SUMMARY
The long noncoding Xist RNA inactivates one X-chromosome in the female mammal. Current models posit that Xist induces silencing as it spreads along X and recruits Polycomb complexes. However, the mechanisms for Xist loading and spreading are currently unknown. Here, we define the nucleation center for Xist RNA and show that YY1 docks Xist particles onto the X chromosome. YY1 is a ‘bivalent’ protein, capable of binding both RNA and DNA through different sequence motifs. Xist’s exclusive attachment to the inactive X is determined by an epigenetically regulated trio of YY1 sites as well as allelic origin. Specific YY1-to-RNA and YY1-to-DNA contacts are required to load Xist particles onto X. YY1 interacts with Xist RNA through Repeat C. We propose that YY1 acts as adaptor between regulatory RNA and chromatin targets.
INTRODUCTION
The diverse functions of long noncoding RNA (ncRNA) are exemplified by X-chromosome inactivation (XCI), the process of dosage compensation in mammals that results in silencing of ~1000 genes on one X-chromosome in the early female embryo (Lyon, 1961; Lucchesi et al., 2005; Wutz and Gribnau, 2007; Payer and Lee, 2008). XCI is regulated by a unique region known as the X-inactivation center (Xic), which sequentially controls the counting of X-chromosomes, the random choice of one of two X-chromosomes for inactivation, and the initiation and spread of silencing along the chromosome. The Xic resides in a 100- to 500-kb X-linked domain with intriguing noncoding genes (Chureau et al., 2002), among which five – Xist, Tsix, Xite, RepA, and Jpx – have been shown to act crucially during XCI (Fig. 1A)(Brockdorff et al., 1992; Brown et al., 1992; Lee and Lu, 1999; Ogawa and Lee, 2003; Zhao et al., 2008; Tian et al., 2010). Recent studies have begun to elucidate complex events surrounding the initiation of XCI, highlighting RNA as central molecules in overcoming challenges presented by this method of dosage compensation (Lee, 2009, 2010).
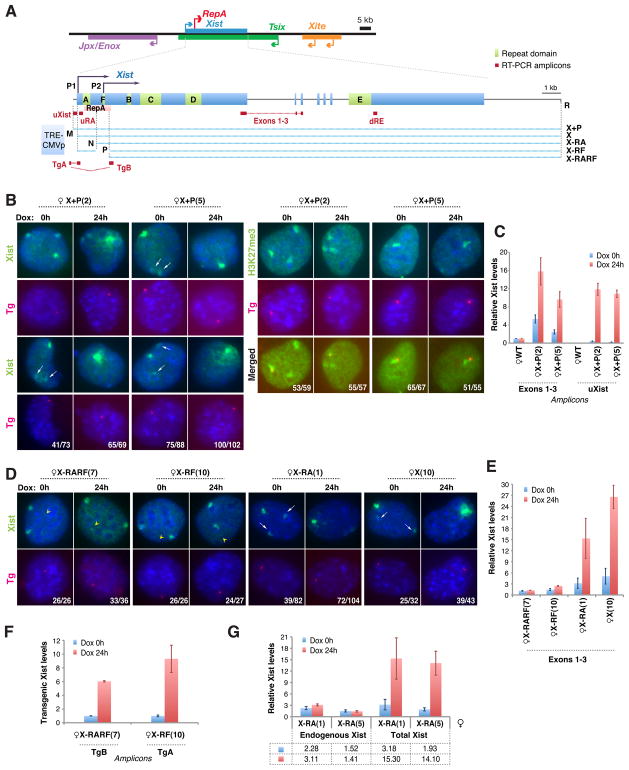
Figure 1. Newly introduced Xist transgenes squelch Xist RNA from Xi in MEFs.
A. Map of Xist and transgenes. Restriction sites used for cloning: M, MluI; R, RsrII; N, NheI; P, PmlI.
B. Xist RNA FISH (left) and H3K27me3 immunostaining (right) in X+P female clones. RNA FISH and immunostaining were followed by DNA FISH using a vector probe (Tg) to confirm transgenic origin of Xist or H3K27me3. Two representative clones shown before and after doxycycline induction (dox, 2μg/ml). Arrows, Xist squelching on Xi in progress. Number of cells with indicated Xist pattern/total cells shown. NOTE: MEF lines are tetraploid due to SV40 Large T-transformation.
C. qRT-PCR of Xist in wildtype female MEF (WT) and two X+P clones. Transgenic RNA quantitated at uXist; total Xist at Exons 1–3. Xist levels normalized to WT (set arbitrarily to 1.0). Averages ± 1 standard deviation (SD) from three independent experiments shown.
D. Serial Xist RNA/Tg DNA FISH in representative clones for four transgenic lines. Arrowheads, transgenic Xist locations. Arrows, Xist squelching in progress. Number of cells with indicated Xist pattern/total cells shown.
E. Xist qRT-PCR measured at Exons 1–3.
F. qRT-PCR of transgenic Xist for X-RF(7) and X-RARF(10). Levels at Dox 0h set to 1.0.
G. qRT-PCR of endogenous (uRA) and total (exons 1–3) Xist in X-RA clones.
A pivotal factor is the 17-kb Xist transcript (Brockdorff et al., 1992; Brown et al., 1992; Clemson et al., 1996; Penny et al., 1996; Marahrens et al., 1997; Wutz et al., 2002), which initiates chromosome-wide silencing as it coats the X and recruits Polycomb complexes (Plath et al., 2003; Silva et al., 2003; Schoeftner et al., 2006; Zhao et al., 2008). In pre-XCI cells, Xist RNA is expressed at basal levels from both X-chromosomes (<5 copies/chromosome)(Sun et al., 2006; Zhao et al., 2008) and is kept in check by Tsix, a 40-kb antisense transcript that antagonizes Xist by blocking its transcriptional induction (Lee and Lu, 1999; Lee, 2000; Sado et al., 2001). When dosage compensation is triggered by cell differentiation, Tsix continues to be expressed from the future active X (Xa), but is downregulated on the future inactive X (Xi) to relieve the first roadblock to Xist activation. Suppression of Tsix on Xi enables RepA RNA to recruit Polycomb complexes to Xist (Sun et al., 2006; Zhao et al., 2008) and renders Xist permissive for activation by Jpx RNA (Tian et al., 2010). Thus, Xist is controlled positively and negatively by several Xic-encoded transcripts that act upon Xist in a requisite allele-specific manner.
These findings underscore one major challenge of the mammalian dosage compensation system: Whereas the fruitfly and roundworm dosage compensation machineries act upon all X-chromosomes in the nucleus (Park et al., 2003; Lucchesi et al., 2005; Pal Bhadra et al., 2006; Laverty et al., 2010; Meyer, 2010), the mammalian machinery must discriminate between two essentially identical X-chromosomes. Furthermore, silencing must occur in phase, so that alleles along one X are coordinately inactivated without affecting X-linked alleles in trans. Conceptually, the mammalian paradigm necessitates an initial decision point (“choice”) in which the cell designates one Xa and one Xi, followed by an execution step in which the Xist-Polycomb silencing machinery is recruited to as yet undefined loci along one X.
Two broad classes of recruiting mechanisms can be envisioned. One postulates that recruitment is regulated locally – that each X-gene or a cluster of genes attracts its own dosage compensation machinery independently of other genes/clusters on the same X. Given strong evidence for the existence of only a single control center (Xic)(Lee et al., 1996; Chureau et al., 2002) and coordinated spread along Xi, this type of mechanism is not favored. More popular is the idea of a primary recruiting center at which Xist particles would load and propagate in a cis-limited manner with the aid of regularly spaced booster elements (Gartler and Riggs, 1983). Because neither recruiting centers nor booster elements have been identified, how Xist binds the X remains elusive. Here, we investigate this problem and discover a developmentally regulated receptor within the Xic that traps the Xist-Polycomb complex. Our model supports the idea of a single nucleation center that loads Xist particles prior to their translocation along Xi.
RESULTS
Squelching of endogenous Xist RNA by newly introduced Xist transgenes
To study Xist RNA localization, we introduced a full-length doxycycline(dox)-inducible Xist transgene (X+P; Fig. 1A) into female mouse embryonic fibroblasts (MEF). RNA fluorescent in situ hybridization (FISH) showed transgene expression and formation of small Xist foci even without dox-induction (Fig. 1B), likely due to inclusion of 180-bp of Xist’s promoter sequence (Pillet et al., 1995; Stavropoulos et al., 2005). [Note: Cells are tetraploid due to SV40 Large T-transformation; two Xi are present]. Dox-induction for 24 hours significantly boosted expression and led to development of large Xist clouds (Fig. 1B). Quantitative RT-PCR (qRT-PCR) indicated that total Xist levels were 2–5 times higher than in wildtype (WT) cells before dox-induction, and increased 2–3 times further upon induction (Fig. 1C; exons1–3). To examine transgenic contributions, we amplified with transgene-specific primers (uXist) and observed >10-fold induction with dox.
Two unusual observations caught our attention. First, the transgene not only formed Xist clouds but was also hypermethylated at H3K27 (H3K27me3) (Fig. 1B). This was unexpected, because previous analyses using a mouse embryonic stem (ES) model showed that the X-chromosome becomes refractory to Xist after the first 3 days of cell differentiation (Wutz and Jaenisch, 2000; Kohlmaier et al., 2004). More surprisingly, ectopic Xist clouds were always more prominent than endogenous clouds. In fact, even before dox induction, the transgene displayed a large Xist cloud and the Xi’s RNA cloud was already suppressed in 56–85% of cells (Fig. 1B). After induction, Xist clouds disappeared from Xi completely in 94–98% of cells (Fig. 1B). Multiple independent clones showed this behavior. We conclude that newly introduced Xist transgenes in MEFs act on the endogenous locus in trans and “squelches” Xist RNA clouds on Xi.
Squelching depends on a 700-bp RNA localization domain around Repeat F
Several mechanisms could underlie squelching. Introduction of homologous sequences could induce RNAi-mediated gene silencing (Wassenegger et al., 1994). Alternatively, the transgene could outcompete endogenous Xist for locus-specific transcription factors (Gill and Ptashne, 1988). Post-transcriptional mechanisms, such as those affecting RNA localization, must also be entertained. To address mechanism, we performed transgene deletional analysis to identify squelching sequences, focusing on Xist’s conserved proximal end. We deleted a 2-kb region spanning Xist’s P1 and P2 promoters (Johnston et al., 1998), Repeat A, and Repeat F (Brockdorff et al., 1992; Brown et al., 1992; Nesterova et al., 2001)(Fig. 1A, X-RARF). In contrast to X+P clones, multiple independent clones of X-RARF did not squelch endogenous Xist (Fig. 1D,E). qRT-PCR showed increased X-RARF expression after dox induction (Fig. 1F), but RNA FISH revealed no RNA accumulation at the X-RARF site (Fig. 1D, arrowhead). These results implied either an RNA localization or stabilization defect in X-RARF. Thus, the deleted 2-kb region is responsible for both squelching and RNA accumulation.
To narrow down required regions, we made smaller deletions and examined multiple independent clones of each (representative clones shown in all analyses below). Transgene X deleted only the Xist promoter and squelched effectively (Fig. 1A,D). The X-RA transgene eliminated the Xist promoter, Repeat A, and RepA RNA (Zhao et al., 2010)(Fig. 1A), but remained squelching-competent (Fig. 1D). By contrast, transgene X-RF deleted a 700-bp region around Repeat F and abolished both squelching and RNA localization (Fig. 1D). Like X-RARF, X-RF induction increased steady state Xist levels (Fig. 1F), but Xist RNA failed to accumulate at the transgene site (Fig. 1D, arrowheads). At the same time, Xist clouds on Xi were spared. There is thus a strong correlation between transgenic Xist accumulation and squelching of endogenous Xist RNA. We conclude that Xist’s promoter, Repeat A, and RepA RNA are not required for squelching and implicate the 700-bp region around Repeat F in both Xist localization and squelching.
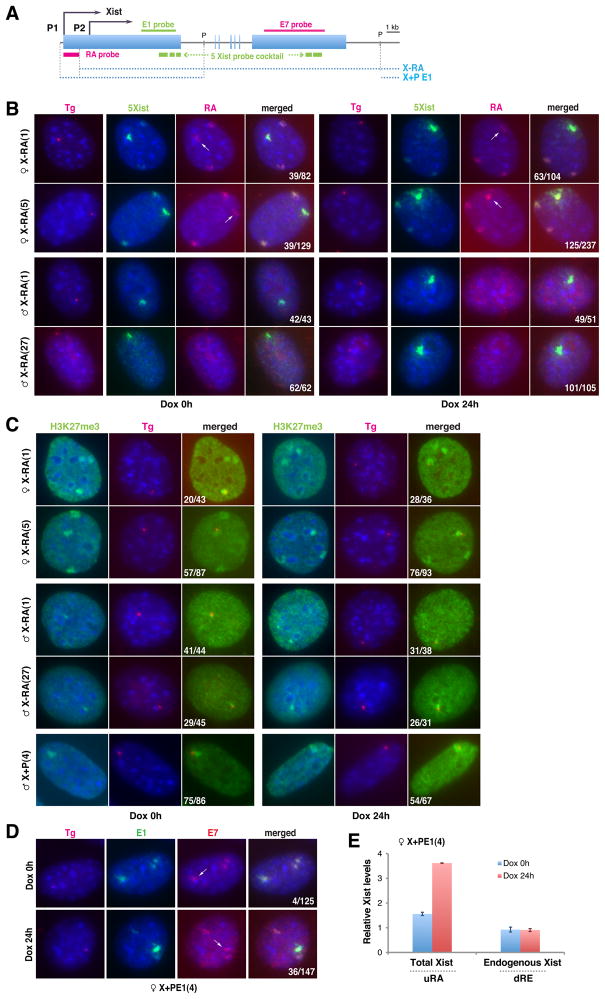
Xist RNA diffuses away from Xi and is attracted to the transgene
We suspected a direct connection between squelching and RNA localization, as squelching occurs when newly introduced transgenes can accumulate Xist RNA. Could the transgene exert trans effects and displace Xist RNA from Xi? Indeed, although Xist clouds faded away on Xi, the stability (or steady state levels) of endogenous Xist RNA was surprisingly not affected (Fig. 1G). To investigate the fate of endogenous Xist RNA, we tracked Xist molecules in squelching-competent X-RA clones. Serial RNA/DNA FISH enabled us to distinguish endogenous versus transgenic RNAs by a Repeat-A probe (Fig. 2A, RA), and X versus transgenic DNA by a vector-specific probe. Intriguingly, endogenous Xist localized not only to Xi but also to the transgenic site (Fig. 2B, arrows). Thus, endogenous Xist RNA trans-migrated between Xi and the homologous ectopic site. This behavior was seen even before dox-induction, demonstrating that high transgene expression is not required to strip Xist from Xi. H3K27me3 enrichment followed Xist accumulation at the transgene site (Fig. 2C). Because X-RA lacks Polycomb-recruiting sequences (Zhao et al., 2008), transgenic H3K27me3 must reflect action of trans-migrated wildtype Xist-Polycomb complexes. Thus, Xist RNA is diffusible and remains stable when not bound to chromatin.
Figure 2. Autosomal transgenes attract Xist RNA way from Xi.
A. Map of Xist, FISH probes, and transgenes. P, PasI.
B. Serial Xist RNA/Tg DNA FISH in X-RA cells ± dox. Tg, transgene insertion site. Endogenous Xist, RA probe (RA); transgenic Xist, 5-Xist-riboprobe mix (5mix). Arrows, trans-migrated endogenous Xist to transgene site.
C. H3K27me3 immunostaining followed by DNA FISH in X-RA cells.
D. RNA FISH of female X+PE1 cells using probes E1 and E7 ± dox. Arrows, trans-migration of endogenous Xist to transgene site.
E. qRT-PCR for total (uRA) and endogenous (dRE) Xist.
Because earlier experiments in male cells had shown that transgenic Xist could not diffuse between X and autosome (Lee et al., 1996; Lee et al., 1999; Wutz and Jaenisch, 2000; Kohlmaier et al., 2004), we examined consequences of introducing our transgenes into male MEF. Consistent with prior reports, RNA/DNA FISH showed that X-RA male cells formed Xist clouds at the transgene site, but the RNA never transmigrated to X (Fig. 2B). Also consistent with previous studies (Plath et al., 2003; Kohlmaier et al., 2004), the Repeat-A-deficient RNA induced H3K27me3 poorly on the autosome in spite of RNA accumulation (Fig. 2C; only H3K27me3 pinpoints at best). However, X+P cells efficiently formed Xist clouds and H3K27me3 foci, further arguing that Xist function is not confined to a developmental time window. Nevertheless, Xist produced from X+P could not bind male Xa. These results demonstrate that, although diffusible, Xist is not promiscuous. The male Xa is resistant to Xist, either because it lacks a receptor for Xist RNA or other accessory factors.
Xist localization requires YY1 protein
To identify the Xist receptor, we designed a “squelching assay”, on the principle that RNA binding sites on Xi and transgene must compete for a limited pool of Xist particles. First, we asked if Xist exon 1 were sufficient to attract RNA in trans and tested transgene X+PE1 (Fig. 2A) in female MEFs by performing RNA FISH using differentially labeled exon 1 and 7 probes that distinguished endogenous from transgenic transcripts. Indeed, exon 1 attracted endogenous Xist RNA, though not as efficiently as full-length transgenes (Fig. 2D, arrows; 22% of cells). As observed in other transgenic lines, Xist RNA remained stable when displaced from Xi in X+PE1 cells (Fig. 2E; S2). Therefore, exon 1 is both necessary and sufficient to squelch endogenous Xist. Receptors for Xist must reside therein.
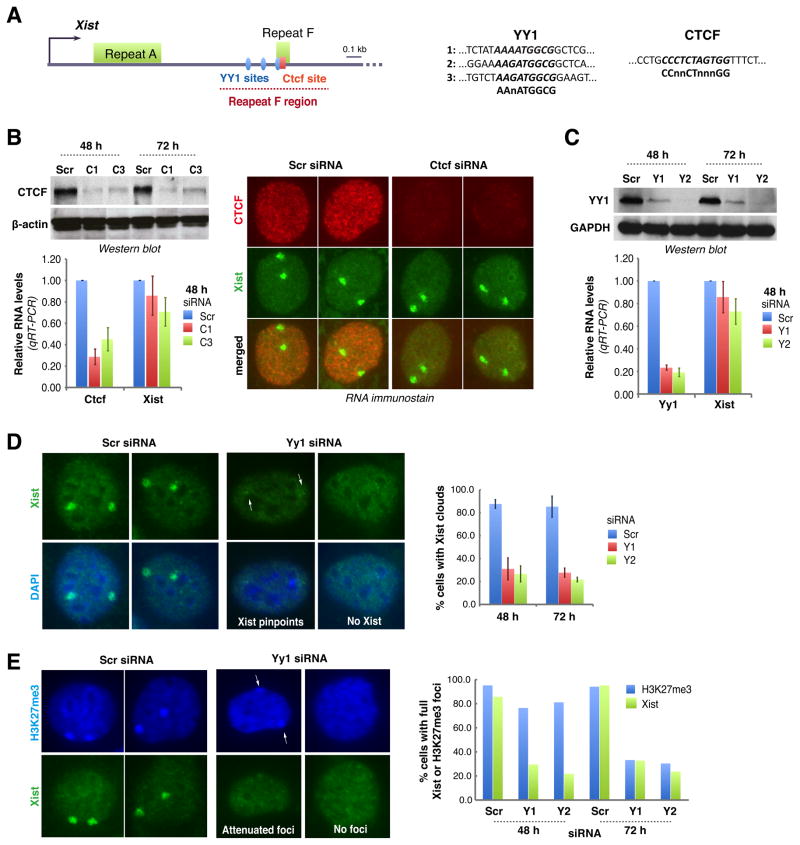
We then searched exon 1 for conserved motifs. Near Repeat F are two potential binding sites for CTCF (Lobanenkov et al., 1990; Essien et al., 2009) and YY1 (Hariharan et al., 1991; Park and Atchison, 1991; Seto et al., 1991; Shi et al., 1991; Kim et al., 2006)(Fig. 3A). These proteins have been implicated in regulation of X-chromosome pairing through binding sites in Tsix/Xite (Donohoe et al., 2007; Xu et al., 2007; Donohoe et al., 2009) and regulation of human XCI through sites upstream of XIST (Hendrich et al., 1993; Pugacheva et al., 2005), but a role in RNA localization had not been suspected. To test whether CTCF is required for Xist localization, we knocked down CTCF in female MEF, but observed no reduction in Xist levels or clouds (Fig. 3B). Thus, CTCF is not needed for Xist binding to Xi.
Figure 3. YY1 is required for Xist localization.
A. Map of the proximal 2-kb region of Xist. One CTCF and three putative YY1 binding sites near Repeat F are shown.
B. Western blot, qRT-PCR, and combined Xist RNA FISH/CTCF immunostain 48 hours after Ctcf knockdown using C1 or C3 siRNA. Averages ± SD of three independent experiments shown.
C. YY1 Western blot and Yy1/Xist qRT-PCR after Yy1 knockdown using Y1 or Y2 siRNA. Averages ± SD from 7 independent experiments shown for qRT-PCR. One representative Western blot shown.
D. Xist FISH after Yy1 knockdown. Cells with pinpoint (arrows) or no Xist were scored negative. Averages ± SD from 206–510 nuclei/sample from three independent experiments.
E. H3K27me3 immunostaining (blue) followed by Xist RNA FISH in Yy1-knockdown cells. Two representative patterns shown. Histogram shows counts (n=62–138).
By contrast, knocking down YY1 (Fig. 3C) resulted in loss of Xist clouds from >70% of cells (Fig. 3D). In cells where Xist was still detectable, RNA signals were pinpoint or severely attenuated (arrows, Fig. 3D). Similar results were obtained for two YY1-specific siRNAs, Y1 and Y2, arguing against off-target effects. Transfection with scrambled siRNA (siRNA-Scr) had no effect on YY1 or Xist. Interestingly, although YY1 knockdown affected Xist localization, it did not affect total RNA levels, agreeing with the assertion that Xist RNA remains stable when displaced from chromatin. Whereas Xist clouds disappeared within 24–48 h of YY1 knockdown, H3K27me3 enrichment persisted up to 48 h and did not disappear from Xi until 72 h (Fig. 3E; 70–80%), consistent with slower kinetics of H3K27me3 turnover. These data demonstrate that YY1 is essential for Xist localization.
A trio of YY1-binding sites serves as nucleation center
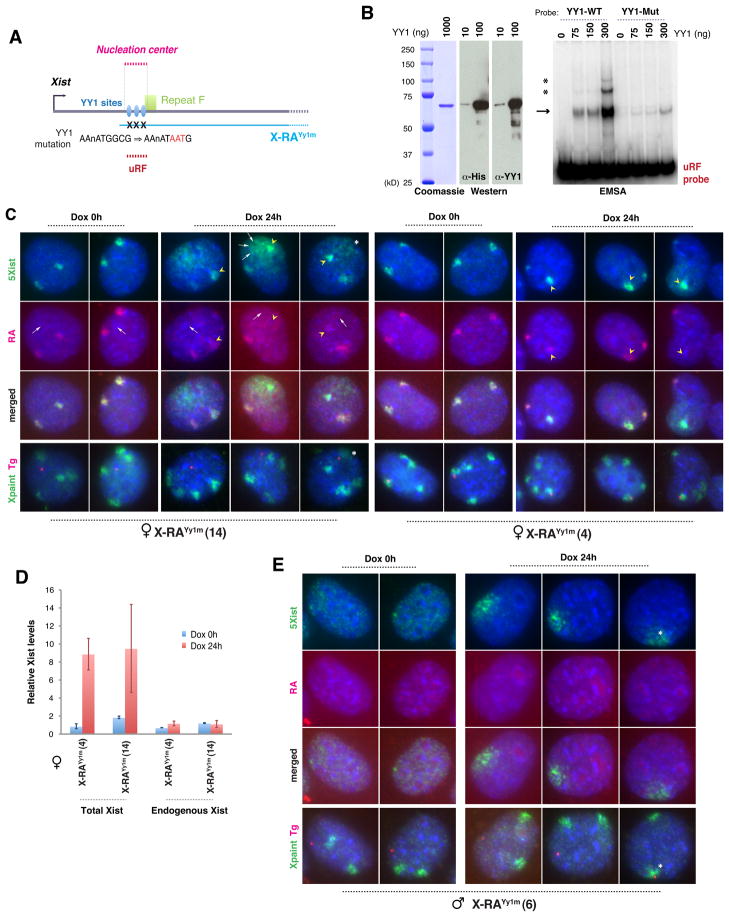
Is YY1 the receptor for Xist particles? To investigate, we examined three conserved elements matching the YY1 consensus, AAnATGGCG, separated by ~100 bp near Repeat F. These elements were previously proposed to bind YY1 based on bioinformatic and ChIP analyses, though direct DNA-protein interactions were not demonstrated (Kim et al., 2006). To test direct binding, we performed electrophoretic mobility shift assays (EMSA) and found that purified recombinant YY1 protein shifted a 280-bp DNA probe containing the trio motif (Fig. 4A,B). Elevating YY1 protein concentration both intensified the shifted band (arrow) and led to appearance of two higher molecular weight species (asterisks) indicative of progressive site occupancy. When the motifs were mutated, YY1 binding was severely attenuated (Fig. 4A,B). Thus, YY1 directly binds the trio motif.
Figure 4. Mutating YY1-binding sites in DNA abolishes Xist RNA loading.
A. Map of proximal Xist, YY1-binding sites, transgenes, and EMSA probe. Site-directed mutation of YY1 sites shown.
B. Left panels: SDS-PAGE, Coomassie staining, and Western blot of purified recombinant His-YY1 protein. Right panel: EMSA using YY1 and a 280-bp uRF probe. WT, wildtype YY1 probe. Mut, mutated YY1 probe. Arrow, YY1-uRF shift. Asterisks, increasing Yy1 occupancy on uRF probe.
C. Two-probe Xist RNA FISH of female X-RAYy1m clones, followed by DNA FISH to locate transgene (Tg) and Xs (X paint). Arrows, transgenic RNA and transgene position. Arrowheads, transmigrated mutated transgenic RNA onto the Xi that is closer. Asterisks, Xa located close to Tg.
D. qRT-PCR of total (Exons 1–3) and endogenous (uRA) Xist in female X-RAYy1m cells.
E. Two-probe Xist RNA FISH followed by DNA FISH in male X-RAYy1m cells. Asterisks, Xa located close to Tg.
To ask if the motif is involved in Xist localization, we performed site-directed mutagenesis of all three YY1 sites on the X-RA transgene (X-RAYy1m; Fig. 4A). X-RA was used because it is both squelching-competent and its RNA can be distinguished from endogenous Xist RNA by RNA FISH using a Repeat A probe. Serial RNA/DNA FISH showed dramatic differences between X-RA (Fig. 2B) and X-RAYy1m clones (Fig. 4C). Before dox induction, RNA was never observed at the X-RAYy1m transgenic site, whereas Xist RNA showed robust accumulation on Xi (Fig. 4C, dox 0h). This result contrasted with obvious squelching in uninduced X-RA clones (Fig. 2B). Dox induction revealed further differences. Transgene expression resulted in a huge burst of RNA around the transgene site, but the RNA seemed to diffuse away (concentration gradient around transgene; Fig. 4C, dox 24h, arrows; 62.9%, n=116). Thus, mutating the YY1-binding sites prevented anchoring of Xist RNA and abolished squelching of Xist RNA from Xi. In wildtype cells, YY1 did not decorate Xi at any time (Fig. S1). We conclude that a trio of YY1-binding sites serves as nucleation center for Xist binding to Xi.
Xist diffuses bidirectionally between Xi and transgene, but Xa is always resistant
Curiously, the two Xi in transgenic cells often did not have equal Xist clouds. The Xi closer to the transgene usually exhibited the larger Xist cloud (Fig. 4C, arrowheads; 49.1%, n=116) and, strangely, this cloud consisted mostly of mutated transgenic rather than Xi-synthesized RNA, as RA-probe signals on proximal Xi were less than on distal Xi. This disparity was observed only after dox-induction. Therefore, transgenic RNA – though it could not bind to its own transgene site in cis – must be able to displace endogenous Xist from the Xi closer to it. This odd finding implied that YY1 must interact with DNA and RNA via different nucleic acid motifs. qRT-PCR showed no change in steady state levels of endogenous or transgenic RNA (Fig. 4D), indicating that both mutated and wildtype Xist molecules are stable even when not chromatin-bound.
At no time did transgenic Xist localize onto Xa, even when Xa was in proximity in female cells (Fig. 4C, asterisk). This was also the case in male MEF clones carrying X-RAYy1m. Prior to dox induction, transgene expression was minimal. Pinpoint nascent Xist transcripts were seen in 68% of cells (n=78), and the rest showed no detectable Xist (Fig. 4E). When induced, transgenic RNA localized poorly around the transgene site (81%, n=74)(Fig. 4E), similar to that observed in X-RAYy1m female cells (Fig. 4C). In males, Xa never attracted Xist RNA even when the transgene locus was close. Xa is therefore always resistant.
Taken together, these data illustrate several crucial points: (i) A cluster of YY1 sites near Repeat F serves nucleation center for Xist binding. (ii) Xist particles are freely diffusible. (iii) Exchange of Xist molecules can occur bidirectionally, from transgene to Xi (Fig. 4D) as well as from Xi to transgene (Fig. 1–2). (iv) While X-RAYy1m transgenes could not strip Xist RNA from Xi, Xi could attract RNA produced by X-RAYy1m. This lack of reciprocity argues that, while YY1 binds the AAnATGGCG motif in DNA, its interaction with Xist RNA does not occur through the corresponding RNA motif, AAnAUGGCG. (v) Xa is refractory to Xist binding, even though Xa also possesses the trio of YY1 sites.
Xi-specific binding of YY1
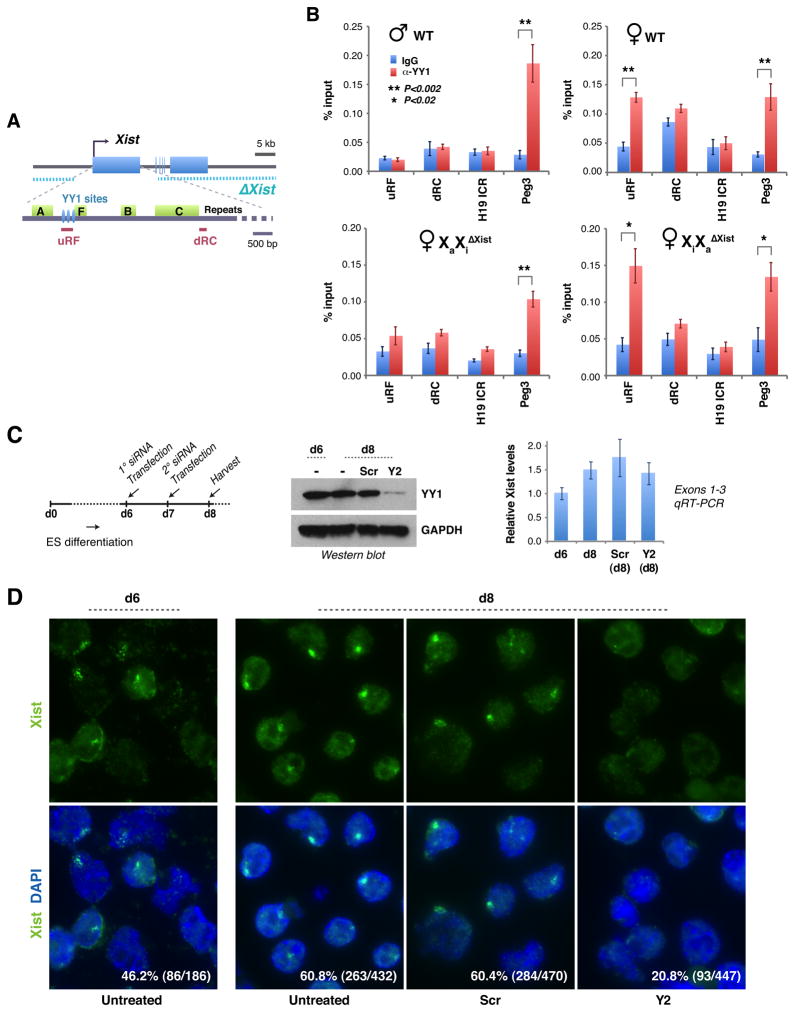
Xa’s immunity implies an epigenetic difference between Xa from Xi. To ask if differential YY1 binding underlies the difference, we examined YY1 binding patterns in vivo by chromatin immunoprecipitation (ChIP) assays using YY1 antibodies and qPCR primers flanking the YY1 sites (Fig. 5A). We observed strong enrichment of YY1 to this region (uRF) in female but not male MEFs (Fig. 5B). The enrichment was comparable to that for intron 1 of Peg3, an imprinted gene known to bind YY1 (Kim et al., 2009). By contrast, no enrichment occurred in a region downstream of the Repeat C (dRC) or in the H19 imprinting control center (ICR). These data demonstrate that YY1 specifically occupies the Repeat F YY1 sites. To distinguish Xa from Xi, we used female MEFs that bear a conditional deletion of Xist exons 1–3 either on Xa (XiXaΔXist) or Xi (XaXiΔXist)(Zhang et al., 2007). ChIP consistently showed enriched YY1 binding to uRF in XiXaΔXist but not in XaXiΔXist. In XiXaΔXist, YY1 could only have bound to Xi, as the uRF region is deleted on Xa. By the same logic, the lack of YY1 enrichment at uRF in XaXiΔXist cells implies that YY1 is not enriched on Xa. Thus, YY1 differentially binds the nucleation center of Xi and Xa. We propose that differential susceptibility of Xa and Xi to Xist is not only the result of allele-specific Xist transcription, but primarily the consequence of allele-specific YY1 occupancy. In differentiating female ES cells, knockdown of YY1 also did not alter the stability of Xist RNA but significantly interfered with Xist localization (Fig. 5C,D). Therefore, we propose that YY1 is crucial for Xist localization during initiation, establishment, and maintenance of XCI.
Figure 5. YY1 binds specifically to Xi.
A. Map of the Xist deletion (Csankovszki et al., 1999; Zhang et al., 2007), ChIP amplicons, and YY1 sites.
B. YY1 ChIP analyses. At least three independent experiments performed for each cell line. Averages ± standard errors (SE) from ≥3 independent experiments shown. Statistical significance, P, determined by the Student t-test (asterisks).
C. YY1 knockdown in differentiating female ES cells (TsixTST/+) via the indicated timeline. Cells were split into siRNA-treated and –untreated samples on day 6 (d6). Western blot showed good knockdown. Xist qRT-PCR showed constant steady state levels; averages ± SD from three independent knockdown experiments shown.
D. Xist RNA FISH after YY1 knockdown in female ES cells. Percentage of Xist+ cells and sample sizes shown.
YY1 is an RNA-binding protein and serves as receptor for Xist
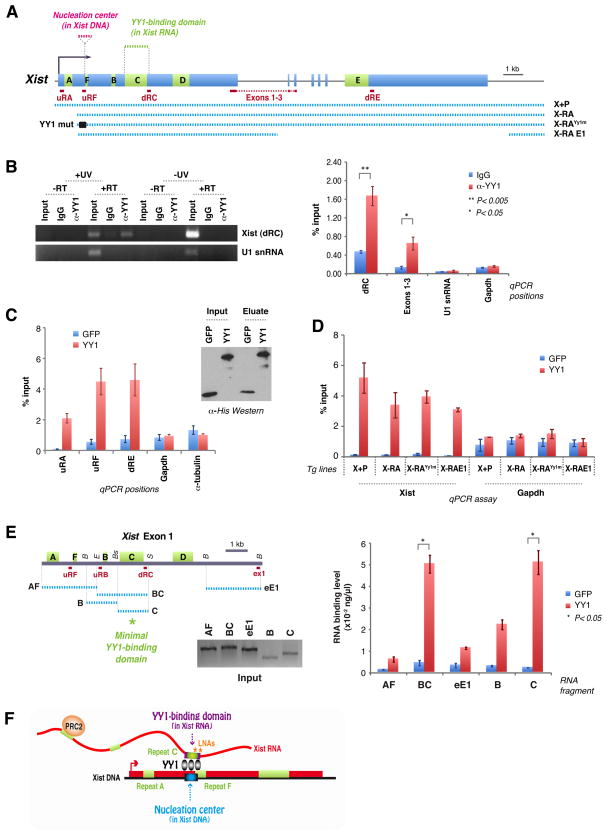
If YY1 serves as docking protein for Xist particles, it must directly interact with Xist RNA. To look for interactions in vivo, we performed RNA immunoprecipitation (RIP) with YY1 antibodies following UV-crosslinking of RNA to protein in MEFs. qRT-PCR of YY1 pulldown material showed significant co-immunoprecipitation of Xist RNA (Fig. 6A,B). The interaction was not detected without UV crosslinking, in RT-negative samples, and when IgG antibodies were used. Moreover, the abundant U1 snRNA was not co-immunoprecipitated. Because UV crosslinking occurs at near-zero Angstrom, the observed pulldown suggests specific and direct Xist-YY1 interaction in vivo.
Figure 6. YY1 is an RNA-binding protein that bridges RNA and chromatin.
A. Map of Xist, transgenes, and RT-PCR amplicons.
B. UV-crosslink RIP of female MEFs, followed by qRT-PCR for Xist (dRC, Exons 1–3) or RNA controls (U1 snRNA, Gapdh). YY1 antibodies or IgG used. 1% input. -UV and -RT controls performed in parallel. Left panel, EtBr-stained gel. Right panel, RT-PCR quantitation. Averages ± SE of 3 independent experiments.
C. RNA pulldown assay using purified His-YY1 or His-GFP (Western blot) and WT female ES RNA. qRT-PCR at 3 different Xist positions (uRF, uRA, dRE) and two controls (Gadph, α-tubulin). Averages of 5 independent experiments ± SE.
D. RNA pulldown assay using RNAs from transgenic lines after dox induction. qRT-PCR performed at dRC. Averages ± SE for 3 independent experiments.
E. RNA pulldown assay using equal molar amounts of in vitro-transcribed RNA fragments AF (2.5kb), BC (2.5kb), eE1 (2.5kb), B (1.2kb), and C (1.8kb) as illustrated in the map. Quantitated by qRT-PCR. 20% of input shown on the gel. P calculated using t-test. B, BamHI; E, EcoRI; Bs, BstBI; S, ScaI. Averages of 2 independent experiments ± SE.
F. Bivalent function of YY1. YY1 contacts Xist RNA and DNA via different sequences. Asterisks, positions of blocking LNAs (Sarma et al., 2010).
To probe its nature, we carried out RNA pulldown assays in vitro using purified recombinant His-tagged YY1 proteins. To ask if YY1 preferentially binds Xist RNA among a complex pool of cellular RNAs, we purified total RNA from female MEFs and quantitated the interaction between YY1 and Xist relative to other RNAs. At multiple qPCR positions (uRF, uRA, dRE), Xist pulldown by YY1 was enriched above background (GFP)(Fig. 6C). Neither Gapdh nor α-tubulin RNA showed enrichment. Therefore, consistent with in vivo RIP, YY1 specifically and directly interacts with Xist in vitro.
Site-directed mutagenesis showed that, although YY1 binds exon 1 DNA via the motif, AAnATGGCG, YY1 cannot bind Xist RNA via the corresponding motif in the RNA (AAnAUGGCG) (Fig. 4). To determine where YY1 binds RNA, we carried out pulldown assays using a panel of mutated transgenic RNAs (Fig. 6A). To isolate transgenic RNAs from endogenous Xist, we introduced the transgenic constructs into male MEFs, induced expression using doxycycline, isolated RNA, and tested the RNA for binding to YY1 in a pulldown assay. All four transgenic RNAs bound YY1 specifically (Fig. 6D, P<0.02 in all cases). The control Gapdh RNA did not demonstrate significant differences between pulldown with YY1 versus GFP. These results show that deleting Repeat A (X-RA) and mutating the clustered YY1 motifs (X-RAYY1m) had no effect on Xist-YY1 interactions, further supporting the notion that YY1 does not bind Xist via AAnAUGGCG.
The ability of X-RAE1 RNA to bind YY1 delimits the interaction domain to the portion of exon 1 downstream of Repeat A (Fig. 6A). To pinpoint Xist RNA’s YY1-binding domain, we generated RNA subfragments, in vitro-transcribed and purified each, and tested them for YY1 binding in a pulldown assay (Fig. 6E). Although several RNA domains showed more binding to YY1 than background (GFP), the difference was strongest and statistically significant only for fragments containing Repeat C, a conserved C-rich element unique to Xist that is repeated 14 times in tandem (Brockdorff et al., 1992; Brown et al., 1992). Repeat C by itself showed 20-fold enrichment (P=0.047). A fragment containing both Repeats B and C showed 10-fold better binding than background (P=0.033). Repeat B might also have some affinity for YY1, as it showed 5-fold enrichment and the difference bordered statistical significance (P= 0.053). Repeat C’s binding to YY1 was especially interesting, given recent observation that locked nucleic acid (LNA) antagomirs against this repeat displace Xist RNA from Xi without affecting RNA stability (Sarma et al., 2010) – a finding that suggested Repeat C as an anchoring sequence to Xi. We propose that Repeat C, and potentially also Repeat B, of Xist RNA make direct contact with YY1, which in turn anchors the Xist particle to Xi via a trio of DNA elements near Repeat F (Fig. 6F). Thus, YY1 is an RNA-binding protein that serves as receptor for the Xist silencing complex on Xi.
DISCUSSION
Here we have elucidated how Xist RNA loads onto Xi and establishes its action in cis. Our work identifies a primary loading site – dubbed the nucleation center – and shows that bound YY1 proteins trap the Xist silencing complex before the RNA promulgates along Xi. A most surprising observation is that Xist is not inherently cis-acting. The RNA freely diffuses, remains stable when displaced from chromatin, and can trans-migrates between any chromosome bearing an open loading site. Importantly, the RNA’s selective action on Xi is not only the result of Xi-specific transcription, but is also due to YY1’s allele-specific binding to the nucleation center of Xi. Even so, YY1 alone cannot specify the Xi fate, as Xist does not nucleate at any other of a large number of genome-wide YY1 sites. We surmise that YY1 and as yet undefined accessory factors – such as lncRNAs like Tsix which are specific to X – may conspire to define the nucleation center.
Our study was initially motivated by “squelching”, a term coined to describe how overexpressed transcription factors indirectly repress gene expression by competing for general transcription machinery (Gill and Ptashne, 1988). Xist squelching is conceptually similar in that supernumerary copies of Xist create direct competition between X-linked and transgene-based binding sites for a limited pool of Xist particles. Why is stripping of Xist RNA from Xi so complete, as indeed transgenes are unlikely to be intrinsically more attractive than Xi? Xist’s preference for transgenes likely arises from the transgene’s multicopy nature, which creates more binding sites and might achieve greater avidity than a single YY1 cluster on Xi. Squelching is thus an RNA migration and localization phenomenon. RNA migration is, however, not directional.
An apparent contradiction may occur between our work and current thinking regarding the nature of Xist RNA and the timing of its action. Many studies have shown that Xist RNA cannot diffuse between chromosomes and operates only during an early developmental time window (Lee et al., 1996; Lee et al., 1999; Wutz and Jaenisch, 2000; Wutz et al., 2002; Kohlmaier et al., 2004; Chow et al., 2007; Jonkers et al., 2008), though work in human transgenic systems have sometimes hinted at partial XIST effects in somatic cells (Clemson et al., 1998; Chow et al., 2007). There are several major differences, however, between prior studies and our work. Previous studies mostly examined male cells, whereas we have examined female cells. Furthermore, previous studies generally introduced the transgenes into ES cells and then investigated their effects in differentiated cell types either ex vivo or in vivo in mice; by contrast, our transgenes were introduced de novo into post-XCI cells. We suggest that our approach bypassed epigenetic modifications (that normally occurs during development) which would have occluded ectopic nucleation sites. Transgenes introduced directly into post-XCI cells as “naked” DNA may retain an open configuration, bind YY1, and attract diffusing Xist particles. Thus, the combination of studying female cells and introducing naked transgene DNA not subjected to the usual developmental programming accounts for our ability to detect squelching and Xist transmigration.
We show that YY1 is bivalent, binding both DNA and RNA. Specific YY1-DNA contacts are required to formulate the nucleation center, and specific YY1-RNA interactions are necessary to load Xist particles (Fig. 6F). YY1’s bivalency bridges regulatory long ncRNA and its chromatin target. Its zinc fingers may mediate the interaction with both DNA and RNA, as some zinc finger proteins can bind RNA as well as DNA in vitro (Iuchi, 2001). Interestingly, although YY1 binds the AAnATGGCG motif on DNA, its interaction with Xist RNA does not occur through the corresponding motif on the RNA. Instead it contacts RNA via Repeat C, a C-rich repeat unique to Xist and one of the best-conserved elements within eutherian Xist/XIST (Brockdorff et al., 1992; Brown et al., 1992). A recent study showed that targeting Repeat C using locked nucleic acids (LNAs) causes rapid Xist displacement from Xi (Sarma et al., 2010). This effect was not observed with LNAs against Repeat B or any other tested sequence. Thus, antagomirs against Repeat C phenocopied the YY1-knockdown. In light of current findings, we suggest that Repeat C LNAs functioned by inhibiting Xist-YY1 interactions and caused release of Xist particles from Xi (Fig. 6F). Repeat A is not required. A human XIST transgene previously shown to localize poorly without the Repeat A region (Chow et al., 2007) actually also deleted three of eight YY1 sites in the human sequence corresponding to the mouse nucleation center. The collective evidence demonstrates that Xist RNA must interact with two proteins for XCI – with EZH2 (PRC2) via Repeat A to form the silencing complex, and with YY1 via Repeat C to load onto the X (Fig. 6F).
Our data have implications for Polycomb regulation. Because the PRC2 subunits, EED, EZH2, SUZ12, and RBAP48, lack sequence-specific DNA binding subunits, cis-acting long ncRNAs have been proposed as locus-specific recruiting tools (Zhao et al., 2008; Lee, 2009, 2010). The concept of YY1 as docking protein is intriguing, given that the related protein, PHO, has been proposed to recruit Polycomb complexes in fruitflies (Ringrose and Paro, 2004; Schwartz and Pirrotta, 2008). Mammalian YY1 has been implicated as a binding partner for PRC2 (Atchison et al., 2003; Wilkinson et al., 2006). This idea has been debated, however, as YY1 has not generally co-purified with PRC2 (Landeira et al., 2010; Li et al., 2010), mutating YY1 sites in HOX-D does not abrogate PRC2 binding (Woo et al., 2010), and YY1 motifs are not enriched near PRC2-binding sites (Mendenhall et al., 2010). Nevertheless, our work demonstrates that YY1 is required for Xist loading and, by inference, for Polycomb recruitment in the context of XCI. XCI may be a special case of PRC2 regulation that involves YY1.
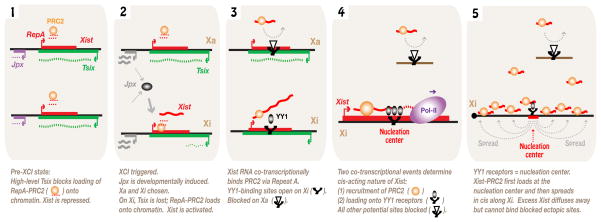
In putting this work in context, we propose that the initiation of XCI and the harnessing of Xist to act strictly in cis result from a series of tightly regulated RNA-protein interactions (Fig. 7). Xist is controlled by two ncRNA switches, Tsix and Jpx, with Tsix blocking Xist expression and Jpx activating it (Tian et al., 2010). In the pre-XCI state, high Tsix and low Jpx expression maintains the activity of both Xs. At the onset of XCI, persistent Tsix expression on the chosen Xa prevents upregulation of Xist (Lee and Lu, 1999). On the chosen Xi, loss of Tsix creates a permissive state for Xist activation by enabling RepA RNA to recruit PRC2 to the Xic and rendering the Xist promoter poised for activation (Zhao et al., 2008). At the same time, the developmentally timed induction of Jpx RNA supplies the required activator for high-level Xist expression. Xist RNA co-transcriptionally recruits PRC2 via its Repeat A motif, but without a mechanism to anchor this complex, Xist-PRC2 freely diffuses away. Our current work shows that a strategically placed nucleation center <1.0 kb downstream of Repeat A traps the Xist-PRC2 complex as the RNA is synthesized and the complex is assembled. Thus, we envision that two co-transcriptional events – the loading of PRC2 and the trapping of the Xist-PRC2 complex by YY1 – account for the cis-acting nature of Xist. Under normal circumstances, Xist cannot act ectopically (though it diffuses) because potential loading sites either lack crucial factors (e.g., YY1) or are blocked by developmentally regulated factors, as exemplified by the allelic equivalent on Xa. Our data support the concept of a single nucleation center within which translocation to chromosome-wide binding sites must originate. Such “spreading elements” cannot function autonomously, as our data suggest that Xist must first engage the nucleation center. How the Xist-PRC2 complex translocates in cis along the X-chromosome remains an open and tantalizing question.
Figure 7. Summary and Model.
Events at the initiation of XCI. Co-transcriptional recruitment of PRC2 and docking onto the YY1-bound nucleation center account for the cis-acting nature of Xist RNA.
EXPERIMENTAL PROCEDURES
Transgene constructs
Transgenes were constructed from an Xist plasmid, pSx9. Xist inserts were generated by PCR and replaced the corresponding region in pSx9 by digesting with SalI and PmlI. All constructs were put into doxycycline-inducible pTRE2hyg (Clontech). 3′ truncations were generated by excising a 13.5-kb PasI transgene fragment. For X-RAYY1m, YY1 sites were altered with QuikChange® Multi Site-Directed Mutagenesis Kit (Stratagene).
Cell lines
XaXiΔXist and XiXaΔXist fibroblasts and TsixTST/+ cells have been described (Zhang et al., 2007; Ogawa et al., 2008). For the tet-inducible system, rt-TA expressing fibroblasts were isolated from 13.5-dpc Rosa26-M2rtTA+/− embryos (Hochedlinger et al., 2005), immortalized with SV-40 large T-antigen, cloned by limiting dilution, and one male and female clone were characterized further. Ploidy was checked by metaphase analysis and X-painting. To generate transgenic MEF lines, 15 μg of linearized DNA was introduced into ~4×106 cells by electroporation (200 V, 1,050 μF), selected in 250 μg/ml hygromycin B, and clones were picked after 2 weeks. Autosomal integration was confirmed by DNA FISH.
RNA FISH, DNA FISH, and immunostaining
Experiments were performed as described (Zhang et al., 2007). Xist RNA was detected using an Xist-riboprobe cocktail unless indicated. RA, E1, E7, and the transgene-specific probe, pSacBII, were labeled by nick translation (Roche). For immunostaining, cells were blocked with PBS, 0.3% Tween20, 3% BSA for 15 minutes before primary antibody incubation. H3K27me3 antibodies were from Active Motif (#39535). DNA FISH combined with RNA FISH or immunostaining was performed as follows: RNA FISH or immunostaining was performed first. Images were captured and their positions recorded on a Nikon Eclipse 90i microscope workstation with Volocity software (Improvision). Slides were then re-fixed in 4% paraformaldehyde, treated with RNaseA to remove RNA signals, and denatured for DNA FISH. After overnight hybridization at 37°C, slides were re-imaged at recorded positions.
Quantitative RT-PCR
Total RNA was isolated using TRIzol® (Invitrogen) and treated with TURBO DNase (Ambion). 500 ng was reverse-transcribed with random primers (Promega) using Superscript® III reverse transcriptase (Invitrogen). Control reactions without reverse transcriptase (-RT) were also prepared. qRT-PCR was performed using iQ SYBR Green Supermix (Bio-Rad) on the CFX96™ system (Bio-Rad). For each primer pair, a standard curve was generated using serial 10-fold dilutions of a plasmid containing the corresponding DNA. Copy numbers of PCR products were determined by comparison to these standard curves. Melting curve analyses showed a single peak for each primer pair, suggesting homogeneity of PCR products. Expression levels were normalized to either α-Tubulin or Gapdh levels. Primer pairs were: uXist F: 5′-TTATGTGGAAGTTCTACATAAACG-3′, R: ACCGCACATCCACGGGAAAC; uRA F: CGGTTCTTCCGTGGTTTCTC, R: GGTAAGTCCACCATACACAC; Exons 1–3 F: GCTGGTTCGTCTATCTTGTGGG, R: CAGAGTAGCGAGGACTTGAAGAG; dRE F: CCCAATAGGTCCAGAATGTC, R: TTTTGGTCCTTTTAAATCTC; Tg-A F: CCGGGACCGATCCAGCCTCC, R: GGTAAGTCCACCATACACAC; Tg-B F: CCGGGACCGATCCAGCCTCC, R: AGCACTGTAAGAGACTATGAACG; α-tubulin F: CTCGCCTCCGCCATCCACCC, R: CTTGCCAGCTCCTGTCTCAC; Gapdh F: ATGAATACGGCTACAGCAACAGG, R: GAGATGCTCAGTGTTGGGGG; Ctcf F: GTAGAAGAACTTCAGGGGGC, R: CTGCTCTAGTGTCTCCACTTC; Yy1 F: CGACGGTTGTAATAAGAAGTTTG, R: ATGTCCCTTAAGTGTGTAG; U1 snRNA F: GGAAATCATACTTACCTGGC, R: AAACGCAGTCCCCCACTACC; uRF-A F: CTCGACAGCCCAATCTTTGTT, R: ACCAACACTTCCACTTAGCC; uRB F: ACTCATCCACCGAGCTACT, R: GATGCCATAAAGGCAAGAAC; ex1 F: GCTGGTTCGTCTATCTTGTGGG, R: CCTGCACTGGATGAGTTACTTG.
siRNA transfection
siRNAs (Integrated DNA Technologies) were sequences were: C1, 5′-CAGAGAAAGTAGTTGGTAA-3′; C3, TGGTCAAGCTTGTAAATAA; Y1, ACAGAAAGGGCAACAATAA; Y2, GCTCAAAGCTAAAACGACA. Control siRNA was purchased from Invitrogen (#12935-200). Cells were transfected with siRNAs at a final concentration 20 nM using Lipofectamine™ RNAiMAX (Invitrogen). For both CTCF and YY1 depletion, transfections were performed twice at 24-hr intervals before cells were collected at indicated timepoints. Knockdown was confirmed with RT-PCR, immunostaining, or Western blotting. Most analyses were performed 48 hrs after transfection when cell growth rates and viabilities were comparable to that of control. CTCF and YY1 antibodies were from Cell Signaling Technology (#2899) and Santa Cruz Biotechnology (sc-7341), respectively.
Chromatin immunoprecipitation (ChIP)
Experiments were performed essentially as described (Takahashi et al., 2000). Approximately 2×106 cells and 2 μg of antibodies were used per ChIP. Before incubating with antibodies, chromatin was treated with 0.2 μg/μl of RNaseA at 37°C for 30 min. Chromatin-antibody complexes were collected with Dynabeads® Protein G (Invitrogen). YY1 antibodies for ChIP were from Santa Cruz (sc-1703). Primer pairs used for qPCR were: uRF-B F: GGGCTGCTCAGAAGTCTAT, R: AAAATCACTGAAAGAAACCAC; dRC F: ACTTTGCATACAGTCCTACTTTACTT, R: GGAAAGGAGACTTGAGAGATGATAC; H19 ICR F: TCGATATGGTTTATAAGAGGTTGG, R: GGGCCACGATATATAGGAGTATGC; Peg3 F: CCCCTGTCTATCCTTAGCG, R: ACTGCACCAGAAACGTCAG.
Electrophoretic mobility shift assay (EMSA)
Recombinant His-YY1 protein was purified as described (Shi et al., 1991) except for protein elution with 250 mM imidazole. For EMSA, 10 fmoles of 5′-end-labeled probes were incubated with 75–300 ng of purified YY1. Binding reactions were carried out for 30 min at room temperature in a final volume of 20μl containing 10 mM Tris-HCl (pH 8.0), 5 mM MgCl2, 0.2 mM ZnCl2, 2mM DTT, 150 mM NaCl, 1 μg poly(dI·dC), 0.1 mg/ml BSA, and 10% glycerol. Complexes were electrophoresed in a 4% acrylamide gel in TBE.
RNA Immunoprecipitation (RIP)
1×107 female MEFs per IP were UV-crosslinked at 254 nm (2000 J/m2) in 10 ml ice-cold PBS and collected by scraping. Cells were incubated in lysis solution (0.5% NP40, 0.5% sodium deoxycholate, 400U/ml RNase Inhibitor (Roche), and protease inhibitor cocktail (Sigma) in PBS pH 7.9) at 4°C for 25 minutes with rotation, followed by DNase treatment (30 U of TURBO DNase, 15 minutes at 37°C). After centrifugation, the supernatant was incubated with 5 μg of IgG or YY1 antibodies immobilized on Dynabeads® Protein G, overnight at 4°C. Beads were washed three times with PBS containing 1% NP40, 0.5% sodium deoxycholate and additional 150mM NaCl (total 300 mM NaCl), and DNase treated (10 U) for 30 min. After washing three more times with the same wash buffer supplemented with 10 mM EDTA, beads were incubated in 100 mM Tris-HCl (pH 7.5), 50 mM NaCl, 10 mM EDTA, 100 μg of Proteinase K (Roche), and 0.5% SDS for 30 min at 55°C. RNA was recovered by phenol-chloroform extraction.
In vitro RNA pulldown assay
2 μg of His-YY1 or His-GFP were immobilized with Dynabeads® His-Tag Isolation and Pulldown (Invitrogen) in PBS with 15 mM β-mercaptoethanol for 2 hrs. 5 μg of total RNA was incubated with protein-bead complexes at room temperature for 1 h in PBS containing 2 mM MgCl2, 0.2 mM ZnCl2, 15 mM β-mercaptoethanol, 100 U/ml RNase Inhibitor, 0.1 mg/ml yeast tRNA (Ambion), 0.05% BSA and 0.2% NP40. RNA was treated with TURBO DNase and renatured by heating and slow cooling. Beads were washed with the same incubation buffer supplemented with additional 150mM NaCl (total 300 mM NaCl). For mutant RNA pulldowns, 500 ng of total RNA from dox-induced transgenic male MEF was used. For RNA fragment pulldowns, each fragment was transcribed in vitro using the MEGAscript® Kit (Ambion). Transcripts were treated with DNase for 1hr at 37°C, TRIzol-purified, and renatured by heating and slow-cooling. 0.5 pmol of RNA and 1μg of protein were used per reaction, and 10% of each pulled-down product was analyzed by qRT-PCR. Standard curves were generated using an Xist plasmid.
Supplementary Material
Acknowledgments
We thank K. Sarma, B. del Rosario, and B. Payer for reading of the manuscript and all lab members for many helpful suggestions. We acknowledge K. Hochedlinger for Rosa26-M2rtTA mice, Y. Shi for His-YY1, B. del Rosario for purified His-GFP. This work was supported by a Korean Research Foundation grant and the MGH ECOR Fund to Y.J., and NIH RO1-GM090278 to J.T.L. J.T.L. is an Investigator of the HHMI.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Atchison L, Ghias A, Wilkinson F, Bonini N, Atchison ML. Transcription factor YY1 functions as a PcG protein in vivo. EMBO J. 2003;22:1347–1358. doi: 10.1093/emboj/cdg124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brockdorff N, Ashworth A, Kay GF, McCabe VM, Norris DP, Cooper PJ, Swift S, Rastan S. The product of the mouse Xist gene is a 15 kb inactive X-specific transcript containing no conserved ORF and located in the nucleus. Cell. 1992;71:515–526. doi: 10.1016/0092-8674(92)90519-i. [DOI] [PubMed] [Google Scholar]
- Brown CJ, Hendrich BD, Rupert JL, Lafreniere RG, Xing Y, Lawrence J, Willard HF. The human XIST gene: analysis of a 17 kb inactive X-specific RNA that contains conserved repeats and is highly localized within the nucleus. Cell. 1992;71:527–542. doi: 10.1016/0092-8674(92)90520-m. [DOI] [PubMed] [Google Scholar]
- Chow JC, Hall LL, Baldry SE, Thorogood NP, Lawrence JB, Brown CJ. Inducible XIST-dependent X-chromosome inactivation in human somatic cells is reversible. Proc Natl Acad Sci U S A. 2007;104:10104–10109. doi: 10.1073/pnas.0610946104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chureau C, Prissette M, Bourdet A, Barbe V, Cattolico L, Jones L, Eggen A, Avner P, Duret L. Comparative sequence analysis of the X-inactivation center region in mouse, human, and bovine. Genome Res. 2002;12:894–908. doi: 10.1101/gr.152902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemson CM, Chow JC, Brown CJ, Lawrence JB. Stabilization and localization of Xist RNA are controlled by separate mechanisms and are not sufficient for X inactivation. J Cell Biol. 1998;142:13–23. doi: 10.1083/jcb.142.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemson CM, McNeil JA, Willard HF, Lawrence JB. XIST RNA paints the inactive X chromosome at interphase: evidence for a novel RNA involved in nuclear/chromosome structure. J Cell Biol. 1996;132:259–275. doi: 10.1083/jcb.132.3.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csankovszki G, Panning B, Bates B, Pehrson JR, Jaenisch R. Conditional deletion of Xist disrupts histone macroH2A localization but not maintenance of X inactivation. Nat Genet. 1999;22:323–324. doi: 10.1038/11887. [DOI] [PubMed] [Google Scholar]
- Donohoe ME, Silva SS, Pinter SF, Xu N, Lee JT. The pluripotency factor Oct4 interacts with Ctcf and also controls X-chromosome pairing and counting. Nature. 2009;460:128–132. doi: 10.1038/nature08098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donohoe ME, Zhang LF, Xu N, Shi Y, Lee JT. Identification of a Ctcf cofactor, Yy1, for the X chromosome binary switch. Mol Cell. 2007;25:43–56. doi: 10.1016/j.molcel.2006.11.017. [DOI] [PubMed] [Google Scholar]
- Essien K, Vigneau S, Apreleva S, Singh LN, Bartolomei MS, Hannenhalli S. CTCF binding site classes exhibit distinct evolutionary, genomic, epigenomic and transcriptomic features. Genome Biol. 2009;10:R131. doi: 10.1186/gb-2009-10-11-r131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gartler SM, Riggs AD. Mammalian X-chromosome inactivation. Annu Rev Genet. 1983;17:155–190. doi: 10.1146/annurev.ge.17.120183.001103. [DOI] [PubMed] [Google Scholar]
- Gill G, Ptashne M. Negative effect of the transcriptional activator GAL4. Nature. 1988;334:721–724. doi: 10.1038/334721a0. [DOI] [PubMed] [Google Scholar]
- Hariharan N, Kelley DE, Perry RP. Delta, a transcription factor that binds to downstream elements in several polymerase II promoters, is a functionally versatile zinc finger protein. Proc Natl Acad Sci USA. 1991;88:9799–9803. doi: 10.1073/pnas.88.21.9799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrich BD, Brown CJ, Willard HF. Evolutionary conservation of possible functional domains of the human and murine XIST genes. Hum Mol Genet. 1993;2:663–672. doi: 10.1093/hmg/2.6.663. [DOI] [PubMed] [Google Scholar]
- Hochedlinger K, Yamada Y, Beard C, Jaenisch R. Ectopic expression of Oct-4 blocks progenitor-cell differentiation and causes dysplasia in epithelial tissues. Cell. 2005;121:465–477. doi: 10.1016/j.cell.2005.02.018. [DOI] [PubMed] [Google Scholar]
- Iuchi S. Three classes of C2H2 zinc finger proteins. Cell Mol Life Sci. 2001;58:625–635. doi: 10.1007/PL00000885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston CM, Nesterova TB, Formstone EJ, Newall AE, Duthie SM, Sheardown SA, Brockdorff N. Developmentally regulated Xist promoter switch mediates initiation of X inactivation. Cell. 1998;94:809–817. doi: 10.1016/s0092-8674(00)81739-0. [DOI] [PubMed] [Google Scholar]
- Jonkers I, Monkhorst K, Rentmeester E, Grootegoed JA, Grosveld F, Gribnau J. Xist RNA is confined to the nuclear territory of the silenced X chromosome throughout the cell cycle. Mol Cell Biol. 2008;28:5583–5594. doi: 10.1128/MCB.02269-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JD, Hinz AK, Bergmann A, Huang JM, Ovcharenko I, Stubbs L, Kim J. Identification of clustered YY1 binding sites in imprinting control regions. Genome Res. 2006;16:901–911. doi: 10.1101/gr.5091406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JD, Kang K, Kim J. YY1’s role in DNA methylation of Peg3 and Xist. Nucleic Acids Res. 2009;37:5656–5664. doi: 10.1093/nar/gkp613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohlmaier A, Savarese F, Lachner M, Martens J, Jenuwein T, Wutz A. A chromosomal memory triggered by Xist regulates histone methylation in X inactivation. PLoS Biol. 2004;2:E171. doi: 10.1371/journal.pbio.0020171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landeira D, Sauer S, Poot R, Dvorkina M, Mazzarella L, Jorgensen HF, Pereira CF, Leleu M, Piccolo FM, Spivakov M, et al. Jarid2 is a PRC2 component in embryonic stem cells required for multi-lineage differentiation and recruitment of PRC1 and RNA Polymerase II to developmental regulators. Nat Cell Biol. 2010;12:618–624. doi: 10.1038/ncb2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laverty C, Lucci J, Akhtar A. The MSL complex: X chromosome and beyond. Curr Opin Genet Dev. 2010;20:171–178. doi: 10.1016/j.gde.2010.01.007. [DOI] [PubMed] [Google Scholar]
- Lee JT. Disruption of imprinted X inactivation by parent-of-origin effects at Tsix. Cell. 2000;103:17–27. doi: 10.1016/s0092-8674(00)00101-x. [DOI] [PubMed] [Google Scholar]
- Lee JT. Lessons from X-chromosome inactivation: long ncRNA as guides and tethers to the epigenome. Genes Dev. 2009;23:1831–1842. doi: 10.1101/gad.1811209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JT. The X as model for RNA’s niche in epigenomic regulation. Cold Spring Harb Perspect Biol. 2010;2:a003749. doi: 10.1101/cshperspect.a003749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JT, Lu N. Targeted mutagenesis of Tsix leads to nonrandom X inactivation. Cell. 1999;99:47–57. doi: 10.1016/s0092-8674(00)80061-6. [DOI] [PubMed] [Google Scholar]
- Lee JT, Lu N, Han Y. Genetic analysis of the mouse X inactivation center defines an 80-kb multifunction domain. Proc Natl Acad Sci U S A. 1999;96:3836–3841. doi: 10.1073/pnas.96.7.3836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JT, Strauss WM, Dausman JA, Jaenisch R. A 450 kb transgene displays properties of the mammalian X-inactivation center. Cell. 1996;86:83–94. doi: 10.1016/s0092-8674(00)80079-3. [DOI] [PubMed] [Google Scholar]
- Li G, Margueron R, Ku M, Chambon P, Bernstein BE, Reinberg D. Jarid2 and PRC2, partners in regulating gene expression. Genes Dev. 2010;24:368–380. doi: 10.1101/gad.1886410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobanenkov VV, Nicolas RH, Adler VV, Paterson H, Klenova EM, Polotskaja AV, Goodwin GH. A novel sequence-specific DNA binding protein which interacts with three regularly spaced direct repeats of the CCCTC-motif in the 5′ flaking sequence of the chicken c-myc gene. Oncogene. 1990;5:1743–1753. [PubMed] [Google Scholar]
- Lucchesi JC, Kelly WG, Panning B. Chromatin remodeling in dosage compensation. Annu Rev Genet. 2005;39:615–651. doi: 10.1146/annurev.genet.39.073003.094210. [DOI] [PubMed] [Google Scholar]
- Lyon MF. Gene action in the X-chromosome of the mouse (Mus musculus L.) Nature. 1961;190:372–373. doi: 10.1038/190372a0. [DOI] [PubMed] [Google Scholar]
- Marahrens Y, Panning B, Dausman J, Strauss W, Jaenisch R. Xist-deficient mice are defective in dosage compensation but not spermatogenesis. Genes Dev. 1997;11:156–166. doi: 10.1101/gad.11.2.156. [DOI] [PubMed] [Google Scholar]
- Mendenhall EM, Koche RP, Truong T, Zhou VW, Issac B, Chi AS, Ku M, Bernstein BE. GC-rich sequence elements recruit PRC2 in mammalian ES cells. PLoS Genet. 2010;6:e1001244. doi: 10.1371/journal.pgen.1001244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer BJ. Targeting X chromosomes for repression. Curr Opin Genet Dev. 2010;20:179–189. doi: 10.1016/j.gde.2010.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nesterova TB, Slobodyanyuk SY, Elisaphenko EA, Shevchenko AI, Johnston C, Pavlova ME, Rogozin IB, Kolesnikov NN, Brockdorff N, Zakian SM. Characterization of the genomic Xist locus in rodents reveals conservation of overall gene structure and tandem repeats but rapid evolution of unique sequence. Genome Res. 2001;11:833–849. doi: 10.1101/gr.174901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa Y, Lee JT. Xite, X-inactivation intergenic transcription elements that regulate the probability of choice. Mol Cell. 2003;11:731–743. doi: 10.1016/s1097-2765(03)00063-7. [DOI] [PubMed] [Google Scholar]
- Ogawa Y, Sun BK, Lee JT. Intersection of the RNA Interference and X-Inactivation Pathways. Science. 2008;320:1336–1341. doi: 10.1126/science.1157676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal Bhadra M, Bhadra U, Birchler JA. Misregulation of sex-lethal and disruption of male-specific lethal complex localization in Drosophila species hybrids. Genetics. 2006;174:1151–1159. doi: 10.1534/genetics.106.060541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park K, Atchison ML. Isolation of a candidate repressor/activator, NF-E1, that binds to the immunoglobulin kappa 3′ enhancer and the immunoglobulin heavy-chain mu E1 site. Proc Natl Acad Sci USA. 1991;88:9804–9808. doi: 10.1073/pnas.88.21.9804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park Y, Mengus G, Bai X, Kageyama Y, Meller VH, Becker PB, Kuroda MI. Sequence-specific targeting of Drosophila roX genes by the MSL dosage compensation complex. Mol Cell. 2003;11:977–986. doi: 10.1016/s1097-2765(03)00147-3. [DOI] [PubMed] [Google Scholar]
- Payer B, Lee JT. X Chromosome Dosage Compensation: How Mammals Keep the Balance. Annu Rev Genet. 2008;42:733–772. doi: 10.1146/annurev.genet.42.110807.091711. [DOI] [PubMed] [Google Scholar]
- Penny GD, Kay GF, Sheardown SA, Rastan S, Brockdorff N. Requirement for Xist in X chromosome inactivation. Nature. 1996;379:131–137. doi: 10.1038/379131a0. [DOI] [PubMed] [Google Scholar]
- Pillet N, Bonny C, Schorderet DF. Characterization of the promoter region of the mouse Xist gene. Proc Natl Acad Sci U S A. 1995;92:12515–12519. doi: 10.1073/pnas.92.26.12515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plath K, Fang J, Mlynarczyk-Evans SK, Cao R, Worringer KA, Wang H, de la Cruz CC, Otte AP, Panning B, Zhang Y. Role of histone H3 lysine 27 methylation in X inactivation. Science. 2003;300:131–135. doi: 10.1126/science.1084274. [DOI] [PubMed] [Google Scholar]
- Pugacheva EM, Tiwari VK, Abdullaev Z, Vostrov AA, Flanagan PT, Quitschke WW, Loukinov DI, Ohlsson R, Lobanenkov VV. Familial cases of point mutations in the XIST promoter reveal a correlation between CTCF binding and pre-emptive choices of X chromosome inactivation. Hum Mol Genet. 2005;14:953–965. doi: 10.1093/hmg/ddi089. [DOI] [PubMed] [Google Scholar]
- Ringrose L, Paro R. Epigenetic regulation of cellular memory by the Polycomb and Trithorax group proteins. Annu Rev Genet. 2004;38:413–443. doi: 10.1146/annurev.genet.38.072902.091907. [DOI] [PubMed] [Google Scholar]
- Sado T, Wang Z, Sasaki H, Li E. Regulation of imprinted X-chromosome inactivation in mice by Tsix. Development. 2001;128:1275–1286. doi: 10.1242/dev.128.8.1275. [DOI] [PubMed] [Google Scholar]
- Sarma K, Levasseur P, Aristarkhov A, Lee JT. Locked nucleic acids reveal sequence requirements and kinetics of Xist RNA localization to the X chromosome. Proc Natl Acad Sci U S A. 2010 doi: 10.1073/pnas.1009785107. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoeftner S, Sengupta AK, Kubicek S, Mechtler K, Spahn L, Koseki H, Jenuwein T, Wutz A. Recruitment of PRC1 function at the initiation of X inactivation independent of PRC2 and silencing. Embo J. 2006;25:3110–3122. doi: 10.1038/sj.emboj.7601187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz YB, Pirrotta V. Polycomb complexes and epigenetic states. Curr Opin Cell Biol. 2008;20:266–273. doi: 10.1016/j.ceb.2008.03.002. [DOI] [PubMed] [Google Scholar]
- Seto E, Shi Y, Shenk T. YY1 is an initiator sequence-binding protein that directs and activates transcription in vitro. Nature. 1991;354:241–245. doi: 10.1038/354241a0. [DOI] [PubMed] [Google Scholar]
- Shi Y, Seto E, Chang LS, Shenk T. Transcriptional repression by YY1, a human GLI-Kruppel-related protein, and relief of repression by adenovirus E1A protein. Cell. 1991;67:377–388. doi: 10.1016/0092-8674(91)90189-6. [DOI] [PubMed] [Google Scholar]
- Silva J, Mak W, Zvetkova I, Appanah R, Nesterova TB, Webster Z, Peters AH, Jenuwein T, Otte AP, Brockdorff N. Establishment of histone h3 methylation on the inactive X chromosome requires transient recruitment of Eed-Enx1 polycomb group complexes. Dev Cell. 2003;4:481–495. doi: 10.1016/s1534-5807(03)00068-6. [DOI] [PubMed] [Google Scholar]
- Stavropoulos N, Rowntree RK, Lee JT. Identification of developmentally specific enhancers for Tsix in the regulation of X chromosome inactivation. Mol Cell Biol. 2005;25:2757–2769. doi: 10.1128/MCB.25.7.2757-2769.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun BK, Deaton AM, Lee JT. A transient heterochromatic state in Xist preempts X inactivation choice without RNA stabilization. Mol Cell. 2006;21:617–628. doi: 10.1016/j.molcel.2006.01.028. [DOI] [PubMed] [Google Scholar]
- Takahashi Y, Rayman JB, Dynlacht BD. Analysis of promoter binding by the E2F and pRB families in vivo: distinct E2F proteins mediate activation and repression. Genes Dev. 2000;14:804–816. [PMC free article] [PubMed] [Google Scholar]
- Tian D, Sun S, Lee JT. The long noncoding RNA, Jpx, is a molecular switch for X chromosome inactivation. Cell. 2010;143:390–403. doi: 10.1016/j.cell.2010.09.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wassenegger M, Heimes S, Riedel L, Sanger HL. RNA-directed de novo methylation of genomic sequences in plants. Cell. 1994;76:567–576. doi: 10.1016/0092-8674(94)90119-8. [DOI] [PubMed] [Google Scholar]
- Wilkinson FH, Park K, Atchison ML. Polycomb recruitment to DNA in vivo by the YY1 REPO domain. Proc Natl Acad Sci U S A. 2006;103:19296–19301. doi: 10.1073/pnas.0603564103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo CJ, Kharchenko PV, Daheron L, Park PJ, Kingston RE. A region of the human HOXD cluster that confers Polycomb-group responsiveness. Cell. 2010;140:99–110. doi: 10.1016/j.cell.2009.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wutz A, Gribnau J. X inactivation Xplained. Curr Opin Genet Dev. 2007;17:387–393. doi: 10.1016/j.gde.2007.08.001. [DOI] [PubMed] [Google Scholar]
- Wutz A, Jaenisch R. A shift from reversible to irreversible X inactivation is triggered during ES cell differentiation. Mol Cell. 2000;5:695–705. doi: 10.1016/s1097-2765(00)80248-8. [DOI] [PubMed] [Google Scholar]
- Wutz A, Rasmussen TP, Jaenisch R. Chromosomal silencing and localization are mediated by different domains of Xist RNA. Nat Genet. 2002;30:167–174. doi: 10.1038/ng820. [DOI] [PubMed] [Google Scholar]
- Xu N, Donohoe ME, Silva SS, Lee JT. Evidence that homologous X-chromosome pairing requires transcription and Ctcf protein. Nat Genet. 2007;39:1390–1396. doi: 10.1038/ng.2007.5. [DOI] [PubMed] [Google Scholar]
- Zhang LF, Huynh KD, Lee JT. Perinucleolar targeting of the inactive X during S phase: evidence for a role in the maintenance of silencing. Cell. 2007;129:693–706. doi: 10.1016/j.cell.2007.03.036. [DOI] [PubMed] [Google Scholar]
- Zhao J, Ohsumi TK, Kung JT, Ogawa Y, Grau DJ, Sarma K, Song JJ, Kingston RE, Borowsky M, Lee JT. Genome-wide identification of polycomb-associated RNAs by RIP-seq. Mol Cell. 2010;40:939–953. doi: 10.1016/j.molcel.2010.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Sun BK, Erwin JA, Song JJ, Lee JT. Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science. 2008;322:750–756. doi: 10.1126/science.1163045. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.