Abstract
Adenoviruses (Ads) hold great promise as gene vectors for diagnostic or therapeutic applications. The native tropism of Ads must be modified to achieve disease site-specific gene delivery by Ad vectors and this should be done in a programmable way and with technology that can realistically be scaled up. To this end, we applied the technologies of designed ankyrin repeat proteins (DARPins) and ribosome display to develop a DARPin that binds the knob domain of the Ad fiber protein with low nanomolar affinity (KD 1.35 nM) and fused this protein with a DARPin specific for Her2, an established cell-surface biomarker of human cancers. The stability of the complex formed by this bispecific targeting adapter and the Ad virion resulted in insufficient gene transfer and was subsequently improved by increasing the valency of adapter–virus binding. In particular, we designed adapters that chelated the knob in a bivalent or trivalent fashion and showed that the efficacy of gene transfer by the adapter–Ad complex increased with the functional affinity of these molecules. This enabled efficient transduction at low stoichiometric adapter-to-fiber ratios. We confirmed the Her2 specificity of this transduction and its dependence on the Her2-binding DARPin component of the adapters. Even the adapter molecules with four fused DARPins could be produced and purified from Escherichia coli at very high levels. In principle, DARPins can be generated against any target and this adapter approach provides a versatile strategy for developing a broad range of disease-specific gene vectors.
Keywords: DARPins, targeted gene delivery, affinity, knob, viral vectors
Introduction
Adenoviruses (Ads) are a family of nonenveloped, double-stranded DNA genome-containing viruses that have been isolated from humans, nonhuman primates and other vertebrates. Studies of Ad biology have identified several Ad serotypes as promising prototypes of gene vectors for gene therapy,1 genetic immunizations,2–4 and molecular–genetic imaging.5,6 To this end, attempts have been made to modify the exterior of human Ad serotype 5 (Ad5) particles to alter the antigenic profile of the virus7 and make it display foreign antigens8,9 and imaging reporters.10,11
In addition, extensive virion-modification studies have been done to alter the native tropism of the virus to make Ad-mediated transgene delivery to desired cell targets both more efficient and target-specific. Those studies focused primarily on modifications to one of the major components of the Ad5 capsid, the fiber protein.12,13 The fiber is a homotrimeric, antenna-shaped protein that associates with another major capsid protein, the penton base, to form the penton capsomers located at the vertices of the icosahedral Ad virions (Fig. 1a). Ad5 achieves entry into a cell by a two-step mechanism. First, the fiber binds to the coxsackie Ad receptor (CAR)14,15; second, the arginine-glycine-aspartic acid (RGD) motif within the penton base protein of the CAR-anchored virion binds to the cellular integrin and triggers Ad internalization.16
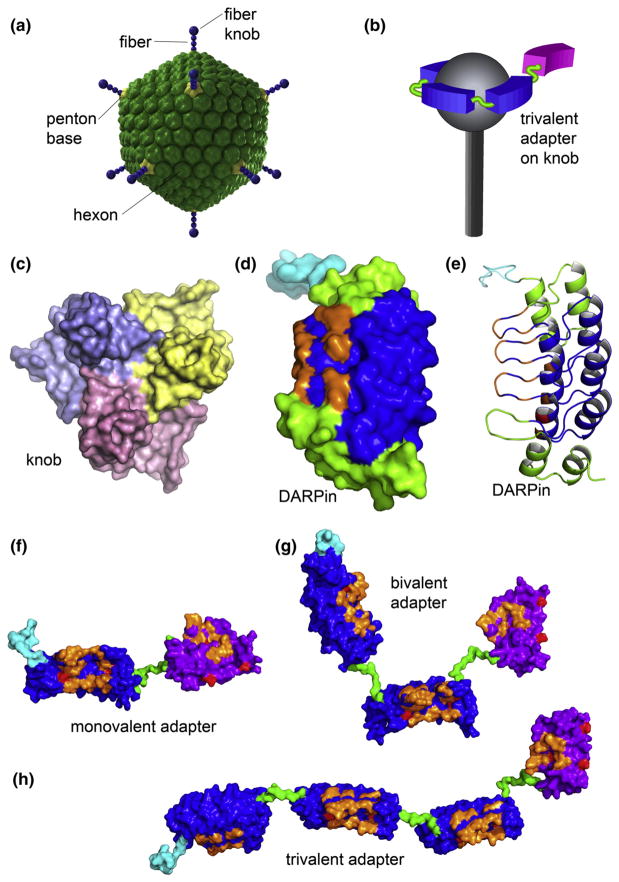
Fig. 1.
The structures of Ad5 and its fiber knob domain, monovalent DARPin, and multivalent DARPin-derived adapters. (a) The icosahedral shell of the Ad5 virion is formed primarily by the hexon protein. Each of the 12 vertices contains the penton capsomer that consists of a trimeric fiber protein anchored within the pentameric penton base. The knob domain located at the end of the fiber binds the primary receptor. (b) The illustration shows the intended arrangement, with the virus fiber protein (gray) bound by three adapter DARPins (blue) connected to a Her2-binding DARPin (magenta) in one polypeptide chain. Please note (c)–(e) and (f)–(h) are drawn to scale. (c) Trimeric knob protein, viewed along the 3-fold symmetry axis from the side distal to the virion surface (PDB ID 1knb). Individual subunits are shown in different colors. (d) and (e) Monovalent DARPin 2E6 model showing the N-terminal His6 tag (cyan), capping repeats (green), and consensus repeats (blue) with seven randomized residues (orange) per repeat for target binding in either space-filling (d) or ribbon representation (e). Each 33 amino acid repeat in the DARPin consists of two antiparallel α-helices and a β-hairpin, resulting in a groove-like binding surface. The capping repeats provide a hydrophilic surface. The resulting DARPins consist of one contiguous polypeptide chain with a randomized binding surface. Bispecific adapter DARPins with varying valency are shown in (f) 2E6-G5, (g) (2E6)2-G5 and (h) (2E6)3-G5. The anti-knob DARPin 2E6 is shown with a blue surface; the anti-HER2 DARPin G5 is shown with a magenta surface, with the randomized residues in orange, the framework mutations in red, the N-terminal His6 tag in cyan and the (Gly4Ser)n linkers in green. The linkers shown between the 2E6 DARPins are (Gly4Ser)2, and (Gly4Ser)3 between 2E6 and G5.
While some Ad serotypes use cell-surface molecules other than the CAR (such as sialic acid, CD46, CD80 and CD86) as their primary receptors,17–19 most of them still use the two-step cell entry mechanism. This mechanistic similarity is paralleled by the evolutionary conservation of the overall structure of the fiber, which consists of three distinct domains.20 The aminoterminal tail domain anchors the fiber within the penton base. The carboxyterminal globular knob domain has two main functions: first, it causes trimerization of the fiber protein, which is essential for the correct assembly of the fiber with the capsid;21 and second, it binds the primary receptor on the cell.22,23 The central shaft domain facilitates binding to the CAR by extending the knob away from the virion.
Attempts to increase the efficacy of Ad5 transduction of CAR-deficient cells (including many types of tumor cells) and to target the virus to alternative receptors have been based on two distinct approaches: (1) genetic modification of the fiber protein; and (2) the use of bispecific molecular adapters capable of cross-linking the virus to the desired receptor. While the former approach yields self-assembling Ad particles with the receptor-targeting ligands being integral parts of the virus, its practicality is limited by the structural compatibility of the ligand and the fiber, as alterations to the fiber protein often disturb its trimerization and assembly with the virus capsid (G. M., N. B. & V. Krasnykh, unpublished results).24 In addition, the functionality of some targeting ligands requires post-translational modifications, such as the formation of disulfide bonds, that are unavailable in the cytoplasm and nucleoplasm of the cells where the synthesis of the fiber and Ad particle assembly occur.25 These limitations are much less of an issue in the latter strategy, in which the production of targeting adapters and the virus assembly process are disengaged and thus mutually independent.
Earlier studies by us and others demonstrated the feasibility of Ad5 targeting with adapters that consisted of peptides, antibodies and their fragments, extracellular domains of CAR (binding to the knob), Gal domain of the blood coagulation factor X (FX) (binding to the hexon) and other proteins.26–35 However, both the efficacy and practicality of these approaches were limited by problems associated with the production of such adapters in sufficient quantities and purity. In addition, the use of adapters derived from soluble CAR required retaining native tropism by the Ad vector,27,28,32 while the in vivo performance of the Ad5 vector targeted with FX-based adapters was compromised by a high concentration of FX in the blood, which resulted in unavoidable replacement of the targeting adapter with FX and the loss of targeting.36
The goal of the present study was to test whether the adapter-based modification of the Ad tropism could be further advanced by developing a novel strategy in which targeting adapters would be assembled with two protein modules, each selected from rationally designed protein libraries and evolved to bind either the virus or the target receptor (Fig. 1b). A design was needed in which these modules could be combined in a multitude of ways to direct the virus to a target receptor of interest. It was particularly important to develop targeting adapters that could be easily prepared in the amounts and with the purity required for subsequent clinical applications. Finally, it was essential to use protein modules that could, in principle, be generated with specificity for any cellular target.
To this end, we applied the technology of designed ankyrin repeat proteins (DARPins),37,38 in which ribosome display37 or phage display39 is used to select target-specific binders from combinatorial libraries of DARPins. DARPins are particularly robust to engineering and, thus, can act as an alternative to antibodies. DARPins are structurally distinct from antibodies; they are built from helical repeats (Fig. 1d and e) and contain a concave protein-interaction surface, which is randomized in the library. These proteins contain no cysteine, can be expressed in soluble form in Escherichia coli at very high levels, and are very stable and resistant to aggregation.40 Here, we report the use of this technology for the development of targeting adapters that bind to the knob domain of the Ad5 fiber (Fig. 1c). By coupling a human epidermal growth factor receptor type 2 (Her2)-specific DARPin to the knob-binding DARPins, we show the ease with which DARPin-based targeting adapters can be assembled, expressed and purified, and we demonstrate that the affinity of such adapters for the Ad5 virion can be increased through affinity maturation and linear multimerization. We show that, by forming complexes with the Ad5 virions, such designed adapters direct the virus to the human cancer-related receptor Her2 on the cell surface, yielding target-specific transduction.
Results
Selection, affinity maturation and characterization of the knob-binding DARPins
The design of a truly targeted Ad vector requires the accomplishment of two distinct tasks: (1) elimination of the native tropism of the virus; and (2) engineering of novel tropism toward the desired molecular target. To these ends, we first chose to develop DARPins that would bind with high affinity to the knob domain of the Ad5 fiber protein whose CAR-binding site was ablated. To facilitate the development of such DARPins, we used E. coli to produce three variants of recombinant Ad5 fiber knob proteins differing in the tags present, all having deletion of the TAYT tetrapeptide within the FG loop. This deletion dramatically decreased the ability of the mutated knob to bind to CAR.41 We verified the similar oligomerization behavior of these proteins as the trimeric, wild-type Ad5 fiber knob by SDS-PAGE in combination with size-exclusion chromatography (SEC) with multi-angle light-scattering (MALS). Both analyses confirmed that all TAYT-deleted knobs were trimers (Supplementary Data Fig. S2) and could thus be used as targets and competitors for the selection of knob-binding DARPins.
Next, we used these knobΔTAYT proteins in the DARPin selection and affinity maturation strategy, which is outlined in Supplementary Data Fig. S3. As an initial set of selected binders had low affinity, we sought to improve affinity by using off-rate selection combined with error-prone PCR on both the pool of initially selected binders and the original DARPin library. In total, 282 DARPins were screened for improved affinity by enzyme-linked immunosorbent assay (ELISA) using crude E. coli extracts from expression cultures at 1/1000 dilution, and the 21 best performing DARPin leads were purified via immobilized metal ion affinity chromatography (IMAC) and then affinity-ranked using an Octet system with the biosensor surface coated with biotinylated knobΔTAYT protein (see Supplementary Data, Methods). The sequencing of several of the most promising DARPin genes showed that amino acid positions that were randomized in the original library38 were identical in all of them but revealed additional mutations that were scattered across the scaffold of the proteins, with three mutations being frequent. These data suggested that the DARPins in this new set were derived from a single progenitor clone. From this pool of binders with the best apparent equilibrium dissociation constants, KD, we chose DARPin 2E6 for further characterization. We determined KD of the DARPin 2E6 using surface plasmon resonance (SPR) measurements with the immobilized biotinylated knobΔTAYT protein to be 1.35 nM (Supplementary Data Fig. S4b). Using SEC-MALS analysis (see below), 2E6 proved to be monomeric without any tendency to aggregate and therefore displayed suitable biophysical properties to serve as a component of the targeting adapter.
We also used SPR to measure the affinity of the DARPin F4 (Supplementary Data Fig. S4a), which was chosen from the initial set of knob binders for use as a low-affinity, knob-binding control. In particular, we wished to test F4 in parallel with 2E6 to correlate the affinity of a given DARPin for the knob with its potency in Ad vector targeting. The KD of DARPin F4 was estimated to be ~400 nM (Supplementary Data Fig. S4a).
We used ELISA to test the ability of the selected DARPins to bind to the fiber contained within the complete Ad virion. In this assay, three types of Ad5 virions containing the wild-type unmodified fiber (Ad5Luc1), the fiber with the mutated knob (Ad5LucFΔTAYT) or no fiber (Ad5LucΔF) were probed with the selected DARPins. As suggested by the binding studies, e.g. ELISA, SPR and SEC at various stages of the selection and affinity maturation process of the selected DARPins to the recombinant purified fiber knob, this assay clearly showed specific, fiber-dependent binding of the DARPin 2E6 to Ad particles (Fig. 2) and thus justified the use of this DARPin as a basis for constructing the virus-anchoring component of bispecific targeting adapters. Notably, DARPin 2E6 showed levels of binding comparable to both the mutated and wild-type virions, suggesting that its binding site on the knob does not involve the FG loop. As expected, the low-affinity early generation DARPin F4 gave rise to lower signals than the high-affinity DARPin 2E6.
Fig. 2.
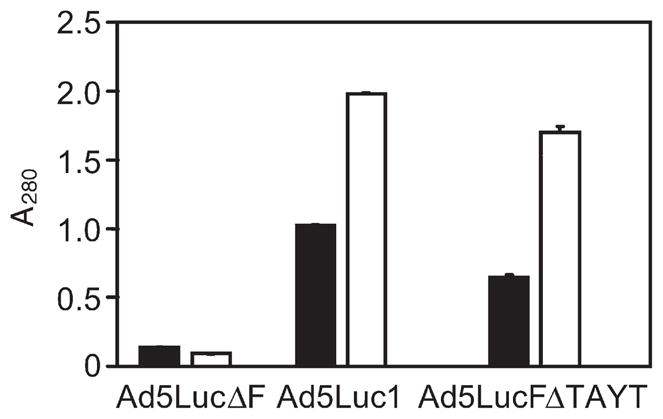
Binding of knob-specific DARPins to virion-associated fibers. DARPins F4 (filled bars) and 2E6 (open bars), immobilized via their biotinylated AviTag to a streptavidin-coated ELISA plate, were probed with the particles of Ad vectors (Ad5lucΔF, fiber-depleted virus; Ad5Luc1, virus containing the wild-type knob; Ad5LucFΔ-TAYT, virus containing the knob with a 4 bp deletion in the FG loop of the knob) shown below the graph. DARPin-bound Ads were detected with an anti-Ad antibody and HRP-labeled secondary antibody. Background determined in casein-coated wells was at an absorbance at 490 nm (A490) of 0.2 and has not been subtracted. Error bars show standard deviation calculated for triplicate data points.
Bispecific, monovalent targeting adapters enabled only suboptimal targeting of the Ad vector
To test the feasibility of Ad vector targeting with the DARPin-derived adapters, we designed the first set of such adapters by fusing the knob-binding DARPin 2E6 with the DARPin G5 described earlier (termed H10-2-G542), which is specific for Her2.
Her2 was chosen as a target for the Ad vector because its over-expression by several types of human tumors has been correlated with the cancer’s aggressiveness and poor prognosis,43,44 thus supporting the development of Her2-targeted therapeutics. Our design strategy was based on our earlier successes with developing bivalent and bispecific DARPin-based fusion proteins, which were expressed in a functional form and at very high levels.45–47 DARPins 2E6 and G5 were fused using a 15 amino acid linker (Gly4Ser)3 that added flexibility to the adapter protein designated 2E6-G5 (Fig. 1f). This design, with generous spacing between the knob-binding and the target-binding domains, was expected to facilitate subsequent cross-linking of the fiber and Her2 through simultaneous engagement of several receptor molecules and knob subunits. As a control, the similar bispecific adapter F4-G5 was designed using the low-affinity, knob-binding DAR-Pin F4. In addition, we fused only the linker itself to the DARPin G5 to make another control molecule, termed Linker-G5, which lacked Ad-binding ability. The resultant proteins were expressed in E. coli and purified as described in Materials and Methods.
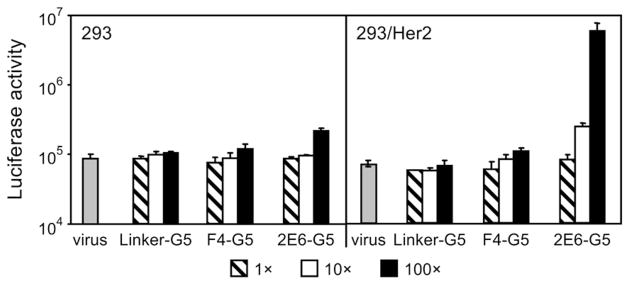
We tested these adapters in an in vitro gene transfer assay in which they were used to target the infection-impaired, firefly luciferase (Fluc)-expressing Ad5 vector containing the fiber with the above described ΔTAYT mutation within the knob. The cells of the two isogenic lines that differed by their Her2 phenotypes, the 293 (Her2-negative) and the 293/Her2 (Her2-expressing, stably transfected with a Her2-expressing plasmid; see Materials and Methods), were used as targets. As expected, regardless of the adapter-to-fiber ratios (defined as the number of adapter molecules/monomeric fiber subunit) used, none of the proteins caused any substantial increase of transduction in the Her2-negative cells when compared to virus alone (Fig. 3). Furthermore, neither the control Linker-G5 protein nor the F4-G5 adapter containing a low-affinity, anti-knob DARPin yielded transduction above the background in 293/Her2 cells. Substantial enhancement of gene transfer to these cells was observed only with the adapter 2E6-G5, which contains the higher affinity knob-binding DARPin 2E6, which yielded a nearly 100-fold increase in reporter expression. However, this augmentation of gene transfer was observed only at the highest tested adapter-to-fiber ratio of 100. This observation suggested that the affinity of the knob-binding DARPin 2E6 was still insufficient to ensure stability of the virus complex with monovalent adapter at lower ratios and made engineering an improved stability of the complex a major focus of subsequent experiments.
Fig. 3.
Adapter-mediated transduction of cultured human cells. Her2-negative 293 cells and Her2-expressing stably transfected 293/Her2 cells were infected with Ad5LucFΔTAYT alone (shown as virus) or preincubated with the adapter fusion proteins (shown below the graph) at adapter-to-fiber ratios of 1, 10 or 100. The virus was added to cells at a multiplicity of infection of 100 VP/cell. Graphs show the activity of luciferase reporter in the lysates of infected cells in relative light units. Error bars show standard deviations calculated for triplicate data points.
Stoichiometry of DARPin-to-knob binding provided a rationale for the design of multivalent targeting adapters
Improvement of adaptor–virus complex stability could potentially be achieved by increasing the avidity within the complex. However, the success of this approach depended on whether the trimeric knob domain of the fiber could actually bind three DARPins. Whether this is possible depends on the location of the epitope, and alternative geometric arrangements within the adapter–knob complex are possible, which could, for instance, result in the binding of just one DARPin to the trimeric knob, as reported for other targets.48 We used three different methods to establish the stoichiometry of 2E6 binding to the knob.
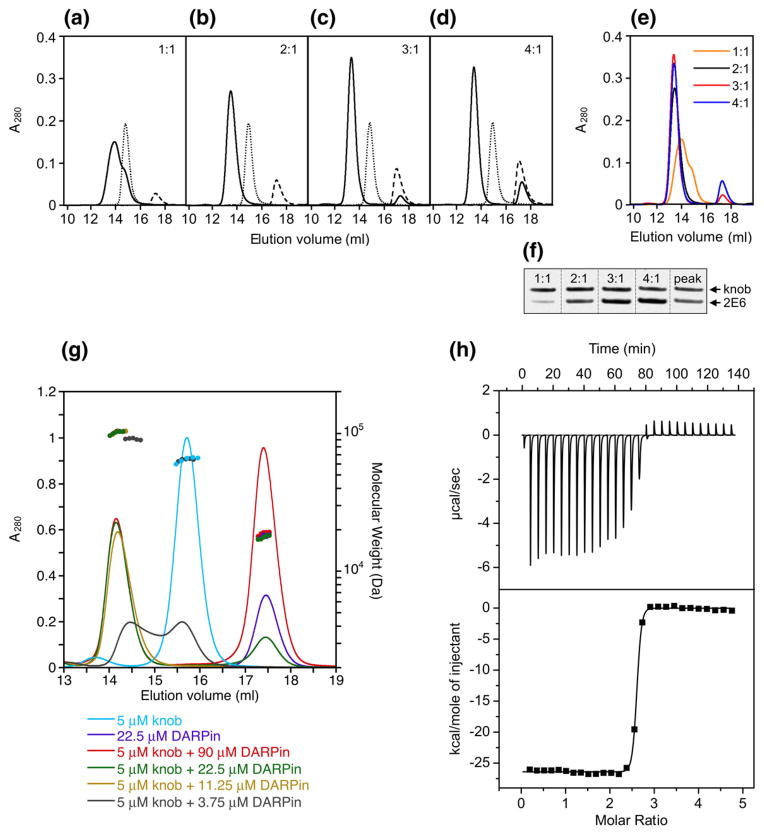
In the first approach, we mixed a fixed amount of the knobΔTAYT protein with DARPin 2E6 at various ratios, resolved these mixtures by SEC, and measured the amounts of the knob and DARPin in the peak corresponding to the knob–DARPin complexes by SDS-PAGE. These experiments showed that the DARPin-binding ability of the knob was exhausted at a 2E6-to-knob ratio of 3 (Fig. 4a–d) because further increase in DARPin quantity did not result in a shift of the corresponding peak position or an increase of the peaks height (Fig. 4e) The measured DARPin-to-knob ratio for complexes collected during chromatography was 2.3 (Fig. 4f).
Fig. 4.
Determination of stoichiometry of DARPin binding to the knob by SEC. (a)–(d) Profiles of mixtures of 2E6 DARPin with the knob protein (one, two, three or four DARPin molar equivalents added per trimeric knob) resolved by SEC. Ratios at which the proteins were mixed are shown in the upper right corner of each panel. Profiles of elution for the knob, DARPin and DARPin-knob mixtures are shown by the dotted, broken and unbroken lines, respectively. (e) Overlay of the curves corresponding to the DARPin-knob mixtures (as in (a)–(d)). (f) SDS-PAGE of mixtures of 2E6 with the knob at indicated ratios, which were used to create a calibration curve to measure the ratio of the proteins in the chromatography peak containing the DARPin–knob complexes. (g) SEC-MALS of DARPin–knob mixtures. The knobΔTAYT protein at a concentration of 5 μM was incubated with different concentrations of DARPin 2E6 (3.75, 11.25, 22.5 and 90 μM). Complex formation was analyzed by size-exclusion chromatography coupled to MALS. The curves shown represent the absorption at 280 nm (peaks; left-hand y-axis). The determined mass of the knobΔTAYT trimer, DARPin 2E6 and the complex are given on top of the corresponding protein peaks (right-hand y-axis). (h) ITC of knobΔTAYT protein (34 μM) with DARPin 2E6 (770 μM stock). Top panel, raw instrumental output (differential power as a function of time). Bottom panel, binding isotherm. The heats measured at each titration step were normalized for the molar concentration and corrected for unspecific heats (symbols). The unbroken line is the best fit according to a binding model assuming identical and independent binding sites. The best fitting parameters are: molar enthalpy (ΔH), −26.5±0.9 kcal mol−1; and number of binding sites (n), 2.5±0.2. Note that the concentrations are so far above KD that a reliable determination of KD is not possible.
Similar experiments were carried out by SEC-MALS (Fig. 4g). This method determines the molecular mass of the DARPin–knob complex independent of its elution profile. At the highest tested DARPin-to-knob ratio of 6:1, a molecular mass of the complex of 99.43 kDa was determined, which was less than 120.21 kDa, the predicted molecular mass of a complex formed by three DARPins (3×18.2 kDa) and one knob trimer (65.61 kDa). This method is independent of potential errors in the concentrations of the stock solutions, because the absolute molar masses of analyzed protein complexes are determined by relating the scattered light intensity to the refractive index (RI) of the protein complex, in turn compared to the RI of the solvent alone. The measured RI is proportional to the concentration of the protein complex, and the specific refractive index increment (dn/dc) used for molar mass determination is known to be constant for proteins and independent of their amino acid composition.49,50
Using a series of DARPins with lower affinity under the same experimental conditions, we saw a correlation between complex size in SEC-MALS and DARPin affinity (Supplementary Data Fig. S5). Using mixtures of DARPins (45 μM) with knobΔ-TAYT protein (10 μM trimer), the stoichiometry of the complexes formed by the DARPins B1 (apparent affinity in the micromolar range), G12 (apparent affinity ~400 nM), 2D2 (apparent affinity ~50 nM) and 2E6 (affinity of 1.35 nM) was determined as 0.45, 0.96, 1.7 and 2.5, respectively. This correlation of the stoichiometry with affinity suggested that under the measurement conditions the occupancy of the knob subunits by the DARPins was not complete and depended on affinity. This finding provided a further rationale to engineer multivalent adapters.
The stoichiometry was examined also by isothermal titration calorimetry (ITC; Fig. 4h), in which the knob (34 μM) was titrated with a stock solution of 770 μM DARPin 2E6. Even at these high concentrations, the knob appeared not to be fully saturated, and the fit yielded a stoichiometry of 2.5±0.2 DARPins per knob.
Taken together, the results of these three independent studies suggested that full occupancy of the Ad5 fibers by monovalent targeting adapters might be difficult to achieve at biologically relevant concentrations, despite the measured affinity of 1.35 nM, prompting us to investigate multivalent binders as a potential solution to this problem.
Linear multimerization of the knob-binding modules increased the stability of the vector–adapter complexes and yielded improved transduction of target cells
We wished to connect two or three knob-binding DARPins with one Her2-binding DARPin within a single molecule (Fig. 1b). Since the exact position of the 2E6-binding site on the mutated knob and the orientation of the knob-bound DARPins with respect to each other were unknown, we used an empirical approach to optimize the length of the linker connecting two adjacent 2E6 DARPins within an adapter. For this purpose, we designed a panel of bivalent derivatives of the type 2E6-Ln-2E6-L3-G5, in which two copies of 2E6 were connected with a flexible linker and n is the number of Gly4Ser repeats in the linker. Six such proteins were designed with one to six copies of the Gly4Ser repeat.
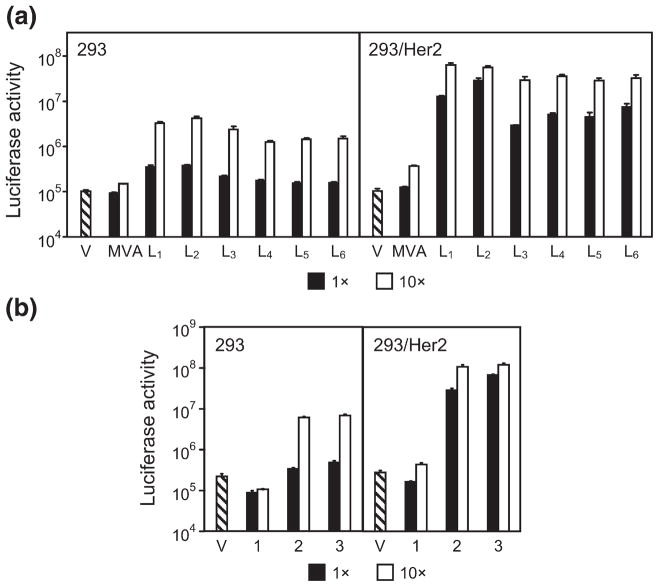
Transduction of cultured cells with the CAR-ablated Ad5LucFΔTAYT vector preincubated with each of these adapters at various adapter-to-fiber ratios yielded several important findings (Fig. 5). First, it showed that compared to the monovalent 2E6-G5, each of the bivalent derivatives drastically improved the efficacy of transduction in Her2-expressing cells (Fig. 5a). For instance, at the lowest adapter-to-fiber ratio of 1, the activity of the vector-expressed transgene was increased 25- to 246-fold, depending on the adapter configuration (linker length L1 – L6). At the same adapter-to-fiber ratio of 1, the increase in transduction of Her2-negative cells was insignificant (1.7- to 4.1-fold). When bivalent adapters were tested at an adapter-to-fiber ratio of 10, transduction of 293/Her2 cells was increased 88- to 192-fold compared to that enabled by the monovalent 2E6-G5 adapter. At this ratio, we noted a significant increase in the transduction of the control 293 cells: compared to the levels of Fluc activity seen with the monovalent 2E6-G5 adapter, the use of bivalent constructs resulted in an 8- to 28-fold increase in nonspecific gene transfer. Despite this increase, the levels of gene transfer for a given adapter–virus complex in target 293/Her2 cells were much higher (12- to 28-fold) than those in nontarget 293 cells. This experiment identified the 2E6-L2-2E6-L3-G5 adapter, which is abbreviated as (2E6)2-G5, as the most efficient mediator of viral infection.
Fig. 5.
Optimization of gene transfer. (a) Bivalent targeting adapters enable improved Ad transduction of Her2-expressing cells. V, virus alone; MVA, monovalent 2E6-G5 adapter, virus in complex with the control monovalent adapter 2E6-G5; L1-L6, virus in complex with one of the six bivalent and bispecific adapters (2E6-Ln-2E6-L3-G5) each containing two copies of 2E6 connected with linkers of various length. L1 denotes Gly4Ser, L2 denotes (Gly4Ser)2, etc. Adapter-to-fiber ratios are shown below the graph. (b) Increased valency of targeting adapter yields improved transduction of Her2-expressing cells. Transgene expression by the vector alone (V) or by the vector in a complex with mono- (1), bi- (2) or trivalent (3) adapters in which the 2E6 DARPin modules are spaced by a (Gly4Ser)2 linker. Transgene expression by CAR binding-ablated Ad in cultured cells is shown as activity of luciferase reporter in the lysates of infected cells displayed in relative light units. Error bars show standard deviations calculated for triplicate data points.
On the basis of these results, we further modified this adapter to include the third copy of the 2E6 DARPin to see whether an additional gain in transduction could be achieved. As shown in Fig. 5b, at an adapter-to-fiber ratio of 1, this trivalent adapter, 2E6-L2-2E6-L2-2E6-L3-G5 (abbreviated as (2E6)3-G5), mediated a 2.4-fold higher transduction of 293/Her2 cells than the bivalent (2E6)2-G5 molecule. In comparison, nonspecific transduction of Her2-negative cells enabled by the (2E6)3-G5 adapter was increased 1.4-fold. This result indicates that the level of gene transfer to 293/Her2 cells at this ratio nearly reached the maximum attainable under the conditions of the experiment, as suggested by the reporter activities seen at the higher adapter-to-fiber ratio of 10. At this higher ratio, we noted essentially the same transduction level of nontarget 293 cells with the trivalent adapter compared to that seen with the bivalent (2E6)2-G5.
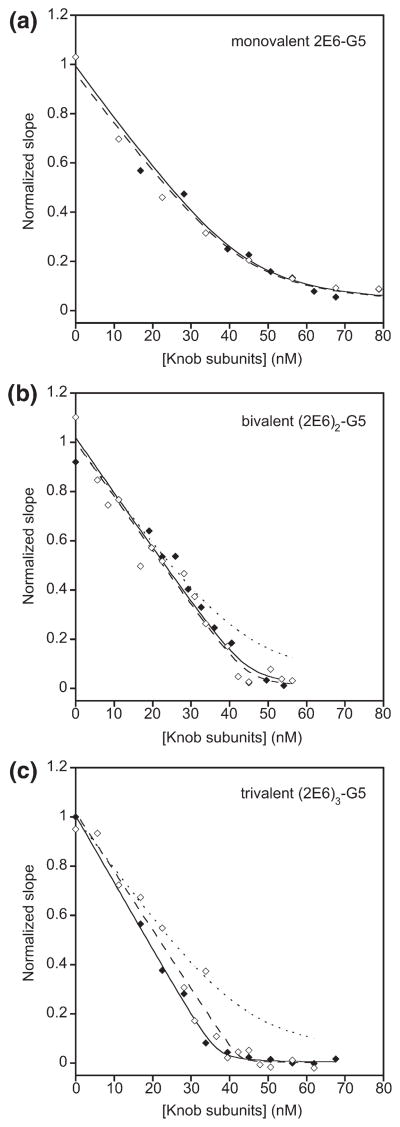
In parallel, we wished to investigate whether the bivalent and trivalent adapters would bind the knob as a chelate; in other words, whether these adapters would show the expected avidity effect of higher valency when binding to a trimeric knob in solution. For this purpose, in an SPR experiment, we coated a biosensor chip with biotinylated knobΔTAYT protein and preincubated tested adapters with increasing amounts of nonbiotinylated knobΔTAYT in solution. The measured initial association rate reflected free adapters in solution at a given inhibitory knob concentration. The rates, plotted as a function of knob concentration, thus allowed determination of the equilibrium between the mono- or multivalent adapters and the knob in solution. When the monovalent adapter 2E6-G5 was titrated with knob, a KD of 2.4 nM was obtained (Fig. 6a), which was in reasonable agreement with the KD of 1.35 nM determined from direct binding kinetics (Supplementary Data Fig. 4b). Titration of bivalent (2E6)2-G5 and trivalent (2E6)3-G5 with knob decreased the linear rate to zero (Fig. 6b and c), with an extremely sharp bend, a tell-tale sign for a KD much tighter than the constant concentration of DARPin used (14–18 nM). The limited sensitivity of the ProteOn instrument required us to do the experiment at these high concentrations, so we could not determine the apparent KD very precisely; instead, we estimated it from simulating the curve and found it to be significantly better than 10−10 M. We could not ascertain, however, whether the trivalent DARPin adapter engaged with two or three knob subunits. Importantly, the curves in Fig. 6b and c are very different from that of the monovalent binder (shown as a dotted line), suggesting that at least bivalent binding to one knob must occur. Thus, chelation of the knob in solution causes this binding improvement of the multivalent DARPins above the monovalent DAR-Pin.
Fig. 6.
Functional affinity of the monovalent, bivalent and trivalent adapters. Competition experiments in solution were performed using SPR technology. Constant concentrations of the corresponding adapter (14–18 nM) were first incubated with increasing amounts of knobΔ-TAYT in solution. Then the binding of free adapter to the knobΔTAYT immobilized on a biosensor chip was measured. The initial on-rates plotted against the total soluble knob subunit concentration and the corresponding fits are shown for the monovalent 2E6-G5 (a), bivalent (2E6)2-G5 (b) and trivalent (2E6)3-G5 (c). In each panel the data obtained in two replicate experiments are depicted with open and filled symbols. The dotted line in (b) and (c) is the fit of the monovalent adapter copied from (a).
We verified also that the bivalent and trivalent adapters show a decreased off-rate from immobilized knob (Supplementary Data Fig. S6). Under these conditions, the off-rate of the bivalent adapter is decreased by ~50-fold compared to the monovalent adapter, while the off-rate of the trivalent adapter is decreased by ~100-fold. Even though we cannot ascertain whether this improved off-rate is due to the chelation of a trimeric knob by a single bi- or trivalent adapter molecule, we note that these data are fully consistent with the measurements in solution illustrated by Fig. 6, which can be explained only as knob chelation by the multivalent adapters.
Her2 specificity of targeted adapter–vector complex was determined by the DARPin ligand
Having demonstrated the key contribution of the tight-binding multivalent knob-specific component of the designed adapters to their desired functionality, we wished to confirm that the DARPin G5, used as a Her2-targeting ligand, determined the specificity of adapter-enabled Ad transduction. To this end, we designed a truncated version of the (2E6)2-G5 adapter, the (2E6)2-L molecule, which lacked the anti-Her2 DARPin, and tested it in combination with the Ad5LucFΔTAYT vector in a gene transfer experiment. Furthermore, to obtain additional evidence of the G5 involvement in Her2 binding and enabling of Ad infection, we used free G5 as an inhibitor of infection. The E2-5 DARPin,38 which has no affinity for Her2, was used as a negative control. Consistent with our design strategy, this experiment confirmed that the Her2-mediated infection was enabled by the G5 ligand within the adapter, as evidenced by the use of the G5-containing adapter but not its G5-lacking analog. This conclusion was supported by the substantial decrease in transduction of 293/Her2 cells by the Ad complex with the (2E6)2-G5 by free DARPin G5 but not by the control DARPin E2-5 (Fig. 7).
Fig. 7.
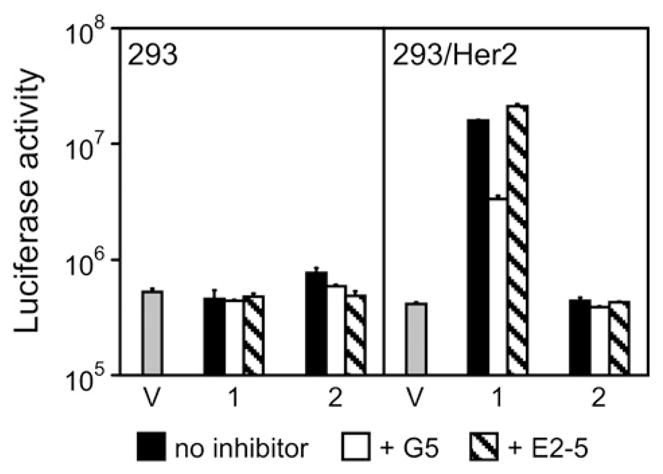
Her2-binding G5 ligand is essential to the specificity of adapter-mediated transduction. Shown is the activity of luciferase reporter in relative light units in the lysates of cells transduced with virus alone (V) or in complex with either the (2E6)2-G5 (1) or its truncated analog (2E6)2-L (2) that lacks the G5 ligand. In addition, cells were preincubated with no inhibitor, free G5, or a free unrelated DARPin (E2-5), as shown below the graph. Error bars show standard deviations calculated for triplicate data points.
Discussion
The goal of this study was to develop a novel, versatile strategy for modification of the natural receptor specificity of Ad5 vector, which could eventually be used to target therapeutic and imaging Ad5-based gene vectors to disease-related biomarkers for the purpose of gene therapy and molecular-genetic imaging.
The argument that is sometimes used against the development of the adapter-mediated targeting strategies is that because these approaches almost always yield complexes formed via noncovalent association of the targeting adapters with the viral particles (VPs), the stability of the resultant complexes might be insufficient under conditions of in vivo gene transfer. Two important considerations make this argument questionable.
First, viruses rely on kinetic stability against unfolding and disassembly of the coat proteins,51,52 and adenovirus is no exception, as none of the protein components of the Ad particle is engaged in covalent interaction with another viral protein. This lack of covalent bonding contributes to the functional plasticity of the particle and enables its stepwise dismantling upon cell entry, which is quintessential for the proper trafficking of the virus inside the cells and the release of its genome into the nucleoplasm. Covalent association of the targeting molecule is not required provided its noncovalent binding is tight enough.
The second counterargument is supported by the recently discovered mechanism of highly efficient (albeit undesirable) transduction of the liver tissue by systemically administered Ad5 vectors. Under these challenging delivery conditions, the virus is able to transduce liver hepatocytes by bonding noncovalently with FX, which functions as a bispecific targeting adapter directing the virus to FX receptors present on hepatocytes.53
With this reasoning in mind, we applied the DARPin technology to develop a protein module that binds with high affinity to the knob domain of the Ad5 fiber protein, which had been mutated to prevent the virus binding to its natural receptor, CAR. To create an adapter that binds to the knob tightly enough for in vivo applications, we designed bi- and trivalent derivatives of the original adapter, where several DARPins chelate the trimeric knob.
Genetic fusion of the selected monovalent, low nanomolar knob-binding DARPin 2E6 with the previously developed Her2-binding DARPin G5 yielded a first-generation targeting adapter that mediated Her2-dependent transduction by the Ad vector containing mutated fiber only at high adapter-to-fiber ratios (Fig. 3). The bi- and trivalent derivatives of the original adapter showed an improved stability of adapter–virus complexes, which yielded significant improvement in transduction efficiency at adapter-to-fiber ratios of 1 and 10 (Fig. 5).
The described approach compares favorably to previously reported adapter-mediated targeting strategies for Ad (G. M., N. B. & V. K., unpublished results).12,13,24–35,54–56 First, its most significant advantage is the ease with which the DARPin-based targeting adapters can be assembled, expressed and purified. For instance, without any optimization of the basic procedure established for individual DARPins, we were able to produce over 50 mg of highly purified trivalent adapter protein from a liter of induced bacterial culture. Assuming a 100% occupancy of Ad fibers with such adapters, this amount is sufficient to target approximately 4× 1016 Ad virions, which is enough to support a midscale phase I gene therapy clinical trial. As described in Materials and Methods, production of these proteins did not require the use of expensive eukaryotic cell cultivation technology or any elaborate purification technique. While bispecific antibodies can be generated in a variety of recombinant formats,57–61 their manufacturing is somewhat challenging. Also, it is more difficult to achieve trivalent binding to the knob using bispecific antibodies and to combine this with another binding site for the cellular target. The tendency of some multivalent antibody constructs to aggregate at high concentrations would make the process of developing antibody-based targeting adapters very challenging. In contrast, the multi-specific DARPin adapters are expressed in the desired functional form, and thus their manufacturing is straightforward.
Second, compared to the targeting adapters derived from soluble CAR proteins,27,28,32 binding of the DARPin-based adapters to the virus does not require wild-type unmodified fiber. This is important because, in our strategy, unavoidable dissociation of some adapter–Ad complexes does not yield infectious CAR-tropic virions that would compromise targeting delivery in vivo. Also, the DARPin-derived adapters compare favorably with the targeting adapters derived from FX, which tend to be displaced from the intravenously administered adapter–Ad complexes with the FX molecules present in the blood at high concentrations, thus resulting in nontargeted virions robustly transducing the liver.36
Third, the growing range of target molecules to which specific DARPins are available,39,46,62–64 and the ease with which DARPin-based adapters can be generated to bind to essentially any target make designing adapters with new specificities straightforward. In addition, chemical modification of DARPin-derived proteins47 with radionuclides might be used to facilitate monitoring of the in vivo distribution of adapter–Ad complexes, and modification with polyethylene glycol to protect these vector complexes from undesired interactions with nontarget tissues.
Fourth, separate production of the Ad vector and the adapter described here makes these processes independent and facilitates preparation of the targeting complexes with desired target specificity and transgene payload from separately produced components. This represents a practical alternative to strategies in which Ad fibers are fused with targeting ligands and new vectors have to be generated for each desired target. While it has been reported to be successful for some fusions,54,55 this could not be achieved in others (G. M., N. B. & V. K., unpublished results).24
As described, the reported transduction strategy might be useful as an efficient method of in vitro transduction, including gene delivery to cell types that are difficult to transfect. This might be useful in high-throughput screening of pharmacological compounds or studies of complex genetic pathways.
Despite the advantages it offers, this strategy will likely need to be further improved to meet the challenges of efficient and targeted gene delivery in vivo. Three factors potentially limiting the in vivo performance of adapter–Ad complexes will have to be addressed: (1) functional affinity of the adapter for the target; (2) functional affinity of the adapter for the fiber knob; and (3) preventing the undesirable FX-mediated transduction of the liver by the targeted complex.
The affinity of the targeting adapter for Her2 could be accomplished by replacing the Her2-targeting ligand G5 within the designed adapter with a DARPin that has a higher affinity. For instance, the Her2-specific DARPin G3, whose affinity for Her2 is 90 pM (compared to 670 pM for G5), could be used.42 To put these affinities into perspective, Wang et al. recently showed that a 60-fold improvement in the affinity of Ad fiber protein for its target receptor (from 10.3 nM to 0.61 nM) yielded a sixfold increase in targeted tumor transduction by the intravenously injected Ad vector.65 Alternatively, if the ligand replacement alone proves insufficient, further gains in affinity could be achieved by incorporating several copies of the ligand.
In addition to attempt to further increase the affinity of the DARPins for the Ad fiber knob by more rounds of directed evolution, additional insight for rational engineering might be obtainable from future structural studies. Such information could be useful in further fine-tuning of the length of the linker connecting individual DARPins within the adapter. An optimized linker design would reduce the probability of undesirable cross-linking of Ad virions by adapters.
The unintended hepatic transduction by the targeted adapter–Ad complex will require modification of the virus hexon protein. This could be accomplished by replacement of all hypervariable regions (HVRs) within the Ad5 hexon with the HVRs of hexons of Ad serotypes that do not bind FX, as described.7,53 Alternatively, the same goal can be achieved by replacement of selected amino acids within the Ad5 HVR5 that are predicted to be involved in FX binding.53,66
Our gene transfer studies showed some level of transduction of nontarget 293 cells by the multivalent adapter–virus complex (Fig. 5). While we could not detect Her2 in the 293 cells by western blotting or flow cytometry (G. M., N. B and V. K., unpublished), the possibility of expression of Her2 by these cells below the detection limits of these methods cannot be ruled out. The fact that Gensler et al. detected Her2 in these cells after immunoprecipitation suggests that there is some level of Her2 expression, which is fairly low.67 This low-level expression of Her2 in 293 cells could be an explanation of the observed transduction. Similarly, an absolute specificity of the G5 DARPin for Her2 cannot be claimed, allowing for some level of off-target binding. However minute these effects could be, they would be amplified dramatically through virus replication in the 293 cells, which would yield a several thousand-fold increase in the number of copies of the reporter transgene at the time of the assay.
In conclusion, in this study we developed a new strategy of altering the natural tropism of the Ad5 virion by modifying the viral particle with rationally designed targeting protein adapters. This strategy is likely to facilitate further development of Ad-based gene vectors for high-throughput assays in vitro and for genetic interventions in humans by allowing for disease site-specific delivery of genes of therapeutic relevance and imaging reporter genes.
Materials and Methods
Expression and purification of recombinant knob proteins
Three variants of the Ad5 fiber knob protein (UniProt P11818, amino acids 387 – 581) were developed, each with a deletion ΔTAYT (amino acids 489 – 492) that ablates binding to the CAR. The knobΔTAYT protein containing an MA(H)6 tag at the N-terminus was expressed in E. coli BL21(DE3)pLysS (Stratagene, La Jolla, CA) using pET20b (Novagen, Madison, WI). The biotinylated knobΔTAYT protein carrying an N-terminal MRGS(H)6 tag followed by an AviTag (Avidity, Aurora, CO) was expressed in E. coli XL1-Blue MRF’ (Stratagene) using the vector pQE30 (Qiagen, Valencia, CA), while the knob version with an N-terminal MA(H)6 tag followed by an AviTag was expressed in E. coli BL21(DE3) (Stratagene) using pET20b. To produce the biotinylated proteins, the host strains were transformed with plasmid pBirA (Avidity) expressing protein biotin ligase. The knob proteins were expressed as described earlier56 and were purified using a combination of IMAC and SEC as described below for DARPins.
Selection of knob-binding DARPins
Selections of knob binders were carried out using the libraries of DARPins containing either two or three randomized repeats37,38 between the flanking repeats. Purified biotinylated knobΔTAYT proteins containing an MRGS(H)6 tag followed by an AviTag at its N-terminus (in the early rounds) or an MA(H)6 tag followed by an AviTag (in the later rounds) were used as targets, the latter target was constructed in order to distinguish the knob from the MRGS(H)6-containing DARPins in assays. Ribosome display was done essentially as described,68,69 but with minor modifications (Supplementary Data, Methods).70 For affinity maturation, error-prone PCR using the nucleoside triphosphate analogs 6-(2-deoxy-β-D-ribofura-nosyl)-3,4-dihydro-8H-pyrimido-[4,5-c]-[1,2]oxazin-7-one triphosphate (dPTP) and 8-oxo-deoxyguanosine triphosphate (8-oxo-dGTP) and off-rate selection was done essentially as described,42,71 using recently summarized considerations for optimal affinity maturation.72
Expression and purification of DARPins
DARPins were expressed either in a 96-well format in E. coli XL1-Blue essentially as described,39 or in larger suspension cultures as follows. Cells were grown in 50 –500 ml of 2YT medium containing 1% (w/v) glucose and 100 μg/ml ampicillin to an absorbance at 600 nm (A600 nm) of 0.8 – 1.0, at which point protein expression was induced with 1 mM isopropylthiogalactoside (IPTG) and growth continued for 3 – 5 h at 37 °C. Cells were harvested by centrifugation and resuspended in 8 ml of TBS-W (50 mM Tris, 400 mM NaCl, 20 mM imidazole and 10% (v/v) glycerol, pH 7.4) additionally containing 1 mg/ml lysozyme per pellet of a 100 ml culture. Cells were disrupted by sonication or with a high-pressure cell disruption system (Constant Systems Ltd., Northants, UK), and the lysate was cleared by centrifugation at 15,000g for 20 min at 4 °C. The cleared lysate was passed over a 500 μl Ni2+-nitrilotriacetic acid (NTA-agarose) column (Qiagen, Valencia, CA) equilibrated with TBS-W. After washing with 20 ml of TBS-W buffer, the bound DARPins were eluted by TBS-W containing 250 mM imidazole. The proteins were dialyzed twice for 2 h against phosphate-buffered saline (PBS; 137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 2 mM KH2PO4, pH 7.4). Some of these preparations were further purified using SEC on a Superdex 200 10/300 column (GE Healthcare Biosciences, Pittsburg, PA).
Genes encoding mono- and multivalent adapters tagged with MRGS(H)6 at their N-termini were assembled in pET20b and expressed in E. coli BL21(DE3)pLysS, and the proteins were purified using the combination of IMAC and SEC described above.
Biotinylated DARPins were expressed in E. coli XL1-Blue, which contained pBirA, using derivatives of pAT224,37 pBD001 and pBD002 (see Supplementary Data, Methods and Fig. S1). This expression yielded DARPins with an N-terminal AviTag and a C-terminal (His)6 tag. The biotinylated proteins were expressed and purified using IMAC, as described above.
Nomenclature
In multivalent proteins (Fig. 1f–h), DARPins were separated by flexible (Gly4Ser)n linkers, which are abbreviated as Ln where n is the number of (Gly4Ser) repeats; for instance, 2E6-L3-G5 refers to the anti-knob DARPin 2E6 separated by (Gly4Ser)3 from the anti-Her2 DARPin G5. Several bivalent proteins of the form 2E6-Ln-2E6-L3-G5 have been tested. We abbreviated the final monovalent construct 2E6-L3-G5 as 2E6-G5 (Fig. 1f), the final bivalent construct 2E6-L2-2E6-L3-G5 as (2E6)2-G5 (Fig. 1g), and the final trivalent construct 2E6-L2-2E6-L2-2E6-L3-G5 as (2E6)3-G5 (Fig. 1h).
Affinity measurements
For the analysis of monovalent 2E6-G5, bivalent (2E6)2-G5 and trivalent (2E6)3-G5 adapters binding to the knob in solution, SPR measurements were done with a ProteOn XPR36 instrument (Bio-Rad Laboratories, Hercules. CA). In each case, one channel of a NeutrAvidin NLC sensor chip (Bio-Rad) was immobilized with the biotinylated knobΔTAYT protein (~200 RU) in the vertical orientation of the fluidic system at a flow rate of 25 μl/min. PBS containing 0.005% (v/v) Tween 20 was used both as the running buffer and for incubations. The corresponding adapter (14 – 18 nM) was first incubated with various amounts of nonbiotinylated knobΔTAYT (0 – 180 nM knob, expressed as the concentration of subunits) for 1 h at room temperature. Then the mixtures were injected in the horizontal orientation of the fluidic system at a flow rate of 30 μl/min and the interaction between the free adapter and biotinylated knobΔTAYT immobilized on the chip was measured for 12 min at 20 °C. At least two independent experiments were done with each adapter.
All sensorgrams were processed using the integrated ProteOn Manager software (Bio-Rad), and the signal of an uncoated channel reference interspot was subtracted from the sensorgrams. The initial slopes of the association curves in the linear phase were plotted against the concentrations of knob in solution (expressed as the concentration of subunits) and KD values were obtained with KaleidaGraph software (Synergy Software, Reading, PA) as described.73,74
Size-exclusion chromatography combined with multiangle light-scattering (SEC-MALS) of knob–DARPin complexes
The mass of each DARPin–target complex was determined using a liquid chromatography system (Agilent LC1100, Agilent Technologies, Santa Clara, CA) coupled to an Optilab rEX refractometer (Wyatt Technology, Santa Barbara, CA) and a miniDAWN three-angle light-scattering detector (Wyatt Technology). For protein separation, a 24 ml Superdex 200 10/30 column (GE Healthcare Biosciences, Pittsburg, PA) was run at 0.5 ml/min in PBS. For the determination of the stoichiometry of the DARPin 2E6-knob complex, 50 μl of solution containing 5 μM trimeric knobΔTAYT protein premixed with various concentrations of DARPin (3.75 – 90 μM) was injected. Analysis of the data was done with the ASTRA software (version 5.2.3.15; Wyatt Technology).
Determination of stoichiometry of DARPin–knob complexes by SEC and SDS-PAGE
Samples comprising either the DARPin 2E6 (50–200 μg) or the knobΔTAYT protein (200 μg), or mixtures of both proteins at molar ratios of 1, 2, 3 and 4 were run on a Superdex 10/300 column exactly as described above. To determine the DARPin/knob stoichiometry in a complex formed at a DARPin/knob mixing molar ratio of 4, a fraction corresponding to the relevant peak of absorbance was collected. Duplicate aliquots of this fraction were then separated on a 4% – 12% (w/v) gradient polyacrylamide/SDS gel side by side with duplicate aliquots of DARPin/knob mixtures prepared at ratios of 1, 2, 3 and 4, which were used as quantitative standards. After separation, the gel was stained with GelCode Blue Stain Reagent (Pierce Protein Research Products, Rockford, IL), destained, and scanned at 800 nm in an Odyssey scanner (Odyssey, Lincoln, NB). The intensity of the bands corresponding to the knob and DARPin 2E6 in standard mixtures was measured and used to establish a linear relationship between the A800 of a band and the amount of protein in that band. This relationship was used to calculate the DARPin/knob stoichiometry in the fraction corresponding to the complex peak collected during SEC.
Isothermal titration calorimetry (ITC)
ITC experiments were performed on a VP-ITC instrument (MicroCal, Inc., Northampton, MA). The calorimeter was calibrated according to the manufacturer’s instruction. The protein samples were prepared in, and thoroughly dialyzed against, the same batch of buffer to minimize artifacts due to minor differences in buffer composition. The concentration of the tested proteins was determined after the samples underwent dialysis and were passed through a 0.22 μm pore-size filter. The sample cell (1.4 ml) was loaded with 34 μM knob protein and the concentration of DARPin 2E6 in the syringe was 770 μM. The titration experiment at 25 °C consisted of 27 injections, each of 10 μl volume and 10 s duration, with a 5 min interval between injections. The stirring rate was 300 rpm. Raw data were integrated, corrected for nonspecific heats, normalized for concentration, and analyzed according to a binding model assuming a set of identical and independent binding sites.75
Cells and cell culture
The human embryonal kidney 293 cell line was purchased from the American Type Culture Collection (ATCC; Manassas, VA); the cell derivatives stably expressing either the wild-type Ad5 fiber (denoted 293/F28) or human Her2 (denoted 293/Her2) were developed by us and described in our earlier reports and the cells were grown in monolayer cultures as described in those reports.54,55
Viruses
The genome of the human Ad serotype 5 vector Ad5Luc1 is identical with the wild-type Ad5 genome except for the E1 region, which is replaced with a firefly luciferase Fluc gene, whose transcription is driven by an immediate-early promoter of cytomegalovirus.76 The wild type fiber gene in the Ad5Luc1 genome was replaced with the gene encoding for a ΔTAYT-mutant fiber, and its complete deletion yielded the genomes of Ad5LucFΔ-TAYT and Ad5LucΔF, respectively. The genomes of Ad5LucFΔTAYT and Ad5LucΔF were assembled in rescue plasmid vectors using homologous DNA recombination in E. coli BJ5183, as described.77 To rescue the viruses, these genomes were excised from the plasmids by restriction endonuclease cleavage and were used to transfect the wild-type fiber-expressing 293/F28 cells. Incorporation of the wild-type fiber expressed by the 293/F28 cells into the progeny virions made them infectious and their continuous propagation possible. The last cycle of amplification was performed in the 293 cells, which yielded the final preparations of Ad5LucFΔTAYT and Ad5LucΔF, containing the mutated fibers or no fibers, respectively.
All viruses were purified by equilibrium centrifugation in CsCl gradients and their particle titers were determined using established methods.54,55
Test of DARPins binding to Ad virions
The wells of a streptavidin-coated 96-well plate were pre-absorbed with purified biotinylated DARPins (500 nmol/well) and blocked with casein. The amount of viruses added was 1010 VPs per well. The DARPin-bound Ad virions were detected with anti-Ad rabbit polyclonal antibodies (ATCC; Manassas, VA) followed by polyclonal swine anti-rabbit immunoglobulin antibody horseradish peroxidase (HRP) conjugate (Dako, Glostrup, Denmark), both used at a 1:1000 dilution. After addition of o-phenylenediamine dihydrochloride (Sigma-Aldrich, St. Louis, MO), color development was stopped with 1 M HCl. The A490 of the plates was read in a DTX880 multimode detector (Beckman-Coulter, Palo Alto, CA) and the absorbance measured in the wells containing casein was used to determine background signals.
Gene transfer
We infected 105 cells seeded in the wells of 24-well plates at a multiplicity of infection of 100 VP/cell with the virus alone or the virus incubated for 1 h at room temperature with a targeting adapter or a control protein. Then the virus-containing medium was aspirated and replaced with fresh medium, and the cells were incubated in a 5% (v/v) CO2 atmosphere at 37 °C for 24 h to allow for transgene expression. At this point, the medium was removed, and the cells were washed with PBS and lysed in Reporter Lysis Buffer (Promega, Madison, WI). We measured the Fluc activity in these lysates using Promega’s luciferase assay system with the Reporter Lysis Buffer kit and a tube luminometer (Sirius; Berthold Detection Systems, Pforzheim, Germany) as described.55
Supplementary Material
Acknowledgments
We thank Christian Hess and Andreas Bosshart for their help with the large-scale protein production. We thank Dr Annemarie Honegger and David Bier for help with preparing images. We are grateful to the personnel of the DNA Analysis Facility at The University of Texas MD Anderson Cancer Center for their help with DNA sequencing. We thank Sue Moreau for editing this manuscript.
This work was supported by Public Health Service grants R01 CA116621 and R01 CA128807 (to V.K.). Additional support was provided by the University Cancer Foundation at The University of Texas M.D. Anderson Cancer Center (to V.K.), the University of Zurich (to A.P and B.D.), the Schweizerische Nationalfonds (SNF) grants 31-128671/1 (to A.P.) and 31-115982 (to I. J.), and the Cancer Center Support grant P30 CA16672.
Abbreviations used
- Ad
adenovirus
- CAR
coxsackie Ad receptor
- FX
blood coagulation factor X
- DARPins
designed ankyrin repeat proteins
- Her2
human epidermal growth factor receptor type 2
- MALS
multi-angle light-scattering
- IMAC
immobilized metal ion affinity chromatography
- SPR
surface plasmon resonance
- Fluc
firefly luciferase
- RI
refractive index
- ITC
isothermal titration calorimetry
Footnotes
Supplementary data to this article can be found online at doi:10.1016/j.jmb.2010.10.040
References
- 1.Amalfitano A, Parks RJ. Separating fact from fiction: assessing the potential of modified adenovirus vectors for use in human gene therapy. Curr Gene Ther. 2002;2:111–133. doi: 10.2174/1566523024605618. [DOI] [PubMed] [Google Scholar]
- 2.Barouch DH, Pau MG, Custers JH, Koudstaal W, Kostense S, Havenga MJ, et al. Immunogenicity of recombinant adenovirus serotype 35 vaccine in the presence of pre-existing anti-Ad5 immunity. J Immunol. 2004;172:6290–6297. doi: 10.4049/jimmunol.172.10.6290. [DOI] [PubMed] [Google Scholar]
- 3.Kobinger GP, Feldmann H, Zhi Y, Schumer G, Gao G, Feldmann F, et al. Chimpanzee adenovirus vaccine protects against Zaire Ebola virus. Virology. 2006;346:394–401. doi: 10.1016/j.virol.2005.10.042. [DOI] [PubMed] [Google Scholar]
- 4.Sullivan NJ, Geisbert TW, Geisbert JB, Xu L, Yang ZY, Roederer M, et al. Accelerated vaccination for Ebola virus haemorrhagic fever in non-human primates. Nature. 2003;424:681–684. doi: 10.1038/nature01876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burton JB, Johnson M, Sato M, Koh SB, Mulholland DJ, Stout D, et al. Adenovirus-mediated gene expression imaging to directly detect sentinel lymph node metastasis of prostate cancer. Nat Med. 2008;14:882–888. doi: 10.1038/nm.1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kishimoto H, Zhao M, Hayashi K, Urata Y, Tanaka N, Fujiwara T, et al. In vivo internal tumor illumination by telomerase-dependent adenoviral GFP for precise surgical navigation. Proc Natl Acad Sci USA. 2009;106:14514–14517. doi: 10.1073/pnas.0906388106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Roberts DM, Nanda A, Havenga MJ, Abbink P, Lynch DM, Ewald BA, et al. Hexon-chimaeric adenovirus serotype 5 vectors circumvent pre-existing anti-vector immunity. Nature. 2006;441:239–243. doi: 10.1038/nature04721. [DOI] [PubMed] [Google Scholar]
- 8.McConnell MJ, Danthinne X, Imperiale MJ. Characterization of a permissive epitope insertion site in adenovirus hexon. J Virol. 2006;80:5361–5370. doi: 10.1128/JVI.00256-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Worgall S, Krause A, Rivara M, Hee KK, Vintayen EV, Hackett NR, et al. Protection against P. aeruginosa with an adenovirus vector containing an OprF epitope in the capsid. J Clin Invest. 2005;115:1281–1289. doi: 10.1172/JCI23135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Le LP, Everts M, Dmitriev IP, Davydova JG, Yamamoto M, Curiel DT. Fluorescently labeled adenovirus with pIX-EGFP for vector detection. Mol Imaging. 2004;3:105–116. doi: 10.1162/15353500200404100. [DOI] [PubMed] [Google Scholar]
- 11.Li J, Le L, Sibley DA, Mathis JM, Curiel DT. Genetic incorporation of HSV-1 thymidine kinase into the adenovirus protein IX for functional display on the virion. Virology. 2005;338:247–258. doi: 10.1016/j.virol.2005.04.005. [DOI] [PubMed] [Google Scholar]
- 12.Krasnykh VN, Douglas JT, van Beusechem VW. Genetic targeting of adenoviral vectors. Mol Ther. 2000;1:391–405. doi: 10.1006/mthe.2000.0062. [DOI] [PubMed] [Google Scholar]
- 13.Nicklin SA, Wu E, Nemerow GR, Baker AH. The influence of adenovirus fiber structure and function on vector development for gene therapy. Mol Ther. 2005;12:384–393. doi: 10.1016/j.ymthe.2005.05.008. [DOI] [PubMed] [Google Scholar]
- 14.Bergelson JM, Cunningham JA, Droguett G, Kurt-Jones EA, Krithivas A, Hong JS, et al. Isolation of a common receptor for Coxsackie B viruses and adenoviruses 2 and 5. Science. 1997;275:1320–1323. doi: 10.1126/science.275.5304.1320. [DOI] [PubMed] [Google Scholar]
- 15.Tomko RP, Xu R, Philipson L. HCAR and MCAR: the human and mouse cellular receptors for subgroup C adenoviruses and group B coxsackie-viruses. Proc Natl Acad Sci USA. 1997;94:3352–3356. doi: 10.1073/pnas.94.7.3352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wickham TJ, Mathias P, Cheresh DA, Nemerow GR. Integrins alpha v beta 3 and alpha v beta 5 promote adenovirus internalization but not virus attachment. Cell. 1993;73:309–319. doi: 10.1016/0092-8674(93)90231-e. [DOI] [PubMed] [Google Scholar]
- 17.Arnberg N, Edlund K, Kidd AH, Wadell G. Adenovirus type 37 uses sialic acid as a cellular receptor. J Virol. 2000;74:42–48. [PMC free article] [PubMed] [Google Scholar]
- 18.Gaggar A, Shayakhmetov DM, Lieber A. CD46 is a cellular receptor for group B adenoviruses. Nat Med. 2003;9:1408–1412. doi: 10.1038/nm952. [DOI] [PubMed] [Google Scholar]
- 19.Short JJ, Pereboev AV, Kawakami Y, Vasu C, Holterman MJ, Curiel DT. Adenovirus serotype 3 utilizes CD80 (B7.1) and CD86 (B7.2) as cellular attachment receptors. Virology. 2004;322:349–359. doi: 10.1016/j.virol.2004.02.016. [DOI] [PubMed] [Google Scholar]
- 20.Chroboczek J, Ruigrok RW, Cusack S. Adenovirus fiber. Curr Top Microbiol Immunol. 1995;199:163–200. doi: 10.1007/978-3-642-79496-4_10. [DOI] [PubMed] [Google Scholar]
- 21.Novelli A, Boulanger PA. Assembly of adenovirus type 2 fiber synthesized in cell-free translation system. J Biol Chem. 1991;266:9299–9303. [PubMed] [Google Scholar]
- 22.Hong JS, Engler JA. Domains required for assembly of adenovirus type 2 fiber trimers. J Virol. 1996;70:7071–7078. doi: 10.1128/jvi.70.10.7071-7078.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Novelli A, Boulanger PA. Deletion analysis of functional domains in baculovirus-expressed adenovirus type 2 fiber. Virology. 1991;185:365–376. doi: 10.1016/0042-6822(91)90784-9. [DOI] [PubMed] [Google Scholar]
- 24.Wickham TJ, Tzeng E, Shears LL, II, Roelvink PW, Li Y, Lee GM, et al. Increased in vitro and in vivo gene transfer by adenovirus vectors containing chimeric fiber proteins. J Virol. 1997;71:8221–8229. doi: 10.1128/jvi.71.11.8221-8229.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Magnusson MK, Hong SS, Henning P, Boulanger P, Lindholm L. Genetic retargeting of adenovirus vectors: functionality of targeting ligands and their influence on virus viability. J Gen Med. 2002;4:356–370. doi: 10.1002/jgm.285. [DOI] [PubMed] [Google Scholar]
- 26.Chen CY, May SM, Barry MA. Targeting adenoviruses with factor x-single-chain antibody fusion proteins. Hum Gene Ther. 2010;21:739–749. doi: 10.1089/hum.2009.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dmitriev I, Kashentseva E, Rogers BE, Krasnykh V, Curiel DT. Ectodomain of coxsackievirus and adenovirus receptor genetically fused to epidermal growth factor mediates adenovirus targeting to epidermal growth factor receptor-positive cells. J Virol. 2000;74:6875–6884. doi: 10.1128/jvi.74.15.6875-6884.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ebbinghaus C, Al-Jaibaji A, Operschall E, Schoffel A, Peter I, Greber UF, Hemmi S. Functional and selective targeting of adenovirus to high-affinity Fc receptor I-positive cells by using a bispecific hybrid adapter. J Virol. 2001;75:480–489. doi: 10.1128/JVI.75.1.480-489.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Haisma HJ, Kamps GK, Bouma A, Geel TM, Rots MG, Kariath A, Bellu AR. Selective targeting of adenovirus to αvβ3 integrins, VEGFR2 and Tie2 endothelial receptors by angio-adenobodies. Int J Pharm. 2010;391:155–161. doi: 10.1016/j.ijpharm.2010.02.032. [DOI] [PubMed] [Google Scholar]
- 30.Hong SS, Galaup A, Peytavi R, Chazal N, Boulanger P. Enhancement of adenovirus-mediated gene delivery by use of an oligopeptide with dual binding specificity. Hum Gene Ther. 1999;10:2577–2586. doi: 10.1089/10430349950016627. [DOI] [PubMed] [Google Scholar]
- 31.Korokhov N, Mikheeva G, Krendelshchikov A, Belousova N, Simonenko V, Krendelshchikova V, et al. Targeting of adenovirus via genetic modification of the viral capsid combined with a protein bridge. J Virol. 2003;77:12931–12940. doi: 10.1128/JVI.77.24.12931-12940.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pereboev AV, Nagle JM, Shakhmatov MA, Triozzi PL, Matthews QL, Kawakami Y, et al. Enhanced gene transfer to mouse dendritic cells using adenoviral vectors coated with a novel adapter molecule. Mol Ther. 2004;9:712–720. doi: 10.1016/j.ymthe.2004.02.006. [DOI] [PubMed] [Google Scholar]
- 33.Reynolds PN, Zinn KR, Gavrilyuk VD, Balyasnikova IV, Rogers BE, Buchsbaum DJ, et al. A targetable, injectable adenoviral vector for selective gene delivery to pulmonary endothelium in vivo. Mol Ther. 2000;2:562–578. doi: 10.1006/mthe.2000.0205. [DOI] [PubMed] [Google Scholar]
- 34.Watkins SJ, Mesyanzhinov VV, Kurochkina LP, Hawkins RE. The ‘adenobody’ approach to viral targeting: specific and enhanced adenoviral gene delivery. Gene Ther. 1997;4:1004–1012. doi: 10.1038/sj.gt.3300511. [DOI] [PubMed] [Google Scholar]
- 35.Wickham TJ, Segal DM, Roelvink PW, Carrion ME, Lizonova A, Lee GM, Kovesdi I. Targeted adenovirus gene transfer to endothelial and smooth muscle cells by using bispecific antibodies. J Virol. 1996;70:6831–6838. doi: 10.1128/jvi.70.10.6831-6838.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen CY, Barry MA. FX domain fused to single-chain antibodies improve oncolytic effect of Ad5 In vitro and In vivo. 13th Annual Meeting of American Society of Gene and Cell Therapy; Washington, DC, USA. 2010. [Google Scholar]
- 37.Binz HK, Amstutz P, Kohl A, Stumpp MT, Briand C, Forrer P, et al. High-affinity binders selected from designed ankyrin repeat protein libraries. Nat Biotechnol. 2004;22:575–582. doi: 10.1038/nbt962. [DOI] [PubMed] [Google Scholar]
- 38.Binz HK, Stumpp MT, Forrer P, Amstutz P, Plückthun A. Designing repeat proteins: well-expressed, soluble and stable proteins from combinatorial libraries of consensus ankyrin repeat proteins. J Mol Biol. 2003;332:489–503. doi: 10.1016/s0022-2836(03)00896-9. [DOI] [PubMed] [Google Scholar]
- 39.Steiner D, Forrer P, Plückthun A. Efficient selection of DARPins with sub-nanomolar affinities using SRP phage display. J Mol Biol. 2008;382:1211–1227. doi: 10.1016/j.jmb.2008.07.085. [DOI] [PubMed] [Google Scholar]
- 40.Wetzel SK, Settanni G, Kenig M, Binz HK, Plückthun A. Folding and unfolding mechanism of highly stable full-consensus ankyrin repeat proteins. J Mol Biol. 2008;376:241–257. doi: 10.1016/j.jmb.2007.11.046. [DOI] [PubMed] [Google Scholar]
- 41.Roelvink PW, Mi Lee G, Einfeld DA, Kovesdi I, Wickham TJ. Identification of a conserved receptor-binding site on the fiber proteins of CAR-recognizing adenoviridae. Science. 1999;286:1568–1571. doi: 10.1126/science.286.5444.1568. [DOI] [PubMed] [Google Scholar]
- 42.Zahnd C, Wyler E, Schwenk JM, Steiner D, Lawrence MC, McKern NM, et al. A designed ankyrin repeat protein evolved to picomolar affinity to Her2. J Mol Biol. 2007;369:1015–1028. doi: 10.1016/j.jmb.2007.03.028. [DOI] [PubMed] [Google Scholar]
- 43.Slamon DJ, Clark GM, Wong SG, Levin WJ, Ullrich A, McGuire WL. Human breast cancer: correlation of relapse and survival with amplification of the HER-2/neu oncogene. Science. 1987;235:177–182. doi: 10.1126/science.3798106. [DOI] [PubMed] [Google Scholar]
- 44.Slamon DJ, Godolphin W, Jones LA, Holt JA, Wong SG, Keith DE. Studies of the HER-2/neu proto-oncogene in human breast and ovarian cancer. Science. 1989;244:707–712. doi: 10.1126/science.2470152. [DOI] [PubMed] [Google Scholar]
- 45.Theurillat JP, Dreier B, Nagy-Davidescu G, Seifert B, Behnke S, Zurrer-Hardi U, et al. Designed ankyrin repeat proteins: a novel tool for testing epidermal growth factor receptor 2 expression in breast cancer. Mod Path. 2010;23:1289–1297. doi: 10.1038/modpathol.2010.103. [DOI] [PubMed] [Google Scholar]
- 46.Winkler J, Martin-Killias P, Plückthun A, Zangemeister-Wittke U. EpCAM-targeted delivery of nanocomplexed siRNA to tumor cells with designed ankyrin repeat proteins. Mol Cancer Ther. 2009;9:2674–2683. doi: 10.1158/1535-7163.MCT-09-0402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zahnd C, Kawe M, Stumpp MT, de Pasquale C, Tamaskovic R, Nagy-Davidescu G, et al. Efficient tumor targeting with high-affinity designed ankyrin repeat proteins: effects of affinity and molecular size. Cancer Res. 2010;70:1595–1605. doi: 10.1158/0008-5472.CAN-09-2724. [DOI] [PubMed] [Google Scholar]
- 48.Veesler D, Dreier B, Blangy S, Lichière J, Tremblay D, Moineau S, Spinelli S, Tegoni M, Plückthun A, Campanacci V, Cambillau C. Crystal structure of a DARPin neutralizing inhibitor of lactococcal phage TP901-1: comparison of DARPin and camelid VHH binding mode. J Biol Chem. 2009;384:30718–30726. doi: 10.1074/jbc.M109.037812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wen J, Arakawa T. Refractive index of proteins in aqueous sodium chloride. Anal Biochem. 2000;280:327–329. doi: 10.1006/abio.2000.4524. [DOI] [PubMed] [Google Scholar]
- 50.Wen J, Arakawa T, Philo JS. Size-exclusion chromatography with on-line light-scattering, absorbance, and refractive index detectors for studying proteins and their interactions. Anal Biochem. 1996;240:155–166. doi: 10.1006/abio.1996.0345. [DOI] [PubMed] [Google Scholar]
- 51.Zlotnick A, Stray SJ. How does your virus grow? Understanding and interfering with virus assembly. Trends Biotechnol. 2003;21:536–542. doi: 10.1016/j.tibtech.2003.09.012. [DOI] [PubMed] [Google Scholar]
- 52.Forrer P, Chang C, Ott D, Wlodawer A, Plückthun A. Kinetic stability and crystal structure of the viral capsid protein SHP. J Mol Biol. 2004;344:179–193. doi: 10.1016/j.jmb.2004.09.030. [DOI] [PubMed] [Google Scholar]
- 53.Alba R, Bradshaw AC, Parker AL, Bhella D, Waddington SN, Nicklin SA, et al. Identification of coagulation factor (F)X binding sites on the adenovirus serotype 5 hexon: effect of mutagenesis on FX interactions and gene transfer. Blood. 2009;114:965–971. doi: 10.1182/blood-2009-03-208835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Belousova N, Korokhov N, Krendelshchikova V, Simonenko V, Mikheeva G, Triozzi PL, et al. Genetically targeted adenovirus vector directed to CD40-expressing cells. J Virol. 2003;77:11367–11377. doi: 10.1128/JVI.77.21.11367-11377.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Belousova N, Mikheeva G, Gelovani J, Krasnykh V. Modification of adenovirus capsid with a designed protein ligand yields a gene vector targeted to a major molecular marker of cancer. J Virol. 2008;82:630–637. doi: 10.1128/JVI.01896-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Krasnykh VN, Mikheeva GV, Douglas JT, Curiel DT. Generation of recombinant adenovirus vectors with modified fibers for altering viral tropism. J Virol. 1996;70:6839–6846. doi: 10.1128/jvi.70.10.6839-6846.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Plückthun A, Pack P. New protein engineering approaches to multivalent and bispecific antibody fragments. Immunotechnology. 1997;3:83–105. doi: 10.1016/s1380-2933(97)00067-5. [DOI] [PubMed] [Google Scholar]
- 58.Presta LG. Molecular engineering and design of therapeutic antibodies. Curr Opin Immunol. 2008;20:460–470. doi: 10.1016/j.coi.2008.06.012. [DOI] [PubMed] [Google Scholar]
- 59.Müller D, Kontermann RE. Recombinant bispecific antibodies for cellular cancer immunotherapy. Curr Opin Mol Ther. 2007;9:319–326. [PubMed] [Google Scholar]
- 60.Marvin JS, Zhu Z. Recombinant approaches to IgG-like bispecific antibodies. Acta Pharmacol Sin. 2005;26:649–658. doi: 10.1111/j.1745-7254.2005.00119.x. [DOI] [PubMed] [Google Scholar]
- 61.Fischer N, Leger O. Bispecific antibodies: molecules that enable novel therapeutic strategies. Pathobiology. 2007;74:3–14. doi: 10.1159/000101046. [DOI] [PubMed] [Google Scholar]
- 62.Amstutz P, Koch H, Binz HK, Deuber SA, Plückthun A. Rapid selection of specific MAP kinase-binders from designed ankyrin repeat protein libraries. Protein Eng Des Sel. 2006;19:219–229. doi: 10.1093/protein/gzl004. [DOI] [PubMed] [Google Scholar]
- 63.Milovnik P, Ferrari D, Sarkar CA, Plückthun A. Selection and characterization of DARPins specific for the neurotensin receptor 1. Protein Eng Des Sel. 2009;22:357–366. doi: 10.1093/protein/gzp011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Schweizer A, Roschitzki-Voser H, Amstutz P, Briand C, Gulotti-Georgieva M, Prenosil E, et al. Inhibition of caspase-2 by a designed ankyrin repeat protein: specificity, structure, and inhibition mechanism. Structure. 2007;15:625–636. doi: 10.1016/j.str.2007.03.014. [DOI] [PubMed] [Google Scholar]
- 65.Wang H, Liu Y, Li Z, Tuve S, Stone D, Kalyushniy O, et al. In vitro and in vivo properties of adenovirus vectors with increased affinity to CD46. J Virol. 2008;82:10567–10579. doi: 10.1128/JVI.01308-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Alba R, Bradshaw AC, Coughlan L, Denby L, McDonald RA, Waddington SN, et al. Biodistribution and retargeting of FX-binding ablated adenovirus serotype 5 vectors. Blood. 2010;116:2656–2664. doi: 10.1182/blood-2009-12-260026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gensler M, Buschbeck M, Ullrich A. Negative regulation of HER2 signaling by the PEST-type protein-tyrosine phosphatase BDP1. J Biol Chem. 2004;279:12110–12116. doi: 10.1074/jbc.M309527200. [DOI] [PubMed] [Google Scholar]
- 68.Zahnd C, Amstutz P, Plückthun A. Ribosome display: selecting and evolving proteins in vitro that specifically bind to a target. Nat Methods. 2007;4:269–279. doi: 10.1038/nmeth1003. [DOI] [PubMed] [Google Scholar]
- 69.Hanes J, Plückthun A. In vitro selection and evolution of functional proteins by using ribosome display. Proc Natl Acad Sci USA. 1997;94:4937–4942. doi: 10.1073/pnas.94.10.4937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Dreier B, Plückthun A. Ribosome display, a technology for selecting and evolving proteins from large libraries. In: Park D, editor. PCR Protocols. 3. Humana Press; New York, NY: 2010. [DOI] [PubMed] [Google Scholar]
- 71.Luginbühl B, Kanyo Z, Jones RM, Fletterick RJ, Prusiner SB, Cohen FE, et al. Directed evolution of an anti-prion protein scFv fragment to an affinity of 1 pM and its structural interpretation. J Mol Biol. 2006;363:75–97. doi: 10.1016/j.jmb.2006.07.027. [DOI] [PubMed] [Google Scholar]
- 72.Zahnd C, Sarkar CA, Plückthun A. Computational analysis of off-rate selection experiments to optimize affinity maturation by directed evolution. Protein Eng Des Sel. 2010;23:175–184. doi: 10.1093/protein/gzp087. [DOI] [PubMed] [Google Scholar]
- 73.Hanes J, Jermutus L, Weber-Bornhauser S, Bosshard HR, Plückthun A. Ribosome display efficiently selects and evolves high-affinity antibodies in vitro from immune libraries. Proc Natl Acad Sci USA. 1998;95:14130–14135. doi: 10.1073/pnas.95.24.14130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zahnd C, Spinelli S, Luginbühl B, Amstutz P, Cambillau C, Plückthun A. Directed in vitro evolution and crystallographic analysis of a peptide binding scFv antibody with low picomolar affinity. J Biol Chem. 2004;279:18870–18877. doi: 10.1074/jbc.M309169200. [DOI] [PubMed] [Google Scholar]
- 75.Jelesarov I, Bosshard HR. Isothermal titration calorimetry and differential scanning calorimetry as complementary tools to investigate the energetics of biomolecular recognition. J Mol Recognit. 1999;12:3–18. doi: 10.1002/(SICI)1099-1352(199901/02)12:1<3::AID-JMR441>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 76.Krasnykh V, Belousova N, Korokhov N, Mikheeva G, Curiel DT. Genetic targeting of an adenovirus vector via replacement of the fiber protein with the phage T4 fibritin. J Virol. 2001;75:4176–4183. doi: 10.1128/JVI.75.9.4176-4183.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Chartier C, Degryse E, Gantzer M, Dieterle A, Pavirani A, Mehtali M. Efficient generation of recombinant adenovirus vectors by homologous recombination in Escherichia coli. J Virol. 1996;70:4805–4810. doi: 10.1128/jvi.70.7.4805-4810.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.