Abstract
Papillomaviruses establish persistent infection in the dividing, basal epithelial cells of the host. The viral genome is maintained as a circular, double-stranded DNA, extrachromosomal element within these cells. Viral genome amplification occurs only when the epithelial cells differentiate and viral particles are shed in squames that are sloughed from the surface of the epithelium. There are three modes of replication in the papillomavirus life cycle. Upon entry, in the establishment phase, the viral genome is amplified to a low copy number. In the second, maintenance phase, the genome replicates at a constant copy number in synchrony with the cellular DNA in dividing cells. And finally, in the vegetative or productive phase, the viral DNA is amplified to a high copy number in differentiated cells and is destined to be packaged in viral capsids. This review discussed the cis elements and protein factors required for each stage of papillomavirus replication.
I. Background
A. Papillomavirus associated disease
Papillomaviruses are ubiquitous, epitheliotrophic viruses that cause warts or papillomas. There are hundreds of different human and animal papillomavirus types that have coevolved with their hosts over millions of years. Each virus is species specific and replicates persistently in a specific type of cutaneous or mucosal epithelium. Papillomaviruses cause a spectrum of disease ranging from clinically inapparent infections, through a variety of benign lesions such as common warts, anogenital warts, and laryngeal papillomatosis (Lacey, 2005) to malignant carcinomas. In humans, persistent infection with high-oncogenic risk HPVs is responsible for virtually all cervical cancer (Parkin et al, 2005; Walboomers et al, 1999), and is associated with a subset of head and neck cancers (Gillison and Lowy, 2004).
B. Life Cycle
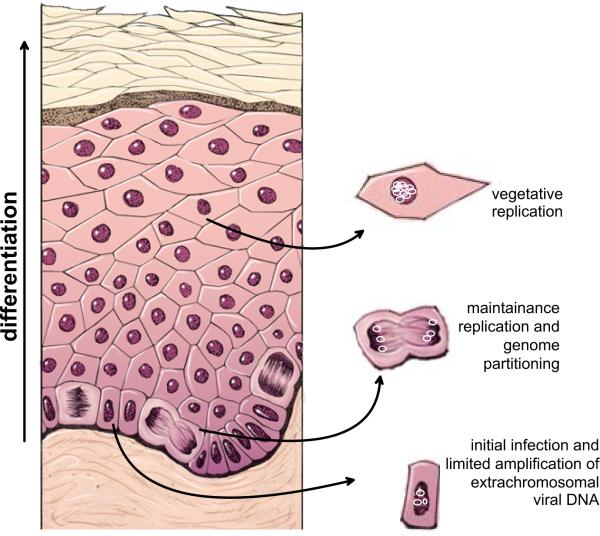
All papillomaviruses have similar life cycles that are tightly linked to differentiation of the host epithelium (see Figure 1). In a normal epithelium, only cells in the lower basal layer are mitotically active. After cell division, one daughter cell is pushed up to replenish the overlying differentiated layers, which are eventually shed from the surface of the epithelium. To initiate infection, papillomaviruses must access the dividing basal cells and it is thought that they do so through microabrasions or wounds. Certain cells in the epithelium, such as those in the bulge of the hair follicle, or the transformation zone of the cervix or epithelial stem cells, may be more susceptible to, or more likely to support, long term persistent infection.
Figure 1. Modes of Replication in the Life cycle of Papillomaviruses.
The diagram shows a stratified epithelium. The lowermost basal layer provides the germinal cells necessary for renewal of the epithelium. Papillomaviruses infect these cells through microabrasions and the viral DNA undergoes a limited amplification after delivery to the nucleus. The viral genome is maintained and partitioned in a regulated manner in these dividing cells. They provide a reservoir of infected cells and, as they divide, individual infected daughter cells are pushed upwards and begin the differentiation process. These differentiated cells support vegetative replication resulting in a great amplification of the viral genome. Eventually, the cells at the outermost layer, containing virion particles, are sloughed from the epithelium.
Papillomaviruses must target a site of epithelial wounding and it has been proposed that laminin 5, secreted by migrating keratinocytes, acts as a transient receptor to trap the virions (Culp et al, 2006). Viral particles also adsorb to membrane-associated heparan sulfate proteoglycans (Giroglou et al, 2001; Joyce et al, 1999; Shafti-Keramat et al, 2003) before transfer to more specific receptors such as the candidate receptor, 6 integrin (Evander et al, 1997; McMillan et al, 1999). Viral particles enter the cell by either calveolae or clathrin-mediated endocytic pathways (Bousarghin et al, 2003; Day et al, 2003; Hindmarsh and Laimins, 2007; Smith et al, 2007). After exiting the endosomes, the L2 minor capsids protein transports the viral DNA into the nucleus to the subnuclear promyelocytic leukemia protein (PML) bodies (Day et al, 2004). Upon infection, many DNA viruses localize to, initiate transcription at, and often disrupt, these bodies, leading to the hypothesis that they are involved in antiviral defense (Negorev and Maul, 2001). In contrast, the presence of the PML protein enhances early transcription of papillomaviruses (Day et al, 2004).
After transportation to the nucleus, the viral genome is amplified to a copy number that may range from 10 to 50 copies per cell, and the infected cell is driven to proliferate to provide a pool of infected basal cells to form the basis of the papilloma. The viral genomes are maintained in the nucleus of the dividing basal cells as extrachromosomal replicating elements that replicate in synchrony with the host cellular DNA in a cell cycle dependent manner. Papillomavirus infections are usually long-lived and persistent and the continually dividing basal cells serve as a reservoir of infected cells for the continual progressive vertical differentiation that occurs during the maturation of the epithelium. Therefore, papillomaviruses need a robust mechanism to retain their extrachromosomal genomes within the nucleus of continually dividing cells to ensure that the infection is sustainable.
Viral genome amplification, late capsid protein synthesis and virion assembly occur in the upper, terminally differentiated cells of the epithelium. Virions are assembled in the superficial differentiated layers and are found throughout the nuclei, frequently organized into paracrystalline arrays. These virus containing cells are destined to be sloughed from the epidermis. The strategy of restricting viral DNA amplification and synthesis of large amounts of viral antigens to the superficial layers of an epithelium ensures long-term persistent infection and is important for immune evasion by the virus. However, this strategy necessitates that the virus amplifies its DNA in cells that would normally have withdrawn from the cell cycle are undergoing differentiation. To achieve this, the papillomaviruses must disrupt the normal differentiation process and sustain cells in an S-phase like state so that the virus has available the enzymes to replicate its own DNA. An unintended result of cell cycle disregulation by a subset of “high risk” viruses is the absence of crucial cell cycle checkpoints, which can lead to genetic instability and malignant progression.
C. Genome Organization and Expression
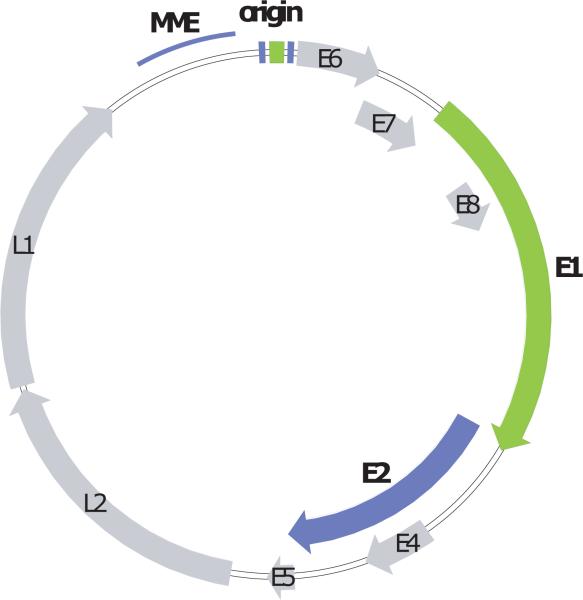
Papillomaviruses have circular, double-stranded DNA genomes of approximately 7000 to 8000bp (see Figure 2). The coding region is divided into early and late regions. The early region contains open reading frames designated E1 through E8, which, for the most part, are expressed in the lower, less differentiated layers of a papilloma. The late region encodes the capsid antigens, L1 and L2, which are expressed in the more superficial, differentiated cells. All viral RNA species are transcribed from one strand and undergo extensive alternative splicing. A region of approximately 500 to 1000bp, located upstream from the coding region, is called the Long Control Region (LCR) or Upstream Regulatory Region (URR). This region contains the replication origin, sequences required for genome maintenance and transcriptional enhancers and promoters.
Figure 2. Papillomavirus genome.
The early and late open reading frames, are indicated as E1–E8 and L1–L2, respectively. The replication elements MME (minichromosome maintenance element) and origin are shown in the LCR region. The E1 and E2 replication proteins are highlighted in green and purple, respectively, with the corresponding binding sites shown in the origin region.
D. Function of Viral Proteins
Papillomaviruses appear to be simple viruses that encode only a few genes, but each viral protein interacts with and regulates a multitude of cellular proteins. Overall, the functions of the viral proteins contribute to initiating an efficient, persistent infection, to evading the host immune system and to producing large amounts of progeny virus.
Basal cells act as a reservoir of cells for the continual renewal of the epithelium and their division is normally regulated by growth factors. Immediately after virus infection, the newly infected cell is driven to proliferate independently from these cellular factors in order to establish a pool of infected basal cells. The E5 protein is a membrane protein that promotes proliferation of infected basal cells by inducing constitutive, ligand-independent activation of growth factor receptors. E5 also interferes with Golgi acidification, resulting in increased cycling and activation of growth factor receptors (reviewed in (DiMaio and Mattoon, 2001). E5 down-regulates surface expression of major histocompatibility complex (MHC) class I molecules, helping the virus to evade the host immune system (Ashrafi et al, 2006; Zhang et al, 2003).
The E6 and E7 proteins also induce cellular proliferation and delay differentiation of the host cells to sustain them in an S-phase like state to ensure the availability of host DNA synthesis proteins for viral DNA replication. The E7 proteins of many “high risk” HPVs accomplish this by binding and inactivating the cellular pRB protein, resulting in expression of S-phase genes. However, this aberrant state induces proteins, such as p53, to arrest the growth of abnormal cells. In turn, the E6 protein binds to the p53 protein and targets it for degradation, thus enabling the cells to remain in a sustained S-phase-like state. The continual division of cells without checkpoints leads to genetic instability and, eventually, a malignant phenotype (reviewed in(Wise-Draper and Wells, 2008)). It is not clear how the “low risk” papillomaviruses fulfill this same requirement as they do not inactivate pRb and p53 functions; they most likely use alternative interactions to accomplish the same goal. For example, the “low risk” HPV E7 proteins bind a unique pRb and calmodulin-binding scaffolding protein called p600, (DeMasi et al, 2005; Huh et al, 2005) and both “low- and high-risk” HPVs are able to target the pRB family member p130 for degradation (Zhang et al, 2006). “Low risk” E6 proteins disrupt the actin cytoskeleton and bind a number of cellular proteins, including ERC-55, the focal adhesion protein paxillin, the E3 ubiquitin ligase E6AP, and the clathrin adaptor complex AP-1 (reviewed in (Wise-Draper and Wells, 2008). E5, E6 and E7 are also important for immune evasion and they act by down regulating interferon responsive genes and MHC Class I surface expression (reviewed in (Woodworth, 2002).
E1 is the primary replication initiator protein. It binds and unwinds the viral replication origin in concert with the E2 protein. The E2 protein is the major transcriptional regulator of the virus and both activates and represses transcription from viral promoters. Additionally, the E2 protein maintains and partitions the replicating extrachromosomal genome. This review will focus on the replication associated functions of these proteins.
The E4 protein, usually expressed from a spliced mRNA species as E1^E4, is synthesized at very high levels in the differentiated layers of the papilloma. E4 interferes with the cell cycle and may also be important for sustaining an S-phase like state in cells that are amplifying viral DNA (Davy et al, 2002). E4 also increases the egress of virions by weakening the viral laden cornified envelopes (Brown et al, 2006) and by disrupting keratin filaments (Doorbar et al, 1991).
Papillomavirus virions form non-enveloped icosahedral structures of 55–60nm diameter containing the genome packaged in a nucleohistone complex (Favre et al, 1977). L1 is the major capsid antigen and can self-assemble into virus like particles (Kirnbauer et al, 1992; Zhou et al, 1991). L2, the minor capsid protein, is important late in infection for packaging the viral genome into the capsid (Day et al, 1998; Zhao et al, 1998) and at early stages of infection to transport the viral DNA to the nucleus to establish a permissive infection (Day et al, 2004).
E. Modes of Viral DNA Replication
The papillomavirus life cycle requires three different modes of viral DNA replication which can be defined as “Establishment”, “Maintenance” and “Amplification”. Upon initial infection, the incoming viral genome is transported to the nucleus where it undergoes a limited amplification to a low copy number to establish the infection. In the second stage of replication, these genomes are replicated and maintained at a constant copy number in mitotically active basal cells. And finally, as the infected cells differentiate and mature, the genome is vegetatively amplified to a large copy number to provide progeny genomes to be encapsidated in virion particles. Some of these steps are very well understood, but others still remain elusive.
II Replication Initiation
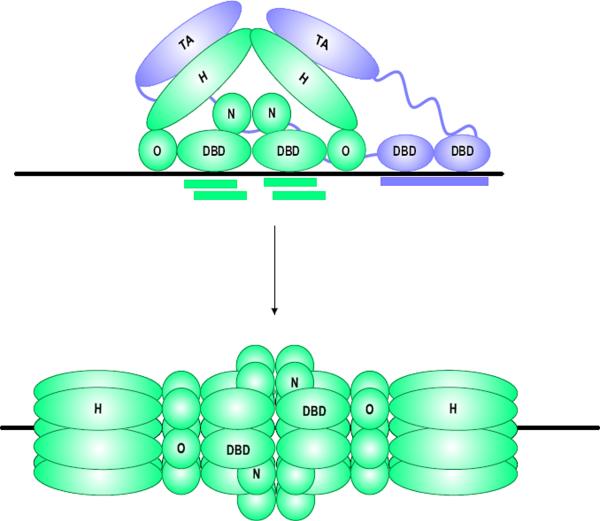
Initiation of papillomavirus DNA replication requires the replication origin, which minimally contains an E1 and E2 binding site (Mohr et al, 1990; Ustav and Stenlund, 1991; Ustav et al, 1991), and the viral E1 and E2 proteins. E1 is the primary replication protein; it is an ATP-dependent helicase that specifically binds, melts and unwinds the viral replication origin to allow access of the cellular replication proteins. Initially, E1 cooperatively binds to the origin with the E2 protein but E1 then assembles into double hexamers that encircle the DNA and bidirectionally unwind the origin.
A. The E1 Initiator Protein
The papillomavirus E1 proteins are 70-kD ATP-dependent helicases that bind specifically to the origin to initiate replication (Yang et al, 1993). They consist of four domains; an N-terminal domain, a sequence specific DNA binding domain, an oligomerization domain and a helicase domain (see Figure 3A). The replication origin contains binding sites for E1 and the loading protein, E2 (Ustav et al, 1991). As shown in Figure 4, A dimer of E1 and a dimer of E2 cooperatively bind to their adjacent sites with the N-terminal domain of E2 forming an interaction with the helicase domain of E1 (Sarafi and McBride, 1995; Sedman et al, 1997). After loading, E2 is dissociated from the complex and E1 coverts into a double hexameric ring helicase that encircles the DNA (Sanders and Stenlund, 1998; Schuck and Stenlund, 2005; Sedman and Stenlund, 1998; Titolo et al, 2000). E1 has the ability to bind DNA both specifically and non-specifically via two different DNA binding regions. Site-specific DNA binding is provided by the DNA binding domain (DBD) (Chen and Stenlund, 1998; Sarafi and McBride, 1995), which recognizes and binds to two pairs of E1 binding sites (E1 BS) in the replication origin. The helicase domain also bind DNA, but with low sequence specificity. This activity is required for the ability of the E1 helicase domain to contact the DNA sequences flanking the E1 BS, including a region that has been termed the A/T-rich region (Schuck and Stenlund, 2006). In the initial binding complex, the interaction of the E2 protein with the helicase domain masks this non-specific DNA binding function, but when E2 is disassociated, the non-specific DNA binding region is available to interact with DNA (Stenlund, 2003). An X-ray crystal structure of the BPV-1 E1 helicase, bound to DNA, has shown that single-stranded DNA is threaded through the hexamer channel and so the replication origin must be melted as E1 converts from a dimer to a double hexamer (Enemark and Joshua-Tor, 2006).
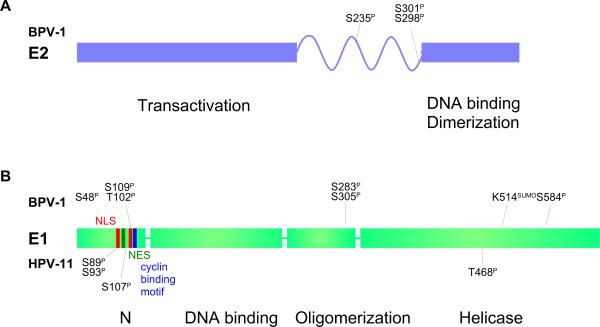
Figure 3. Domain structure of the E1 and E2 proteins.
A. The full-length E2 proteins of all papillomaviruses consist of two conserved domains linked by a less conserved hinge region. The N-terminal domain is important for transcriptional regulation and interaction with the E1 protein. The C-ternminal domain is a specific DNA binding and dimerization domain. Phosphorylation sites mapped in BPV-1 E2 are indicated (Lehman and Botchan, 1998; McBride et al, 1989).
B. The E1 proteins consist of four domains. The N-terminal domain is important for intracellular localization and contains both nuclear localization signals (NLS) and nuclear export signals (NES). The next domain is an origin specific DNA binding domain, followed by an oligomerization domain. The C-terminal domain contains the helicase function. Phosphorylation sites mapped in BPV-1 are indicated above and those mapped in HPV11 are indicated below. See text for references.
Figure 4. Initiation of DNA Replication.
Initially, dimers of the E1 and E2 proteins co-operative together to bind to their specific binding sites on the replication origin. The transactivation domain of E2 interacts with the helicase domain of E1. This complex converts to a double E1 hexamer that encircles the origin. In the absence of E2, the helicase domain can make additional, non-specific interactions with DNA surrounding the origin.
Compared to most known cellular helicases, virally encoded helicase proteins such as E1 and SV40 T antigen are unique in their ability to cause melting, as well as unwinding, of the DNA helix. For E1, a beta hairpin (also found in T antigen) pries apart the DNA strands to melt the helix (Castella et al, 2006; Liu et al, 2007). When the double hexamer has formed, each subunit of the helicase domain projects its beta hairpin into the channel of the helicase in a right-handed staircase pattern. Each hairpin contacts adjacent nucleotides of single stranded DNA in a sequential pattern and ATP hydrolysis and ADP release causes the helicase to move along these nucleotides in what has been termed a coordinated escort mechanism (Enemark and Joshua-Tor, 2006). Each of the hexamers translocates bidirectionally with a 3' to 5' polarity to unwind the origin. The oligomerization domain provides a rigid collar around the DNA to help stabilize the interactions between the helicase domains (Enemark and Joshua-Tor, 2006). The structure of the E1 DNA binding domain and the hexameric helicase are shown in Figure 5D and 5E, respectively.
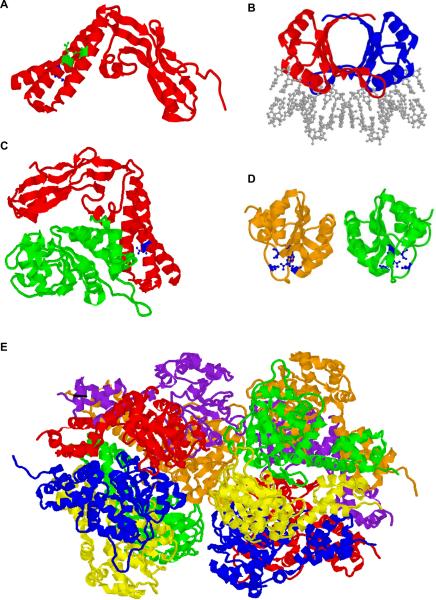
Figure 5. Structures of Papillomavirus E1 and E2 Protein Domains and Complexes.
A. The transactivation domain (residues 1–201) of the HPV-16 E2 protein from the pdb file 1DTO (Antson et al, 2000). Residues R37 and I73 are shown in green and residue D39 is shown in blue.
B. A dimer of the DNA binding/dimerization domain of BPV-1 E2 bound to DNA (residues 326–410) from the pdb file 2BOP (Hegde et al, 1992). The DNA is shown in gray.
C. The transactivation domain of HPV-18 E2 (residues 1–215; red) in complex with a c-terminal domain of HPV18 E1 (residues 428–631; green) from the pdb file 1TUE (Abbate et al, 2004). E2 residue D43 (equivalent to BPV-1 D39 in panel A) is shown in blue.
D. A dimer of the BPV-1 E1 DNA binding domain (residues 159–303) from the pdb file 1FO8 (Enemark et al, 2000). Residues shown in blue are important for DNA contact.
E. Double hexameric ring structure of the BPV-1 E1 helicase domain (residues 306–577) from the pdb file 2GXA (Enemark and Joshua-Tor, 2006). Each E1 monomer is shown in a different color.
The N-terminal domain of E1 is only required for replication in vivo (Ferran and McBride, 1998; Sun et al, 1998) and likely plays a role in regulating intracellular localization. The E1 protein shuttles from the nucleus to the cytoplasm and the N-terminal domain contains both nuclear import and export signals that are regulated by cyclin A/E-Cdk2 (Deng et al, 2004; Hsu et al, 2007; Lentz et al, 1993; Ma et al, 1999). A conserved caspase cleavage site located between residues 46 and 49 in this domain is also important for vegetative DNA replication (Moody et al, 2007) (see below).
In addition to origin recognition, melting and unwinding, E1 recruits and interacts with many cellular replication factors. Viral DNA synthesis requires replication protein A (RPA) to stabilize single stranded DNA, topoisomerase I to relieve tortional stress, and the host DNA polymerase α primase to prime replication (Clower et al, 2006; Conger et al, 1999; Han et al, 1999; Kuo et al, 1994; Loo and Melendy, 2004; Masterson et al, 1998; Melendy et al, 1995; Muller et al, 1994). Also recruited are Proliferating Cell Nuclear Antigen (PCNA), Replication Factor C (RFC) and Polymerase δ (Kuo et al, 1994).
B. The E2 Loading Factor
The papillomavirus E2 proteins regulate transcription, as well as being required for viral DNA replication. The E2 gene encodes multiple proteins that are the result of expression from different promoters and/or alternative RNA splicing. The full-length E2 protein is a polypeptide of between 350 and 500 amino acids in length, and is often referred to as E2-TA or E2 transactivator. E2-TA regulates the activity of viral promoters by binding to multiple 12bp palindromic sequences in E2-specific enhancers within the LCR. As shown in Figure 3A, E2 consists of two conserved domains; a 200 amino acid N-terminal domain, which is important for transcriptional activation and repression, interaction with the E1 protein, as well as many cellular factors, and a C-terminal domain of approximately 100 amino acids that has sequence-specific DNA binding and dimerization properties (reviewed in (McBride and Myers, 1997). The region between these domains varies in sequence and in length and is not well conserved among different genera of viruses. However, within each papillomavirus genus, the E2 hinge region contains auxiliary elements that are only conserved among related viruses. For example, there are elements important for regulated E2 degradation in BPV-1 E2 (Penrose and McBride, 2000), for nuclear localization in HPV-11 E2 (Lai et al, 1999; Zou et al, 2000), and for mitotic chromosome binding (Poddar et al, 2008) and transcriptional regulation in HPV-8 E2 (Steger et al, 2002).
The DNA binding domain of the E2 proteins forms a dimeric beta-barrel structure that positions two alpha recognition helices to contact the consensus binding site (Hegde et al, 1992). A crystal structure of the BPV-1 E2 DNA binding domain is shown in Figure 5B. The consensus binding site motif with the highest affinity has the sequence ACCGN4CGGT, with a preference for AT nucleotides in the center for many HPVs (Androphy et al, 1987; Bedrosian and Bastia, 1990; Li et al, 1989). The E2 protein also binds to the motif ACCN6GGT and binding to sites AACN6GGT, ACCN6GTT and ACAN5CGGT has also been observed (Li et al, 1989; Newhouse and Silverstein, 2001). Some viruses contain few consensus E2 sites (the genital HPVs have four conserved sites) and others contain many more (the BPV-1 genome has 17 binding sites). These sites are important for viral transcriptional regulation, initiation of replication and for viral genome partitioning and maintenance. Figure 6 shows the position of E2 binding sites in the LCR of several different papillomaviruses.
Figure 6. E1 Binding Origin Sequences.
The palindromic E1 binding origin sequence from a range of papillomaviruses is shown. The genus to which each virus belongs is shown to the right.
The E2-TA protein recruits many basic cellular transcription factors and coactivators to activate viral transcription. E2 also represses transcription from viral promoters when the E2 binding sites overlap essential promoter elements, such as the TATA box or Sp1 binding sites (Bernard et al, 1989; Cripe et al, 1987; Thierry and Yaniv, 1987). However, repression also requires a function of the transactivation domain in addition to DNA binding (Dowhanick et al, 1995; Goodwin et al, 1998; Soeda et al, 2006; Thierry and Yaniv, 1987).
The transactivation domain of the E2 proteins is also well characterized and forms a cashew shaped structure consisting of a bundle of three alpha helices in the N-terminal half linked to a beta sheet region in the C-terminal half by a region that has been termed the fulcrum (Antson et al, 2000). The structure of the E2 transactivation domain in complex with the helicase domain of E1 has also been solved (Abbate et al, 2004). Representative structures are shown in Figure 5. The regions of the transactivation domain required for transcriptional regulation and replication are well defined and separated (Abroi et al, 1996; Brokaw et al, 1996; Sakai et al, 1996). For example, E2 residues arginine 37 and isoleucine 73, located on the face of two adjacent helices in the N-terminal portion of the transactivation domain, are important for transcriptional regulation (Antson et al, 2000) whereas glutamate at position 39 on the opposite side of the domain is key for interaction with arginine 454 in the helicase domain of E1 (Abbate et al, 2004).
The E2 transactivation domain has also been implicated in self-interaction and looping of DNA containing E2 binding sites and looped E2-DNA structures have been observed by electron microscopy for BPV-1, HPV-16 and HPV-11 E2 proteins (Antson et al, 2000; Hernandez-Ramon et al, 2008; Knight et al, 1991; Sim et al, 2008). However, the precise interaction between N-terminal domains of different E2 proteins, at least in the observed crystal structures, is not consistent. In the X-ray crystal structure of the HPV16 E2 transactivation domain, intermolecular interactions were observed between residues arginine 37 and isoleucine 73, which are crucial for the transcriptional activation function (Antson et al, 2000). In contrast, a crystal structure of the BPV-1 N-terminal domain revealed a disulphide bond between cysteine residues 57, which interfered with the E1 interaction face of the domain (Sanders et al, 2007). A different monomer-monomer interface was also observed in the protein crystal lattice obtained for the HPV-11 E2 protein (Wang et al, 2004). Thus, self-interaction of the transactivation domains may be important for E2 function, but the interactions observed in X-ray structures could also be the result of non-specific crystal packaging interfaces (Janin et al, 2007).
Several E2 functions are important for initiation of viral DNA replication. As described above, E2 helps load the E1 helicase by co-operatively binding to adjacent binding motifs in the origin and by increasing the specificity of E1 DNA binding by masking its non-specific binding activity (Stenlund, 2003). E2 also enhances replication by alleviating repression by nucleosomes to allow the E1/E2 complex to bind to the origin (Li and Botchan, 1994). Furthermore, E2 interacts with RPA, a single-stranded binding protein required for DNA replication (Li and Botchan, 1993) and with Replication Factor C, RF/C (Wu et al, 2006).
C. The replication Origin
The papillomavirus replication origin contains an E1 binding site, several E2 binding sites and an A/T rich region (Mohr et al, 1990; Ustav et al, 1993; Ustav et al, 1991). The E1 binding sites consists of an 18bp palindrome that contains multiple overlapping recognition sequences for the E1 protein (Chen and Stenlund, 2001; Holt et al, 1994; Titolo et al, 2003). E1 is a monomer in solution but binds to the origin as a dimer, along with a dimer of the E2 protein. The E1 binding sites consist of overlapping hexanucleotide sequences with a consensus ATTGTT, which are separated by three nucleotides from the adjacent half of the palindrome (Auster and Joshua-Tor, 2004). The sequence is not highly conserved but, in most cases, the thymidines at position 2 and 5 are invariant. The sequences of a series of E1 binding sites are shown in Figure 7.
Figure 7. Map of the E1 and E2 binding sites in the Long Control Regions in a Range of Papillomaviruses.
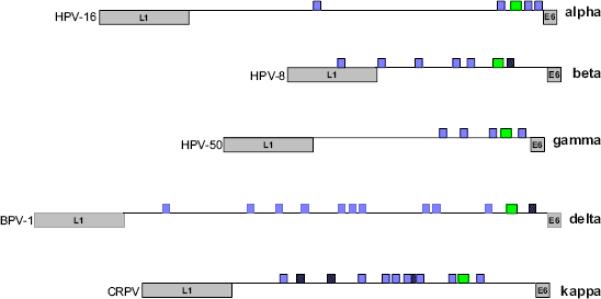
The LCRs of papillomaviruses from five different genera are shown. The end of the L1 and beginning of the E6 open reading frames are indicated. The E1 binding site is indicated as a green box. E2 binding sites that match the consensus ACCN6GGT are shown in purple. E2 sites that deviate from this consensus are shown in hatched purple.
For most papillomaviruses, at least one E2 binding site is required for transient replication and additional sites greatly stimulate replication (Akgul et al, 2003; Chiang et al, 1992; Lee et al, 1997; McShan and Wilson, 1997b; Remm et al, 1992; Russell and Botchan, 1995; Sun et al, 1996). In some cases, the E1 binding site is dispensable and two E2 binding sites can suffice (Lu et al, 1995; Sverdrup and Khan, 1995). Presumably, in the latter case the E2 protein can load the E1 helicase efficiently enough in the absence of specific E1 binding. It has also been shown that, at high levels, the E1 protein can support replication in the absence of the E2 protein (Gopalakrishnan and Khan, 1995). Again, presumably at higher concentrations the E1 protein is less dependent on E2 for loading. Although these studies provide insight into the mechanisms of viral replication, it is almost certain that in the actual viral life cycle initiation of DNA replication in the initial amplification stage requires both the E1 and E2 proteins, and their cognate binding sites. HPV-11 E2 can be observed as a large disk/ring-shaped protein particle bound to the three E2 sites adjacent to the HPV-11 origin. Inclusion of the fourth, upstream E2 site results in DNA molecules containing loops from this site to the promoter proximal sites (Sim et al, 2008).
D. Regulation of Replication Initiation
There are many ways in which papillomavirus replication can be regulated. The mechanisms described in this section could regulate initiation of DNA synthesis in the establishment phase but may also affect maintenance replication and vegetative viral DNA amplification. Additional regulatory mechanisms that are more likely to be specific for the latter modes are described later in the article.
After initial infection, the papillomavirus DNA is delivered to the nucleus in complex with the L2 minor capsid protein (Day et al, 2004) and, similar to many other viruses, localizes adjacent to the PML nuclear domains. L2 also recruits E2 to these domains (Day et al, 1998) and presumably this site is a privileged site for viral DNA replication. Transient replication studies with HPV-11, show that the E1 and E2 proteins accumulate in replication foci that are often associated with PML nuclear domains (Swindle et al, 1999). E1 localization is also regulated by sumoylation (Rangasamy and Wilson, 2000), which may regulate intranuclear localization (Rangasamy et al, 2000) and E1 access to the nuclear replication domains. Although not well understood, it is likely that localization to the subnuclear sites of viral replication is tightly regulated.
The E1 proteins are extensively regulated by posttranslational modifications and at least ten phosphorylation sites have been identified in the BPV-1 E1 protein (Lentz et al, 2006). It appears that many of these phosphorylation events regulate nucleocytoplasmic shuttling of the E1 proteins. The N-terminal domain of the E1 protein contains both a bipartite nuclear localization signal (NLS) (Lentz et al, 1993) and a nuclear export signal (NES) (Rosas-Acosta and Wilson, 2008) and localization appears to be regulated by phosphorylation (Bian et al, 2007; Deng et al, 2004; Hsu et al, 2007; Yu et al, 2007a). The cell cycle kinases that modify the cellular MCM helicase complex, cyclin E/cdk2 and cyclin A/cdk2, also phosphorylate the E1 proteins from several papillomaviruses (Cueille et al, 1998; Lin et al, 2000; Ma et al, 1999). The E1 proteins are also modified by CK2 (Lentz, 2002; McShan and Wilson, 1997a), mitogen-activated protein kinases (Yu et al, 2007a), protein kinases A and C (Zanardi et al, 1997) and p34cdc2 (Lentz et al, 1993).
Initiation of replication in the establishment phase is dependent on the levels of the E1 and E2 proteins. Levels of both proteins can be regulated at the transcriptional and translational levels, but there is strong evidence that turnover of both proteins is tightly regulated. The BPV-1 E1 protein is quite short-lived and is a target of the ubiquitin ligase APC (Mechali et al, 2004). However, when it is bound to cyclin E-cyclin-dependent kinase 2 (Cdk2) before the start of DNA synthesis, E1 becomes resistant to ubiquitin-mediated degradation. Thus, APC controls the levels of E1, and this might be important to limit the level of amplificational replication and/or to maintain a constant low copy number of the viral genome during the maintenance phase of replication. The half-life of the E2 protein is also regulated by CK2 phosphorylation (Penrose et al, 2004), which in turn regulates the levels of replication and genome copy number. The mechanism will be described in more detail below.
Both the E1 and E2 open reading frames encode truncated products that can potentially negatively regulate replication. For example, some papillomaviruses encode a truncated E1 protein, designated E1-M or E1 modulator, which consists of the N-terminal domain of the E1 protein (Lusky and Botchan, 1986; Rotenberg et al, 1989). However, although earlier studies indicated that the BPV-1 E1-M protein might modulate maintenance replication (Berg et al, 1986), subsequent studies showed that it was non-essential (Hubert and Lambert, 1993). Most papillomaviruses also encode truncated E2 protein products. For example, BPV-1 encodes two E2 “repressor” proteins. One, E2-TR, is expressed from an internal promoter and initiation codon (Lambert et al, 1987) and the other is encoded by a spliced message that fuses 11 amino acids from the E8 open reading frame to the C-terminal domain of E2 (Choe et al, 1989; Lambert et al, 1990). All E2 proteins contain the DNA binding and dimerization domain and so the shorter forms of the protein can antagonize the activities of the E2-TA protein, either by competing for binding to the DNA sites or by forming heterodimers (Barsoum et al, 1992; Lambert et al, 1987; Lim et al, 1998). Similar truncated E2 proteins that can regulate both transient amplification replication and stable episomal maintenance are encoded by the HPVs. For example, an E1M^E2C protein encoded by HPV-11 (Chiang et al, 1992), an HPV-31 E8^E2C protein (Stubenrauch et al, 2000) and a CRPV E9^E2C protein (Jeckel et al, 2003) all regulate replication of their respective viruses. The HPV-31 E8^E2 protein has been studied in some detail and it seems that the E8 residues contribute specifically to this function and it is not simply due to direct interference with E2 DNA binding domain functions (Zobel et al, 2003). Of note is the fact that the BPV1 E2 binding sites adjacent to the origin have relatively weak affinity for the E2 protein compared to other sites (Li et al, 1989) and binding is only efficient in the presence of the E1 protein (Sarafi and McBride, 1995). Since the E2 transactivation domain is required for efficient co-operative binding with E1 (Winokur and McBride, 1996), this would preclude binding of the shorter E2-TR proteins to these sites. This mechanism could allow the E2 repressor proteins to regulate E2-TA binding at other sites (e.g. for transcription) without interfering with replication.
Papillomavirus replication is also regulated by the p53 protein. P53 inhibits transient amplificational replication of BPV-1 and HPV-11 but does not affect stable maintenance replication (Ilves et al, 2003). An intact DNA-binding and oligomerization domain of p53 is necessary for repression, while the N-terminal transcription activation and C-terminal regulatory domains are dispensible (Ilves et al, 2003). Replication inhibition can be mediated by direct binding of p53 to the HPV16 E2 protein (Brown et al, 2008)or, as in the case of HPV-8, by competitively binding to a p53 site adjacent to the origin which overlaps an E2 site (Akgul et al, 2003). This inhibition may be important to limit the amount of replication in the newly infected basal cells. The papillomavirus E6 proteins bind, and in some cases degrade, the p53 protein. In turn, the E2 protein modulates transcriptional expression of the E6/E7 genes. These components could form a regulatory loop to balance and limit replication (Ilves et al, 2003).
Papillomavirus replication is also regulated by cellular factors such as the TATA-binding protein (TBP), Yin-yang 1 (YY1) and the CCAAT displacement protein (CDP). TBP prevents formation of the HPV 11 E1-E2 complex at the origin of replication by antagonizing E2 binding to the origin (Hartley and Alexander, 2002) and YY1 can inhibit replication by sequestering the E2 protein (Lee et al, 1998; O'Connor et al, 1996). CDP represses transcription and replication of several papillomaviruses (Narahari et al, 2006; O'Connor et al, 2000; Pattison et al, 1997) by binding to a site that overlaps the E1 binding site in the replication origin of many papillomaviruses. Binding of the E2 proteins to the origin region helps to relieve repression by displacing CDP (Narahari et al, 2006). Since CDP is expressed in undifferentiated but not in differentiated cells (Ai et al, 1999), keratinocyte differentiation would alleviate repression repression and permit vegetative genome amplification. Clearly there is much interplay between regulation of transcription and replication in the early stages of the papillomavirus life cycle. Cellular proteins inhibit replication both directly and indirectly by modulating expression of the E1 and E2 replication proteins. In many cases, the viral E6 and E7 proteins are equipped to counteract the effects of these cellular repressors and the E2 protein itself can alleviate repression. At a certain level, the E2 protein downregulates E6 and E7 expression and so all of these processes can reach equilibrium.
III Maintenance Replication
After the initial amplification step, the viral genome must be maintained in the dividing basal cells to sustain a persistent infection. The genomes can be maintained indefinitely at a constant copy number as extrachromosomal elements. The genomes are very difficult to detect by in situ techniques in the basal cells of a papilloma and thus there are estimated to be less than twenty copies per cell (Evans et al, 2003). A few cell lines that have been isolated from clinical lesions also maintain the viral genome extrachromosomally (Bedell et al, 1991; Stanley et al, 1989), as do keratinocytes transfected with the viral genome (Flores et al, 1999; Frattini et al, 1996; Meyers et al, 1997). The genome replicates in these cells in culture at about 50 to 200 copies per cell.
As described above, initiation of viral DNA replication requires the E1 and E2 proteins and the origin of replication. However, plasmids containing the minimal origin will only replicate transiently and the replicated DNA is lost with time. Long-term, stable replication of BPV-1 requires multiple E2 DNA binding sites in cis to the origin (Piirsoo et al, 1996). This observation was pivotal in identifying that the E2 protein had a separate function in viral genome maintenance. It was subsequently shown that E2 partitions and maintains the viral genomes by attaching them to mitotic chromosomes (Ilves et al, 1999; Skiadopoulos and McBride, 1998). A model of E2-mediated viral genome partitioning is shown in Figures 8A and 8B.
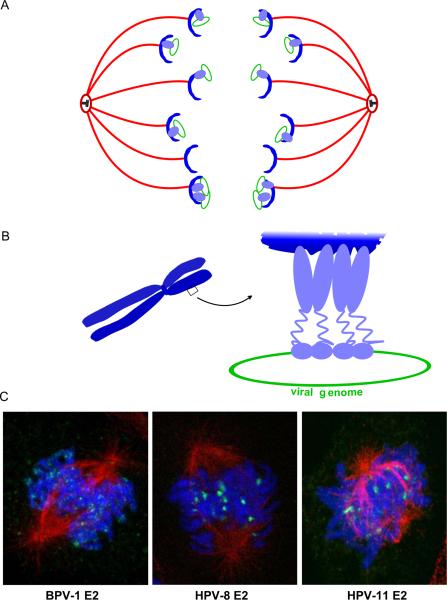
Figure 8. Mechanism of Papillomavirus Genome Partitioning.
A. Model for partitioning of papillomavirus genomes. Cellular chromosomes (blue crescents) are partitioned by attachment to the spindle in mitotis. The viral E2 protein (purple) binds to the viral genome and tethers it to mitotic chromosomes, thus hitchhiking on the cellular chromosomes.
B. Model for chromosomal tethering by the BPV-1 E2 protein. The E2 DNA binding domain binds to sites in the viral genome while the transactivation domain interacts with proteins, such as Brd4, on mitotic chromosomes.
C. Binding patterns of three different E2 proteins, as detected by immunofluorescence, on mitotic chromosomes. BPV-1 E2 forms small speckles on the arms of all chromosomes. HPV-8 E2 is observed in fewer, larger speckles that bind adjacent to the centromere of acrocentric chromosomes. HPV-11 E2 is also observed binding to the pericentromeric region of chromosomes, but only in certain fixation conditions (Oliveira et al, 2006). The E2 proteins are shown in green, cellular DNA in blue and the mitotic spindle in red.
It can be argued that a specific viral genome partitioning mechanism is not required when there are sufficient copies of the viral genome. In this scenario, each daughter cell is likely to randomly contain at least one genome, which can be amplified to a level established by a copy number control mechanism. However, compared to cell lines, the viral genome copy number in the basal cells of papillomas is virtually undetectable (Evans et al, 2003). Furthermore, it has been shown that the E2 protein tethers the viral genomes to mitotic chromosomes. This ensures that the viral genomes are partitioned to daughter cells in approximately equal numbers and guarantees that they are retained in the nucleus after cell division. Furthermore, this strategy of faithfully maintaining and partitioning viral genomes by tethering them to host mitotic chromosomes using a viral DNA binding protein is common to many persistent DNA viruses (McBride et al, 2006).
A. Role of the E1 Protein in Maintenance Replication
In general, it has been assumed that initiation of replication of the viral DNA in the maintenance phase occurs in an E1 and E2-dependent manner, similar to that described for the establishment phase of replication. However, it is also feasible that viral DNA synthesis could be initiated by cellular proteins in the maintenance phase and that viral functions are only required for retention and partitioning of the viral genomes. Evidence that E1 might not be required for maintenance replication comes from a study with a BPV-1 genome encoding a temperature sensitive E1 protein (Kim and Lambert, 2002). It was observed that while the E1 function was required for establishment of the genomes, they could be maintained as extrachromosomal elements at the non-permissive temperature, thus, indicating that E1 was not absolutely required. Other evidence that the modes of replication initiation might be different comes from the observation that p53 can inhibit amplificational replication but does not seem to repress stable episomal maintenance (Ilves et al, 2003). In general, the requirements for initiation of replication may be less stringent than previously presumed as recent studies have revealed that cellular origins may be a passive result of local chromatin structure rather than well defined sequence elements (Gilbert, 2004).
B. The E2 Tethering Protein
The first clue to the precise role of E2 in genome partitioning was the observation that both the BPV-1 E2 protein and the viral genomes are observed as small speckles over the arms of all mitotic chromosomes (Skiadopoulos and McBride, 1998; Zheng et al, 2005). Further analysis showed that E2 binds mitotic chromosomes through protein-protein interactions mediated by the transactivation domain. The DNA binding domain binds to multiple E2 binding sites in the viral genome (in the MME) and tethers it to the condensed chromosomes (Bastien and McBride, 2000; Ilves et al, 1999). In cells expressing E2, plasmids containing at least eight E2 binding sites are maintained and segregated and are observed bound to mitotic chromosomes (Ilves et al, 1999; Piirsoo et al, 1996). Examples of the the pattern of mitotic chromosome binding of different E2 proteins are shown in Figure 8.
C. Genome Partitioning in Different Papillomaviruses
It was initially assumed that all papillomaviruses would maintain their genomes using a conserved mechanism similar to BPV-1. However, characterization of E2 tethering in other papillomaviruses has revealed that, although most E2 proteins associate with mitotic chromosomes, the E2 chromosomal target varies for different papillomaviruses. A study of the mitotic chromosomal binding pattern of E2 proteins from thirteen different papillomaviruses showed that most E2 proteins, from very different genera and host species, were found bound to cellular mitotic chromosomes. However, the binding pattern varied among different E2 proteins (Oliveira et al, 2006). For example, BPV-1 E2 is detected in many small speckles over the arms of all chromosomes but HPV-8 E2 binds primarily to the peri-centromeric region of a subset of chromosomes. The binding target of the alpha-papillomaviruses, which infect the mucosal epithelia and include the cancer associated “high-risk” HPVs, has been more controversial. Using conditions similar to those used for the BPV-1 studies, the alpha papillomaviruses E2 proteins are only found in association with mitotic chromosomes in early (prophase) and late (telophase) stages of mitosis (Donaldson et al, 2007; Gammoh et al, 2006; Oliveira et al, 2006). In theory, the association of the E2 protein/viral genome complex with telophase chromatin would be sufficient to ensure that the genomes are partitioned and maintained in the nucleus. However, under certain fixation conditions, the alpha E2 proteins are observed in a pericentromeric pattern on metaphase chromosomes (Oliveira et al, 2006) and so may have a target similar to that of the beta-papillomaviruses (as exemplified by HPV-8 E2). The E2 proteins of the alpha-papillomaviruses may need a specialized cellular environment or additional cellular or viral factors to stabilize their association with mitotic chromosomes. Notably, stable extrachromosomal genome replication of these viruses also requires E6 and E7 gene functions (Park and Androphy, 2002; Thomas et al, 1999). Unfortunately, the E2 proteins are very difficult to detect when expressed from replicating viral genomes and so it is still not clear whether the localization observed reflects the mechanism of genome partitioning used by these viruses.
A different study of the alpha-papillomavirus HPV11 E2 protein demonstrated that both the N-terminal and C-terminal E2 domains, when fused to GFP, could independently associate with the mitotic spindle rather than the chromosomes (Van Tine et al, 2004). This localization was found to be dynamic, similar to many cellular chromosomal passenger proteins, and by late stages of mitosis the C-terminal domain of E2 was observed associated with the central spindle microtubules and by cytokinesis was located in the midbody (Dao et al, 2006). A similar study reports the colocalization of a small percentage of the BPV-1 E2 protein with the central spindle microtubules, in complex with the mitotic kinesin-like protein, MKlp (Yu et al, 2007b). Whether this microtubule localization of E2 is involved in genome partitioning remains to be determined as genomes tethered to E2 through this interaction would ultimately be stranded in the cytoplasm of cells.
D. Other Viral Tethering Proteins
Remarkably, the strategy of maintaining and partitioning extrachromosomal viral genomes by tethering them to cellular chromosomes is a common strategy among diverse DNA viruses that cause persistent infection. This has been shown for the gamma herpesviruses Epstein-Barr virus (EBV), Kaposi's sarcoma associated Herpesvirus (KSHV), Herpesvirus saimiri (HVS) and murine gamma herpesvirus-68 (MHV-68). Each virus encodes a DNA binding protein that binds specifically to repeated sites in the viral DNA and tethers the genome to the cellular mitotic chromosomes. The best studied of these herpesvirus tethering proteins are EBNA-1 and LANA, from EBV and KHSV, respectively. It has been proposed that EBNA-1 interacts with mitotic chromosomes in two different ways. The nucleolar proliferation antigen, p40, otherwise known as EBP2, is the major chromosomal protein target for EBNA-1 (Wu et al, 2000). However, the regions of EBNA-1 that bind mitotic chromosomes have also been proposed to be AT hooks that directly interact with the cellular DNA (Sears et al, 2004). LANA interacts with a number of different mitotic chromosome binding proteins such as histone H1, DEK, Methyl CpG binding protein, Brd2, Brd4 and a H2A/H2B histone dimer (Barbera et al, 2006; Cotter and Robertson, 1999; Krithivas et al, 2002; Platt et al, 1999; You et al, 2006). These targets are summarized in Table 1.
TABLE 1.
Chromosomal Targets of Viral Tethering Proteins
| Virus | Tethering Protein | Putative Target or Accessory factor | Ref |
|---|---|---|---|
| Papillomaviruses | |||
| BPV-1 | E2 | Brd4 | (Baxter et al, 2005; You et al, 2004) |
| HPV-8 | E2 | rDNA | (Poddar et al, 2008) |
| HPV-11 | E2 | Mitotic spindle | (Van Tine et al, 2004) |
| BPV-1, HPV-16 | E2 | ChLR1 | (Parish et al, 2006) |
| Herpesviruses | |||
| Epstein-Barr Virus, EBV |
EBNA-1 | hEBP2 (p40) A-T hook |
(Kapoor and Frappier, 2003) (Sears et al, 2004) |
| Kaposi's sarcoma associated herpesvirus (HHV-8) | LANA | Brd2 Brd4 Histone H1 Histones H2A/B DEK, MeCBP NuMA |
(Platt et al, 1999; Viejo-Borbolla et al, 2005) (You et al, 2006) (Cotter and Robertson, 1999) (Barbera et al, 2006) (Krithivas et al, 2002) (Si et al, 2008) |
| Herpesvirus saimiri | ORF 73 | MeCP2 | (Calderwood et al, 2004; Griffiths and Whitehouse, 2007) |
The EBNA-1 and LANA tethering proteins have many features in common with E2. In addition to genome tethering, each protein regulates viral transcription and is involved in initiation of viral DNA replication (although the herpesviruses have no E1 counterpart and use cellular proteins to initiate replication). Despite having no sequence homology, the DNA binding domains of E2 and EBNA (Bochkarev et al, 1995) and the putative structure of LANA (Grundhoff and Ganem, 2003) form similar dimeric anti-parallel beta-barrel DNA binding structures.
E. The Role of Cis Elements in Genome Partitioning
The LCR of BPV-1 contains 11 E2 binding sites (Li et al, 1989); at least eight of which are required for genome maintenance and have been mapped to an element designated the Minichromosome Maintenance Element or MME (Piirsoo et al, 1996). Four E2 sites are sufficient for E2-mediated plasmid maintenance in yeast (Brannon et al, 2005). However, yeast undergoes a “closed mitosis” in which the nuclear membrane does not break down and this might result in less stringent requirements for plasmid maintenance. The herpesvirus tethering proteins, EBNA-1 and LANA, also bind to large numbers of tandem binding sites in their respective viral genomes to link them to the host chromosomes.
Most other papillomaviruses contain only four consensus E2 binding sites and it is not yet clear whether this would be sufficient for stable genome maintenance using a mechanism similar to BPV-1. Fewer sites may suffice if these viruses use different chromosomal targets and mechanisms to partition their genomes. But these genomes may also contain additional elements that bind cellular or viral proteins and stabilize the tethering complex. It is likely that for many of the papillomaviruses, the E2 binding sites will not be sufficient for stable genome maintenance. One potential candidate cis elements is a Matrix attachment region (MAR). These DNA sequences anchor chromatin fibers to the nuclear matrix and generate transcriptionally active domains. MARs have been mapped in papillomavirus genomes in vitro (Tan et al, 1998). There is also precedent that these elements could be important for viral genome maintenance; plasmids containing cellular MAR elements are stably maintained in cells over many cell generations by attachment to cellular mitotic chromosomes (Baiker et al, 2000) via an interaction with the scaffold attachment protein SAF-A (Jenke et al, 2002).
Several studies have shown that sub-genomic fragments of HPV DNA can replicate autonomously in yeast in an E1 and E2 independent manner (see Section V.D. below). A recent study has shown that similar multiple sub-genomic fragments of HPV16 DNA can also replicate and/or persist in mammalian cells, in the absence of E1 and E2, further implicating the role of cellular factors in binding to viral cis elements (Pittayakhajortwut and Angeletti, 2008). Sequences such as MAR elements, HMG, Topoisomerase II, Telomere-repeat binding factor and CENP-B binding sites have been identified in the HPV16 genome and could be potential candidates for binding to cellular genome maintenance factors (Pittayakhajortwut and Angeletti, 2008).
Another potential mechanism of regulation is methylation of the E2 binding sites. E2 is unable to bind to its consensus sequence when it contains a methylated CpG dinucleotide (Kim et al, 2003; Thain et al, 1996). In replicating cells maintaining the HPV16 gemone as an episome, the E2 binding sites, especially binding site 2, were often methylated. In contrast, in differentiated cells the genomes become hypomethylated (Kim et al, 2003). This may also indicate that the E2 binding sites are not crucial for genome maintenance.
One of the difficulties in designing a papillomavirus segregation assay is the multifunctional nature of the E2 protein. Because E2 is also important for replication initiation and transcriptional activation, it is necessary to separate these functions to study the role of E2 in genome maintenance. Analysis of the roles of individual E2 binding sites on long-term replication of the viral genome is difficult because the sites are also required for replication initiation and for transcriptional regulation of the virus, including regulation of expression of the E1 and E2 proteins themselves. In HPV31, mutation of each E2 site, except BS2, resulted in integration of the viral genome (Stubenrauch et al, 1998b). To date, the only successful partitioning assay is for BPV1 (Abroi et al, 2004).
F. Papillomavirus Chromosomal Tethering Targets
Disruption of the chromosomal interaction of the papillomavirus E2/genome tethering complex could have great therapeutic potential and so an important goal has been to identify and characterize the cellular chromosomal targets that mediate viral genome segregation. Table 1 lists chromosomal targets for a range of papillomavirus E2 proteins as well as proposed targets for other viral tethering proteins.
1. Brd4
The best characterized chromosomal target, to date, is the cellular protein, Brd4. Brd4 is a major component of the tethering complex of BPV-1, and probably a subset of other papillomaviruses (Baxter et al, 2005; McPhillips et al, 2006; You et al, 2004). Brd4 is a double bromodomain protein that binds to acetylated histone tails of histones H3 and H4 and, in many cell types, remains bound to chromatin through mitosis (Dey et al, 2003). Brd4 is a member of the BET family of double bromodomain proteins (Florence and Faller, 2001) that “read” the histone code by binding to acetylated residues. Brd4 has been isolated in complex with the replication factor, RFC (Maruyama et al, 2002) and as part of the mouse transcriptional Mediator complex (Jiang et al, 1998). It is also a component of P-TEFb (Jang et al, 2005) and stimulates RNA polymerase II-dependent transcriptional elongation.
In many cells, Brd4 is detected as a diffuse coat around mitotic chromosomes and it has been hypothesized to play a role in epigenetic memory (Dey et al, 2003). In the presence of the BPV-1 E2 protein, both E2 and Brd4 colocalize on mitotic chromosomes in punctuate spots (McPhillips et al, 2005) and analysis of mutated E2 proteins shows that the mitotic chromosome binding activity of E2 correlates strongly with Brd4 interaction (Baxter et al, 2005). Two highly conserved residues in the transactivation domain, R37 and I73, previously shown to be crucial for transcriptional activation, are critical for Brd4 binding in all E2 proteins tested (Baxter et al, 2005; McPhillips et al, 2006; Senechal et al, 2007). In fact, interaction of E2 with Brd4 is also crucial for E2-mediated transcriptional regulation in all E2 proteins analyzed (Ilves et al, 2006; McPhillips et al, 2006; Schweiger et al, 2006; Wu et al, 2006).
The dimerization of E2 through the E2 C-terminal DNA binding domain promotes efficient binding of E2 to Brd4 and to mitotic chromosomes (Cardenas-Mora et al, 2008). Furthermore, novel E2 “single chain heterodimer” proteins containing one transactivation domain linked to a dimeric DNA binding domain binds Brd4 much less efficiently than the wildtype E2 protein (Kurg et al, 2006). In support of this, an E2-Brd4 heterotetramer, with two Brd4 peptides sandwiched between two E2 N-terminal domains, was recently observed in a crystallographic structure of the HPV16 E2 transactivation domain in complex with a C-terminal 20 amino acid peptide from Brd4 (Abbate et al, 2006). Dimerization of the E2 protein through the C-terminal domain would greatly stabilize such a complex. Cooperative binding of E2 and Brd4 into higher order complexes could explain why the Brd4-E2 complex is observed as punctuate speckles on mitotic chromosomes (McPhillips et al, 2005), whereas in the absence of E2 it forms a diffuse coating (Dey et al, 2000).
When exogenously expressed, the C-terminal E2 binding region of Brd4 interferes with the E2-Brd4 interaction, the association of E2 and viral genomes with mitotic chromosomes, BPV-1-induced cellular transformation and viral genome maintenance (You et al, 2004; You et al, 2005). E2 is unable to maintain plasmids containing E2 binding sites in Saccharomyces cerevisiae but this function can be reconstituted with Brd4 expression (Brannon et al, 2005). Thus, the E2-Brd4 association is pivotal to BPV-1 maintenance replication. However, while Brd4 is clearly an important component of the tethering complex for several papillomaviruses, it is not clear that it is the key link between the E2/genome complex and the cellular chromosomes. Brd4 is not a stable tether, interacting only with histones that are acetylated (Dey et al, 2003) and most of which become deacetylated during mitosis (Kruhlak et al, 2001). Perhaps because of this, the interaction of E2 and Brd4 is not passive and E2 actively stabilizes the interaction of Brd4 with chromatin and redistributes it into punctate dots on the mitotic chromosomes (McPhillips et al, 2005). Thus, the E2/genome complex does not passively hitchhike on mitotic chromosomes but instead modifies and stabilizes the Brd4-chromatin interaction to ensure the stable transmission of the viral genome. It has also not been conclusively shown that disruption of the E2-Brd4 interaction directly impacts viral genome maintenance in mammalian cells. Inhibition of the E2-Brd4 interaction by either downregulation of Brd4 by siRNA, or by expression of the dominant negative CTD of Brd4 results in loss of viral genomes, but it is not clear whether this is due to inhibition of the transcriptional function of E2, which also requires Brd4, or whether it is due to a direct inhibition of chromosomal tethering (Schweiger et al, 2006; You et al, 2004; You et al, 2005). A peptide from Brd4, corresponding to the E2 interacting region, was also able to displace HPV16 genomes from mitotic chromosomes (Abbate 2006), but it has yet to be shown that alpha HPV16 E2 protein binds to mitotic chromosomes in a similar pattern.
Brd4 is clearly an important part of the BPV-1 E2 tethering complex but it does not seem to be involved in genome tethering of all papillomaviruses (McPhillips et al, 2006). The E2 proteins of some viruses (EEPV, ROPV, HPV-1a) show complete colocalization of E2 and Brd4 on mitotic chromosomes as is seen for BPV1, others some show partial colocalization and others show none (McPhillips et al, 2006). Furthermore, mutations in E2 that abrogate Brd4 binding (in R37 and I73 residues) do not affect mitotic chromosomal localization for E2 proteins from viruses such as HPV8 and HPV31 (McPhillips et al, 2006). Consistent with these findings, an HPV31 genome containing a mutation in E2 residue 73 (I73L), which disrupts the interaction of E2 and Brd4 (Senechal et al, 2007), is maintained as a stable episome and can be amplified in differentiated keratinocytes (Stubenrauch et al, 1998a). In CRPV, however, E2 mutations that abrogate the Brd4 interaction have a detrimental effect on the induction of tumors (a domestic rabbit model resulting in abortive infection) suggesting that in CRPV the E2-Brd4 interaction is important for some aspects of pathogenesis (Jeckel et al, 2002). Notably, the CRPV E2 protein interacts with Brd4 much more efficiently that the alpha papillomavirus E2 proteins and so may have a more significant role in the CRPV life cycle (McPhillips et al, 2006). In a different study, HPV11 E2 was observed bound to mitotic microtubules but also did not colocalize with Brd4 (Dao et al, 2006). Nevertheless, all E2 proteins tested interact with Brd4, albeit with different binding efficiencies. This is because Brd4 is required for E2-mediated transcription, but not genome partitioning, of all papillomaviruses.
2. rDNA Loci
Oliveira et al. observed that certain E2 proteins, as exemplified by HPV-8, associate with the pericentromeric region of mitotic chromosomes rather than being distributed over the chromosome arms (Oliveira et al, 2006). Further investigation has shown the peri-centromeric binding target to be the ribosomal RNA gene loci on the short arms of acrocentric chromosomes (Poddar et al, 2008). This interaction does not require the Brd4 protein and is an attractive target for chromosomal tethering for several reasons. The tandemly-arranged repeating rDNA units (400 per diploid cell) greatly increase the local concentration of theE2 binding target. Also, the rDNA loci have specialized mechanisms for replication, cohesion, condensation and segregation (D'Amours et al, 2004; Freeman et al, 2000; Kobayashi et al, 1998; Sullivan et al, 2004; Torres-Rosell et al, 2005) and RNA polymerase I transcription factors remain bound throughout mitosis (McStay, 2006). The resulting, unique chromatin structure could provide unique targets on the host chromosomes. Furthermore, the rDNA genes are sequestered in the nucleoli in interphase, which would leave the E2 protein free to participate in viral transcription and replication until the nucleolar envelope breaks down in early mitosis. Notably, the cellular target of the Epstein-Barr virus tethering protein, EBNA-1 is a protein termed p40 nucleolar proliferation antigen, or hEBP2, and is involved in pre-RNA processing (Shire et al, 1999). hEBP2 is nucleolar in interphase (Chatterjee et al, 1987) but is distributed along condensed chromosomes in mitosis (Wu et al, 2000). The C-terminal domain of the LANA tethering protein of HHV-8 has also been shown to bind to pericentromeric and telomeric regions of certain metaphase chromosomes (Kelley-Clarke et al, 2007), some of which the authors speculate might be acrocentric chromosomes. In certain conditions, the E2 proteins from the alpha-genus, are also observed binding to pericentromeric regions of chromosomes (Oliveira et al, 2006). Thus, the rDNA loci might be an attractive tethering target for several papillomaviruses.
3. ChLR1
Another cellular protein that is involved in papillomavirus genome partitioning is ChLR1, a DNA helicase involved in sister chromatid cohesion (Parish et al, 2006). E2 interacts with and colocalizes with ChLR1, but only at early stages of mitosis. Furthermore, BPV-1 genomes that encode an E2 protein which is unable to interact with ChLR1 are not maintained extrachromosomally and downregulation of hChLR1 results in viral genome loss. Therefore, hCHLR1 may be important for loading, but not maintaining, the E2/genome complex on mitotic chromosomes.
4. Mitotic Spindle
In a different study, several of the alpha-papillomavirus E2 proteins have been observed to associate with the mitotic spindle (Van Tine et al, 2004). Both the N-terminal and C-terminal domains of HPV-11 E2 can associate with the microtubules and a short sequence in the DNA binding domain important for this interaction has been mapped (Dao et al, 2006). Notably, this region is not well conserved in BPV-1 E2. The E2 proteins have also been observed to bind to the centrosome (Donaldson et al, 2007; Van Tine et al, 2004). Whether this localization reflects the tethering function of E2, or the site of its proteasomal degradation, (Fabunmi et al, 2000) remains to be determined.
G. Replication Licensing
Maintenance papillomavirus DNA replication is coupled to that of the host cell and, on average, each genome is replicated once per cell cycle to give an overall constant copy number. This copy number could be maintained if replication depends on the availability of a limiting factor; genomes would replicate randomly and replication would cease when this factor is exhausted. An alternative scenario is that each genome is licensed and is only replicated once per cell cycle. Early studies indicated that BPV-1 was subject to replication licensing in mouse cells (Botchan et al, 1986), but further studies showed that when cells were isolated after S-phase a portion of the genomes had undergone multiple rounds of replication indicating a random choice mechanism (Gilbert and Cohen, 1987; Ravnan et al, 1992). A more recent study analyzed the mode of replication of the alpha papillomaviruses, HPV-16 and HPV-31. However, the results were mixed in that the different genomes seemed to have different modes of replication and this could change in different cell types (Hoffmann et al, 2006). One hypothesis is that the mode of replication is actually determined by the host cell and that lines derived from infection of transit amplifying cells give rise to random choice viral replication whereas infection of keratinocyte stem cells results in genomes that undergo replication licensing (Hoffmann et al, 2006). The genomes of Epstein-Barr virus (EBV) are also maintained as episomes and each genome is replicated in an ordered once per cell cycle fashion (Yates and Guan, 1991). However, latent EBV replication is initiated by the cellular ORC and MCM proteins, which are normally licensed in each cell cycle. Since papillomaviruses encode their own initiator protein, it is less likely that E1-dependent replication would be subject to licensing.
H. Regulation of Genome Copy Number and Partitioning
It is not well understood how papillomavirus genome copy number is maintained and regulated. For BPV-1, E2 protein levels correlate with genome copy number and greatly increased amounts of E2 protein and viral genomes are observed tethered to the host chromosomes when the half-life of the E2 protein is increased (Penrose and McBride, 2000; Skiadopoulos and McBride, 1998). Mutation of a CK2 phosphorylation site in the BPV-1 E2 protein greatly reduces protein degradation by the proteasome and results in a protein with a much longer half-life and a virus with greatly increased genome copy number (McBride and Howley, 1991). However, the E2 CK2 phosphorylation site is conserved only among the delta papillomaviruses, which cause fibropapillomas. Nevertheless, the levels of all E2 proteins are observed to dramatically decrease in mitosis (Bellanger et al, 2001). Since the E2-genome complex must be tethered to the host chromosomes throughout the length of mitosis, it is very likely that these processes are tightly regulated during persistent infection. Expression of the E1 protein of BPV-1 interferes with E2 protein-mediated tethering of the viral DNA to mitotic chromosomes, introducing yet another level of regulation to papillomavirus genome partitioning (Voitenleitner and Botchan, 2002).
The truncated E2 repressor proteins can also regulate genome copy number and may be important to limit runaway replication. BPV-1 containing mutations that eliminate expression of the E2-TR protein replicate at greatly increased copy number (Lambert et al, 1990; Riese, II et al, 1990). In HPV31, the E8^E2 protein also regulates genome copy number (Zobel et al, 2003). The E2 repressor proteins contain the DNA binding and dimerization domain of E2 and could potentially regulate replication either by directly forming heterodimers with the full-length E2 protein or by competing for binding to sites in the viral genome (Lim et al, 1998).
IV. Vegetative Replication
The third stage of viral replication is vegetative DNA replication, where viral genomes are amplified to a high copy number destined to be packaged in the capsids of progeny virions. Vegetative DNA replication occurs only in the differentiating layers of a papilloma and less is known about this mode of replication. Vegetative DNA replication will take place in organotypic raft cultures and in xenografts of mice (McBride et al, 2000; Meyers et al, 1992), but usually requires that genomes are maintained extrachromosomally at earlier stages of the infection. This makes it difficult to separate the requirements for the three modes of replication.
By studying replicative intermediates, some studies have reported that there is a switch from bidirectional theta replication in the maintenance stage to a rolling circle mode in the vegetative stage (Dasgupta et al, 1992; Flores and Lambert, 1997). The rolling circle mechanism is an efficient method for generating large amounts of viral DNA; for example, bacteriophage lambda replicates in a bidirectional theta mode early in infection, and then switches to the sigma rolling circle mode at late times. This leads to long concatamers of DNA that are cut into unit genome lengths for packaging (reviewed by (Narajczyk et al, 2007)). Similarly, herpesviruses such as Epstein Barr virus have two modes of replication and initiation switches from the latent “plasmid” origin (oriP) to the lytic origin (oriLyt) for vegetative replication (Hammerschmidt and Sugden, 1988). Vegetative replication of EBV yields intermediates of large head to tail concatemeric molecules, which are subsequently cleaved and packaged as described for lambda, and it is assumed that they are generated by a rolling circle mechanism (reviewed in (Tsurumi et al, 2005). However, initiator proteins involved in rolling circle replication usually have an associated nuclease activity and rolling circle replication (RCR) motifs (Ilyina and Koonin, 1992). Notably, the Rep initiator protein of Adeno-Associated virus (AAV), which initiates rolling circle replication, has a DNA binding domain structure very related to the SV40 T antigen and the papillomavirus E1 protein (Hickman et al, 2004). However, while the RCR motif regions of the latter two proteins are structurally homologous to the analogous region in Rep, the catalytic RCR residues have not been conserved. Therefore, it remains unclear whether papillomaviruses switch to a different replication mechanism for vegetative replication.
Vegetative replication is triggered in differentiated cells and this may be due to an increase in the levels of the E1 and E2 replication proteins. Certainly in BPV-1 infected papillomas, cells undergoing genome amplification contain greatly increased levels of the E2 protein (Burnett et al, 1990; Penrose and McBride, 2000) and BPV-1 containing cells which spontaneously amplify the viral genome also contain high levels of E2 (Burnett et al, 1990). An example of this colocalization is shown in figure 9. To date, immunological detection of the E1 protein has not been possible but mRNA species that are predicted to express both the E1 and E2 proteins are induced at this stage of infection in HPV31 (Ozbun and Meyers, 1998). E1 transcripts are expressed from both early and late promoters, but only the early promoter is repressed by the E2 protein. Therefore, a switch of transcription to the late promoter would allow E1 to be expressed to high levels that were not possible from genomes being maintained in the basal cells.
Figure 9. Vegetative DNA amplification in a BPV-1 Papilloma.
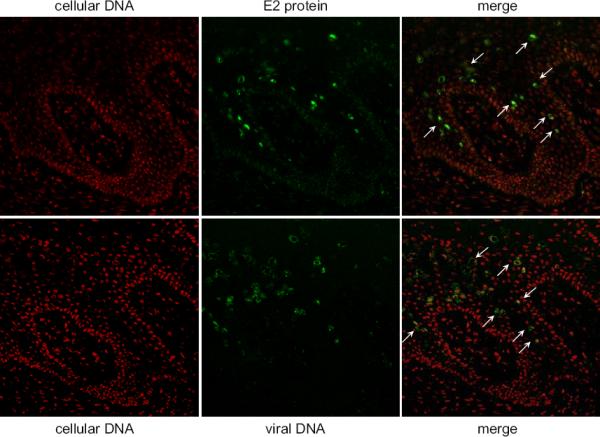
E2-specific immunofluorescence and BPV DNA-specific fluorescent in situ hybridization were performed on serial sections of bovine wart tissue. E2 protein expression is shown in green in the top panels and viral DNA is shown in green in the bottom panels. In both cases cellular DNA is counterstained in red. Cells containing high levels of both E2 protein and viral DNA are indicate by arrows.
Moody et al. recently demonstrated that HPV infection could specifically activate caspases upon differentiation and, in turn, the caspases cleaved the E1 protein in the N-terminal domain at a highly conserved DxxD motif located between residues 46 and 49 (Moody et al, 2007). Mutation of this motif inhibited genome amplification indicating that E1 cleavage might also be important for regulating vegetative replication.
V Other Aspects of Papillomavirus Replication
A. Establishment of a replication competent environment
For all modes of replication, the papillomaviruses rely on the host replication machinery. Initial genome amplification and maintenance takes place in the cycling basal cells and so the virus has available the host DNA replication factors it requires. Nevertheless, the regulatory activities of the E5, E6 and E7 proteins are likely to enhance replication, both directly and indirectly. Both the E6 and E7 proteins of HPV-16 and HPV-31 are required for long term maintenance of the viral genome (Park and Androphy, 2002; Thomas et al, 1999). E6 and E7 could indirectly enhance replication by promoting cellular proliferation and division but they could also interfere with inhibitory factors such as TBP, CDP, YY1 and p53 (Hartley and Alexander, 2002; Ilves et al, 2003; Narahari et al, 2006).
The cell cycle regulatory functions of the E6 and E7 proteins are absolutely required for vegetative amplification in differentiated cells as these cells would normally have withdrawn from the cell cycle and the E6 and E7 proteins are required to maintain or induce an pseudo S-phase like state so that the cellular replication machinery is available. In addition, the expression of the E1^E4 protein can cause a G2 arrest and so expression of both proteins in the same cell results in an pseudo S-phase like environment conducive to viral genome amplification. At high expression levels, which is also observed in cells amplifying the genome, E2 can also result in G2 arrest (Frattini et al, 1997). This cell cycle arrest could allow sustained synthesis of viral DNA and be important for vegetative replication.
B. Differences in Replication Strategies of Different Papillomaviruses
Papillomaviruses have coevolved with their hosts over millions of years and individual HPV types have existed since the evolutionary origin of humans (Bernard et al, 2006). Each virus is epitheliotropic but infects either mucosal or cutaneous epithelia, usually at a specific site of the body. Although each virus has a very similar genomic organization and the viral proteins are relatively well conserved, infection by different papillomaviruses results in a wide spectrum of pathologies. These can range from clinically inapparent infections, through a diverse variety of benign papillomas and warts, to a subset of infections that can progress to malignant carcinomas. These pathological differences are most likely due to different viral types, different epithelial host cells and the immune response of the host. However, in each case the virus has a similar strategy in that it must establish a persistent infection in the dividing cells of the epithelium and restrict vegetative growth to the more differentiated cells that are destined to be sloughed from the epithelium. The timing of productive replication, however, differs among the papillomaviruses. Some, such as the cutaneous mu viruses, trigger vegetative viral DNA amplification as soon as the host cells leave the basal layer while others, such as the mucosal alpha viruses, postpone productive replication until later stages of differentiation (Peh et al, 2002).
Similarly, it is becoming apparent that different papillomaviruses use different chromosomal targets for partitioning the viral genomes and the chromosome binding pattern of E2 seems to be characteristic of each evolutionary group. Notably, all E2 proteins studied to date interact with the Brd4 protein to regulate viral transcription (McPhillips et al, 2006; Schweiger et al, 2006), but only a subset of viruses also use Brd4 as a chromosomal tether in mitosis (McPhillips et al, 2006). For regulation of transcription and initiation of DNA replication, the E2 proteins must interact precisely with many cellular protein complexes that tightly regulate these processes. For genome partitioning, the E2 protein must simply tether the viral genome to a stable chromosomal target. This could be a cellular protein, such as Brd4, which is also used for other E2 functions or could be a completely different target used only for mitotic partitioning.
C. Comparison of Papillomavirus DNA Replication with Cellular DNA replication and Replication of Other Viruses
The replicon model first proposed by Jacob and Brenner proposed that replication was initiated by an initiator protein binding to a genetic element designated the replicator (Jacob and BRENNER, 1963). In higher eukaryotes, the initiator is actually a complex of proteins that is assembled on the replicator in a regulated manner. The replicator element is recognized by ORC, the origin recognition complex, followed by Cdt1 and CDC6, which function to recruit and load the helicase, MCM. The assembled MCM complex must be activated by cyclin dependent kinases (reviewed in (Masai et al, 2005). While many viruses utilize the cellular replication machinery, they also often encode their own initator proteins. For papillomaviruses, the specific DNA binding domain of E1 is analogous to ORC and the helicase function is analogous to the cellular helicase MCM. The loading functions of Cdt1 and CDC6 are carried out by the E2 protein.
Other viruses also encode initiator proteins. The large T antigen of SV40 initiates replication in a very similar manner to E1. Both proteins have limited sequence homology (Mansky et al, 1997) but extensive structural homology in the helicase and DNA binding domains (Enemark et al, 2000; Enemark and Joshua-Tor, 2006; Li et al, 2003; Luo et al, 1996; Meinke et al, 2006; Meinke et al, 2007). However, SV40 has no counterpart to E2 and the double hexameric helicase of T antigen assembles onto the replication origin in the absence of a cellular or viral loading factor (Borowiec et al, 1990). In permissive cells, SV40 is a lytic virus that undergoes runaway replication and does not establish its genome as a persistent episome. Therefore, the role of E2 in papillomavirus regulation may be to regulate runaway replication as well as to maintain and segregate the viral genome.
Another virus, Epstein-Barr virus (EBV), establishes long-term persistent infections similar to the papillomaviruses. EBV replicates in synchrony with the cellular DNA and does not encode a viral helicase initiator protein (Kirchmaier and Sugden, 1998; Yates and Guan, 1991). Instead, replication is regulated by assembly of ORC, MCM and other cellular replication factors (Chaudhuri et al, 2001; Schepers et al, 2001). EBV encodes a single protein, EBNA-1, that is required for maintenance replication and segregation of the viral genome. EBNA-1, binds to the latent replication origin, oriP (Rawlins et al, 1985; Yates et al, 1985), which contains a family of repeats (FR) and a dyad symmetry (DS) element. The FR element is essential for stable episomal maintenance and contains multiple EBNA-1 binding sites. It functions in concert with the DS element, which contains phased EBNA1 sites adjacent to telomere repeat factor (TRF) binding sites, which together function as an efficient origin of DNA replication initiation (Deng et al, 2003; Deng et al, 2002). Thus the function of EBNA is analogous to that of the E2 protein.
D. Papillomavirus Replication in Saccharomyces cerevisiae
There are many genetic systems and screens in yeast that enable the study of plasmid maintenance in a much more practical and efficient manner than in mammalian cells. Several studies have shown that papillomavirus genomes will replicate stably in yeast (Angeletti et al, 2002; Chattopadhyay et al, 2005; Kim et al, 2005). Papillomavirus genomes, and sub-genomic fragments, are replicated and maintained through sequences that can substitute for autonomously replicating sequence (ARS) and centromere (CEN) elements, even in the absence of the E1 and E2 proteins. Although this does not directly parallel the requirements for papillomavirus replication in mammalian cells, the elements identified in these studies may be important for viral genome replication in addition to E1 and E2-dependent elements.
In a different type of study, yeast plasmids containing an ARS element and E2 binding sites could be maintained in yeast in the presence of the BPV-1 E2 protein and the mammalian Brd4 protein (Brannon et al, 2005). Notably, S. cerevisiae do express a Brd4 family member, Bdf1, but this is not sufficient for E2-dependent maintenance. Bdf1 does not contain a region homologous to the C-terminal region of Brd4 that interacts with E2, but a fusion protein of Bdf1-Brd4 can support E2-plasmid maintenance.
E. Anti-viral replication therapies
Many antiviral therapies for HPV-associated disease have focused on the replication proteins (reviewed in (Fradet-Turcotte and Archambault, 2007)). Small molecule indandione inhibitors, designed to inhibit interaction between the N-terminal domain of E2 and the helicase domain of E1, inhibit HPV-11 replication (White et al, 2003). Polyamides have been designed to inhibit specific E1 and E2 DNA binding function (Schaal et al, 2003). An other attractive target is the enzymatic activities of the E1 ATPase/helicase and several pharmaceutical companies have candidates for these inhibitors (reviewed in (Fradet-Turcotte and Archambault, 2007). There is also great interest in designing therapeutics that would disrupt the tethering of the viral genome to mitotic chromosomes, which is predicted to cure persistent papillomavirus infection.
F. Papillomavirus-based vectors
Gene therapy vectors that replicate extrachromosomally are advantageous compared to those that integrate randomly into the host genome as they will not result in insertional mutagenesis and are not susceptible to positional effects, yet will persist long term in dividing cells. One of the first descriptions of the concept of viral based vectors was described by Rogers in 1966 in a paper entitled “Shope Papillomavirus: a passenger in man and its significance to the potential control of the host genome” (Friedmann, 2001; Rogers, 1966). However, the actual use of papillomavirus-based vectors was not realized until the 1980s (Sarver et al, 1981). The first vectors consisted of the early region of BPV-1 linked to an expression cassette for the foreign gene. However, these early vectors were not widely adopted because they transformed cells, had a limited host range and transcription of the foreign gene often caused the plasmid to integrate (Waldenstrom et al, 1992). This was likely due to interference between the viral regulatory elements and the insertion of a heterologous enhancer and promoter sequences in the compact papillomavirus genomes. More recently vectors have been developed based on the modular elements required for replication. For example, newer BPV-1 based vectors contain only the LCR and the E1 and E2 genes (Mannik et al, 2002; Ohe et al, 1995) and are maintained in transgenic mice for several generations (Mannik et al, 2003). Human papillomavirus-based vectors have also been explored and shown to be able to replicate, at least transiently, in human cell lines and in the lungs of mice (Gadi et al, 1999; Sverdrup et al, 1999). However, at this point these studies have shown mainly “proof of principle” of papillomavirus based vectors and they have not yet been widely or practically used to express foreign genes. More detailed knowledge of the mechanisms of human papillomavirus replication and genome maintenance should enable the development of more successful vectors.
Acknowledgements
The author's research is supported by the Intramural Research Program of the NIAID, NIH
Reference List
- 1.Abbate EA, Berger JM, Botchan MR. The X-ray structure of the papillomavirus helicase in complex with its molecular matchmaker E2. Genes Dev. 2004;18:1981–1996. doi: 10.1101/gad.1220104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abbate EA, Voitenleitner C, Botchan MR. Structure of the papillomavirus DNA-tethering complex E2:Brd4 and a peptide that ablates HPV chromosomal association. Mol.Cell. 2006;24:877–889. doi: 10.1016/j.molcel.2006.11.002. [DOI] [PubMed] [Google Scholar]
- 3.Abroi A, Ilves I, Kivi S, Ustav M. Analysis of chromatin attachment and partitioning functions of bovine papillomavirus type 1 E2 protein. J.Virol. 2004;78:2100–2113. doi: 10.1128/JVI.78.4.2100-2113.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Abroi A, Kurg R, Ustav M. Transcriptional and replicational activation functions in the BPV1 E2 protein are encoded by different structural determinants. J.Virol. 1996;70:6169–6179. doi: 10.1128/jvi.70.9.6169-6179.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ai W, Toussaint E, Roman A. CCAAT displacement protein binds to and negatively regulates human papillomavirus type 6 E6, E7, and E1 promoters. J.Virol. 1999;73:4220–4229. doi: 10.1128/jvi.73.5.4220-4229.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Akgul B, Karle P, Adam M, Fuchs PG, Pfister HJ. Dual role of tumor suppressor p53 in regulation of DNA replication and oncogene E6-promoter activity of epidermodysplasia verruciformis-associated human papillomavirus type 8. Virology. 2003;308:279–290. doi: 10.1016/s0042-6822(02)00133-2. [DOI] [PubMed] [Google Scholar]
- 7.Androphy EJ, Lowy DR, Schiller JT. Bovine papillomavirus E2 trans-activating gene product binds to specific sites in papillomavirus DNA. Nature. 1987;325:70–73. doi: 10.1038/325070a0. [DOI] [PubMed] [Google Scholar]
- 8.Angeletti PC, Kim K, Fernandes FJ, Lambert PF. Stable replication of papillomavirus genomes in Saccharomyces cerevisiae. J.Virol. 2002;76:3350–3358. doi: 10.1128/JVI.76.7.3350-3358.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Antson AA, Burns JE, Moroz OV, Scott DJ, Sanders CM, Bronstein IB, Dodson GG, Wilson KS, Maitland NJ. Structure of the intact transactivation domain of the human papillomavirus E2 protein. Nature. 2000;403:805–809. doi: 10.1038/35001638. [DOI] [PubMed] [Google Scholar]
- 10.Ashrafi GH, Brown DR, Fife KH, Campo MS. Down-regulation of MHC class I is a property common to papillomavirus E5 proteins. Virus Research. 2006;120:208–211. doi: 10.1016/j.virusres.2006.02.005. [DOI] [PubMed] [Google Scholar]
- 11.Auster AS, Joshua-Tor L. The DNA-binding domain of human papillomavirus type 18 E1. Crystal structure, dimerization, and DNA binding. J.Biol.Chem. 2004;279:3733–3742. doi: 10.1074/jbc.M311681200. [DOI] [PubMed] [Google Scholar]
- 12.Baiker A, Maercker C, Piechaczek C, Schmidt SB, Bode J, Benham C, Lipps HJ. Mitotic stability of an episomal vector containing a human scaffold/matrix-attached region is provided by association with nuclear matrix. Nat.Cell Biol. 2000;2:182–184. doi: 10.1038/35004061. [DOI] [PubMed] [Google Scholar]
- 13.Barbera AJ, Chodaparambil JV, Kelley-Clarke B, Joukov V, Walter JC, Luger K, Kaye KM. The nucleosomal surface as a docking station for Kaposi's sarcoma herpesvirus LANA. Science. 2006;311:856–861. doi: 10.1126/science.1120541. [DOI] [PubMed] [Google Scholar]
- 14.Barsoum J, Prakash SS, Han P, Androphy EJ. Mechanism of action of the papillomavirus E2 repressor: repression in the absence of DNA binding. J.Virol. 1992;66:3941–3945. doi: 10.1128/jvi.66.6.3941-3945.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bastien N, McBride AA. Interaction of the papillomavirus E2 with mitotic chromosomes. Virology. 2000;270:124–134. doi: 10.1006/viro.2000.0265. [DOI] [PubMed] [Google Scholar]
- 16.Baxter MK, McPhillips MG, Ozato K, McBride AA. The mitotic chromosome binding activity of the papillomavirus E2 protein correlates with interaction with the cellular chromosomal protein, Brd4. J.Virol. 2005;79:4806–4818. doi: 10.1128/JVI.79.8.4806-4818.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bedell MA, Hudson JB, Golub TR, Turyk ME, Hosken M, Wilbanks GD, Laimins LA. Amplification of human papillomavirus genomes in vitro is dependent on epithelial differentiation. J.Virol. 1991;65:2254–2260. doi: 10.1128/jvi.65.5.2254-2260.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bedrosian CL, Bastia D. The DNA-binding domain of HPV-16 E2 protein interaction with the viral enhancer: Protein-induced DNA bending and role of the nonconserved core sequence in binding site affinity. Virology. 1990;174:557–575. doi: 10.1016/0042-6822(90)90109-5. [DOI] [PubMed] [Google Scholar]
- 19.Bellanger S, Demeret C, Goyat S, Thierry F. Stability of the human papillomavirus type 18 E2 protein is regulated by a proteasome degradation pathway through its amino-terminal transactivation domain. J.Virol. 2001;75:7244–7251. doi: 10.1128/JVI.75.16.7244-7251.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Berg L, Lusky M, Stenlund A, Botchan MR. Repression of bovine papilloma virus replication is mediated by a virally encoded trans-acting factor. Cell. 1986;46:753–762. doi: 10.1016/0092-8674(86)90351-x. [DOI] [PubMed] [Google Scholar]
- 21.Bernard BA, Bailly C, Lenoir MC, Darmon M, Thierry F, Yaniv M. The human papillomavirus type 18 (HPV18) E2 gene product is a repressor of the HPV18 regulatory region in human keratinocytes. J.Virol. 1989;63:4317–4324. doi: 10.1128/jvi.63.10.4317-4324.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bernard HU, Calleja-Macias IE, Dunn ST. Genome variation of human papillomavirus types: phylogenetic and medical implications. Int.J.Cancer. 2006;118:1071–1076. doi: 10.1002/ijc.21655. [DOI] [PubMed] [Google Scholar]
- 23.Bian XL, Rosas-Acosta G, Wu YC, Wilson VG. Nuclear import of bovine papillomavirus type 1 E1 protein is mediated by multiple alpha importins and is negatively regulated by phosphorylation near a nuclear localization signal. J.Virol. 2007;81:2899–2908. doi: 10.1128/JVI.01850-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bochkarev A, Barwell JA, Pfuetzner RA, Furey WJ, Edwards AM, Frappier L. Crystal structure of the DNA-binding domain of the Epstein-Barr virus origin-binding protein EBNA 1. Cell. 1995;83:39–46. doi: 10.1016/0092-8674(95)90232-5. [DOI] [PubMed] [Google Scholar]
- 25.Borowiec JA, Dean FB, Bullock PA, Hurwitz J. Binding and unwinding--how T antigen engages the SV40 origin of DNA replication. Cell. 1990;60:181–184. doi: 10.1016/0092-8674(90)90730-3. [DOI] [PubMed] [Google Scholar]
- 26.Botchan M, Berg L, Reynolds J, Lusky M. The bovine papillomavirus replicon. Ciba Found.Symp. 1986;120:53–67. doi: 10.1002/9780470513309.ch5. [DOI] [PubMed] [Google Scholar]
- 27.Bousarghin L, Touze A, Sizaret PY, Coursaget P. Human papillomavirus types 16, 31, and 58 use different endocytosis pathways to enter cells. J.Virol. 2003;77:3846–3850. doi: 10.1128/JVI.77.6.3846-3850.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brannon AR, Maresca JA, Boeke JD, Basrai MA, McBride AA. Reconstitution of papillomavirus E2-mediated plasmid maintenance in Saccharomyces cerevisiae by the Brd4 bromodomain protein. Proc.Natl.Acad.Sci.U.S.A. 2005;102:2998–3003. doi: 10.1073/pnas.0407818102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brokaw JL, Blanco M, McBride AA. Amino acids critical for the functions of the bovine papillomavirus type 1 E2 transactivator. J.Virol. 1996;70:23–29. doi: 10.1128/jvi.70.1.23-29.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brown C, Kowalczyk AM, Taylor ER, Morgan IM, Gaston K. p53 represses human papillomavirus type 16 DNA replication via the viral E2 protein. Virol.J. 2008;5:5. doi: 10.1186/1743-422X-5-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brown DR, Kitchin D, Qadadri B, Neptune N, Batteiger T, Ermel A. The human papillomavirus type 11 E1--E4 protein is a transglutaminase 3 substrate and induces abnormalities of the cornified cell envelope. Virology. 2006;345:290–298. doi: 10.1016/j.virol.2005.09.048. [DOI] [PubMed] [Google Scholar]
- 32.Burnett S, Strom AC, Jareborg N, Alderborn A, Dillner J, Moreno-Lopez J, Pettersson U, Kiessling U. Induction of bovine papillomavirus E2 gene expression and early region transcription by cell growth arrest: correlation with viral DNA amplification and evidence for differential promoter induction. J.Virol. 1990;64:5529–5541. doi: 10.1128/jvi.64.11.5529-5541.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Calderwood MA, Hall KT, Matthews DA, Whitehouse A. The herpesvirus saimiri ORF73 gene product interacts with host-cell mitotic chromosomes and self-associates via its C terminus. J.Gen.Virol. 2004;85:147–153. doi: 10.1099/vir.0.19437-0. [DOI] [PubMed] [Google Scholar]
- 34.Cardenas-Mora J, Spindler JE, Jang MK, McBride AA. Dimerization of the Papillomavirus E2 Protein is Required for Efficient Mitotic Chromosome Association and Brd4 Binding. Journal of Virology. 2008 doi: 10.1128/JVI.00772-08. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Castella S, Bingham G, Sanders CM. Common determinants in DNA melting and helicase-catalysed DNA unwinding by papillomavirus replication protein E1. Nucleic Acids Res. 2006;34:3008–3019. doi: 10.1093/nar/gkl384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chatterjee A, Freeman JW, Busch H. Identification and partial characterization of a Mr 40,000 nucleolar antigen associated with cell proliferation. Cancer Res. 1987;47:1123–1129. [PubMed] [Google Scholar]
- 37.Chattopadhyay A, Schmidt MC, Khan SA. Identification of a 450-bp region of human papillomavirus type 1 that promotes episomal replication in Saccharomyces cerevisiae. Virology. 2005;340:133–142. doi: 10.1016/j.virol.2005.06.029. [DOI] [PubMed] [Google Scholar]
- 38.Chaudhuri B, Xu H, Todorov I, Dutta A, Yates JL. Human DNA replication initiation factors, ORC and MCM, associate with oriP of Epstein-Barr virus. Proc.Natl.Acad.Sci.U.S.A. 2001;98:10085–10089. doi: 10.1073/pnas.181347998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen G, Stenlund A. Characterization of the DNA-binding domain of the bovine papillomavirus replication initiator E1. J.Virol. 1998;72:2567–2576. doi: 10.1128/jvi.72.4.2567-2576.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen G, Stenlund A. The E1 initiator recognizes multiple overlapping sites in the papillomavirus origin of DNA replication. J.Virol. 2001;75:292–302. doi: 10.1128/JVI.75.1.292-302.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chiang CM, Dong G, Broker TR, Chow LT. Control of human papillomavirus type 11 origin of replication by the E2 family of transcription regulatory proteins. J.Virol. 1992;66:5224–5231. doi: 10.1128/jvi.66.9.5224-5231.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Choe J, Vaillancourt P, Stenlund A, Botchan M. Bovine papillomavirus type 1 encodes two forms of a transcriptional repressor: Structural and functional analysis of new viral cDNAs. J.Virol. 1989;63:1743–1755. doi: 10.1128/jvi.63.4.1743-1755.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Clower RV, Fisk JC, Melendy T. Papillomavirus E1 protein binds to and stimulates human topoisomerase I. J.Virol. 2006;80:1584–1587. doi: 10.1128/JVI.80.3.1584-1587.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Conger KL, Liu JS, Kuo SR, Chow LT, Wang TS. Human papillomavirus DNA replication. Interactions between the viral E1 protein and two subunits of human dna polymerase alpha/primase. J.Biol.Chem. 1999;274:2696–2705. doi: 10.1074/jbc.274.5.2696. [DOI] [PubMed] [Google Scholar]
- 45.Cotter MA, Robertson ES. The latency-associated nuclear antigen tethers the Kaposi's sarcoma- associated herpesvirus genome to host chromosomes in body cavity-based lymphoma cells. Virology. 1999;264:254–264. doi: 10.1006/viro.1999.9999. [DOI] [PubMed] [Google Scholar]
- 46.Cripe TP, Haugen TH, Turk JP, Tabatabai F, Schmid PG, 3d., Dürst M, Gissmann L, Roman A, Turek LP. Transcriptional regulation of the human papillomavirus- 16 E6–E7 promoter by a keratinocyte-dependent enhancer, and by viral E2 trans-activator and repressor gene products: implications for cervical carcinogenesis. EMBO J. 1987;6:3745–3753. doi: 10.1002/j.1460-2075.1987.tb02709.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cueille N, Nougarede R, Mechali F, Philippe M, Bonne-Andrea C. Functional interaction between the bovine papillomavirus virus type 1 replicative helicase E1 and cyclin E-Cdk2. J.Virol. 1998;72:7255–7262. doi: 10.1128/jvi.72.9.7255-7262.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Culp TD, Budgeon LR, Marinkovich MP, Meneguzzi G, Christensen ND. Keratinocyte-secreted laminin 5 can function as a transient receptor for human papillomaviruses by binding virions and transferring them to adjacent cells. J.Virol. 2006;80:8940–8950. doi: 10.1128/JVI.00724-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.D'Amours D, Stegmeier F, Amon A. Cdc14 and condensin control the dissolution of cohesin-independent chromosome linkages at repeated DNA. Cell. 2004;117:455–469. doi: 10.1016/s0092-8674(04)00413-1. [DOI] [PubMed] [Google Scholar]
- 50.Dao LD, Duffy A, Van Tine BA, Wu SY, Chiang CM, Broker TR, Chow LT. Dynamic localization of the human papillomavirus type 11 origin binding protein E2 through mitosis while in association with the spindle apparatus. J.Virol. 2006;80:4792–4800. doi: 10.1128/JVI.80.10.4792-4800.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dasgupta S, Zabielski J, Simonsson M, Burnett S. Rolling-circle replication of a high-copy BPV-1 plasmid. J.Mol.Biol. 1992;228:1–6. doi: 10.1016/0022-2836(92)90485-3. [DOI] [PubMed] [Google Scholar]
- 52.Davy CE, Jackson DJ, Wang Q, Raj K, Masterson PJ, Fenner NF, Southern S, Cuthill S, Millar JB, Doorbar J. Identification of a G(2) arrest domain in the E1 wedge E4 protein of human papillomavirus type 16. J.Virol. 2002;76:9806–9818. doi: 10.1128/JVI.76.19.9806-9818.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Day PM, Baker CC, Lowy DR, Schiller JT. Establishment of papillomavirus infection is enhanced by promyelocytic leukemia protein (PML) expression. Proc.Natl.Acad.Sci.U.S.A. 2004;101:14252–14257. doi: 10.1073/pnas.0404229101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Day PM, Lowy DR, Schiller JT. Papillomaviruses infect cells via a clathrin-dependent pathway. Virology. 2003;307:1–11. doi: 10.1016/s0042-6822(02)00143-5. [DOI] [PubMed] [Google Scholar]
- 55.Day PM, Roden RB, Lowy DR, Schiller JT. The papillomavirus minor capsid protein, L2, induces localization of the major capsid protein, L1, and the viral transcription/replication protein, E2, to PML oncogenic domains. J.Virol. 1998;72:142–150. doi: 10.1128/jvi.72.1.142-150.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.DeMasi J, Huh KW, Nakatani Y, Munger K, Howley PM. Bovine papillomavirus E7 transformation function correlates with cellular p600 protein binding. Proc.Natl.Acad.Sci.U.S.A. 2005;102:11486–11491. doi: 10.1073/pnas.0505322102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Deng W, Lin BY, Jin G, Wheeler CG, Ma T, Harper JW, Broker TR, Chow LT. Cyclin/CDK regulates the nucleocytoplasmic localization of the human papillomavirus E1 DNA helicase. J.Virol. 2004;78:13954–13965. doi: 10.1128/JVI.78.24.13954-13965.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Deng Z, Atanasiu C, Burg JS, Broccoli D, Lieberman PM. Telomere repeat binding factors TRF1, TRF2, and hRAP1 modulate replication of Epstein-Barr virus OriP. J.Virol. 2003;77:11992–12001. doi: 10.1128/JVI.77.22.11992-12001.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Deng Z, Lezina L, Chen CJ, Shtivelband S, So W, Lieberman PM. Telomeric proteins regulate episomal maintenance of Epstein-Barr virus origin of plasmid replication. Mol.Cell. 2002;9:493–503. doi: 10.1016/s1097-2765(02)00476-8. [DOI] [PubMed] [Google Scholar]
- 60.Dey A, Chitsaz F, Abbasi A, Misteli T, Ozato K. The double bromodomain protein Brd4 binds to acetylated chromatin during interphase and mitosis. Proc.Natl.Acad.Sci.U.S.A. 2003;100:8758–8763. doi: 10.1073/pnas.1433065100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dey A, Ellenberg J, Farina A, Coleman AE, Maruyama T, Sciortino S, Lippincott-Schwartz J, Ozato K. A bromodomain protein, MCAP, associates with mitotic chromosomes and affects G(2)-to-M transition. Mol.Cell Biol. 2000;20:6537–6549. doi: 10.1128/mcb.20.17.6537-6549.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.DiMaio D, Mattoon D. Mechanisms of cell transformation by papillomavirus E5 proteins. Oncogene. 2001;20:7866–7873. doi: 10.1038/sj.onc.1204915. [DOI] [PubMed] [Google Scholar]
- 63.Donaldson MM, Boner W, Morgan IM. TopBP1 regulates human papillomavirus type 16 E2 interaction with chromatin. J.Virol. 2007;81:4338–4342. doi: 10.1128/JVI.02353-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Doorbar J, Ely S, Sterling J, McLean C, Crawford L. Specific interaction between HPV-16 E1–E4 and cytokeratins results in collapse of the epithelial cell intermediate filament network. Nature. 1991;352:824–827. doi: 10.1038/352824a0. [DOI] [PubMed] [Google Scholar]
- 65.Dowhanick JJ, McBride AA, Howley PM. Suppression of cellular proliferation by the papillomavirus E2 protein. J.Virol. 1995;69:7791–7799. doi: 10.1128/jvi.69.12.7791-7799.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Enemark EJ, Chen G, Vaughn DE, Stenlund A, Joshua-Tor L. Crystal structure of the DNA binding domain of the replication initiation protein E1 from papillomavirus. Mol.Cell. 2000;6:149–158. [PubMed] [Google Scholar]
- 67.Enemark EJ, Joshua-Tor L. Mechanism of DNA translocation in a replicative hexameric helicase. Nature. 2006;442:270–275. doi: 10.1038/nature04943. [DOI] [PubMed] [Google Scholar]
- 68.Evander M, Frazer IH, Payne E, Qi YM, Hengst K, McMillan NA. Identification of the alpha6 integrin as a candidate receptor for papillomaviruses. J.Virol. 1997;71:2449–2456. doi: 10.1128/jvi.71.3.2449-2456.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Evans MF, Aliesky HA, Cooper K. Optimization of biotinyl-tyramide-based in situ hybridization for sensitive background-free applications on formalin-fixed, paraffin-embedded tissue specimens. BMC.Clin.Pathol. 2003;3:2. doi: 10.1186/1472-6890-3-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Fabunmi RP, Wigley WC, Thomas PJ, DeMartino GN. Activity and regulation of the centrosome-associated proteasome. Journal of Biological Chemistry. 2000;275:409–413. doi: 10.1074/jbc.275.1.409. [DOI] [PubMed] [Google Scholar]
- 71.Favre M, Breitburd F, Croissant O, Orth G. Chromatin-like structures obtained after alkaline disruption of bovine and human papillomaviruses. J.Virol. 1977;21:1205–1209. doi: 10.1128/jvi.21.3.1205-1209.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ferran MC, McBride AA. Transient viral DNA replication and repression of viral transcription are supported by the C-terminal domain of the bovine papillomavirus type 1 E1 protein. J.Virol. 1998;72:796–801. doi: 10.1128/jvi.72.1.796-801.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Florence B, Faller DV. You bet-cha: a novel family of transcriptional regulators. Front Biosci. 2001;6:D1008–D1018. doi: 10.2741/florence. [DOI] [PubMed] [Google Scholar]
- 74.Flores ER, Allen-Hoffmann BL, Lee D, Sattler CA, Lambert PF. Establishment of the human papillomavirus type 16 (HPV-16) life cycle in an immortalized human foreskin keratinocyte cell line. Virology. 1999;262:344–354. doi: 10.1006/viro.1999.9868. [DOI] [PubMed] [Google Scholar]
- 75.Flores ER, Lambert PF. Evidence for a switch in the mode of human papillomavirus type 16 DNA replication during the viral life cycle. J.Virol. 1997;71:7167–7179. doi: 10.1128/jvi.71.10.7167-7179.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Fradet-Turcotte A, Archambault J. Recent advances in the search for antiviral agents against human papillomaviruses. Antiviral Therapy. 2007;12:431–451. [PMC free article] [PubMed] [Google Scholar]
- 77.Frattini MG, Hurst SD, Lim HB, Swaminathan S, Laimins LA. Abrogation of a mitotic checkpoint by E2 proteins from oncogenic human papillomaviruses correlates with increased turnover of the p53 tumor suppressor protein. EMBO J. 1997;16:318–331. doi: 10.1093/emboj/16.2.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Frattini MG, Lim HB, Laimins LA. In vitro synthesis of oncogenic human papillomaviruses requires episomal genomes for differentiation-dependent late expression. Proc.Natl.Acad.Sci.U.S.A. 1996;93:3062–3067. doi: 10.1073/pnas.93.7.3062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Freeman L, ragon-Alcaide L, Strunnikov A. The condensin complex governs chromosome condensation and mitotic transmission of rDNA. J.Cell Biol. 2000;149:811–824. doi: 10.1083/jcb.149.4.811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Friedmann T. Stanfield Rogers: insights into virus vectors and failure of an early gene therapy model. Mol.Ther. 2001;4:285–288. doi: 10.1006/mthe.2001.0454. [DOI] [PubMed] [Google Scholar]
- 81.Gadi VK, Zou N, Liu JS, Cheng S, Broker TR, Sorscher EJ, Chow LT. Components of human papillomavirus that activate transcription and support plasmid replication in human airway cells. Am.J.Respir.Cell Mol.Biol. 1999;20:1001–1006. doi: 10.1165/ajrcmb.20.5.3479. [DOI] [PubMed] [Google Scholar]
- 82.Gammoh N, Grm HS, Massimi P, Banks L. Regulation of human papillomavirus type 16 E7 activity through direct protein interaction with the E2 transcriptional activator. J.Virol. 2006;80:1787–1797. doi: 10.1128/JVI.80.4.1787-1797.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Gilbert DM. In search of the holy replicator. Nat.Rev.Mol.Cell Biol. 2004;5:848–855. doi: 10.1038/nrm1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Gilbert DM, Cohen SN. Bovine papilloma virus plasmids replicate randomly in mouse fibroblasts throughout S phase of the cell cycle. Cell. 1987;50:59–68. doi: 10.1016/0092-8674(87)90662-3. [DOI] [PubMed] [Google Scholar]
- 85.Gillison ML, Lowy DR. A causal role for human papillomavirus in head and neck cancer. Lancet. 2004;363:1488–1489. doi: 10.1016/S0140-6736(04)16194-1. [DOI] [PubMed] [Google Scholar]
- 86.Giroglou T, Florin L, Schafer F, Streeck RE, Sapp M. Human papillomavirus infection requires cell surface heparan sulfate. J.Virol. 2001;75:1565–1570. doi: 10.1128/JVI.75.3.1565-1570.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Goodwin EC, Naeger LK, Breiding DE, Androphy EJ, DiMaio D. Transactivation-competent bovine papillomavirus E2 protein is specifically required for efficient repression of human papillomavirus oncogene expression and for acute growth inhibition of cervical carcinoma cell lines. J.Virol. 1998;72:3925–3934. doi: 10.1128/jvi.72.5.3925-3934.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Gopalakrishnan V, Khan SA. E1 protein of human papillomavirus type 1a is sufficient for initiation of viral DNA replication. PNAS temp. 1995;91:9597–9601. doi: 10.1073/pnas.91.20.9597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Griffiths R, Whitehouse A. Herpesvirus saimiri episomal persistence is maintained via interaction between open reading frame 73 and the cellular chromosome-associated protein MeCP2. J.Virol. 2007;81:4021–4032. doi: 10.1128/JVI.02171-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Grundhoff A, Ganem D. The latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus permits replication of terminal repeat-containing plasmids. J.Virol. 2003;77:2779–2783. doi: 10.1128/JVI.77.4.2779-2783.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hammerschmidt W, Sugden B. Identification and characterization of oriLyt, a lytic origin of DNA replication of Epstein-Barr virus. Cell. 1988;55:427–433. doi: 10.1016/0092-8674(88)90028-1. [DOI] [PubMed] [Google Scholar]
- 92.Han Y, Loo YM, Militello KT, Melendy T. Interactions of the papovavirus DNA replication initiator proteins, bovine papillomavirus type 1 E1 and simian virus 40 large T antigen, with human replication protein A. J.Virol. 1999;73:4899–4907. doi: 10.1128/jvi.73.6.4899-4907.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Hartley KA, Alexander KA. Human TATA binding protein inhibits human papillomavirus type 11 DNA replication by antagonizing E1-E2 protein complex formation on the viral origin of replication. J.Virol. 2002;76:5014–5023. doi: 10.1128/JVI.76.10.5014-5023.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Hegde RS, Grossman SR, Laimins LA, Sigler PB. Crystal structure at 1.7 A of the bovine papillomavirus-1 E2 DNA- binding domain bound to its DNA target. Nature. 1992;359:505–512. doi: 10.1038/359505a0. [DOI] [PubMed] [Google Scholar]
- 95.Hernandez-Ramon EE, Burns JE, Zhang W, Walker HF, Allen S, Antson AA, Maitland NJ. Dimerization of the human papillomavirus type 16 E2 N terminus results in DNA looping within the upstream regulatory region. J.Virol. 2008;82:4853–4861. doi: 10.1128/JVI.02388-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Hickman AB, Ronning DR, Perez ZN, Kotin RM, Dyda F. The nuclease domain of adeno-associated virus rep coordinates replication initiation using two distinct DNA recognition interfaces. Mol.Cell. 2004;13:403–414. doi: 10.1016/s1097-2765(04)00023-1. [DOI] [PubMed] [Google Scholar]
- 97.Hindmarsh PL, Laimins LA. Mechanisms regulating expression of the HPV 31 L1 and L2 capsid proteins and pseudovirion entry. Virol.J. 2007;4:19. doi: 10.1186/1743-422X-4-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Hoffmann R, Hirt B, Bechtold V, Beard P, Raj K. Different modes of human papillomavirus DNA replication during maintenance. J.Virol. 2006;80:4431–4439. doi: 10.1128/JVI.80.9.4431-4439.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Holt SE, Schuller G, Wilson VG. DNA binding specificity of the bovine papillomavirus E1 protein is determined by sequences contained within an 18-base-pair inverted repeat element at the origin of replication. J.Virol. 1994;68:1094–1102. doi: 10.1128/jvi.68.2.1094-1102.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Hsu CY, Mechali F, Bonne-Andrea C. Nucleocytoplasmic shuttling of bovine papillomavirus E1 helicase downregulates viral DNA replication in S phase. J.Virol. 2007;81:384–394. doi: 10.1128/JVI.01170-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Hubert WG, Lambert PF. The 23-kilodalton E1 phosphoprotein of bovine papillomavirus type 1 is nonessential for stable plasmid replication in murine C127 cells. J.Virol. 1993;67:2932–2937. doi: 10.1128/jvi.67.5.2932-2937.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Huh KW, DeMasi J, Ogawa H, Nakatani Y, Howley PM, Munger K. Association of the human papillomavirus type 16 E7 oncoprotein with the 600-kDa retinoblastoma protein-associated factor, p600. Proc.Natl.Acad.Sci.U.S.A. 2005;102:11492–11497. doi: 10.1073/pnas.0505337102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Ilves I, Kadaja M, Ustav M. Two separate replication modes of the bovine papillomavirus BPV1 origin of replication that have different sensitivity to p53. Virus Res. 2003;96:75–84. doi: 10.1016/s0168-1702(03)00174-6. [DOI] [PubMed] [Google Scholar]
- 104.Ilves I, Kivi S, Ustav M. Long-term episomal maintenance of bovine papillomavirus type 1 plasmids is determined by attachment to host chromosomes, which Is mediated by the viral E2 protein and its binding sites. J.Virol. 1999;73:4404–4412. doi: 10.1128/jvi.73.5.4404-4412.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Ilves I, Maemets K, Silla T, Janikson K, Ustav M. Brd4 is involved in multiple processes of the bovine papillomavirus type 1 life cycle. J.Virol. 2006;80:3660–3665. doi: 10.1128/JVI.80.7.3660-3665.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Ilyina TV, Koonin EV. Conserved sequence motifs in the initiator proteins for rolling circle DNA replication encoded by diverse replicons from eubacteria, eucaryotes and archaebacteria. Nucleic Acids Res. 1992;20:3279–3285. doi: 10.1093/nar/20.13.3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Jacob F, BRENNER S. On the regulation of DNA synthesis in bacteria: the hypothesis of the replicon. C.R.Hebd.Seances Acad.Sci. 1963;256:298–300. [PubMed] [Google Scholar]
- 108.Jang MK, Mochizuki K, Zhou M, Jeong HS, Brady JN, Ozato K. The bromodomain protein Brd4 is a positive regulatory component of P-TEFb and stimulates RNA polymerase II-dependent transcription. Mol.Cell. 2005;19:523–534. doi: 10.1016/j.molcel.2005.06.027. [DOI] [PubMed] [Google Scholar]
- 109.Janin J, Rodier F, Chakrabarti P, Bahadur RP. Macromolecular recognition in the Protein Data Bank. Acta Crystallogr.D.Biol.Crystallogr. 2007;63:1–8. doi: 10.1107/S090744490603575X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Jeckel S, Huber E, Stubenrauch F, Iftner T. A transactivator function of cottontail rabbit papillomavirus e2 is essential for tumor induction in rabbits. J.Virol. 2002;76:11209–11215. doi: 10.1128/JVI.76.22.11209-11215.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Jeckel S, Loetzsch E, Huber E, Stubenrauch F, Iftner T. Identification of the E9/E2C cDNA and functional characterization of the gene product reveal a new repressor of transcription and replication in cottontail rabbit papillomavirus. J.Virol. 2003;77:8736–8744. doi: 10.1128/JVI.77.16.8736-8744.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Jenke BH, Fetzer CP, Stehle IM, Jonsson F, Fackelmayer FO, Conradt H, Bode J, Lipps HJ. An episomally replicating vector binds to the nuclear matrix protein SAF-A in vivo. EMBO Rep. 2002;3:349–354. doi: 10.1093/embo-reports/kvf070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Jiang YW, Veschambre P, Erdjument-Bromage H, Tempst P, Conaway JW, Conaway RC, Kornberg RD. Mammalian mediator of transcriptional regulation and its possible role as an end-point of signal transduction pathways. Proc Natl.Acad.Sci.U.S.A. 1998;95:8538–8543. doi: 10.1073/pnas.95.15.8538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Joyce JG, Tung JS, Przysiecki CT, Cook JC, Lehman ED, Sands JA, Jansen KU, Keller PM. The L1 major capsid protein of human papillomavirus type 11 recombinant virus-like particles interacts with heparin and cell-surface glycosaminoglycans on human keratinocytes. J.Biol.Chem. 1999;274:5810–5822. doi: 10.1074/jbc.274.9.5810. [DOI] [PubMed] [Google Scholar]
- 115.Kapoor P, Frappier L. EBNA1 partitions Epstein-Barr virus plasmids in yeast cells by attaching to human EBNA1-binding protein 2 on mitotic chromosomes. J.Virol. 2003;77:6946–6956. doi: 10.1128/JVI.77.12.6946-6956.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Kelley-Clarke B, Ballestas ME, Komatsu T, Kaye KM. Kaposi's sarcoma herpesvirus C-terminal LANA concentrates at pericentromeric and peri-telomeric regions of a subset of mitotic chromosomes. Virology. 2007;357:149–157. doi: 10.1016/j.virol.2006.07.052. [DOI] [PubMed] [Google Scholar]
- 117.Kim K, Angeletti PC, Hassebroek EC, Lambert PF. Identification of cis-acting elements that mediate the replication and maintenance of human papillomavirus type 16 genomes in Saccharomyces cerevisiae. J.Virol. 2005;79:5933–5942. doi: 10.1128/JVI.79.10.5933-5942.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Kim K, Garner-Hamrick PA, Fisher C, Lee D, Lambert PF. Methylation patterns of papillomavirus DNA, its influence on E2 function, and implications in viral infection. J.Virol. 2003;77:12450–12459. doi: 10.1128/JVI.77.23.12450-12459.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Kim K, Lambert PF. E1 protein of bovine papillomavirus 1 is not required for the maintenance of viral plasmid DNA replication. Virology. 2002;293:10–14. doi: 10.1006/viro.2001.1305. [DOI] [PubMed] [Google Scholar]
- 120.Kirchmaier AL, Sugden B. Rep*: a viral element that can partially replace the origin of plasmid DNA synthesis of Epstein-Barr virus. J.Virol. 1998;72:4657–4666. doi: 10.1128/jvi.72.6.4657-4666.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Kirnbauer R, Booy F, Cheng N, Lowy DR, Schiller JT. Papillomavirus L1 major capsid protein self-assembles into virus-like particles that are highly immunogenic. Proc.Natl.Acad.Sci.U.S.A. 1992;89:12180–12184. doi: 10.1073/pnas.89.24.12180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Knight JD, Li R, Botchan M. The activation domain of the bovine papillomavirus E2 protein mediates association of DNA-bound dimers to form DNA loops. Proc.Natl.Acad.Sci.USA. 1991;88:3204–3208. doi: 10.1073/pnas.88.8.3204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Kobayashi T, Heck DJ, Nomura M, Horiuchi T. Expansion and contraction of ribosomal DNA repeats in Saccharomyces cerevisiae: requirement of replication fork blocking (Fob1) protein and the role of RNA polymerase I. Genes Dev. 1998;12:3821–3830. doi: 10.1101/gad.12.24.3821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Krithivas A, Fujimuro M, Weidner M, Young DB, Hayward SD. Protein interactions targeting the latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus to cell chromosomes. J.Virol. 2002;76:11596–11604. doi: 10.1128/JVI.76.22.11596-11604.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Kruhlak MJ, Hendzel MJ, Fischle W, Bertos NR, Hameed S, Yang XJ, Verdin E, Bazett-Jones DP. Regulation of global acetylation in mitosis through loss of histone acetyltransferases and deacetylases from chromatin. J.Biol.Chem. 2001;276:38307–38319. doi: 10.1074/jbc.M100290200. [DOI] [PubMed] [Google Scholar]
- 126.Kuo SR, Liu JS, Broker TR, Chow LT. Cell-free replication of the human papillomavirus DNA with homologous viral E1 and E2 proteins and human cell extracts. J.Biol.Chem. 1994;269:24058–24065. [PubMed] [Google Scholar]
- 127.Kurg R, Tekkel H, Abroi A, Ustav M. Characterization of the functional activities of the bovine papillomavirus type 1 E2 protein single-chain heterodimers. J.Virol. 2006;80:11218–11225. doi: 10.1128/JVI.01127-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Lacey CJ. Therapy for genital human papillomavirus-related disease. J.Clin.Virol. 2005;32:S82–S90. doi: 10.1016/j.jcv.2004.10.020. [DOI] [PubMed] [Google Scholar]
- 129.Lai MC, Teh BH, Tarn WY. A human papillomavirus E2 transcriptional activator. The interactions with cellular splicing factors and potential function in pre-mRNA processing. J.Biol.Chem. 1999;274:11832–11841. doi: 10.1074/jbc.274.17.11832. [DOI] [PubMed] [Google Scholar]
- 130.Lambert PF, Monk BC, Howley PM. Phenotypic analysis of bovine papillomavirus type 1 E2 repressor mutants. J.Virol. 1990;64:950–956. doi: 10.1128/jvi.64.2.950-956.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Lambert PF, Spalholz BA, Howley PM. A transcriptional repressor encoded by BPV-1 shares a common carboxy-terminal domain with the E2 transactivator. Cell. 1987;50:69–78. doi: 10.1016/0092-8674(87)90663-5. [DOI] [PubMed] [Google Scholar]
- 132.Lee D, Kim H, Lee Y, Choe J. Identification of sequence requirement for the origin of DNA replication in human papillomavirus type 18. Virus Res. 1997;52:97–108. doi: 10.1016/s0168-1702(97)00114-7. [DOI] [PubMed] [Google Scholar]
- 133.Lee KY, Broker TR, Chow LT. Transcription factor YY1 represses cell-free replication from human papillomavirus origins. J.Virol. 1998;72:4911–4917. doi: 10.1128/jvi.72.6.4911-4917.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Lehman CW, Botchan MR. Segregation of viral plasmids depends on tethering to chromosomes and is regulated by phosphorylation. Proc Natl.Acad.Sci.U.S.A. 1998;95:4338–4343. doi: 10.1073/pnas.95.8.4338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Lentz MR. A carboxyl-terminal serine of the bovine papillomavirus E1 protein is phosphorylated in vivo and in vitro. Virus Res. 2002;83:213–219. doi: 10.1016/s0168-1702(02)00003-5. [DOI] [PubMed] [Google Scholar]
- 136.Lentz MR, Pak D, Mohr I, Botchan MR. The E1 replication protein of bovine papillomavirus type 1 contains an extended nuclear localization signal that includes a p34cdc2 phosphorylation site. J.Virol. 1993;67:1414–1423. doi: 10.1128/jvi.67.3.1414-1423.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Lentz MR, Stevens SM, Jr., Raynes J, Elkhoury N. A phosphorylation map of the bovine papillomavirus E1 helicase. Virol.J. 2006;3:13. doi: 10.1186/1743-422X-3-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Li D, Zhao R, Lilyestrom W, Gai D, Zhang R, DeCaprio JA, Fanning E, Jochimiak A, Szakonyi G, Chen XS. Structure of the replicative helicase of the oncoprotein SV40 large tumour antigen. Nature. 2003;423:512–518. doi: 10.1038/nature01691. [DOI] [PubMed] [Google Scholar]
- 139.Li R, Botchan MR. The acidic transcriptional activation domains of VP16 and p53 bind the cellular replication protein A and stimulate in vitro BPV-1 DNA replication. Cell. 1993;73:1207–1221. doi: 10.1016/0092-8674(93)90649-b. [DOI] [PubMed] [Google Scholar]
- 140.Li R, Botchan MR. Acidic transcription factors alleviate nucleosome-mediated repression of DNA replication of bovine papillomavirus type 1. Proc.Natl.Acad.Sci.USA. 1994;91:7051–7055. doi: 10.1073/pnas.91.15.7051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Li R, Knight J, Bream G, Stenlund A, Botchan M. Specific recognition nucleotides and their DNA context determine the affinity of E2 protein for 17 binding sites in the BPV- 1 genome. Genes Dev. 1989;3:510–526. doi: 10.1101/gad.3.4.510. [DOI] [PubMed] [Google Scholar]
- 142.Lim DA, Gossen M, Lehman CW, Botchan MR. Competition for DNA binding sites between the short and long forms of E2 dimers underlies repression in bovine papillomavirus type 1 DNA replication control. J.Virol. 1998;72:1931–1940. doi: 10.1128/jvi.72.3.1931-1940.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Lin BY, Ma T, Liu JS, Kuo SR, Jin G, Broker TR, Harper JW, Chow LT. HeLa cells are phenotypically limiting in cyclin E/CDK2 for efficient human papillomavirus DNA replication. J.Biol.Chem. 2000;275:6167–6174. doi: 10.1074/jbc.275.9.6167. [DOI] [PubMed] [Google Scholar]
- 144.Liu X, Schuck S, Stenlund A. Adjacent residues in the E1 initiator beta-hairpin define different roles of the beta-hairpin in Ori melting, helicase loading, and helicase activity. Mol.Cell. 2007;25:825–837. doi: 10.1016/j.molcel.2007.02.009. [DOI] [PubMed] [Google Scholar]
- 145.Loo YM, Melendy T. Recruitment of replication protein A by the papillomavirus E1 protein and modulation by single-stranded DNA. J.Virol. 2004;78:1605–1615. doi: 10.1128/JVI.78.4.1605-1615.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Lu JZ, Sun YN, Rose RC, Bonnez W, McCance DJ. Two E2 binding sites (E2BS) alone or one E2BS plus an A/T-rich region are minimal requirements for the replication of the human papillomavirus type 11 origin. J.Virol.1993.Dec. 1995;67:7131–7139. doi: 10.1128/jvi.67.12.7131-7139.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Luo X, Sanford DG, Bullock PA, Bachovchin WW. Solution structure of the origin DNA-binding domain of SV40 T-antigen. Nat.Struct.Biol. 1996;3:1034–1039. doi: 10.1038/nsb1296-1034. [DOI] [PubMed] [Google Scholar]
- 148.Lusky M, Botchan MR. A bovine papillomavirus type 1-encoded modulator function is dispensable for transient viral replication but is required for establishment of the stable plasmid state. J.Virol. 1986;60:729–742. doi: 10.1128/jvi.60.2.729-742.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Ma T, Zou N, Lin BY, Chow LT, Harper JW. Interaction between cyclin-dependent kinases and human papillomavirus replication-initiation protein E1 is required for efficient viral replication. Proc.Natl.Acad.Sci.U.S.A. 1999;96:382–387. doi: 10.1073/pnas.96.2.382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Mannik A, Piirsoo M, Nordstrom K, Ustav E, Vennstrom B, Ustav M. Effective generation of transgenic mice by Bovine papillomavirus type 1 based self-replicating plasmid that is maintained as extrachromosomal genetic element in three generations of animals. Plasmid. 2003;49:193–204. doi: 10.1016/s0147-619x(03)00012-x. [DOI] [PubMed] [Google Scholar]
- 151.Mannik A, Runkorg K, Jaanson N, Ustav M, Ustav E. Induction of the bovine papillomavirus origin “onion skin”-type DNA replication at high E1 protein concentrations in vivo. J.Virol. 2002;76:5835–5845. doi: 10.1128/JVI.76.11.5835-5845.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Mansky KC, Batiza A, Lambert PF. Bovine papillomavirus type 1 E1 and simian virus 40 large T antigen share regions of sequence similarity required for multiple functions. J.Virol. 1997;71:7600–7608. doi: 10.1128/jvi.71.10.7600-7608.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Maruyama T, Farina A, Dey A, Cheong J, Bermudez VP, Tamura T, Sciortino S, Shuman J, Hurwitz J, Ozato K. A Mammalian bromodomain protein, Brd4, interacts with replication factor C and inhibits progression to S phase. Mol.Cell Biol. 2002;22:6509–6520. doi: 10.1128/MCB.22.18.6509-6520.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Masai H, You Z, Arai K. Control of DNA replication: regulation and activation of eukaryotic replicative helicase, MCM. IUBMB.Life. 2005;57:323–335. doi: 10.1080/15216540500092419. [DOI] [PubMed] [Google Scholar]
- 155.Masterson PJ, Stanley MA, Lewis AP, Romanos MA. A C-terminal helicase domain of the human papillomavirus E1 protein binds E2 and the DNA polymerase alpha-primase p68 subunit. J.Virol. 1998;72:7407–7419. doi: 10.1128/jvi.72.9.7407-7419.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.McBride AA, Dlugosz A, Baker CC. Production of Infectious BPV-1 Virions from Cloned Viral DNA Using An Organotypic Raft /Xenograft Technique. Proc Natl.Acad.Sci.U.S.A. 2000;97:5534–5539. doi: 10.1073/pnas.97.10.5534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.McBride AA, Bolen JB, Howley PM. Phosphorylation sites of the E2 transcriptional regulatory proteins of bovine papillomavirus type 1. J.Virol. 1989;63:5076–5085. doi: 10.1128/jvi.63.12.5076-5085.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.McBride AA, Howley PM. Bovine papillomavirus with a mutation in the E2 serine 301 phosphorylation site replicates at a high copy number. J.Virol. 1991;65:6528–6534. doi: 10.1128/jvi.65.12.6528-6534.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.McBride AA, Myers G. The E2 proteins: an update. In: Myers G, et al., editors. Human Papillomaviruses 1997. Los Alamos National Laboratory; Los Alamos: 1997. pp. III54–III99. [Google Scholar]
- 160.McBride AA, Oliveira JG, McPhillips MG. Partitioning viral genomes in mitosis: same idea, different targets. Cell Cycle. 2006;5:1499–1502. doi: 10.4161/cc.5.14.3094. [DOI] [PubMed] [Google Scholar]
- 161.McMillan NA, Payne E, Frazer IH, Evander M. Expression of the alpha6 integrin confers papillomavirus binding upon receptor-negative B-cells. Virology. 1999;261:271–279. doi: 10.1006/viro.1999.9825. [DOI] [PubMed] [Google Scholar]
- 162.McPhillips MG, Oliveira JG, Spindler JE, Mitra R, McBride AA. Brd4 is required for e2-mediated transcriptional activation but not genome partitioning of all papillomaviruses. J.Virol. 2006;80:9530–9543. doi: 10.1128/JVI.01105-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.McPhillips MG, Ozato K, McBride AA. Interaction of bovine papillomavirus E2 protein with Brd4 stabilizes its association with chromatin. J.Virol. 2005;79:8920–8932. doi: 10.1128/JVI.79.14.8920-8932.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.McShan GD, Wilson VG. Casein kinase II phosphorylates bovine papillomavirus type 1 E1 in vitro at a conserved motif. J.Gen.Virol. 1997a;78:171–177. doi: 10.1099/0022-1317-78-1-171. [DOI] [PubMed] [Google Scholar]
- 165.McShan GD, Wilson VG. Reconstitution of a functional bovine papillomavirus type 1 origin of replication reveals a modular tripartite replicon with an essential AT-rich element. Virology. 1997b;237:198–208. doi: 10.1006/viro.1997.8793. [DOI] [PubMed] [Google Scholar]
- 166.McStay B. Nucleolar dominance: a model for rRNA gene silencing. Genes Dev. 2006;20:1207–1214. doi: 10.1101/gad.1436906. [DOI] [PubMed] [Google Scholar]
- 167.Mechali F, Hsu CY, Castro A, Lorca T, Bonne-Andrea C. Bovine papillomavirus replicative helicase E1 is a target of the ubiquitin ligase APC. J.Virol. 2004;78:2615–2619. doi: 10.1128/JVI.78.5.2615-2619.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Meinke G, Bullock PA, Bohm A. Crystal structure of the simian virus 40 large T-antigen origin-binding domain. J.Virol. 2006;80:4304–4312. doi: 10.1128/JVI.80.9.4304-4312.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Meinke G, Phelan P, Moine S, Bochkareva E, Bochkarev A, Bullock PA, Bohm A. The crystal structure of the SV40 T-antigen origin binding domain in complex with DNA. PLoS.Biol. 2007;5:e23. doi: 10.1371/journal.pbio.0050023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Melendy T, Sedman J, Stenlund A. Cellular factors required for papillomavirus DNA replication. J.Virol. 1995;69:7857–7867. doi: 10.1128/jvi.69.12.7857-7867.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Meyers C, Frattini MG, Hudson JB, Laimins LA. Biosynthesis of human papillomavirus from a continuous cell line upon epithelial differentiation. Science. 1992;257:971–973. doi: 10.1126/science.1323879. [DOI] [PubMed] [Google Scholar]
- 172.Meyers C, Mayer TJ, Ozbun MA. Synthesis of infectious human papillomavirus type 18 in differentiating epithelium transfected with viral DNA. J.Virol. 1997;71:7381–7386. doi: 10.1128/jvi.71.10.7381-7386.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Mohr IJ, Clark R, Sun S, Androphy EJ, MacPherson P, Botchan MR. Targeting the E1 replication protein to the papillomavirus origin of replication by complex formation with the E2 transactivator. Science. 1990;250:1694–1699. doi: 10.1126/science.2176744. [DOI] [PubMed] [Google Scholar]
- 174.Moody CA, Fradet-Turcotte A, Archambault J, Laimins LA. Human papillomaviruses activate caspases upon epithelial differentiation to induce viral genome amplification. Proc.Natl.Acad.Sci.U.S.A. 2007;104:19541–19546. doi: 10.1073/pnas.0707947104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Muller F, Seo YS, Hurwitz J. Replication of bovine papillomavirus type 1 origin-containing DNA in crude extracts and with purified proteins. J.Biol.Chem. 1994;269:17086–17094. [PubMed] [Google Scholar]
- 176.Narahari J, Fisk JC, Melendy T, Roman A. Interactions of the cellular CCAAT displacement protein and human papillomavirus E2 protein with the viral origin of replication can regulate DNA replication. Virology. 2006;350:302–311. doi: 10.1016/j.virol.2006.01.047. [DOI] [PubMed] [Google Scholar]
- 177.Narajczyk M, Baranska S, Wegrzyn A, Wegrzyn G. Switch from theta to sigma replication of bacteriophage lambda DNA: factors involved in the process and a model for its regulation. Molecular Genetics and Genomics. 2007;278:65–74. doi: 10.1007/s00438-007-0228-y. [DOI] [PubMed] [Google Scholar]
- 178.Negorev D, Maul GG. Cellular proteins localized at and interacting within ND10/PML nuclear bodies/PODs suggest functions of a nuclear depot. Oncogene. 2001;20:7234–7242. doi: 10.1038/sj.onc.1204764. [DOI] [PubMed] [Google Scholar]
- 179.Newhouse CD, Silverstein SJ. Orientation of a novel DNA binding site affects human papillomavirus- mediated transcription and replication. J.Virol. 2001;75:1722–1735. doi: 10.1128/JVI.75.4.1722-1735.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.O'Connor MJ, Stunkel W, Koh CH, Zimmermann H, Bernard HU. The differentiation-specific factor CDP/Cut represses transcription and replication of human papillomaviruses through a conserved silencing element. J.Virol. 2000;74:401–410. [PMC free article] [PubMed] [Google Scholar]
- 181.O'Connor MJ, Tan SH, Tan CH, Bernard HU. YY1 represses human papillomavirus type 16 transcription by quenching AP-1 activity. J.Virol. 1996;70:6529–6539. doi: 10.1128/jvi.70.10.6529-6539.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Ohe Y, Zhao D, Saijo N, Podack ER. Construction of a novel bovine papillomavirus vector without detectable transforming activity suitable for gene transfer. Hum. Gene Ther. 1995;6:325–333. doi: 10.1089/hum.1995.6.3-325. [DOI] [PubMed] [Google Scholar]
- 183.Oliveira JG, Colf LA, McBride AA. Variations in the association of papillomavirus E2 proteins with mitotic chromosomes. Proc. Natl. Acad. Sci. U.S.A. 2006;103:1047–1052. doi: 10.1073/pnas.0507624103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Ozbun MA, Meyers C. Human papillomavirus type 31b E1 and E2 transcript expression correlates with vegetative viral genome amplification. Virology. 1998;248:218–230. doi: 10.1006/viro.1998.9285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Parish JL, Bean AM, Park RB, Androphy EJ. ChlR1 is required for loading papillomavirus E2 onto mitotic chromosomes and viral genome maintenance. Mol. Cell. 2006;24:867–876. doi: 10.1016/j.molcel.2006.11.005. [DOI] [PubMed] [Google Scholar]
- 186.Park RB, Androphy EJ. Genetic analysis of high-risk e6 in episomal maintenance of human papillomavirus genomes in primary human keratinocytes. J. Virol. 2002;76:11359–11364. doi: 10.1128/JVI.76.22.11359-11364.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Parkin DM, Bray F, Ferlay J, Pisani P. Global cancer statistics, 2002. CA Cancer J. Clin. 2005;55:74–108. doi: 10.3322/canjclin.55.2.74. [DOI] [PubMed] [Google Scholar]
- 188.Pattison S, Skalnik DG, Roman A. CCAAT displacement protein, a regulator of differentiation-specific gene expression, binds a negative regulatory element within the 5′ end of the human papillomavirus type 6 long control region. J. Virol. 1997;71:2013–2022. doi: 10.1128/jvi.71.3.2013-2022.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Peh WL, Middleton K, Christensen N, Nicholls P, Egawa K, Sotlar K, Brandsma J, Percival A, Lewis J, Liu WJ, Doorbar J. Life cycle heterogeneity in animal models of human papillomavirus- associated disease. J. Virol. 2002;76:10401–10416. doi: 10.1128/JVI.76.20.10401-10416.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Penrose KJ, Garcia-Alai M, Prat-Gay G, McBride AA. CK2 phosphorylation-induced conformational switch triggers degradation of the papillomavirus E2 protein. J. Biol. Chem. 2004;279:22430–22439. doi: 10.1074/jbc.M314340200. [DOI] [PubMed] [Google Scholar]
- 191.Penrose KJ, McBride AA. Proteasome-mediated degradation of the papillomavirus E2-TA protein is regulated by phosphorylation and can modulate viral genome copy number. J. Virol. 2000;74:6031–6038. doi: 10.1128/jvi.74.13.6031-6038.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Piirsoo M, Ustav E, Mandel T, Stenlund A, Ustav M. Cis and trans requirements for stable episomal maintenance of the BPV-1 replicator. EMBO J. 1996;15:1–11. [PMC free article] [PubMed] [Google Scholar]
- 193.Pittayakhajortwut D, Angeletti PC. Analysis of cis-elements that facilitate extrachromosomal persistence of human papillomavirus genomes. Virology. 2008;374:304–314. doi: 10.1016/j.virol.2008.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Platt GM, Simpson GR, Mittnacht S, Schulz TF. Latent nuclear antigen of Kaposi's sarcoma-associated herpesvirus interacts with RING3, a homolog of the Drosophila female sterile homeotic (fsh) gene. J. Virol. 1999;73:9789–9795. doi: 10.1128/jvi.73.12.9789-9795.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Poddar A, Reed SC, McPhillips MG, McBride AA. The Human Papillomavirus Type 8 E2 Tethering Protein Targets the Ribosomal DNA Loci of Host Mitotic Chromosomes. 2008 doi: 10.1128/JVI.01936-08. manuscript submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Rangasamy D, Wilson VG. Bovine papillomavirus E1 protein is sumoylated by the host cell Ubc9 protein. J. Biol. Chem. 2000;275:30487–30495. doi: 10.1074/jbc.M003898200. [DOI] [PubMed] [Google Scholar]
- 197.Rangasamy D, Woytek K, Khan SA, Wilson VG. SUMO-1 modification of bovine papillomavirus E1 protein is required for intranuclear accumulation. J. Biol. Chem. 2000;275:37999–38004. doi: 10.1074/jbc.M007777200. [DOI] [PubMed] [Google Scholar]
- 198.Ravnan J-B, Gilbert GM, Ten Hagen KG, Cohen SN. Random-choice replication of extrachromosomal bovine papillomavirus (BPV) molecules in heterogeneous clonally-derived BPV-infected cell lines. J. Virol. 1992;66:6946–6952. doi: 10.1128/jvi.66.12.6946-6952.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Rawlins DR, Milman G, Hayward SD, Hayward GS. Sequence-specific DNA binding of the Epstein-Barr virus nuclear antigen (EBNA-1) to clustered sites in the plasmid maintenance region. Cell. 1985;42:859–868. doi: 10.1016/0092-8674(85)90282-x. [DOI] [PubMed] [Google Scholar]
- 200.Remm M, Brain R, Jenkins JR. The E2 binding sites determine the efficiency of replication for the origin of human papillomavirus type 18. Nucleic. Acids. Res. 1992;20:6015–6021. doi: 10.1093/nar/20.22.6015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Riese DJ, II, Settleman J, Neary K, DiMaio D. Bovine papillomavirus E2 repressor mutant displays a high- copy-number phenotype and enhanced transforming activity. J. Virol. 1990;64:944–949. doi: 10.1128/jvi.64.2.944-949.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Rogers S. Shope Papilloma Virus - A Passenger in Man and Its Significance to Potential Control of Host Genome. Nature. 1966;212:1220–1222. doi: 10.1038/2121220a0. [DOI] [PubMed] [Google Scholar]
- 203.Rosas-Acosta G, Wilson VG. Identification of a nuclear export signal sequence for bovine papillomavirus E1 protein. Virology. 2008;373:149–162. doi: 10.1016/j.virol.2007.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Rotenberg MO, Chow LT, Broker TR. Characterization of rare human papillomavirus type 11 mRNAs coding for regulatory and structural proteins, using the polymerase chain reaction. Virology. 1989;172:489–497. doi: 10.1016/0042-6822(89)90191-8. [DOI] [PubMed] [Google Scholar]
- 205.Russell J, Botchan MR. cis-Acting components of human papillomavirus (HPV) DNA replication: linker substitution analysis of the HPV type 11 origin. J. Virol. 1995;69:651–660. doi: 10.1128/jvi.69.2.651-660.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Sakai H, Yasugi T, Benson JD, Dowhanick JJ, Howley PM. Targeted mutagenesis of the human papillomavirus type 16 E2 transactivation domain reveals separable transcriptional activation and DNA replication functions. J. Virol. 1996;70:1602–1611. doi: 10.1128/jvi.70.3.1602-1611.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Sanders CM, Sizov D, Seavers PR, Ortiz-Lombardia M, Antson AA. Transcription activator structure reveals redox control of a replication initiation reaction. Nucleic Acids Res. 2007;35:3504–3515. doi: 10.1093/nar/gkm166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Sanders CM, Stenlund A. Recruitment and loading of the E1 initiator protein: an ATP-dependent process catalysed by a transcription factor. EMBO J. 1998;17:7044–7055. doi: 10.1093/emboj/17.23.7044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Sarafi TR, McBride AA. Domains of the BPV-1 E1 replication protein required for origin-specific DNA binding and interaction with the E2 transactivator. Virology. 1995;211:385–396. doi: 10.1006/viro.1995.1421. [DOI] [PubMed] [Google Scholar]
- 210.Sarver N, Gruss P, Law MF, Khoury G, Howley PM. Bovine papilloma virus deoxyribonucleic acid: a novel eucaryotic cloning vector. Mol. Cell. Biol. 1981;1:486–496. doi: 10.1128/mcb.1.6.486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Schaal TD, Mallet WG, McMinn DL, Nguyen NV, Sopko MM, John S, Parekh BS. Inhibition of human papilloma virus E2 DNA binding protein by covalently linked polyamides. Nucleic Acids Res. 2003;31:1282–1291. doi: 10.1093/nar/gkg206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Schepers A, Ritzi M, Bousset K, Kremmer E, Yates JL, Harwood J, Diffley JF, Hammerschmidt W. Human origin recognition complex binds to the region of the latent origin of DNA replication of Epstein-Barr virus. EMBO J. 2001;20:4588–4602. doi: 10.1093/emboj/20.16.4588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Schuck S, Stenlund A. Assembly of a double hexameric helicase. Mol. Cell. 2005;20:377–389. doi: 10.1016/j.molcel.2005.09.020. [DOI] [PubMed] [Google Scholar]
- 214.Schuck S, Stenlund A. Surface mutagenesis of the bovine papillomavirus E1 DNA binding domain reveals residues required for multiple functions related to DNA replication. J. Virol. 2006;80:7491–7499. doi: 10.1128/JVI.00435-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Schweiger MR, You J, Howley PM. Bromodomain protein 4 mediates the papillomavirus e2 transcriptional activation function. J. Virol. 2006;80:4276–4285. doi: 10.1128/JVI.80.9.4276-4285.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Sears J, Ujihara M, Wong S, Ott C, Middeldorp J, Aiyar A. The amino terminus of Epstein-Barr Virus (EBV) nuclear antigen 1 contains AT hooks that facilitate the replication and partitioning of latent EBV genomes by tethering them to cellular chromosomes. J. Virol. 2004;78:11487–11505. doi: 10.1128/JVI.78.21.11487-11505.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Sedman J, Stenlund A. The papillomavirus E1 protein forms a DNA-dependent hexameric complex with ATPase and DNA helicase activities. J. Virol. 1998;72:6893–6897. doi: 10.1128/jvi.72.8.6893-6897.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Sedman T, Sedman J, Stenlund A. Binding of the E1 and E2 proteins to the origin of replication of bovine papillomavirus. J. Virol. 1997;71:2887–2896. doi: 10.1128/jvi.71.4.2887-2896.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Senechal H, Poirier GG, Coulombe B, Laimins LA, Archambault J. Amino acid substitutions that specifically impair the transcriptional activity of papillomavirus E2 affect binding to the long isoform of Brd4. Virology. 2007;358:10–17. doi: 10.1016/j.virol.2006.08.035. [DOI] [PubMed] [Google Scholar]
- 220.Shafti-Keramat S, Handisurya A, Kriehuber E, Meneguzzi G, Slupetzky K, Kirnbauer R. Different heparan sulfate proteoglycans serve as cellular receptors for human papillomaviruses. J. Virol. 2003;77:13125–13135. doi: 10.1128/JVI.77.24.13125-13135.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Shire K, Ceccarelli DF, Avolio-Hunter TM, Frappier L. EBP2, a human protein that interacts with sequences of the Epstein-Barr virus nuclear antigen 1 important for plasmid maintenance. J. Virol. 1999;73:2587–2595. doi: 10.1128/jvi.73.4.2587-2595.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Si H, Verma SC, Lampson M, Cai Q, Robertson ES. KSHV Encoded LANA can Interact with the Nuclear Mitotic Apparatus Protein to Regulate Genome Maintenance and Segregation. J. Virol. 2008 doi: 10.1128/JVI.00342-08. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Sim J, Ozgur S, Lin BY, Yu JH, Broker TR, Chow LT, Griffith J. Remodeling of the human papillomavirus type 11 replication origin into discrete nucleoprotein particles and looped structures by the E2 protein. J. Mol. Biol. 2008;375:1165–1177. doi: 10.1016/j.jmb.2007.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Skiadopoulos MH, McBride AA. Bovine papillomavirus type 1 genomes and the E2 transactivator protein are closely associated with mitotic chromatin. J. Virol. 1998;72:2079–2088. doi: 10.1128/jvi.72.3.2079-2088.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Smith JL, Campos SK, Ozbun MA. Human papillomavirus type 31 uses a caveolin 1- and dynamin 2-mediated entry pathway for infection of human keratinocytes. J. Virol. 2007;81:9922–9931. doi: 10.1128/JVI.00988-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Soeda E, Ferran MC, Baker CC, McBride AA. Repression of HPV16 early region transcription by the E2 protein. Virology. 2006;351:29–41. doi: 10.1016/j.virol.2006.03.016. [DOI] [PubMed] [Google Scholar]
- 227.Stanley MA, Browne HM, Appleby M, Minson AC. Properties of a non-tumorigenic human cervical keratinocyte cell line. Int. J. Cancer. 1989;43:672–676. doi: 10.1002/ijc.2910430422. [DOI] [PubMed] [Google Scholar]
- 228.Steger G, Schnabel C, Schmidt HM. The hinge region of the human papillomavirus type 8 E2 protein activates the human p21(WAF1/CIP1) promoter via interaction with Sp1. J. Gen. Virol. 2002;83:503–510. doi: 10.1099/0022-1317-83-3-503. [DOI] [PubMed] [Google Scholar]
- 229.Stenlund A. E1 initiator DNA binding specificity is unmasked by selective inhibition of non-specific DNA binding. EMBO J. 2003;22:954–963. doi: 10.1093/emboj/cdg091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Stubenrauch F, Colbert AM, Laimins LA. Transactivation by the E2 protein of oncogenic human papillomavirus type 31 is not essential for early and late viral functions. J. Virol. 1998a;72:8115–8123. doi: 10.1128/jvi.72.10.8115-8123.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Stubenrauch F, Hummel M, Iftner T, Laimins LA. The E8E2C protein, a negative regulator of viral transcription and replication, is required for extrachromosomal maintenance of human papillomavirus type 31 in keratinocytes. J.Virol. 2000;74:1178–1186. doi: 10.1128/jvi.74.3.1178-1186.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Stubenrauch F, Lim HB, Laimins LA. Differential requirements for conserved E2 binding sites in the life cycle of oncogenic human papillomavirus type 31. J.Virol. 1998b;72:1071–1077. doi: 10.1128/jvi.72.2.1071-1077.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Sullivan M, Higuchi T, Katis VL, Uhlmann F. Cdc14 phosphatase induces rDNA condensation and resolves cohesin-independent cohesion during budding yeast anaphase. Cell. 2004;117:471–482. doi: 10.1016/s0092-8674(04)00415-5. [DOI] [PubMed] [Google Scholar]
- 234.Sun Y, Han H, McCance DJ. Active domains of human papillomavirus type 11 E1 protein for origin replication. J.Gen.Virol. 1998;79:1651–1658. doi: 10.1099/0022-1317-79-7-1651. [DOI] [PubMed] [Google Scholar]
- 235.Sun YN, Lu JZ, McCance DJ. Mapping of HPV-11 E1 binding site and determination of other important cis elements for replication of the origin. Virology. 1996;216:219–222. doi: 10.1006/viro.1996.0050. [DOI] [PubMed] [Google Scholar]
- 236.Sverdrup F, Khan SA. Two E2 binding sites alone are sufficient to function as the minimal origin of replication of human papillomavirus type 18 DNA. J.Virol. 1995;69:1319–1323. doi: 10.1128/jvi.69.2.1319-1323.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Sverdrup F, Sheahan L, Khan S. Development of human papillomavirus plasmids capable of episomal replication in human cell lines. Gene Ther. 1999;6:1317–1321. doi: 10.1038/sj.gt.3300957. [DOI] [PubMed] [Google Scholar]
- 238.Swindle CS, Zou N, Van Tine BA, Shaw GM, Engler JA, Chow LT. Human papillomavirus DNA replication compartments in a transient DNA replication system. J.Virol. 1999;73:1001–1009. doi: 10.1128/jvi.73.2.1001-1009.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Tan SH, Bartsch D, Schwarz E, Bernard HU. Nuclear matrix attachment regions of human papillomavirus type 16 point toward conservation of these genomic elements in all genital papillomaviruses. J.Virol. 1998;72:3610–3622. doi: 10.1128/jvi.72.5.3610-3622.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Thain A, Jenkins O, Clarke AR, Gaston K. CpG methylation directly inhibits binding of the human papillomavirus type 16 E2 protein to specific DNA sequences. J.Virol. 1996;70:7233–7235. doi: 10.1128/jvi.70.10.7233-7235.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Thierry F, Yaniv M. The BPV1-E2 trans-acting protein can be either an activator or a repressor of the HPV18 regulatory region. EMBO J. 1987;6:3391–3397. doi: 10.1002/j.1460-2075.1987.tb02662.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Thomas JT, Hubert WG, Ruesch MN, Laimins LA. Human papillomavirus type 31 oncoproteins E6 and E7 are required for the maintenance of episomes during the viral life cycle in normal human keratinocytes. Proc.Natl.Acad.Sci.U.S.A. 1999;96:8449–8454. doi: 10.1073/pnas.96.15.8449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Titolo S, Brault K, Majewski J, White PW, Archambault J. Characterization of the minimal DNA binding domain of the human papillomavirus e1 helicase: fluorescence anisotropy studies and characterization of a dimerization-defective mutant protein. J.Virol. 2003;77:5178–5191. doi: 10.1128/JVI.77.9.5178-5191.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Titolo S, Pelletier A, Pulichino AM, Brault K, Wardrop E, White PW, Cordingley MG, Archambault J. Identification of domains of the human papillomavirus type 11 E1 helicase involved in oligomerization and binding to the viral origin. J.Virol. 2000;74:7349–7361. doi: 10.1128/jvi.74.16.7349-7361.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Torres-Rosell J, Machin F, Farmer S, Jarmuz A, Eydmann T, Dalgaard JZ, Aragon L. SMC5 and SMC6 genes are required for the segregation of repetitive chromosome regions. Nat.Cell Biol. 2005;7:412–419. doi: 10.1038/ncb1239. [DOI] [PubMed] [Google Scholar]
- 246.Tsurumi T, Fujita M, Kudoh A. Latent and lytic Epstein-Barr virus replication strategies. Rev.Med.Virol. 2005;15:3–15. doi: 10.1002/rmv.441. [DOI] [PubMed] [Google Scholar]
- 247.Ustav E, Ustav M, Szymanski P, Stenlund A. The bovine papillomavirus origin of replication requires a binding site for the E2 transcriptional activator. Proc Natl.Acad.Sci.U.S.A. 1993;90:898–902. doi: 10.1073/pnas.90.3.898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Ustav M, Stenlund A. Transient replication of BPV-1 requires two viral polypeptides encoded by the E1 and E2 open reading frames. EMBO J. 1991;10:449–457. doi: 10.1002/j.1460-2075.1991.tb07967.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Ustav M, Ustav E, Szymanski P, Stenlund A. Identification of the origin of replication of bovine papillomavirus and characterization of the viral origin recognition factor E1. EMBO J. 1991;10:4321–4329. doi: 10.1002/j.1460-2075.1991.tb05010.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Van Tine BA, Dao LD, Wu SY, Sonbuchner TM, Lin BY, Zou N, Chiang CM, Broker TR, Chow LT. Human papillomavirus (HPV) origin-binding protein associates with mitotic spindles to enable viral DNA partitioning. Proc Natl.Acad.Sci.U.S.A. 2004;101:4030–4035. doi: 10.1073/pnas.0306848101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Viejo-Borbolla A, Ottinger M, Bruning E, Burger A, Konig R, Kati E, Sheldon JA, Schulz TF. Brd2/RING3 interacts with a chromatin-binding domain in the Kaposi's Sarcoma-associated herpesvirus latency-associated nuclear antigen 1 (LANA-1) that is required for multiple functions of LANA-1. J.Virol. 2005;79:13618–13629. doi: 10.1128/JVI.79.21.13618-13629.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Voitenleitner C, Botchan M. E1 protein of bovine papillomavirus type 1 interferes with E2 protein- mediated tethering of the viral DNA to mitotic chromosomes. J.Virol. 2002;76:3440–3451. doi: 10.1128/JVI.76.7.3440-3451.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Walboomers JM, Jacobs MV, Manos MM, Bosch FX, Kummer JA, Shah KV, Snijders PJ, Peto J, Meijer CJ, Munoz N. Human papillomavirus is a necessary cause of invasive cervical cancer worldwide. J.Pathol. 1999;189:12–19. doi: 10.1002/(SICI)1096-9896(199909)189:1<12::AID-PATH431>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 254.Waldenstrom M, Schenstrom K, Sollerbrant K, Hansson L. Replication of bovine papillomavirus vectors in murine cells. Gene. 1992;120:175–181. doi: 10.1016/0378-1119(92)90091-3. [DOI] [PubMed] [Google Scholar]
- 255.Wang Y, Coulombe R, Cameron DR, Thauvette L, Massariol MJ, Amon LM, Fink D, Titolo S, Welchner E, Yoakim C, Archambault J, White PW. Crystal structure of the E2 transactivation domain of human papillomavirus type 11 bound to a protein interaction inhibitor. J.Biol.Chem. 2004;279:6976–6985. doi: 10.1074/jbc.M311376200. [DOI] [PubMed] [Google Scholar]
- 256.White PW, Titolo S, Brault K, Thauvette L, Pelletier A, Welchner E, Bourgon L, Doyon L, Ogilvie WW, Yoakim C, Cordingley MG, Archambault J. Inhibition of human papillomavirus DNA replication by small molecule antagonists of the E1–E2 protein interaction. J.Biol.Chem. 2003;278:26765–26772. doi: 10.1074/jbc.M303608200. [DOI] [PubMed] [Google Scholar]
- 257.Winokur PL, McBride AA. The transactivation and DNA binding domains of the BPV-1 E2 protein have different roles in cooperative origin binding with the E1 protein. Virology. 1996;221:44–53. doi: 10.1006/viro.1996.0351. [DOI] [PubMed] [Google Scholar]
- 258.Wise-Draper TM, Wells SI. Papillomavirus E6 and E7 proteins and their cellular targets. Front Biosci. 2008;13:1003–1017. doi: 10.2741/2739. [DOI] [PubMed] [Google Scholar]
- 259.Woodworth CD. HPV innate immunity. Front Biosci. 2002;7:d2058–d2071. doi: 10.2741/A898. [DOI] [PubMed] [Google Scholar]
- 260.Wu H, Ceccarelli DF, Frappier L. The DNA segregation mechanism of Epstein-Barr virus nuclear antigen 1. EMBO Rep. 2000;1:140–144. doi: 10.1093/embo-reports/kvd026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Wu SY, Lee AY, Hou SY, Kemper JK, Erdjument-Bromage H, Tempst P, Chiang CM. Brd4 links chromatin targeting to HPV transcriptional silencing. Genes Dev. 2006;20:2383–2396. doi: 10.1101/gad.1448206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Yang L, Mohr I, Fouts E, Lim DA, Nohaile M, Botchan M. The E1 protein of bovine papillomavirus 1 is an ATP-dependent DNA helicase. Proc.Natl.Acad.Sci.USA. 1993;90:5086–5090. doi: 10.1073/pnas.90.11.5086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Yates JL, Guan N. Epstein-Barr virus-derived plasmids replicate only once per cell cycle and are not amplified after entry into cells. J.Virol. 1991;65:483–488. doi: 10.1128/jvi.65.1.483-488.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Yates JL, Warren N, Sugden B. Stable replication of plasmids derived from Epstein-Barr virus in various mammalian cells. Nature. 1985;313:812–815. doi: 10.1038/313812a0. [DOI] [PubMed] [Google Scholar]
- 265.You J, Croyle JL, Nishimura A, Ozato K, Howley PM. Interaction of the bovine papillomavirus E2 protein with Brd4 tethers the viral DNA to host mitotic chromosomes. Cell. 2004;117:349–360. doi: 10.1016/s0092-8674(04)00402-7. [DOI] [PubMed] [Google Scholar]
- 266.You J, Schweiger MR, Howley PM. Inhibition of E2 binding to Brd4 enhances viral genome loss and phenotypic reversion of bovine papillomavirus-transformed cells. J.Virol. 2005;79:14956–14961. doi: 10.1128/JVI.79.23.14956-14961.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.You J, Srinivasan V, Denis GV, Harrington WJ, Jr., Ballestas ME, Kaye KM, Howley PM. Kaposi's sarcoma-associated herpesvirus latency-associated nuclear antigen interacts with bromodomain protein Brd4 on host mitotic chromosomes. J.Virol. 2006;80:8909–8919. doi: 10.1128/JVI.00502-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Yu JH, Lin BY, Deng W, Broker TR, Chow LT. Mitogen-activated protein kinases activate the nuclear localization sequence of human papillomavirus type 11 E1 DNA helicase to promote efficient nuclear import. J.Virol. 2007a;81:5066–5078. doi: 10.1128/JVI.02480-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Yu T, Peng YC, Androphy EJ. Mitotic kinesin-like protein 2 binds and colocalizes with papillomavirus E2 during mitosis. J.Virol. 2007b;81:1736–1745. doi: 10.1128/JVI.01638-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Zanardi TA, Stanley CM, Saville BM, Spacek SM, Lentz MR. Modulation of bovine papillomavirus DNA replication by phosphorylation of the viral E1 protein. Virology. 1997;228:1–10. doi: 10.1006/viro.1996.8375. [DOI] [PubMed] [Google Scholar]
- 271.Zhang B, Chen W, Roman A. The E7 proteins of low- and high-risk human papillomaviruses share the ability to target the pRB family member p130 for degradation. Proc.Natl.Acad.Sci.U.S.A. 2006;103:437–442. doi: 10.1073/pnas.0510012103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Zhang BY, Li P, Wang E, Brahmi Z, Dunn KW, Blum JS, Roman A. The E5 protein of human papillornavirus type 16 perturbs MHC class II antigen maturation in human foreskin keratinocytes treated with interferon-gamma. Virology. 2003;310:100–108. doi: 10.1016/s0042-6822(03)00103-x. [DOI] [PubMed] [Google Scholar]
- 273.Zhao KN, Sun XY, Frazer IH, Zhou J. DNA packaging by L1 and L2 capsid proteins of bovine papillomavirus type 1. Virology. 1998;243:482–491. doi: 10.1006/viro.1998.9091. [DOI] [PubMed] [Google Scholar]
- 274.Zheng PS, Brokaw J, McBride AA. Conditional mutations in the mitotic chromosome binding function of the bovine papillomavirus type 1 E2 protein. J.Virol. 2005;79:1500–1509. doi: 10.1128/JVI.79.3.1500-1509.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Zhou J, Sun XY, Stenzel DJ, Frazer IH. Expression of vaccinia recombinant HPV 16 L1 and L2 ORF proteins in epithelial cells is sufficient for assembly of HPV virion-like particles. Virology. 1991;185:251–257. doi: 10.1016/0042-6822(91)90772-4. [DOI] [PubMed] [Google Scholar]
- 276.Zobel T, Iftner T, Stubenrauch F. The papillomavirus E8-E2C protein represses DNA replication from extrachromosomal origins. Mol.Cell Biol. 2003;23:8352–8362. doi: 10.1128/MCB.23.22.8352-8362.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Zou N, Lin BY, Duan F, Lee KY, Jin G, Guan R, Yao G, Lefkowitz EJ, Broker TR, Chow LT. The hinge of the human papillomavirus type 11 E2 protein contains major determinants for nuclear localization and nuclear matrix association. J.Virol. 2000;74:3761–3770. doi: 10.1128/jvi.74.8.3761-3770.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]