Abstract
Integral membrane proteins are integrated cotranslationally into the membrane of the endoplasmic reticulum in a process mediated by the Sec61 translocon. Transmembrane α-helices in a translocating polypeptide chain gain access to the surrounding membrane through a lateral gate in the wall of the translocon channel [van den Berg B, et al. (2004) Nature 427:36–44; Zimmer J, et al. (2008) Nature 455:936–943; Egea PF, Stroud RM (2010) Proc Natl Acad Sci USA 107:17182–17187]. To clarify the nature of the membrane-integration process, we have measured the insertion efficiency into the endoplasmic reticulum membrane of model hydrophobic segments containing nonproteinogenic aliphatic and aromatic amino acids. We find that an amino acid’s contribution to the apparent free energy of membrane-insertion is directly proportional to the nonpolar accessible surface area of its side chain, as expected for thermodynamic partitioning between aqueous and nonpolar phases. But unlike bulk-phase partitioning, characterized by a nonpolar solvation parameter of 23 cal/(mol·Å2), the solvation parameter for transfer from translocon to bilayer is 6–10 cal/(mol·Å2), pointing to important differences between translocon-guided partitioning and simple water-to-membrane partitioning. Our results provide compelling evidence for a thermodynamic partitioning model and insights into the physical properties of the translocon.
Keywords: flexizyme, hydrophobicity, nonproteinogenic amino acid, transmembrane helix
In eukaryotic cells, membrane proteins destined for the plasma membrane and the various compartments along the endo- and exocytic pathways are synthesized by endoplasmic reticulum (ER)-bound ribosomes and cotranslationally integrated into the ER membrane in a process mediated by the Sec61 translocon complex; the homologous SecYEG translocon mediates membrane-protein integration into the inner membrane of prokaryotes (1, 2). Subsequent to membrane integration, membrane proteins fold and oligomerize in the ER and are then moved further along the secretory pathway by vesicular transport.
During the membrane-integration step, hydrophobic segments in the translocating nascent polypeptide chain exit the Sec61 translocon through a lateral gate and become embedded in the surrounding lipid bilayer (3–5). Cotranslational, translocon-mediated integration of transmembrane α-helices into the ER membrane sets the stage for all subsequent folding and oligomerization events and hence represents a critical step in the maturation of membrane proteins.
In previous studies, we have provided quantitative data on the propensities of the 20 natural amino acids to promote the integration of transmembrane helices into the ER membrane and have shown that they depend both on hydrophobicity and on position within the helix (3, 6). Although the partitioning of transmembrane helices between the Sec61 translocon and the lipid membrane bears strong similarities to partitioning of solutes between water and lipid membranes, translocon-to-bilayer partitioning may not be equivalent to water-to-bilayer partitioning (7). Insights into the differences between the two partitioning processes might be revealed if the physicochemical properties of the translocon could be probed chemically. However, given the somewhat idiosyncratic collection of proteinogenic amino acids used in nature, it has hitherto not been possible to vary side-chain chemistry in the systematic fashion required to unravel fully the physicochemical basis for translocon-mediated membrane partitioning. In order to probe the membrane-integration mechanism in greater detail, we have taken advantage of a suppressor tRNA-based technique to introduce nonproteinogenic aliphatic and aromatic amino acids into a model hydrophobic segment and measure their apparent free energies of membrane insertion. The results show not only that translocon/membrane partitioning is quantitatively different from simple water/membrane partitioning, but also reveal a physicochemical asymmetry between the cytoplasmic and lumenal ends of a transmembrane helix, which is likely related to the structure of the translocon.
Results
Experimental Approach.
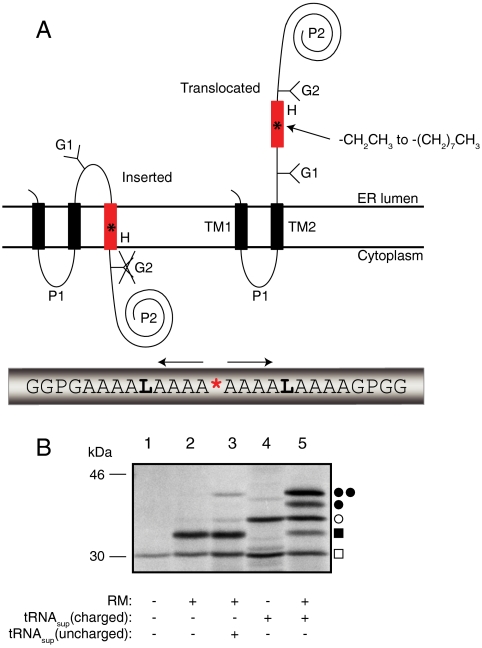
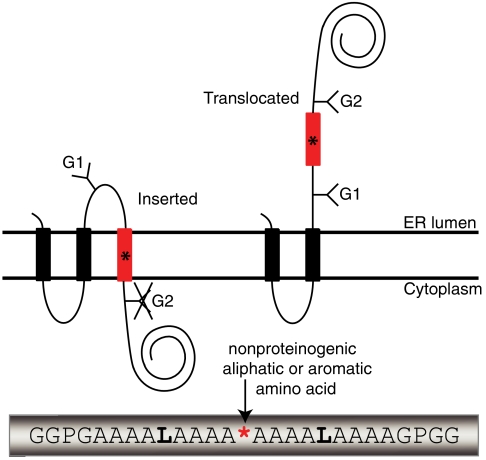
The basic approach is illustrated in Fig. 1. A suppressor tRNA (tRNAsup) is charged with the desired nonproteinogenic amino acid using the Flexizyme system (8, 9) and is then added to an in vitro translation system programmed with an mRNA encoding an engineered version of the well-characterized membrane-protein leader peptidase (Lep). The Lep construct has two N-terminal transmembrane helices (TM1, TM2) and contains a hydrophobic test segment (H segment) flanked by two canonical Asn-X-Thr acceptors sites for N-linked glycosylation (G1, G2). A UAG stop codon that can be recognized by the charged tRNAsup serves to position the nonproteinogenic amino acid in the H segment.
Fig. 1.
(A) Model H segments are introduced into the Lep host protein between two engineered acceptor sites for N-linked glycosylation (G1, G2). A typical H segment is shown, including a nonproteinogenic amino acid (*) and GGPG…GPGG flanking regions. The fractions of membrane-inserted and noninserted H segment are determined by quantitation of radiolabeled singly and doubly glycosylated forms of the protein. (B) In vitro translation of a Lep construct with the H segment AAAALAAAAAAAAALAAAX (X = Me-Trp) in the absence and presence of charged or uncharged tRNAsup and dog pancreas RMs. □, Truncated Lep protein resulting from termination at the UAG stop codon in the H segment; ▪, truncated Lep protein glycosylated on the G1 site; ○, full-length, nonglycosylated Lep protein; ●, full-length Lep protein glycosylated on the G1 site; ●●, full-length Lep protein glycosylated on the G1 and G2 sites. The degree of suppression in the absence of added tRNAsup is negligible (lane 2), and is very low (4%) with added uncharged tRNAsup (lane 3). Suppression is > 50% in the presence of charged tRNAsup (lanes 4 and 5). The amounts of sample loaded in -RM and +RM lanes were adjusted to give roughly equal signals.
When the translation reaction is carried out in the presence of ER-derived dog pancreas rough microsomes (RMs), TM1 and TM2 insert into the ER membrane as shown (10). If the H segment is recognized as a transmembrane segment by the translocon and inserted into the membrane, only the G1 acceptor site is accessible to the lumenal oligosaccharyl transferase enzyme and receives a glycan moiety; if, in contrast, the H segment is translocated across the membrane, both G1 and G2 become glycosylated (11). The apparent free energy of membrane insertion of a given H segment is calculated as ΔGapp = -RT ln(f1/f2), where R is the gas constant, T the absolute temperature (298 K), f1 the amount of singly glycosylated molecules, and f2 the amount of doubly glycosylated molecules (6). The H segments analyzed here have the composition GGPG-[1X,nL,(18-n)A]-GPGG, where X is the nonproteinogenic amino acid and n is chosen such that, for any given X, -1 kcal/mol ≤ ΔGapp ≤ +1 kcal/mol. The GGPG…GPGG flanks are included in order to break any regular secondary structure and thereby insulate the H segment from the influences of the surrounding sequence and to prevent it from shifting in position along the membrane normal (6).
Membrane-Insertion Characteristics of Nonpolar Side Chains.
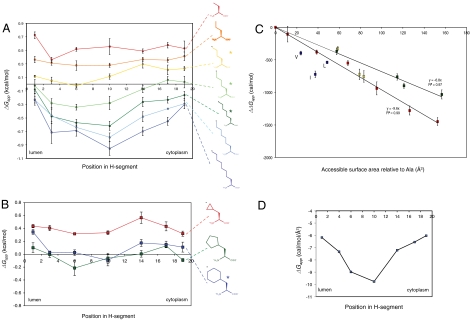
Because hydrophobicity is central to transmembrane-helix integration into the ER membrane, we first asked how nonpolar surface area correlates with propensity for membrane insertion. The most straightforward way to determine the relation between nonpolar surface area and ΔGapp is to choose amino acids with linear alkyl side chains for X. We therefore scanned amino acids with linear side chains containing two to eight carbons along an H segment with n = 2 leucines. The results are shown in Fig. 2A and listed in Table S1. Three important conclusions can be drawn: (i) ΔGapp varies in a regular fashion with the length of the side chain; (ii) the difference in ΔGapp between the terminal and central positions of the side chain increases with the length of the side chain; and (iii) for the longer side chains, there is a noticeable asymmetry in the curves when one compares positions close to the N-terminal end with those close to the C-terminal end of the H segment (especially evident comparing positions 6 and 14, Fig. 2 A and D). Cyclic aliphatic and nonpolar aromatic side chains yield similar results, Figs. 2B and 3A.
Fig. 2.
(A) Apparent free energy of insertion (ΔGapp) for 19-residue-long H segments carrying a single nonproteinogenic amino acid with a linear alkyl side chain (shown on the right) in the indicated positions. The H segments all have the sequence AAAALAAAAAAAAALAAAA with the Ala in the indicated position replaced by the nonproteinogenic amino acid. The lumenal, N-terminal end of the H segment is on the left and the cytoplasmic end on the right. Error bars show standard deviations; * indicates side chains for which the ΔGapp value for position 6 is significantly smaller than the value for position 14 (two-sided t test, p < 0.01), i.e., that have a significant asymmetry in the ΔGapp profile. (B) ΔGapp for cyclic alkyl side chains. The H segments all have the sequence AAAALAAAAAAAAALAAAA with the Ala in the indicated position replaced by the nonproteinogenic amino acid. (C) ΔGapp decreases in proportion to the increase in ASA of the nonproteinogenic amino acid. ΔΔGapp is measured relative to the AAAALAAAAAAAAALAAAA H segment and the nonproteinogenic amino acid is in the middle position (position 10) in the H segment. The change in ASA is calculated relative to a model AAAALAAAAAAAAALAAAA α-helix. Red data points are for linear alkyl side chains, yellow for cyclic alkyl side chains (cyclopropyl, cyclopentyl, cyclohexyl), green for aromatic side chains lacking polar groups (Phe, 1-naphthyl, 2-naphthyl, biphenyl), and blue for the branched natural amino acids Leu, Ile, and Val. The average change in ΔGapp is -9.6 cal/(mol·Å2) for the linear alkyl side chains and -6.8 cal/(mol·Å2) for the nonpolar aromatic side chains. (D) Average change in ΔGapp per square angstrom (Å2) of accessible surface area as a function of position in the H segment (calculated from the data in A using linear regression as in C).
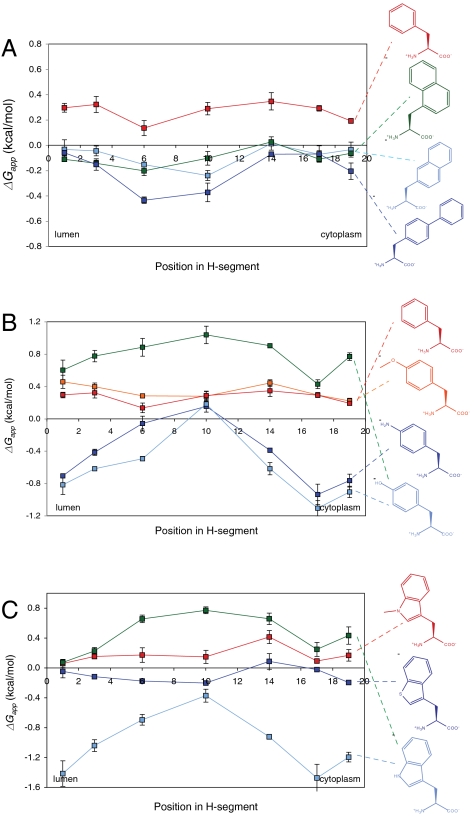
Fig. 3.
ΔGapp for different aromatic amino acids. The H segments used in A have the sequence AAAALAAAAAAAAALAAAA, with the Ala in the indicated position replaced by the nonproteinogenic amino acid. In B, the H segments used for Phe, Me-Tyr, and Tyr (green trace) also have this sequence, whereas the sequence used for Tyr (light-blue trace) and the aniline side chain is AAAALALAAAAALALAAAA. In C, AAAALAAAAAAAAALAAAA is used for Trp (green trace), Me-Trp and the benzothiophene side chain, and AAAALALAAAAALALAAAA is used for Trp (light-blue trace).
As seen in Fig. 2C, there is a strict correlation between the increase in side-chain accessible surface area (ASA) and ΔGapp that holds for aliphatic side chains with at least up to three times the ASA of the largest natural aliphatic amino acids. For the middle position in the H segment, ΔGapp decreases by approximately 10 cal/(mol·Å2), whereas the decrease is smaller when the side-chain is closer to the ends of the H segment, Fig. 2D. A similar relation between ASA and ΔGapp holds for the nonpolar aromatic side chains, but the decrease in ΔGapp is only about 7 cal/(mol·Å2) for the middle position of the H segment in this case, Fig. 2C.
Except for the slight asymmetry in the curves, these results are those expected if membrane insertion of the H segment is driven by thermodynamic partitioning into a lipid bilayer environment. First, similar to our results, the partitioning free energy of nonpolar compounds between aqueous buffer and a bilayer-mimetic solvent is proportional to ASA and can be characterized by the atomic solvation parameter σ. For partitioning between buffer and nonpolar solvents such as n-octanol, σ is approximately 23–25 cal/(mol·Å2) for aliphatic side chains and approximately 16 cal/(mol·Å2) for nonpolar aromatic side chains (12, 13). The corresponding values for translocon/bilayer partitioning in the middle of the membrane, approximately 10 and 7 cal/(mol·Å2), are 2.5-fold smaller, but the value for nonpolar aromatic side chains is approximately 70% of the value for aliphatic side chains in both cases. Second, large aliphatic compounds are expected to partition preferentially into the center of the bilayer (14), as has been shown experimentally for n-hexane (15), and we see the same trend in our data, Fig. 2D.
Membrane-Insertion Characteristics of Polar Aromatic Side Chains.
We also tested a selection of polar aromatic side chains, some containing non-carbon atoms, Fig. 3. As found previously, Tyr and Trp behave differently compared to non-polar aromatic residues such as Phe in that they promote membrane insertion significantly better when located near the ends of the H segment than when in the middle (3, 6); the same behavior is now seen for the aniline side chain but not for methylated tyrosine, methylated tryptophan, or the benzothiophene side chain. This observation points to the hydrogen-bonding ability of the Tyr, Trp, and aniline side chains (and the lack thereof in the Phe, methyl-Tyr, methyl-Trp, and benzothiophene side chains) as a critical distinguishing factor. A study of the interactions of Trp analogs with and without H-bonding ability showed that the “aromaticity” of Trp was the dominant cause of the preferential partitioning of Trp into lipid bilayer interfaces (16). Our results indicate that H bonding becomes important when polar aromatic residues are inserted in locations below the membrane-water interface.
Transmembrane Asymmetry.
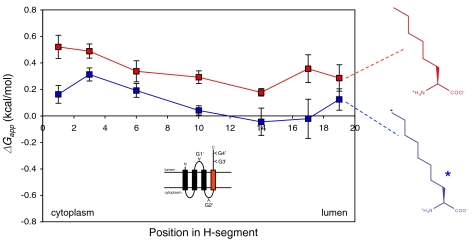
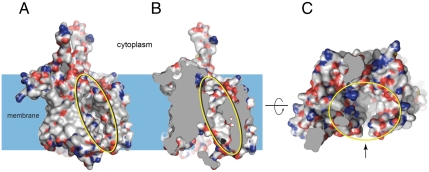
Finally, what could be the cause of the slight transbilayer asymmetry in the ΔGapp curves in Fig. 2A? We asked whether the asymmetry correlates with the N-to-C-terminal polarity of the H segment or with the orientation of the H segment relative to the membrane. To invert the membrane orientation of H segments containing hexyl and octyl side chains in different positions, we inserted an additional GGPG-[7L,12A]-GPGG transmembrane segment between TM2 and the H segment, Fig. 4. Comparing Figs. 4 and 2A, there is a general trend that the membrane-insertion efficiency is higher when the alkyl side chain is in the lumenal half of the H segment (i.e., the asymmetry in the ΔGapp curves correlates with the membrane orientation of the H segment), suggesting that it reflects an asymmetry in either the ER membrane or the Sec61 translocon. Although data are scarce, the ER is thought not to have a strong asymmetry in lipid composition between the two leaflets (17, 18). Therefore, it is more likely that the origin of the asymmetry is to be sought in the translocon. Although we cannot know the exact nature of the translocon asymmetry until a high-resolution structure of a functionally open translocon becomes available, the recent structure of a translocon with a partially open lateral gate (5) reveals the nascent-chain conduit as a deep canyon lined mainly by apolar residues and with scattered polar groups projecting from the canyon walls, Fig. 5. It does not seem unreasonable that the complex shape and physicochemical heterogeneity of the translocon channel might underlie the observed asymmetry in the ΔGapp curves.
Fig. 4.
ΔGapp for Ncyt-Clum orientated linear hexyl and octyl side chains. The GGPG…GPGG flanked H segments are inserted C terminal to an added third transmembrane helix (GGPGALAALALAALAALALAALAGPGG) in the Lep host protein and acceptor sites for N-linked glycosylation are engineered into the construct, as shown in the cartoon. If the H segment does not insert into the membrane, only the G1′ site will receive a glycan; if it does insert, the G3′ and G4′ sites will also be modified. The H segments all have the composition [X,18A] and have the nonproteinogenic amino acid X in the indicated position. The cytoplasmic, N-terminal end of the H segment is on the left and the lumenal on the right. The ΔGapp value for position 3 is significantly larger than the value for position 17 (two-sided t test, p < 0.01) for the octyl side chain (indicated by *).
Fig. 5.
Different views of the nascent-chain conduit in Pyrococcus furiosus SecYEβ (5) whose structure reveals a partially open (“primed”) translocon. (A) View along the membrane plane, showing the partially open lateral gate (yellow oval). (B) Same view as in A but with the front of the molecule removed to show the rear wall of the channel. (C) View of the upper parts of the channel (yellow oval) seen from the cytoplasmic end. The lateral gate is indicated by the arrow. Carbon atoms are displayed in white, oxygen atoms in red, nitrogen atoms in blue, and sulfur atoms in yellow.
Discussion
Our results provide compelling evidence for the thermodynamic partitioning model of translocon-mediated integration of transmembrane helices into the ER membrane (3, 6, 19–21). In its simplest version, this model pictures the transmembrane helix as equilibrating between the translocon channel and the surrounding membrane. As judged from the available X-ray structures of prokaryotic homologs of the Sec61 translocon (4, 5, 22, 23), the channel is quite narrow and lined by a mixture of polar and apolar amino acids, providing an environment that is less polar than aqueous buffer. Likewise, the ER membrane, which, like all biological membranes, has a high protein content, may offer an environment that is less apolar than a pure lipid bilayer (24). Indeed, the solvation parameters for partitioning of aliphatic and nonpolar aromatic side chains obtained here [approximately 10 and 7 cal/(mol·Å2)] are a factor 2.5 smaller than found in classical solute transfer experiments (13, 25–28), suggesting that simple water-to-lipid partitioning measurements do not capture the full complexity of translocon-to-membrane partitioning (7). The asymmetry in the ΔGapp curves, Figs. 2 and 4, and effects on the hydrophobicity threshold for H segment insertion caused by mutations in the Sec61 translocon (29) support the notion that our measurements report on partitioning between the translocon channel and the surrounding lipid, although we cannot completely rule out more complicated models where the H segment can also explore the membrane-water interface region in the vicinity of the translocon during the membrane-insertion step.
Little is known about the energetics of the passage of nascent membrane-protein chains through the Sec61 translocon. Is the chain pushed steadily through the translocon by the ribosome? Or does the chain diffuse through the translocon aided by accessory proteins, such as BiP, which give directionality via a Brownian ratchet mechanism (30, 31)? If the chain is driven steadily through the translocon by the ribosome, Schow et al. (7) have suggested that there are only two independent equilibria that need be considered: one between translocon and bilayer (ΔGtbi) and one between water and bilayer (ΔGwbi). The first equilibrium process determines whether a transmembrane helix enters the membrane and the second determines if it stays there. Schow et al. (7) further estimated that ΔGwbi ≈ 2.6ΔGtbi, and suggested that the discrepancy could be explained by assuming that the translocon/bilayer solvation parameter σtbi = σwbi/2.6 = 8.8 cal/(mol·Å2). This value is remarkably close to the value for σtbi (approximately 10 cal/(mol·Å2)) reported here. Although the agreement may be fortuitous, it does suggest a possible route to understanding quantitatively the physical chemistry of translocon-to-membrane partitioning.
Materials and Methods
Enzymes and Chemicals.
All enzymes were purchased from Fermentas, except Phusion DNA polymerase from Finnzyme and SP6 RNA polymerase from Promega. The QuikChange™ Site-Directed Mutagenesis kit and deoxyribonucleotides were from Stratagene, and the Megashortscript™ T7 kit was from Ambion Inc. The plasmid pGEM1, the rabbit reticulocyte lysate system, and the RNasin were from Promega. Oligonucleotides were from Eurofins MWG Operon. All chemicals were from Sigma-Aldrich, except DMSO from J.T. Baker Chemicals, ethanol from Kemetyl, and [35S]-methionine from PerkinElmer.
DNA Manipulations.
For cloning, a modified version of the Escherichia coli lepB gene in a pGEM1 vector was used (6). It harbors an engineered test segment (H segment) in the coding region of the P2 domain between a Spe1 cleavage site in codons 226–227 and a Kpn1 cleavage site in codon 253 (WT lepB codon positions) as well as two glycosylation acceptor sites for N-linked glycosylation at codons 96–98 (G1: Asn-Ser-Thr) and codons 258–260 (G2: Asn-Ala-Thr). The sequences of the H segment’s flanking regions are QETKENGIRLSETSGGPG-(H segment)-GPGGVPGQQNATWIVPP (Spe1 and Kpn1 cleavage sites underlined). The introduction of amber stop codons (TAG) into the termini of the H-segment encoding sequence was done by site-directed mutagenesis using Pfu Turbo polymerase. To introduce amber stop codons in or near the center of the H segment, double-stranded oligonucleotides encoding the H segment with the amber stop (including also GGPG/GPGG) and flanked by N-terminal Spe1 and C-terminal Kpn1 sticky ends were first generated by annealing of two pairs of complementary oligonucleotides with overlapping overhangs (each 18–45 nucleotides long), followed by annealing of the pairs via the complementary overhangs and cloning into the lepB gene between the Spe1 and Kpn1 cleavage sites (6).
For Lep constructs with a Ncyt-Clum orientated H segment, a large part of the P2 domain was replaced by the corresponding part of the P2 domain of a Lep construct where all positive charges after codon 179 until 30 codons upstream of the end of the protein were replaced by alanines, and a new H segment with composition GGPG-ALAALALAALAALALAALA-GPGG was introduced between Apa1 and Mfe1 cleavage sites, located in codons -71 and -45 relative to the first codon of the original H segment. The construct further has two additional glycosylation acceptor sites for N-linked glycosylation, one (G2′: Asn-Ala-Thr) 16–18 codons downstream of the new H segment and the other (G4′: Asn-Ser-Thr) 7–9 codons downstream of G3′ (the G3′ site is the same as the G2 site in the original construct with Nout-Cin orientated H segment). The gene fragment of the replacing P2 domain was generated by PCR amplification and inserted between a Xho1 cleavage site (lepB codons 180 and 181) and a Sma1 cleavage site directly downstream of the 3′ end of the lepB gene.
Preparation of Flexible tRNA Aminoacylation Ribozyme [Dinitrobenzyl Flexizyme (dFx); Enhanced Flexizyme ( eFx)] and Microbacteriophage L5 (ML)-Derived  [
[ (tRNAsup)].
(tRNAsup)].
Preparations were done using the same protocol. First, double-stranded DNA templates encoding the RNA species and an N-terminal T7 promotor sequence (8, 32) were generated by PCR extension of annealed overlapping oligonuclotides. DNA templates were then amplified by PCR using primers complementary to both ends of the templates, followed by phenol/chloroform extraction and ethanol precipitation. The DNA was used in a second step for transcription by T7 polymerase using the Ambion Megashortscript™ T7 kit, and the RNA product was isopropanol precipitated and purified over 12% denaturing PAGE. After cutting out the RNA band, RNA was eluted for 2 h in 0.3 M NaCl, ethanol precipitated, and dissolved in dH2O (70–250 μM final concentration).
Materials for the Synthesis and Characterization of Amino Acid Derivatives.
All experiments dealing with air- and moisture-sensitive compounds were conducted under an atmosphere of dry argon. For TLC analysis, Merck precoated plates (silica gel 60 F254, Art 5715, 0.25 mm) were used. For flash column chromatography, silica gel 60 (Merck Art 7734, 70–230 mesh) was used. For silica gel preparative TLC preparation, Merck precoated plates (silica gel 60 F254, Art 5744, 0.5 mm) were used. Cyanomethylester (CME) and 3,5-dinitrobenzylester derivatives were prepared using a previously described procedure (8, 33). For general procedures of synthesis and characterization of compounds, see SI Text.
Acylation of  .
.
Flexizyme (dFx in all cases, except for aromatic amino acids where eFx was used) and  (tRNAsup), each 250 μM in 71 mM Hepes-K buffer, pH 7.5 (total volume 7 μL), were heated at 95 °C for 2 min and cooled to room temperature over 5 min. One microliter MgCl2 (200 mM for dFx and 3 M for eFx reactions) and 2 μL of amino acid 3,5-dinitrobenzyl ester (25 mM, nonaromatic amino acids) or amino acid CME (25 mM, aromatic amino acids) were added. The reaction was carried out for 2 h on ice and then stopped with 40 μM AcONa (0.3 M, pH 5.2). RNA was ethanol precipitated twice, once with a 0.1 M ethanolic AcONa solution (pH 5.2) and once with 95% ethanol. More detailed information can be found elsewhere (8).
(tRNAsup), each 250 μM in 71 mM Hepes-K buffer, pH 7.5 (total volume 7 μL), were heated at 95 °C for 2 min and cooled to room temperature over 5 min. One microliter MgCl2 (200 mM for dFx and 3 M for eFx reactions) and 2 μL of amino acid 3,5-dinitrobenzyl ester (25 mM, nonaromatic amino acids) or amino acid CME (25 mM, aromatic amino acids) were added. The reaction was carried out for 2 h on ice and then stopped with 40 μM AcONa (0.3 M, pH 5.2). RNA was ethanol precipitated twice, once with a 0.1 M ethanolic AcONa solution (pH 5.2) and once with 95% ethanol. More detailed information can be found elsewhere (8).
Expression in Vitro and Quantification of Membrane-Insertion Efficiency.
All constructs were transcribed for 60 min at 37 °C using a standard SP6 polymerase transcription protocol (34). Resulting mRNA was translated for 80 min at 30 °C in rabbit reticulocyte lysate (40–60 pg mRNA per µL translation mix) in the presence of [35S]-Met (370 pCi/μL translation mix; 1 Ci = 37 GBq), dog pancreas RMs (50 pL/μL translation mix), amino acid mix (each 75 μM/μL translation mix), amino acid-tRNAsup (3.5 pmol/μL translation mix), and RNasin (3 units per µL translation mix). Translation products were analyzed by SDS-PAGE. Gels were visualized on a Fuji FLA-3000 PhosphoImager using the Image Reader 8.1j software and quantified using ImageGauge V 3.45 and the Qtiplot 0.9.3-rc2 softwares (35). The degree of membrane integration of each H segment was calculated as an apparent equilibrium constant between the membrane-integrated and nonintegrated forms: Kapp = f1/f2, where f1 is the fraction of singly and f2 the fraction of doubly glycosylated Lep molecules, and the results were then converted to apparent free energies, ΔGapp = -RT ln Kapp. All reported ΔGapp values are averages of three or four independent measurements. The degree of suppression of the UAG stop codon obtained in the in vitro translation system varied between 50% and 75% for the different aminoacylated tRNAsup species, and was between 2% and 6% (all positions shown in Fig. 2 were tested) when an uncharged tRNAsup was present in the translation mix.
Accessible Surface Area Calculations.
Model helices of the H segments without flanking GGPG/GPGG sequences were generated using MacPyMol and, if needed, the side chains of the nonnatural amino acids manually modeled at the appropriate position in the helix. The accessible surface area was then calculated using Naccess version 2.1.1 (copyright S. Hubbard and J. Thornton 1992–1996) with a rolling probe size of 1.4 Å.
Supplementary Material
Acknowledgments.
We thank Drs. Kálmán Szabó and Erik Lindahl, Stockholm University, for technical advice, and Dr. Berhard Dobberstein, Heidelberg University, for kindly providing dog pancreas rough microsomes. This work was supported by grants from the European Research Council (ERC-2008-AdG 232648), the Swedish Foundation for Strategic Research, the Swedish Research Council, and the Swedish Cancer Foundation (G.v.H.), the Swedish Foundation for International Cooperation in Research and Higher Education and Henrik Granholms Stiftelse (I.M.N.), the Japan Science and Technology Agency, Strategic International Cooperative Program (H.S.), and the US National Institutes of Health (S.H.W.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
See Author Summary on page 12571.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1100120108/-/DCSupplemental.
References
- 1.Rapoport TA. Protein translocation across the eukaryotic endoplasmic reticulum and bacterial plasma membranes. Nature. 2007;450:663–669. doi: 10.1038/nature06384. [DOI] [PubMed] [Google Scholar]
- 2.White SH, von Heijne G. How translocons select transmembrane helices. Annu Rev Biophys. 2008;37:23–42. doi: 10.1146/annurev.biophys.37.032807.125904. [DOI] [PubMed] [Google Scholar]
- 3.Hessa T, et al. Molecular code for transmembrane-helix recognition by the Sec61 translocon. Nature. 2007;450:1026–1030. doi: 10.1038/nature06387. [DOI] [PubMed] [Google Scholar]
- 4.van den Berg B, et al. X-ray structure of a protein-conducting channel. Nature. 2004;427:36–44. doi: 10.1038/nature02218. [DOI] [PubMed] [Google Scholar]
- 5.Egea PF, Stroud RM. Lateral opening of a translocon upon entry of protein suggests the mechanism of insertion into membranes. Proc Natl Acad Sci USA. 2010;107:17182–17187. doi: 10.1073/pnas.1012556107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hessa T, et al. Recognition of transmembrane helices by the endoplasmic reticulum translocon. Nature. 2005;433:377–381. doi: 10.1038/nature03216. [DOI] [PubMed] [Google Scholar]
- 7.Schow EV, et al. Arginine in membranes: The connection between molecular dynamics simulations and translocon-mediated insertion experiments. J Membr Biol. 2011;239:35–48. doi: 10.1007/s00232-010-9330-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Murakami H, Ohta A, Ashigai H, Suga H. A highly flexible tRNA acylation method for non-natural polypeptide synthesis. Nat Methods. 2006;3:357–359. doi: 10.1038/nmeth877. [DOI] [PubMed] [Google Scholar]
- 9.Xiao H, Murakami H, Suga H, Ferre-D’Amare AR. Structural basis of specific tRNA aminoacylation by a small in vitro selected ribozyme. Nature. 2008;454:358–361. doi: 10.1038/nature07033. [DOI] [PubMed] [Google Scholar]
- 10.Johansson M, Nilsson I, von Heijne G. Positively charged amino acids placed next to a signal sequence block protein translocation more efficiently in Escherichia coli than in mammalian microsomes. Mol Gen Genet. 1993;239:251–256. doi: 10.1007/BF00281625. [DOI] [PubMed] [Google Scholar]
- 11.Sääf A, Wallin E, von Heijne G. Stop-transfer function of pseudo-random amino acid segments during translocation across prokaryotic and eukaryotic membranes. Eur J Biochem. 1998;251:821–829. doi: 10.1046/j.1432-1327.1998.2510821.x. [DOI] [PubMed] [Google Scholar]
- 12.Wimley WC, Creamer TP, White SH. Solvation energies of amino acid sidechains and backbone in a family of host-guest pentapeptides. Biochemistry. 1996;35:5109–5124. doi: 10.1021/bi9600153. [DOI] [PubMed] [Google Scholar]
- 13.Karplus PA. Hydrophobicity regained. Protein Sci. 1997;6:1302–1307. doi: 10.1002/pro.5560060618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Marqusee JA, Dill KA. Solute partitioning into chain molecule interphases: Monolayers, bilayer membranes, and micelles. J Chem Phys. 1986;85:434–444. [Google Scholar]
- 15.White SH, King GI, Cain JE. Location of hexane in lipid bilayers determined by neutron diffraction. Nature. 1981;290:161–163. [Google Scholar]
- 16.Yau WM, Wimley WC, Gawrisch K, White SH. The preference of tryptophan for membrane interfaces. Biochemistry. 1998;37:14713–14718. doi: 10.1021/bi980809c. [DOI] [PubMed] [Google Scholar]
- 17.Buton X, Morrot G, Fellmann P, Seigneuret M. Ultrafast glycerophospholipid-selective transbilayer motion mediated by a protein in the endoplasmic reticulum membrane. J Biol Chem. 1996;271:6651–6657. doi: 10.1074/jbc.271.12.6651. [DOI] [PubMed] [Google Scholar]
- 18.van Meer G, Voelker DR, Feigenson GW. Membrane lipids: Where they are and how they behave. Nat Rev Mol Cell Biol. 2008;9:112–124. doi: 10.1038/nrm2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Heinrich S, Mothes W, Brunner J, Rapoport T. The Sec61p complex mediates the integration of a membrane protein by allowing lipid partitioning of the transmembrane domain. Cell. 2000;102:233–244. doi: 10.1016/s0092-8674(00)00028-3. [DOI] [PubMed] [Google Scholar]
- 20.Gumbart J, Chipot C, Schulten K. Free-energy cost for translocon-assisted insertion of membrane proteins. Proc Natl Acad Sci USA. 2011;108:3596–3601. doi: 10.1073/pnas.1012758108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rychkova A, Vicatos S, Warshel A. On the energetics of translocon-assisted insertion of charged transmembrane helices into membranes. Proc Natl Acad Sci USA. 2010;107:17598–17603. doi: 10.1073/pnas.1012207107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zimmer J, Nam Y, Rapoport TA. Structure of a complex of the ATPase SecA and the protein-translocation channel. Nature. 2008;455:936–943. doi: 10.1038/nature07335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tsukazaki T, et al. Conformational transition of Sec machinery inferred from bacterial SecYE structures. Nature. 2008;455:988–991. doi: 10.1038/nature07421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Johansson AC, Lindahl E. The role of lipid composition for insertion and stabilization of amino acids in membranes. J Chem Phys. 2009;130:185101. doi: 10.1063/1.3129863. [DOI] [PubMed] [Google Scholar]
- 25.Chothia C. The nature of the accessible and buried surfaces in proteins. J Mol Biol. 1976;105:1–12. doi: 10.1016/0022-2836(76)90191-1. [DOI] [PubMed] [Google Scholar]
- 26.Eisenberg D, McLachlan AD. Solvation energy in protein folding. Nature. 1986;319:199–203. doi: 10.1038/319199a0. [DOI] [PubMed] [Google Scholar]
- 27.Hermann RB. Use of solvent cavity area and number of packed solvent molecules around a solute in regard to hydrocarbon solubilities and hydrophobic interactions. Proc Natl Acad Sci USA. 1977;74:4144–4145. doi: 10.1073/pnas.74.10.4144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reynolds JA, Gilbert DB, Tanford C. Empirical correlation between hydrophobic free energy and aqueous cavity surface area. Proc Natl Acad Sci USA. 1974;71:2925–2927. doi: 10.1073/pnas.71.8.2925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Junne T, Kocik L, Spiess M. The hydrophobic core of the Sec61 translocon defines the hydrophobicity threshold for membrane integration. Mol Biol Cell. 2010;21:1662–1670. doi: 10.1091/mbc.E10-01-0060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Simon SM, Peskin CS, Oster GF. What drives the translocation of proteins? Proc Natl Acad Sci USA. 1992;89:3770–3774. doi: 10.1073/pnas.89.9.3770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liebermeister W, Rapoport TA, Heinrich R. Ratcheting in post-translational protein translocation: A mathematical model. J Mol Biol. 2001;305:643–656. doi: 10.1006/jmbi.2000.4302. [DOI] [PubMed] [Google Scholar]
- 32.Ohta A, Murakami H, Higashimura E, Suga H. Synthesis of polyester by means of genetic code reprogramming. Chem Biol. 2007;14:1315–1322. doi: 10.1016/j.chembiol.2007.10.015. [DOI] [PubMed] [Google Scholar]
- 33.Saito H, Kourouklis D, Suga H. An in vitro evolved precursor tRNA with aminoacylation activity. EMBO J. 2001;20:1797–1806. doi: 10.1093/emboj/20.7.1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nilsson I, Johnson AE, von Heijne G. How hydrophobic is alanine? J Biol Chem. 2003;278:29389–29393. doi: 10.1074/jbc.M212310200. [DOI] [PubMed] [Google Scholar]
- 35.Hedin LE, et al. Membrane insertion of marginally hydrophobic transmembrane helices depends on sequence context. J Mol Biol. 2010;396:221–229. doi: 10.1016/j.jmb.2009.11.036. [DOI] [PubMed] [Google Scholar]