Abstract
Primary cilia regulate polarized protein trafficking in photoreceptors, which are dynamic and highly compartmentalized sensory neurons of retina. The ciliary protein Cep290 modulates cilia formation and is frequently mutated in syndromic and non-syndromic photoreceptor degeneration. However, the underlying mechanism of associated retinopathy is unclear. Using the Cep290 mutant mouse rd16 (retinal degeneration 16), we show that Cep290-mediated photoreceptor degeneration is associated with aberrant accumulation of its novel interacting partner Rkip (Raf-1 kinase inhibitory protein). This effect is phenocopied by morpholino-mediated depletion of cep290 in zebrafish. We further demonstrate that ectopic accumulation of Rkip leads to defective cilia formation in zebrafish and cultured cells, an effect mediated by its interaction with the ciliary GTPase Rab8A. Our data suggest that Rkip prevents cilia formation and is associated with Cep290-mediated photoreceptor degeneration. Furthermore, our results indicate that preventing accumulation of Rkip could potentially ameliorate such degeneration.
Keywords: Centrosome, Eye, Neurodegeneration, Neurons, Photoreceptors, cep290, Cilia, Ciliopathies, Retina, Retinal Degeneration
Introduction
Primary cilia are microtubule-based organelles that are generated from mother centrioles (also called basal body) by an evolutionarily conserved process called intraflagellar transport (IFT)2 (1, 2). IFT and other ciliary proteins, such as Bardet-Biedl syndrome proteins, retinitis pigmentosa GTPase regulator, and nephronophthisis-associated proteins facilitate cilia formation and maintenance by cooperating with small GTPases such as Rab8A (3–8).
Cilia regulate diverse processes, including visual perception and signal transduction (9–11). Defects in ciliary function are associated with a wide range of disorders, collectively called ciliopathies (12), such as Bardet-Biedl syndrome, Joubert syndrome, and Meckel-Gruber syndrome. These disorders are characterized by multiple clinical features including mental retardation, renal malfunction, and polydactyly. Photoreceptor degeneration, leading to retinitis pigmentosa (RP) and childhood onset Leber congenital amaurosis, is commonly observed in ciliopathies (13). However, the mechanism by which ciliary dysfunction results in photoreceptor degeneration is not clear.
Vertebrate photoreceptors are highly polarized sensory neurons whose apical component is compartmentalized into inner segment (IS) and outer segment (OS). A connecting cilium extends from the basal body in the distal IS, extends to form axoneme in the OS, and elaborates into stacks of lamellar membranous discs which comprise the OS (14–16). The connecting cilium is the structural equivalent of the transition zone (TZ) of primary cilia and is involved in the docking and sorting of cargo proteins destined for the axoneme. OS discs, the primary sensory apparatus of the retina, undergo periodic shedding and renewal in response to light (17–20). It is estimated that during disc renewal, ∼2000 molecules of opsin are transported to each OS in human retina (19–21). Additionally, light-dependent translocation of arrestin, transducin, and recoverin occurs via the TZ (22–24). This transport may be governed by IFT or diffusion (25–28). Macromolecular protein complexes at the TZ of cilia govern the stringently regulated transport of proteins (29, 30). Consistently, the TZ-associated proteins are also involved in cilia-dependent retinal degenerative diseases (31).
The ciliary protein Cep290 localizes primarily to the TZ of photoreceptors and regulates ciliogenesis and photoreceptor viability (32–35). Cep290 mutations are associated with severe and syndromic disorders such as Joubert and Meckel-Gruber syndromes (35, 36), as well as tissue restricted disease, such as Leber congenital amaurosis (37). Previously, we showed that in-frame deletion of 299 amino acids in Cep290 is associated primarily with retinal degeneration in the rd16 (retinal degeneration 16) mouse (32). We have termed this domain, “deleted in rd16 domain” (DRD; or DSD, deleted in sensory dystrophy). The rd16 mice exhibit rod and cone dysfunction by postnatal day (P) 12. The truncated Cep290 protein (ΔCep290) with a deletion of the DRD is still expressed in the rd16 mouse (32). Here, we show that Cep290, specifically the DRD interacts with Raf-1 kinase inhibitory protein (Rkip), a novel ciliary protein in photoreceptors. We also provide evidence of the involvement of aberrant accumulation of Rkip in the manifestation of Cep290-associated photoreceptor degeneration.
EXPERIMENTAL PROCEDURES
Antibodies
Cep290 and Rab8A antibodies (rabbit) have been characterized (32, 38). Mouse anti-RAB8A was purchased from BD Biosciences. Rabbit polyclonal and mouse monoclonal antibodies against Rkip and anti-myc antibodies were purchased from Invitrogen. Anti-γ-tubulin and mouse anti-FLAG antibodies were procured from Sigma. Antibody against GFP was purchased from Torrey Pines Biolabs and anti-Pcm-1 was purchased from Novus Biologicals.
Plasmids and shRNA
RKIP and RAB8A cDNAs were amplified from human lymphocytes and cloned into pGEX4T1 (GE Healthcare), eGFP-C1, mCherry-C1, mECFP-C1 (from Clontech), and pQCXIP-mCherry (kindly provided by Dr. Ben Margolis, University of Michigan). Human CEP290 was amplified from human retinal RNA (a kind gift of Dr. Anand Swaroop, National Eye Institute) by RT-PCR followed by cloning into pEGFP-C1 vector. The construct was sequence verified and used in zebrafish rescue experiments. The FLAG-CEP290 plasmid DNA was a generous gift of Dr. Joseph Gleeson (University of California, San Diego, CA). Site-directed mutagenesis was performed using the Stratagene QuikChange kit. The CEP290 shRNA construct (pLKO.1-CEP290) was purchased from Sigma (5′-AAATTAAGATGCTCACCGAACTCGA-3′).
Cell Culture and Transfections
COS7, HEK293T, and hTERT-RPE1 cells were cultured as indicated in ATCC guidelines. DNA was transfected using Arrest-in reagent (Open Biosystems) or FuGENE 6 (Roche Applied Science). For generating stable cell clones, we employed retrovirus-mediated infection using standard methods. For cilia induction, cells were cultured for 24 h with 10% serum and then serum-deprived (2% serum) for 48 h before fixation. Cilia growth was assessed by positive acetylated α-tubulin staining.
GST Pulldown, Immunoprecipitations (IP), and Tandem Mass Spectrometry Analysis
GST pulldown analysis was carried out as described (39). Briefly, in vitro translation was performed using the [35S]methionine in vitro translated reaction mix (PROMEGA TnT Quick kit). 35S signal was analyzed with STORM 840 (GE Healthcare) and GST pulldown input was analyzed by Coomassie Brilliant Blue staining. Immunoprecipitations were performed, as described (39). The precipitated proteins were resolved by two-dimensional gel electrophoresis and stained with SYPRO Ruby (Invitrogen). The protein spots that were detected specifically in the immunoprecipitates from WT mouse retina and not in rd16 retinal extracts were analyzed by tandem mass spectrometry (Michigan Proteome Consortium, University of Michigan). For GTP/GDP loading of retinal extracts, ∼300 μg of bovine or mouse proteins lysates were prepared in buffer containing 0.1 mm non-hydrolyzable GDP or GTP analogs and 30 mm MgCl2.
Rkip Quantification in Immunoblots
For measurement of integrated densities of Rkip immunoreactive bands, blot films were scanned and the area of each band was calculated manually in pixels. The mean gray value of each selected band × the area represent the integrated density. The measurements were done using Image J software (NIH, Bethesda).
Immunofluorescence and Immunogold EM
Immunofluorescence of fixed frozen retinal sections and of cultured cells was performed using Leica SP5, Olympus FV500, or Olympus FV1000 confocal microscopes, as previously described (40). The excitation wavelength for pericentrin staining in mCherry-overexpressing cells was 650 nm so that it does not coincide with the mCherry staining. However, in some figures, it is represented as red color. DAPI or Hoechst were used as nuclear counterstains. Immunogold staining using anti-Rkip antibody was performed using 20-nm gold particles, as described (35).
Zebrafish Manipulations
The translation blocking cep290-morpholino (MO; 5′-GCCGCAGGCATTCTTCAGGTCAGCT-3′), and standard negative control (SNC) morpholino were purchased from Gene Tools Inc. For rescue and overexpression experiments, human mRNA was transcribed with appropriate primers using mMESSAGE mMACHINE kit (Ambion). Injections, phenotypic analysis, and staining of Kupffer vesicles (KV) cilia in zebrafish embryos were performed as described (6).
RESULTS
Cep290 Interacts with Rkip
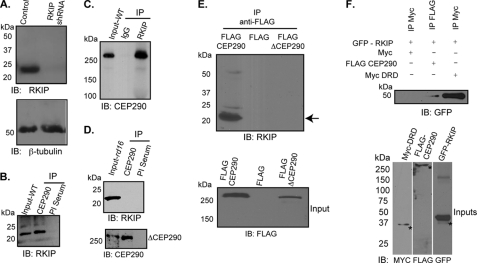
To delineate the pathogenic mechanism of Cep290-mediated photoreceptor degeneration, we first sought to identify Cep290-interacting proteins in mammalian retina. To this end, we performed immunoprecipitation (IP) of Cep290 from P10 wild type (WT) and rd16 retinal extracts followed by two-dimensional gel electrophoresis and tandem mass spectrometry (MS/MS) of the precipitated proteins. To identify the interactions mediated by the Cep290-DRD, we specifically focused on the proteins that were precipitated in WT but not in the rd16 retina and had cut off of three unique peptides (supplemental Fig. S1). Our analysis revealed four different peptides of Rkip (Table 1), which belongs to a family of phosphatidylethanolamine-binding proteins (41). Interestingly, Rkip localizes to centrosomes in dividing cells and was identified as one of the abundant proteins in the mouse photoreceptor ciliary proteome (42, 43). We therefore validated CEP290-Rkip association by performing immunoblot analysis of the precipitated proteins using WT and rd16 retinas. We first validated the specificity of the anti-Rkip antibody. We generated hTERT-RPE1 cells that stably express RKIP-shRNA or control shRNA (scrambled). Immunoblot analysis of cell extracts revealed that although the control cells expressed Rkip protein, no immunoreactive band was detected in RKIP-shRNA-treated cells (Fig. 1A). Equal loading of protein extracts was conformed by immunoblotting of the same extracts using anti-β-tubulin antibody (Fig. 1A, lower panel). The anti-Cep290 and anti-Rkip antibodies could, respectively, co-immunoprecipitate Rkip and Cep290 from WT mouse retina (Fig. 1, B and C). IP using rd16 retina did not reveal Rkip-immunoreactive bands (Fig. 1D); however, we could detect anti-Rkip immunoreactive bands at high exposures (data not shown). These results indicate that, although other domains may also be involved, the Cep290-DRD mediates a majority of Cep290 association with Rkip.
TABLE 1.
Mouse (M. musculus; accession number AAG25635) Rkip peptides identified by MS/MS analysis of Cep290-immunoprecipitated proteins
| Sequences | |
|---|---|
| 1. | 48NRPSSISWDGLDPG62 |
| 2. | 63LYTLVLTDPDAPSR76 |
| 3. | 94GNDISSGTVLSDYVGSGPPS113 |
| 4. | 120YVWLVYEQEQPLS132 |
FIGURE 1.
Cep290 interacts with Rkip. A, immunoblot (IB) analysis of protein extracts from control and RKIP-shRNA expressing hTERT-RPE1 cells was performed using anti-Rkip (upper panel) or anti-β-tubulin (lower panel) antibodies. Apparent molecular mass is denoted in kDa. B and C, IP was performed with antibodies to Cep290 (B) or Rkip (C) using mouse retinal extracts. IP with preimmune (PI) serum or normal rabbit IgG was used as negative control. Input represents 10% of the amount of protein used for IP. D, IP using anti-Cep290 antibody (upper panel) from rd16 mouse retina revealed no Rkip-immunoreactive band. Lower panel shows the efficacy of IP using the Cep290 antibody, as ΔCep290 can be immunoprecipitated from rd16 mouse retina. E, COS7 cells were transiently transfected with constructs encoding FLAG epitope, Flag-Cep290, or Flag-ΔCep290. Protein extracts were subjected to IP using anti-FLAG antibody and immunoblotting using anti-Rkip or anti-FLAG antibodies. Arrow indicates the Rkip-immunoreactive band in the FLAG-Cep290 expressing cell extracts. Lower panel shows that FLAG-CEP290 and FLAG-ΔCEP290 proteins were expressed in the cells. F, lysates from COS7 cells transiently transfected with GFP-Rkip, Myc-DRD, and Flag-CEP290 were subjected to IP using the indicated antibodies followed by immunoblotting with anti-GFP antibody. Asterisks represent specific immunoreactive bands. Lower panel shows input samples of the different extracts used for IP.
We then performed transient transfection of COS7 cells using constructs expressing FLAG-Cep290, FLAG-ΔCep290, or FLAG epitope (empty vector). Protein extracts were subjected to IP using anti-FLAG antibody followed by immunoblotting using anti-Rkip antibody. Our analysis revealed Rkip-immunoreactive bands in protein extracts of cells expressing full-length FLAG-Cep290 but not FLAG-ΔCep290 or FLAG epitope alone (Fig. 1E). These results indicate that deletion of the DRD results in tremendously reduced or undetectable association of the mutant Cep290 protein with Rkip. To further validate whether the DRD of Cep290 directly mediates its interaction with Rkip, we performed co-transfection and co-IP experiments using COS7 cells and constructs encoding GFP-Rkip and FLAG-Cep290 or Myc-DRD (Fig. 1F). We found that IP using anti-Myc or anti-FLAG antibody pulls down GFP-Rkip, as determined by immunoblotting using the anti-GFP antibody. In a control experiment, Myc tag or FLAG tag alone did not interact with Rkip. To assess whether CEP290-DRD and Rkip can also interact in vitro, we used purified recombinant GST-Rkip in a GST pulldown assay with in vitro translated 35S-DRD. We found that GST-Rkip could pull down radiolabeled DRD, although GST moiety alone could not associate with the DRD (data not shown).
Rkip Co-localizes with Cep290 at the Connecting Cilium (Transition Zone) of Photoreceptors
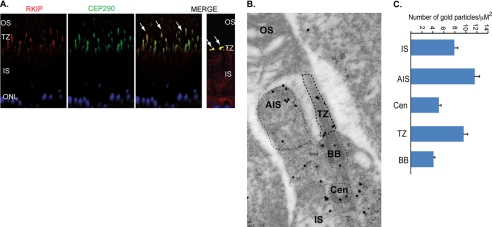
We next investigated the localization of Rkip in mouse retina. In adult mouse retina, we found that Rkip co-localizes with Cep290 in the TZ, which is the region between the basal body and the axoneme (Fig. 2A). In photoreceptors, Rkip staining also co-localizes with acetylated α-tubulin, a ciliary marker (supplemental Fig. S2A). To further investigate the subcellular localization of Rkip in mouse retina, we performed immunogold labeling of Rkip. Our analysis revealed predominant staining of Rkip in the apical inner segment, TZ, and to a lesser extent at the basal body and centriole of photoreceptors (Fig. 2B). Quantitative analysis further revealed that a majority of Rkip staining is present in the apical inner segment and transition zone (Fig. 2C). In cultured hTERT-RPE1 cells, Rkip shows expression in the cytosol and also near the basal body, as determined by co-localization with γ-tubulin (supplemental Fig. S2B). Such localization at the base of the cilium in hTERT-RPE1 cells as opposed to a low level basal body staining in photoreceptors may represent cell type-specific differences in the recruitment of Rkip.
FIGURE 2.
Rkip primarily localizes to cilia. A, retinal sections from WT mice were stained with anti-Cep290 (green) and anti-Rkip (red) antibodies. Merge image shows co-localization of Rkip and Cep290 at the TZ (arrows). Right panel in the merged image shows a magnified image of the co-localization of Rkip and Cep290 at the junction between the inner and outer segments (ONL, outer nuclear layer). B, immunogold labeling of Rkip in mouse photoreceptors reveals broad expression of Rkip in photoreceptors with predominant staining in the apical inner segment (AIS) and TZ. CEN, centriole; BB, basal body. C, histogram shows quantitative analysis of the labeling in the different regions of the photoreceptors. Electron micrographs were analyzed for immunogold particles in a defined square area of the different regions (AIS, BB, and TZ). The number of gold particles counted across various sections was plotted as average number of particles in the region counted. At least 10 cells in each section across 10 different sections were used for analyzing the gold particle density.
Rkip Levels Are Up-regulated in rd16 Mouse Retina
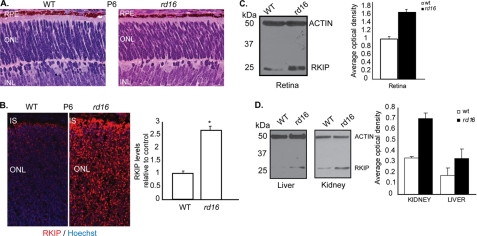
As Rkip cannot associate with ΔCep290 in rd16 retina, we tested whether Rkip expression and/or localization are altered in the rd16 photoreceptors. To this end, we utilized retinal sections from postnatal day 6 mice. This stage were selected so that effect on Rkip expression or localization could be analyzed prior to onset of photoreceptor degeneration in the rd16 mice (32) (see Fig. 3A for comparison at P6). We detected diffuse but predominant staining of Rkip in the IS of P6 WT retina. Surprisingly, in P6 rd16 retina, there was ∼2.5-fold increase in the levels of endogenous Rkip, as compared with WT (Fig. 3B). Similar results were obtained with retinal sections from P5 or P10 rd16 mice (data not shown). Immunoblot analysis of retinal extracts of WT and rd16 mice using the anti-Rkip antibody revealed similar findings. The rd16 retinal protein extract exhibited an almost 1.8-fold increase in the optical density of the Rkip-immunoreactive band compared with wild type age-matched controls (Fig. 3C). We also detected an increase in Rkip intensity in liver and kidney homogenates of rd16 mice compared with WT mice (Fig. 3D). Such a phenomenon could result from up-regulation of gene expression, protein translation, or protein stability. We detected less than a 2-fold change in the expression of Rkip mRNA in a quantitative real time reverse transcriptase-PCR (RT-PCR) assay in the rd16 retina (data not shown). Moreover, an increase in Rkip levels did not require new protein synthesis because knockdown of Cep290 in hTERT-RPE1 cells resulted in higher Rkip levels even in the presence of cycloheximide, a protein synthesis inhibitor (data not shown). These studies indicate that the accumulation of Rkip is likely due to increased stability.
FIGURE 3.
Cep290 regulates intracellular levels of Rkip. A, histological analysis of retinal sections of P6 WT and rd16 mice. RPE, retinal pigment epithelium; ONL, outer nuclear layer; INL, inner nuclear layer. B, increased endogenous levels of Rkip were detected in P6 rd16 retinas by immunofluorescence using anti-Rkip antibody (red). Nuclei are stained with Hoechst (blue). Right panel shows graphical representation of the Rkip signal in arbitrary fluorescent units. Data from three different animals in each group are represented mean ± S.D. (*, p = 0.03; paired t test). C, immunoblot analysis revealed increased Rkip levels in P6 rd16 retinal lysates. The histogram shows quantitative analysis of the optical density of the Rkip-immunoreactive band relative to actin (control). Data represent average ± S.D. of at least three independent experiments (*, p = 0.002; paired t test). D, immunoblot analysis and quantification of Rkip levels in P6 rd16 kidney and liver lysates. Actin was used as loading control. Lower panel shows statistically significant differences in Rkip levels in kidney (p = 0.00003) and liver (p = 0.001).
Expression of Cep290-DRD Partially Rescues cep290 Knockdown Phenotype
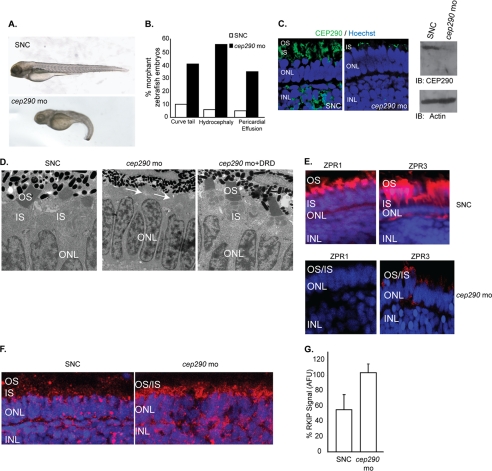
We next tested whether expressing the Cep290-DRD may partly rescue the photoreceptor degeneration phenotype observed in cep290-knockdown embryos. We and others have shown that morpholino-mediated knockdown of ciliary proteins, including cep290, results in measurable phenotypes in zebrafish embryos (6, 35, 40, 44, 45). Consistent with previous reports (35), injection of a translation-blocking morpholino against cep290 resulted in tail extension anomalies, pericardial effusion, hydrocephaly, and microphthalmia at 4 days post-fertilization (dpf) in ∼60% of zebrafish embryos. In contrast, when embryos were treated with a SNC morpholino, only 5% were defective (Fig. 4, A and B). Moreover, the observed phenotypes can be rescued by co-injection of mRNA encoding human CEP290, indicating that these are specific phenotypes and not related to MO toxicity (data not shown). Down-regulation of the Cep290 protein levels was confirmed by immunofluorescence and immunoblot analysis of 4 dpf control and cep290-knockdown embryos using anti-Cep290 antibody (Fig. 4C). We then analyzed retinal morphology of control and cep290-knockdown zebrafish embryos at 4 dpf using transmission electron microscopy. Depletion of cep290 resulted in severe photoreceptor defects with failure to form OS in defective embryos (Fig. 4D, middle panel). Commensurate with this, we detected significant loss of expression of rod and cone photoreceptor-specific marker proteins Zpr3 and Zpr1, respectively (Fig. 4E). Moreover, we detected an ∼1.8-fold increase in Rkip levels in the cep290-knockdown zebrafish embryo retina (Fig. 4, F and G).
FIGURE 4.
Depletion of cep290 in zebrafish is associated with morphological anomalies and retinal defects. A and B, injection of cep290-MO and not a SNC-MO results in ciliary defects in zebrafish embryos, including curly tail, pericardial effusion, hydrocephaly, and microphthalmia (quantified in B). C, left panel, retinal cryosections from zebrafish embryos injected with cep290-mo or SNC were stained with anti-Cep290 antibody (green). Nuclei are stained with Hoechst (blue). ONL, outer nuclear layer; INL, inner nuclear layer. Right panel shows immunoblot (IB) analysis of lysates of zebrafish injected with SNC or cep290-MO using anti-Cep290 or actin antibody (loading control). D, transmission EM of retinal sections from 4 dpf zebrafish embryos injected with SNC or cep290-MO or co-injected with cep290-MO and mRNA encoding the DRD (cep290-mo+DRD) was performed to detect the effect on photoreceptor development. Arrows indicate loss of photoreceptor OS due to knockdown of cep290. E, effect of loss of cep290 on the expression of photoreceptor-specific proteins ZPR1 and ZPR3 (red) was examined by immunofluorescence analysis of zebrafish embryos treated with SNC or cep290-MO. Nuclei were stained with Hoechst (blue). F, SNC or cep29-MO-treated zebrafish retinas were analyzed for Rkip expression by immunofluorescence using anti-Rkip antibody. Histogram in panel G depicts the up-regulation of Rkip levels following depletion of cep290 as compared with controls. The data are representative of three independent experiments with at least 20 embryos analyzed in each experiment. AFU, arbitrary fluorescent units.
We then assessed whether expression of the DRD of Cep290 can partially rescue the cep290-MO-associated phenotype in zebrafish. Our analysis revealed that co-injection of mRNA encoding the DRD and cep290-morpholino results in partial yet significant rescue of the photoreceptor phenotype, as determined by development of photoreceptor OS (Fig. 4D, and supplemental Fig. S3) and expression of photoreceptor marker proteins (data not shown). We also sometimes observed swelling of photoreceptor nuclei and in the inner nuclear layer in some sections from retinas expressing cep290-MO and the DRD. Although this was not a consistent occurrence, we consider that such phenomena may represent inner retinal remodeling in response to the expression of the DRD and formation of photoreceptor OS.
Overexpression of Rkip Results in Ciliary Defects in Zebrafish
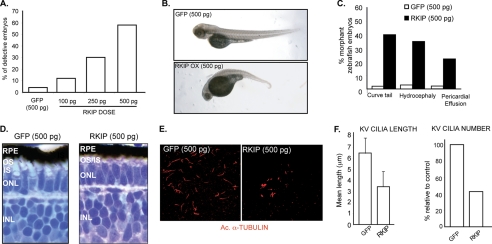
If Cep290-dependent developmental phenotypes in zebrafish are mediated by Rkip, then overexpression of Rkip should recapitulate the cep290-knockdown phenotype. To test this hypothesis, we overexpressed human Rkip in zebrafish embryos and assessed developmental phenotypes. Injection of different doses of mRNA encoding Rkip resulted in a dose-dependent effect on the development of zebrafish embryos as compared with control embryos injected with mRNA encoding GFP. Approximately 40% of embryos injected with Rkip-encoding mRNA (500 pg) displayed curly tail and hydrocephaly and 20% of the embryos exhibited pericardial effusion (Fig. 5, A–C). Histological analysis of retinas of Rkip-overexpressing embryos (500 pg of RKIP mRNA) revealed perturbed photoreceptor outer segment development at 4 dpf (Fig. 5D).
FIGURE 5.
Increase in Rkip levels results in cilia-related defects in zebrafish embryos. A, indicated doses of RKIP mRNA and maximum dose (500 pg) of GFP mRNA were injected into zebrafish embryos. Total number of defective embryos (morphants) was assessed at 4 dpf and quantified. B, representative pictures of embryos with overexpression of Rkip or GFP (control). C, ectopic expression of Rkip results in ciliary defects in zebrafish embryos. Injection of 500 pg of Rkip mRNA results in tail extension abnormalities, hydrocephaly, and pericardial effusion. As a control, we injected 500 pg of GFP mRNA. D, ectopic expression of Rkip affects photoreceptor morphology as evidenced by analysis of toluidine blue-stained plastic-embedded retinas at 4 dpf. E, overexpression of Rkip affects KV cilia formation and extension. KV cilia were analyzed by immunostaining with the ciliary marker acetylated α-tubulin (Ac α-tubulin; red). F represents quantification of length (left) and number (right) of KV cilia. Data represent mean ± S.D. of three independent experiments (p < 0.001).
Rkip Levels Are Critical for Cilia Formation
As disruption of CEP290 results in aberrant ciliogenesis (34), we examined whether overexpression of RKIP affects cilia assembly in zebrafish embryos. Injection of mRNA encoding RKIP (500 pg) into wild type zebrafish embryos resulted in defective ciliogenesis, as determined by acetylated α-tubulin staining of the KV (Fig. 5E). Numbers of cilia in the KV were decreased by 60%; and cilia length was reduced by 45–50% compared with control cells overexpressing GFP (Fig. 5F).
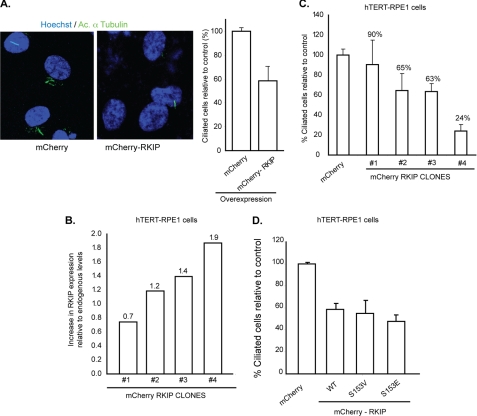
To further validate the role of Rkip in regulating ciliogenesis, we generated hTERT-RPE1 cells stably expressing recombinant mCherry-Rkip or mCherry alone. Analysis of these cells revealed a dose-dependent effect of Rkip levels on ciliogenesis (Fig. 6, A–C). Although a 1.9-fold increase in the levels of mCherry-Rkip over endogenous Rkip resulted in ∼76% decrease in the number of ciliated cells compared with controls, a 0.7-fold increase in mCherry-Rkip levels was associated with only a 10% decrease in the number of ciliated cells. Similar results were obtained using GFP-Rkip (data not shown). We did not detect a significant difference in the number of Rkip-overexpressing cells undergoing cell division compared with controls, as revealed by staining with Ki-67, a cell cycle progression marker (data not shown). Rkip has been shown to regulate spindle checkpoint in a Raf-1 kinase and MAP kinase pathway-dependent manner. Previous studies showed that phosphorylation of Rkip at residue Ser-153, is critical for its association with centrosomes and its function as a regulator of MAPK signaling cascade (46). We therefore, asked whether Rkip-mediated regulation of ciliogenesis also depends upon its phosphorylation. Transient overexpression of mCherry-Rkip S153E (that mimics the phosphorylated state of Rkip) or phosphorylation-deficient (Rkip S153V) mutant revealed a similar effect on cilia biogenesis as observed with WT mCherry-Rkip (Fig. 6D). These results indicate that phosphorylation of Rkip at Ser-153 does not affect its ability to reduce cilia formation.
FIGURE 6.
Increased levels of Rkip affect cilia formation in hTERT-RPE1 cells. A, hTERT-RPE1 cells stably expressing mCherry-Rkip or mCherry alone (control) were assessed for cilia growth by immunofluorescence using acetylated (Ac) α-tubulin (green). Histogram shows quantitative analysis of the number of ciliated cells. Data represent mean ± S.D. (p < 0.05). B and C, the levels of mCherry-Rkip were analyzed from four different clones of hTERT-RPE1 cells with stable expression of mCherry-Rkip and a ratio over the level of endogenous Rkip expression was calculated. The different clones were cultured in ciliogenesis conditions (serum starvation) and the number of ciliated cells was assessed by the acetylated α-tubulin immunofluorescence signal. All clones displayed a dose-dependent effect of overexpression of Rkip on ciliogenesis (C). Data represent mean ± S.D. of more than 100 cells analyzed in three independent experiments. D, number of ciliated hTERT-RPE1 cells was assessed after overexpression of mCherry-tagged constitutively phosphorylated (S153E) or non-phosphorylated (S153V) mutants of Rkip. Cilia growth was assessed by acetylated α-tubulin staining. As control, we overexpressed mCherry tag alone. Data represent mean ± S.D. of more than 100 cells analyzed in each group on three independent experiments.
Rkip Modulates Ciliary Localization of Rab8A
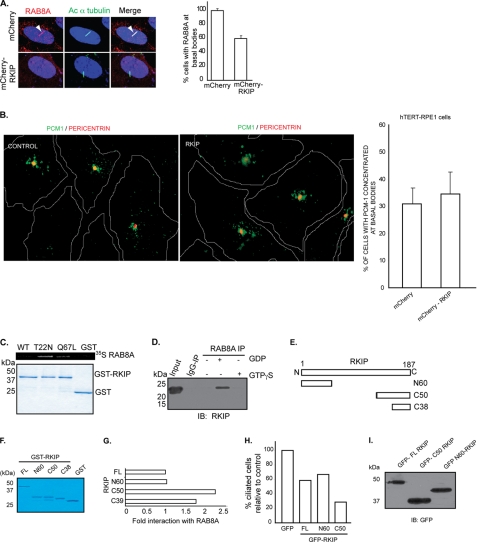
To investigate the mechanism by which accumulation of Rkip mediates cilia assembly, we assessed whether components of cilia formation are recruited to the basal bodies. Because depletion of CEP290 in human RPE cells results in defective targeting of Rab8A to the primary cilium (34), we analyzed whether overexpression of Rkip also affects Rab8A localization. Immunofluorescence analysis of hTERT-RPE1 cells revealed mislocalization of Rab8A in about 40% of the cells that stably express mCherry-Rkip compared with those expressing mCherry alone (Fig. 7A). It was also shown that depletion of CEP290 affects Pcm-1 distribution in cultured cells (33); however, our analysis did not reveal a significant alteration in the distribution of Pcm-1 in vitro (Fig. 7B) or of other ciliary proteins, including Ift88/Polaris and retinitis pigmentosa GTPase regulator (data not shown). These results suggest that Rkip modulates the localization of a distinct set of proteins required for cilia assembly and maintenance, at least in cultured cell lines.
FIGURE 7.
Overexpression of Rkip affects Rab8A localization in cells. A, confocal immunofluorescence of mCherry-Rkip or mCherry alone hTERT-RPE1 stably overexpressing cells stained with anti-Rab8A antibody (represented in red). Rab8A positive staining at cilia (arrow) was assessed by co-localization with acetylated (Ac) α-tubulin (represented in green). There is a considerable decrease in the number of cells with Rab8A at the cilia in Rkip-overexpressing cells (right panel; histogram). Data represent mean ± S.D. (p < 0.005). B, overexpression of Rkip does not seem to affect Pcm-1 distribution in hTERT-RPE1 cells. Localization of Pcm-1 around the basal bodies was assessed by co-localization with Pericentrin (represented as red) in hTERT-RPE1 cells stably expressing mCherry-Rkip (Rkip) or mCherry alone (control). No major difference was observed between the two groups, as shown in the right panel. Data represent mean ± S.D. from more than 150 cells analyzed in each group in three independent experiments. C, GST pulldown assay was performed using GST-Rkip and in vitro translated 35S-Rab8A WT, GDP-locked (T22N), and GTP-locked (Q67L) mutants. Interaction was assessed by autoradiography (upper panel). Lower panel shows Coomassie staining of the proteins used in the assay. D, bovine retinal lysates (∼300 mg), prepared in the presence of GDP or GTP non-hydrolyzable analogs were subjected to IP using Rab8A antibody or IgG (control). Precipitated proteins were analyzed by SDS-PAGE and immunoblotting (IB) using anti-Rkip antibody. E, schematic representation of different deleted variants of Rkip utilized in the GST pulldown assay. F, Coomassie Blue-stained gel of the Rkip variants after purification from E. coli. G, quantitative analysis of the interaction between Rkip (full-length; FL and deleted variants) and 35S-Rab8A-T22N in GST pulldown assay. Data represent mean of three independent experiments. H, quantitative analysis of the number of ciliated hTERT-RPE1 cells after transient overexpression of different GFP-Rkip variants or GFP alone (control). Data represent mean of three independent experiments (n >200). I, lysates from transientely transfected hTERT-RPE1 cells with GFP-tagged variants of Rkip were analyzed by SDS-PAGE and immunoblotting with GFP antibody.
To delineate how Rkip modulates Rab8A localization, we tested the potential interaction of Rkip with Rab8A. Using GST pulldown assay, we found that Rkip interacts preferentially with the GDP-locked mutant of Rab8A (T22N) (Fig. 7C). Little interaction was detected with the GTP-locked mutant of Rab8A (Q67L). We further validated Rkip-Rab8A association in bovine retinal lysates. We found that Rkip preferentially associates with Rab8A in the presence of GDP substrate as compared with GTPγS (Fig. 7D).
We then sought to identify the minimal domain of Rkip required for its association with Rab8A. We expressed and purified recombinant GST-fused fragments of Rkip from Escherichia coli (Fig. 7, E and F). These fragments include amino-terminal (N) 60 amino acids and carboxyl-terminal (C) 50 and 38 (C38) amino acids. Although we could not detect a domain of Rkip that does not interact with Rab8A in the GST pulldown assay using in vitro synthesized 35S-RAB8A T22N, we found that the Rkip-C38 and Rkip-C50 show, respectively, 1.7- and 2.2-fold greater interaction with Rab8A compared with full-length (FL) Rkip or Rkip-N60 (Fig. 7G). Other domains, N139, C127, and C93, also showed similar association as the FL-Rkip (data not shown). As C50 exhibited maximal association with Rkip, we tested whether overexpression of this domain should result in a more severe effect on cilia formation than FL-Rkip or Rkip-N60. Overexpression of GFP-Rkip-C50 in hTERT-RPE1 cells resulted in 60% fewer ciliated cells compared with those expressing GFP alone (Fig. 7H). As predicted, expression of FL-Rkip or Rkip-N60 resulted in a 35–40% decrease in the number of ciliated cells. The different domains of Rkip were expressed at similar levels in hTERT-RPE1 cells, as determined by immunoblot analysis of cell lysates using anti-GFP antibody (Fig. 7I).
DISCUSSION
Development of targeted therapy depends upon delineating molecular mechanisms of underlying diseases and identification of a majority of patient population. Cep290 is an ideal candidate for such analyses. Mutations in CEP290 are a frequent cause of Leber congenital amaurosis and are also associated with numerous syndromic ciliary disorders. We previously reported that the rd16 mouse carries a hypomorphic allele of Cep290 and predominantly exhibits retinal degeneration due to ciliary dysfunction (32). We now show that pathogenesis of Cep290-dependent retinopathy can be partly attributed to aberrant endogenous levels of the Rkip protein. Although other proteins or factors may also be involved, our findings suggest that selective degradation of proteins may act as a key intermediate in the manifestation of retinal degeneration in Cep290 and other ciliopathies.
The GTPase Rab8A is a critical component of the protein trafficking machinery of photoreceptors (47). Remarkable studies have shown that overexpression of the GDP-locked variant of Rab8A (Rab8A-T22N) results in mistrafficking of rhodopsin and photoreceptor degeneration (47). Whereas the Rab8A T22N mutant disrupts cilia formation, expression of the GTP-locked variant Rab8A Q67L promotes ciliogenesis (3). As Rkip interacts preferably with the GDP-bound form of Rab8A in the retina and accumulation of Rkip due to Cep290 mutation is associated with retinopathy, we propose that the Rkip-Rab8A (GDP) complex may need to dissociate for the release of Rab8A-GDP and subsequent conversion to Rab8A-GTP for appropriate ciliary transport. Degradation of Rkip may facilitate the release of Rab8A-GDP. The molecular mechanisms underlying Rkip degradation and release of Rab8A are still under investigation. Cep290 seems to play a major role in this process as even hypomorphic Cep290 mutation in the rd16 mouse can result in aberrant accumulation of Rkip and early onset severe retinal degeneration.
Rkip is a multifunctional protein involved in diverse processes, including tumor suppression, cell cycle regulation, and Raf-1/MAPK signaling cascade (48). Inhibition of Rkip results in increased cell proliferation by affecting spindle checkpoint, resulting in chromosomal aberrations and cancer progression (43). This effect seems to be mediated by its action on the Raf-1 and MAP kinase pathways (49). In contrast, ablation of Rkip in mice does not seem to affect normal development and maintenance although mild defects in olfaction and reproduction were observed (50). We postulate that there are cell type-specific roles of Rkip that may or may not be interdependent on its phosphorylation status or action on MAP kinase pathway. The regulation of mitotic progression by Rkip is also correlated with its ability to inhibit prostate cancer progression. As tumor progression has recently been shown to be associated with ciliary function (10, 51), further studies will be important to delineate the molecular mechanisms that unify such diverse functions of Rkip in distinct cell types.
Our immunolocalization studies reveal an interesting localization of Rkip to the apical inner segment and the TZ of photoreceptor cilia. Remarkable studies have shown that the apical plasma membrane and the periciliary ridge complex can act as docking sites for rhodopsin-containing vesicles during vectorial transport to the OS (52). The concentrated signal of Rkip at these sites and its ability to interact with Rab8A suggests its involvement in the regulation of docking and transport of membrane protein-containing vesicles in photoreceptors. Multiple other proteins, including the Usher syndrome proteins, also localize to these structures and are predicted to provide structural and functional support to the privileged membrane domain of photoreceptors (53, 54). Rkip may also play a role in regulating the function of these proteins. Further studies are needed to test such functions of Rkip in photoreceptors and other cilia.
The extent of dysfunction of a particular organ in the disease may depend upon the relative load of ciliary function, including protein trafficking and signal transduction. As Cep290 mutations are associated with multisystem disorders, one would predict that accumulation of Rkip and subsequent mislocalization of Rab8A and/or other signaling moieties should result in the dysfunction of the cilia and homeostasis in multiple organ systems. Although we detected an increase in Rkip levels in extra-retinal tissues, such as kidney, defects in rd16 mice appear to be limited to sensory neurons (32, 55). We reckon that Cep290-DRD in rd16 mouse still retains its ability to interact at low levels with Rkip and that such an interaction may be sufficient to retain the function of other organ systems. A hypothesis that remains to be tested is that loss of function mutations in Cep290, as seen in syndromic ciliopathies, may completely abolish its interaction with Rkip and subsequently result in relatively higher levels of Rkip and subsequent mislocalization of key signaling proteins in other tissues as opposed to the predominantly retinal (or sensory) defects observed with hypomorphic CEP290 mutations.
It should be noted that aberrant protein degradation has been implicated in the pathogenesis of some rhodopsin mutations (56) and in neurodegenerative disorders, such as Alzheimer and Parkinson diseases (57). In these disorders, the mutant protein undergoes misfolding and forms toxic aggregates in the cytosol (58, 59). We provide evidence for the aberrant accumulation of a protein in photoreceptors that seems to function downstream of the mutated disease protein Cep290. Previous studies showed that Bardet-Biedl syndrome-associated disease manifestation could also be correlated to defects in protein degradation pathways and developmental signaling cascades (60). It will be interesting to identify and characterize other such proteins whose levels are up-regulated in retina and other cell types due to mutations in ciliary proteins.
Our findings that Rkip levels are up-regulated in Cep290-associated photoreceptor degeneration can be clinically significant because of its potential role as a prognostic marker. Rkip and other such proteins also provide possible therapeutic targets in the management of retinal and other neurodegenerative disorders. Additional comprehensive analysis of ciliary function of Rkip should allow development of optimal therapeutic modalities.
Supplementary Material
Acknowledgments
We thank Dr. Anand Swaroop and Dr. Jan Klysik for critical discussions, Dr. Peter Hitchcock for help with zebrafish manipulations, Steve Lentz and Alice Hackett for assistance in microscopy, and Stephen Atkins for help with protein purification.
This work was supported, in whole or in part, by National Institutes of Health Grants EY007961 and 5P60 DK20572 to the Michigan Diabetes Research and Training Center, the Foundation for Fighting Blindness, and Midwest Eye Banks and Transplantation Center, and Vision Core Facilities Grant EY07003.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3.
- IFT
- intraflagellar transport
- Rkip
- Raf-1 kinase inhibitory protein
- SNC
- standard negative control
- RPE
- retinal pigment epithelium
- IS
- inner segment
- OS
- outer segment
- TZ
- transition zone
- DRD
- deleted in rd16 domain
- dpf
- days post-fertilization
- GTPγS
- guanosine 5′-3-O-(thio)triphosphate
- IP
- immunoprecipitation
- FL
- full-length
- KV
- Kupffer vesicles
- P
- postnatal day
- MO
- morpholino.
REFERENCES
- 1. Rosenbaum J. L., Witman G. B. (2002) Nat. Rev. Mol. Cell Biol. 3, 813–825 [DOI] [PubMed] [Google Scholar]
- 2. Sorokin S. (1962) J. Cell Biol. 15, 363–377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Nachury M. V., Loktev A. V., Zhang Q., Westlake C. J., Peränen J., Merdes A., Slusarski D. C., Scheller R. H., Bazan J. F., Sheffield V. C., Jackson P. K. (2007) Cell 129, 1201–1213 [DOI] [PubMed] [Google Scholar]
- 4. Seo S., Baye L. M., Schulz N. P., Beck J. S., Zhang Q., Slusarski D. C., Sheffield V. C. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 1488–1493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hodges M. E., Scheumann N., Wickstead B., Langdale J. A., Gull K. (2010) J. Cell Sci. 123, 1407–1413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ghosh A. K., Murga-Zamalloa C. A., Chan L., Hitchcock P. F., Swaroop A., Khanna H. (2010) Hum. Mol. Genet. 19, 90–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Murga-Zamalloa C. A., Atkins S. J., Peranen J., Swaroop A., Khanna H. (2010) Hum. Mol. Genet. 19, 3591–3598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Pazour G. J., Baker S. A., Deane J. A., Cole D. G., Dickert B. L., Rosenbaum J. L., Witman G. B., Besharse J. C. (2002) J. Cell Biol. 157, 103–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Rosenbaum J. L., Cole D. G., Diener D. R. (1999) J. Cell Biol. 144, 385–388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wong S. Y., Seol A. D., So P. L., Ermilov A. N., Bichakjian C. K., Epstein E. H., Jr., Dlugosz A. A., Reiter J. F. (2009) Nat. Med. 15, 1055–1061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Yoder B. K. (2006) Dev. Cell 10, 541–542 [DOI] [PubMed] [Google Scholar]
- 12. Gerdes J. M., Davis E. E., Katsanis N. (2009) Cell 137, 32–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Adams N. A., Awadein A., Toma H. S. (2007) Ophthalmic Genet. 28, 113–125 [DOI] [PubMed] [Google Scholar]
- 14. Besharse J. C. (1986) The Retina: A Model for Cell Biological Studies Part I, Academic Press, New York [Google Scholar]
- 15. Kennedy B., Malicki J. (2009) Dev. Dyn. 238, 2115–2138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Young R. W. (1968) J. Ultrastruct. Res. 23, 462–473 [DOI] [PubMed] [Google Scholar]
- 17. Besharse J. C., Hollyfield J. G. (1976) J. Exp. Zool. 198, 287–302 [DOI] [PubMed] [Google Scholar]
- 18. Besharse J. C., Hollyfield J. G., Rayborn M. E. (1977) Science 196, 536–538 [DOI] [PubMed] [Google Scholar]
- 19. LaVail M. M. (1973) J. Cell Biol. 58, 650–661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Besharse J. C., Hollyfield J. G., Rayborn M. E. (1977) J. Cell Biol. 75, 507–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Williams D. S. (2002) Vision Res. 42, 455–462 [DOI] [PubMed] [Google Scholar]
- 22. Sokolov M., Lyubarsky A. L., Strissel K. J., Savchenko A. B., Govardovskii V. I., Pugh E. N., Jr., Arshavsky V. Y. (2002) Neuron 34, 95–106 [DOI] [PubMed] [Google Scholar]
- 23. Arshavsky V. Y. (2003) Sci. STKE 2003, PE43. [DOI] [PubMed] [Google Scholar]
- 24. Strissel K. J., Lishko P. V., Trieu L. H., Kennedy M. J., Hurley J. B., Arshavsky V. Y. (2005) J. Biol. Chem. 280, 29250–29255 [DOI] [PubMed] [Google Scholar]
- 25. Marszalek J. R., Liu X., Roberts E. A., Chui D., Marth J. D., Williams D. S., Goldstein L. S. (2000) Cell 102, 175–187 [DOI] [PubMed] [Google Scholar]
- 26. Nair K. S., Hanson S. M., Mendez A., Gurevich E. V., Kennedy M. J., Shestopalov V. I., Vishnivetskiy S. A., Chen J., Hurley J. B., Gurevich V. V., Slepak V. Z. (2005) Neuron 46, 555–567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Besharse J. C., Baker S. A., Luby-Phelps K., Pazour G. J. (2003) Adv. Exp. Med. Biol. 533, 157–164 [PubMed] [Google Scholar]
- 28. Calvert P. D., Schiesser W. E., Pugh E. N., Jr. (2010) J. Gen. Physiol. 135, 173–196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Murga-Zamalloa C. A., Swaroop A., Khanna H. (2009) J. Genet. 88, 399–407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Nachury M. V., Seeley E. S., Jin H. (2010) Annu. Rev. Cell Dev. Biol. 26, 59–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hildebrandt F., Benzing T., Katsanis N. (2011) N. Engl. J. Med. 364, 1533–1543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Chang B., Khanna H., Hawes N., Jimeno D., He S., Lillo C., Parapuram S. K., Cheng H., Scott A., Hurd R. E., Sayer J. A., Otto E. A., Attanasio M., O'Toole J. F., Jin G., Shou C., Hildebrandt F., Williams D. S., Heckenlively J. R., Swaroop A. (2006) Hum. Mol. Genet. 15, 1847–1857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kim J., Krishnaswami S. R., Gleeson J. G. (2008) Hum. Mol. Genet. 17, 3796–3805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Tsang W. Y., Bossard C., Khanna H., Peränen J., Swaroop A., Malhotra V., Dynlacht B. D. (2008) Dev. Cell 15, 187–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sayer J. A., Otto E. A., O'Toole J. F., Nurnberg G., Kennedy M. A., Becker C., Hennies H. C., Helou J., Attanasio M., Fausett B. V., Utsch B., Khanna H., Liu Y., Drummond I., Kawakami I., Kusakabe T., Tsuda M., Ma L., Lee H., Larson R. G., Allen S. J., Wilkinson C. J., Nigg E. A., Shou C., Lillo C., Williams D. S., Hoppe B., Kemper M. J., Neuhaus T., Parisi M. A., Glass I. A., Petry M., Kispert A., Gloy J., Ganner A., Walz G., Zhu X., Goldman D., Nurnberg P., Swaroop A., Leroux M. R., Hildebrandt F. (2006) Nat. Genet. 38, 674–681 [DOI] [PubMed] [Google Scholar]
- 36. Valente E. M., Silhavy J. L., Brancati F., Barrano G., Krishnaswami S. R., Castori M., Lancaster M. A., Boltshauser E., Boccone L., Al-Gazali L., Fazzi E., Signorini S., Louie C. M., Bellacchio E., Bertini E., Dallapiccola B., Gleeson J. G. (2006) Nat. Genet. 38, 623–625 [DOI] [PubMed] [Google Scholar]
- 37. den Hollander A. I., Koenekoop R. K., Yzer S., Lopez I., Arends M. L., Voesenek K. E., Zonneveld M. N., Strom T. M., Meitinger T., Brunner H. G., Hoyng C. B., van den Born L. I., Rohrschneider K., Cremers F. P. (2006) Am. J. Hum. Genet. 79, 556–561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Peränen J., Auvinen P., Virta H., Wepf R., Simons K. (1996) J. Cell Biol. 135, 153–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Khanna H., Hurd T. W., Lillo C., Shu X., Parapuram S. K., He S., Akimoto M., Wright A. F., Margolis B., Williams D. S., Swaroop A. (2005) J. Biol. Chem. 280, 33580–33587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Khanna H., Davis E. E., Murga-Zamalloa C. A., Estrada-Cuzcano A., Lopez I., den Hollander A. I., Zonneveld M. N., Othman M. I., Waseem N., Chakarova C. F., Maubaret C., Diaz-Font A., MacDonald I., Muzny D. M., Wheeler D. A., Morgan M., Lewis L. R., Logan C. V., Tan P. L., Beer M. A., Inglehearn C. F., Lewis R. A., Jacobson S. G., Bergmann C., Beales P. L., Attié-Bitach T., Johnson C. A., Otto E. A., Bhattacharya S. S., Hildebrandt F., Gibbs R. A., Koenekoop R. K., Swaroop A., Katsanis N. (2009) Nat. Genet. 41, 739–745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Simister P. C., Banfield M. J., Brady R. L. (2002) Acta Crystallogr. D Biol. Crystallogr. 58, 1077–1080 [DOI] [PubMed] [Google Scholar]
- 42. Liu Q., Tan G., Levenkova N., Li T., Pugh E. N., Jr., Rux J. J., Speicher D. W., Pierce E. A. (2007) Mol. Cell Proteomics 6, 1299–1317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Eves E. M., Shapiro P., Naik K., Klein U. R., Trakul N., Rosner M. R. (2006) Mol. Cell 23, 561–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Leitch C. C., Zaghloul N. A., Davis E. E., Stoetzel C., Diaz-Font A., Rix S., Alfadhel M., Al-Fadhel M., Lewis R. A., Eyaid W., Banin E., Dollfus H., Beales P. L., Badano J. L., Katsanis N. (2008) Nat. Genet. 40, 443–448 [DOI] [PubMed] [Google Scholar]
- 45. Ross A. J., May-Simera H., Eichers E. R., Kai M., Hill J., Jagger D. J., Leitch C. C., Chapple J. P., Munro P. M., Fisher S., Tan P. L., Phillips H. M., Leroux M. R., Henderson D. J., Murdoch J. N., Copp A. J., Eliot M. M., Lupski J. R., Kemp D. T., Dollfus H., Tada M., Katsanis N., Forge A., Beales P. L. (2005) Nat. Genet. 37, 1135–1140 [DOI] [PubMed] [Google Scholar]
- 46. Zeng L., Imamoto A., Rosner M. R. (2008) Expert Opin. Ther. Targets 12, 1275–1287 [DOI] [PubMed] [Google Scholar]
- 47. Deretic D., Huber L. A., Ransom N., Mancini M., Simons K., Papermaster D. S. (1995) J. Cell Sci. 108, 215–224 [DOI] [PubMed] [Google Scholar]
- 48. Odabaei G., Chatterjee D., Jazirehi A. R., Goodglick L., Yeung K., Bonavida B. (2004) Adv. Cancer Res. 91, 169–200 [DOI] [PubMed] [Google Scholar]
- 49. Klysik J., Theroux S. J., Sedivy J. M., Moffit J. S., Boekelheide K. (2008) Cell Signal. 20, 1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Theroux S., Pereira M., Casten K. S., Burwell R. D., Yeung K. C., Sedivy J. M., Klysik J. (2007) Brain Res. Bull. 71, 559–567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Han Y. G., Kim H. J., Dlugosz A. A., Ellison D. W., Gilbertson R. J., Alvarez-Buylla A. (2009) Nat. Med. 15, 1062–1065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Peters K. R., Palade G. E., Schneider B. G., Papermaster D. S. (1983) J. Cell Biol. 96, 265–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Yang J., Liu X., Zhao Y., Adamian M., Pawlyk B., Sun X., McMillan D. R., Liberman M. C., Li T. (2010) PLoS Genet. 6, e1000955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Maerker T., van Wijk E., Overlack N., Kersten F. F., McGee J., Goldmann T., Sehn E., Roepman R., Walsh E. J., Kremer H., Wolfrum U. (2008) Hum. Mol. Genet. 17, 71–86 [DOI] [PubMed] [Google Scholar]
- 55. McEwen D. P., Koenekoop R. K., Khanna H., Jenkins P. M., Lopez I., Swaroop A., Martens J. R. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 15917–15922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Kaushal S., Khorana H. G. (1994) Biochemistry 33, 6121–6128 [DOI] [PubMed] [Google Scholar]
- 57. Rubinsztein D. C. (2006) Nature 443, 780–786 [DOI] [PubMed] [Google Scholar]
- 58. Sung C. H., Davenport C. M., Nathans J. (1993) J. Biol. Chem. 268, 26645–26649 [PubMed] [Google Scholar]
- 59. Noorwez S. M., Sama R. R., Kaushal S. (2009) J. Biol. Chem. 284, 33333–33342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Gerdes J. M., Liu Y., Zaghloul N. A., Leitch C. C., Lawson S. S., Kato M., Beachy P. A., Beales P. L., DeMartino G. N., Fisher S., Badano J. L., Katsanis N. (2007) Nat. Genet. 39, 1350–1360 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.