Abstract
Sex in mammals is determined in the foetal gonad by the presence or absence of the Y chromosome gene Sry, which controls whether bipotential precursor cells differentiate into testicular Sertoli cells or ovarian granulosa cells1. This pivotal decision in a single gonadal cell type ultimately controls sexual differentiation throughout the body. Sex determination can be viewed as a battle for primacy in the foetal gonad between a male regulatory gene network in which Sry activates Sox9 and a female network involving Wnt/β-catenin signaling (Supplemental Fig. 1)2. In females the primary sex-determining decision is not final: loss of the FOXL2 transcription factor in adult granulosa cells can reprogramme granulosa cells into Sertoli cells2. Here we show that sexual fate is also surprisingly labile in the testis: loss of the DMRT1 transcription factor3 in mouse Sertoli cells, even in adults, activates Foxl2 and reprogrammes Sertoli cells into granulosa cells. In this environment, theca cells form, oestrogen is produced, and germ cells appear feminized. Thus Dmrt1 is essential to maintain mammalian testis determination, and competing regulatory networks maintain gonadal sex long after the foetal choice between male and female. Dmrt1 and Foxl2 are conserved throughout vertebrates4,5 and Dmrt1-related sexual regulators are conserved throughout metazoans3. Antagonism between Dmrt1 and Foxl2 for control of gonadal sex may therefore extend beyond mammals. Reprogramming due to loss of Dmrt1 also may help explain the etiology of human syndromes linked to DMRT1, including disorders of sexual differentiation6 and testicular cancer7.
Human chromosome 9p deletions removing DMRT1 are associated with XY male-to-female sex reversal, and Dmrt1 homologues determine sex in several non-mammalian vertebrates8,9,10. In mice, Dmrt1 is expressed and required in both germ cells and Sertoli cells of the testis11,12,13. XY Dmrt1 null mutant mice are born as males with testes, although these gonads later undergo abnormal differentiation14; hence the role of Dmrt1 in mammalian sex determination has been unclear. Here we examine Dmrt1 mutant testes during postnatal development, asking whether loss of Dmrt1 causes postnatal feminization in mice.
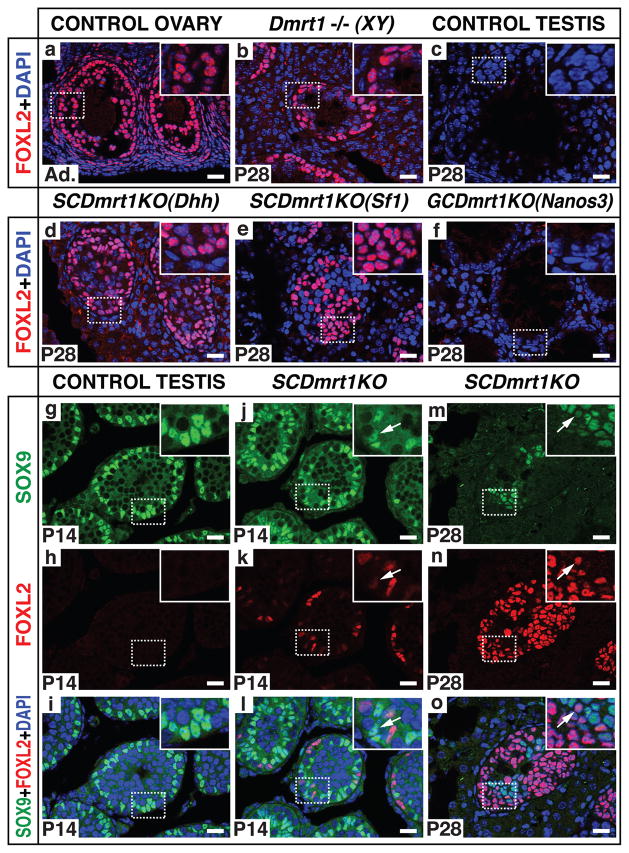
We first examined gonads of Dmrt1 null mutant males (Dmrt1−/−) for the presence of FOXL2, a female-specific transcription factor expressed in granulosa cells and theca cells15,16, the two somatic cell types of the ovarian follicle (Fig. 1a). Four weeks after birth, abundant FOXL2-positive cells were present within mutant seminiferous tubules (Fig. 1b), which in control testes contain only germ cells and Sertoli cells (Fig 1c). To establish the origin of the FOXL2-positive cells, we deleted Dmrt1 either in germ cells (using Nanos3-cre) or in Sertoli cells (using Dhh-cre or Sf1-cre) (Supplemental Fig. 2a–l; Supplemental Table 1). Loss of Dmrt1 in foetal Sertoli cells (SCDmrt1KO) but not in foetal germ cells (GCDmrt1KO) induced FOXL2 expression (Fig. 1d–f). SCDmrt1KO gonads retained small numbers of germ cells, which appeared to arrest in meiotic prophase based on SYCP3 localization (Supplemental Fig. 3). These results demonstrate that DMRT1 expression in Sertoli cells prevents FOXL2 expression and suggest that Dmrt1 mutant testes become feminized during the first postnatal month.
Figure 1. DMRT1 maintains SOX9 and suppresses FOXL2 expression in postnatal Sertoli cells.
(a–c) FOXL2 expression detected by immunofluorescence (IF) in adult granulosa and theca cells of control ovary (a) and intratubular cells of Dmrt1 null testis at postnatal day 28 (P28) (b) but not in control testis (c). (d–f) FOXL2 is robustly expressed when Dmrt1 is mutated in foetal Sertoli cells with Dhh-cre (d) or Sf1-cre (e) but not when Dmrt1 is mutated in foetal germ cells with Nanos3-cre (f). (g–o) Timing of FOXL2 expression. FOXL2 is absent from control testis at P14 (g–i). Cells expressing FOXL2 or FOXL2 and SOX9 (arrowheads) are present in SCDmrt1KO testis at P14 (j–l). FOXL2-positive cells are abundant in SCDmrt1KO testis at P28 and most cells no longer express SOX9 (m–o). Scale bars: 20 mm.
Next we examined the timing of FOXL2 induction. At postnatal day 7 (P7), SCDmrt1KO testes had seminiferous tubules in which all Sertoli cells expressed SOX9 normally (Supplemental Fig. 2m–r), but at P14 some intratubular cells co-expressed SOX9 and FOXL2 or lacked SOX9 and strongly expressed FOXL2 (Fig. 1g–l). By P28 few SOX9-positive cells remained and most intratubular cells strongly expressed FOXL2 (Fig. 1m–o). Histologic analysis of mutant gonads is shown in Supplemental Fig. 4. These results show that foetal loss of Dmrt1 causes postnatal Sertoli cells to lose the male-promoting SOX9 and instead express the female-promoting FOXL2.
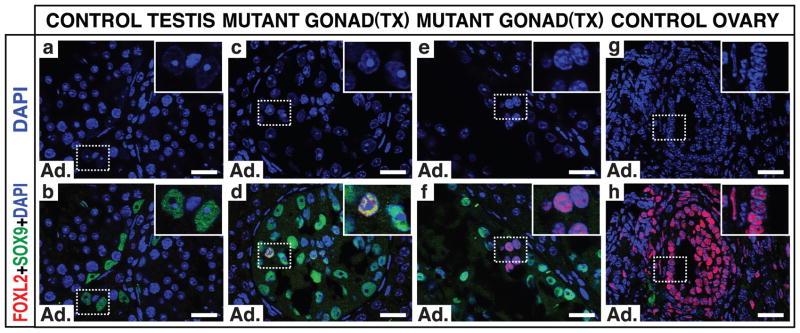
Loss of Foxl2 in the adult ovary can lead to transdifferentiation of granulosa cells to Sertoli cells2, so we asked whether loss of Dmrt1 in the adult testis activates Foxl2 and causes the reciprocal sex transformation, from Sertoli to granulosa. Indeed, one month after deletion of Dmrt1 in adult males (using a tamoxifen-inducible cre transgene), we observed cells with typical Sertoli cell features including tripartite nucleoli but expressing both SOX9 and FOXL2 (Fig. 2a–d), as well as cells with typical granulosa cell nuclear morphology that lacked SOX9 and strongly expressed FOXL2 (Fig. 2e–h). Thus antagonism between DMRT1 and FOXL2 continues into adulthood and Sertoli cell fate remains plastic even after terminal differentiation.
Figure 2. Sertoli-to-granulosa transdifferentiation in the adult testis.
Expression of FOXL2 and SOX9 one month after tamoxifen injection into Dmrt1flox/flox adult males (8 weeks and older) carrying inducible ubiquitous cre transgene UBC-cre/ERT2. (a,b) Sertoli cells in control testis express SOX9 but not FOXL2. (c–f) Mutant testis has Sertoli-like cells expressing SOX9 or SOX9 and FOXL2 (inset, d) and granulosa-like cells expressing only FOXL2 (inset, f). (g,h) FOXL2-positive cells in control ovary have DAPI morphology similar to FOXL2 single-positive cells of mutant testis. FOXL2-positive cells in mutant testis resemble granulosa cells: they lack the tripartite nucleoli of Sertoli cells, have smaller and more rounded nuclei, and have more punctate DAPI staining. UBC-cre/ERT2 also deletes Dmrt1 in germ cells, causing precocious meiosis12; after one month germ cells are nearly absent. Scale bars: 20 μm.
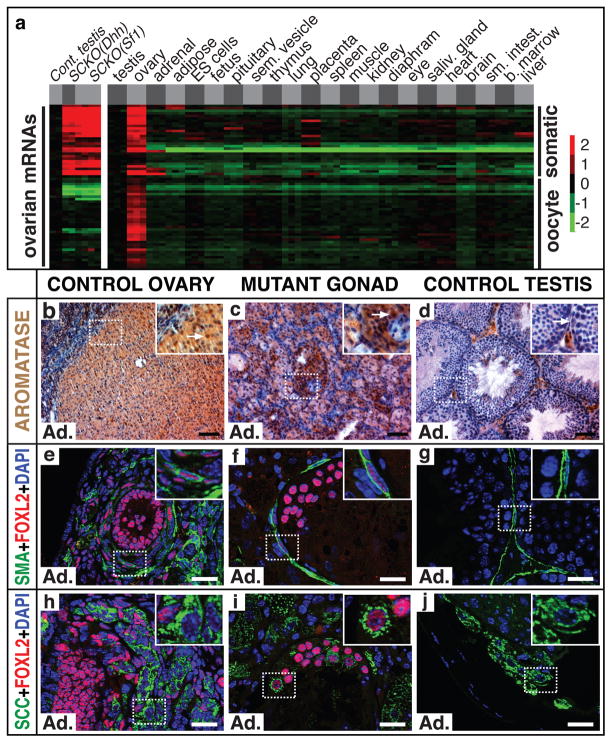
To further evaluate the transformation of mutant gonads, we compared the mRNA profile of control and mutant P28 testes; 5030 mRNAs were expressed >8-fold differently across this dataset or a dataset comparing testis to 21 other tissues including ovary; (Supplemental Fig. 5a). We calculated Pearson correlation coefficients for expression of these 5030 mRNAs in mutant gonads relative to each tissue and found that the mutant gonad most closely resembled ovary (Supplemental Fig. 5b; average R=0.75). Many mRNAs with decreased expression in mutant gonads also were low in other tissues, likely reflecting a lack of male germ cells, which comprise much of the testis mass. Also, some mRNAs elevated in mutant gonads were elevated in other tissues. Therefore, to specifically evaluate ovary-enriched mRNAs, we used bioGPS (biogps.gnf.org; SI) to identify 65 mRNAs with expression closely correlated to Foxl2 and then compared their expression in ovary relative to the other 21 tissues (Fig. 3a; Supplemental Fig. 6). This comparison confirmed that these mRNAs are highly ovary-enriched. About 40% were elevated in mutant gonads relative to control testes; about 80% of the remainder were oocyte-enriched. Thus loss of Dmrt1 causes large changes in mRNA expression, including induction of multiple ovary-enriched mRNAs. mRNA profiling of Dmrt1 mutant gonads perinatally and at P9 did not reveal apparent feminization17,18, consistent with the observation that FOXL2 expression starts at ~P14.
Figure 3. Feminization of SCDmrt1KO XY gonads.
(a) Expression of ovary-enriched mRNAs with expression profiles similar to Foxl2 (see SI). mRNAs labeled “somatic” were enriched in ovarian somatic cells; those labeled “oocyte” were enriched in female germ cells. See Supplemental Fig. 6 for higher resolution image. (b–d) IHC detection of CYP19A1/Aromatase expression in follicles of control adult ovary (a) and in adult XY SCDmrt1KO gonad (b) but only in interstitial Leydig cells of control testis (c). Arrows indicate Aromatase-positive granulosa cells in ovary and mutant gonad and negative Sertoli cell in control testis. Scale bars: 50 μm. (e–g) IF detection of smooth muscle actin (SMA) and FOXL2. Ovarian theca cells (inset, e) are elongated cells expressing both proteins; similar cells are present in mutant gonads (f); peritubular myoid cells in control testes express SMA and not FOXL2 (g). Scale bars: 20 μm. (h–j) IF detection of cells coexpressing FOXL2 in the nucleus and steroidogenic enzyme CYP11A1/SCC at high levels in the cytoplasm in control ovary (h) and XY Dmrt1KO gonads (i). SCC-positive cells in control testis (j) are interstitial Leydig cells. Mutant gonads were SCDmrt1KO(Dhh). Ad.=adult. Scale bars: 20μm.
Further analysis of the mRNA profiling data identified highly elevated expression (>5-fold, p<0.001) of many mRNAs expressed in granulosa cells and required for ovarian development or function. These included Foxl2, Nr5a2/Lrh1, Wnt4, LH receptor (Lhcgr), prolactin receptor (Prlr), FSH receptor (Fshr), follistatin (Fst), Sfrp4, Igfbp5, Inhbb, Inha, and Lnfg (Supplemental Table 2). Foxl2os, a noncoding RNA transcribed from the opposite strand of the Foxl2 coding region, also was highly over-expressed and has been suggested as a positive regulator of Foxl219. We confirmed elevated expression in mutant gonads of LRH1, a transcription factor expressed only in granulosa cells within the ovary20 and absent from the testis (Supplemental Fig. 7a–f). Nr5a2/Lrh1 is likely a direct target of DMRT1 regulation, based on binding of DMRT1 to its promoter proximal sequences in vivo (Supplemental Fig. 7g). Based on mRNA and protein expression data and changes in cellular morphology, we conclude that loss of Dmrt1 in testes reprogrammes Sertoli cells into granulosa cells.
Granulosa cells produce oestrogens, which are essential for ovarian development in many vertebrates; in mammals oestrogen signaling also acts with FOXL2 to repress Sox9 transcription in adult granulosa cells2. HSD17β1 and CYP19A1/Aromatase are enzymes critical for oestrogen synthesis, and mRNAs for both enzymes were elevated in mutant gonads (Supplemental Fig. 8). Aromatase protein is robustly expressed in granulosa cells and was strongly expressed in mutant gonads (Fig. 3b–d). Consistent with these enzyme changes, oestradiol was elevated in serum of adult mutants relative to control adult males (SI). Although expression of the androgenic enzyme Hsd17β3 was not affected in mutant gonads (Supplemental Fig. 8), androgen levels were reduced based on severely decreased seminal vesicle weight, a sensitive indicator of androgen activity (350 +/− 52 mg vs 182 +/− 36 mg; n = 3, P=0.01).
Theca cells are induced during follicle growth in the ovary, likely in response to granulosa cell signals21, and together with granulosa cells and oocytes they comprise the functional unit of the ovary. Because mutant gonads contained apparently functional granulosa cells, we asked whether theca cells also formed. Theca cells have spindle-shaped nuclei and express both FOXL2 and smooth muscle actin (SMA) (Fig. 3e). Adult mutant gonads contained cells closely resembling theca cells and expressing both proteins (Fig. 3f). The theca-like cells likely derive either from granulosa cells or peritubular myoid cells (which also are elongated and express SMA; Figure 3g). However, since seminiferous tubule integrity was lost prior to formation of these cells (Fig. 3f; Supplemental Fig. 9) they could potentially derive from interstitial cells that invaded the tubule remnants. We also observed intratubular cells strongly expressing the steroidogenic enzyme SCC (Fig. 3h–j); these cells resembled luteinized granulosa cells of the ovary (Fig. 3h), suggesting that granulosa cells in the mutant gonad are responsive to gonadotropins. We therefore tested the effect of exogenous gonadotropin stimulation; treated mutants, but not controls, had additional luteinized granulosa cells and germ cells with oocyte-like nuclear morphology that expressed the oocyte-specific proteins MATER and ZP2 (Supplemental Fig. 10). This result suggests that both somatic cells and germ cells are feminized in mutant gonads.
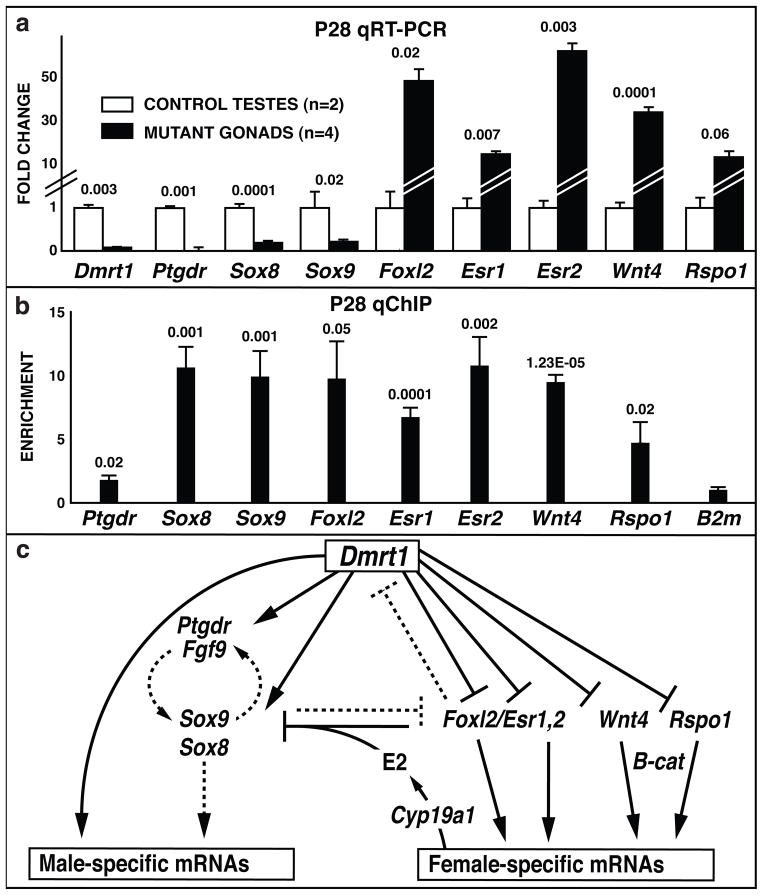
The preceding results indicate that DMRT1 is essential for postnatal sex maintenance. DMRT1 is a sequence-specific transcriptional regulator, capable of activating or repressing transcription of target genes18,22. To help find targets of DMRT1 regulation with potential roles in sex maintenance we examined expression of known foetal sex-determining genes in mutant gonads at P28 by qRT-PCR (Fig. 4a). Among masculinizing genes, Ptgdr, Sox9, and Sox8, which acts redundantly with Sox923,24, were reduced. Among feminizing genes, Foxl2, Esr1, Esr2, Wnt4 and Rspo1 were elevated. We assayed binding of DMRT1 to DNA of P28 testes by quantitative chromatin immunoprecipitation (qChIP), guided by genome-wide ChIP data from P9 testes (ChIP-chip18 and ChIP-seq [unpublished]). DMRT1 bound both upstream and downstream of Sox9 and upstream of Sox8, and bound weakly near Ptgdr. DMRT1 bound strongly near Foxl2, Esr1, Esr2, Wnt4 and Rspo1 (Fig. 4b). All of the DMRT1-associated regions contained at least one close match to the DMRT1 DNA binding consensus18,22.
Figure 4. DMRT1 regulation of postnatal gene expression.
(a) qRT-PCR analysis of sex-determining genes at P28. Significance of expression changes is indicated (Students t-test). Mutant gonads were SCDmrt1KO(Sf1); SCDmrt1KO(Dhh) mutant gonads and equivalent expression changes. (b) qChIP analysis of DMRT1 DNA binding in P28 testes. Significance of enrichment relative to B2m (Students t-test) is shown. (c) Model for regulation by postnatal sex maintenance by DMRT1. Proposed direct regulation based on ChIP and mRNA expression data is indicated by solid lines; indirect or potential regulation is indicated by dashed lines. (Model adapted from Veitia2).
Based on mRNA and protein expression data and ChIP analysis, we propose a model for postnatal sex maintenance (Fig. 4b) in which DMRT1 maintains male fates by repressing multiple female-promoting genes and activating male-promoting genes. Sox9 is dispensable for testis differentiation after sex determination24,25, suggesting that other critical male regulators remain to be found; Sox8 is a clear candidate based on its redundancy with Sox923,24. We find that DMRT1 represses Foxl2, which is known to maintain postnatal ovarian fate. FOXL2 also represses Dmrt12; thus antagonism between these sex-specific transcriptional regulators may be central to sex maintenance in both sexes throughout reproductive life. Wnt4 and Rspo1 also are prime candidates for postnatal sex maintenance based on their requirement in ovarian determination in the foetus26,27. Indeed, P28 mutant gonads had elevated nuclear β-Catenin in somatic cells, as in ovaries, but control testes did not, indicating active Wnt/β-catenin signaling in the mutant gonads (Supplemental Fig. 11). Functional analysis of Wnt4, Rspo1 and other known foetal sex regulators will be important to establish their roles in sex maintenance. The analysis presented here demonstrates that deletion of Dmrt1 during foetal development induces postnatal feminization of the testis, causing male-to-female primary sex reversal. Moreover, deletion of Dmrt1 in adults can reprogramme differentiated Sertoli cells into apparent granulosa cells. Why are Dmrt1 mutants feminized only after birth? Another male-promoting gene may act redundantly with Dmrt1 prior to P14, masking its function; alternatively, the testis may lack potential feminizing activity from genes such as Foxl2 prior to P14. Why are Dmrt1 mutant mice born male, whereas human 9p deletions removing DMRT1 can cause XY feminization at birth? The human sex reversal may reflect failure to maintain male sex determination, and the longer human gestation may permit testis-to-ovary reprogramming before birth. Alternatively, human testes may have potential feminizing activity earlier or may lack masculinizing genes redundant with DMRT1. Our results may provide insights into the aetiology of human gonadal disorders, including gonadoblastoma and granulosa cell tumors of the testis. Moreover, because many genes implicated in this study are evolutionarily conserved, similar mechanisms may control adult sex-switching in fish and may maintain sexual fate in the adult gonads of other vertebrates or even in other phyla.
Methods Summary
Mouse breeding
Dmrt1 mutant and control males were generated as described12; tissue-specific Cre recombinase strains are in Supplemental Table 1. Adult wild type or Dmrt1flox/flox females were used as controls. Mice were mixed C57BL/6J, 129S1, and FVB genetic background. Protocols were approved by the Institutional Animal Care and Use Committee.
Immunofluorescence (IF) and immunohistochemistry (IHC)
IF and IHC were performed as described12. Antibodies are listed in Supplemental Table 3. Analyses included at least two biological replicates.
Tamoxifen treatment
Tamoxifen-inducible deletion of Dmrt1 in adult males was as described12. Testes were harvested one to two months post-treatment.
mRNA expression analysis
mRNA expression profiling and data analysis were as described13 except total testis RNA was isolated from 4-week-old animals using TRIzol reagent (Invitrogen #15596-026). Additional detail is in Supplemental Methods.
qRT-PCR
qRT-PCR was as described12. qRT-PCR primers are listed in Supplemental Table 4.
Chromatin immunoprecipitation
ChIP followed by either microarray (ChIP-chip) or qPCR analysis (qChIP) were as described18. Gene-specific primers used for qChIP are in Supplemental Table 4.
Methods
Mouse breeding
Conditional Dmrt1 mutant and control males were generated as described12; tissue-specific Cre recombinase strains are in Supplemental Table 1. Adult wild type or Dmrt1flox/flox females were used as controls. Mice were of mixed C57BL/6J, 129S1, and FVB genetic background. Protocols were approved by the University of Minnesota Institutional Animal Care and Use Committee.
Immunofluorescence (IF) and immunohistochemistry (IHC)
Both IF and IHC were performed as described12. Antibodies are listed in Supplemental Table 3. Analyses included a minimum of two biological replicates.
Tamoxifen treatment
Tamoxifen-inducible deletion of Dmrt1 in adult males was performed as previously described12. Testes were harvested one to two months following treatment.
mRNA expression analysis
mRNA expression profiling and data analysis were performed as described13 except total testis RNA was isolated from 4-week-old animals using TRIzol reagent (Invitrogen #15596-026). Affymetrix Mouse Genome 439 2.0 arrays were normalized by GC-RMA normalization28 using GeneData Refiner. The Raw .cel files and the normalized data are deposited in GEO29 as GSE27261. GSE9954 was obtained from the GEO database. The arrays with the highest sample IDs were removed from the tissue dataset to select 22 tissue types, each with three experimental replicates. When multiple probe sets were mapped to the same gene symbol, these values were averaged to obtain one value for each gene symbol. Direct Pearson Correlation R-values were calculated using all array data following reduction to gene symbols, and these values are shown in Figure 2b.
Each experiment in our dataset was divided by the average expression value from control testis tissue. GSE9954 data were separately divided by the average signal obtained from the GSE9954 testis samples. This was done separately for each dataset to determine how samples from each dataset differed from a baseline “testis” expression state. Cluster 3.0 software30 was used to: i) log base 2 transform the data; ii) filter the dataset for genes that showed at least three observations with abs(val) >= 3 (8-fold) which resulted in 5030 genes passing the filter using both datasets combined; and iii) cluster the data on the gene-axis using average linkage hierarchial clustering. The experimental axis was defined by order of decreasing correlation to the mutant testes calculated as described above. Javatreeview Software31 was used to generate heatmap images.
qRT-PCR
qRT-PCR was performed as described12. qRT-PCR primers are listed in Supplemental Table 4.
Chromatin immunoprecipitation
ChIP followed by either microarray (ChIP-chip) or qPCR analysis (qChIP) were performed as described18. Gene specific primers used for qChIP are in Supplemental Table 4.
Oestadiol assays
Serum oestradiol was assayed using a clinical electrochemiluminescence immunoassay (Roche Estradiol II, 03000079 122) according to manufacturer’s instructions. Three of three males assayed had levels below the detection limit, whereas two of three females had measurable oestradiol (5.0 and 19.7 pg/dl). Two of three SCDmrt1KO(Dhh) mutant males had measurable oestradiol (5.6 and 21.2 pg/dl).
Gonadotropin treatment
6–8 week old mutant males, control males, and control females were treated with 5 units of pregnant mare serum by intraperitoneal injection and gonads were harvested 48 hours later.
Supplementary Material
Acknowledgments
We thank Katarina Hatzi, Anna Minkina, Aiden Peterson, the U of MN Mouse Genetics Laboratory, and Chris Small for technical assistance, Jurrien Dean, Reiner Veitia, and Ken-ichirou Morohashi for antibodies, David Greenstein and Anne Marie Weber-Main for comments on the manuscript, Carlos Manivel histology, Michael Steffes and Deanna Gabrielson for oestradiol analysis, and the U of MN Supercomputing Institute for computational resources. This work was funded by the NIH (GM59152), the Minnesota Medical Foundation, and a postdoctoral fellowship from the NSF (to CKM).
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Author Contributions C.K.M. performed mouse breeding and analysis of protein and mRNA expression; M.W.M. performed ChIP analysis; A.S. performed bioinformatic analysis; C.K.M., D.Z. and V.J.B. designed the study, analyzed data, and wrote the paper; M.D.G. provided mRNA profiling expertise; all authors discussed the results and edited the paper.
Author Information mRNA expression profiling data have been deposited at GEO (GSE27261) and can be reviewed at http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?token=zlctbiugsamymtc&acc=GSE27261.
References
- 1.Koopman P, Gubbay J, Vivian N, Goodfellow P, Lovell-Badge R. Male development of chromosomally female mice transgenic for Sry. Nature. 1991;351:117–121. doi: 10.1038/351117a0. [DOI] [PubMed] [Google Scholar]
- 2.Uhlenhaut NH, et al. Somatic sex reprogramming of adult ovaries to testes by FOXL2 ablation. Cell. 2009;139:1130–1142. doi: 10.1016/j.cell.2009.11.021. S0092-8674(09)01433-0 [pii] [DOI] [PubMed] [Google Scholar]
- 3.Raymond CS, et al. Evidence for evolutionary conservation of sex-determining genes. Nature. 1998;391:691–695. doi: 10.1038/35618. [DOI] [PubMed] [Google Scholar]
- 4.Loffler KA, Zarkower D, Koopman P. Etiology of ovarian failure in blepharophimosis ptosis epicanthus inversus syndrome: FOXL2 is a conserved, early-acting gene in vertebrate ovarian development. Endocrinology. 2003;144:3237–3243. doi: 10.1210/en.2002-0095. [DOI] [PubMed] [Google Scholar]
- 5.Raymond CS, Kettlewell JR, Hirsch B, Bardwell VJ, Zarkower D. Expression of Dmrt1 in the genital ridge of mouse and chicken embryos suggests a role in vertebrate sexual development. Dev Biol. 1999;215:208–220. doi: 10.1006/dbio.1999.9461. S0012-1606(99)99461-7 [pii] [DOI] [PubMed] [Google Scholar]
- 6.Tannour-Louet M, et al. Identification of de novo copy number variants associated with human disorders of sexual development. PLoS One. 2010;5:e15392. doi: 10.1371/journal.pone.0015392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Turnbull C, et al. Variants near DMRT1, TERT and ATF7IP are associated with testicular germ cell cancer. Nat Genet. 2010;42:604–607. doi: 10.1038/ng.607. ng.607 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yoshimoto S, et al. A W-linked DM-domain gene, DM-W, participates in primary ovary development in Xenopus laevis. Proc Natl Acad Sci U S A. 2008;105:2469–2474. doi: 10.1073/pnas.0712244105. 0712244105 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Smith CA, et al. The avian Z-linked gene DMRT1 is required for male sex determination in the chicken. Nature. 2009;461:267–271. doi: 10.1038/nature08298. nature08298 [pii] [DOI] [PubMed] [Google Scholar]
- 10.Matsuda M, et al. DMY is a Y-specific DM-domain gene required for male development in the medaka fish. Nature. 2002;417:559–563. doi: 10.1038/nature751. nature751 [pii] [DOI] [PubMed] [Google Scholar]
- 11.Kim S, Bardwell VJ, Zarkower D. Cell type-autonomous and non-autonomous requirements for Dmrt1 in postnatal testis differentiation. Dev Biol. 2007;307:314–327. doi: 10.1016/j.ydbio.2007.04.046. S0012-1606(07)00869-X [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Matson CK, et al. The mammalian doublesex homolog DMRT1 is a transcriptional gatekeeper that controls the mitosis versus meiosis decision in male germ cells. Dev Cell. 2010;19:612–624. doi: 10.1016/j.devcel.2010.09.010. S1534-5807(10)00428-4 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Krentz AD, et al. The DM domain protein DMRT1 is a dose-sensitive regulator of fetal germ cell proliferation and pluripotency. Proc Natl Acad Sci U S A. 2009;106:22323–22328. doi: 10.1073/pnas.0905431106. 0905431106 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Raymond CS, Murphy MW, O’Sullivan MG, Bardwell VJ, Zarkower D. Dmrt1, a gene related to worm and fly sexual regulators, is required for mammalian testis differentiation. Genes Dev. 2000;14:2587–2595. doi: 10.1101/gad.834100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schmidt D, et al. The murine winged-helix transcription factor Foxl2 is required for granulosa cell differentiation and ovary maintenance. Development. 2004;131:933–942. doi: 10.1242/dev.00969. dev.00969 [pii] [DOI] [PubMed] [Google Scholar]
- 16.Uda M, et al. Foxl2 disruption causes mouse ovarian failure by pervasive blockage of follicle development. Hum Mol Genet. 2004;13:1171–1181. doi: 10.1093/hmg/ddh124. ddh124 [pii] [DOI] [PubMed] [Google Scholar]
- 17.Fahrioglu U, Murphy MW, Zarkower D, Bardwell VJ. mRNA expression analysis and the molecular basis of neonatal testis defects in Dmrt1 mutant mice. Sex Dev. 2007;1:42–58. doi: 10.1159/000096238. SXD2007001001042 [pii] [DOI] [PubMed] [Google Scholar]
- 18.Murphy MW, et al. Genome-wide analysis of DNA binding and transcriptional regulation by the mammalian Doublesex homolog DMRT1 in the juvenile testis. Proc Natl Acad Sci U S A. 2010;107:13360–13365. doi: 10.1073/pnas.1006243107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cocquet J, Pannetier M, Fellous M, Veitia RA. Sense and antisense Foxl2 transcripts in mouse. Genomics. 2005;85:531–541. doi: 10.1016/j.ygeno.2005.01.007. S0888-7543(05)00021-2 [pii] [DOI] [PubMed] [Google Scholar]
- 20.Duggavathi R, et al. Liver receptor homolog 1 is essential for ovulation. Genes Dev. 2008;22:1871–1876. doi: 10.1101/gad.472008. 22/14/1871 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Orisaka M, Tajima K, Tsang BK, Kotsuji F. Oocyte-granulosa-theca cell interactions during preantral follicular development. J Ovarian Res. 2009;2:9. doi: 10.1186/1757-2215-2-9. 1757-2215-2-9 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Murphy MW, Zarkower D, Bardwell VJ. Vertebrate DM domain proteins bind similar DNA sequences and can heterodimerize on DNA. BMC Mol Biol. 2007;8:58. doi: 10.1186/1471-2199-8-58. 1471-2199-8-58 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chaboissier MC, et al. Functional analysis of Sox8 and Sox9 during sex determination in the mouse. Development. 2004;131:1891–1901. doi: 10.1242/dev.01087. dev.01087 [pii] [DOI] [PubMed] [Google Scholar]
- 24.Barrionuevo F, et al. Testis cord differentiation after the sex determination stage is independent of Sox9 but fails in the combined absence of Sox9 and Sox8. Dev Biol. 2009;327:301–312. doi: 10.1016/j.ydbio.2008.12.011. S0012-1606(08)01414-0 [pii] [DOI] [PubMed] [Google Scholar]
- 25.Chang H, et al. Wt1 negatively regulates beta-catenin signaling during testis development. Development. 2008;135:1875–1885. doi: 10.1242/dev.018572. dev.018572 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vainio S, Heikkila M, Kispert A, Chin N, McMahon AP. Female development in mammals is regulated byWnt-4 signalling. Nature. 1999;397:405–409. doi: 10.1038/17068. [DOI] [PubMed] [Google Scholar]
- 27.Parma P, et al. R-spondin1 is essential in sex determination, skin differentiation and malignancy. Nat Genet. 2006;38:1304–1309. doi: 10.1038/ng1907. ng1907 [pii] [DOI] [PubMed] [Google Scholar]
- 28.Wu J, Irazarry RA, Gentleman R, Martinez-Murillo, Spencer F. A model-based background adjustment for oligonucleotide expression arrays. Journal of the American Statistical Association. 2004;99:909–917. [Google Scholar]
- 29.Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.de Hoon MJ, Imoto S, Nolan J, Miyano S. Open source clustering software. Bioinformatics. 2004;20:1453–1454. doi: 10.1093/bioinformatics/bth078. bth078 [pii] [DOI] [PubMed] [Google Scholar]
- 31.Saldanha AJ. Java Treeview--extensible visualization of microarray data. Bioinformatics. 2004;20:3246–3248. doi: 10.1093/bioinformatics/bth349. bth349 [pii] [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.