Abstract
Motivation: Chimeric DNA sequences often form during polymerase chain reaction amplification, especially when sequencing single regions (e.g. 16S rRNA or fungal Internal Transcribed Spacer) to assess diversity or compare populations. Undetected chimeras may be misinterpreted as novel species, causing inflated estimates of diversity and spurious inferences of differences between populations. Detection and removal of chimeras is therefore of critical importance in such experiments.
Results: We describe UCHIME, a new program that detects chimeric sequences with two or more segments. UCHIME either uses a database of chimera-free sequences or detects chimeras de novo by exploiting abundance data. UCHIME has better sensitivity than ChimeraSlayer (previously the most sensitive database method), especially with short, noisy sequences. In testing on artificial bacterial communities with known composition, UCHIME de novo sensitivity is shown to be comparable to Perseus. UCHIME is >100× faster than Perseus and >1000× faster than ChimeraSlayer.
Contact: robert@drive5.com
Availability: Source, binaries and data: http://drive5.com/uchime.
Supplementary information: Supplementary data are available at Bioinformatics online.
1 INTRODUCTION
1.1 Background
Current sequencing technologies often require DNA samples to be amplified using the polymerase chain reaction (PCR). Amplification produces chimeric sequences that stem from two or more original sequences (the parents of the chimera). The most common mechanism is incomplete template extension, when a partially extended sequence from one sequence reanneals to another parent in the next cycle of PCR. The resulting chimeras are often difficult to identify during downstream analysis (Ashelford et al., 2005). This problem is particularly acute in population studies that sequence a single region, such as the bacterial 16S ribosomal RNA gene (16S) or the fungal Internal Transcribed Spacer (ITS) region, to estimate diversity or find differences between populations, e.g. between diseased and control samples. In the case of 16S, published studies report that curated databases may contain up to 46% chimeric sequences (Ashelford et al., 2005, 2006; Huber et al., 2004). Factors including sequence similarity, number of PCR cycles and relative abundance of gene-specific PCR templates influence chimera formation (Acinas et al., 2005; Haas et al, 2011; Lahr and Katz, 2009; Thompson et al., 2002; Wang and Wang, 1996, 1997). While chimeras with two segments (bimeras) are most common, chimeras with >2 segments (multimeras) may form at comparable rates and account for a significant fraction of the unique sequences in an amplified sample (Lahr and Katz, 2009).
1.2 Previous work
Previous chimera detection methods include CHIMERA_CHECK (Maidak et al., 1999), Pintail (Ashelford et al., 2005), Mallard (Ashelford et al., 2006), Bellerophon (Huber et al., 2004), ChimeraChecker (Nilsson et al., 2010), ChimeraSlayer (Haas et al., 2011) and Perseus (Quince et al., 2011). Pintail and Mallard are 16S-specific programs that use a reference database of trusted chimera-free reference sequences. The query sequence is aligned to all (Pintail) or all pairs (Mallard) of reference sequences. Evolutionary distance is computed in a sliding window across the query sequence and variations in distance are compared with the known rate variability in the 16S gene, with larger variations indicating a chimera. ChimeraChecker is an ITS-specific method using BLAST (Altschul et al., 1997) to search a reference database for taxonomic anomalies. If, for example, the closest match to the ITS1 region is different from the closest match to the ITS2 region, the query is flagged as potentially chimeric. ChimeraSlayer searches a multiple alignment of chimera-free reference sequences and constructs three-way alignments with candidate parents. ChimeraSlayer was shown to be more sensitive than earlier methods (Haas et al., 2011). Although ChimeraSlayer is presented as a 16S-specific method, it would likely perform well with another sequence type if a reference multiple alignment is available. Perseus is designed to detect chimeras in 454 pyrosequencing reads that have been filtered by the AmpliconNoise algorithm (Quince et al., 2011). Assuming that a chimera has undergone fewer rounds of amplification than its parents, the query is compared with all pairs of sequences having higher abundance. The closest pair is selected, and its three-way alignment with the query sequence is made. Supervised learning is employed to determine the parameters of the model.
1.3 UCHIME
To improve speed and accuracy of chimera detection, we created a new algorithm, UCHIME. In our tests, UCHIME achieved higher sensitivity than the best previous method based on a reference database (ChimeraSlayer), while maintaining lower or comparable error rates. In particular, UCHIME has much better performance on short, noisy sequences and on multimeras. The algorithm has no explicit dependencies on any one region and should perform well on different sequence types. UCHIME can use a trusted reference database of non-chimeric sequences (like ChimeraSlayer) and also offers a de novo mode (like Perseus). UCHIME does not require a multiple alignment of the reference database. UCHIME reports a score for each sequence, allowing the user to trade sensitivity for specificity by adjusting the minimum score threshold used to discriminate chimeras from biological sequences. No training is required as we have found the UCHIME score parameters to be robust when presented with different types of input data. The default score threshold gave good sensitivity with low error rates (0–3%) on our tests.
2 METHODS
2.1 UCHIME algorithm
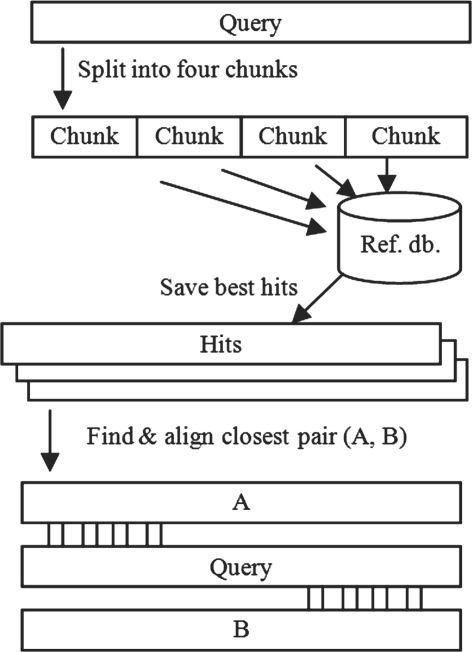
The UCHIME algorithm is illustrated in Figure 1. The query sequence is divided into four non-overlapping segments (chunks), each of which is used to search a reference database, which is assumed to be chimera free. The best matches to each chunk are noted, and the two best candidate parents are identified from matches to all chunks. A three-way multiple alignment of the query to these two candidates is constructed. If a pair of segments extracted from these two candidates has identity ≥0.8% closer to the query sequence than either candidate alone, a score is computed from the alignment and a chimera is reported if the score exceeds a predetermined threshold. In reference mode, the user provides a database of trusted sequences. In de novo mode, the database is constructed on the fly using a strategy similar to Perseus: sequences are considered in the order of decreasing abundance, and candidate parents must have abundance at least 2× that of the query sequence, assuming chimeras are less abundant than their parents because they undergo fewer rounds of amplification. Sequences not classified as chimeric are added to the reference database.
Fig. 1.
UCHIME schematic. The query sequence is divided into four chunks, each of which is used to search the reference database. The best few hits to each chunk are saved, and the closest two sequences are found by calculating smoothed identity with the query. A three-way chimeric alignment is constructed, and a chimera is reported if its score [Equation (2)] exceeds a preset threshold.
2.2 Chimeric alignments and models
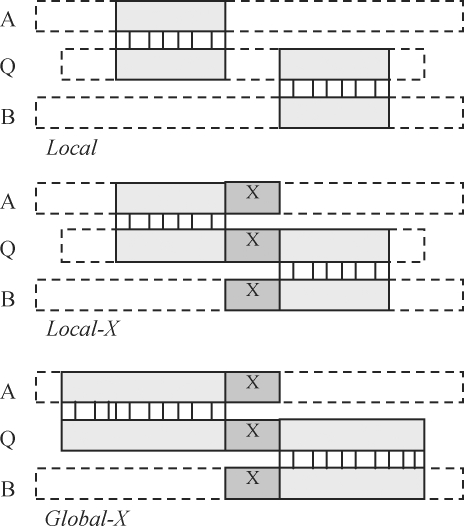
UCHIME searches for a chimeric alignment between a query sequence (Q) and two candidate parents (A and B). We identify three types of alignment as shown in Figure 2, which we call local, local-X and global-X, respectively. We aggressively reduce the number of chimeric alignments that are forwarded to the classification stage because with a given error rate, false positives increase with the number of classifications, while the number of true positives is at most one. We limit the number of classifications by (i) searching for global-X alignments, as fewer global-X alignments usually exist compared with local or local-X; (ii) examining only two candidate parents; and (iii) discarding models having distance to the closest parent (divergence) <0.8%, as classification is harder when differences are small and a failure to detect a chimera with very small divergence only rarely degrades experimental results. If parents or close proxies (step-parents) are present in the reference database, then it is usually possible to construct a chimeric alignment. However, the existence of a chimeric alignment is not sufficient to reliably classify a sequence as an amplification artifact. Chimeric alignments may alternatively be explained by (i) chance biological similarity, e.g. in fast-evolving regions; (ii) convergent evolution due to similar selection pressure in different lineages; (iii) naturally occurring chimeras due to biological processes such as lateral gene transfer; (iv) sequencer error; or (v) poor-quality alignments. One might naively expect that a global-X search would fail to find most multimeras, but in practice global-X proved to have surprisingly good sensitivity and was more effective for finding multimeras than other approaches we have tried, including local-X, which is available as an option in UCHIME. The effectiveness of global-X may be explained by the fact that many multimeras resemble noisy bimeras, and UCHIME is tolerant of noise. All results reported here were obtained using global-X search unless otherwise stated.
Fig. 2.
Chimeric alignments. We identify three types of chimeric alignment between a query sequence Q and two candidate parents A and B: local, local-X and global-X. A chimeric alignment has two non-overlapping segments of Q, one of which is closer to A than to B by some measure of evolutionary distance while the other is closer to B than to A. In a local chimeric alignment, these two segments can be non-contiguous and may only cover a part of Q. In a local-X alignment, the segments are contiguous with an intervening crossover segment (X) which is identical in Q, A and B. A global-X alignment is a special case of a local-X alignment that covers all of Q, but not necessarily all of A or B.
2.3 Scoring function
In a typical chimeric alignment, most columns are identities q=a=b, where q, a and b are letters from Q, A and B, respectively. A column in which at least one sequence differs from the other two is called a diff. Diffs can be considered as votes for or against the model (Fig. 3). For example, a diff q=a, q≠b increases the distance d(Q,B) while leaving d(Q,A) unchanged. If such a diff is found in the segment that is closer to A, it can be regarded as a ‘yes’ vote supporting the model; if it is found in the segment that is closer to B then it contradicts the model and is regarded as a ‘no’ vote. A diff in which all three sequences differ or in which a=b, q≠a, q≠b increases the distance of Q to both A and B and is regarded as an ‘abstain’ vote that neither supports nor contradicts the model. Let Yg, Ng and Ag be the total number of yes, no and abstain votes in segment g of the model, where g is L (left) or R (right). If YL>NL and YR>NR, the alignment is chimeric and the model is closer to Q than A or B alone. The number of diffs may be very small in more challenging cases. For example, in a 16S experiment using 200 nt reads, clusters of radius ~3% might be used in an attempt to identify species (Stackebrandt and Goebel, 1994). It would then be important to identify chimeras with divergences as low as ~2%, which could have a few as four diffs with their closest parents. In such cases, the small amount of evidence available should increase the uncertainty of the classification. UCHIME uses a numerical score for discrimination, as follows. Each segment is assigned a score:
| (1) |
Intuitively, this can be understood as a generalization of the ratio Y/N, which must be >1 for the alignment to be chimeric. The β parameter (which should be ≥1 and is set to 8 by default) gives a no vote a higher weight than a yes vote, and the n parameter (which should be >0 and is set to 1.4 by default) acts as a pseudocount prior (Durbin et al., 1998) on the number of no votes. A positive value of n reduces H, especially when Y is small; this models increased uncertainty with reduced evidence. Abstain votes also lower the score as they indicate noise or the use of a step-parent, either of which should increase uncertainty. The query is classified as a chimera if:
| (2) |
Here, h is the minimum score threshold (0.28 by default). This score is ad hoc; i.e. was not derived from a theoretical model. It was chosen because it is conceptually simple, fast to compute, has only two tunable parameters (β and n) plus an adjustable threshold (h) and was found to perform well empirically.
Fig. 3.
Chimeric alignment showing diffs and votes. This figure shows a region from an alignment generated by UCHIME. Diffs and votes are annotated. The ‘Model’ row indicates the three segments of the alignment which are closer to A, the crossover (X) and closer to B, respectively. Diffs are ‘A’ = diff with Q closer to A in the A segment, ‘a’ = diff with Q closer to A in the B segment, and similarly for ‘B’ and ‘b’. A ‘p’ diff indicates that the parents agree but are different from Q. Votes are ‘+’ (yes), ‘!’ (no) and ‘0’ (abstain), indicating whether the corresponding diff supports or contradicts the model.
2.4 Parent selection and alignment construction
Candidate parents are found by (i) splitting the query into subsequences (chunks); (ii) using each chunk to search the database; and (iii) saving the best few hits to each chunk. We have found any reasonable procedure to be effective for this stage. More difficult is to reduce the number of candidates in order to suppress the false positives caused by attempting to classify too many models. UCHIME selects the best two candidates according to the following procedure. A pair-wise alignment is computed between the query Q and each candidate parent P. The identity between P and Q is smoothed over a window (default size 32). For each position in Q, the highest value for the smoothed identity among the parents is recorded. The best candidate is then identified as the one with most positions having highest smoothed identity. Note that this does not require the positions to be contiguous. This can be effective in the case of a multimera where multiple disjoint segments are derived from a single parent sequence, which may occur when a sequence is highly abundant in the sample. The positions in which the best candidate has highest smoothed identity are removed from Q, and the second candidate is identified in the same way from the remaining positions. UCHIME then constructs a star multiple alignment (Altschul, 1989), i.e. one that preserves the pair-wise alignments of Q to the two candidate parents. Following ChimeraSlayer, columns in the three-way alignment containing a gap or adjacent to a column containing a gap are discarded as these tend to occur in regions that are less reliably aligned. Diffs are identified in the remaining columns. Finally, dynamic programming on the vector of diffs is used to find segments of a global-X or local-X labeling of the alignment that maximizes H.
2.5 De novo mode and abundance skew
In de novo mode, UCHIME starts with an empty reference database. Sequences are considered in the order of decreasing abundance. If a sequence is classified as chimeric, it is discarded; otherwise it is added to the reference database. Candidate parents are required to have abundance at least λ times that of the query sequence, on the assumption that a chimera has undergone fewer rounds of amplification and will therefore be less abundant than its parents. The parameter λ is called the abundance skew, and by default λ=2, assuming at least one more round of amplification for the parents.
2.6 Training and validation datasets
Three test datasets were used in this work. (i) SIM2 is a selected subset of the simulated bimeras and control sequences used to train and evaluate ChimeraSlayer. (ii) MOCK is the Uneven datasets used to evaluate Perseus (Quince et al., 2011). They are derived from pyrosequencing reads of ‘mock’ communities, i.e. experimentally mixed DNAs of known composition. These reads were processed by AmpliconNoise (Quince et al., 2011), which attempts to remove sequencing error and generates a set of predicted sequences for the amplicons. Sequences in this set were classified as biological or chimeric by comparing them to reference sequences for the species in each community, and chimera detection algorithms were assessed by their success in reproducing this classification. (iii) SIMM is a new set of simulated m-meras created for this work. SIM2 and SIMM were used to compare the performance of the reference database mode of UCHIME with ChimeraSlayer, MOCK was used to compare the de novo mode of UCHIME with Perseus. The parameters of UCHIME were trained on SIM2; the score threshold h was set to a value giving an average error rate over the whole SIM2 dataset lower than the error rate of ChimeraSlayer on the same data. UCHIME was trained by an exhaustive search over manually selected pairs (β,n). The optimal pair (β+,n+) was identified by maximizing the area under a receiver operating characteristic curve (Mason and Graham, 2002). Given β+ and n+, an optimal score threshold h+ is determined by (i) specifying a maximum desired error rate or minimum desired sensitivity and (ii) maximizing sensitivity or minimizing error rate, respectively. After training, the sensitivity of UCHIME averaged over all SIM2 sets was 70.6% with an error rate of 0.49%, compared with 54.6% sensitivity and 0.62% errors for ChimeraSlayer.
2.7 Creation of the SIMM dataset
In order to test UCHIME on multimeras, we implemented CHSIM, a simulator capable of creating m-meras with any number of segments. Input to CHSIM is a set of chimera-free parent sequences. In each iteration of the simulation, a preset number of chimeras (default 100) are created at crossovers where parents have an identical k-mer; in this experiment, we used k=10. Crossover points are selected at random, weighted by the frequency of the k-mer in the set of parent sequences. This biases crossovers to occur between similar sequences in regions of higher sequence similarity, as presumably happens in real experiments. Non-homologous crossovers are permitted, and exactly one occurred in the simulations used to create SIMM (ch646_m4_90_95). At the end of each iteration, chimeras are added to the pool of parent sequences, allowing multimeras to form when one or two existing chimeras cross over. To create the SIMM dataset, parents were the set of 86 reference sequences for species in the Uneven sets of MOCK. These have length ~250 nt and cover the V2 hypervariable region of the 16S gene. These relatively short parents were chosen to model the short sequences obtained by current sequencing technologies, which can be more challenging for chimera detection algorithms owing to the smaller number of diffs needed to cause divergences that are experimentally relevant (Haas et al., 2011). Several simulations were performed using the same set of parent sequences with different random number seeds. Segments in a chimera were required to be unique to one parent, otherwise an m-mera may be identical to an (m−1)-mera. Chimeras with m>4 were found to be very rare due to the short sequence length. Chimeras with m=2, 3 and 4 in three divergence ranges (90–95%, 95–97% and 97–99%) were identified, for a total of nine bins, each containing 100 simulated chimeras.
2.8 Program versions
Unless otherwise stated, UCHIME results were obtained using the USEARCH v4.2.52. Perseus results were obtained using v1.24 of the AmpliconNoise package. MAFFT v6.853 (Katoh and Toh, 2008) was used by Perseus to create alignments. The reference database used for both ChimeraSlayer and UCHIME was the ‘gold’ set in http://sourceforge.net/projects/microbiomeutil/files/, version 2011-11-02. Unless otherwise stated, Perseus results were obtained using PerseusD v1.24, a variant of the original Perseus algorithm that follows UCHIME by only testing parents that have been classified as non-chimeric and are at least twice as abundant as the query. For a comparison of Perseus with PerseusD, see the Supplemental Material.
3 RESULTS
3.1 Assessment on SIM2
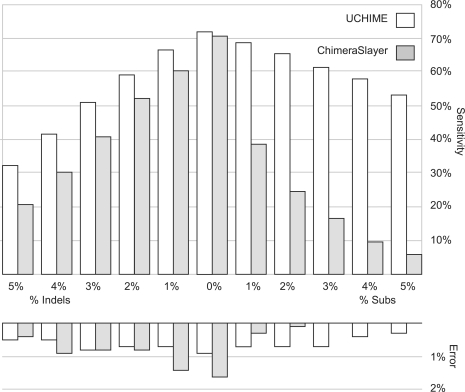
The SIM2 dataset contains simulated bimeras and control sequences with lengths 200, 300 and full-length (FL). Bimeras are created by selecting two random segments of the control sequences. Ten additional sets are provided for each length in which from 1% to 5% of sites were mutated by introducing simulated substitutions or indels, respectively. These mutations model cases where reference sequences are diverged from the true parents due to biological variation, sequencing error or other factors. Results are presented in Table 1, Supplementary Table S1 and in Figure 4, which shows sensitivity and specificity on the length 200 sets, which are the shortest and therefore most difficult. As seen in Figure 4, UCHIME has higher sensitivity on all length 200 sets, with increasing improvement at higher mutation rates. The sensitivity of ChimeraSlayer falls rapidly as substitutions are introduced, even at the relatively low rate of 1%, while the sensitivity of UCHIME degrades only slightly. At a substitution rate of 2%, which is well within the range observed for 16S genes within strains of a single bacterial species, the sensitivity of ChimeraSlayer drops by more than half (from 71% to 25%), compared with a reduction of only 8% for UCHIME (from 72% to 66%).
Table 1.
Performance of UCHIME and ChimeraSlayer (CS) on the SIM2 benchmark
| Length | Mutations | CS Sens. (Err.) | UCHIME Sens. (Err.) |
|---|---|---|---|
| FL | None | 90.3 (1.0) | 90.8 (0.5) |
| FL | 1% indels. | 83.6 (0.9) | 94.3 (0.3) |
| FL | 1% subs. | 87.4 (0.4) | 90.4 (0.2) |
| 300 nt | None | 77.5 (1.9) | 81.3 (1.9) |
| 300 nt | 1% indels. | 66.6 (1.9) | 76.4 (1.3) |
| 300 nt | 1% subs. | 55.5 (0.4) | 78.5 (1.0) |
| 200 nt | None | 70.7 (1.6) | 72.7 (0.9) |
| 200 nt | 1% indels. | 60.4 (1.4) | 66.6 (0.6) |
| 200 nt | 1% subs. | 38.6 (0.3) | 69.6 (0.6) |
Sensitivity (Sens.) and error rate (Err.) are shown for selected subsets of the SIM2 benchmark: lengths 200, 300 and FL (full-length genes) with no added mutations and with 1% substitutions and indels respectively, as indicated in the Mutations column. For full results, see Supplementary Table S1. UCHIME has higher sensitivity on all these subsets; both programs have similar error rates in the range ~0.5% to ~2%. UCHIME is more tolerant of noise, especially with substitutions in short sequences where the sensitivity is improved from 38.6% to 69.6% (200 nt) and from 55.5% to 78.5% (300 nt). Values are given in percentages.
Fig. 4.
Performance of UCHIME and ChimeraSlayer on length 200 tests in SIM2. These results show that UCHIME has higher sensitivity than ChimeraSlayer on all length 200 sets, with increasing improvement at higher mutation rates, especially when substitutions are present. The UCHIME error rate is <1% on all sets.
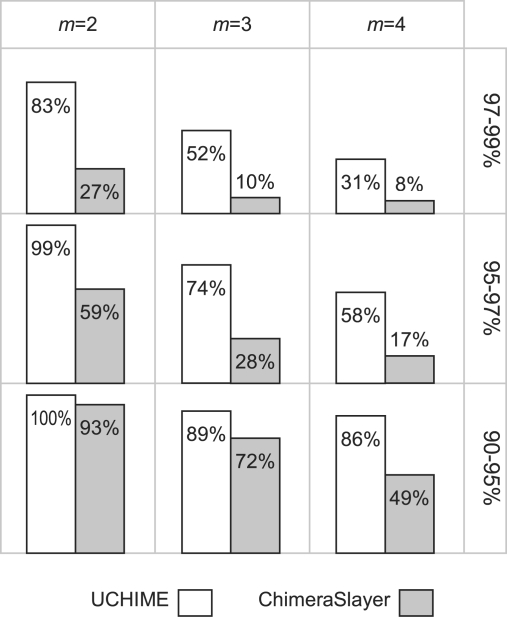
3.2 Assessment on SIMM
The SIMM dataset contains 900 simulated chimeras of length ~250 nt divided into nine bins by divergence and the number of segments. As in SIM2, 10 additional sets were created by adding from 1% to 5% substitutions or indels. Results are presented in Supplementary Table S2 and Figure 5, which shows sensitivity on the set with 1% substitutions as we consider this level of noise to be reasonably realistic. (While indel errors are relatively common in pyrosequencing, this is mainly due to homopolymers which can be handled in a preprocessing step, e.g. by truncating runs of identical letters). Error rates are not shown since both programs find no false positives in the parent sequences. Again we observe that UCHIME has greatly improved sensitivity compared with ChimeraSlayer, especially to chimeras with small divergence and/or larger numbers of segments. Similar trends are seen in the other sets (Supplementary Table S2).
Fig. 5.
Sensitivity on the SIMM set with 1% substitutions. UCHIME has higher sensitivity than ChimeraSlayer on all subsets, especially to chimeras with small divergence and larger numbers of segments. In the 3×3 grid shown in the figure, columns indicate the number of segments (m) in an m-mera and rows correspond to divergence ranges.
3.3 UCHIME and PerseusD compared on MOCK
Results on the MOCK sets are shown in Table 2. UCHIME is shown to have similar sensitivities and error rates to Perseus. Given that UCHIME was trained entirely on a very different dataset (SIM2) in reference database mode with no separate training of the de novo mode, we interpret these results as demonstrating that the UCHIME algorithm is highly robust when presented with new types of data.
Table 2.
Performance of UCHIME and PerseusD on the MOCK datasets
| Set | GoodSeqs | Chimeras | Sensitivity |
Errors |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PerseusD (%) | UCdn (%) | UCref (%) | PerseusD | UCdn | UCref | |||||||
| Uneven1 | 94 | 898 | 93 | 95 | 89 | 1 | 0 | 2 | ||||
| Uneven2 | 77 | 742 | 93 | 93 | 86 | 0 | 1 | 2 | ||||
| Uneven3 | 75 | 925 | 93 | 94 | 91 | 0 | 2 | 1 | ||||
|
m=2 |
m=3 |
m=4 |
||||||||||
| N | PerseusD | UCdn | UCref | N | PerseusD | UCdn | UCref | N | PerseusD | UCdn | UCref | |
| Uneven1 | 816 | 761 | 780 | 737 | 81 | 73 | 68 | 61 | 1 | 0 | 1 | 1 |
| Uneven2 | 669 | 619 | 624 | 578 | 71 | 66 | 60 | 57 | 2 | 1 | 1 | 1 |
| Uneven3 | 843 | 797 | 806 | 782 | 82 | 64 | 64 | 59 | 0 | 0 | 0 | 0 |
Input sequences are denoised amplicons and abundances predicted by AmpliconNoise (Quince et al., 2011). These results show that de novo UCHIME (UCdn) with default parameters (obtained by training on SIM2) has similar sensitivities and error rates to PerseusD. In reference database mode (UCref), using the microbiome utils gold reference database, UCHIME performance is similar, reflecting the fact that many of the 16S sequences in the communities are found in the gold database. N is the number of chimeric sequences and m is the number of segments in the chimera. GoodSeqs and Chimeras are the total numbers of biological sequences and chimeras, respectively, found using a separate reference database of 16S sequences for species in the communities (see Quince et al., 2011 for details).
3.4 Computational resources
All the tested programs have modest memory requirements, needing at most 50 Mb to complete the tests reported here. Execution times required to execute UCHIME, ChimeraSlayer and Perseus on a representative dataset are shown in Table 3. Two versions of UCHIME are tested: a stand-alone program and a second implementation of the UCHIME algorithm in the USEARCH package (Edgar, 2010). The stand-alone version of UCHIME is more than 100× faster than Perseus in de novo mode and more than 1000× faster than ChimeraSlayer in reference database mode, with a further order of magnitude achieved by the USEARCH version.
Table 3.
Execution times
| Program | Mode | Time (h:min:s) |
|---|---|---|
| usearch –uchime | de novo | 0:02 |
| UCHIME | de novo | 0:13 |
| PerseusD | de novo | 32:06 |
| usearch –uchime | ref. db. | 0:34 |
| UCHIME | ref. db. | 13:19 |
| ChimeraSlayer | ref. db. | 4:28:28 |
Elapsed time required to execute two implementations of the UCHIME algorithm compared with ChimeraSlayer and Perseus on the Uneven1 subset of the MOCK data, which has 1124 sequences. The ChimeraSlayer reference database (5181 sequences) was used for both UCHIME and ChimeraSlayer. The stand-alone UCHIME program is tested and also an implementation of the same algorithm in the USEARCH package (Edgar, 2010). A single-core, 1 GHz 32-bit i86 Linux computer with 1 GB RAM was used.
4 DISCUSSION
Chimeric sequence identification poses a challenging problem in algorithm design, especially with short reads, where the available evidence is often limited to very small numbers of observed differences. UCHIME achieves a significant improvement in detection accuracy over previous methods that use a reference database, and performs comparably to a state-of-the art de novo method designed specifically for pyrosequencing, despite the fact that UCHIME was not designed or trained for this particular type of data. UCHIME achieves much faster execution speeds than previous programs. Our results show that UCHIME has robust performance when presented with different types of 16S data and is tolerant of simulated noise, suggesting that UCHIME is likely to perform well with other types of data, e.g. the fungal ITS region or reads from novel sequencing technologies.
UCHIME requires either a database with adequate coverage of the phylogenetic diversity in the input sequences (reference mode), or an estimate of unique amplicon sequences and their abundances (de novo mode). Construction of a suitable reference database and robust estimation of amplicon sequences and their abundances (denoising) are both challenging problems that are discussed in more detail in the Supplementary Material.
Although we regard the experiments reported here as informative for comparing algorithms, realism is hard to achieve both in simulations and in mock communities, so results may not be predictive of sensitivity and error rates that would be achieved on experimental data from environmental samples.
The emerging interest in characterizing the effects of members of the rare biosphere in a range of clinical and environmental contexts, combined with the rapid decrease in sequencing cost, challenges us to improve the efficiency of sequence analysis so that that computational cost does not become a limiting factor. UCHIME meets this challenge for an essential step in many experiments, offering a unique combination of accuracy and speed that will be of great value to biologists.
Supplementary Material
ACKNOWLEDGEMENTS
R.C.E. thanks Henrik Flyvbjerg for helpful discussions.
Funding: National Institutes of Health (grant U54-HG004969 to B.J.H.); Engineering and Physical Sciences Research Council Career Acceleration Fellowship (EP/H003851/1 to C.Q.); National Institutes of Health (grant HG004872 and HHMI to R.K., in part).
Conflict of Interest: none declared.
REFERENCES
- Acinas S.G., et al. PCR-induced sequence artifacts and bias: insights from comparison of two 16S rRNA clone libraries constructed from the same sample. Appl. Environ. Microbiol. 2005;71:8966–8969. doi: 10.1128/AEM.71.12.8966-8969.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul S.F. Trees, stars and multiple biological sequence alignment. SIAM J. Appl. Math. 1989;49:197–209. [Google Scholar]
- Altschul S.F., et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashelford K.E., et al. At least 1 in 20 16S rRNA sequence records currently held in public repositories is estimated to contain substantial anomalies. Appl. Environ. Microbiol. 2005;71:7724–7736. doi: 10.1128/AEM.71.12.7724-7736.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashelford K.E., et al. New screening software shows that most recent large 16S rRNA gene clone libraries contain chimeras. Appl. Environ. Microbiol. 2006;72:5734–5741. doi: 10.1128/AEM.00556-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durbin R., et al. Biological Sequence Analysis. Cambridge, UK: Cambridge University Press; 1998. [Google Scholar]
- Edgar R.C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- Haas B.J., et al. Chimeric 16S rRNA sequence formation and detection in Sanger and 454-pyrosequenced PCR amplicons. Genome Res. 2011;21:494–504. doi: 10.1101/gr.112730.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber T., et al. Bellerophon: a program to detect chimeric sequences in multiple sequence alignments. Bioinformatics. 2004;20:2317–2319. doi: 10.1093/bioinformatics/bth226. [DOI] [PubMed] [Google Scholar]
- Katoh K., Toh H. Recent developments in the MAFFT multiple sequence alignment program. Brief. Bioinformatics. 2008;9:286–298. doi: 10.1093/bib/bbn013. [DOI] [PubMed] [Google Scholar]
- Lahr D.J., Katz L.A. Reducing the impact of PCR-mediated recombination in molecular evolution and environmental studies using a new-generation high-fidelity DNA polymerase. Biotechniques. 2009;47:857–866. doi: 10.2144/000113219. [DOI] [PubMed] [Google Scholar]
- Maidak B.L., et al. A new version of the RDP (Ribosomal Database Project) Nucleic Acids Res. 1999;27:171–173. doi: 10.1093/nar/27.1.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason S.J., Graham N.E. Areas beneath the relative operating characteristics (ROC) and relative operating levels (ROL) curves: statistical significance and interpretation. Q. J. Meteorol. Soc. 2002;128:2145–2166. [Google Scholar]
- Nilsson R., et al. An open source chimera checker for the fungal ITS region. Mol. Ecol. Res. 2010;10:1076–1081. doi: 10.1111/j.1755-0998.2010.02850.x. [DOI] [PubMed] [Google Scholar]
- Quince C., et al. Removing noise from pyrosequenced amplicons. BMC Bioinformatics. 2011;12:38. doi: 10.1186/1471-2105-12-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stackebrandt E., Goebel B.M. A place for DNA-DNA reassociation and 16S rRNA sequence analysis in the present species definition in Bacteriology. Int. J. Syst. Bacteriol. 1994;44:846–849. [Google Scholar]
- Thompson J.R., et al. Heteroduplexes in mixed-template amplifications: formation, consequence and elimination by ‘reconditioning PCR’. Nucleic Acids Res. 2002;30:2083–2088. doi: 10.1093/nar/30.9.2083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G.C., Wang Y. The frequency of chimeric molecules as a consequence of PCR co-amplification of 16S rRNA genes from different bacterial species. Microbiology. 1996;142(Pt 5):1107–1114. doi: 10.1099/13500872-142-5-1107. [DOI] [PubMed] [Google Scholar]
- Wang G.C., Wang Y. Frequency of formation of chimeric molecules as a consequence of PCR coamplification of 16S rRNA genes from mixed bacterial genomes. Appl. Environ. Microbiol. 1997;63:4645–4650. doi: 10.1128/aem.63.12.4645-4650.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.