Summary
Somatic cell nuclear transfer and transcription factor-based reprogramming revert adult cells to an embryonic state, and yield pluripotent stem cells that can generate all tissues. These two reprogramming methods reset genomic methylation, an epigenetic modification of DNA that influences gene expression, by different mechanisms and kinetics, leading us to hypothesize that the resulting pluripotent stem cells might have different properties. Here we observe that low passage induced pluripotent stem cells (iPSC) derived by factor-based reprogramming harbor residual DNA methylation signatures characteristic of their somatic tissue of origin, which favors their differentiation along lineages related to the donor cell, while restricting alternative cell fates. Such an “epigenetic memory” of the donor tissue could be reset by differentiation and serial reprogramming, or by treatment of iPSC with chromatin-modifying drugs. In contrast, the differentiation and methylation of nuclear transfer-derived pluripotent stem cells were more similar to classical embryonic stem cells than were iPSC, consistent with more effective reprogramming. Our data demonstrate that factor-based reprogramming can leave an epigenetic memory of the tissue of origin that may influence efforts at directed differentiation for applications in disease modeling or treatment.
Introduction
Direct reprogramming of somatic cells with the transcription factors Oct4, Sox2, Klf4, and c-Myc1 yields induced pluripotent stem cells (iPSC) with striking similarity to embryonic stem cells from fertilized embryos (fESC). Like fESC, iPSC form teratomas, differentiated tumors with tissues from all three embryonic germ layers, and when injected into murine blastocysts contribute to all tissues, including the germ line. iPSC from mouse embryo fibroblasts generate “all-iPSC mice” following injection into tetraploid blastocysts2, thereby satisfying the most stringent criterion of pluripotency3. Embryonic tissues are the most efficiently reprogrammed, producing iPSC that are nearly identical to fESC. In contrast, reprogramming from accessible adult tissues, most applicable for modeling diseases and generating therapeutic cells, is inefficient and limited by barriers related to the differentiation state and age of the donor’s cells4,5,6. Aged cells have higher levels of Ink4/Arf, which limits the efficiency and fidelity of reprogramming5. Moreover, terminally differentiated blood cells reprogram less efficiently than blood progenitors6. As with cloning by nuclear transfer in frogs and mice, the efficiency and yield of reprogrammed genomes declines with increasing age and differentiation status of the donor cell7, and varies with the methylation state of the donor nucleus8.
Different tissues show variable susceptibility to reprogramming. Keratinocytes reprogram more readily than fibroblasts9, and iPSC from stomach or liver cells harbor fewer integrated proviruses than fibroblasts, suggesting they require lower levels of the reprogramming factors to achieve pluripotency10. When differentiated into neurospheres, iPSC from adult tail-tip fibroblasts retain more teratoma-forming cells than iPSC from embryonic fibroblasts, again indicating heterogeneity based on the tissue of origin11. Moreover, cells can exist in intermediate states of reprogramming that interconvert with continuous passage or treatment with chromatin-modifying agents12,13. Although generic iPSC are highly similar to fESC, in practice iPSC generated from various tissues may harbor significant differences, both functional and molecular.
Transcription factor reprogramming differs markedly from nuclear transfer, particularly with regard to DNA demethylation, which commences immediately upon transfer of a somatic nucleus into ooplasm14, but occurs over days to weeks during the derivation of iPSC13. Because demethylation is a slow and inefficient process in factor-based reprogramming, we postulated that residual methylation might leave iPSC with an “epigenetic memory,” and that methylation might be more effectively erased by nuclear transfer. Here we compare the differentiation potential and genomic methylation of pluripotent stem cells (iPSC, ntESC, and fESC), and find evidence that iPSC indeed retain a methylation signature of their tissue of origin.
Results
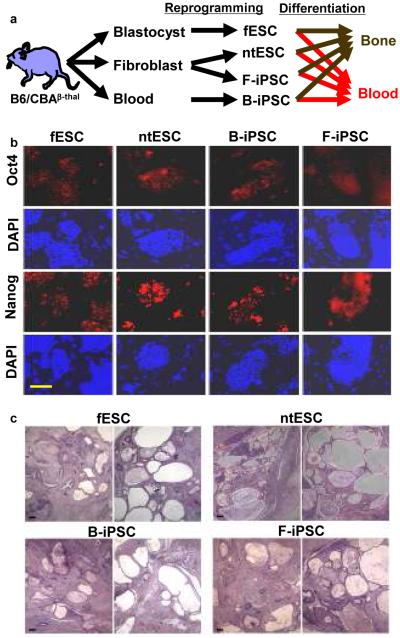
Initially we sought to compare the in vivo engraftment potential of hematopoietic stem cells derived from fESC, ntESC, and iPSC in a mouse model of thalassemia. However, even in vitro we observed strikingly different blood-forming potential; thus, we focused here instead on understanding this phenomenon. Our initial set of pluripotent stem cells were derived from the hybrid C57BL/6 x CBA (B6/CBAF1) strain carrying a deletion in the beta-globin locus15, which is otherwise irrelevant to this study (Fig. 1a). We isolated fESC cells from naturally fertilized embryos and derived ntESC cells from nuclei of dermal fibroblasts8. We infected early bone marrow cells (Kit+, Lin−, CD45+) or dermal fibroblasts from aged mice with retroviral vectors carrying Oct4, Sox2, Klf4, and Myc, and selected blood-derived and fibroblast-derived iPSC colonies (B-iPSC, F-iPSC). Hematopoietic progenitors and fibroblasts yielded a comparable frequency of reprogrammed colonies (0.02%), which consistent with prior reports5, was lower than the yield from fibroblasts of a juvenile mouse (0.1%). We characterized the fESC, ntESC, and iPSC lines for expression of Oct4 and Nanog by immunohistochemistry, and demonstrated multi-lineage differentiation potential in teratomas (Fig. 1b, c; Supplementary Fig. 1). By criteria typically applied to human samples and appropriate for a therapeutic model3, all stem cell lines manifest pluripotency.
Figure 1. Pluripotent stem cells and their characterization.
a, Experimental schema. fESC, ntESC, F-iPSC, and B-iPSC were derived from B6/CBA F1 mice by reprogramming and/or cell culture, characterized for pluripotency by criteria applied to human cells, followed by differentiation analysis for osteogenic or hematopoietic lineages. b, Expression of pluripotency markers NANOG and OCT4 by immunohistochemistry. 4,6-Diamidino-2-phenylindole (DAPI) staining for total cell content. Feeder fibroblasts serve as internal negative controls. c, Teratoma analysis: tumor histology from indicated cell lines shows highly cystic structures consisting of differentiated elements of all three embryonic germ layers. Scale bar, 200μm.
Differentiation of pluripotent stem cells
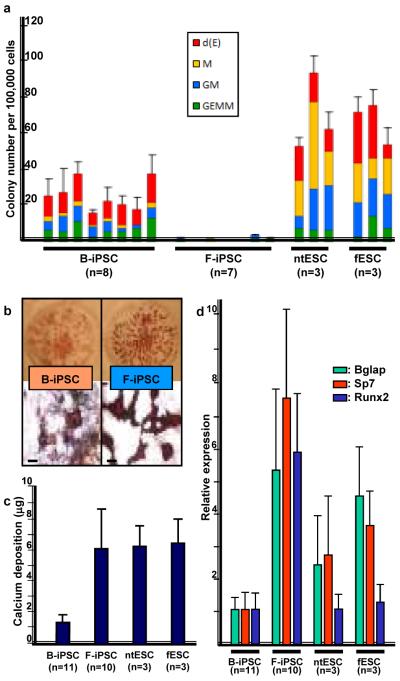
To test blood potential, we differentiated multiple pluripotent stem cell clones into embryoid bodies (EBs), dissociated cells, and assayed for hematopoietic colony forming cells16. All pluripotent cells generated comparable EBs but markedly different numbers of hematopoietic colonies. Consistently, blood-derived B-iPSC yielded more hematopoietic colonies than F-iPSC (Fig. 2a). Hematopoietic colony formation from ntESC and fESC were higher than the iPSC lines.
Figure 2. Differentiation of cell lines.
a, Hematopoietic colony number per 100,000 EB cells differentiated from indicated cell lines. b, Alizarin Red staining of osteogenic cultures of B-iPSC (left) and F-iPSC (right). Top: 3 cm culture dish; Bottom: stained colonies; scale bar, 500 μm. c, Quantification of elemental calcium by inductively coupled plasma – atomic emission spectroscopy49 in 5×105 cells after osteogenic differentiation of indicated cell lines. d, Q-PCR of osteogenic genes, Bglap, Sp7, and Runx2 in indicated cell lines after osteogenic differentiation. Gene expression was normalized to Actin. n=number of independent clones tested. Error bars=s.d.
We then tested differentiation into osteoblasts, a mesenchymal lineage that can be derived from fibroblasts17,18. By alizarin red staining, a marker of osteogenic cells19, F-iPSC produced more sharply defined osteogenic colonies (Fig. 2b), deposited more elemental calcium (Fig. 2c), and showed higher expression of three osteoblast-associated genes (Fig. 2d) than B-iPSC. By these criteria, F-iPSC show enhanced osteogenic potential, reflecting a propensity to differentiate towards a mesenchymal lineage. In contrast, ntESC cells behaved comparably to fESC in hematopoietic and osteogenic assays.
DNA methylation of pluripotent stem cells
We hypothesized that the different pluripotent cells might harbor different patterns of genomic DNA methylation; thus, we performed Comprehensive High-throughput Array-based Relative Methylation (CHARM) analysis, which interrogates ~4.6 million CpG sites, including almost all CpG islands and nearby sequences termed shores20,21, but does not assess non-CpG methylation. We determined the number of differentially methylated regions (DMRs) between pair-wise comparisons, using a threshold area cutoff of 2, corresponding to a 5% false discovery rate (FDR22; Supplementary Table 1a). By this analysis, ntESC were most similar to fESC (only 229 DMRs), whereas F-iPSC differed most extensively (5304 DMRs). Relative to fESC, hypermethylated DMRs predominated for F-iPSC (3349=63%) and B-iPSC (516=74%). Highlighting their functional differences, 5202 DMRs were identified between B-iPSC and F-iPSC. We confirmed the results of CHARM analysis by bisulfite pyrosequencing of multiple loci (Supplementary Fig. 2).
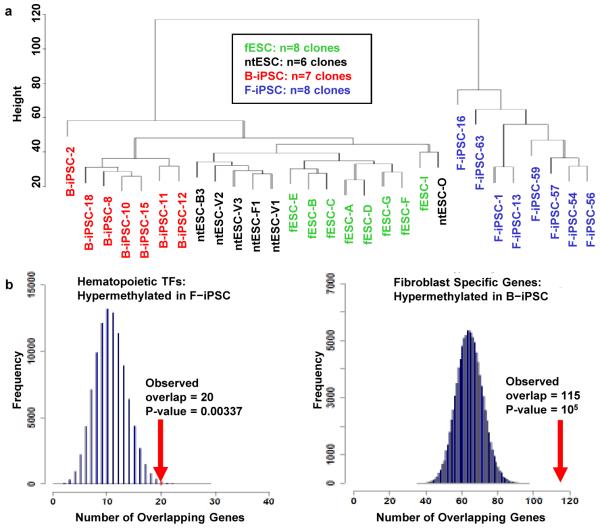
Unsupervised hierarchical clustering of DMRs between B-iPSC and F-iPSC easily distinguished iPSC from ntESC and fESC, which cluster together (Fig. 3a). B-iPSC cluster nearer to ntESC and fESC than do F-iPSC, which represent a strikingly separate cluster. These data indicate that the methylation patterns of ntESC are more like fESC than are either iPSC.
Figure 3. Analysis of methylation in stem cell lines.
a, Cluster dendrogram using probes from DMRs that distinguish B-iPSC and F-iPSC. Cell clones are described in Supplemental Table 5. b, Enrichment of DMRs for hematopoiesis and fibroblast-related transcription factors in B-iPSC and F-iPSC, relative to chance (blue histogram; 100,000 random permutations). Left panel: 20 of 74 hematopoiesis-related transcription factors overlap DMRs hypermethylated in F-IPSC (p=0.0034); Right panel: 115 of 764 fibroblast-specific genes overlap DMRs hypermethylated in B-iPSC (p=10−5).
Several lines of evidence support a mechanistic link between differential methylation and hematopoietic propensity of iPSC lines. First, literature survey of genes for the top 24 DMRs that distinguish B-iPSC and F-iPSC links 11 to hematopoiesis and 3 to osteogenesis (Supplementary Table 2). Of the 11 hematopoietic loci, 10 are hypermethylated in F-iPSC relative to B-iPSC. Second, of 74 hematopoietic transcription factors23, 20 are in or near DMRs that are hypermethylated in F-iPSC versus B-iPSC, twice that predicted by chance (p=0.0034; Fig. 3b left panel, Supplementary Fig. 3a, and Supplementary Table 3). Similarly, of 764 fibroblast-specific genes, 115 are hypermethylated in B-iPSC, twice that predicted by chance (p=10−5; Fig 3b right panel). Given the correlation between methylation and transcriptional silencing24, our data suggest that iPSC harbor epigenetic marks antagonistic to cell lineages distinct from the donor cell type.
We asked whether DMRs that distinguish B-iPSC from fESC might allow us to identify their hematopoietic lineage of origin. In a separate CHARM experiment 25, we examined genome-wide methylation in highly purified multipotent and lineage-specific hematopoietic progenitors. Comparing DMRs in B-iPSC to those that define hematopoietic progenitors, we observed that B-iPSC cluster alongside Common Myeloid Progenitors (CMP) and distant from Common Lymphoid Progenitors (CLP; Supplementary Fig. 4a and Supplementary Table 4), which is notable given that B-iPSC were derived from Kit+, lineage-negative myeloid marrow precursors. Next, we asked whether the tissue of origin (bone marrow vs fibroblast) could be identified by the methylation state of tissue specific DMRs in F-iPSC, B-iPSC, and Bl-iPSC (a B lymphocyte-derived iPSC line described below). Using DMRs that distinguish fibroblast and bone marrow, and examining methylation in iPSCs and somatic cells from two different genetic backgrounds (B6CBA and B6129), we found that F-iPSC cluster alongside fibroblasts, and distant from bone marrow (Supplementary Fig. 4b). Similarly, the hematopoietic-derived B-iPSC and Bl-iPSC grouped with somatic cells from bone marrow. Thus, residual methylation indicates the tissue of origin of iPSC, and for blood-derivatives even their precise lineage, further supporting the phenomenon of epigenetic memory in iPSC.
Reprogrammed state of iPSC and ntESC
We postulated that the differing methylation signatures of B-iPSC, F-iPSC, and ntESC reflect disparate reprogramming, and confirmed this by two independent computational analyses. First, we overlapped DMRs that distinguish B-iPSC, F-iPSC, and ntESC from fESC with genes specifically expressed in undifferentiated murine fESC26. By this analysis, ntESC showed the fewest DMRs at loci corresponding to the most highly expressed fESC-specific genes, and B-iPSC showed fewer DMRs at these loci than F-iPSC (Supplementary Fig. 5a). Second, we overlapped DMRs with the DNA binding locations for seven transcription factors that compose a core protein network of pluripotency27, and found the fewest DMRs at core transcription factor binding sites in ntESC, and less overlap in B-iPSC than in F-iPSC (Supplementary Table 5). These analyses indicate that F-iPSC harbor more residual methylation than B-iPSC at loci directly linked to the gene expression and pluripotency networks of fESC, whereas ntESC show the least differential methylation and appear closest to fESC at these critical loci.
Further analysis of Oct4 and Nanog indicates that although both are detected by immunohistochemistry in B-iPSC and F-iPSC (Fig. 1b), Oct4 mRNA is fully expressed from a demethylated promoter in both types of iPSC, whereas Nanog mRNA is sub-optimally expressed from a promoter that retains considerable methylation in F-iPSC (Supplementary Fig. 6). When assessed by blastocyst chimerism, B-iPSC contribute to all tissues, including the germ line, whereas F-iPSC contribute only poorly (Supplementary Fig. 7a), although they can be found in SSEA1+ germ cells of the gonadal ridge (Supplementary Fig. 7b). Thus, while both B-iPSC and F-iPSC generate robust multi-lineage teratomas, satisfying criteria for pluripotency typically applied to human cells3, broader functional assessments available in the mouse system confirm their differential degree of reprogramming. In this comparison of iPSC derived from accessible tissues of aged adult mice, bone marrow yields stem cells with superior features of pluripotency, but neither iPSC is equivalent to ntESC or fESC.
Stringently-defined pluripotent stem cells
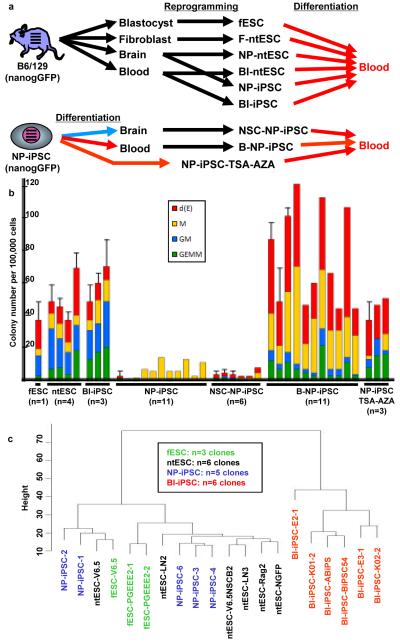
To determine if blood-forming potential differs among cell lines that satisfy more stringent criteria for pluripotency, we analyzed lines derived from a uniform genetic background (B6/129F1) that all express a Nanog-eGFP reporter gene28, and for which pluripotency was demonstrated by blastocyst chimerism and transmission through the germ line (Fig. 4a, upper schema; Supplementary Table 6). These studies involve “secondary” iPSC lines derived from neural progenitor cells (NP-iPSC29) and B-lymphocytes (Bl-iPSC30) of mice chimerized with iPSC carrying proviruses that express doxycycline-inducible reprogramming factors from identical proviral integration sites. NP-iPSC and Bl-iPSC were compared to ntESC generated from neural progenitor cells (NP-ntESC8), blood progenitor cells (B-ntESC31), and fibroblasts (F-ntESC31, 32), as well as fESC.
Figure 4. Stringently-defined pluripotent stem cells and their characterization.
a, Experimental schema. Four horizontal lines indicate integrated proviruses carrying dox-inducible reprogramming factors in some experiments. Characteristics of individual clones in all subsequent panels can be found in Supplemental Table 5. b, Hematopoietic colony number per 100,000 EB cells differentiated from indicated cell lines. n=number of independent clones tested. Error bars=s.d., added for clones repeated three or more times. c, Cluster dendrogram using probes from DMRs that distinguish Bl-iPSC and NP-iPSC.
All cell lines were differentiated into embryoid bodies and assayed for hematopoietic colony forming activity16. Across multiple clones, we observed higher blood forming potential of iPSC derived from B lymphocytes (Bl-iPSC) than from neural progenitors (NP-iPSC; Fig. 4b). In contrast, we observed that ntESC, regardless of tissue origin (fibroblasts, neural progenitors, or T-cells), and fESC displayed an equivalently robust blood forming potential (Fig. 4b). In this independent set of iPSC lines, qualified as pluripotent by stringent criteria, we again observed consistent differences in blood formation, with blood derivatives showing more robust hematopoiesis in vitro than neural derivatives.
Resetting differentiation propensity
Finally, we asked whether we could rescue the poor blood-forming potential of NP-iPSC by differentiation into hematopoietic lineages16,33, followed by a tertiary round of reprogramming back to pluripotency by doxycycline induction of the endogenous reprogramming factors (Fig. 4a, lower schema). As a control, we differentiated NP-iPSCs into neural stem cells8, followed by tertiary reprogramming to pluripotency. Resulting iPSC clones were selected for expression of the Nanog-eGFP reporter and shown to express Oct4 and Nanog by immunohistochemistry (Supplementary Fig. 8) and to chimerize murine blastocysts (Supplementary Fig. 9). The tertiary blood-derived B-NP-iPSC showed higher hematopoietic colony-forming potential than the tertiary NSC-NP-iPSC (Fig. 4b), and generated larger hematopoietic colonies with more cells per colony (Supplementary Fig. 10a, b). These data indicate that the poor blood-forming potential of secondary NP-iPSC can be enhanced by differentiation into hematopoietic progeny, followed by tertiary reprogramming. In contrast, tertiary reprogramming via neural intermediates yields iPSC that retain poor hematopoietic potential.
The reduced blood potential of NP-iPSC might be explained by residual epigenetic marks that restrict blood fates or a lack of epigenetic marks that enable blood formation. We sought to determine whether treatment of NP-iPSC with pharmacologic modulators of gene expression and DNA methylation might reactivate latent hematopoietic potential. We treated NP-iPSC in vitro with Trichostatin A (TSA), a potent inhibitor of histone deacetylase34, and 5-azacytidine (AZA), a methylation-resistant cytosine analogue35. After 18 days of drug treatment, the resulting cells displayed higher blood forming activity (NP-iPSC-TSA-AZA; Fig. 4b). For unclear reasons, tertiary reprogramming through blood intermediates or drug treatment of NP-iPSC produced altered ratios of colony sub-types, perhaps suggesting different efficiencies of lineage reprogramming.
Methylation in secondary and tertiary iPSC
We used CHARM to examine the methylome of the germ-line competent pluripotent stem cells, the tertiary reprogrammed B-NP-iPSC and NSC-NP-iPSC, and the drug-treated NP-iPSC (Fig. 4a). In pair-wise comparisons (Supplementary Table 1b), the NP-iPSC showed only a small number of DMRs relative to fESC (553), fewer than the numbers of DMRs distinguishing ntESC from fESC (679), indicating that selection using the Nanog-GFP reporter and derivation from young donor tissue yields more equivalently reprogrammed cells. Despite equivalent Nanog-GFP expression, B lymphocyte-derived Bl-iPSC harbored more DMRs (1485) relative to fESC than did the NP-iPSC. Cluster dendrogram analysis, employing the most variable DMRs that distinguish Bl-iPSC and NP-iPSC, showed NP-iPSC to be more similar to fESC than are Bl-iPSC, which represent a distinct cluster (Fig. 4c). These data suggest that neural progenitors are more completely reprogrammed to an ESC-like state than blood donor cells. Cluster dendrogram analysis failed to distinguish among NP-iPSC, ntESC, and fESC, but assessment of the overlap of DMRs with loci for highly expressed ESC-specific genes26 and core pluripotency transcription factor binding sites27 indicated differences among these three pluripotent cell types, and reveal that ntESC have the fewest DMRs affecting these critical loci (Supplementary Fig. 5b).
Relative to fESC, hypermethylated DMRs predominated in NP-iPSC and Bl-iPSC (417 (75%) and 1423 (96%), respectively; Supplementary Table 1b), confirming that even when pluripotency is documented by stringent criteria, iPSC retain residual methylation. By analysis of overlapping DMRs, Bl-iPSC cluster with progenitors of the lymphoid lineage (CLP) rather than the myeloid lineage (Supplementary Fig. 4a; Supplementary Table 425). To illustrate this point, the Gcnt2 gene, which encodes the enzyme responsible for the blood group I antigen, and Gata2, a regulator of hematopoiesis and erythropoiesis, are both hypermethylated and transcriptionally silent in the lymphoid lineage25. Bl-iPSC showed hypermethylation at these loci relative to fESC, whereas the myeloid-derived B-iPSC did not (Supplementary Fig. 11). Thus, a methylation signature correctly identifies the blood lineage of origin of B-lymphocyte derived iPSC.
Furthermore, we found that neural-related genes tended to be differentially methylated between Bl-iPSC and NP-iPSC (Supplementary Fig. 3b). Treatment of NP-iPSC with TSA and AZA enhances blood-forming potential and increases hypomethylated DMRs (626; Supplementary Table 1c). We found significant overlap between these DMRs and genes enriched in mouse hematopoietic stem cells (MSigDG signature STEMCELL_HEMATOPOIETIC_UP; Supplementary Fig. 3c), suggesting that drug treatment erases inhibitory methylation signatures at hematopoietic loci.
We compared DMRs in iPSCs with high hematopotietic potential (B-NP-iPSC and NP-iPSC-TSA-AZA) to those with low hematopoietic potential (NP-iPSC and NSC-NP-iPSC), and found that B-NP-iPSC and NP-iPSC-TSA-AZA harbored higher gene-body methylation of Wnt3 (Supplementary Fig. 12), a gene which along with its homologue Wnt3a plays a major role in blood development from fESC36. The blood-deficient NP-iPSC and NSC-NP-iPSC lines lacked gene body methylation. While promoter methylation is repressive, gene body methylation is seen in active genes37. When iPSC were differentiated into embryoid bodies, the blood-prone NP-iPSC-TSA-AZA showed higher levels of Wnt3/3a expression than the blood-deficient NP-iPSC (Supplementary Fig. 13a). Interestingly, supplementation of the culture media with Wnt3a during embryoid body differentiation restored blood-forming potential in the blood-deficient NP-iPSC and NSC-NP-iPSC lines, but had little effect on the already robust hematopoietic potential of B-NP-iPSC (Supplementary Fig. 13b). Albeit preliminary, these data correlate differential gene body methylation and expression of the Wnt3 locus with enhanced blood-forming potential in iPSC lines.
Discussion
Here we demonstrate that iPSC retain an epigenetic memory of their tissue of origin. Our data reveal several important principles that relate to the technical limitations inherent in the process of reprogramming, and which in practice influence the differentiation propensity of specific isolates of iPSC.
First, tissue source influences the efficiency and fidelity of reprogramming9,10,11. From aged mice, blood cells were reprogrammed more closely to fESC than dermal fibroblasts, which yielded only incompletely reprogrammed cells. Neural progenitor-derived iPSC were most similar to fESC, consistent with evidence that such cells can be reprogrammed with fewer transcription factors38. Whereas neural progenitors are not readily accessible, iPSC can be generated by direct reprogramming of human blood39.
Second, analysis of DNA methylation reveals substantial differences between iPSC and embryo-derived ESC (ntESC and fESC). iPSC derived from non-hematopoietic cells (neural progenitors and fibroblasts) retain residual methylation at loci required for hematopoietic fate, which manifests as reduced blood-forming potential in vitro. Residual methylation signatures link iPSC to their tissue of origin, and even discriminate between the myeloid and lymphoid origins of blood-derived iPSC. Prior studies reporting residual hypermethylation in iPSC37,22 did not establish a link between DMRs at specific loci, tissue of origin, and altered differentiation potential. While residual methylation is mostly repressive, we have shown for Wnt3 that residual gene body methylation in blood-derived iPSC is associated with enhanced blood potential. Interestingly, the poor blood potential of neural progenitor-derived iPSC, which lack this epigenetic mark and express lower levels of endogenous Wnt3, can be enhanced by supplementing differentiating cultures with exogenous Wnt3a cytokine, indicating that manipulating culture conditions can overcome epigenetic barriers.
Third, the differentiation propensity and methylation profile of iPSC can be reset. When blood-deficient neural progenitor-derived iPSC (NP-iPSC) are differentiated into blood and then reprogrammed to pluripotency, their blood-forming potential is markedly increased. Alternatively, treatment of NP-iPSC with chromatin-modifying compounds increases blood-forming potential and is associated with reduced methylation at hematopoietic loci. For some applications, epigenetic memory of the donor cell may be advantageous, as directed differentiation to specific tissue fates remains a challenge.
Fourth, nuclear transfer-derived ESC are more faithfully reprogrammed than most iPSC generated from adult somatic tissues. Like the immediate and rapid demethylation of the sperm pronucleus following fertilization, somatic nuclei are rapidly demethylated by nuclear transfer into ooplasm, prompting speculation that the egg harbors an active demethylase14. In contrast, demethylation is a late phenomenon in factor-based reprogramming, and likely occurs passively13. Studying how ooplasm erases methylation might identify biochemical functions that would enhance factor-based reprogramming. Failure to demethylate pluripotency genes is associated with intermediate or partial states of reprogramming12,29,40, and knock-down of the maintenance methyltranferase DNMT1 or treatment with the demethylating agent 5-AZA can convert intermediate states to full pluripotency. Demethylation appears passage dependent13, and reprogramming efficiency correlates with the rate of cell division and the passage number41. In our experiments, we compared pluripotent stem cells of comparable low passage number (Supplemental Table 5), but continued serial passage may homogenize the differentiation potential of pluripotent cell types.
The mRNA expression program of iPSC and fESC are strikingly similar42. Minor differences in mRNA and microRNA expression have been reported40, but removal of transgenes reduces the differences43. The Dlk1-Dio3 locus, whose expression correlates with capacity to generate “all-iPSC” mice44, is not differentially methylated and expressed in at least some iPSC lines that manifest epigenetic memory (our unpublished observations). Thus even the most stringently-defined iPSC might retain epigenetic memory. Importantly, differences between iPSC and fESC may not manifest until differentiation, when the specific loci that retain residual epigenetic marks are expressed, influencing cell fates. Methylation is but one molecular feature of “epigenetic memory” in iPSC. Faulty restoration of bivalent domains, which mark developmental loci with both active and repressive histone modifications 45, and loss of pioneer factors, which in fESC and iPSC occupy enhancers of genes expressed only in differentiated cells46, represent two other potential mechanisms.
Although ideal, generic iPSC may be functionally and molecularly indistinguishable from fESC, we have shown in practice that even rigorously selected iPSC can retain epigenetic marks characteristic of the donor cell that influence differentiation propensity. Epigenetic differences are unlikely to be essential features of iPSC, but rather reflect stochastic variations associated with the technical challenges of achieving complete reprogramming. Given that we lack reporter genes for selecting human iPSC, and cannot qualify their pluripotency by assaying embryo chimerism, the behavior of human cells will likely be influenced by epigenetic memory. Human ESC can also manifest variable differentiation potential47. These data highlight our limited understanding of the epigenetic heterogeneity of pluripotent stem cells, and the need for improved methods to ensure reprogramming of somatic cells to a fully naïve, “ground state” of pluripotency48.
METHODS SUMMARY
ES cells and iPSCs were cultured in ESC media containing 15% FBS, and 1,000 U/ml of LIF. For the reprogramming of somatic cells, retrovirus expressing Oct4, Sox2, Klf4, and Myc were introduced. For the somatic cells containing inducible reprogramming factors, the media was supplemented with 2 ng/ml of doxycycline. For DNA and RNA isolation, fESC or iPSCs were trypsinized and re-plated onto new tissue culture dishes for 45 minutes to remove feeder cells, and nucleic acids were extracted from the non-adherent cell suspension. Genomic DNA methylation analysis and pyrosequencing were performed by previously published methods20,21.
Full Methods and any associated references are available in the online version of the paper at www.nature.com/nature.
Supplementary Material
Acknowledgements
G.Q.D is an investigator of the Howard Hughes Medical Institute and was supported by NIH grants RO1-DK70055 and RO1-DK59279, and special funds received under the American Recovery and Reinvestment Act (RC2-HL102815). K.K. was supported by NIH (K99HL093212-01), LLS (3567-07), and Cooley’s Anemia Foundation. A.P.F was funded by NIH grants R37CA054358 and P50HG003233. I.L.W. was funded by NIH grants R01AI047457, R01AI047458, CA86065, and HL099999, and the Thomas and Stacey Siebel Foundation. L.I.R.E. was supported by Special Fellow Career Development award from the Leukemia and Lymphoma Society. J.S. is supported by a fellowship from the California Institute for Regenerative Medicine (T1-00001).
APPENDIX
METHODS
Tissue culture
Bl-iPSC and NP-iPSC have been described30,50,51. The cells were cultured in standard ES maintenance media.
Generation of B-iPSC and F-iPSC
B-iPSC were generated from bone marrow cells collected from one-year-old B6CBAF1 mice. Early progenitor cells (lin−, CD45+, and cKit+) were sorted by FACS (HemNeoFlow Facility at the Dana Farber Cancer Institute) and stained with lineage-specific antibodies (B220;RA3-6B2, CD19;1D3, CD3;145-2c11, CD4;GK1.5, CD8;536.7, Ter119;ter119, Gr-1;RB6-8C5), CD45 specific antibody (30-F11), and cKit antibody (2B8). 105 sorted cells were infected with retrovirus generated from pMXOct4, pMXSox2, pMXKlf41, and pEYK3.1cMyc52 in 6 well dishes with 0.5 ml of each viral supernatant (total 2 ml per well), and spun at 2500 rpm at 20C for 90 minutes (BenchTop Centrifuge, BeckmanCoulter, Allegra-6R). cMyc was cloned into pEYK3.1 containing two loxp sites to enable removal of the cMyc by Cre treatment. The reprogramming factor-infected cells were plated on to irradiated OP9 feeder cells in 10 cm tissue culture dish in IMDM media (Invitrogen) supplemented with 10% FBS, 1x penicillin/streptomycin/glutamine (Invitrogen), VEGF (R&D Systems, 40ng/ml), Flt (R&D Systems, 100 ng/ml), TPO (R&D Systems, 100ng/ml), and SCF (R&D Systems, 40ng/ml) on day 0. The media were changed on day 2. Cells were collected by media centrifugation and returned to culture during media changes. On day 5, cultured cells were trypsinized and replated on to four 10 cm dishes pre-coated with irradiated mouse embryonic fibroblast in ES maintenance media. Media were changed daily until ES-like colonies were observed. F-iPSC were generated from tail tip fibroblasts of one-year-old B6CBAF1 mice. 106 fibroblast cells were plated onto all wells of a 6 well plate and spin-infected with the four viral supernatants, as for the generation of B-iPSC. Cells were cultured further in DMEM media (Invitrogen) supplemented with 15% FBS, 1x penicillin/streptomycin/glutamine (Invitrogen). On day 5, the cultured cells were trypsinized, and replated in four 10 cm dishes on irradiated mouse embryonic fibroblast with ES maintenance media. Media was changed every day until ES-like colonies were observed.
Differentiation of iPSC to hematopoietic and osteogenic lineages
Hematopoietic colony forming activity of d6 EBs differentiated from pluripotent stem cells was measured in methylcellulose medium with IL3, IL6, Epo, and SCF (M3434, StemCell Tech.) as described16. Hematopoietic colony type was determined on day 10. Colony identity was confirmed by leukostain analysis of cytospin preparation of the methylcellulose colonies. Osteogenic differentiation was performed by culturing pluripotent stem cells in 15 μl hanging drops (800 cells/drop) in ES differentiation media16. Embryoid bodies (EB) from hanging drops were collected at 2 days, transferred to a 10 cm dish of non-tissue culture grade plastic with 10−6 M of retinoic acid, and cultured for 3 days on the shaker (50 rpm) in an incubator. EBs were equally distributed among 3 wells of a 6-well tissue culture dish, and cultured in αMEM media supplemented with 10 % FBS, 1x penicillin/streptomycin/glutamine (Invitrogen), 2 nM triiodothyronine, 1x insulin/transferrin/triacostatin A (Gibco, #51300-044). The media were changed every other day. On day 11, one well of each sample was used to measure calcium concentration, osteogenic gene expression (RNA isolation), and for Alizarin Red staining. For Alizarin Red staining, cells were washed with PBS and fixed with 4% paraformaldehyde for 5 minutes at 20C. Fixed cells were incubated for 15 minutes in Alizarin Red staining solution (Alizarin Red (Sigma, A5533) 2% in H2O, pH 4-4.3 adjusted with NH4OH, filtered with 0.45uM membrane), and washed with Tris-HCl, pH4.0. Elemental calcium concentrations were measured by inductively coupled plasma – atomic emission spectroscopy (ICP-AES, HORIBA Jobin Yvon Activa-M)49 at the Center for Materials Science and Engineering at MIT, and three measurements were conducted to obtain mean and standard deviation values. To measure ionized calcium, cells were treated with 5% HNO3 (for dissolution of calcium molecules) and 10% HClO4 acid solutions (to remove organic compounds) in a cell culture flask and then briefly sonicated for 10 min. The solution was incubated for > 3 hrs on the titer plate shaker. The obtained values were converted to calcium concentration using a reference solution made by Fluka (Calcium Standard for AAS, TraceCERT®), and normalized by 5 × 105 initiated cells.
Quantitative RT-PCR Analysis
The expression levels of osteogenic genes (Runx2, Sp7, and Bglap) were quantified by real-time RT-PCR with Quantifast SYBR Green RT-PCR kit (Qiagen, Hilden, Germany). Total RNAs (2 ug) were reverse-transcribed in a volume of 20 ul by using the SuperScript III First-Strand Synthesis System (Invitrogen, Carlsbad, California, USA), and the resulting cDNA was diluted into a total volume of 500 ul. 5 ul of this synthesized cDNA solution was used for analysis. For osteogenic genes, each reaction was performed in a 25 ul volume using the Quantifast SYBR Green RT-PCR kit (Qiagen, Hilden, Germany). The conditions were programmed as follows: initial denaturation at 95C for 5 min followed by 40 cycles of 10 s at 95C and 30 s at 60C, then 1 min at 95C, 30 s at 55C, and 30 sec at 95C. For pluripotent genes, each reaction was performed in a 25 ul volume using using the Brilliant SYBR Green QPCR master mix kit (Stratagene, Cedar Creek, TX, USA). The conditions were programmed as follows: initial denaturation at 95C for 10 min followed by 40 cycles of 30 s at 95C, 1 min at 55C, and 1 min at 72C, then 1 min at 95C, 30 s at 55C, and 30 sec at 95C. Primers used in the quantitative RT-PCR are listed in Supplementary Table 7. All of the samples were duplicated, and the PCR reaction was performed using an Mx3005P (Stratagene, Cedar Creek, TX, USA), which can detect the amount of synthesized signals during each PCR cycle. The relative amounts of the mRNAs were determined using the MxPro program (Stratagene, Cedar Creek, TX, USA). The amount of PCR product was normalized to a percentage of the expression level of b-Actin. The RT-PCR products of Oct4, Nanog, and b-Actin were also evaluated on 0.8% agarose gels after staining with ethidium bromide. The cycle numbers of the PCR were reduced in order to optimize the difference in band intensities (Oct4, Nanog, and b-Actin were 29, 33, and 28, respectively) (Supplementary Table 7).
DNA methylation analysis
5ug of genomic DNA from each sample was fractionated, digested with McrBC, gel purified, labeled and hybridized to a CHARM microarray as previously described20, 21. For each probe, the averaged methylation values across the same cell type were computed and converted to percent methylation (p). p was used to find regions of differential methylation (Δp) for each pairwise cell type comparison and the absolute area of each region was calculated by multiplying the number of probes by Δp. For data analysis, we used area value 2 as the cutoff to define differentially methylated regions (DMRs). Our previous study indicated that this cutoff corresponds to 5% false discovery rate (Doi et al. unpublished data). Bisulfite pyrosequencing analysis of individual regions was performed as previously described21. Primer sequences are provided in Supplementary Table 7.
Teratoma and chimera analysis
Teratomas were assessed by injecting 106 undifferentiated cells into the subcutaneous tissue above the rear haunch of Rag2/γC immunodeficient mice (Taconic), and teratoma formation was monitored for 3 months post injection. Collected tumors were processed by the Pathology Core of the Dana-Farber/Harvard Cancer Center. Chimera analysis of pluripotent cells was conducted by injecting GFP+ or GFP− cells into blastocysts isolated from C57BL/6 (GFP− or GFP+) embryos, which were collected at the two-cell stage. The fertilized embryo was collected from the oviduct and cultured in KSOM media (Specialty Media). A mouse strain expressing GFP from the human ubiquitin promoter (Jackson Laboratory) was used to ensure maximum expression in various tissues, and enabled injected cells to be distinguished from host cells. The reconstituted blastocysts were implanted into 2.5 day pseudopregnant CD1 females. Chimeras were allowed to develop to adulthood to gauge skin chimerism and germ cell transmission, or were dissected at embryonic day 12.5 to isolate gonad, liver, heart, and MEF for flow analysis. Gonads were stained with SSEA1 antibody (Hybridoma Bank) for 1 hour, and treated with APC-conjugated mouse IgM antibody (BD Pharmingen, #550676) to detect SSEA1 positive germ cells by flow cytometry (LSRII, BD Biosciences, Hematology/Oncology Flow Cytometry Core Facility of Children’s Hospital Boston).
Generation of NSC-NP-iPSC, B-NP-iPSC, and NP-iPSC-TSA-AZA
Neural Progenitor (NP) iPSC harboring integrated proviruses carrying the four reprogramming factrors29 were differentiated to neural stem cells (NSC) as described53. Reprogramming factors in cultured NSC were induced by doxycycline, and colonies expressing GFP from the nanog-reporter were selected to yield NSC-NP-iPSC. NP-iPSC from blood lineages (B-NP-iPSC) were obtained by differentiating the NP-iPSC via EB for 6 days, infecting with HoxB4ERT retrovirus54, and co-culturing on OP9 in the presence of 4-hydroxytamoxifen (4-HT) to enable isolation of hematopoietic cells16. Day 15 hematopoietic cells were harvested, stained with CD45+ (BD Pharmingen, #557659), and sorted for hematopoietic cells. Only minimal hematopoietic colonies were observed on OP9 culture in the absence of 4-HT. Harvested CD45+ hematopoietic cells were induced by doxycycline and colonies expressing GFP from the nanog-reporter were selected to yield B-NP-iPSC. Methylcellulose hematopoietic colony analysis was conducted in the absence of 4-HT as a negative control. The dissociated EBs (2×105 cells) from NP-iPSC were infected with HoxB4-ERT virus and then plated on methylcellulose media. Only 1.7 +/− 1.2 colonies (n=3) were formed in the absence of hydroxytamoxifen, which indicates the limited functional HoxB4 expression in the absence of 4-HT. NP-iPSC-TSA-AZA cells were isolated by treating cells for 9 days with Trichostatin A (TSA, 100nM) and 5-azacytidine (AZA, 1mM), in 3-day cycles: drug treatment occurred on two consecutive days, followed by one day of non-treatment. Undifferentiated colonies were recovered to conduct methylcellulose analysis. Wnt3a (R&D System, 1324-WN-002/CF, 40ng/ml) was added to EB culture media between day 2 and 4, and hematopoietic potential was tested by plating on methylcellulose media as described above.
Gene Enrichment analysis
We took a permutation approach to assess the enrichment of hematopoiesis and fibroblast related genes in DMRs. Gene lists were derived from MSigDB (http://www.broadinstitute.org/gsea/msigdb/index.jsp) for Supplementary Figure 3c, and cell-type signatures are described in Cahan et al.23 for all other enrichment analyses. To identify cell-type signatures, gene expression profiles of more than 80 distinct cell types were downloaded from Gene Expression Omnibus, normalized, and searched for sets of genes that exhibit cell type-specific expression patterns, using the template matching method described in Pavlidis et al.55. Enrichment P-values were calculated as the number of times that a random selection of genes from the 13,931 profiled met or exceeded the observed overlap based on 100,000 random selections. The number of randomly selected genes was the same as the number of genes in the DMR list. Fig. 3b (left panel): 20/74 hematopoiesis-related transcription factors are among the 1,997 genes hypermethylated in F-iPSC vs B-iPSC (P-value=0.00337). Fig. 3b (right panel): 115/562 fibroblast-specific genes are among the 1,589 genes hypermethylated in B-iPSC vs F-iPSC (P-value=0.00001). Supplementary Figure 3A: 12/130 liver-specific genes are among the 1,321 differentially methylated in F-iPSC vs B-iPSC (P-value=0.58178). Supplementary Figure 3B: 250/1764 neural-specific genes are among the 1,805 differentially methylated in Bl-iPSC vs. NP-iPSC (P-value=0.05813). Supplementary Figure 3C: 63/526 genes up-regulated in hematopoietic stem cells are among the 1,133 genes hypomethylated in NP-iPSC-TSA-AZA vs NP-iPSC (P-value=0.00116).
Method Reference
- 50.Kirov G, et al. Variation in the protocadherin gamma A gene cluster. Genomics. 2003;82:433–440. doi: 10.1016/s0888-7543(03)00167-8. [DOI] [PubMed] [Google Scholar]
- 51.Markoulaki S, et al. Transgenic mice with defined combinations of drug-inducible reprogramming factors. Nat Biotechnol. 2009;27:169–171. doi: 10.1038/nbt.1520. Epub 2009 Jan 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Koh EY, Chen T, Daley GQ. Novel retroviral vectors to facilitate expression screens in mammalian cells. Nucleic Acids Res. 2002;30:e142. doi: 10.1093/nar/gnf142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Conti L, et al. Niche-independent symmetrical self-renewal of a mammalian tissue stem cell. PLoS Biol. 2005;3:e283. doi: 10.1371/journal.pbio.0030283. Epub 2005 Aug 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schiedlmeier B, et al. HOXB4’s road map to stem cell expansion. Proc Natl Acad Sci U S A. 2007;104:16952–16957. doi: 10.1073/pnas.0703082104. Epub 12007 Oct 16916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pavlidis P, Noble WS. Analysis of strain and regional variation in gene expression in mouse brain. Genome biology. 2001:2. doi: 10.1186/gb-2001-2-10-research0042. RESEARCH0042. [DOI] [PMC free article] [PubMed] [Google Scholar]
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Author Contributions K.K. and G.Q.D. conceived the experimental plan. K.K., A.D., B.W., K.N., Z.R., H.J., L.E., A.Y., A.T., KC.C., H.H., S.MF., O.N., TJ.Y., R.I., N.J., J.S., and J.H. performed the experiments. P.C., J.K., and M.J.A. performed statistical analysis. A.D., B.W., and A.F. performed CHARM and guided analysis of methylation. K.K., A.D., B.W., K.N., P.C., J.K., H.J., T.Y., A.P.F., and G.Q.D. wrote the manuscript.
References
- 1.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. Epub 2006 Aug 2010. [DOI] [PubMed] [Google Scholar]
- 2.Zhao XY, et al. iPS cells produce viable mice through tetraploid complementation. Nature. 2009;461:86–90. doi: 10.1038/nature08267. [DOI] [PubMed] [Google Scholar]
- 3.Daley GQ, et al. Broader implications of defining standards for the pluripotency of iPSCs. Cell Stem Cell. 2009;4:200–201. doi: 10.1016/j.stem.2009.02.009. author reply 202. [DOI] [PubMed] [Google Scholar]
- 4.Marion RM, et al. A p53-mediated DNA damage response limits reprogramming to ensure iPS cell genomic integrity. Nature. 2009;460:1149–1153. doi: 10.1038/nature08287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li H, et al. The Ink4/Arf locus is a barrier for iPS cell reprogramming. Nature. 2009;460:1136–1139. doi: 10.1038/nature08290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eminli S, et al. Differentiation stage determines potential of hematopoietic cells for reprogramming into induced pluripotent stem cells. Nat Genet. 2009;41:968–976. doi: 10.1038/ng.428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gurdon JB, Melton DA. Nuclear reprogramming in cells. Science. 2008;322:1811–1815. doi: 10.1126/science.1160810. [DOI] [PubMed] [Google Scholar]
- 8.Blelloch R, et al. Reprogramming efficiency following somatic cell nuclear transfer is influenced by the differentiation and methylation state of the donor nucleus. Stem Cells. 2006;24:2007–2013. doi: 10.1634/stemcells.2006-0050. Epub 2006 May 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Maherali N, et al. A high-efficiency system for the generation and study of human induced pluripotent stem cells. Cell stem cell. 2008;3:340–345. doi: 10.1016/j.stem.2008.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Aoi T, et al. Generation of pluripotent stem cells from adult mouse liver and stomach cells. Science. 2008;321:699–702. doi: 10.1126/science.1154884. Epub 2008 Feb 2014. [DOI] [PubMed] [Google Scholar]
- 11.Miura K, et al. Variation in the safety of induced pluripotent stem cell lines. Nat Biotechnol. 2009;27:743–745. doi: 10.1038/nbt.1554. [DOI] [PubMed] [Google Scholar]
- 12.Chan EM, et al. Live cell imaging distinguishes bona fide human iPS cells from partially reprogrammed cells. Nat Biotechnol. 2009 doi: 10.1038/nbt.1580. [DOI] [PubMed] [Google Scholar]
- 13.Mikkelsen TS, et al. Dissecting direct reprogramming through integrative genomic analysis. Nature. 2008;454:49–55. doi: 10.1038/nature07056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Santos F, Hendrich B, Reik W, Dean W. Dynamic reprogramming of DNA methylation in the early mouse embryo. Dev Biol. 2002;241:172–182. doi: 10.1006/dbio.2001.0501. [DOI] [PubMed] [Google Scholar]
- 15.Skow LC, et al. A mouse model for beta-thalassemia. Cell. 1983;34:1043–1052. doi: 10.1016/0092-8674(83)90562-7. [DOI] [PubMed] [Google Scholar]
- 16.Kyba M, Perlingeiro RC, Daley GQ. HoxB4 confers definitive lymphoid-myeloid engraftment potential on embryonic stem cell and yolk sac hematopoietic progenitors. Cell. 2002;109:29–37. doi: 10.1016/s0092-8674(02)00680-3. [DOI] [PubMed] [Google Scholar]
- 17.Bourne S, Polak JM, Hughes SP, Buttery LD. Osteogenic differentiation of mouse embryonic stem cells: differential gene expression analysis by cDNA microarray and purification of osteoblasts by cadherin-11 magnetically activated cell sorting. Tissue Eng. 2004;10:796–806. doi: 10.1089/1076327041348293. [DOI] [PubMed] [Google Scholar]
- 18.Wdziekonski B, Villageois P, Dani C. Differentiation of mouse embryonic stem cells and of human adult stem cells into adipocytes. Curr Protoc Cell Biol. 2007 doi: 10.1002/0471143030.cb2304s34. Chapter 23, Unit 23.24. [DOI] [PubMed] [Google Scholar]
- 19.Hood RC, Neill WM. A modification of alizarin red S technic for demonstrating bone formation. Stain Technol. 1948;23:209–218. doi: 10.3109/10520294809106251. [DOI] [PubMed] [Google Scholar]
- 20.Irizarry RA, et al. Comprehensive high-throughput arrays for relative methylation (CHARM) Genome Res. 2008;18:780–790. doi: 10.1101/gr.7301508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Irizarry RA, et al. The human colon cancer methylome shows similar hypo- and hypermethylation at conserved tissue-specific CpG island shores. Nat Genet. 2009;41:178–186. doi: 10.1038/ng.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Doi A, et al. Differential methylation of tissue- and cancer-specific CpG island shores distinguishes human induced pluripotent stem cells, embryonic stem cells and fibroblasts. Nat Genet. 2009 doi: 10.1038/ng.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cahan P, Li H, Collins JJ, Daley QG. Tissue-specific gene expression and autoregulatory loops. 2010 manuscript in preparation. [Google Scholar]
- 24.Turker MS. Gene silencing in mammalian cells and the spread of DNA methylation. Oncogene. 2002;21:5388–5393. doi: 10.1038/sj.onc.1205599. [DOI] [PubMed] [Google Scholar]
- 25.Ji H, et al. A comprehensive methylome map of myeloid and lymphoid commitment in hematopoietic progenitors. 2010 doi: 10.1038/nature09367. Submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Perez-Iratxeta C, et al. Study of stem cell function using microarray experiments. FEBS Lett. 2005;579:1795–1801. doi: 10.1016/j.febslet.2005.02.020. [DOI] [PubMed] [Google Scholar]
- 27.Kim J, Chu J, Shen X, Wang J, Orkin SH. An extended transcriptional network for pluripotency of embryonic stem cells. Cell. 2008;132:1049–1061. doi: 10.1016/j.cell.2008.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wernig M, et al. In vitro reprogramming of fibroblasts into a pluripotent ES- cell-like state. Nature. 2007;448:318–324. doi: 10.1038/nature05944. Epub 2007 Jun 2006. [DOI] [PubMed] [Google Scholar]
- 29.Wernig M, et al. A drug-inducible transgenic system for direct reprogramming of multiple somatic cell types. Nat Biotechnol. 2008;26:916–924. doi: 10.1038/nbt1483. Epub 2008 Jul 2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hanna J, et al. Direct reprogramming of terminally differentiated mature B lymphocytes to pluripotency. Cell. 2008;133:250–264. doi: 10.1016/j.cell.2008.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brambrink T, Hochedlinger K, Bell G, Jaenisch R. ES cells derived from cloned and fertilized blastocysts are transcriptionally and functionally indistinguishable. Proc Natl Acad Sci U S A. 2006;103:933–938. doi: 10.1073/pnas.0510485103. Epub 2006 Jan 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Eggan K, et al. Hybrid vigor, fetal overgrowth, and viability of mice derived by nuclear cloning and tetraploid embryo complementation. Proc Natl Acad Sci U S A. 2001;98:6209–6214. doi: 10.1073/pnas.101118898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nadin BM, Goodell MA, Hirschi KK. Phenotype and hematopoietic potential of side population cells throughout embryonic development. Blood. 2003;102:2436–2443. doi: 10.1182/blood-2003-01-0118. Epub 2003 Jun 2412. [DOI] [PubMed] [Google Scholar]
- 34.Eden S, Hashimshony T, Keshet I, Cedar H, Thorne AW. DNA methylation models histone acetylation. Nature. 1998;394:842. doi: 10.1038/29680. [DOI] [PubMed] [Google Scholar]
- 35.Chiu CP, Blau HM. 5-Azacytidine permits gene activation in a previously noninducible cell type. Cell. 1985;40:417–424. doi: 10.1016/0092-8674(85)90155-2. [DOI] [PubMed] [Google Scholar]
- 36.Lengerke C, et al. BMP and Wnt specify hematopoietic fate by activation of the Cdx-Hox pathway. Cell stem cell. 2008;2:72–82. doi: 10.1016/j.stem.2007.10.022. [DOI] [PubMed] [Google Scholar]
- 37.Ball MP, et al. Targeted and genome-scale strategies reveal gene-body methylation signatures in human cells. Nat Biotechnol. 2009;27:361–368. doi: 10.1038/nbt.1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim JB, et al. Oct4-induced pluripotency in adult neural stem cells. Cell. 2009;136:411–419. doi: 10.1016/j.cell.2009.01.023. [DOI] [PubMed] [Google Scholar]
- 39.Loh YH, et al. Generation of induced pluripotent stem cells from human blood. Blood. 2009;113:5476–5479. doi: 10.1182/blood-2009-02-204800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chin MH, et al. Induced pluripotent stem cells and embryonic stem cells are distinguished by gene expression signatures. Cell stem cell. 2009;5:111–123. doi: 10.1016/j.stem.2009.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hanna J, et al. Direct cell reprogramming is a stochastic process amenable to acceleration. Nature. 2009;462:595–601. doi: 10.1038/nature08592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Okita K, Ichisaka T, Yamanaka S. Generation of germline-competent induced pluripotent stem cells. Nature. 2007;448:313–317. doi: 10.1038/nature05934. Epub 2007 Jun 2006. [DOI] [PubMed] [Google Scholar]
- 43.Soldner F, et al. Parkinson’s disease patient-derived induced pluripotent stem cells free of viral reprogramming factors. Cell. 2009;136:964–977. doi: 10.1016/j.cell.2009.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stadtfeld M, et al. Aberrant silencing of imprinted genes on chromosome 12qF1 in mouse induced pluripotent stem cells. Nature. 465:175–181. doi: 10.1038/nature09017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bernstein BE, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 46.Xu J, et al. Transcriptional competence and the active marking of tissue-specific enhancers by defined transcription factors in embryonic and induced pluripotent stem cells. Genes Dev. 2009;23:2824–2838. doi: 10.1101/gad.1861209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Osafune K, et al. Marked differences in differentiation propensity among human embryonic stem cell lines. Nat Biotechnol. 2008;26:313–315. doi: 10.1038/nbt1383. [DOI] [PubMed] [Google Scholar]
- 48.Silva J, et al. Promotion of reprogramming to ground state pluripotency by signal inhibition. PLoS Biol. 2008;6:e253. doi: 10.1371/journal.pbio.0060253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nomlru T, Kaneco S, Tanaka T, I to S, Kawaguchi H. Determination of Calcium Content in Cells by Inductively Coupled Plasma Spectrometry Individual BiologicalAtomic Emission. Anal. Chem. 1994;66:3000–3004. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.