Abstract
The dicotyledon seedling undergoes organ-specific photomorphogenic development when exposed to light. The cotyledons open and expand, the apical hook opens, and the hypocotyl ceases to elongate. Using the large and easily dissected seedlings of soybean (Glycine max ‘Williams 82’), we show that genes involved in photosynthesis and its regulation dominate transcripts specific to the cotyledon, even in etiolated seedlings. Genes for cell wall biosynthesis and metabolism are expressed at higher levels in the hypocotyl, while examination of genes expressed at higher levels in the hook region (including the shoot apical meristem) reveals genes involved in cell division and protein turnover. The early transcriptional events in these three organs in response to a 1-h treatment of far-red light are highly distinctive. Not only are different regulatory genes rapidly regulated by light in each organ, but the early-responsive genes in each organ contain a distinctive subset of known light-responsive cis-regulatory elements. We detected specific light-induced gene expression for the root phototropism gene RPT2 in the apical hook and also phenotypes in Arabidopsis (Arabidopsis thaliana) rpt2 mutants demonstrating that the gene is necessary for normal photomorphogenesis in the seedling apex. Significantly, expression of the RPT2 promoter fused to a β-glucuronidase reporter gene shows differential expression across the hook region. We conclude that organ-specific, light-responsive transcriptional networks are active early in photomorphogenesis in the aerial parts of dicotyledon seedlings.
Photomorphogenic responses control a wide range of important developmental events throughout the lifetime of plants, including seed germination, deetiolation, shade avoidance, and flowering (Monte et al., 2007; Josse et al., 2008). Seedling photomorphogenesis (or deetiolation) is the phenomenon whereby a dark-grown seedling, which features an elongated hypocotyl, closed cotyledons, an apical hook, and undifferentiated chloroplasts, displays an inhibition of hypocotyl elongation, opening of cotyledons and apical hook, and chloroplast maturation after it is exposed to light. Photomorphogenesis is thus both a developmental process and a response to environmental stimuli. The timing of deetiolation is of key importance to the survival of plants. Early opening of the hook and cotyledons while still in the soil will lead to the damage of young embryos. A delayed deetiolation response may result in late initiation of photosynthesis and depletion of nutrients, ultimately limiting the ability of the seedling to survive.
The phytochrome family mediates photomorphogenesis in response to red and far-red (FR) light. In Arabidopsis (Arabidopsis thaliana), the five phytochromes (phyA–phyE) perform overlapping yet distinct physiological functions. In particular, phyA mediates the response of etiolated seedlings to FR, while phyB to phyE are largely responsible for the response to larger doses of continuous red light in etiolated seedlings (Sharrock and Quail, 1989; Clack et al., 1994; Devlin et al., 1998; Quail, 2002). The photosensory activity of the phytochromes results from their capacity to undergo light-induced, reversible switching between the biologically inactive Pr form and the biologically active Pfr form (Borthwick et al., 1952; Smith, 2000; Quail, 2002). The active Pfr form is translocated from the cytoplasm to the nucleus (Fankhauser and Chen, 2008), where it interacts with transcription factors that likely trigger changes in downstream gene expression and, subsequently, morphological changes (Ni et al., 1998; Quail, 2002).
Many regulatory factors in phytochrome signaling have been reported in recent years (Franklin et al., 2005; Monte et al., 2007; Josse et al., 2008; Sharrock, 2008), greatly increasing our understanding of molecular and cellular mechanisms of photomorphogenesis. However, another important feature of seedling photomorphogenesis, the cellular specificity of photoreceptors and photomorphogenic responses at the molecular level, is poorly understood. In seedling photomorphogenesis, for example, different organs display distinct responses to the light stimulus. During deetiolation, cell expansion occurs in the cotyledon and the concave side of apical hook, while inhibition of cell growth is observed in the hypocotyl and the convex side of apical hook. Questions such as how the same light signal triggers distinct, even opposite, responses in different tissues and organs, and whether there is coordination or communication between organs, have been of interest in the field (Bou-Torrent et al., 2008). As early as in 1995, some light-responsive genes, such as β-TUBULIN1 and SUPPRESSOR OF PHYA, were shown to be regulated organ specifically (Leu et al., 1995; Zhu et al., 2008). As the expression microarray became a standard tool for global expression analysis, the organ-specific light response began to be examined by profiling light-responsive gene expression in individual organs such as cotyledon, hypocotyl, root, and shoot apex (Ma et al., 2005; López-Juez et al., 2008). However, our knowledge of the tissue-specific regulation of light signaling and thus the regulation of photomorphogenesis at the cellular level is far from complete. For example, the apical hook plays a key role in early seedling establishment by protecting cotyledons and the meristematic primordia in the etiolated seedling during soil penetration. The timing of hook opening, therefore, is critical to the survival of the young seedling. Despite its distinct photomorphogenic behavior and importance to seedling survival, the light response of the apical hook has only recently begun to be explored (Li et al., 2004; Khanna et al., 2007). A global study of transcriptional responses to light in the apical hook has not yet been performed, to our knowledge, perhaps due to the significant difficulty of extracting sufficient RNA from this small organ in Arabidopsis seedlings.
We argue that our knowledge of the photomorphogenic control of plant development can be refined by examining the response of the transcriptome to light thoroughly in seedlings at the organ level. We performed an organ-specific expression profiling study with soybean (Glycine max ‘Williams 82’), which permits accurate expression profiling of multiple tissues thanks to its large etiolated seedlings, without resorting to RNA amplification. The response to a short FR treatment was studied in order to identify the early regulatory events as well as to eliminate the effects of photosynthesis. In this work, gene expression in the cotyledon, apical hook (including the apical meristem), and hypocotyl was compared between seedlings treated with continuous far-red light (FRc) for 1 h and dark-grown seedlings using microarrays. FRc-responsive genes were identified and their regulation by FRc was shown to be organ specific. To identify organ-specific FRc responses and to investigate the relationship between organ-specific expression and light-induced expression, a single-channel analysis was performed with the microarray data to identify genes showing expression that was significantly stronger in individual organs. Both analyses were selectively verified by quantitative real-time reverse transcription (QRT)-PCR. The deetiolation responses in FRc of Arabidopsis mutants carrying defects in an ortholog of one of the identified genes, the root phototropism gene RPT2, were examined to study the function of the gene in photomorphogenesis.
RESULTS
Global Expression Analysis Identified 27 Genes as Early FR-Responsive Genes in Soybean Seedling Photomorphogenesis
We conducted an expression profiling experiment to identify the organ-specific gene regulation triggered in the early stage of seedling deetiolation. In order to remove the potential complications of (1) signaling via multiple photoreceptor pathways and (2) gene expression responses to the initiation of photosynthesis, nonphotosynthetic FRc was given for 1 h to induce the changes in gene expression associated with deetiolation via the phyA signaling pathway (Tepperman et al., 2001). The expression profiles of the cotyledon, apical hook, and hypocotyl of soybean seedlings were compared between FRc-treated seedlings and dark controls using the soybean cDNA microarray described by Vodkin et al. (2004; Fig. 1). Four independent biological replicates, each containing the tissue of a single seedling, were used in separate hybridizations. To control for labeling differences, the dyes used for labeling the cohybridized samples (Cy3 and Cy5) were swapped in the biological replicates. Microarray data were preprocessed and normalized by Global Locally Weighted Scatterplot Smoothing and then statistically analyzed using the rank product method (Breitling et al., 2004). Genes with mRNAs differentially expressed in FR versus dark as identified by an absolute fold change of greater than 2-fold, and which were statistically significant after applying a false discovery rate (FDR) correction of 5%, were defined as “FR-responsive genes.” In total, microarray analysis identified 27 FR-responsive genes according to these criteria, including four genes identified as differentially expressed between the cotyledon plus/minus FR samples, 15 genes identified as responding in apical hook, and 10 genes identified in hypocotyl (Table I). Intriguingly, only two of the genes fulfilled these criteria in multiple tissues (Glyma02g04170 is responsive to FRc in both hook and hypocotyl, and Glyma06g14170 is FRc responsive in cotyledon and hook).
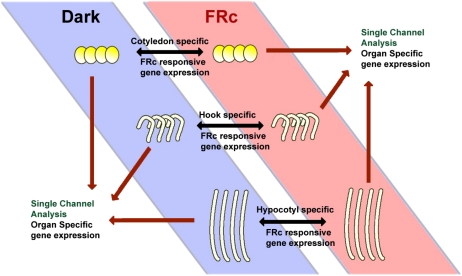
Figure 1.
Two approaches to identify differential gene expression. To identify the genes regulated by FRc in the hook, hypocotyl, and cotyledon, RNA samples from the three dark-grown soybean organs were directly compared with RNA samples from FRc-treated plant organs by hybridizing both samples on the same array (represented by the black two-headed arrows). To identify gene expression that was organ specific, regardless of light responsiveness, single-channel data from each array was extracted (represented by the single-headed dark red arrows), which represented the expression profile of an organ in a specific light treatment. Single-channel profiles of all three organs with the same light treatment were then compared with each other to identify the genes that are highly expressed in one organ compared with the other two. [See online article for color version of this figure.]
Table I. FRc-responsive genes identified by microarray analysis.
| Soybean Identifier | Organ | Log2 Fold Changea | FDR |
| % | |||
| Glyma02g42500 | Cotyledon | −1.86 | 0 |
| Glyma13g37320 | Cotyledon | 1.04 | 0 |
| Glyma01g38590 | Cotyledon | 1.22 | 0 |
| Glyma06g14170 | Cotyledon | 1.38 | 0 |
| Glyma11g03850 | Hook | −1.31 | 0 |
| Glyma08g28730 | Hook | −1.12 | 0 |
| Glyma15g16190 | Hook | −1.1 | 0 |
| Glyma11g36210 | Hook | −1.09 | 0 |
| Glyma07g21150 | Hook | −1.02 | 0 |
| Glyma02g04170 | Hook | −1.01 | 0 |
| Glyma18g05720 | Hook | 1.02 | 0 |
| Glyma11g08850 | Hook | 1.02 | 0 |
| Glyma08g45310 | Hook | 1.02 | 1.3 |
| Glyma05g34870 | Hook | 1.07 | 2.1 |
| Glyma02g13930 | Hook | 1.1 | 2 |
| Glyma09g05180 | Hook | 1.27 | 0 |
| Glyma08g45300 | Hook | 1.4 | 0 |
| Glyma06g14170 | Hook | 1.73 | 0 |
| Glyma20g28890 | Hook | 1.89 | 0 |
| Glyma02g04170 | Hypocotyl | −1.16 | 0 |
| Glyma12g13290 | Hypocotyl | 1.07 | 5 |
| Glyma17g03350 | Hypocotyl | 1.09 | 3.3 |
| Glyma20g38030 | Hypocotyl | 1.12 | 3.8 |
| Glyma15g09150 | Hypocotyl | 1.18 | 0 |
| Glyma10g33370 | Hypocotyl | 1.18 | 0 |
| Glyma07g09220 | Hypocotyl | 1.21 | 0 |
| Glyma16g03280 | Hypocotyl | 1.23 | 0 |
| Glyma11g01350 | Hypocotyl | 1.3 | 0 |
| Glyma10g12060 | Hypocotyl | 1.34 | 0 |
Base 2 logarithm of fold change (FR/dark). Positive log2 fold change value means up-regulation of the gene, while negative log2 fold change value means down-regulation.
An Organ-Specific Gene Expression Pattern in Response to FRc
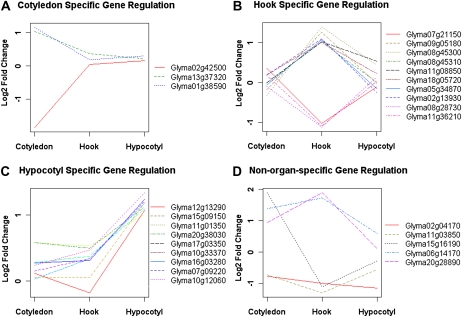
With the previously described criteria (FDR < 0.05 and fold change > 2), we found that only two genes out of the 27 FRc-responsive genes meet these criteria in more than one organ. The early FR regulation of most genes detected thus seems to be organ specific. However, it is possible that some of those 27 FRc-responsive genes are also induced or repressed by FRc in a second organ but fail to pass the arbitrary cutoff due to array noise and/or differences in statistical power. Therefore, the fold changes of the 27 FRc-regulated genes were compared across all three organs using replicated microarray data to get a clear picture of the organ specificity of FR gene response (Fig. 2). In addition to the organ where the gene was identified to be FR responsive, if the gene is also responsive in another organ with a fold change greater than 1.5 (equal to a log2 fold change of 0.6), the gene is considered to be regulated by FRc in more than one organ. Such stringent parameters for calling organ-specific gene regulation allow us to identify organ-specific gene regulation with confidence. According to these criteria, 81.5% (22 of 27) of the FR-responsive genes are regulated by FRc specifically in only one organ (Fig. 2, A–C). Ten genes were specifically regulated only in the apical hook (Fig. 2B), nine genes were regulated specifically in the hypocotyl (Fig. 2C), while only three genes were shown to be specifically regulated in the cotyledon (Fig. 2A). Some genes that are regulated in more than one organ were also identified (Fig. 2D). Overall, these results support the hypothesis that many light-triggered mRNA-level changes are likely to be tissue specific during the early stage of the transcriptional signaling cascade. The organ specificity of transcriptional light responses is a possible reason for discrepancies between the FRc-responsive genes identified in this experiment and those identified in whole-seedling Arabidopsis experiments (Tepperman et al., 2001). For example, the Arabidopsis ortholog of Glyma09g05180 (AT4G02570), which was up-regulated specifically in the apical hook by 2.41-fold in our experiment, was not found to be significantly regulated by FRc in either of the two previously published whole-seedling Arabidopsis microarray experiments (Tepperman et al., 2001; Ghassemian et al., 2006).
Figure 2.
Organ-specific regulation of mRNA levels in response to FRc. The expression level changes of genes derived from the microarray experiment were plotted as logarithm of fold change to base 2 (hence, value 0 means no change, while value +/−1 means up- or down- regulation by 2-fold) across cotyledon, hook, and hypocotyl. The majority of the genes show a FRc-induced change in expression only in one organ, cotyledon (A), apical hook (B), or hypocotyl (C), while a smaller number of genes are not regulated organ specifically (D). [See online article for color version of this figure.]
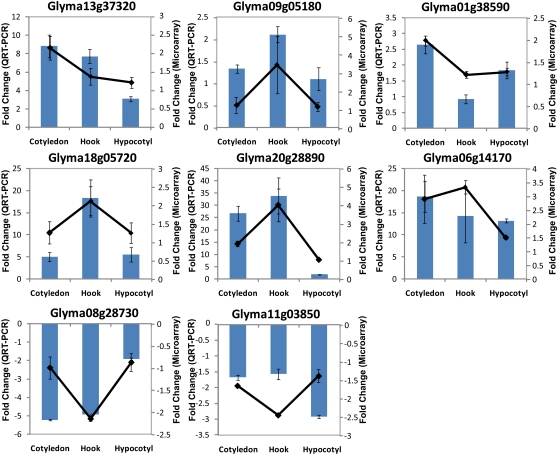
Because false-positive results from arrays of the type used for this study have been reported (Woo et al., 2004), QRT-PCR was performed to confirm the gene regulation patterns. Gene expression was monitored in 1-h FRc-treated samples versus dark controls by means of an independently conducted, replicated, and controlled set of QRT-PCR experiments. Five reference gene candidates, ubiquitin (Glyma20g27950), α-tubulin (Glyma04g09350), β-tubulin (Glyma04g02610), histone H3.2 minor (Glyma14g40590), and phosphoenolpyruvate carboxylase (Glyma13g36670), were chosen based on microarray data and literature (Tuteja et al., 2004). These five candidates were tested for their stability in expression level across three organs in both FRc and dark conditions using QRT-PCR. Histone H3.2 minor was chosen to be the reference gene because it was most consistently expressed in different organs in response to either treatment among the five candidates (Supplemental Fig. S1). Eight genes that display distinct organ-specific regulation patterns were chosen from the 27 FR-responsive genes identified by the microarray for QRT-PCR analysis. The mRNA levels of genes were measured with three biological replicates where each biological replicate was a pool of eight seedlings. Three technical replicates were performed for each biological replicate sample. The results of these highly replicated QRT-PCR experiments thus allow greater statistical power. The data were first converted to replicated expression values normalized with the internal reference gene, and then a fold change (FR/dark) was calculated, using the ΔΔCT method with experiment-determined amplification efficiency incorporated (Livak and Schmittgen, 2001). QRT-PCR results are in agreement with the organ-specific regulation of mRNA levels revealed by microarray analysis for all but one of the genes tested (Fig. 3), although the magnitude of the change observed is generally greater from the QRT-PCR. We interpret this as being the result of background hybridization to the microarray limiting the observable fold change. In the case of Glyma11g03850, a discrepancy between microarray data and QRT-PCR results was observed (Fig. 3). Although the direction of change in expression level in response to light is consistent between the QRT-PCR and microarray results for Glyma11g03850, the organ specificity of mRNA regulation is switched from hook specific to hypocotyl specific. Since the QRT-PCR was performed as an entirely separate experiment with an independently collected plant sample set, the discrepancy may be due to variation in the dissection of hook and hypocotyl between the two batches of samples, which, in the case of a gene whose expression is confined to a small group of cells (e.g. the region where hypocotyl starts and apical hook ends), could create a difference in the observed regulation pattern. Overall, the QRT-PCR results support the organ-specific regulation pattern revealed by the microarray data and hence suggest that light-responsive organ-specific transcriptional regulation early in the transcriptional cascade is part of the mechanism underlying the tissue-specific photomorphogenesis.
Figure 3.
FR-responsive mRNA expression patterns measured by microarray and by QRT-PCR. In each graph, the absolute fold change derived from microarray data (black lines, right axis) and QRT-PCR (columns, left axis) were plotted for cotyledon, hook, and hypocotyl. Positive fold change indicates higher mRNA levels in FRc compared with darkness, while negative fold change signifies lower mRNA levels in response to FRc. Error bars represent se of all biological replicates. [See online article for color version of this figure.]
Functional Annotation of the Soybean cDNA Microarray Sheds Light on the Roles of Organ-Specific FRc-Responsive Genes
To allow further investigation of the biological meaning of the microarray data, functional annotation was generated for the array probes. Functional assignments of the 27 genes of interest are listed in Table II. We identified Arabidopsis orthologs for 21 out of 27 FR-responsive genes, among which 16 genes have well-annotated functions while the other five have poorly known functions. Among the well-annotated genes, some were previously reported to be involved in light signal transduction, such as RPT2 (Glyma18g05720 [Arabidopsis ortholog AT2G30520]; Sakai et al., 2000; Inada et al., 2004) and ATHB-2 (Glyma11g03850 [Arabidopsis ortholog AT4G16780]; Ohgishi et al., 2001). Some other genes are known as downstream effectors for light response, such as chalcone synthase (Glyma11g01350 [Arabidopsis ortholog AT5G13930]), which is involved in the generation of protective anthocyanin pigments in response to light (Batschauer et al., 1991; Kubasek et al., 1992, 1998), and early light-inducible protein (Glyma20g28890 [Arabidopsis ortholog AT3G22840]). Some other genes are involved in protein regulation and modification, such as the ubiquitin-dependent protein catabolic process (Glyma09g05180 [Arabidopsis ortholog AT4G02570] and Glyma20g38030 [Arabidopsis ortholog AT1G09100]) and protein phosphorylation/dephosphorylation (Glyma12g13290 [Arabidopsis ortholog AT4G28400]), which are two known mechanisms of controlling protein activity in the light signaling pathway (Wei and Deng, 2003; Monte et al., 2007). Two genes (Glyma02g04170 and Glyma02g42500) that encode proteins with the domain with unknown function DUF231, including the freezing tolerance regulator Eskimo1 (ESK1; Xin et al., 2007), were both down-regulated by FRc. Six genes of the identified 27 FR-responsive genes (22%) do not have a clear Arabidopsis homolog identifiable by TBLASTX with an E-value cutoff of 1E-6. The annotation of identified FR-responsive genes agrees well with our current knowledge of photomorphogenesis, confirming that our microarray experiment led to the identification of photomorphogenic regulators and suggesting that not all photomorphogenesis-related genes in soybean have orthologs in Arabidopsis.
Table II. Annotation of identified FRc-responsive genes.
| Soybean Identifier | Arabidopsis Homolog | Annotation |
| Glyma02g42500 | AT3G55990 | Encodes ESK1 (Eskimo1), a member of a large gene family of DUF231 domain proteins whose members encode a total of 45 proteins of unknown function; ESK1 functions as a negative regulator of cold acclimation; mutations in the ESK1 gene provide strong freezing tolerance |
| Glyma13g37320 | AT4G28290 | Unknown protein |
| Glyma01g38590 | AT3G26300 | CYP71B34 (cytochrome P450, family 71, subfamily B, polypeptide 34); oxygen binding |
| Glyma06g14170 | AT5G24460 | Hydrolase |
| Glyma11g03850 | AT4G16780 | ATHB-2 (ARABIDOPSIS THALIANA HOMEOBOX PROTEIN 2); DNA binding/transcription factor |
| Glyma08g28730 | AT1G67265 | DVL3/RTFL21 (ROTUNDIFOLIA-LIKE21) |
| Glyma15g16190 | No hits | No hits |
| Glyma11g36210 | AT5G10180 | AST68 (sulfate transporter 2.1) |
| Glyma07g21150 | AT2G26500 | Cytochrome b6f complex subunit (petM) |
| Glyma02g04170 | AT1G60790 | Similar to unknown protein; contains InterPro domain protein of unknown function DUF231, plant (InterPro:IPR004253) |
| Glyma18g05720 | AT2G30520 | RPT2 (ROOT PHOTOTROPISM2) |
| Glyma11g08850 | AT4G35680 | Similar to unknown protein; contains InterPro domain protein of unknown function DUF241, plant (InterPro:IPR004320) |
| Glyma08g45310 | No hits | No hits |
| Glyma05g34870 | AT1G14870 | Identical to uncharacterized protein; At1g14870 contains InterPro domain Asp and Asn hydroxylation site (InterPro:IPR000152); contains InterPro domain protein of unknown function Cys-rich (InterPro:IPR006461) |
| Glyma02g13930 | No hits | No hits |
| Glyma09g05180 | AT4G02570 | ATCUL1 (CULLIN1) |
| Glyma08g45300 | No hits | No hits |
| Glyma20g28890 | AT3G22840 | ELIP1 (EARLY LIGHT-INDUCABLE PROTEIN); chlorophyll binding |
| Glyma12g13290 | AT4G28400 | Protein phosphatase 2C, putative (PP2C, putative) |
| Glyma17g03350 | No hits | No hits |
| Glyma20g38030 | AT1G09100 | RPT5B (26S PROTEASOME AAA-ATPASE SUBUNIT RPT5B); ATPase/calmodulin |
| Glyma15g09150 | AT1G63310 | Similar to oxidoreductase, acting on NADH or NADPH |
| Glyma10g33370 | AT2G33360 | Similar to unknown protein |
| Glyma07g09220 | AT4G28940 | Catalytic |
| Glyma16g03280 | No hits | No hits |
| Glyma11g01350 | AT5G13930 | ATCHS/CHS/TT4 (CHALCONE SYNTHASE); naringenin-chalcone synthase |
| Glyma10g12060 | AT5G06900 | CYP93D1 (cytochrome P450, family 93, subfamily D, polypeptide 1); oxygen binding |
Single-Channel Analysis of the Microarray Data Reveals an Organ-Specific Gene Expression Pattern
Spotted microarray data can be analyzed in multiple dimensions in order to compare samples across multiple microarrays, in addition to comparing samples physically hybridized into the same array (Dhaubhadel et al., 2007). To address whether the FR-responsive genes were also genes that are expressed in an organ-specific pattern regardless of light response, a single-channel approach was taken to reanalyze the microarray data. The gene expression profile of one organ (either the Cy3 or Cy5 channel in one hybridization array) was compared with that of other organs (profiled in the Cy3 or Cy5 channel from a second array) in either FRc-treated or dark control samples (Fig. 1). The microarray data were normalized and processed as described in “Materials and Methods.” Statistical analysis was performed again using the rank product method with FDR cutoff of 5% and a greater fold change, 4-fold cutoff, to minimize the influence of between-array noise and to identify organ-specific expression with confidence. Transcripts that are 4-fold or more abundant in one organ relative to the other two organs and show statistical significance were considered to be organ-specific transcripts.
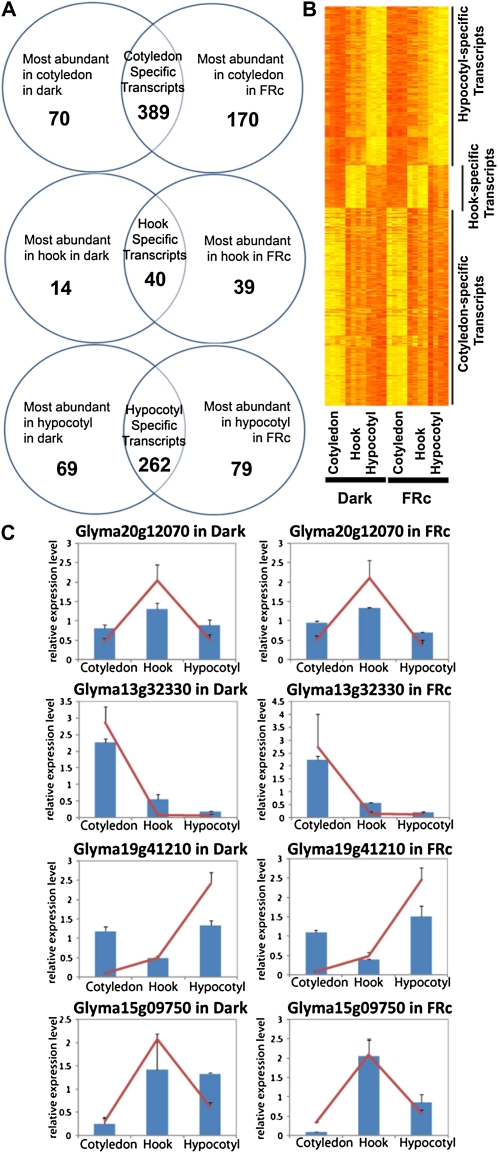
The analysis revealed that in etiolated seedlings, 459 genes are expressed at statistically significant, 4-fold higher levels in the cotyledon with respect to apical hook and hypocotyl. These 459 genes represent the cotyledon-specific transcripts in 7-d-old etiolated seedlings. In seedlings treated with FRc for 1 h, 559 transcripts are more abundant in the cotyledon compared with apical hook and hypocotyl, which represent the cotyledon-specific transcripts in the seedlings exposed to 1 h of FRc. Comparing the two gene lists led to identification of the overlapping set of 389 transcripts, henceforth referred to as “cotyledon-specific genes” in this study, which represent genes with 4-fold higher mRNA levels in cotyledon irrespective of light conditions (Fig. 4, A and B; Supplemental Fig. S2A; Supplemental Table S1). With the same process, 40 “hook-specific genes” and 262 “hypocotyl-specific genes” were identified (Fig. 4, A and B; Supplemental Fig. S2, B and C; Supplemental Tables S2 and S3). We expected to see more cotyledon-specific genes than hook-specific genes and hypocotyl-specific genes, because the cotyledon is distinct in morphology, function, and organogenesis from the other two organs in question. The result agrees well with our expectation. Some of the organ-specific transcripts were selected and confirmed by replicated QRT-PCR experiments. (The gene Glyma15g09750 was just short of the 4-fold criterion for hook specific [fold change of hook versus hypocotyl as 3.64-fold] but was included because of its potential biological significance as an ARF6-like gene.) The QRT-PCR results agreed well with those from the microarray (Fig. 4C), confirming the reproducibility of the single-channel analysis method.
Figure 4.
Identification and QRT-PCR confirmation of organ-specific transcripts. A, Organ-specific transcripts in dark-grown etiolated seedlings and seedlings treated with 1 h of FRc were identified by microarray using a single-channel approach and the rank product method. The intersection of these two groups represents the transcripts that are consistently organ specific in this experiment, used hereafter as the organ-specific gene lists. B, Heat map showing normalized expression values of organ-specific genes across three organs in two different light conditions with four biological replicates per organ per light condition. Yellow indicates high expression, while orange indicates low expression. C, Confirmation of organ-specific expression by QRT-PCR. In each graph, the relative expression levels (normalized to the mean expression level in all three organs) derived from the microarray (red lines) and QRT-PCR data (blue columns) were plotted for cotyledon, hook, and hypocotyl. The error bars represent se of all biological replicates.
We investigated whether Gene Ontology (GO) terms were overrepresented in the genes that were significantly more abundant in each organ. For this analysis, we included all genes whose mRNA levels are significantly higher with respect to the other two organs with FDR < 5% (without the 4-fold cutoff criterion), allowing greater statistical power for the detection of overrepresented GO terms. GO term annotations were assigned to soybean genes by determining the GO annotations of their Arabidopsis orthologs using The Arabidopsis Information Resource (TAIR; Huala et al., 2001). The percentage of genes annotated with a given GO term in the organ-specific genes was compared with the percentage of genes annotated with the same GO term in the complete probe set of the soybean cDNA microarray, and the statistical significance of any difference was assessed by means of the hypergeometric distribution. Raw P values calculated using the hypergeometric method were then submitted to FDR control for multiple-testing error with FDR < 5%, to find the statistically significant GO terms. The overrepresented GO terms in cotyledon-specific genes include chloroplast thylakoid membrane (GO:0009535), chloroplast ribulose bisphosphate carboxylase complex (GO:0009573), oxygen-evolving complex (GO:0009654), Cys protease inhibitor activity (GO:0004869), and cytochrome b6f complex (GO:0009512). Taken together, these terms indicate an expected predominance of photosynthesis-associated genes that are specific to the cotyledon (Table III). The higher expression levels of photosynthesis- and chloroplast-related genes in the cotyledons are observed even before the seedlings are exposed to a light signal. This suggests that components of the photosynthetic machinery are already expressed in a cotyledon-specific manner in the darkness. Hook-specific genes are enriched with GO terms related to cell division and protein turnover (Table III). This could indicate that active cell division has a role in hook opening or subsequent development, especially considering that the meristemic tissue is closely adjacent to the apical hook. The most significant GO term in hypocotyl-specific genes was “cell wall-related genes,” which agrees well with an expected importance of cell wall synthesis activity in hypocotyls in either elongating etiolated seedlings or during seedling deetiolation, where the elongation of the hypocotyl is inhibited (Table III). Overall, the significantly overrepresented GO terms identified in the organ-specific genes agree well with our current knowledge of seedling photomorphogenesis.
Table III. Overrepresented GO terms in organ-specific genes.
GO annotation was determined for each transcript by determining the GO annotation for its Arabidopsis homolog (E-value cutoff of 1e-6) using TAIR. P was calculated by the hypergeometric method. FDR was controlled to 0.05.
| Organ | GO Term | GO Identifier | Organ-Specific Genes with the GO Identifier | Whole Probe Sets with the GO Identifier | P |
| % | |||||
| Cotyledon | Chloroplast thylakoid membrane | GO:0009535 | 7.32 | 0.74 | 3.04E-18 |
| Chloroplast ribulose bisphosphate carboxylase complex | GO:0009573 | 0.91 | 0.03 | 1.03E-04 | |
| Oxygen-evolving complex | GO:0009654 | 1.22 | 0.09 | 2.08E-04 | |
| Cys protease inhibitor activity | GO:0004869 | 0.91 | 0.05 | 6.33E-04 | |
| Cytochrome b6f complex | GO:0009512 | 0.61 | 0.02 | 2.25E-03 | |
| Hook | Biological process | GO:0008150 | 9.72 | 1.10 | 1.41E-05 |
| Endomembrane system | GO:0012505 | 16.67 | 5.74 | 7.68E-04 | |
| Ubiquitin ligase complex | GO:0000151 | 4.17 | 0.33 | 1.71E-03 | |
| DNA primase activity | GO:0003896 | 1.39 | 0.00 | 2.74E-03 | |
| Hypocotyl | Cell wall | GO:0005618 | 3.90 | 0.32 | 5.00E-08 |
| Metabolic process | GO:0008152 | 3.90 | 1.07 | 9.22E-04 | |
| Oxidoreductase activity | GO:0016491 | 2.16 | 0.40 | 2.28E-03 | |
We next compared the set of genes regulated by FR in an organ-specific manner and the set of genes expressed in an organ-specific pattern to investigate whether the FR-responsive genes are also expressed in an organ-specific pattern. The result shows that there is little overlap between the two gene sets, with three exceptions. Glyma15g16190 and Glyma07g21150 are expressed in high abundance in the apical hook of etiolated seedlings (Supplemental Table S2), meanwhile they are down-regulated by FRc specifically in the apical hook. Glyma01g38590 is highly expressed in cotyledon and induced by FRc in the cotyledon. However, the remainder of the FR-responsive genes identified (24 out of 27) were not shown to be expressed at significantly higher levels in the organ where they show organ-specific regulation, suggesting that there is no strong correlation between organ-specific gene expression and organ-specific transcriptional responses to light.
cis-Regulatory Elements Identified in the Promoters of Organ-Specific Genes
The regulatory nucleotide sequence in gene promoters plays a key role in the transcriptional response of plants to light stimuli (Hudson and Quail, 2003). A number of cis-regulatory elements have been characterized, and for light-responsive elements their specific binding to trans-regulatory factors in the phytochrome-mediated light signaling pathway is known in many cases, such as GATA, G-box, I-box, and the CCA1 binding motif (Donald and Cashmore, 1990; Wang et al., 1997; Teakle et al., 2002). To search for potential organ-specific light-responsive cis-regulatory elements, two enumerative approach-based motif-finding tools, Sift and Elefinder, were used to identify overrepresented promoter motifs in the microarray-identified coregulated gene sets (a coregulated gene set is a group of genes regulated by FRc in the same manner [e.g. the same organ]). Sift was developed for identifying overrepresented promoter motifs in coregulated Arabidopsis genes sampled from the Arabidopsis Affymetrix array (Hudson and Quail, 2003). In this study, we used an updated version of Sift, which allows the detection of motifs including degenerate nucleotides and with more rigorous statistics, with promoter sequences from the now-completed genome of soybean (Schmutz et al., 2010). Elefinder is a new program similar to Sift, but rather than detecting new motifs, it is designed to detect previously characterized motifs that are overrepresented (Hudson, 2005). Both tools are available at http://stan.cropsci.uiuc.edu/tools.php. The abundance of a motif in promoters of coregulated genes was compared with the abundance of the same motif in promoters of all the genes presented on the microarray, using both Elefinder and Sift. Motifs that are significantly more abundant in coregulated gene sets with respect to the rest of the microarray were determined by first calculating a P value using the hypergeometric method and then correcting for multiple tests by using FDR < 5%.
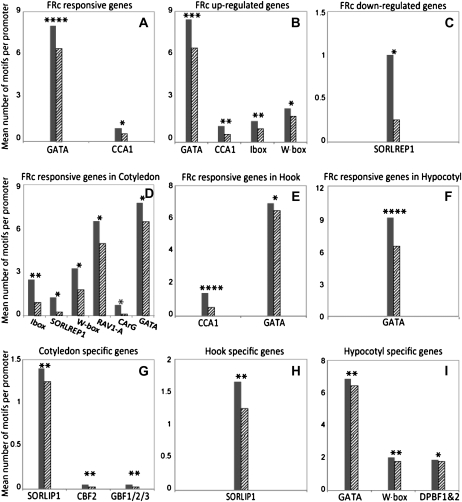
We investigated the overrepresented motifs in the coregulated genes identified by the microarray experiment at multiple levels by testing sets of promoters from FR-responsive genes, FR up-regulated genes, FR down-regulated genes, organ-specific FR-responsive genes, and organ-specific genes. Figure 5 shows the significantly overrepresented known regulatory motifs identified in those groups by Elefinder. Significantly overrepresented motifs in the 27 FR-responsive genes include the formerly characterized GATA binding site and CCA1 binding site, which are known light-responsive motifs (Fig. 5A; Wang et al., 1997; Teakle et al., 2002). FR up-regulated genes and FR down-regulated genes have distinct overrepresented motifs in their promoters (Fig. 5, B and C), suggesting that the same light signal is transduced through two separate pathways leading to positive and negative regulation of downstream effectors. In the promoters of FR up-regulated genes, the GATA binding motif and CCA1 binding motif are again overrepresented, as well as another formerly described light-responsive motif, I-box (Donald and Cashmore, 1990). In the promoters of FR down-regulated genes, SORLREP1, first reported as a light-repressed motif by Hudson and Quail (2003), is the most overrepresented motif (Fig. 5C). Motif analysis also revealed that the most over-represented motif in FRc-induced genes is distinct in different organs. The I-box was most overrepresented in the promoters of cotyledon FR-responsive genes, the CCA1 binding motif in the hook, and the GATA binding site in the hypocotyl (Fig. 5, D–F). This suggests that distinct interactions between transcription factors and cis-regulatory elements occur in different organs and tissues in response to light stimulus.
Figure 5.
Overrepresented motifs involved in organ-specific and FRc-responsive gene expression. Known cis-regulatory motifs were detected in promoters of organ-specific and FRc-responsive genes using the Elefinder software. Motifs that were overrepresented in genes identified in this study as FRc responsive or organ specific to a statistically significant degree were identified using a hypergeometric distribution-based algorithm. This analysis was conducted with the following gene lists: genes that are responsive (in either direction) to 1 h of FRc (A); genes that are up-regulated by 1 h of FRc (B); genes that are down-regulated by 1 h of FRc (C); genes that are responsive to 1 h of FRc in cotyledons (D); genes that are responsive to 1 h of FRc in the apical hook (E); genes that are responsive to 1 h of FRc in the hypocotyl (F); genes that are expressed at higher levels in cotyledons compared with the apical hook and hypocotyl (G); genes that are expressed at higher levels in the apical hook compared with cotyledons and hypocotyl (H); and genes that are expressed at higher levels in the hypocotyl compared with cotyledons and apical hook (I). In each graph, the mean number of motifs per promoter of the genes in the coregulated gene set (gray bars) was compared with the mean number of motifs per promoter of the soybean cDNA microarray probe set (hatched bars).
The significant motifs identified in organ-specific FR-responsive promoters could either be involved in FR-induced photomorphogenesis or simply overrepresented in promoters of genes expressed in the organ in question. To address this issue, we prepared the list of overrepresented cis-regulatory motifs in genes from the “organ-specific” gene lists compared with the motifs associated with organ-specific FR regulation. In cotyledons and hook, organ-specific gene expression and organ-specific FR gene regulation are associated with different motifs (Fig. 5); therefore, the light-responsive motifs identified in cotyledon and hook are likely to be truly light responsive. Note that SORLIP1, a motif known to be overrepresented in light-induced genes (Hudson and Quail, 2003), appears in this experiment to be specific to cotyledon-expressed genes and not to light-induced genes. The most overrepresented light-responsive motif in hypocotyl, the GATA motif, is also observed to be present at significantly higher levels in hypocotyl-specific gene promoters (Fig. 5I), while the mean number of GATA motifs per promoter in hypocotyl FR-responsive genes is higher than that in hypocotyl-specific genes. The hitherto undescribed differentiation of known light-regulatory motifs into cotyledon, hook, and hypocotyl has several implications for the mechanism of photomorphogenesis in different tissues.
In addition to searching for known motifs in coregulated gene groups using Elefinder, we also analyzed all 6-, 7-, 8-, and 9-mers in those promoters using Sift (Hudson and Quail, 2003; Walley et al., 2007) to search for novel cis-regulatory motifs. A new motif, TGNGCNANT, was identified as significantly overrepresented in FR up-regulated gene promoters. Another motif, CNACGTGG, which shares strong similarity with the known light-responsive element G-box, was identified as significantly overrepresented in the cotyledon-specific gene expression (Table IV). No other motifs were identified as significant using the FDR cutoff at 5%. The fact that most of the motifs identified as significant by Elefinder were not detected by Sift is likely a result of the relatively low statistical power of Sift when corrected for false discovery, since Sift examines millions of putative motifs while Elefinder examines only hundreds of known motifs.
Table IV. Novel FRc-responsive and organ-specific cis-regulatory elements revealed by Sift.
| Coregulated Gene Set | Motif Sequence | Coregulated Gene Set Containing the Motif | Reference Gene Set Containing the Motif | P | FDR |
| % | |||||
| FRc-positive regulated genes | TGNGCNANT | 80.0 | 20.6 | 2.15E-08 | <0.05 |
| Cotyledon-specific genes | CNACGTGG | 13.0 | 4.3 | 1.28E-10 | <0.001 |
Identification of New Signaling Factors in FRc-Regulated Photomorphogenesis
The microarray experiment allowed the identification of genes that are regulated by light in an organ-specific manner. We then investigated whether any genes identified can be shown to play an organ-specific role in FRc-induced photomorphogenesis. Because the apical hook has a critical role in seedling deetiolation but the mechanism underlying hook opening is poorly understood, we investigated genes involved in the hook-specific transcriptional regulation.
Glyma18g05720 is up-regulated in response to FRc, with a larger fold change in apical hook than in cotyledon or hypocotyl, as indicated by the microarray and confirmed by QRT-PCR (Table I; Figs. 2B and Fig. 3). Glyma18g05720 encodes a likely ortholog of the Arabidopsis RPT2 (Sakai et al., 2000; Inada et al., 2004); hence, the protein encoded by this transcript is referred to as GmRPT2L (for Glycine max RPT2-like) hereafter. GmRPT2L shares 66.4% amino acid identity and 89.5% similarity with Arabidopsis RPT2. RPT2 was first reported as a positive regulator downstream of PHOT1 in the blue light signaling pathway of root phototropism (Motchoulski and Liscum, 1999; Sakai et al., 2000) and later shown to mediate light-induced stomatal opening by associating with PHOT1 (Inada et al., 2004).
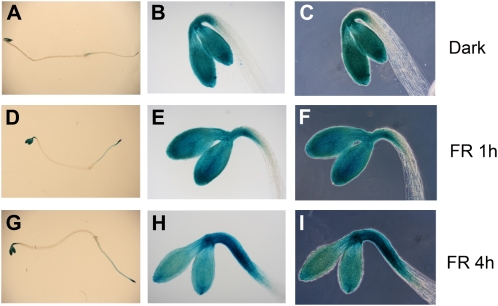
To better understand the role of RPT2 in FRc-induced photomorphogenesis, we studied the spatial expression pattern of RPT2 during deetiolation. The GUS-reporter system was used to test the expression pattern of pRPT2:GUS (Inada et al., 2004) in 1-h FRc-treated Arabidopsis seedlings as well as dark-grown seedlings (Fig. 6, A–F). In darkness, the promoter of RPT2 drives GUS expression in the concave side of the hook and the root tip as well as in cotyledons (Fig. 6, A–C). The spatial expression pattern of RPT2:GUS in 1-h FRc-treated seedlings is very similar to that in the etiolated seedlings (Fig. 6, D–F). Because 1 h is unlikely to be long enough for the distribution of the GUS protein to change significantly, we further examined the GUS expression pattern in seedlings treated with 4 h of FRc. The GUS activity in the hook expanded from the concave side of the hook to the convex side in 4-h FRc-treated seedlings (Fig. 6, G–I), which agrees well with the observed induction of RPT2 in the apical hook overall in response to FRc.
Figure 6.
Histochemical localization of GUS expression in transgenic Arabidopsis plants carrying pRPT2:GUS. Three-day-old dark-grown seedlings carrying a construct with the promoter of the RPT2 gene driving expression of the GUS gene are shown after histochemical staining for GUS. Seedlings are shown without light treatment or after 1 or 4 h of treatment with FR. A to C, Dark-grown seedlings showing GUS staining in the whole seedling under bright-field illumination (staining in the apical hook, cotyledons, and root; A), the seedling apex under bright-field illumination (staining the concave region of the apical hook and the cotyledons; B), and the same view as B under phase-contrast illumination (C). D to F, The same views as A to C in seedlings after treatment with 1 h of FRc, showing a similar pattern to the dark-grown seedlings as the apical hook begins to open. G to I, The same views as A to C in seedlings after treatment with 4 h of FRc, showing loss of the differential expression gradient across the apical hook.
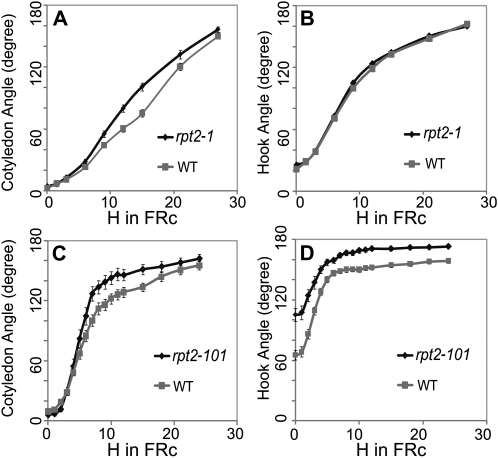
To investigate any role of RPT2 in FRc-induced photomorphogenesis, we exploited the available Arabidopsis mutant resources to obtain two ethyl methanesulfonate-induced alleles of rpt2, rpt2-1 (null mutant; Sakai et al., 2000) and the previously undescribed rpt2-101 (where a G-to-A transition at position 115 in the open reading frame causes a G39R substitution). Seed of rpt2-101 was obtained as a TILLING line from the Arabidopsis Biological Resource Center at Ohio State University with accession number CS91521 (Till et al., 2003). Seeds of these lines were first grown in darkness for 3 d and then given 10 μmol m−2 s−1 FR light for 24 or 27 h. After transfer to FRc, the angles of cotyledons and apical hooks were measured every 3 h for rpt2-1 and once per hour for the first 12 h followed by once every 3 h for rpt2-101 (since rpt2-101 displays a more rapid opening curve compared with rpt2-1). The background accessions of the two mutant lines (Landsberg erecta for rpt2-1 and Big Mama [Torii et al., 1996] for rpt2-101) were also included in the assay as controls. This result suggests that although the mRNA for RPT2 is regulated by FR more strongly in the apical hook than in the other two organs, the light-induced hook opening rate of rpt2 mutants is similar to that of the wild type. However, rpt2-101 showed an altered hook angle in darkness (Fig. 7, B and D). For both rpt2 mutants, cotyledon opening is significantly faster in the mutants compared with their corresponding wild type (Fig. 7, A and C). RPT2 is thus necessary for normal photomorphogenesis in FRc. The more rapid opening of the cotyledon in the mutants may indicate a negative regulatory role of RPT2 in seedling photomorphogenesis.
Figure 7.
Angle of the cotyledon and apical hook in rpt2 mutants compared with their background accession (WT) during growth under FRc. Three-day-old dark-grown seedlings of rpt2-1 (A and B) and rpt2-101 (C and D) seedlings and their corresponding background accessions were transferred to FRc. The angle between the cotyledons (A and C) and the hook angle (measured as the angle between the cotyledon axis and the hypocotyl; B and D) of both mutants and the background accession were measured at 3-h intervals as well as at 1-h intervals in the most rapidly changing part of the curve. The same seedlings were measured repeatedly under dim green safelight and then returned to the FRc treatment. The error bars represent se.
DISCUSSION
Spatial Specificity of Gene Regulation in Photomorphogenesis
Using microarrays, it is now possible to study the role of spatial specificity of gene regulation in photomorphogenesis, as others have done (Ma et al., 2005; López-Juez et al., 2008). In our study, we were able to use the large seedling size and genomic resources available for soybean to show that several genes are expressed at significantly higher levels in cotyledon, hook, or hypocotyl. When the genes responsive to FRc within 1 h are considered, some genes show stronger responses in one organ than in other organs. We identified more organ-specific genes than FRc-regulated genes in this experiment, which is not surprising considering the large differences in the biological roles the three organs play. The cotyledons showed a larger number of organ-specific genes, consistent with the special role played by the cotyledon in energy supply. Hook and hypocotyl cells have similar fates in seedling development; thus, fewer genes are expected to be specific to one of these two tissues. Such organ-specific expression and regulation of gene expression provides a reasonable explanation for the mechanism of organ-specific photomorphogenic responses.
In our study, we identified 27 genes that were regulated in cotyledon, hook, and hypocotyl by 1 h of FRc. Seven genes were repressed and 20 genes were induced. A previous study (Tepperman et al., 2001) performed on Arabidopsis whole seedlings using an Affymetrix assay identified 56 genes induced and six genes repressed by 1 h of FRc via phyA. Among the 56 genes, transcription factors were significantly enriched. We identified a similar trend of repression versus induction but a smaller number of regulated genes and fewer transcription factors. A comparison of our regulated gene list with the Arabidopsis gene list based on orthologous genes showed the following: (1) two out of the 27 genes, RPT2 and ATHB2, were also reported to be regulated in the same direction by FRc in the Arabidopsis Affymetrix data; and (2) the majority of the 27 genes were not identified as being regulated in the Arabidopsis Affymetrix data. A few differences in experimental approaches may have contributed to this. First, a biological replicate in our microarray experiment contained tissues from a single seedling to enhance confidence in true positives by intentionally allowing between-individual variance to be measured. However, this could lead to false negatives due to higher noise, especially for low-expression genes such as transcription factors. In contrast, Tepperman et al. (2001) used a pool of many Arabidopsis seedlings as a single biological replicate. Second, Tepperman et al. (2001) used an Affymetrix array, while in this study a mechanically spotted array was used. Overall, the level of noise and the resolution provided by the array differ substantially between the two experiments. Moreover, Tepperman et al. (2001) reported 62 early-responsive genes, of which 21 are transcription factors. Of these 21 transcription factor genes, only 11 are represented by soybean orthologs in the soybean cDNA microarray. Six of these 11 genes showed consistent repression/induction by FRc greater than 1.6-fold in our array data (thus, we consider them to be confirmed in both species), while the others showed smaller fold changes, possibly due to array noise, as described earlier. Two of these six transcription factors were reported in our study as significantly responsive genes (RPT2-like and HAT4/ATHB2-like). Therefore, the correlation of the two gene lists may be greater than apparent at first sight. The discrepancies were most likely caused by the fold change/FDR cutoff, due to the sensitivity of the microarray and the stringency of the statistical methods applied. That our study identified some FR-regulated genes that were not reported to be responsive in the study by Tepperman et al. (2001) might be due to the difference in spatial resolution (organ level versus the whole seedling). Although the fundamental mechanisms of photomorphogenesis are likely conserved among dicots, timing and magnitude differences in light-responsive gene regulation between Arabidopsis and soybean might exist and might also account for differences in the significant gene lists.
The difference in overrepresented cis-regulatory motifs in different organs provides more insight into the mechanism of organ specification, suggesting that organ-specific interaction of transcription factors and cis-regulatory motifs occurs, even for motifs already known to mediate light responses. This finding suggests that photomorphogenic regulatory networks vary in different organs and tissues. Such distinctive, tissue-specific regulatory networks provide a mechanism for the organ-specificity of seedling photomorphogenesis at the tissue level. SORLIP1, which was reported to be a motif overrepresented in light-induced genes, appears in this experiment to be specific to cotyledon-expressed genes and not to light-induced genes. It is hard to distinguish between a cotyledon-specific motif and a light-inducible, photosynthesis-related motif in this context. The FR-regulated genes described here are regulated 2-fold within 1 h at the organ-specific level, but the data set in which SORLIP1 was discovered consists of genes induced by FR within 24 h in the whole seedlings (Hudson and Quail, 2003). Thus, this motif could be correlated with genes that are light responsive beyond the 1-h time point.
The Apical Hook and the Role of RPT2
The apical hook displays distinct morphological behavior in deetiolation. As a result of the interplay of light stimulus and hormonal regulation, the apical hook shows localized cell expansion, leading to unfolding of the hook. However, apical hooks were not included in the previous organ-specific light response studies (Ma et al., 2005; López-Juez et al., 2008), presumably due to the extremely small size of the hook in Arabidopsis seedlings. In our study, we took advantage of the relatively large soybean seedlings to show a list of genes regulated by FRc in the apical hook in dicot seedlings, to our knowledge for the first time. However, the apical hook region we dissected was still necessarily heterogeneous, limited by how visually distinguishable the organs in question are, the need for a sample of large enough size for effective RNA isolation, the need to rapidly process the samples to avoid RNA degradation and touch responses, and the maintenance of consistent light conditions. First, the concave and convex sides of the hooks play very different, almost opposing, roles during hook opening. For the apical hook to open, the concave side of the hook has to show active growth, primarily by cell elongation, while the convex side of the hook has to show slower or no cell wall expansion. Cambial cell division may also be asymmetric. Second, the hook section of the seedling as used in this study contains the shoot apical meristem and also likely some hypocotyl tissue. Since the shoot apical meristem and leaf primordia, with many cells undergoing cell fate determination and rapid expansion, were included in apical hook tissue, the “hook” sample contains the most rapid regions of cell division. This provides an explanation of why the largest number of FRc-responsive genes were detected in the apical hook region and that the overrepresented GO terms for the hook-specific genes are related to cell division and protein turnover. Also, the junction of the apical hook and hypocotyl is not a clearly defined line that could be seen visually. Thus, the hook sample may contain a variable number of cells where gene regulation responsible for the hypocotyl elongation is active. This may explain the discrepancy between QRT-PCR and microarray results for gene Glyma11g03850 (ATHB2-like), whose organ specificity is flipped between the hook and hypocotyl in these experiments.
To further investigate whether organ-specific transcriptional regulation can influence photomorphogenesis, we investigated Arabidopsis mutants in a gene with a known role in light-regulated cell elongation (RPT2/Glyma18g05720) to determine whether hook-specific developmental defects were present. This mutant has a known role in tropic responses but no previously known effects on phytochrome-mediated, nondirectional photomorphogenic responses. A missense mutant allele of rpt2 showed an altered hook angle in darkness (Fig. 7D). The missense mutation in this gene may be affecting other systems as a result of gain of function; alternatively, this mutation may have a dominant negative effect over redundant similar genes. Both the missense and stop codon alleles displayed faster cotyledon opening (Fig. 7, A and C). This result indicates that this gene does play a role in the morphogenesis of the apical zone but that it is likely more important for cotyledon angle than for hook angle. Therefore, while a gene expression profile in the apical hook does not always predict a mutant phenotype in that structure, a previously undescribed phenotype for rpt2 was observed, which is consistent with a role of this gene in photomorphogenesis of the apical zone during deetiolation. Since cotyledon opening and hook opening are both rapid responses in the apical area mediated largely by cell expansion, it is likely that these responses are related and is possible that the cotyledon-opening response is the more sensitive of the two to perturbation of regulatory factors. Alternately, it is also possible that a signal causing cotyledon opening originates in the hook and is transported to the base of the cotyledons, thus affecting cotyledon opening. Such intercellular signaling, induced by phytochrome, between different tissues has been reported in tobacco (Nicotiana tabacum; Bischoff et al., 1997). A careful examination of the spatial gene expression pattern of RPT2 in the seedlings at the developmental stages concerned in this study was thus necessary.
Therefore, we examined the spatial expression pattern of RPT2 with Arabidopsis transgenic plants carrying the pRPT2:GUS reporter system. In dark-grown seedlings, the GUS signal was observed most strongly in the hook, cotyledons, and roots (Fig. 6, A–C). The signal in the hook was asymmetric, with a strong bias toward the concave side of the hook. After giving etiolated seedlings 1 h of FRc, no strong change was observed (Fig. 6, D–F), although the microarray and QRT-PCR suggested a major induction of RPT2 in the hook. This is not unexpected, because the microarray and QRT-PCR assay examined the mRNA level of RPT2, while the GUS assay examined a combination of transcriptional and translational regulation of RPT2. Therefore, 1 h is likely too short for the GUS protein levels to change significantly, even though the transcriptional regulation likely occurs as early as 1 h. We thus also examined the GUS expression pattern of RPT2 after 4 h of FRc treatment. The region strongly stained by GUS expanded from the hook concave region to the hook convex region, as well as in the direction of hypocotyl, in 4-h FRc-treated seedlings (Fig. 6, G–I). This agrees well with the induction of RPT2 in response to FRc in the apical hook observed by microarray and QRT-PCR.
Given the evidence described above, we postulate that RPT2 is expressed preferentially in the concave side of the hook, cotyledons, and root in dark-grown seedlings. When stimulated by FRc, RPT2 is significantly induced by light in the apical zone of the seedlings, with an expansion of the region of expression from the concave side of the hook to the whole hook (Figs. 2B, 3, and 6, G–I). The spatial expression and light induction of RPT2 in the apical zone agrees well with its proposed role as a signaling factor in cotyledon and hook opening. Since mutants in RPT2 open their cotyledons more quickly, RPT2 may function to fine-tune the speed of cotyledon opening to prevent damage to seedlings caused by premature opening of the cotyledons. The signal of pRPT2:GUS is strongest in the concave side of the hook in dark-grown seedlings and the gradient disappears in FRc, consistent with a role in the suppression of cell expansion in the concave region of the hook in darkness. However, the hook opening of one allele of the rpt2 mutant is comparable to that of the wild type. A potential explanation is that the function of RPT2 in hook opening is redundant and therefore more robust to perturbation. It is also possible that RPT2 does not control hook opening but rather functions as a sensor of the hook-opening process to signal the subsequent cotyledon opening. RPT2 has already been shown to be involved in the blue light responses of root phototropism and stomatal opening in the leaf (Sakai et al., 2000; Inada et al., 2004). This multiplicity of roles suggests that RPT2 acts downstream of multiple photoreceptors to control differential cell expansion responses but plays little role in determining organ development. Instead, spatiotemporal changes in RPT2 expression are likely interpreted within the organellar and/or developmental context.
Novel Regulatory Factors in Photomorphogenesis
Several more genes of interest were identified from the organ-specific regulated gene list and the organ-specific expressed gene list, further study of which may lead to a better understanding of seedling deetiolation. Glyma02g42500 encodes a protein similar to ESK1/AT3G55990 (67% identity and 83% similarity), which is a negative regulator of cold tolerance in Arabidopsis (Xin and Browse, 1998; Xin et al., 2007; Table II). ESK1 contains a conserved DUF231. In our study, Glyma02g42500 was repressed significantly by 1 h of FRc (Table I), indicating that cold tolerance was induced by FRc. The possible adaptive value of cross talk between the cold tolerance and light regulation responses has been reported (Franklin and Whitelam, 2007). An increased FR-red ratio (as a result of FR being preferentially transmitted from low-angle sunlight) may indicate a shorter daylength and longer twilight period as winter approaches (Franklin and Whitelam, 2007). Therefore, induction of cold tolerance genes by FRc may help plants prepare for winter. In the case of germination and seedling establishment, as our study suggests, a higher FR-red ratio may be an indicator that germination is in a period of low-angle sunlight where cold tolerance is required for the plants to survive. Interestingly, another gene containing a DUF231 domain, Glyma02g04170, was also down-regulated by 1 h of FRc in our study (Tables I and II). The significant selection advantage to be gained by early germination and establishment (e.g. canopy penetration) could have led to the development of interaction between cold acclimation and light adaptation responses.
Another interesting finding is that Glyma09g05180, regulated by FRc specifically in the apical hook (Table I; Figs. 2B and 3), encodes a CULLIN1-like protein (CUL1-like). The soybean CUL1-like protein shares 82.7% identity and 96.1% similarity with AtCUL1 (AT4G02570) by protein-protein Smith-Waterman alignment. CUL1 is a key subunit of the ubiquitin protein ligase (E3) complex SCF (for SKP1-CUL1-F-box), which specifies the substrate proteins for 26S proteasome in the ubiquitin/26S proteasome pathway (Pintard et al., 2004). SCF complex-associated protein degradation controls the turnover of important regulatory proteins in light signaling, including the light receptors (Dieterle et al., 2001; Harmon and Kay, 2003; Wei and Deng, 2003; Franklin et al., 2005). An Arabidopsis mutation in CUL1 was reported to display hypersensitivity to FR and a delay in phyA degradation in response to FR (Quint et al., 2005). In addition, the SCF complex was also shown to degrade AUX/IAA proteins under auxin stimuli (Gray et al., 2001). Auxin is known to play a key role in apical hook maintenance (Liscum and Reed, 2002; Vandenbussche et al., 2010; Zádníková et al., 2010). Therefore, CUL1 may be involved in the apical hook opening processes, either by targeting photomorphogenic regulators for degradation or recruiting auxin-responsive factors for degradation. It may also act as a cross talk point for the hormone pathway and the light signaling pathway in the apical hook opening process.
CONCLUSION
Taken together, our data demonstrate that even the transcriptional responses to FRc that occur within 1 h of illumination are organ specific and developmentally regulated. The expressions of several genes show a light response that is specific to one or more organs. In addition, many genes show tissue-specific expression during photomorphogenesis. Thus, even studies of early signal transduction events in phytochrome signaling should be informed by the knowledge that the transcriptional networks and cascades mediating photomorphogenesis are likely to be distinct in different plant tissues. We have demonstrated that organ-specific profiling can be helpful in predicting the morphogenic roles of genes involved in spatially controlled developmental processes and that organ-specific genes regulated by a light stimulus can be correlated with a distinct subset of known cis-regulatory elements.
MATERIALS AND METHODS
Plant Material and Light Treatment
For the microarray experiments, etiolated soybean seedlings (Glycine max ‘Williams 82’) were first surface sterilized in a solution of 5% Clorox and 0.1% Tween 20 for 10 min and then grown hydroponically in water, with sterile glass beads as the solid matrix, in transparent Magenta boxes at 20°C in darkness for 8 d. Seedlings were irradiated with FRc (peak at 733 nm) generated by Snap-Lite light-emitting diode arrays (Quantum Devices). The FRc irradiation was given at 24.7 μmol m−2 s−1 for 1 h. FRc-treated samples were harvested and immediately frozen in liquid nitrogen at 1 h after the start of irradiation. Dark control samples were harvested at 0 h, before the beginning of the irradiation. Plant material for the QRT-PCR experiment was prepared under the same conditions except that seedlings were grown in compost (Sunshine Mix LC1). For the phenotypic study of rpt2, two lines, rpt2-1 (Sakai et al., 2000) and rpt2-101 (CS91521; from the Arabidopsis [Arabidopsis thaliana] TILLING project; Till et al., 2003) were used. All genotypes were verified with PCR amplification with gene-specific primers and sequencing through the putative single-nucleotide polymorphism region. rpt2-1 was compared with its background accession, Landsberg erecta (CS20 from the Arabidopsis Biological Resource Center), and rpt2-101 was compared with its background accession Big Mama (CS89540). The seeds of mutants and their controls were surface sterilized by means of chlorine gas sterilization, then planted on Murashige and Skoog agar plates, and stratified for 5 d in darkness at 4°C. The plates were then treated with 120 μmol m−2 s−1 white light for 3 h to synchronize germination. Seeds were kept in the dark for 72 h to germinate, then treated with 10 μmol m−2 s−1 FR light for 24 h (for rpt2-1 the treatment continued to 27 h). Photographs of seedlings were taken repeatedly of the same plants either once per hour or once per 3 h under green safelight (cool fluorescent light through color effect filters 119, 116, and 101 [Lee filters]; Supplemental Fig. S3) for the measurement of cotyledon opening and hook opening. At least 30 seedlings of each genotype were included in each replicate of the experiment to increase the statistical power of the measurement. The mutant and its corresponding control lines were planted on the same plate for reliable comparison, and at least two plates were included in each comparison for better replication. Experiments were repeated at least twice to verify the results.
RNA Extraction
For the microarray experiments, each seedling was frozen immediately in liquid nitrogen and transferred to RNAlater-ICE (Ambion), which was precooled to −80°C. The purpose of this step was to fix samples while at the same time softening the samples to facilitate dissection. Frozen, fixed seedlings were then dissected into cotyledon, hypocotyl, and apical hook regions (roots were discarded). RNA was isolated from the three different parts of an individual seedling in quadruplicate following the pine tree method (Chang et al., 1993) except for some minor modifications: the homogenization was performed using Ultra-Turrax T8 Homogenizers (IKA) at top speed for 1 min, and phase lock gel (Eppendorf) was used to facilitate the chloroform-isoamyl alcohol extraction. For QRT-PCR, pools of eight seedlings were harvested in triplicate from both dark control and 1-h FR-treated samples and immediately cryofrozen in liquid nitrogen. Seedlings were dissected into cotyledon, hypocotyl, and apical hook regions on dry ice, and then the same modified pine tree method was used for RNA isolation.
Microarray
Pairwise comparison of FRc versus dark was performed with the 18K soybean cDNA arrays (Vodkin et al., 2004). Each set of the 18K soybean cDNA array contains two slides, 18kA and 18kB; together, they present a low-redundancy set of approximately 36,000 sequenced cDNAs. Three two-color pairwise comparisons, one for each organ, were carried out between FRc-treated samples and dark control samples in quadruplicate (Fig. 1). Each biological replicate was a total RNA sample extracted from one dissected organ from one single seedling. By examining the expression profile of a single seedling, we wanted to detect gene regulation events consistent among all individuals. By contrast, significant changes due to a small number of responsive individuals in a pool of seedlings could cause false significant fold change of gene expression level. A dye swap was included in the experimental design to control for the possible bias caused by labeling methods. The 3DNA Array Detection Array Kit (Genisphere) was used for cDNA synthesis, labeling, and microarray hybridization. The 3DNA Array Detection Array 900 Kit was used for labeling RNA samples from apical hook and hypocotyl, and the Array 50 Kit was used for the cotyledon samples, because the Array 900 Kit affords better sensitivity for samples where mRNA is limiting (due to the limited amount of total RNA one can extract from the apical hook or hypocotyl from a single seedling) at the expense of higher noise levels. Arrays were scanned with a Packard ScanArray Express scanner (Perkin-Elmer Life Sciences) to generate the false color array images. Images were then processed with Genepix Pro 4.0 (MDS Analytical Technologies) to generate Genepix Results files (which contain general information on image acquisition and analysis as well as the raw data of channel F635 [for Cy5] and channel F532 [for Cy3] extracted from each individual feature). F635 median and F532 median were used for the data analysis: a Perl script merge.pl was obtained courtesy of Min Li and Steven Clough (U.S. Department of Agriculture-Agricultural Research Service and University of Illinois) and adapted to extract F635 median and F532 median values from the GPR files and perform data preprocessing, including removing empty/blank spots and low-expression features. Data were normalized using Global Locally Weighted Scatterplot Smoothing in the MAANOVA package, part of the Bioconductor package for the R computing language and environment, to generate normalized relative expression matrices (R Development Core Team, 2009). Expression levels of each gene were then compared between dark control and FR-treated samples in individual organs in order to determine which genes showed statistically significant changes by using the rank product analysis method (Breitling et al., 2004) at a FDR (Benjamini and Hochberg, 1995) cutoff of 5%. The above-described analysis methods were automated using an in-house Perl script, which carries out the entire data analysis pipeline automatically from GPR files to rank product statistical analysis. In order to refine our analysis to those transcripts showing strong changes in expression that are likely to be biologically significant, only genes showing expression changes greater than 2-fold that were also statistically significant were defined as “FR-responsive genes.”
Organ-specific gene expression was determined using an alternative approach with single-channel data (F635 median or F532 median). Single-channel information representing median gene expression levels from cotyledon, apical hook, and hypocotyl were compared across organs in both dark-grown seedlings and FRc-treated seedlings (Fig. 1). Raw data were first preprocessed to remove empty/blank spots and flagged data. Normalization across all channels was then performed by first multiplying each channel by a specific constant to make the mean intensity the same for each individual channel and then converting to base 2 log values. Another script was written and used to remove spots with expression value less than the negative control before submitting the data for differential expression analysis. Genes that are significantly more highly expressed in one organ compared with the other two organs were defined as organ-specific genes by the rank product method (FDR = 5%, fold change cutoff of 4-fold).
Microarray Annotation
The 18k cDNA soybean microarrays are supplied with annotation derived from a BLASTX search against the nonredundant protein database using 5′ and 3′ sequences of the cDNA clones (cutoff E value of 10E-6; Vodkin et al., 2004). However, many of these annotations are outdated or absent. A combination of the annotation of soybean chromosome scale assembly (ftp.jgi-psf.org/pub/JGI_data/phytozome/v5.0/Gmax/annotation/initialRelease/Glyma1.cDNA.fa.gz; Schmutz et al., 2010) and the soybean gene index (Quackenbush et al., 2001) followed by BLASTX of these sequences against the TAIR database (www.arabidopsis.org) and the plant protein database (http://www.ncbi.nlm.nih.gov/) was used to generate additional annotation for the 18k cDNA microarray (Supplemental Table S4). The new method was able to provide information on protein function of 5,327 out of 7,950 previously “unknown genes” from the previous annotation spreadsheet supplied with the soybean microarray.
QRT-PCR
Primers for QRT-PCR were designed using Primer Express version 2.0 (Applied Biosystems) based on the soybean EST sequences corresponding to cDNA microarray probes and were then used to search against the JGI soybean chromosome scale assembly (Soybean Genome Project, Department of Energy Joint Genome Institute) to ensure the specificity of the primers. Four control genes derived from array probes, ubiquitin (Glyma20g27950), α-tubulin (Glyma04g09350), β-tubulin (Glyma04g02610), and histone H3.2 minor (Glyma14g40590), were chosen from the microarray as candidate reference genes because they showed relatively constant expression between different light conditions in the microarray data (data not shown). Phosphoenolpyruvate carboxylase (Glyma13g36670) was chosen as an additional candidate reference gene, specifically for organ-specific gene verification, because it has been shown to be expressed constantly across different tissue types (Tuteja et al., 2004). These five candidates were tested for their stability in expression level across three organs in both FRc and dark conditions using QRT-PCR. Histone H3.2 minor was chosen to be the reference gene because it was most consistently expressed in different organs in both experimental conditions among the five candidates (Supplemental Fig. S1). Residual genomic DNA was removed from RNA samples using TURBO DNA-free (Ambion). First-strand cDNA synthesis was accomplished using SuperScript III Reverse Transcriptase (Invitrogen). QRT-PCR was performed using Brilliant SYBR Green QPCR Master Mix (Stratagene) and the Mx3000P QPCR system (Stratagene). Amplification efficiencies of all tested genes including reference candidates were determined by dilution series. QRT-PCR products were analyzed using 3% agarose gel electrophoresis to ensure specific amplification of a single product. Data were analyzed using the ΔΔCT method with the experimentally determined amplification efficiency incorporated (Livak and Schmittgen, 2001). Additionally, analysis with multiple reference genes was performed using the geNorm method (Vandesompele et al., 2002). geNorm analysis and results from the ΔΔCT method were directly comparable (data not shown).
Promoter Motif Analysis
Promoter motif analysis was performed using promoter motif analysis tools, Sift and Elefinder, to search for overrepresented cis-regulatory elements in the promoters of coregulated genes (a coregulated gene set is a group of genes regulated by FRc in the same manner [e.g. same organ or same direction] identified by the microarray experiments; Hudson and Quail, 2003). The abundance of motifs in the promoters of coregulated genes was compared with the abundance of motifs in the promoters of all probes in the soybean cDNA microarray using the hypergeometric distribution combined with FDR control at 5% to identify overrepresented motifs. The extraction of promoter sequences of soybean genes was facilitated by the Soybean Genome Project Glyma 1.0 gene set and the soybean chromosome-scale assembly (Soybean Genome Project, Department of Energy Joint Genome Institute). The EST sequences of cDNA microarray probes were used to search against Glyma 1.0 predicted gene models by BLASTN (Supplemental Table S4; E-value cutoff of 1e-10, identity ≥ 95%). The resultant top hits are the corresponding predicted gene models of cDNA microarray probes (named the 31k set herein, as it contains about 31,000 transcripts). An in-house Perl script was used to extract 2-kb upstream repeat-masked genomic sequence from the starting site of each mRNA in the 31k set. The promoter sequences of coregulated genes were compared with the promoter sequences of the 31k set using Sift (Hudson and Quail, 2003) to identify any significantly overrepresented motifs with the size of six to nine nucleotides, and Elefinder (http://stan.cropsci.uiuc.edu/tools.php) was used to search for significantly overrepresented motifs that have been reported before.
GUS Staining Assay
The Arabidopsis pRPT2:GUS line was produced and kindly provided by Dr. Tatsuya Sakai (Inada et al., 2004). The seeds were first surface sterilized by chlorine gas sterilization, planted on half-strength Murashige and Skoog agar plates without Suc, and stratified for 4 d in darkness at 4°C. Seeds were then treated with 120 μmol m−2 s−1 white light for 2 h to synchronize germination. Seeds were kept in darkness for 72 h at 20°C to germinate and then treated with 20 μmol m−2 s−1 FR light, while the dark controls were maintained in darkness. After the FRc treatment, GUS staining was performed with FRc-treated seedlings and dark controls with a protocol previously described (Campisi et al., 1999). The staining was stopped after 3 h. Photomicrographs were taken in whole-mount bright-field and phase-contrast illumination.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Expression levels of candidate reference genes.
Supplemental Figure S2. Identification of organ-specific genes.
Supplemental Figure S3. The light spectrum of green safelight.
Supplemental Table S1. Cotyledon-specific gene expression.
Supplemental Table S2. Hook-specific gene expression.
Supplemental Table S3. Hypocotyl-specific gene expression.
Supplemental Table S4. Annotation of the 18K soybean cDNA microarray probes.
Acknowledgments
We thank Dr. Tatsuya Sakai for the kind gift of the rpt2-1 seeds and pRPT2:GUS seeds, Dr. Lila Vodkin for providing the soybean cDNA microarray and for useful discussion about the data analysis, Dr. Jeremy Schmutz for providing prerelease versions of the soybean genome assembly, Dr. Karen Hudson for critical reading of the manuscript, and Dr. Steve Clough for suggestions on data analysis and for generously sharing Perl scripts.
References
- Batschauer A, Ehmann B, Schäfer E. (1991) Cloning and characterization of a chalcone synthase gene from mustard and its light-dependent expression. Plant Mol Biol 16: 175–185 [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B 57: 289–300 [Google Scholar]
- Bischoff F, Millar AJ, Kay SA, Furuya M. (1997) Phytochrome-induced intercellular signalling activates cab:luciferase gene expression. Plant J 12: 839–849 [Google Scholar]
- Borthwick HA, Hendricks SB, Parker MW, Toole EH, Toole VK. (1952) A reversible photoreaction controlling seed germination. Proc Natl Acad Sci USA 38: 662–666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bou-Torrent J, Roig-Villanova I, Martínez-García JF. (2008) Light signaling: back to space. Trends Plant Sci 13: 108–114 [DOI] [PubMed] [Google Scholar]
- Breitling R, Armengaud P, Amtmann A, Herzyk P. (2004) Rank products: a simple, yet powerful, new method to detect differentially regulated genes in replicated microarray experiments. FEBS Lett 573: 83–92 [DOI] [PubMed] [Google Scholar]
- Campisi L, Yang Y, Yi Y, Heilig E, Herman B, Cassista AJ, Allen DW, Xiang H, Jack T. (1999) Generation of enhancer trap lines in Arabidopsis and characterization of expression patterns in the inflorescence. Plant J 17: 699–707 [DOI] [PubMed] [Google Scholar]
- Chang S, Puryear J, Cairney J. (1993) A simple and efficient method for isolating RNA from pine trees. Plant Mol Biol Rep 11: 113–116 [Google Scholar]
- Clack T, Mathews S, Sharrock RA. (1994) The phytochrome apoprotein family in Arabidopsis is encoded by five genes: the sequences and expression of PHYD and PHYE. Plant Mol Biol 25: 413–427 [DOI] [PubMed] [Google Scholar]
- Devlin PF, Patel SR, Whitelam GC. (1998) Phytochrome E influences internode elongation and flowering time in Arabidopsis. Plant Cell 10: 1479–1487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhaubhadel S, Gijzen M, Moy P, Farhangkhoee M. (2007) Transcriptome analysis reveals a critical role of CHS7 and CHS8 genes for isoflavonoid synthesis in soybean seeds. Plant Physiol 143: 326–338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieterle M, Zhou YC, Schäfer E, Funk M, Kretsch T. (2001) EID1, an F-box protein involved in phytochrome A-specific light signaling. Genes Dev 15: 939–944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donald RG, Cashmore AR. (1990) Mutation of either G box or I box sequences profoundly affects expression from the Arabidopsis rbcS-1A promoter. EMBO J 9: 1717–1726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fankhauser C, Chen M. (2008) Transposing phytochrome into the nucleus. Trends Plant Sci 13: 596–601 [DOI] [PubMed] [Google Scholar]
- Franklin KA, Larner VS, Whitelam GC. (2005) The signal transducing photoreceptors of plants. Int J Dev Biol 49: 653–664 [DOI] [PubMed] [Google Scholar]
- Franklin KA, Whitelam GC. (2007) Light-quality regulation of freezing tolerance in Arabidopsis thaliana. Nat Genet 39: 1410–1413 [DOI] [PubMed] [Google Scholar]
- Ghassemian M, Lutes J, Tepperman JM, Chang HS, Zhu T, Wang X, Quail PH, Lange BM. (2006) Integrative analysis of transcript and metabolite profiling data sets to evaluate the regulation of biochemical pathways during photomorphogenesis. Arch Biochem Biophys 448: 45–59 [DOI] [PubMed] [Google Scholar]
- Gray WM, Kepinski S, Rouse D, Leyser O, Estelle M. (2001) Auxin regulates SCF(TIR1)-dependent degradation of AUX/IAA proteins. Nature 414: 271–276 [DOI] [PubMed] [Google Scholar]
- Harmon FG, Kay SA. (2003) The F box protein AFR is a positive regulator of phytochrome A-mediated light signaling. Curr Biol 13: 2091–2096 [DOI] [PubMed] [Google Scholar]
- Huala E, Dickerman AW, Garcia-Hernandez M, Weems D, Reiser L, LaFond F, Hanley D, Kiphart D, Zhuang M, Huang W, et al. (2001) The Arabidopsis Information Resource (TAIR): a comprehensive database and Web-based information retrieval, analysis, and visualization system for a model plant. Nucleic Acids Res 29: 102–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson M. (2005) Elefinder. http://stan.cropsci.uiuc.edu/cgi-bin/elefinder/soyelefinder_array.cgi (June 22, 2011)
- Hudson ME, Quail PH. (2003) Identification of promoter motifs involved in the network of phytochrome A-regulated gene expression by combined analysis of genomic sequence and microarray data. Plant Physiol 133: 1605–1616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inada S, Ohgishi M, Mayama T, Okada K, Sakai T. (2004) RPT2 is a signal transducer involved in phototropic response and stomatal opening by association with phototropin 1 in Arabidopsis thaliana. Plant Cell 16: 887–896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Josse EM, Foreman J, Halliday KJ. (2008) Paths through the phytochrome network. Plant Cell Environ 31: 667–678 [DOI] [PubMed] [Google Scholar]
- Khanna R, Shen Y, Marion CM, Tsuchisaka A, Theologis A, Schäfer E, Quail PH. (2007) The basic helix-loop-helix transcription factor PIF5 acts on ethylene biosynthesis and phytochrome signaling by distinct mechanisms. Plant Cell 19: 3915–3929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubasek WL, Shirley BW, McKillop A, Goodman HM, Briggs W, Ausubel FM. (1992) Regulation of flavonoid biosynthetic genes in germinating Arabidopsis seedlings. Plant Cell 4: 1229–1236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leu WM, Cao XL, Wilson TJ, Snustad DP, Chua NH. (1995) Phytochrome A and phytochrome B mediate the hypocotyl-specific downregulation of TUB1 by light in Arabidopsis. Plant Cell 7: 2187–2196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Johnson P, Stepanova A, Alonso JM, Ecker JR. (2004) Convergence of signaling pathways in the control of differential cell growth in Arabidopsis. Dev Cell 7: 193–204 [DOI] [PubMed] [Google Scholar]
- Liscum E, Reed JW. (2002) Genetics of Aux/IAA and ARF action in plant growth and development. Plant Mol Biol 49: 387–400 [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- López-Juez E, Dillon E, Magyar Z, Khan S, Hazeldine S, de Jager SM, Murray JA, Beemster GT, Bögre L, Shanahan H. (2008) Distinct light-initiated gene expression and cell cycle programs in the shoot apex and cotyledons of Arabidopsis. Plant Cell 20: 947–968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L, Sun N, Liu X, Jiao Y, Zhao H, Deng XW. (2005) Organ-specific expression of Arabidopsis genome during development. Plant Physiol 138: 80–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monte E, Al-Sady B, Leivar P, Quail PH. (2007) Out of the dark: how the PIFs are unmasking a dual temporal mechanism of phytochrome signalling. J Exp Bot 58: 3125–3133 [DOI] [PubMed] [Google Scholar]
- Motchoulski A, Liscum E. (1999) Arabidopsis NPH3: a NPH1 photoreceptor-interacting protein essential for phototropism. Science 286: 961–964 [DOI] [PubMed] [Google Scholar]
- Ni M, Tepperman JM, Quail PH. (1998) PIF3, a phytochrome-interacting factor necessary for normal photoinduced signal transduction, is a novel basic helix-loop-helix protein. Cell 95: 657–667 [DOI] [PubMed] [Google Scholar]
- Ohgishi M, Oka A, Morelli G, Ruberti I, Aoyama T. (2001) Negative autoregulation of the Arabidopsis homeobox gene ATHB-2. Plant J 25: 389–398 [DOI] [PubMed] [Google Scholar]
- Pintard L, Willems A, Peter M. (2004) Cullin-based ubiquitin ligases: Cul3-BTB complexes join the family. EMBO J 23: 1681–1687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quackenbush J, Cho J, Lee D, Liang F, Holt I, Karamycheva S, Parvizi B, Pertea G, Sultana R, White J. (2001) The TIGR Gene Indices: analysis of gene transcript sequences in highly sampled eukaryotic species. Nucleic Acids Res 29: 159–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quail PH. (2002) Phytochrome photosensory signalling networks. Nat Rev Mol Cell Biol 3: 85–93 [DOI] [PubMed] [Google Scholar]
- Quint M, Ito H, Zhang W, Gray WM. (2005) Characterization of a novel temperature-sensitive allele of the CUL1/AXR6 subunit of SCF ubiquitin-ligases. Plant J 43: 371–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team (2009) R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna: http://www.R-project.org (June 22, 2011) [Google Scholar]
- Sakai T, Wada T, Ishiguro S, Okada K. (2000) RPT2: a signal transducer of the phototropic response in Arabidopsis. Plant Cell 12: 225–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmutz J, Cannon SB, Schlueter J, Ma J, Mitros T, Nelson W, Hyten DL, Song Q, Thelen JJ, Cheng J, et al. (2010) Genome sequence of the palaeopolyploid soybean. Nature 463: 178–183 [DOI] [PubMed] [Google Scholar]
- Sharrock RA. (2008) The phytochrome red/far-red photoreceptor superfamily. Genome Biol 9: 230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharrock RA, Quail PH. (1989) Novel phytochrome sequences in Arabidopsis thaliana: structure, evolution, and differential expression of a plant regulatory photoreceptor family. Genes Dev 3: 1745–1757 [DOI] [PubMed] [Google Scholar]
- Smith H. (2000) Phytochromes and light signal perception by plants: an emerging synthesis. Nature 407: 585–591 [DOI] [PubMed] [Google Scholar]
- Teakle GR, Manfield IW, Graham JF, Gilmartin PM. (2002) Arabidopsis thaliana GATA factors: organisation, expression and DNA-binding characteristics. Plant Mol Biol 50: 43–57 [DOI] [PubMed] [Google Scholar]
- Tepperman JM, Zhu T, Chang HS, Wang X, Quail PH. (2001) Multiple transcription-factor genes are early targets of phytochrome A signaling. Proc Natl Acad Sci USA 98: 9437–9442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Till BJ, Reynolds SH, Greene EA, Codomo CA, Enns LC, Johnson JE, Burtner C, Odden AR, Young K, Taylor NE, et al. (2003) Large-scale discovery of induced point mutations with high-throughput TILLING. Genome Res 13: 524–530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torii KU, Mitsukawa N, Oosumi T, Matsuura Y, Yokoyama R, Whittier RF, Komeda Y. (1996) The Arabidopsis ERECTA gene encodes a putative receptor protein kinase with extracellular leucine-rich repeats. Plant Cell 8: 735–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuteja JH, Clough SJ, Chan WC, Vodkin LO. (2004) Tissue-specific gene silencing mediated by a naturally occurring chalcone synthase gene cluster in Glycine max. Plant Cell 16: 819–835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandenbussche F, Petrásek J, Zádníková P, Hoyerová K, Pesek B, Raz V, Swarup R, Bennett M, Zazímalová E, Benková E, et al. (2010) The auxin influx carriers AUX1 and LAX3 are involved in auxin-ethylene interactions during apical hook development in Arabidopsis thaliana seedlings. Development 137: 597–606 [DOI] [PubMed] [Google Scholar]
- Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F. (2002) Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 3: H0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vodkin LO, Khanna A, Shealy R, Clough SJ, Gonzalez DO, Philip R, Zabala G, Thibaud-Nissen F, Sidarous M, Strömvik MV, et al. (2004) Microarrays for global expression constructed with a low redundancy set of 27,500 sequenced cDNAs representing an array of developmental stages and physiological conditions of the soybean plant. BMC Genomics 5: 73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walley JW, Coughlan S, Hudson ME, Covington MF, Kaspi R, Banu G, Harmer SL, Dehesh K. (2007) Mechanical stress induces biotic and abiotic stress responses via a novel cis-element. PLoS Genet 3: 1800–1812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ZY, Kenigsbuch D, Sun L, Harel E, Ong MS, Tobin EM. (1997) A Myb-related transcription factor is involved in the phytochrome regulation of an Arabidopsis Lhcb gene. Plant Cell 9: 491–507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei N, Deng XW. (2003) The COP9 signalosome. Annu Rev Cell Dev Biol 19: 261–286 [DOI] [PubMed] [Google Scholar]
- Woo Y, Affourtit J, Daigle S, Viale A, Johnson K, Naggert J, Churchill G. (2004) A comparison of cDNA, oligonucleotide, and Affymetrix GeneChip gene expression microarray platforms. J Biomol Tech 15: 276–284 [PMC free article] [PubMed] [Google Scholar]
- Xin Z, Browse J. (1998) eskimo1 mutants of Arabidopsis are constitutively freezing-tolerant. Proc Natl Acad Sci USA 95: 7799–7804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin Z, Mandaokar A, Chen J, Last RL, Browse J. (2007) Arabidopsis ESK1 encodes a novel regulator of freezing tolerance. Plant J 49: 786–799 [DOI] [PubMed] [Google Scholar]
- Zádníková P, Petrásek J, Marhavy P, Raz V, Vandenbussche F, Ding Z, Schwarzerová K, Morita MT, Tasaka M, Hejátko J, et al. (2010) Role of PIN-mediated auxin efflux in apical hook development of Arabidopsis thaliana. Development 137: 607–617 [DOI] [PubMed] [Google Scholar]
- Zhu D, Maier A, Lee JH, Laubinger S, Saijo Y, Wang H, Qu LJ, Hoecker U, Deng XW. (2008) Biochemical characterization of Arabidopsis complexes containing CONSTITUTIVELY PHOTOMORPHOGENIC1 and SUPPRESSOR OF PHYA proteins in light control of plant development. Plant Cell 20: 2307–2323 [DOI] [PMC free article] [PubMed] [Google Scholar]