3 group (Figure 14B) [18]. Removal of this functional group precludes hydrogen-bonding contacts in the minor groove, and this absence significantly reduces extension efficiencies for both the dZ:T and dZ:dF base pairs. In both cases, the lower efficiencies are caused by 100-fold reductions in Vmax rather than in the Km for the next correct nucleotide [18]. Finally, it is important to note that the extension efficiencies for all of these non-natural base pairs (dF:dQ versus dQ:dF and dF:dZ versus dZ:dF) are asymmetrical. In all cases, the difference results from perturbations in Vmax values rather than an effect on the Km for the next correct nucleotide. Collectively, these results re-iterate that correlations between the kinetics of primer elongation and shape complementary between non-natural base pairs do not exist. Instead, the major regulator for extension appears to be the presence and position of functional groups that line the minor groove of DNA.
To investigate if this mechanism is universal amongst all DNA polymerase, five other polymerases including Pol α, Pol β, T7, Taq, and HIV-RT were tested for their ability to extend nucleic acid containing various non-hydrogen bonding nucleoside isoteres [18]. These studies identified two broad classes of enzyme activities that are influenced by the presence or absence of hydrogen-bonding groups present in the minor groove. The first group of DNA polymerases include the E. coli Klenow fragment, Taq, and HIV-RT as they efficiently extend beyond nonpolar base pairs containing dQ but not dZ. These data suggest that specific interactions between key amino acids of each polymerase with functional groups present in the minor groove are necessary for extension. The second group comprised of the eukaryotic polymerases, pol α and pol β, as well as the high fidelity bacteriophage T7 DNA polymerase, failed to extend beyond any base pair containing a non-natural nucleobase. These results indicate that these enzymes may need hydrogen bonding groups at both minor groove sites of DNA for elongation to occur.
Collectively, these results support a model in which minor groove interactions are used by DNA polymerases as a mechanism that proofreads the fidelity of the formed base pair. The absence of correct hydrogen-bonding contacts within the minor-groove is viewed by the polymerase as an incorrect base-pair which then prevent subsequent nucleotide incorporation event [18]. An alternative explanation is that the lack of minor-groove interactions prevents polymerase translocation to the next template position that precludes the incorporation of the next correct nucleotide. In either case, the mechanism invoking the role of proper minor groove contacts in primer elongation can be easily extrapolated to explain why natural mispairs such as G:T or A:C are poorly extended [67–69].
Re-Evaluating the Importance of Shape Complementary during Primer Extension
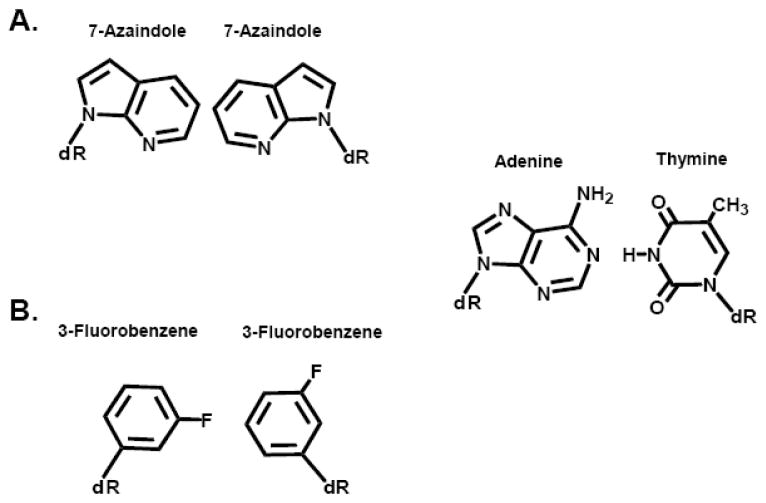
Romesberg and colleagues used several different classes of non-natural nucleotides to probe the underlying mechanism accounting for the inefficient extension of non-natural base pairs. Their collective studies reveal an inverse correlation between the efficiency of forming a non-natural base pair with is ability to be extended. In general, base pairs between large aromatic and hydrophobic nucleobases are enzymatically synthesized with high catalytic efficiencies due to hydrophobic packing interactions. However, efficient primer elongation is significantly limited even though these base pairs are thermodynamically stable. One exceptional example is the inability of the E. coli Klenow fragment to extend the self pair formed between 7-azaindole although the polymerase synthesizes the self-pair with a moderate efficiency of ~105 M−1min−1 [26] (Figure 15A). The inability to extend the 7-azaindole self pair is proposed to reflect an intercalated structure at the primer terminus that causes mispositioning of the 3′-OH in the polymerase’s active site which prevents phosphoryl transfer [70, 71].
Figure 15.
(A) Chemical structure of the self-pair formed between 7-azaindole. (B) Chemical structure of the self pair formed between 3-fluorobenzene. The natural base pair of adenine: thymine is provided as a reference.
To evaluate this mechanism, the Romesberg group measured the extension kinetics of a series of non-natural nucleosides composed of a simple phenyl ring containing fluoro, bromo, cyano, and/or methyl substituents [72]. The judicious placement of these substituent groups allowed the shape, hydrophobicity, and electronic properties of the nucleobase to be systematically varied. Of several nucleotides tested, one intriguing example is 3-fluorobenzene monophosphate (d3FB) (Figure 15B) as it is incorporated opposite itself and then extended by the E. coli Klenow fragment with a rate that is only 100-fold slower than a natural base pair [72]. These results are noteworthy since they demonstrate that large aromatic surface areas are not essential for enzymatically forming a non-natural base pair. Furthermore, the lack of an extended aromatic surface area makes the d3FB self-pair too small to form a distorted primer terminus via inappropriate intercalation of one nucleobase stacked over the other. Instead, the bases interact in an edge-to-edge manner that resembles the conformation of natural base pairs [73]. In this regard, it is intriguing that the specific meta-fluoro substitution pattern of d3FB is optimal for elongation as similarly positioned methyl, bromo, and cyano substituents show reduced extension efficiencies despite possessing an “in-plane” primer-terminus [73]. These data argue that the specific dipole-dipole alignments of the fluoro groups within the 3FB self-pair are just as important as the proper shape of the non-natural base pair for efficient synthesis and extension.
Attempts to rapidly identify other extendable base pairs were performed by screening 3,600 possible self-pairing and heteropairing combinations [74]. One noteworthy combination is the base pair formed between d5SICS and dMMO2 which is synthesized and extended by the E. coli Klenow fragment with remarkable efficiency in either strand context (Figure 16). This polymerase forms the d5SICS:dMMO2 heteropair with a second-order rate constant of 4.7*107 M−1min−1 while other A-family members such as the T7 and Taq DNA polymerases are slightly less efficient at ~106 M−1min−1. However, the efficiency for extending this mispair is low as these polymerases extend this base pair with second-order rate constants approaching 104 M−1min−1 and are 100-fold lower than for their formation. B-family polymerases such as Thermococcus litoralis and Thermococcus 9°N-7 are similar as they form the d5SICS:dMMO2 heteropair with high efficiencies of ~107 M−1min−1 but have difficulties in extending it (104 M−1min−1). Only the X-family DNA polymerase, pol β is more proficient at extending the mispair than forming it. In fact, pol β elongates the non-natural mispair with the same efficiency as elongating a correct Watson-Crick pair (~105 M−1min−1) and 100-fold more efficient than extending a G:T mispair (~103 M−1min−1).
Figure 16.

Structure of the base pairing combination of dMMO2 and d5SICS.
Is Extension of a Non-Natural Base Pair a Two-Polymerase Affair?
As indicated earlier, the Romesberg group demonstrated that the E. coli Klenow fragment forms the 7-azaindole self pair with moderate efficiency but is unable to extend it [26]. In contrast, pol β is more proficient at extending the 7-azaindole self pair but is unable to form it [75]. As a result, combining the activities of these two polymerases allows for the complete replication of the non-natural base pair under in vitro reaction conditions [75]. Thus, the use of two distinct DNA polymerases provides a way to circumvent obstacles associated with expanding the genetic code.
Similar results were reported by Delaney et al. testing the ability of various non hydrogen-bonding nucleotides to be efficiently replicated under in vivo conditions [20]. In these experiments, the non-natural nucleobases dF and dQ were inserted into a phage genome and subsequently transformed into E. coli. Genetic assays demonstrate that these two analogs were by-passed with moderate efficiencies under normal growth conditions. However, the by-pass efficiency increased significantly under damage response (SOS) conditions induced by treatment with UV light, implying that error-prone DNA polymerase are required for the complete replication of the non-natural base pair. Despite the likely involvement of translesion DNA polymerases, however, both isoteres were replicated with high fidelity as dF directed the incorporation of dAMP while dQ directed the incorporation of dTMP.
While these data re-iterate that Watson-Crick hydrogen bonds are not needed for high-fidelity replication of a base pair, they also argue that the complete and efficient replication of a non-natural base pair is a “two polymerase” affair. Under these conditions, one polymerase forms the mispair while another is responsible for extending beyond it. This mechanism is similar to models explaining how certain DNA lesions such as thymine dimers and other cross-linked adducts are efficiently bypassed under cellular conditions [76–78]. In these models, high fidelity DNA polymerases dissociate or stall at the thymine dimer. At this point, pol η is recruited to the site of DNA damage and incorporates one or more dAMPs opposite the lesion. Extension beyond the lesion is catalyzed by pol ζ which incorporates only one or two nucleotides beyond the thymine dimer. After this, DNA synthesis resumes by the high fidelity DNA polymerase. The mechanism for by-passing a non-natural nucleobase in the template would proceed in a similar fashion. However, upon encountering a modified nucleobase, the high fidelity DNA polymerase would incorporate a natural dNMP opposite it. Since high-fidelity polymerases are reluctant to extend beyond the non-natural basepair, polymerase switching would occur such that pol ζ or another error-prone DNA polymerase would bind to the mispair and extend it.
The Kinetics of Exonuclease Proofreading
All of the aforementioned studies monitoring the incorporation and extension of non-natural nucleotides have been performed using DNA polymerases lacking exonuclease-proofreading activity. Exonuclease proofreading is essential for maintaining genomic fidelity by removing inappropriately misinserted nucleotides before they are extended. For a complete discussion of the exonucleolytic pathway, please refer to the article by Linda Reha-Krantz presented in this Special Issue. For the purposes of our discussion, we condense this multistep pathway into four simple yet distinct steps: mispair recognition, formation of the exonuclease complex, hydrolysis of the phosphate ester bond followed by movement of the corrected primer into the polymerase’s active site. The dynamics of this pathway have been extensively studies with natural mispairs such as A:C and G:G mismatches [79–82]. However, the sections below examine how various non-natural nucleobases have been used to study the molecular details of the first two steps of the exonucleolytic pathway - recognition of the mispair and subsequent movement of the misaligned primer into the exonuclease site to form the exonuclease complex.
Early studies using wild-type E. coli Klenow fragment demonstrated that the isosteric pairs of A:dF and A:T are edited at very different rates [83]. Despite the fact that both base pairs are nearly identical in shape and size, dAMP is excised ~40-fold more efficiently when paired opposite dF than T. Similarly, a terminal dZMP opposite template T is excised at a ~30-fold faster rate compared to the excision of a terminal dAMP opposite T. At first glance, it is not surprising that these non-natural nucleotides are excised with higher catalytic efficiencies since the lack of hydrogen bonding interactions may distort the geometry of the non-natural pair. However, it is intriguing that the excision efficiencies for C:dF and T:dF base pairs are similar to that for an A:dF pair since the former have suboptimal geometrical alignments compared to the later which closely resembles a canonical A:T base pair. To account for this unexpected behavior, a model was proposed invoking the overall stability of the primer-terminus as being the chief determinant of the editing rate since the measured proofreading rates correlated well with the thermodynamic melting temperatures of the DNA substrates. This suggests that melting of the DNA and/or partitioning of the primer into the exonuclease site is more facile as a consequence of an unstable terminal base pair.
To further investigate this mechanism, pre-steady-state kinetic techniques were used to more accurately measure rate constants for kinetic steps encompassing mispair recognition and hydrolysis using the wild-type bacteriophage T4 polymerase [84]. These studies tested the generally accepted kinetic model for fidelity positing that the rate constant for dNMP excision should be inversely correlated with the rate constant for dNMP incorporation. Put another way, the more difficult it is to form a mispair, the easier it should be to excise it and vice versa. Indeed, this correlation exists with natural pairs as dAMP is rapidly incorporated opposite T (kpol ~100 sec−1) and slowly excised (kexo ~1 sec−1) when paired opposite this complementary partner. The excision of kinetically favored base pairs formed with non-natural nucleotides such as 5-NI nucleotide and 5-Ph nucleotide provide an excellent model to test the validity of this model. For example, 5-NI nucleotide is inserted opposite an abasic site ~100-times faster than dAMP (compare 130 sec−1 versus 0.15 sec−1, respectively). Furthermore, the rate constant of 10 sec−1 for the excision of 5-NI nucleotide opposite an abasic site is slower than the rate constant of ~30 sec−1 measured for the excision of dAMP. This difference was proposed to reflect the higher stability of 5-NI nucleotide: abasic site pair compared to the dAMP:abasic site mispair. Additional studies reveal that the larger non-natural nucleotide, 5-Ph nucleotide, is excised from opposite an abasic site with a 10-fold slower rate constant compared to 5-NI nucleotide. The slower rate of excision is consistent with a model invoking enhanced stability due to an increase base-stacking potential caused by large pi-stacking surface areas coupled with shape complementary. Collectively, these studies indicate that the relative stability of the terminal base pair is one of the predominant factors that influences whether it will be excised or extended.
Conclusions and Future Directions
In describing their proposed structure of DNA, [85] Watson and Crick wrote “It has not escaped our notice that the specific pairing we have postulated immediately suggests a possible copying mechanism for the genetic material.” Indeed, their structure not only provided a biophysical explanation on how genetic material is stably stored, it immediately evoked a mechanism explaining how genetic information is copied by DNA polymerases and transmitted by RNA polymerases. Perhaps the most obvious feature is the intuitive relationship between hydrogen bond acceptor-donor pairs and nucleic acid stability that immediately infers a simple yet elegant way for polymerases to faithfully copy nucleic acid. In fact, early models aimed at understanding how spontaneous mutations occur and how DNA damaging agents compromise the fidelity of polymerization have relied heavily on the concept of altered hydrogen-bonding patterns.
Sixty years later, our knowledge on the mechanism and fidelity of DNA polymerization has grown significantly. Much of this growth comes from the use of non-natural nucleotides as chemical entities to probe the mechanism of DNA polymerization. The results of these research endeavors challenge the existing concept that replication occurs in “2-dimensions” via simple hydrogen-bonding interactions. Instead, several new models have emerged that take into account all of the biophysical features that are subtlety yet elegantly provided in the three-dimensional structure of DNA originally described by Watson and Crick [85].
This review provides a historical perspective that explores some of the pioneering efforts using non-natural nucleotides to probe which biophysical feature is most important for efficient and accurate DNA polymerization. For example, Benner’s work illustrates how polymerases behave when hydrogen-bonding functional groups are permutated to form different pairing combinations. Yokoyama’s research describes how steric encumbrance can be used to influence polymerase fidelity. Work by Kool, Romesberg, and Schultz have challenged the long-standing paradigm that polymerization can only occur through hydrogen-bonding interactions. Their results collectively highlight the contributions of shape-complementarity and the power of desolvation during the polymerization process. In addition, their work demonstrates polymerization efficiency and fidelity are not identical, and that one can be achieved at the expense of the other. This last issue is perhaps best illustrated by the independent efforts of the Kutcha and Berdis/Lee laboratories. By “de-evolving” natural nucleotides, Kuchta and co-workers have developed chemical probes to interrogate how DNA polymerases balance the positive and negative features of a nucleotide to maintain fidelity. Taking a slightly different approach, Berdis and coworkers have “evolved” a series of non-natural nucleotides that are selectively incorporated opposite various non-coding DNA lesions. Their research underscores the influence of electronic configurations of the nucleobase as a positive effector during translesion DNA synthesis.
A cursor overview of this chapter evokes two important questions. Namely, does a unified mechanism for polymerization exist, and if so, which model is correct? At this point, the data argue against a unified mechanism. This statement is not meant to imply that any of the aforementioned models is incorrect. Instead, it is more appropriate to say that each model is incomplete since each of the chemical probes used to study DNA synthesis suffers from limitations that are inherent within their design. These limitations are caused by the remarkably schizophrenic nature of the natural nucleotides that act as substrates for DNA polymerases. As described earlier, each natural nucleobase possesses a variety of different biophysical properties including hydrogen-bonding potential, π-electron density, hydrophobic nature, and defined shapes and sizes. As such, it is unlikely that only one parameter is solely responsible for the high catalytic efficiency associated with their utilization. Indeed, it is more likely that DNA polymerases use all of these features during DNA synthesis but in an unequal manner during each kinetic step along the polymerization pathway. For example, shape complementarity may be more important during the initial binding of a nucleotide to the polymerase:DNA complex while desolvation and the subsequent formation of hydrogen-bonding interactions play more important roles during kinetic steps encompassing conformational changes and phosphoryl transfer, respectively.
A second argument against a unified model for polymerization reflects the diversity of DNA polymerases that perform different cellular functions. Low-fidelity polymerases such as viral replicases appear to use hydrogen-bonding interactions as the preeminent force during replication [34]. Indeed, certain low fidelity polymerase such as pol kappa [86], pol eta [87], and pol beta [88] do not incorporate dFMP, and these negative results suggest that these polymerases indeed require hydrogen-bonding interactions for polymerization. With viral polymerases, the reliance on hydrogen-bonding recognition introduces a greater frequency of mutations that plays an important role in their survival and adaptability through evolutionary processes. In contrast, high-fidelity polymerases involved in chromosomal replication appear to use a combination of shape complementarity [13], desolvation [24], and π-electron stacking interactions [59] to distinguish between correct versus incorrect nucleotides to maintain replicative fidelity.
In addition, it is clear that DNA polymerases behave differently when replicating normal versus damaged nucleic acid. High-fidelity polymerases, for example, tend to misreplicate damaged DNA while DNA polymerases that are classified as “error-prone” generally incorporate the correct dNTP opposite the DNA lesion (reviewed in [89] and [90]). Comparing the structures of various high fidelity and error-prone DNA polymerases has provided some insight into the structural nuances between the two classes of enzymes that explain their distinct behavior. However, it is amazing that even members of error-prone DNA polymerase family use mutually different mechanisms for replicating damaged DNA. One mechanism favors the use of shape complementarity and is based upon the available structures of Dpo4, an error-prone DNA polymerase from S. solfataricus whose active site is significantly larger than that of high fidelity DNA polymerases [91]. The other mechanism, based on the structure of Rev1, involves hydrogen-bonding interactions in which the incoming dNTP “pairs” with an amino acid in the active site rather than with the templating nucleobase [92]. The different tactics used by these two DNA polymerases makes it clear that a unified mechanism for polymerization is unlikely to exist. In this case, the use of non-natural nucleotides has expanded our appreciation of the subtleties that various DNA polymerases display during replication.
Indeed, comprehensive structure-activity relationships are just beginning to develop by integrating the results of studies using non-natural nucleotides with available structural information on various DNA polymerases [51, 59, 93]. To date, the E. coli Klenow fragment is the only DNA polymerase that has been studied with the several divergent classes of non-natural nucleotide described in this review. Thus, other high, medium, and low fidelity DNA polymerases must be studied with the various classes of nucleotide analogs to expand upon this limited data set. In addition, structural studies of DNA polymerase bound with nucleic acid containing non-natural nucleobases are needed to augment these kinetic studies [94]. This field of structural biology will undoubtedly provide remarkable insight into how DNA polymerases utilize non-natural nucleotides, and what features of the substrate and polymerase can be altered to modulate the efficiency of incorporation and extension.
While non-natural nucleotides are valuable tools for understanding the mechanism of DNA polymerization, their greatest utility has yet to be fully realized. In particular, the ability of non-natural nucleotides to be effectively incorporated into DNA applications toward applied research efforts in the biomedical sciences. One example has been attempts to expand nature’s genetic code by introducing a new “third” base pair (reviewed in [27]). Expanding the genetic code of an organism from four potential pairing combinations to eight would provide additional coding information for the synthesis of novel proteins that could contain an array of non-natural amino acids [27]. This technology would expand the repetoire of functional groups present on the amino acid side chains to increase protein stability [95] and the chemical diversity of enzymes [96]. Significant progress in these areas has been made by the Schultz [97–99] and Yokoyama [100–102] laboratories toward developing in vitro and in vivo transcription/translation systems with non-natural base-pairs.
Finally, non-natural nucleotides will play important roles in a variety of biomedical applications. Novel base-pairing partners have already been developed for use as universal primers for polymerase chain reaction (PCR) [103], biosensors [104], and nanowires [105]. In addition, these non-natural nucleotides can serve as substrates for DNA polymerases during the misreplication of damaged DNA [48–51]. The ability to selectively insert a non-natural nucleotide opposite various DNA lesions will provide useful tools to detect DNA lesions. This feature could be important in detecting the physiological effects of DNA damaging agents under in vitro or in vivo conditions.
Footnotes
Abbreviations used in this review include the following: dNTP, deoxynucleoside triphosphate; A, adenine; C, cytidine; G, guanine; T, thymine; dAMP, adenosine-2′-deoxyriboside monophosphate; dCMP, cytosine-2′-deoxyriboside; dGMP, guanosine-2′-deoxyriboside dTMP, thymine-2′-deoxyriboside; dF, 2,4-difluorotoluene; dFMP, 2,4-difluorotoluene monophosphate; dPMP, pyrene 2′-deoxyriboside monophosphate; 5-NI nucleotide, 5-nitro-indolyl-2′-deoxyriboside triphosphate; 5-Nap nucleotide, 5-napthyl-indolyl-2′deoxyriboside triphosphate; 5-Ph nucleotide, 5-phenyl-indolyl-2′-deoxyriboside triphosphate; 5-CE-nucleotide, 5-cyclohexene-indolyl-2′deoxyriboside triphosphate; 5-CH-nucleotide, 5-cyclohexyl-indolyl-2′deoxyriboside triphosphate; dQ, 9-methyl-1H-imidazo-[4,5-b]pyridine; dZ, 4-methylbenzimidazole nucleoside; d3FB, 3-fluorobenzene 2′deoxyriboside.
Other base-pairing interactions such as Hoogsteen and wobble base pairs have been identified and extensively characterized (reviewed in [1]).
Incorporation opposite thymine is evaluated since the 5-substituted indolyl deoxynucleotides are predicted to be complementary partners since they mimic the core structure of dATP.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Cheng YK, Pettitt BM. Stabilities of double- and triple-strand helical nucleic acids. Prog Biophys Mol Biol. 1992;58:225–257. doi: 10.1016/0079-6107(92)90007-s. [DOI] [PubMed] [Google Scholar]
- 2.Egli M, Pallan PS. Insights from crystallographic studies into the structural and pairing properties of nucleic acid analogs and chemically modified DNA and RNA oligonucleotides. Annu Rev Biophys Biomol Struct. 2007;36:281–305. doi: 10.1146/annurev.biophys.36.040306.132556. [DOI] [PubMed] [Google Scholar]
- 3.Goodman MF, Fygenson KD. DNA polymerase fidelity: from genetics toward a biochemical understanding. Genetics. 1998;148:1475–1482. doi: 10.1093/genetics/148.4.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kunkel TA, Bebenek K. DNA replication fidelity. Annu Rev Biochem. 2000;69:497–529. doi: 10.1146/annurev.biochem.69.1.497. [DOI] [PubMed] [Google Scholar]
- 5.Mitra R, Pettitt BM, Rame GL, Blake RD. The relationship between mutation rates for the (C-G)-->(T-A) transition and features of T-G mispair structures in different neighbor environments, determined by free energy molecular mechanics. Nucleic Acids Res. 1993;21:6028–6037. doi: 10.1093/nar/21.25.6028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stano NM, Jeong YJ, Donmez I, Tummalapalli P, Levin MK, Patel SS. DNA synthesis provides the driving force to accelerate DNA unwinding by a helicase. Nature. 2005;435:370–373. doi: 10.1038/nature03615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Piccirilli JA, Krauch T, Moroney SE, Benner SA. Enzymatic incorporation of a new base pair into DNA and RNA extends the genetic alphabet. Nature. 1990;343:33–37. doi: 10.1038/343033a0. [DOI] [PubMed] [Google Scholar]
- 8.Horlacher J, Hottiger M, Podust VN, Hubscher U, Benner SA. Recognition by viral and cellular DNA polymerases of nucleosides bearing bases with nonstandard hydrogen bonding patterns. Proc Natl Acad Sci U S A. 1995;92:6329–6333. doi: 10.1073/pnas.92.14.6329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lutz MJ, Held HA, Hottiger M, Hubscher U, Benner SA. Differential discrimination of DNA polymerase for variants of the non-standard nucleobase pair between xanthosine and 2,4-diaminopyrimidine, two components of an expanded genetic alphabet. Nucleic Acids Res. 1996;24:1308–1313. doi: 10.1093/nar/24.7.1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ishikawa M, Hirao I, Yokoyama S. Chemical synthesis of novel base pairs and their enzymatic incorporation into DNA. Nucleic Acids Symp Ser. 1999:125–126. doi: 10.1093/nass/42.1.125. [DOI] [PubMed] [Google Scholar]
- 11.Ishikawa M, Hirao I, Yokoyama S. Synthesis of 3-(2-deoxy-β-D-ribofuranosyl)pyridin-2-one and 2-amino-6-(N,N-dimethylamino)-9-(2-deoxy-β-D-ribofuranosyl)purine derivatives for an unnatural base pair. Tetrahedron Letters. 2000;41:3931–3934. [Google Scholar]
- 12.Fujiwara T, Kimoto M, Sugiyama H, Hirao I, Yokoyama S. Synthesis of 6-(2-thienyl)purine nucleoside derivatives that form unnatural base pairs with pyridin-2-one nucleosides. Bioorg Med Chem Lett. 2001;11:2221–2223. doi: 10.1016/s0960-894x(01)00415-2. [DOI] [PubMed] [Google Scholar]
- 13.Moran S, Ren RX, Kool ET. A thymidine triphosphate shape analog lacking Watson-Crick pairing ability is replicated with high sequence selectivity. Proc Natl Acad Sci U S A. 1997;94:10506–10511. doi: 10.1073/pnas.94.20.10506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kool ET. Active site tightness and substrate fit in DNA replication. Annu Rev Biochem. 2002;71:191–219. doi: 10.1146/annurev.biochem.71.110601.135453. [DOI] [PubMed] [Google Scholar]
- 15.Kool ET. Hydrogen bonding, base stacking, and steric effects in dna replication. Annu Rev Biophys Biomol Struct. 2001;30:1–22. doi: 10.1146/annurev.biophys.30.1.1. [DOI] [PubMed] [Google Scholar]
- 16.Kool ET. Replication of non-hydrogen bonded bases by DNA polymerases: a mechanism for steric matching. Biopolymers. 1998;48:3–17. doi: 10.1002/(SICI)1097-0282(1998)48:1<3::AID-BIP2>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 17.Morales JC, Kool ET. Efficient replication between non-hydrogen-bonded nucleoside shape analogs. Nat Struct Biol. 1998;5:950–954. doi: 10.1038/2925. [DOI] [PubMed] [Google Scholar]
- 18.Morales JC, Kool ET. Functional hydrogen-bonding map of the minor groove binding tracks of six DNA polymerases. Biochemistry. 2000;39:12979–12988. doi: 10.1021/bi001578o. [DOI] [PubMed] [Google Scholar]
- 19.Guckian KM, Krugh TR, Kool ET. Solution structure of a DNA duplex containing a replicable difluorotoluene-adenine pair. Nat Struct Biol. 1998;5:954–959. doi: 10.1038/2930. [DOI] [PubMed] [Google Scholar]
- 20.Delaney JC, Henderson PT, Helquist SA, Morales JC, Essigmann JM, Kool ET. High-fidelity in vivo replication of DNA base shape mimics without Watson-Crick hydrogen bonds. Proc Natl Acad Sci U S A. 2003;100:4469–4473. doi: 10.1073/pnas.0837277100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim TW, Brieba LG, Ellenberger T, Kool ET. Functional evidence for a small and rigid active site in a high fidelity DNA polymerase: probing T7 DNA polymerase with variably sized base pairs. J Biol Chem. 2006;281:2289–2295. doi: 10.1074/jbc.M510744200. [DOI] [PubMed] [Google Scholar]
- 22.Mizukami S, Kim TW, Helquist SA, Kool ET. Varying DNA base-pair size in subangstrom increments: evidence for a loose, not large, active site in low-fidelity Dpo4 polymerase. Biochemistry. 2006;45:2772–2778. doi: 10.1021/bi051961z. [DOI] [PubMed] [Google Scholar]
- 23.Sintim HO, Kool ET. Remarkable sensitivity to DNA base shape in the DNA polymerase active site. Angew Chem Int Ed Engl. 2006;45:1974–1979. doi: 10.1002/anie.200504296. [DOI] [PubMed] [Google Scholar]
- 24.Petruska J, Sowers LC, Goodman MF. Comparison of nucleotide interactions in water, proteins, and vacuum: model for DNA polymerase fidelity. Proc Natl Acad Sci U S A. 1986;83:1559–1562. doi: 10.1073/pnas.83.6.1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu Y, Ogawa AK, Berger M, McMinn DL, Schultz PG, Romesberg FE. Efforts toward expansion of the genetic alphabet: Optimization of interbase hydrphobic interactions. J Am Chem Soc. 2000;122:7621–7632. [Google Scholar]
- 26.Berger M, Wu Y, Ogawa AK, McMinn DL, Schultz PG, Romesberg FE. Universal bases for hybridization, replication and chain termination. Nucleic Acids Res. 2000;28:2911–2914. doi: 10.1093/nar/28.15.2911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mendel D, Cornish VW, Schultz PG. Site-directed mutagenesis with an expanded genetic code. Annu Rev Biophys Biomol Struct. 1995;24:435–462. doi: 10.1146/annurev.bb.24.060195.002251. [DOI] [PubMed] [Google Scholar]
- 28.Leconte AM, Matsuda S, Hwang GT, Romesberg FE. Efforts towards expansion of the genetic alphabet: pyridone and methyl pyridone nucleobases. Angew Chem Int Ed Engl. 2006;45:4326–4329. doi: 10.1002/anie.200601272. [DOI] [PubMed] [Google Scholar]
- 29.Rose GD, Wolfenden R. Hydrogen bonding, hydrophobicity, packing, and protein folding. Annu Rev Biophys Biomol Struct. 1993;22:381–415. doi: 10.1146/annurev.bb.22.060193.002121. [DOI] [PubMed] [Google Scholar]
- 30.Raschke TM. Water structure and interactions with protein surfaces. Curr Opin Struct Biol. 2006;16:152–159. doi: 10.1016/j.sbi.2006.03.002. [DOI] [PubMed] [Google Scholar]
- 31.Kincaid K, Beckman J, Zivkovic A, Halcomb RL, Engels JW, Kuchta RD. Exploration of factors driving incorporation of unnatural dNTPS into DNA by Klenow fragment (DNA polymerase I) and DNA polymerase alpha. Nucleic Acids Res. 2005;33:2620–2628. doi: 10.1093/nar/gki563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moore CL, Zivkovic A, Engels JW, Kuchta RD. Human DNA primase uses Watson-Crick hydrogen bonds to distinguish between correct and incorrect nucleoside triphosphates. Biochemistry. 2004;43:12367–12374. doi: 10.1021/bi0490791. [DOI] [PubMed] [Google Scholar]
- 33.Ramirez-Aguilar KA, Kuchta RD. Herpes simplex virus-1 primase: a polymerase with extraordinarily low fidelity. Biochemistry. 2004;43:9084–9091. doi: 10.1021/bi049335+. [DOI] [PubMed] [Google Scholar]
- 34.Ramirez-Aguilar KA, Moore CL, Kuchta RD. Herpes simplex virus 1 primase employs watson-crick hydrogen bonding to identify cognate nucleoside triphosphates. Biochemistry. 2005;44:15585–15593. doi: 10.1021/bi0513711. [DOI] [PubMed] [Google Scholar]
- 35.Beckman J, Kincaid K, Hocek M, Spratt T, Engels J, Cosstick R, Kuchta RD. Human DNA polymerase alpha uses a combination of positive and negative selectivity to polymerize purine dNTPs with high fidelity. Biochemistry. 2007;46:448–460. doi: 10.1021/bi061243s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Patro JN, Urban M, Kuchta RD. Role of the 2-amino group of purines during dNTP polymerization by human DNA polymerase alpha. Biochemistry. 2009;48:180–189. doi: 10.1021/bi801823z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Naegeli H. Roadblocks and detours during DNA replication: mechanisms of mutagenesis in mammalian cells. Bioessays. 1994;16:557–564. doi: 10.1002/bies.950160809. [DOI] [PubMed] [Google Scholar]
- 38.Loeb LA. Cancer cells exhibit a mutator phenotype. Adv Cancer Res. 1998;72:25–56. doi: 10.1016/s0065-230x(08)60699-5. [DOI] [PubMed] [Google Scholar]
- 39.Saito I. Chemistry of drug-induced DNA lesions. Toxicol Lett. 1993;67:3–15. doi: 10.1016/0378-4274(93)90042-v. [DOI] [PubMed] [Google Scholar]
- 40.Otsuka C, Sanadai S, Hata Y, Okuto H, Noskov VN, Loakes D, Negishi K. Difference between deoxyribose- and tetrahydrofuran-type abasic sites in the in vivo mutagenic responses in yeast. Nucleic Acids Res. 2002;30:5129–5135. doi: 10.1093/nar/gkf666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Boiteux S, Guillet M. Abasic sites in DNA: repair and biological consequences in Saccharomyces cerevisiae. DNA Repair (Amst) 2004;3:1–12. doi: 10.1016/j.dnarep.2003.10.002. [DOI] [PubMed] [Google Scholar]
- 42.Kroeger KM, Goodman MF, Greenberg MM. A comprehensive comparison of DNA replication past 2-deoxyribose and its tetrahydrofuran analog in Escherichia coli. Nucleic Acids Res. 2004;32:5480–5485. doi: 10.1093/nar/gkh873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Miller H, Grollman AP. Kinetics of DNA polymerase I (Klenow fragment exo-) activity on damaged DNA templates: effect of proximal and distal template damage on DNA synthesis. Biochemistry. 1997;36:15336–15342. doi: 10.1021/bi971927n. [DOI] [PubMed] [Google Scholar]
- 44.Berdis AJ. Dynamics of translesion DNA synthesis catalyzed by the bacteriophage T4 exonuclease-deficient DNA polymerase. Biochemistry. 2001;40:7180–7191. doi: 10.1021/bi0101594. [DOI] [PubMed] [Google Scholar]
- 45.Strauss BS. The ‘A rule’ of mutagen specificity: a consequence of DNA polymerase bypass of non-instructional lesions? Bioessays. 1991;13:79–84. doi: 10.1002/bies.950130206. [DOI] [PubMed] [Google Scholar]
- 46.Matray TJ, Kool ET. A specific partner for abasic damage in DNA. Nature. 1999;399:704–708. doi: 10.1038/21453. [DOI] [PubMed] [Google Scholar]
- 47.Reineks EZ, Berdis AJ. Evaluating the contribution of base stacking during translesion DNA replication. Biochemistry. 2004;43:393–404. doi: 10.1021/bi034948s. [DOI] [PubMed] [Google Scholar]
- 48.Zhang X, Lee I, Berdis AJ. Evaluating the contributions of desolvation and base-stacking during translesion DNA synthesis. Org Biomol Chem. 2004;2:1703–1711. doi: 10.1039/b401732c. [DOI] [PubMed] [Google Scholar]
- 49.Zhang X, Lee I, Berdis AJ. The use of nonnatural nucleotides to probe the contributions of shape complementarity and pi-electron surface area during DNA polymerization. Biochemistry. 2005;44:13101–13110. doi: 10.1021/bi050585f. [DOI] [PubMed] [Google Scholar]
- 50.Zhang X, Lee I, Zhou X, Berdis AJ. Hydrophobicity, shape, and pi-electron contributions during translesion DNA synthesis. J Am Chem Soc. 2006;128:143–149. doi: 10.1021/ja0546830. [DOI] [PubMed] [Google Scholar]
- 51.Vineyard D, Zhang X, Donnelly A, Lee I, Berdis AJ. Optimization of non-natural nucleotides for selective incorporation opposite damaged DNA. Org Biomol Chem. 2007;5:3623–3630. doi: 10.1039/b712480e. [DOI] [PubMed] [Google Scholar]
- 52.Loakes D. Survey and summary: The applications of universal DNA base analogues. Nucleic Acids Res. 2001;29:2437–2447. doi: 10.1093/nar/29.12.2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bergstrom DE, Wang G, Toma JD, Gerry N, Nichols R, Andrews P. Re-engineering DNA: design, synthesis, and properties of modified nucleic acids. Nucleic Acids Symp Ser. 1993:11–12. [PubMed] [Google Scholar]
- 54.Zhang P, Johnson WT, Klewer D, Paul N, Hoops G, Davisson VJ, Bergstrom DE. Exploratory studies on azole carboxamides as nucleobase analogs: thermal denaturation studies on oligodeoxyribonucleotide duplexes containing pyrrole-3-carboxamide. Nucleic Acids Res. 1998;26:2208–2215. doi: 10.1093/nar/26.9.2208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hoops GC, Zhang P, Johnson WT, Paul N, Bergstrom DE, Davisson VJ. Template directed incorporation of nucleotide mixtures using azole-nucleobase analogs. Nucleic Acids Res. 1997;25:4866–4871. doi: 10.1093/nar/25.24.4866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Paul N, Nashine VC, Hoops G, Zhang P, Zhou J, Bergstrom DE, Davisson VJ. DNA polymerase template interactions probed by degenerate isosteric nucleobase analogs. Chem Biol. 2003;10:815–825. doi: 10.1016/j.chembiol.2003.08.008. [DOI] [PubMed] [Google Scholar]
- 57.Loakes D, Brown DM, Linde S, Hill F. 3-Nitropyrrole and 5-nitroindole as universal bases in primers for DNA sequencing and PCR. Nucleic Acids Res. 1995;23:2361–2366. doi: 10.1093/nar/23.13.2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Smith CL, Simmonds AC, Hamilton AL, Martin DL, Lashford AG, Loakes D, Hill F, Brown DM. Use of 5-nitroindole-2′-deoxyribose-5′-triphosphate for labelling and detection of oligonucleotides. Nucleosides Nucleotides. 1998;17:555–564. doi: 10.1080/07328319808005198. [DOI] [PubMed] [Google Scholar]
- 59.Sheriff A, Motea E, Lee I, Berdis AJ. Mechanism and dynamics of translesion DNA synthesis catalyzed by the Escherichia coli Klenow fragment. Biochemistry. 2008;47:8527–8537. doi: 10.1021/bi800324r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Seeman NC. From genes to machines: DNA nanomechanical devices. Trends Biochem Sci. 2005;30:119–125. doi: 10.1016/j.tibs.2005.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lee JF, Stovall GM, Ellington AD. Aptamer therapeutics advance. Curr Opin Chem Biol. 2006;10:282–289. doi: 10.1016/j.cbpa.2006.03.015. [DOI] [PubMed] [Google Scholar]
- 62.Benner SA. Expanding the genetic lexicon: incorporating non-standard amino acids into proteins by ribosome-based synthesis. Trends Biotechnol. 1994;12:158–163. doi: 10.1016/0167-7799(94)90076-0. [DOI] [PubMed] [Google Scholar]
- 63.Hohsaka T, Sisido M. Incorporation of non-natural amino acids into proteins. Curr Opin Chem Biol. 2002;6:809–815. doi: 10.1016/s1367-5931(02)00376-9. [DOI] [PubMed] [Google Scholar]
- 64.Wang SX, Mure M, Medzihradszky KF, Burlingame AL, Brown DE, Dooley DM, Smith AJ, Kagan HM, Klinman JP. A crosslinked cofactor in lysyl oxidase: redox function for amino acid side chains. Science FIELD Full Journal Title:Science (New York, NY) 1996;273:1078–1084. doi: 10.1126/science.273.5278.1078. [DOI] [PubMed] [Google Scholar]
- 65.Deamer D. A giant step towards artificial life? Trends Biotechnol. 2005;23:336–338. doi: 10.1016/j.tibtech.2005.05.008. [DOI] [PubMed] [Google Scholar]
- 66.Kent S. Novel forms of chemical protein diversity -- in nature and in the laboratory. Curr Opin Biotechnol. 2004;15:607–614. doi: 10.1016/j.copbio.2004.10.003. [DOI] [PubMed] [Google Scholar]
- 67.Joyce CM, Sun XC, Grindley ND. Reactions at the polymerase active site that contribute to the fidelity of Escherichia coli DNA polymerase I (Klenow fragment) J Biol Chem. 1992;267:24485–24500. [PubMed] [Google Scholar]
- 68.Creighton S, Huang MM, Cai H, Arnheim N, Goodman MF. Base mispair extension kinetics. Binding of avian myeloblastosis reverse transcriptase to matched and mismatched base pair termini. J Biol Chem. 1992;267:2633–2639. [PubMed] [Google Scholar]
- 69.Mendelman LV, Petruska J, Goodman MF. Base mispair extension kinetics. Comparison of DNA polymerase alpha and reverse transcriptase. J Biol Chem. 1990;265:2338–2346. [PubMed] [Google Scholar]
- 70.Brotschi C, Mathis G, Leumann CJ. Bipyridyl- and biphenyl-DNA: a recognition motif based on interstrand aromatic stacking. Chemistry. 2005;11:1911–1923. doi: 10.1002/chem.200400858. [DOI] [PubMed] [Google Scholar]
- 71.Zahn A, Brotschi C, Leumann CJ. pentafluorophenyl-phenyl interactions in biphenyl-DNA. Chemistry. 2005;11:2125–2129. doi: 10.1002/chem.200401128. [DOI] [PubMed] [Google Scholar]
- 72.Matsuda S, Leconte AM, Romesberg FE. Minor groove hydrogen bonds and the replication of unnatural base pairs. J Am Chem Soc. 2007;129:5551–5557. doi: 10.1021/ja068282b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Matsuda S, Fillo JD, Henry AA, Rai P, Wilkens SJ, Dwyer TJ, Geierstanger BH, Wemmer DE, Schultz PG, Spraggon G, Romesberg FE. Efforts toward expansion of the genetic alphabet: structure and replication of unnatural base pairs. J Am Chem Soc. 2007;129:10466–10473. doi: 10.1021/ja072276d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Leconte AM, Hwang GT, Matsuda S, Capek P, Hari Y, Romesberg FE. Discovery, characterization, and optimization of an unnatural base pair for expansion of the genetic alphabet. J Am Chem Soc. 2008;130:2336–2343. doi: 10.1021/ja078223d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Anthony YW, Ogawa K, McMinn Dustin L, Liu Jianquan, Schultz Peter G, Romesberg Floyd E. Efforts toward the Expansion of the Genetic Alphabet: Information Storage and Replication with Unnatural Hydrophobic Base Pairs. J Amer Chem Soc. 2000;122:3274–3287. [Google Scholar]
- 76.McCulloch SD, Kokoska RJ, Chilkova O, Welch CM, Johansson E, Burgers PM, Kunkel TA. Enzymatic switching for efficient and accurate translesion DNA replication. Nucleic Acids Res. 2004;32:4665–4675. doi: 10.1093/nar/gkh777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Plosky BS, Woodgate R. Switching from high-fidelity replicases to low-fidelity lesion-bypass polymerases. Curr Opin Genet Dev. 2004;14:113–119. doi: 10.1016/j.gde.2004.02.002. [DOI] [PubMed] [Google Scholar]
- 78.Friedberg EC, Lehmann AR, Fuchs RP. Trading places: how do DNA polymerases switch during translesion DNA synthesis? Mol Cell. 2005;18:499–505. doi: 10.1016/j.molcel.2005.03.032. [DOI] [PubMed] [Google Scholar]
- 79.Carver TE, Jr, Hochstrasser RA, Millar DP. Proofreading DNA: recognition of aberrant DNA termini by the Klenow fragment of DNA polymerase I. Proc Natl Acad Sci U S A. 1994;91:10670–10674. doi: 10.1073/pnas.91.22.10670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Johnson AA, Johnson KA. Exonuclease proofreading by human mitochondrial DNA polymerase. J Biol Chem. 2001;276:38097–38107. doi: 10.1074/jbc.M106046200. [DOI] [PubMed] [Google Scholar]
- 81.Miller H, Perrino FW. Kinetic mechanism of the 3′-->5′ proofreading exonuclease of DNA polymerase III. Analysis by steady state and pre-steady state methods. Biochemistry. 1996;35:12919–12925. doi: 10.1021/bi960326d. [DOI] [PubMed] [Google Scholar]
- 82.Strauss BS, Sagher D, Acharya S. Role of proofreading and mismatch repair in maintaining the stability of nucleotide repeats in DNA. Nucleic Acids Res. 1997;25:806–813. doi: 10.1093/nar/25.4.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Morales JC, Kool ET. Importance of terminal base pair hydrogen-bonding in 3′-end proofreading by the Klenow fragment of DNA polymerase I. Biochemistry. 2000;39:2626–2632. doi: 10.1021/bi992173a. [DOI] [PubMed] [Google Scholar]
- 84.Zhang X, Lee I, Berdis AJ. A potential chemotherapeutic strategy for the selective inhibition of promutagenic DNA synthesis by nonnatural nucleotides. Biochemistry. 2005;44:13111–13121. doi: 10.1021/bi050584n. [DOI] [PubMed] [Google Scholar]
- 85.Watson JD, Crick FH. Molecular structure of nucleic acids; a structure for deoxyribose nucleic acid. Nature. 1953;171:737–738. doi: 10.1038/171737a0. [DOI] [PubMed] [Google Scholar]
- 86.Wolfle WT, Washington MT, Kool ET, Spratt TE, Helquist SA, Prakash L, Prakash S. Evidence for a Watson-Crick hydrogen bonding requirement in DNA synthesis by human DNA polymerase kappa. Mol Cell Biol. 2005;25:7137–7143. doi: 10.1128/MCB.25.16.7137-7143.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Washington MT, Helquist SA, Kool ET, Prakash L, Prakash S. Requirement of Watson-Crick hydrogen bonding for DNA synthesis by yeast DNA polymerase eta. Mol Cell Biol. 2003;23:5107–5112. doi: 10.1128/MCB.23.14.5107-5112.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kirby TW, DeRose EF, Beard WA, Wilson SH, London RE. A thymine isostere in the templating position disrupts assembly of the closed DNA polymerase beta ternary complex. Biochemistry. 2005;44:15230–15237. doi: 10.1021/bi0511742. [DOI] [PubMed] [Google Scholar]
- 89.Green CM, Lehmann AR. Translesion synthesis and error-prone polymerases. Adv Exp Med Biol. 2005;570:199–223. doi: 10.1007/1-4020-3764-3_7. [DOI] [PubMed] [Google Scholar]
- 90.Lehmann AR. New functions for Y family polymerases. Mol Cell. 2006;24:493–495. doi: 10.1016/j.molcel.2006.10.021. [DOI] [PubMed] [Google Scholar]
- 91.Ling H, Boudsocq F, Woodgate R, Yang W. Crystal structure of a Y-family DNA polymerase in action: a mechanism for error-prone and lesion-bypass replication. Cell. 2001;107:91–102. doi: 10.1016/s0092-8674(01)00515-3. [DOI] [PubMed] [Google Scholar]
- 92.Nair DT, Johnson RE, Prakash L, Prakash S, Aggarwal AK. Rev1 employs a novel mechanism of DNA synthesis using a protein template. Science. 2005;309:2219–2222. doi: 10.1126/science.1116336. [DOI] [PubMed] [Google Scholar]
- 93.Hwang GT, Romesberg FE. Unnatural substrate repertoire of A, B, and X family DNA polymerases. J Am Chem Soc. 2008;130:14872–14882. doi: 10.1021/ja803833h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zahn KE, Belrhali H, Wallace SS, Doublie S. Caught bending the A-rule: crystal structures of translesion DNA synthesis with a non-natural nucleotide. Biochemistry. 2007;46:10551–10561. doi: 10.1021/bi7008807. [DOI] [PubMed] [Google Scholar]
- 95.Hodgson DR, Sanderson JM. The synthesis of peptides and proteins containing non-natural amino acids. Chem Soc Rev. 2004;33:422–430. doi: 10.1039/b312953p. [DOI] [PubMed] [Google Scholar]
- 96.Kiick KL, Saxon E, Tirrell DA, Bertozzi CR. Incorporation of azides into recombinant proteins for chemoselective modification by the Staudinger ligation. Proc Natl Acad Sci U S A. 2002;99:19–24. doi: 10.1073/pnas.012583299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Chin JW, Cropp TA, Chu S, Meggers E, Schultz PG. Progress toward an expanded eukaryotic genetic code. Chem Biol. 2003;10:511–519. doi: 10.1016/s1074-5521(03)00123-6. [DOI] [PubMed] [Google Scholar]
- 98.Anderson JC, Wu N, Santoro SW, Lakshman V, King DS, Schultz PG. An expanded genetic code with a functional quadruplet codon. Proc Natl Acad Sci U S A. 2004;101:7566–7571. doi: 10.1073/pnas.0401517101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Ryu Y, Schultz PG. Efficient incorporation of unnatural amino acids into proteins in Escherichia coli. Nat Methods. 2006;3:263–265. doi: 10.1038/nmeth864. [DOI] [PubMed] [Google Scholar]
- 100.Takai K, Yokoyama S. Roles of 5-substituents of tRNA wobble uridines in the recognition of purine-ending codons. Nucleic Acids Res. 2003;31:6383–6391. doi: 10.1093/nar/gkg839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Kobayashi T, Sakamoto K, Takimura T, Sekine R, Kelly VP, Kamata K, Nishimura S, Yokoyama S. Structural basis of nonnatural amino acid recognition by an engineered aminoacyl-tRNA synthetase for genetic code expansion. Proc Natl Acad Sci U S A. 2005;102:1366–1371. doi: 10.1073/pnas.0407039102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Hino N, Okazaki Y, Kobayashi T, Hayashi A, Sakamoto K, Yokoyama S. Protein photo-cross-linking in mammalian cells by site-specific incorporation of a photoreactive amino acid. Nat Methods. 2005;2:201–206. doi: 10.1038/nmeth739. [DOI] [PubMed] [Google Scholar]
- 103.Johnson SC, Sherrill CB, Marshall DJ, Moser MJ, Prudent JR. A third base pair for the polymerase chain reaction: inserting isoC and isoG. Nucleic Acids Res. 2004;32:1937–1941. doi: 10.1093/nar/gkh522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Ting R, Thomas JM, Lermer L, Perrin DM. Substrate specificity and kinetic framework of a DNAzyme with an expanded chemical repertoire: a putative RNaseA mimic that catalyzes RNA hydrolysis independent of a divalent metal cation. Nucleic Acids Res. 2004;32:6660–6672. doi: 10.1093/nar/gkh1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Bang GS, Cho S, Kim BG. A novel electrochemical detection method for aptamer biosensors. Biosens Bioelectron. 2005;21:863–870. doi: 10.1016/j.bios.2005.02.002. [DOI] [PubMed] [Google Scholar]