Abstract
Sig1R (Sigma-1receptor) is a 25-kDa protein structurally unrelated to other mammalian proteins. Sig1R is present in brain, liver, and heart and is overexpressed in cancer cells. Studies using exogenous sigma ligands have shown that Sig1R interacts with a variety of ion channels, but its intrinsic function and mechanism of action remain unclear. The human ether-à-gogo related gene (hERG) encodes a cardiac channel that is also abnormally expressed in many primary human cancers, potentiating tumor progression through the modulation of extracellular matrix adhesive interactions. We show herein that sigma ligands inhibit hERG current density and cell adhesion to fibronectin in K562 myeloid leukemia cells. Heterologous expression in Xenopus oocytes demonstrates that Sig1R potentiates hERG current by stimulating channel subunit biosynthesis. Silencing Sig1R in leukemic K562 cells depresses hERG current density and cell adhesion to fibronectin by reducing hERG membrane expression. In K562 cells, Sig1R silencing does not modify hERG mRNA contents but reduces hERG mature form densities. In HEK cells expressing hERG and Sig1R, both proteins co-immunoprecipitate, demonstrating a physical association. Finally, Sig1R expression enhances both channel protein maturation and stability. Altogether, these results demonstrate for the first time that Sig1R controls ion channel expression through the regulation of subunit trafficking activity.
Keywords: Adhesion, Leukemia, Membrane Trafficking, Myeloid Cell, Potassium Channels, hERG, Sigma Receptors
Introduction
Sig1R (Sigma-1receptor) is a 25-kDa protein anchored to ER, mitochondria, nucleus, and plasma membranes (1–3). The structure is unrelated to other mammalian proteins (4) and presents two putative transmembrane segments (5). Sig1Rs are discretely distributed in the brain and peripheral tissues such as liver, kidney, heart, ovaries, and testis (2). Interestingly, Sig1Rs are overexpressed in numerous cancer cell types and interact with cell cycle and apoptosis pathways (6–8). The protein binds a large panel of exogenous compounds such as antipsychotics, opioids, and psychostimulants (2). It also interacts with endogenous steroids (9) and endogenous hallucinogenic tryptamines (10). However, in the absence of any characterized high affinity endogenous ligand, the classification of this protein as a classical receptor remains controversial. Sig1Rs participate in nociception, cardiac activity, memory, drug addiction, apoptosis, cell cycle, and immune response, but the primary molecular mechanism governed by Sig1Rs remains elusive (2). A recent breakthrough was introduced by Hayashi and Su (11), demonstrating that Sig1Rs physically associate with the chaperone Bip at the mitochondria-associated ER4 membrane where they regulate calcium fluxes through inositol 1,4,5-triphosphate receptors. The emerging concept of the Sig1R as an interorganelle signaling modulator, activated either by ligands or cell stress, was then proposed (1). On the other hand, exogenous sigma ligands inhibit ion channels from different molecular families including voltage-dependent K+ channels (Kv), voltage-dependent Ca2+ channels, voltage-dependent Na+ channels, volume-regulated chloride channels, NMDA, or acid-sensing ion channels (1, 5, 7, 10, 12–15). Thus, Sig1Rs could exert its physiological role through the regulation of ion channels. Nevertheless, the constitutive activity of Sig1Rs on ion channels remains to be thoroughly addressed.
The involvement of abnormally expressed channels in the multiple facets of cancer development has been recently demonstrated (16, 17). We showed in this context that Sig1Rs modulate cell apoptosis resistance and cell cycle through the regulation of Kv1.3 and volume-regulated Cl− channels in lung cancer and leukemia cells (7, 8). hERG encodes a voltage-dependent K+ channel that regulates cardiac repolarization (18, 19). In a series of recent studies, the team of Arcangeli (20, 21) has proposed hERG as a biological marker of leukemia and several solid tumors. hERG forms membrane multi-protein signaling complexes with ECM receptors (integrins) and growth factor receptors (VEGF) to control adhesion, migration, differentiation, invasive process, and chemotherapy resistance of cancer cells. We investigate in the present study the putative links between hERG and Sig1R in a chronic myeloid cell line (K562), HEK 293 cells, and Xenopus oocytes. Using both electrophysiological and biochemical approaches, we demonstrate that the expression of Sig1R increases hERG current density through a regulation of channel subunit maturation and stability.
MATERIALS AND METHODS
The K562 cell line was obtained from Dr. S. Brown (Cambridge, UK) and cultured in RPMI 1640 medium supplemented with 5% FBS, 1 mm sodium pyruvate, 2 mm l-glutamine, 50 units/ml penicillin, and 50 μg/ml streptomycin. HEK 293 cells were cultured in DMEM supplemented with 10% FBS, 1 mm sodium pyruvate, 2 mm l-glutamine, 50 units/ml penicillin, and 50 μg/ml streptomycin.
Chemicals and Reagents
Unless otherwise stated, all cell culture media, supplements, and antibiotics were purchased from Invitrogen. All other chemicals are from Sigma. Igmesine is a kind gift of Dr. F. Roman (Pfizer, Fresnes, France).
Animals
Female Xenopus laevis were anesthetized in 0.2% MS222 (tricaine methanesulfonate), according to the procedure recommended by our ethics committee. The surgery consisted in the removal of roughly five ovarian lobes containing oocytes. Following the surgery, the animals were kept in cold tap water to recover from anesthesia, monitored for 3 h, and finally replaced in their aquarium.
Preparation of cRNA
cRNAs were prepared from hERG1 cDNA (kind gift of Dr. G. Robertson, Wisconsin University) or Sig1R cDNA, using a T7 or SP6 transcription kit (Ambion, Huntingdon, UK). cRNA concentration and integrity were estimated from a formamide/formaldehyde agarose gel in MOPS buffer.
Patch Clamp Experiments
K562 cells were prepared as described previously (7). The external solution was 45 mm KCl, 90 mm NaCl, 2 mm MgCl2, 1 mm CaCl2, and 10 mm Hepes (pH adjusted to 7.4 with HCl, 285 mosm/liter). Soft glass patch electrodes (Brand, Wertheim, Germany) were made on a horizontal pipette puller (P-97; Sutter Instrument Co., Novato, CA) to achieve a final resistance ranging from 3 to 5 mΩ. The internal solution was 130 mm potassium aspartate, 2 mm MgCl2, 1 mm CaCl2, 10 mm EGTA, 10 mm Hepes, 2 mm ATP, and 100 μm GTP (pH adjusted to 7.2 with KOH, 290 mosm/liter). Electrical signals were amplified with an Axopatch 200B amplifier (Molecular Device, Foster City, CA) and acquired with a DIGIDATA 1440 interface and pCLAMP 10.2 software (Axon Instruments). K+ currents were recorded at a 10-kHz sampling frequency and filtered at 2 kHz. Sigma ligands were added to external solutions that were administered in the vicinity of the cell under study through the use of a gravity-feed system (rate, 2 ml/min).
Double Electrode Voltage Clamp Experiments
Xenopus oocytes were maintained in modified Barth's saline medium adjusted to [K+] = 90 mm (substituted for Na+ to magnify K+ driving force at −120 mV) for activation and in normal modified Barth's saline medium ([K+] = 1 mm) for inactivation experiments. In the latter case, a three-pulse protocol was used: after a 1-s depolarizing step to 40 mV to fully activate hERG, 10-ms conditioning prepulses from 40 to −140 mV were applied, before repolarizing to +50 mV, where tail current values were recorded.
Adhesion Experiments
Fibronectin (FN) (Roche Applied Science) diluted to 40 μg/ml was coated overnight at room temperature onto a Nunc MaxiSorp flat-bottomed Immunoplate (Thermo Fisher, Langenselbold, Germany). The surface was then blocked with 40 mg/ml BSA for 1 h at room temperature. The plate was washed once with RPMI (without FBS). K562 cells were harvested and resuspended at 106 cells/ml in RPMI (without FBS). They were incubated for 1 h in the presence or absence of 10 μm E-4031 (a potent HERG channel blocker, Enzo Life Sciences, Lausen, Switzerland) or 10 μm igmesine. 105 cells were then plated in each well, and the plate was incubated for 2 h at 37 °C, 5% CO2. Medium and unbound cells were removed by three PBS+ washes. 0.2% (w/v) crystal violet, 20% (v/v) methanol solution was added and incubated for 20 min. The plate was washed thrice by immersion in water and dried before the addition of lysis buffer (75 mm NaCl, 25 mm Tris, pH 7.4, 10% SDS). Lysis was conducted under intermittent agitation for 20 min before reading absorbance at 540 nm on an iEMS plate reader (Labsystems, Helsinki, Finland).
Western Blot Experiments
Oocytes were homogenized in 20 μl/oocyte of 250 mm sucrose, 20 mm Tris-HCl, pH 7.4, containing 0.5 mm Pefabloc® (Roche Applied Science) and kept on ice at all times. The homogenate was prepared as described previously (22). The proteins were blotted onto PVDF (Millipore, Molsheim, France) membranes, using a Biometra semi-dry transfer system (Whatman, Göttingen, Germany). hERG was detected in the samples using a 1:2500 dilution of a polyclonal anti-hERG1(CT) pan antibody (Alexis, San Diego, CA) and a peroxidase coupled-anti-rabbit IgG secondary antibody (1:80000 dilution). Sig1R was detected using a 1:1000 dilution of a polyclonal Sigma receptor antibody from Santa Cruz Biotechnology (Heidelberg, Germany) and anti-goat IgG secondary antibody (1:40000). Both signals were visualized using ECL reagent (Pierce) on a Fusion FX-7 image acquisition system (Vilber Lourmat, Torcy, France). For K562 cells, total membrane proteins were isolated as follows: the cells were briefly washed with cold PBS, and then cell lysis was done in TE buffer (10 mm Tris, 1 mm EDTA, pH 7.4) containing 0.5 mm Pefabloc for 15 min on ice. The homogenate was centrifuged successively at 350 × g, 1,400 × g, and 3,000 × g for 10 min at 4 °C, and the pellet was discarded after each centrifugation. The final supernatant was ultracentrifuged at 255,000 × g for 2 h at 4 °C. The protein pellet was solubilized in a suitable volume of lysis buffer, and the proteins were assayed as previously mentioned. Sig1R was detected as described above. For control of loading, we used either anti-actin (1:2000), anti-α-tubulin (1:50,000), or anti-calnexin (1/1000) antibodies (Sigma). Densitometric analysis of the data were performed with Image J analysis software (National Institutes of Health), and the results were corrected for protein loading by normalization for α-tubulin, actin, or calnexin expression.
shRNA Transduction
Lentiviral particles were obtained from Sigma (MISSION® shRNA lentiviral transduction particles). On day 1, K562 cells were plated in a 6-well plate at a density of 20,000 cells/well in complete medium. On day 2, medium was removed, and cells were incubated in complete medium containing 8 μg/ml of hexadimethrin bromide (Aldrich) and transduced at a multiplicity of infection of 5. Clones SHC002V (nontarget shRNA) and SHVRC-TRCN0000061011 (shSig1R targeted) were used for transduction. On day 3, a new transduction round was applied. On day 6, puromycin (0.5 mg/ml) was added in fresh medium to start selection of transduced cells.
Co-immunoprecipitation
On day 1, cell culture dishes were placed on ice and washed three times with ice-cold PBS+ (PBS with 0.5 mm Ca2+). Then ice-cold lysis buffer (150 mm NaCl, 0.5 mm EDTA, 50 mm Tris-HCl, pH 8.00, 1% dodecyl-β-d-maltoside) was added (1 ml for two 100-mm dishes). For experiments using Sig1R ligand, the cells were incubated for 30 min with 10–5 m igmesine prior to lysis. The cells were scraped off, and tubes were placed for 2 h on a low speed rotator at 4 °C. In the meantime, anti-mouse IgG (whole molecule)-agarose beads were incubated in PBS (2% BSA). Then cell lysate was cleared by 10 min of centrifugation at 13,000 rpm. Supernatant was placed in a fresh tube kept on ice, and total protein concentration was determined. 6 mg were used for each sample and incubated in 2 ml (final volume) of 150 mm NaCl, 0.5 mm EDTA, 50 mm Tris-HCl, pH 8.00, and 0.5% dodecyl-β-d-maltoside. Anti-Myc tag IgG (1/500) (Euromedex, France) was added to the suspension and incubated at 4 °C for 1 h on the low speed rotator. 20 μl of saturated beads were then added to each 2-ml sample and agitated by rotation at 4 °C overnight. Next day, the tubes were centrifuged, and the supernatants were removed. The beads were washed in PBS with 0.5% DMM for 5 min and then washed four times for 5 min in PBS with 0.1% DMM. Finally, the beads were resuspended in 20 μl of 2× loading buffer. The samples were heated at 95 °C for 3 min and were run on a 7–12% SDS-PAGE. hERG was probed with 1:2500 dilution of a polyclonal anti-hERG1(CT) pan antibody as described above. Sig1R was probed with 1:1000 dilution of a rabbit anti-Sig1R polyclonal antibody (Santa Cruz, Heidelberg, Germany) and detected with an anti-rabbit IgG-peroxidase (1:80000 dilution). The signal was visualized as described above.
Flow Cytometry
K562 cells were incubated for 20 min at 4 °C in a solution of PBS, 3% FBS, 2 mm EDTA containing an rabbit antibody directed against an external loop of hERG (Alomone Labs, Jerusalem, Israel). After washing in PBS, 3% FBS, 2 mm EDTA, the cells were stained for 20 min at 4 °C with AlexaFluor 488 conjugated anti-rabbit IgG (Santa Cruz Biotechnologies). The cells were analyzed with a FASCalibur flow cytometer and Cell Quest pro software (Becton Dickinson, Bedford, MA). The use of an appropriate isotypic control allowed eliminating nonspecific signal in the population.
hERG-transduced HEK 293
The coding sequence of hERG1 was subcloned into the mammalian expression vector pPRIhygro to generate pPRIhygro-hERG1. pPRIhygro is derived from pPRIpu (23), where puromycin resistance gene was exchanged for hygromycin resistance gene (sequences of pPRIpu and pPRIhygro are available on request). Highly pure recombinant plasmids were obtained by anion exchange chromatography (NucleobondAx, Macherey-Nagel, Dären, Germany) and were used to stably transduce HEK 293 cells. For transduction experiments, HEK293 cells were seeded at 30–40% density in 100-mm dishes in DMEM supplemented with 10% FCS. To generate retroviruses, 293T cells were transfected the following day with 10 μg of an empty pPRIhygro plasmid or the pPRIhygro-hERG1 construct and with 5 μg of pCMV-VSVG and 5 μg of pCMV-gag-pol plasmids using the classic calcium phosphate transfection technique. 6 h after transfection, the cells were washed, and fresh medium was added. Replication-defective retroviruses were recovered in the culture medium between 24 and 72 h post-transfection. These retroviral supernatants were filtered through sterile 0.45-μm filters and then added directly to HEK 293 cells in the presence of 4 μg/ml hexadimethrin bromide to enhance retroviral transduction efficiency. On day 6, hygromycin (100 ng/ml) was added in fresh medium to start selection of transduced cells. Western analyses were performed to check correct expression of hERG1.
HERG/cmycSig1R-transfected HEK 293
Similarly, cmyc-Sig1R cDNA (c-Myc tag added in phase at the N-terminal part of the protein) (7) was subcloned in pPRIPu vector. 10 μg of pPRIPu-cmyc-Sig1R cDNA was transfected in HEK stably expressing hERG1 using the classic calcium phosphate transfection technique. As control, a cmyc-eGFP cDNA sequence was subcloned in pPRIPu vector and transfected in the same way (all of the cDNA sequences are available on request). Obtention of stable HEK cells expressing hERG/cmycSig1R and hERG/cmyc-eGFP was achieved by puromycin selection.
Real Time PCR
Measurements were performed with a Light Cycler 1.5 (Roche Applied Science) using SYBR green I dye detection according to the manufacturer's recommendations. cDNA, synthesized from 4 μg of total RNA using random primers and Superscript III (Invitrogen), was added to a reaction mixture (Faststart DNA SYBR green I; Roche Applied Science) with appropriate primers at 0.5 μm each. The relative mRNA abundance was calculated using a standard curve method. Expression levels were normalized to the levels of the constitutively expressed 36B4 ribosomal protein mRNA. Oligonucleotides used for PCR were: hERG1up, 5′-TGAGGGCATTAGCTGGTCTAACT-3′; hERG1dw, 5′-GCAGTAAATAGCAGAAAAGTCCTTGA-3′; h36B4up, 5′-AATCCCTGACGCACCGCCGTGATG-3′; and h36B4dw, 5′-TGGGTTGTTTTCCAGGTGCCCTCG-3′.
Pulse-Chase Experiments
To analyze hERG maturation, HEK293 cells expressing GFP or sigma1R and hERG grown in 6-cm diameter dishes were incubated for 1 h in methionine/cysteine-free DMEM, then incubated for 10 min in the presence of 100 μCi [35S]methionine (PerkinElmer Life Sciences, Waltham, MA), and chased for increasing amounts of time (up to 3 h) in the presence of DMEM containing 10% FBS as described previously (24). The cells were lysed in radioimmune precipitation assay buffer containing protease inhibitors and the resulting protein G-Sepharose clarified lysate immunoprecipitated with anti-hERG antibodies (Enzo Life Sciences, mAb A12) for 3 h on ice. Protein G-Sepharose beads were then added for 45 min. After five washes, immunoprecipitated material was resolved by SDS-PAGE. Immunoprecipitated hERG was then visualized by fluorography on x-ray films. To monitor hERG stability, a similar experimental approach was undertaken except that the pulse with [35S]methionine was for 1 h, and the chase lasted up to 8 h. Biological replicates were performed in duplicate, and technical replicates were in triplicate. Quantitation was performed by scanning densitometry.
RESULTS
Sigma Ligands Inhibit hERG Current and Adhesion to FN in K562 Cells
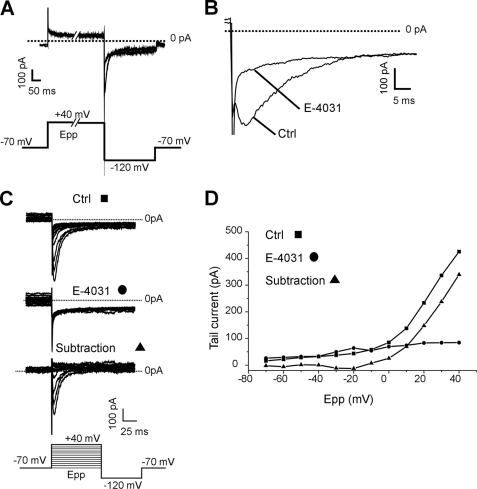
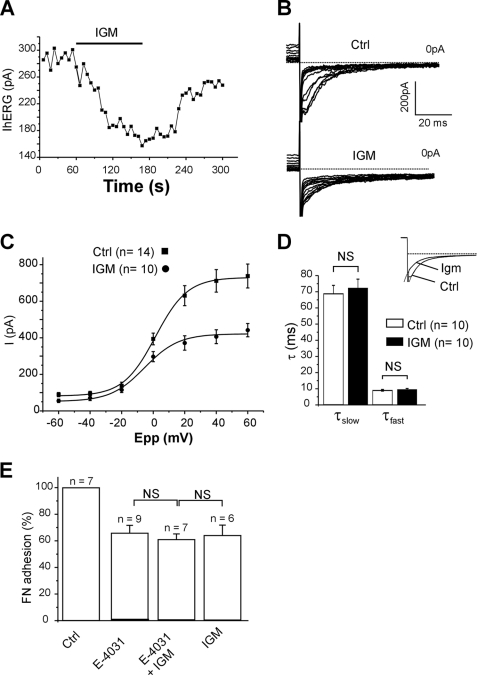
Patch clamp experiments were performed in the whole cell variant of the whole cell technique. In high extracellular K+ concentration, fast repolarizations at −120 mV following a 2-s prepulse at 40 mV gave rise to rapidly inactivating inward tail currents, fully abrogated by perfusion of the specific hERG inhibitor E-4031 (1 μm) (25) (Fig. 1, A and B). Amplitude of tail currents was dependent on prepulse amplitude, and graphical subtraction revealed that E-4031 inhibited a voltage-dependent conductance with an activation threshold of approximately −20 mV (Fig. 1, C and D). Altogether these data confirmed the presence of functional hERG channels in our K562 cell line (26). To verify a potential interaction between the Sig1R and hERG, we studied the effects of sigma-1 selective ligands, i.e. igmesine and (+)pentazocine (4, 7, 14, 27) on tail currents. Extracellular applications of igmesine or (+)pentazocine reversibly depressed the tail currents recorded at −120 mV and following an activating prepulse at 40 mV. The current was reduced by 40.85 ± 2.83% (n = 10) and 21.19 ± 1.77% (n = 3) for igmesine and (+)pentazocine, respectively (10 μm each). The maximal inhibition occurred within 3 min following the onset of drug applications (Fig. 2A). The effect of igmesine (10 μm) was next studied on hERG I/V plots recorded from −70 to 40 mV. Igmesine (10 μm) produced a dramatic reduction of the maximal current amplitude for potentials ranging from −10 to 40 mV (Fig. 2, B and C), suggesting that the drug mainly affected current density. Fitting steady-state activation plots using a Boltzmann function revealed a nonsignificant 4-mV leftward shift in voltage dependence (−0.47 ± 0.89 mV (n = 14) and −4.4 ± 1.76 mV (n = 10)) in the absence or presence of igmesine, respectively, NS, Mann-Whitney). Igmesine changed neither the fast nor the slow deactivating components of hERG tail current recorded at −120 mV (Fig. 2D). Taken together, these results suggest that Sig1R is functionally linked to hERG channels. We then explored the role of this link on the integrin-dependent cell adhesion to ECM in vitro. K562 cells express a single subtype of FN-selective integrin, i.e. the α5β1 (28). Both E-4031 and igmesine inhibited specific FN-dependent cell adhesion (Fig. 2E). Interestingly, the effects of igmesine on FN adhesion were not additive with those produced by E-4031 (Fig. 2E), strongly suggesting that Sig1Rs modulate, at least in part, FN adhesion through the control of hERG.
FIGURE 1.
Characterization of hERG currents in K562 cells. A, superimposed tail currents recorded at −120 mV following a 2-s prepulse at +40 mV (Epp) to fully activate hERG channels, in the absence (control) or the presence of E4031 (1 μm). B, detail of the tail currents presented in A. C, families of tail currents recorded following prepulses from −70 to 40 mV in the absence (control) or the presence of E-4031 (1 μm). The bottom current traces represent the graphic subtraction. D, corresponding I/V plots. Tail current amplitudes are plotted against prepulse potential (Epp). Ctrl, control.
FIGURE 2.
Sigma ligands depress hERG current and cell adhesion to FN in K562 cells. A, time course of hERG current recorded at −120 mV after a 40-mV prepulse in a K562 cell. Igmesine (IGM, left panel, 10 μm) was applied to the external side of the cells during the lap of time represented by the black bar. B, families of hERG currents recorded at −120 mV following prepulses from −70 to 40 mV in a single cell in the absence (upper panel, control) or the presence of igmesine (10 μm, lower panel, IGM). C, corresponding I/V plots. Tail current amplitudes are represented as a function of prepulse potential (Epp) and fitted with a Boltzmann function. D, histogram showing slow and fast deactivating constants at −120 mV in the absence (control) or the presence of igmesine (IGM, 10 μm). Deactivation was fitted with a double-exponential function. The inset shows representative superimposed tail currents recorded before and during igmesine (Igm) application. The values are the means ± S.E. NS, not significant (Student's t test). E, histogram representing the percentage of K562 cells adhering to FN, in control conditions (100%), in the presence of E-4031 (10 μm), E4031 + igmesine (10 μm each), or igmesine alone (10 μm). The values are the means ± S.E. of six to nine independent experiments. NS, not significant (Student's t test). Ctrl, control.
Sig1R Expression Potentiates hERG Current Density in Xenopus Oocytes
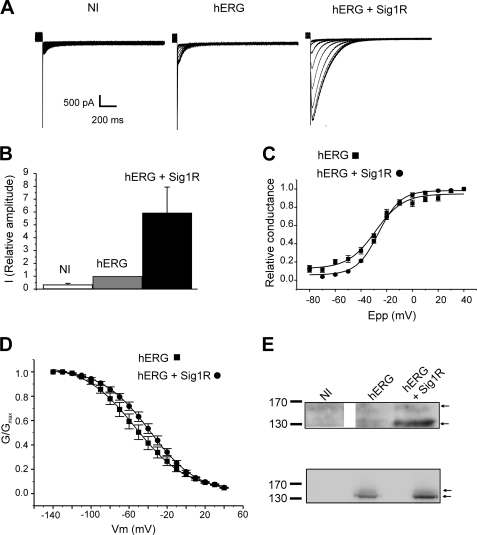
To address the function of Sig1R on hERG in the absence of any exogenous ligand, we next studied the effects of Sig1R expression on hERG currents expressed in Xenopus oocytes. Injection of hERG cRNA gave rise to voltage-dependent tail currents that were absent in water-injected oocytes (Fig. 3A, left and middle panels). Co-injection of Sig1R cRNA with hERG cRNA resulted in a 5-fold increase in current amplitude (Fig. 3, A and B). Sig1R expression did not significantly affect voltage-dependent activation (Fig. 3C and Table 1) or inactivation parameters (Fig. 3D and Table 1), indicating that Sig1Rs mainly modulate hERG current density. To further explore this hypothesis, Western blot analysis from control and injected oocytes were performed using the same cRNA concentration as those used for voltage-clamp experiments shown in Fig. 3A. hERG cRNA injection gave rise to a merely detectable protein expression (Fig. 3E). By contrast, when hERG and Sig1R cRNAs were co-injected, both mature (155 kDa) and immature (135 kDa) hERG glycoforms were clearly resolved (Fig. 3E) (Ficker et al., 2003), correlating the current densities detected in the same conditions (Fig. 3A). The same potentiating effect was observed when hERG and Sig1R cRNA were injected at the same concentrations (Fig. 3E, lower panel).
FIGURE 3.
Sig1R expression stimulates hERG currents in Xenopus oocytes. A, families of tail currents recorded in noninjected (NI), hERG cRNA-injected (hERG; 25 pg/oocyte), and hERG + Sig1R cRNA-injected (hERG + Sig1R; 25 pg and 5 ng/oocyte, respectively) oocytes (representative experiment). The voltage protocol is given in the description of Fig. 1C. B, relative current amplitudes at −120 mV in noninjected, hERG cRNA-injected, and hERG + Sig1R cRNA-injected oocytes (arbitrary units, 1 corresponding to the mean current recorded in hERG cRNA injected oocytes). The values are the means ± S.E. of six independent experiments. C, voltage-dependent activation curves in hERG cRNA-injected (black squares) and hERG + Sig1R-cRNA-injected oocytes (black circles). The plots were fitted with a Boltzmann function. The values are the means ± S.E. of six independent experiments. D, voltage-dependent inactivation curves in hERG cRNA-injected (black squares) and hERG + Sig1R cRNA-injected oocytes (black circles). For the detailed voltage protocols, see “Materials and Methods.” The values are the means ± S.E. of n = 8 independent experiments. E, upper panel, Western blots probed with a hERG antibody in noninjected (NI), hERG cRNA-injected (hERG; 25 pg/oocyte), and hERG + Sig1R cRNA-injected (hERG + Sig1R; 25 pg and 5 ng/oocyte, respectively) oocytes. Lower panel, Western blot of the same experiment using an identical concentration for hERG and Sig1R cRNA (15 ng/oocyte). Each Western blot is representative of three independent experiments.
TABLE 1.
hERG voltage-dependent activation and inactivation parameters in Xenopus oocytes in the absence or presence of Sig1R
Steady-state activation and inactivation were fitted with the Clampfit software using the following Boltzmann function: G/Gmax = 1/(1 + e((V½ − V)/k)). NS, not significant.
| Activation |
Inactivation |
|||
|---|---|---|---|---|
| V½ | Slope (k) | V½ | Slope (k) | |
| hERG (n = 6) | −28.7 ± 0.3 mV | 10.2 ± 2.7 | −67.8 ± 9.1 mV | −22.2 ± 0.1 |
| hERG + Sig1R (n = 8) | −26.8 ± 1.0 mV | 9.2 ± 0.44 | −64.7 ± 6.6 mV | −17.9 ± 0.8 |
| Mann-Whitney | NS | NS | NS | NS |
Sig1R Silencing Modulates hERG Expression and Cell Adhesion to FN in K562 Cells
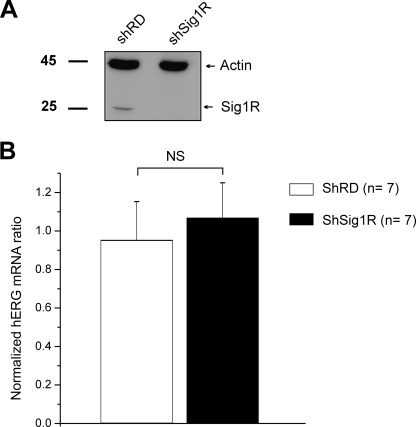
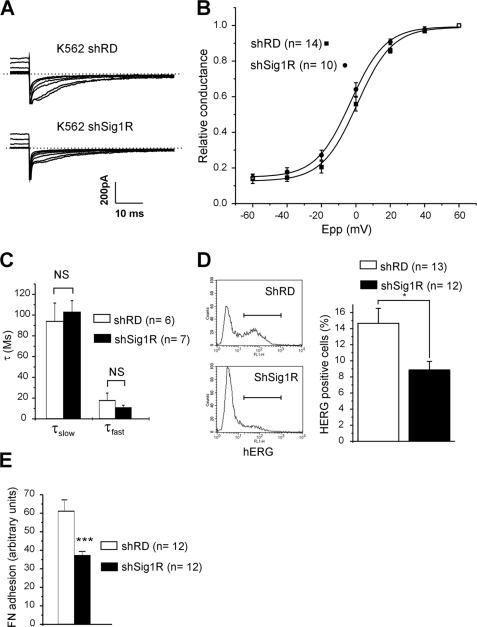
We further explored the function of Sig1R in K562 cells using a shRNA silencing strategy. K562 cells were transduced with lentivirus containing either a random shRNA or a Sig1R-directed shRNA, yielding two cell populations named, respectively, shRD and shSig1R. Western blot experiments revealed a dramatic decrease in Sig1R expression in the shSig1R cell line (Fig. 4A). Quantitative PCR experiments revealed that Sig1R repression had no effect on hERG mRNA contents (Fig. 4B), ruling out any regulation of hERG transcription by Sig1R expression. Patch clamp experiments showed that the mean tail current amplitude recorded in shSig1R cells was clearly reduced when compared with shRD cells (Fig. 5A). At +40 mV, current density was 29.5 ± 1.7 pA/pF in shRD cells (n = 40) and 18.1 ± 1.4 pA/pF in shSig1R cells (n = 45, p < 0.0001, Mann-Whitney). Interestingly, Sig1R extinction neither significantly shifted voltage dependence activation (Fig. 5B and Table 2), nor modified deactivation kinetic time constants (Fig. 5C). The resting potential was not modified by Sig1R silencing (−24.5 ± 3.1 mV (n = 13) and −25.5 ± 3.8 mV (n = 12) for shRD and shSig1R K562, respectively, NS, Mann-Whitney), suggesting that hERG does not participate to the resting potential through a tonic window K+ current.
FIGURE 4.
Sig1R silencing does not alter hERG transcription in K562 cells. A, Western blots probed with an anti-Sig1R or actin antibodies in shRD and shSig1R cells. B, hERG mRNA quantitative expression in shRD and shSig1R K562 cells. hERG mRNA levels were normalized to 36B4 ribosomal protein mRNA. NS, not significant (Student's t test).
FIGURE 5.
Sig1R silencing reduces hERG current density in K562 cells. A, families of hERG currents recorded in response to the voltage protocol described in the description of Fig. 1C. These representative traces were obtained from shRD K562 (K562 shRD, upper panel) and shSig1R K562 cells (K562 shSig1R, lower panel). B, mean activation plots obtained from shRD K562 (black squares) and shSig1R K562 cells (black circles). The plots were fitted using a Boltzmann function. C, deactivation rate constant of hERG current at −120 mV in shRD K562 (white bars) and shSig1R K562 cells (black bars). Deactivation was fitted with a double-exponential function. The values are the means ± S.E., Student's t test. D, left panel, surface expression of hERG by flow cytometry in shRD and shSig1R K562 cells (representative experiment). Right panel, corresponding histogram. *, p < 0.02, Mann-Whitney. E, FN adhesion in shRD (white bar) and shSig1R (black bar) K562 cells. The values are the means ± S.E. of 12 independent experiments. ***, p < 0,001 (Student's t test). In a single experiment, each value is the mean of three distinct wells.
TABLE 2.
hERG kinetic properties in shRD and shSig1R K562 cells
Steady-state activation was fitted with the Clampfit software using the following Boltzmann function: G/Gmax = 1/(1 + e((V½ − V)/k)). Deactivation kinetics were fitted with the following double-exponential function: f(t) = A1e−t/τslow + C1 + A2e−t/τfast + C2. NS, not significant.
| V1/2 | Slope (k) | Deactivation | |
|---|---|---|---|
| shRD K562 | −0.5 ± 1.9 mV (n = 15) | 9.7 ± 0.7 (n = 15) | τslow = 94.1 ± 17.5 ms (n = 6) |
| τfast = 17.8 ± 7.0 ms (n = 6) | |||
| shSig1R K562 | −2.1 ± 1.6 mV (n = 10) | 9.7 ± 1.6 (n = 10) | τslow = 103.0 ± 29.1 ms (n = 7) |
| τfast = 10.9 ± 2.2 ms (n = 7) | |||
| Mann-Whitney | NS | NS | NS |
We then explored the plasma membrane expression of the channel by performing extracellular labeling of hERG in nonpermeabilized K562 cells. Flow cytometry analysis demonstrated that hERG membrane expression was significantly reduced in Sig1R-silenced cells (35.5 ± 0.1%; Fig. 5D), demonstrating that Sig1R regulates hERG current density through the number of ion channel at the plasma membrane. As expected, the decrease of hERG current density and membrane expression was accompanied by an inhibition of K562 cell adhesion to FN in shSig1R compared with the shRD population (≈40%; Fig. 5E).
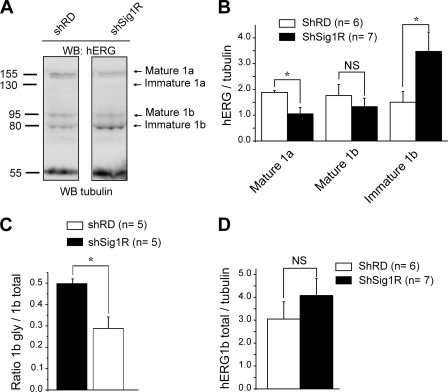
At the protein level, Western blots using a pan-hERG antibody revealed that K562 cells express two different isoforms of hERG, i.e. hERG1a, corresponding to the full-length protein, and a N-terminal-truncated splicing variant hERG1b (29–31). We mainly detected the mature hERG1a isoform, a 155-kDa band representing the fully glycosylated form (mature hERG 1a), whereas the 135-kDa core-glycosylated immature form of hERG1a was seldom detected (Fig. 6A, left panel). By contrast, the hERG1b spliced isoform constantly appeared as two distinct bands, the 95-kDa fully glycosylated mature and the 80-kDa core-glycosylated immature forms (Fig. 6A, left panel). Both hERG mature glycoforms are known to represent the fraction of core-glycosylated channel subunit that has exited the ER to further process through the Golgi and reach the plasma membrane (32, 33). Sig1R silencing strikingly altered the hERG expression pattern. We observed in shSig1R cells a significant decrease in both hERG1a and 1b mature forms. In the same time, SigR silencing induced a dramatic increase in the hERG1b immature form (Figs. 6A, left panel, and 7B). Further analysis indicated that Sig1R silencing significantly reduced the apparent hERG1b maturation, which was quantified as the ratio hERG1bmature/hERG1btotal (33) (Fig. 6C), without significantly modifying the total amount of hERG (Fig. 6D).
FIGURE 6.
Sig1R silencing alters hERG protein expression in K562 and cells. A, Western blots (WB) probed with the anti-pan hERG or anti-tubulin antibodies, performed in shRD and shSig1R K562 cells. B, histogram of the densitometric analysis of mature and immature hERG isoforms in shRD (white bars) and shSig1R K562 cells (black bars). The values correspond to the densitometric ratio hERG/tubulin. C, trafficking efficiency of hERG1b in shRD and shSig1R (black bar) in K562 cells. Trafficking efficiency is calculated as the densitometric ratio (mature)/(mature + immature). D, histogram showing the total amount of hERG1b in shRD and Sig1R K562 cells. *, p < 0,05 (Mann-Whitney). NS, not significant.
FIGURE 7.
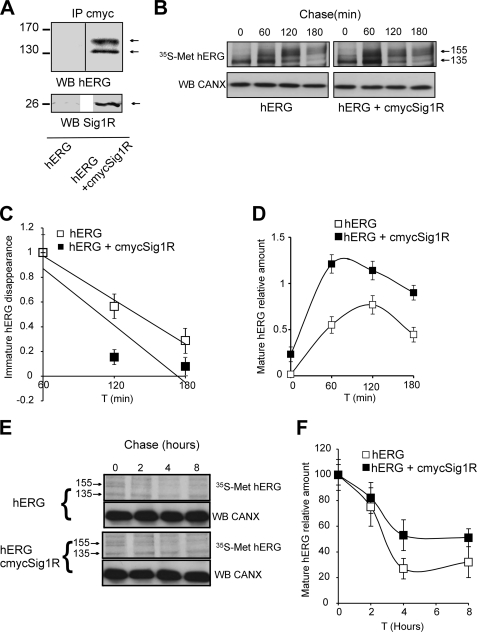
Sig1Rs enhance hERG subunit maturation and stability in HEK cells through a direct interaction. A, immunoprecipitation (IP) of HEK 293 cell lysate proteins with an anti-c-Myc antibody performed in cells transduced with hERG + cmyc-GFP as control (HEK + hERG) or hERG + cmycSig1R (hERG + cmycSig1R) and probed with anti-hERG (upper panel) or anti-Sig1R (lower panel) antibodies. B, 10-min [35S]methionine pulse followed by up to 3-h chase experiments to analyze hERG maturation in the same cells as in A. Radiolabeled hERG was immunoprecipitated and visualized by fluorography following separation by SDS-PAGE. C, quantitation of immature hERG disappearance based on the experiments presented in B. The disappearance rate was determined between 60 and 180 min of chase. Quantitation was performed on nine experimental points obtained in two independent experiments and represented as the means ± S.E. D, quantitation of mature hERG appearance based on the experiments presented in B. Quantitation was performed on nine experimental points obtained in two independent experiments and represented as the means ± S.D. E, 1-h [35S]methionine pulse followed by up to 8-h chase experiments to analyze hERG stability. Radiolabeled hERG was immunoprecipitated and visualized by fluorography following separation by SDS-PAGE. F, quantitation was performed on nine experimental points obtained in two independent experiments and represented as the means ± S.E. based on the experiments presented in E.
The apparent effect of Sig1R on channel maturation was further explored in HEK cells transduced with either hERG + cmycGFP (control experiments) or hERG + cmycSig1R. In a first set of experiments, we successfully co-immunoprecipitated cmycSig1R with hERG, demonstrating a molecular interaction between the two proteins. Interestingly, Sig1R was associated with both mature and immature forms (Fig. 7A). We next studied hERG time course maturation processing by performing pulse-chase experiments. This revealed that maturation of hERG was increased in Sig1R-overexpressing cells (Fig. 7B). Quantitation revealed that newly synthesized immature hERG disappeared faster in Sig1R cells than in control cells (Fig. 7C), thus correlating with the appearance of the mature forms (Fig. 7D). We then analyzed the stability of the mature hERG in both Sig1R expressing and control cells. This revealed that mature hERG, most likely localized at the plasma membrane, was more stable in Sig1R expressing cells than in control cells (Fig. 7E). This apparent stabilization therefore correlated with enhanced biogenesis and current density potentiation in K562 and Xenopus oocytes.
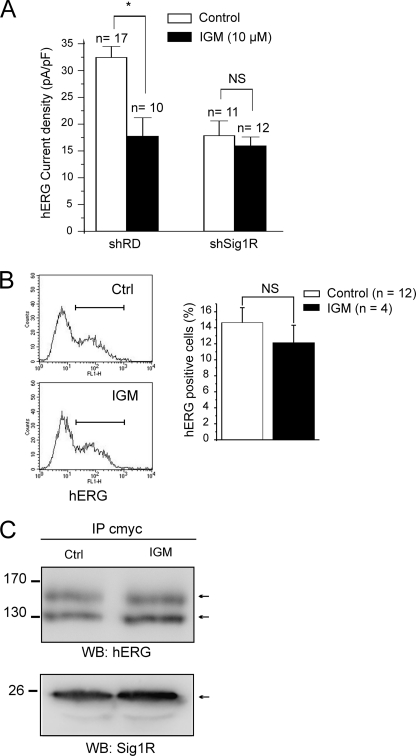
Finally, we addressed the question of the mechanism involved in the sigma ligand-induced inhibition of hERG current. Igmesine (30 min of incubation, 10 μm) significantly reduced hERG current density in shRD K562 cells but produced no effects in shSig1R cells (Fig. 8A), demonstrating a specific effect through Sig1R. However, the same igmesine incubation protocol did not inhibit hERG membrane expression measured by flow cytometry in K562 (Fig. 8B), suggesting that the current inhibition produced by sigma ligands is not the result of an altered membrane channel stability. Using HEK 293 cells stably expressing both hERG and Sig1R, co-IP experiments revealed that igmesine incubation did not reduce the association between hERG and Sig1R (Fig. 8C).
FIGURE 8.
Sigma ligands inhibit hERG current without decreasing hERG membrane expression or altering Sig1R-hERG association. A, effect of cell incubation with igmesine (IGM, 10 μm, 30 min) on hERG current density in shRD and shSig1R K562 cells. *, p < 0.002, Mann-Whitney. B, left panel, effect of with igmesine (IGM, 10 μm, 30 min) on hERG surface expression by flow cytometry in shRD K562 cells. Right panel, corresponding histogram. NS, not significant, Mann-Whitney. C, effect of HEK 293 cell incubation with igmesine (IGM, 10 μm, 30 min) on Sig1R-hERG association. Immunoprecipitation (IP) with an anti-c-Myc antibody of HEK 293 cell lysate proteins. Western blots (WB) were probed with anti-hERG (upper panel) or anti-Sig1R (lower panel) antibodies. The cells were transduced with hERG + cmycSig1R. Representative of three independent experiments. Ctrl, control.
DISCUSSION
The function of Sig1R is poorly understood, and its main endogenous ligand remains unknown. However, the acute effects of sigma ligands on K+, Ca2+, Na+, and Cl− currents suggest a functional link between Sig1Rs and ion channels from different molecular families, a hypothesis supported by several studies showing that Sig1Rs physically bind ion channel subunits (1, 5, 34, 35). We have previously demonstrated that sigma ligands block leukemia and lung cancer cell cycle in the G1 phase through the inhibition of Kv1.3 and volume-regulated Cl− channels (7, 8). In the present study, we have explored the putative link between hERG and Sig1Rs. hERG is a K+ channel involved in cardiac repolarization that is also abnormally expressed in several cancer cells including leukemia. Physically associated with integrins and VEGF receptors, hERG enhances tumor cell progression through the regulation of cell/ECM interaction (20, 21).
Although the ability of sigma ligands to inhibit ion currents is well known, the primary activity of Sig1Rs at the level of ion channels is not understood. To address this question, we performed heterologous co-expression of hERG and Sig1Rs in Xenopus oocytes and observed that Sig1Rs strongly increased current density without altering voltage-dependent activation and inactivation parameters. Accordingly, silencing Sig1Rs in K562 cells resulted in a reduction in current density, whereas other parameters (i.e. open probability and deactivation rate) remained unchanged. Hence, these results demonstrate for the first time that Sig1R expression stimulates ion currents. Our results, however, contrast with a previous report showing that Sig1R accelerated the inactivation rate of Kv1.4 in Xenopus oocyte (5), indicating that interacting modalities may depend on the channel type.
hERG current is mainly regulated by modifications of voltage-dependent and kinetic parameters but also results from the biogenesis/degradation ratio of subunits forming functional membrane channels (32, 36). We show herein that Sig1R expression enhanced hERG protein level in Xenopus oocytes, suggesting that Sig1R stimulates the current density by increasing the number of channels at the plasma membrane. This hypothesis was confirmed by the silencing of Sig1Rs in K562, inducing a decrease in hERG membrane labeling measured by flow cytometry. Accordingly, the level of the mature isoforms of hERG, i.e. hERG1a (full-length isoform) and hERG1b (N-terminally truncated isoform), which co-assemble in heterotetramers to form functional channels (33), was reduced in shK562 cells. Interestingly, Sig1R silencing did not alter hERG mRNA production in K562 cells, but Sig1R co-immunoprecipitated with hERG in HEK cells transduced with both proteins. In a whole, these data reveal that Sig1Rs control the post-translational biogenesis of hERG. Our attention turned therefore on the mechanism involved in the Sig1R-induced current potentiation. The analysis of hERG expression patterns in K562 cells silenced for Sig1R revealed a reduced maturation of both hERG1a and hERG1b isoforms, very similar to the profile described in hERG-expressing CHO cells and treated with Hsp90 inhibitors (32). This analogy suggests that Sig1Rs potentiate ER/Golgi translocation of channel subunits, leading to an increase in the number of functional channels at the plasma membrane. The function of Sig1R in hERG maturation is consistent with its association with the ER-resident 135-kDa channel immature form and was further confirmed by pulse-chase experiments in HEK cells showing that Sig1R expression accelerated hERG rate of maturation. Sig1R can then be proposed as a candidate for the group of proteins controlling hERG trafficking such as Hsp/c70, Hsp90, or the KCNE1 K+ channel β subunit (32, 37). Consistent with a function in hERG folding and/or maturation, in NG108-15 cells, Sig1Rs are associated at the mitochondria-associated ER membrane to the ER chaperone BiP, belonging to the Hsp70 family (11).
Furthermore, we found that Sig1R also associates with the 155-kDa mature form and potentiates its stability. Because the mature form represents the membrane functional channel (38), it can be proposed that Sig1R stimulates hERG current density by synergetically promoting hERG maturation and lowering channel membrane recycling.
Acute application of sigma ligands induced a reversible inhibition of hERG. The overall effect observed corresponds to a reduction in current density, as shown by the inhibition elicited by igmesine in conditions where the tail current represents the maximal activation state (Po = 1) and inactivation is fully removed. This observation is consistent with previous reports showing that sigma ligands mainly inhibit current density associated with Kv1.3, voltage-dependent Na+ channels, volume-regulated Cl− channels, or acid-sensing ion channels (7, 8, 10, 12, 14, 15). It was thereby tempting to postulate that sigma ligands inhibit hERG currents by disrupting Sig1R from hERG subunits, leading to a decrease in the number of hERG surface expression. However, although igmesine decreased current density in K562 cells in a Sig1R-dependent manner, the ligand failed to significantly reduce hERG membrane labeling using the same sigma ligand incubation protocol. Moreover, the direct association between the two proteins expressed in HEK cells was not reduced by igmesine, as shown by co-immunoprecipitation experiments. It can then be concluded that sigma ligands do not inhibit hERG current by disrupting the Sig1R-channel complex and further plasma membrane expression. Sigma ligands are known to provoke a rapid redistribution of sigma receptors localization (39–41). Interestingly, cell treatment with the Sig1R agonist SKF 10,047 provoked the exclusion of Sig1Rs and associated proteins from lipid rafts, likely through a competition with cholesterol at Sig1R sterol-binding sites (42). Sigma ligands may thus induce the redistribution of the Sig1R-hERG complex within the plasma membrane leading to alteration in channel function without modifying plasma membrane expression (43).
Although the exact mechanism by which sigma ligands regulate hERG current will require further investigation, to our knowledge our data unravel the first mechanical and physiological link between Sig1R and ion channel function. The role of Sig1R on channel maturation and trafficking leads to reconsideration of the intrinsic function of sigma receptors in brain or heart but also in cancer. In many tumors, abnormally expressed hERG subunits associate with β1 integrin and VEGF receptors to form channel signaling macrocomplexes involved in cell proliferation, invasiveness, and chemotherapy resistance (20, 29, 44). Understanding the regulation of hERG in cancer cells thus represents a question of paramount importance. In the human leukemic preosteoclastic cell line FLG 29.3, cell binding to FN transiently hyperpolarized membrane potential through hERG activation to potentiate integrin signaling machinery (45, 46). These results indicate that hERG activation is a necessary signaling step in the adhesion process. We show herein that the reduction of hERG current density by either Sig1R silencing or sigma ligands was accompanied by a reduction of the specific K562-cell FN adhesion. Moreover, igmesine and E-4031 produced no additive effects, demonstrating that Sig1Rs modulate cell/ECM interaction through the regulation of hERG channels. In K562 cells expressing both αvβ3 or the native α5β1, depolarization has been shown to enhance FN adhesion (47). Thus, it could be argued that hERG inhibition by either sigma ligands or Sig1R silencing would depolarize cells, potentiating FN adhesion. Nevertheless, E-4031 (47) and Sig1R silencing (our study) did not modify K562 resting potential, indicating that hERG inhibition cannot lead to FN adhesion through depolarization. It can then be proposed that Sig1R inhibition (by either gene silencing or pharmacological inhibition by ligands) or E-4031 leads to a reduced hERG participation to the β1-integrin signaling complex during the FN adhesion process (45). Interestingly, it has been demonstrated that members of the ether-à-gogo channel family could potentiate cancer cell invasiveness in a K+ flux-independent manner, suggesting a mechanism independent of the regulation of membrane resting potential (48).
Altogether, our results unravel the regulating function of Sig1Rs on hERG expression. Sig1R may thus be considered as a new pharmacological target to reduce the activity membrane signaling channel macrocomplexes involved in cancer progression.
Acknowledgments
We thank Drs. Hélène Guizouarn, Isabelle Mus-Veteau, and Said Bendahhou for fruitful discussions about the manuscript; Dr. Céline Feillet for English correction; and Agnès Loubat for kind help during FACS experiments.
This work was supported by the University of Nice-Sophia Antipolis, the CNRS and the Association Ti'Toine Normandie.
- ER
- endoplasmic reticulum
- Kv
- voltage-dependent K+ channels
- FN
- fibronectin
- ECM
- extracellular matrix
- MOPS
- 4-morpholinopropanesulfonic acid
- Sig1R
- Sigma1 receptor
- hERG
- human Ether-à-gogo-related gene.
REFERENCES
- 1. Su T. P., Hayashi T., Maurice T., Buch S., Ruoho A. E. (2010) Trends Pharmacol. Sci. 31, 557–566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Tsai S. Y., Hayashi T., Mori T., Su T. P. (2009) Cent. Nerv. Syst. Agents Med. Chem. 9, 184–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Mavlyutov T. A., Epstein M. L., Andersen K. A., Ziskind-Conhaim L., Ruoho A. E. (2010) Neuroscience 167, 247–255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hanner M., Moebius F. F., Flandorfer A., Knaus H. G., Striessnig J., Kempner E., Glossmann H. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 8072–8077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Aydar E., Palmer C. P., Klyachko V. A., Jackson M. B. (2002) Neuron 34, 399–410 [DOI] [PubMed] [Google Scholar]
- 6. Spruce B. A., Campbell L. A., McTavish N., Cooper M. A., Appleyard M. V., O'Neill M., Howie J., Samson J., Watt S., Murray K., McLean D., Leslie N. R., Safrany S. T., Ferguson M. J., Peters J. A., Prescott A. R., Box G., Hayes A., Nutley B., Raynaud F., Downes C. P., Lambert J. J., Thompson A. M., Eccles S. (2004) Cancer Res. 64, 4875–4886 [DOI] [PubMed] [Google Scholar]
- 7. Renaudo A., L'Hoste S., Guizouarn H., Borgèse F., Soriani O. (2007) J. Biol. Chem. 282, 2259–2267 [DOI] [PubMed] [Google Scholar]
- 8. Renaudo A., Watry V., Chassot A. A., Ponzio G., Ehrenfeld J., Soriani O. (2004) J. Pharmacol. Exp. Ther. 311, 1105–1114 [DOI] [PubMed] [Google Scholar]
- 9. Monnet F. P., Maurice T. (2006) J. Pharmacol. Sci. 100, 93–118 [DOI] [PubMed] [Google Scholar]
- 10. Fontanilla D., Johannessen M., Hajipour A. R., Cozzi N. V., Jackson M. B., Ruoho A. E. (2009) Science 323, 934–937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hayashi T., Su T. P. (2007) Cell 131, 596–610 [DOI] [PubMed] [Google Scholar]
- 12. Soriani O., Le Foll F., Galas L., Roman F., Vaudry H., Cazin L. (1999) Am. J. Physiol. 277, E73–E80 [DOI] [PubMed] [Google Scholar]
- 13. Soriani O., Foll F. L., Roman F., Monnet F. P., Vaudry H., Cazin L. (1999) J. Pharmacol. Exp. Ther. 289, 321–328 [PubMed] [Google Scholar]
- 14. Soriani O., Vaudry H., Mei Y. A., Roman F., Cazin L. (1998) J. Pharmacol. Exp. Ther. 286, 163–171 [PubMed] [Google Scholar]
- 15. Herrera Y., Katnik C., Rodriguez J. D., Hall A. A., Willing A., Pennypacker K. R., Cuevas J. (2008) J. Pharmacol. Exp. Ther. 327, 491–502 [DOI] [PubMed] [Google Scholar]
- 16. Prevarskaya N., Skryma R., Shuba Y. (2010) Trends Mol. Med. 16, 107–121 [DOI] [PubMed] [Google Scholar]
- 17. Fraser S. P., Pardo L. A. (2008) EMBO Rep. 9, 512–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Trudeau M. C., Warmke J. W., Ganetzky B., Robertson G. A. (1995) Science 269, 92–95 [DOI] [PubMed] [Google Scholar]
- 19. Sanguinetti M. C., Jiang C., Curran M. E., Keating M. T. (1995) Cell 81, 299–307 [DOI] [PubMed] [Google Scholar]
- 20. Pillozzi S., Brizzi M. F., Bernabei P. A., Bartolozzi B., Caporale R., Basile V., Boddi V., Pegoraro L., Becchetti A., Arcangeli A. (2007) Blood 110, 1238–1250 [DOI] [PubMed] [Google Scholar]
- 21. Pillozzi S., Masselli M., De Lorenzo E., Accordi B., Cilia E., Crociani O., Amedei A., Veltroni M., D'Amico M., Basso G., Becchetti A., Campana D., Arcangeli A. (2011) Blood 117, 902–914 [DOI] [PubMed] [Google Scholar]
- 22. Guizouarn H., Martial S., Gabillat N., Borgese F. (2007) Blood 110, 2158–2165 [DOI] [PubMed] [Google Scholar]
- 23. Albagli-Curiel O., Lécluse Y., Pognonec P., Boulukos K. E., Martin P. (2007) BMC. Biotechnol. 7, 85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Cameron P. H., Chevet E., Pluquet O., Thomas D. Y., Bergeron J. J. (2009) J. Biol. Chem. 284, 34570–34579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sanguinetti M. C., Jurkiewicz N. K. (1990) J. Gen. Physiol. 96, 195–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Cavarra M. S., del Mónaco S. M., Assef Y. A., Ibarra C., Kotsias B. A. (2007) J. Membr. Biol. 219, 49–61 [DOI] [PubMed] [Google Scholar]
- 27. Roman F. J., Pascaud X., Martin B., Vauché D., Junien J. L. (1990) J. Pharm. Pharmacol. 42, 439–440 [DOI] [PubMed] [Google Scholar]
- 28. Meng X., Cheng K., Krohkin O., Mould A. P., Humphries M. J., Ens W., Standing K., Wilkins J. A. (2005) J. Cell Sci. 118, 4009–4016 [DOI] [PubMed] [Google Scholar]
- 29. Guasti L., Crociani O., Redaelli E., Pillozzi S., Polvani S., Masselli M., Mello T., Galli A., Amedei A., Wymore R. S., Wanke E., Arcangeli A. (2008) Mol. Cell. Biol. 28, 5043–5060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sale H., Wang J., O'Hara T. J., Tester D. J., Phartiyal P., He J. Q., Rudy Y., Ackerman M. J., Robertson G. A. (2008) Circ. Res. 103, e81–e95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Jones E. M., Roti Roti E. C., Wang J., Delfosse S. A., Robertson G. A. (2004) J. Biol. Chem. 279, 44690–44694 [DOI] [PubMed] [Google Scholar]
- 32. Ficker E., Dennis A. T., Wang L., Brown A. M. (2003) Circ. Res. 92, e87–100 [DOI] [PubMed] [Google Scholar]
- 33. Phartiyal P., Sale H., Jones E. M., Robertson G. A. (2008) J. Biol. Chem. 283, 3702–3707 [DOI] [PubMed] [Google Scholar]
- 34. Le Guennec J. Y., Ouadid-Ahidouch H., Soriani O., Besson P., Ahidouch A., Vandier C. (2007) Recent Pat. Anticancer Drug Discov. 2, 189–202 [DOI] [PubMed] [Google Scholar]
- 35. Carnally S. M., Johannessen M., Henderson R. M., Jackson M. B., Edwardson J. M. (2010) Biophys. J. 98, 1182–1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Guo J., Massaeli H., Xu J., Jia Z., Wigle J. T., Mesaeli N., Zhang S. (2009) J. Clin. Invest. 119, 2745–2757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. McDonald T. V., Yu Z., Ming Z., Palma E., Meyers M. B., Wang K. W., Goldstein S. A., Fishman G. I. (1997) Nature 388, 289–292 [DOI] [PubMed] [Google Scholar]
- 38. Petrecca K., Atanasiu R., Akhavan A., Shrier A. (1999) J. Physiol. 515, 41–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Morin-Surun M. P., Collin T., Denavit-Saubié M., Baulieu E. E., Monnet F. P. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 8196–8199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Hayashi T., Su T. P. (2003) J. Pharmacol. Exp. Ther. 306, 726–733 [DOI] [PubMed] [Google Scholar]
- 41. Mavlyutov T. A., Ruoho A. E. (2007) J. Mol. Signal. 2, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Palmer C. P., Mahen R., Schnell E., Djamgoz M. B., Aydar E. (2007) Cancer Res. 67, 11166–11175 [DOI] [PubMed] [Google Scholar]
- 43. Ganapathi S. B., Fox T. E., Kester M., Elmslie K. S. (2010) Am. J. Physiol. Cell Physiol. 299, C74–C86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Pillozzi S., Arcangeli A. (2010) Adv. Exp. Med. Biol. 674, 55–67 [DOI] [PubMed] [Google Scholar]
- 45. Hofmann G., Bernabei P. A., Crociani O., Cherubini A., Guasti L., Pillozzi S., Lastraioli E., Polvani S., Bartolozzi B., Solazzo V., Gragnani L., Defilippi P., Rosati B., Wanke E., Olivotto M., Arcangeli A. (2001) J. Biol. Chem. 276, 4923–4931 [DOI] [PubMed] [Google Scholar]
- 46. Cherubini A., Hofmann G., Pillozzi S., Guasti L., Crociani O., Cilia E., Di Stefano P., Degani S., Balzi M., Olivotto M., Wanke E., Becchetti A., Defilippi P., Wymore R., Arcangeli A. (2005) Mol. Biol. Cell 16, 2972–2983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Vernon-Wilson E. F., Auradé F., Tian L., Rowe I. C., Shipston M. J., Savill J., Brown S. B. (2007) J. Leukoc. Biol. 82, 1278–1288 [DOI] [PubMed] [Google Scholar]
- 48. Downie B. R., Sánchez A., Knötgen H., Contreras-Jurado C., Gymnopoulos M., Weber C., Stühmer W., Pardo L. A. (2008) J. Biol. Chem. 283, 36234–36240 [DOI] [PMC free article] [PubMed] [Google Scholar]