Abstract
Mammalian aging is associated with elevated levels of oxidative damage of DNA, proteins, and lipids as a result of unbalanced prooxidant and antioxidant activities. Accumulating evidence indicates that oxidative stress is a major physiological inducer of aging. p53, the guardian of the genome that is important for cellular responses to oxidative stresses, might be a key coordinator of oxidative stress and aging. In response to low levels of oxidative stresses, p53 exhibits antioxidant activities to eliminate oxidative stress and ensure cell survival; in response to high levels of oxidative stresses, p53 exhibits prooxidative activities that further increase the levels of stresses, leading to cell death. p53 accomplishes these context-dependent roles by regulating the expression of a panel of genes involved in cellular responses to oxidative stresses and by modulating other pathways important for oxidative stress responses. The mechanism that switches p53 function from antioxidant to prooxidant remains unclear, but could account for the findings that increased p53 activities have been linked to both accelerated aging and increased life span in mice. Therefore, a balance of p53 antioxidant and prooxidant activities in response to oxidative stresses could be important for longevity by suppressing the accumulation of oxidative stresses and DNA damage. Antioxid. Redox Signal. 15, 1669–1678.
p53 Is a Critical Tumor Suppressor
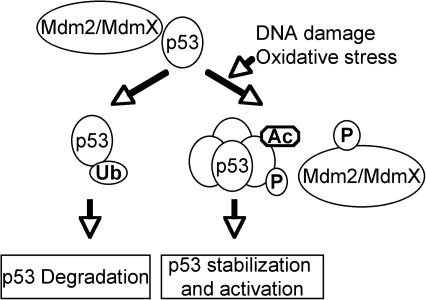
The critical tumor suppressor p53 plays important roles in cell-cycle arrest, apoptosis, senescence, or differentiation in response to various genotoxic and cellular stresses, including oxidative stress (73, 102, 133). As a guardian of the genome, the inactivation of wild-type p53 function by direct gene mutation or disruption of pathways important for p53 activation is a prerequisite for the development of most human cancers (35, 92, 127). As a transcription factor, p53 consists of two N-terminal transactivation domains, a core DNA-binding domain and a C-terminal oligomerization domain (55, 92). Because of its potent activity in inducing apoptosis and senescence, the p53 stability and activity are tightly regulated by posttranslational mechanisms (47, 51, 129). In the absence of stresses, p53 is inactive and unstable because of its interaction with Mdm2 and MdmX, which inactivate p53 and ubiquitinate p53 for proteasome-dependent degradation (Fig. 1). In response to stresses, p53 is modified posttranslationally through phosphorylation, acetylation, methylation, and sumoylation at various sites, disrupting the interaction between p53 and its negative regulators, leading to the activation and stabilization of p53 (68, 85, 104).
FIG. 1.
Activation of p53 in response to DNA damage and oxidative stresses. In the absence of stresses, the negative regulators of p53, such as Mdm2/MdmX, suppress p53 activity and induce its degradation. In response to DNA damage and oxidative stress, p53 and its negative regulators are posttranslationally modified, leading to p53 activation by disrupting the interaction between p53 and its negative regulators.
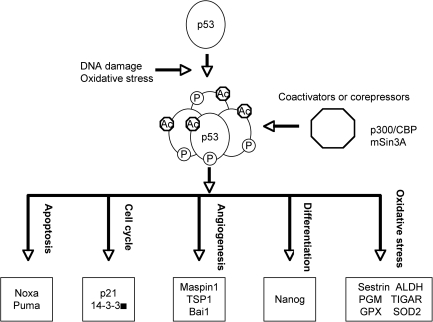
As a transcription factor, p53 can directly regulate the expression of hundreds of genes, products of which mediate various p53-dependent functions (Fig. 2) (43, 53, 69). For example, p21 and 14-3-3σ are responsible for p53-dependent cell-cycle arrest (30, 31, 50); p53 can also induce embryonic stem (ES) cell differentiation by suppressing the expression of Nanog, which is required for the self-renewal of ES cells (64). In response to high levels of DNA damage, p53 induces apoptosis and senescence by upregulating apoptotic genes such as Noxa and Puma (66, 71). These functions of p53 prevent the passage of DNA damage to the daughter cells and thus maintain genomic stability. In response o oxidative stresses, p53 activates the transcription of a number of genes involved in regulating oxidative stress, such as Sestrin, glutathione peroxidase (GPX), aldehyde dehydrogenase (ALDH), and tumor protein 53–induced nuclear protein 1(TP53INP1) (14, 16, 115, 130). p53 can also regulate the cellular oxidative stress levels by modulating glycolysis through inducing the expression of TIGAR (TP53-induced glycolysis and apoptosis regulator) and suppressing the expression of phosphoglycerate mutase (PGM) (9, 58).
FIG. 2.
p53 target genes are mediators of various p53-dependent functions in response to DNA damage and oxidative stresses.
p53 and Aging
Recent studies have functionally linked p53 to aging in various organisms (Fig. 3). The p53 orthologue in Caenorhabditis elegans, Cep-1, is involved in negatively regulating the life span of the worm, because the reduced expression of Cep-1 results in increased longevity (4). Expression of dominant-negative versions of Drosophila melanogaster p53 (Dmp53) in adult neurons extends the life span and increases the genotoxic stress resistance in the fly (8). Because the expression of the dominant-negative Dmp53 does not further increase the life span of flies that are calorie restricted, these findings suggest that p53 is involved in mediating the calorie-restricted life span in flies. However, mutagenesis studies in C. elegans show that certain mutations extending the life span increase activities of p53 and cancer resistance (94). Therefore, increased p53 activities are associated with both accelerated aging and increased life span in C. elegans.
FIG. 3.
Summary of the modulation of p53 effects on the lifespan of various organisms.
A similarly complicated scenario is also observed when studying the roles of p53 in mammalian aging. One mouse model, in which the N-terminus of p53 is truncated, exhibits increased p53 activities and accelerated aging (119). However, because of the large deletion of the genomic DNA upstream of p53 that contains 24 genes (40), it remains unclear whether any of these deleted genes is responsible for these aging phenotypes. The potential involvement of N-terminus–truncated p53 in aging is further supported by the overexpression of the N-terminus–deleted p53 isoform p44 in mice, leading to accelerated aging (72). This study suggests that p44 modulates the life span by inhibiting the PTEN and IGF signal pathways (39, 75, 110). To link p53 to aging in humans, a recent study shows that polymorphism of p53 at codon 72 (arginine-to-proline substitution) reduces p53 activities, correlating with increased life span but also with higher cancer risk in older individuals (120). Therefore, it has been suggested that p53 might suppress cancer at the cost of longevity.
The notion that increased p53 activity induces aging in mice is challenged by recent studies of mouse models with increased p53 activities. For example, mice with a hypomorphic mutation in Mdm2 exhibit increased p53 activity but normal life span (78). In addition, mice with an additional copy of p53 and ARF exhibit an enhanced expression of antioxidant activity and decreased levels of endogenous oxidative stresses, correlating with increased life span (74). Therefore, the increased antioxidant activity of p53 in these transgenic mice prevents the accumulation of oxidative stresses to the high levels required to induce p53-dependent apoptosis and senescence, thus delaying aging in these mice. In summary, the functions of p53 in aging are complex and could be context dependent. In this context, mild and transient activation of p53 in response to a low dosage of oxidative stress could protect cells from oxidative damage. In contrast, persistent activation of p53 in response to high levels of oxidative stresses can result in cell death and organismal aging. In further support of this notion, persistent activation of p53 depletes adult stem cells primarily through p53-dependent apoptosis (64).
Oxidative Stress and Aging
The free radical hypothesis remains the most well-established theory on the mechanism of aging (46). The increased ROS production and a decreased antioxidant capacity are thought to contribute to the aging process by oxidative modification of different macromolecules, such as lipids, proteins, and genomic DNA (12, 20, 25, 62, 63, 65, 96, 109, 117). In the context of DNA, oxidative damage to mitochondrial and nuclear DNA is significantly increased in different tissues in old rats and mice (20, 45, 61, 67, 76, 82, 116). Levels of lipid-peroxidation products are also increased with aging (44, 83, 87, 97, 108, 113, 119, 123). In addition, aging-related oxidative modification of different proteins causes changes in protein structure, enzyme activities, transcriptional activities, and signal-transduction pathways (32, 70, 103, 111, 112, 124), leading to age-related diseases. In summary, the levels of oxidative damage are increased during aging in various organisms, including C. elegans (11, 52, 121), flies (3, 64), and mice (22, 74, 79).
Free radicals are physiologic byproducts of metabolism and are rapidly eliminated by various antioxidant enzymes in cells (23). For example, the antioxidant enzymes, including superoxide dismutase (SOD), catalase, and peroxiredoxins, convert superoxide to hydrogen peroxide and eventually to water (5, 19, 99). SODs catalyze the breakdown of the superoxide anion into oxygen and hydrogen peroxide. Mice lacking SOD2 develop neurologic defects and die soon after birth because of excessive mitochondrial production of ROS (77); mice lacking SOD1 are viable but have numerous pathologies and a reduced life span (98). Catalase converts hydrogen peroxide into water and oxygen (19, 132). Humans and mice deficient in catalase can still efficiently remove H2O2, implying that other enzymes are also involved in this reaction (72, 88). Peroxiredoxins catalyze the reduction of hydrogen peroxide, organic peroxide, and peroxynitrite (99). These enzymes can be divided into there classes: typical 2-cysteine peroxiredoxins, atypical 2-cysteine peroxiredoxins, and 1-cysteine peroxiredoxins (128). Mice lacking peroxiredoxins 1 and 2 have a shortened life span (55, 86). Together, these findings underscore the importance of antioxidant enzymes in preventing aging processes. In further support of this notion, a diet rich in the building-block nutrients of antioxidant enzymes, including cofactors for SOD (manganese, zinc, and copper), show beneficial effects on delaying aging (1, 24, 49, 59, 81, 106).
In further support of the notion that oxidative stress is an inducer of aging, treatment with antioxidants can increase the life spans of various organisms and has a beneficial impact on aging-related diseases (6, 29, 38, 57, 114, 119). A low dose of dietary supplement with antioxidants partially mimics the effects of caloric restriction and delays aging in mice (6), and long-term treatment with free radical scavenging Schisandrin B, a dibenzocyclooctadiene derivative isolated from the fruit of Schisandra chinensis, delays aging-related functional impairment in various organs and improves the survival rate of aging mice (114). A dietary supplement of cysteine, which is required for the synthesis of the primary antioxidant glutathione, has clear benefits in delaying some aspects of aging (29). However, clinical trials have also found no significant beneficial effects of supplementation with antioxidant vitamin E, indicating that not all antioxidants have antiaging activities (55, 107, 125).
p53 and Oxidative Stress
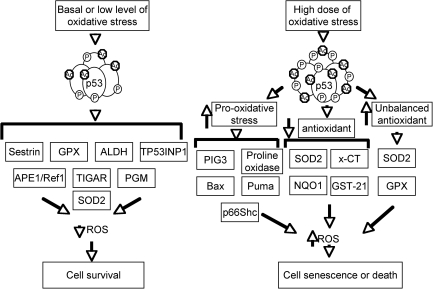
ROS levels have a significant impact on cell growth, survival and development, and tumorigenesis (17). p53 plays key and complex roles in cellular responses to oxidative stresses (84, 100). In response to low levels of oxidative stresses, p53 plays primarily antioxidant roles. In this context, a number of p53 target genes, including Sestrin, glutathione peroxidase (GPX), and aldehyde dehydrogenase (ALDH), are involved in reducing oxidative stresses (Fig. 4). For example, Sestrin protects the cells from hydrogen peroxide–induced damage by generation of peroxiredoxins (14). GPX is a primary antioxidant enzyme that scavenges hydrogen peroxide or organic hydroperoxides (115). Aldehyde dehydrogenase (ALDH) also contributes to the antioxidant function of p53 (130).
FIG. 4.
Context-dependent roles of p53 in cellular responses to oxidative stresses by turning on distinct target genes. At basal or low levels of oxidative stress, p53 regulates the expression of Sestrin, GPX, ALDH, TP53INP1, SOD2, TIGAR, and PGM to eliminate ROS, and therefore, promotes cellular survival. In response to high levels of oxidative stress, p53 induces the expression of prooxidative genes and suppresses the expression of antioxidant genes to increase ROS levels and promote apoptosis. Unbalanced antioxidants can also induce ROS to promote cell death.
p53 can also reduce the intracellular levels of ROS by regulating cellular metabolism. In this context, p53 induces the expression of TIGAR (TP53-induced glycolysis and apoptosis regulator), which slows glycolysis and promotes the production of NAPDH to decrease ROS levels (9). In addition, p53 suppresses the expression of phosphoglycerate mutase (PGM), leading to a decrease of pyruvate required for oxidative respiration in mitochondria and thus reduced ROS production (10, 74).
In response to high levels of oxidative stress, p53 exhibits prooxidative activities by turning on prooxidative genes such as PIG3 and proline oxidase (27, 95). Overexpression of these genes leads to higher levels of oxidative stress. In addition, p53 induces the expression of BAX and PUMA, which induce apoptosis through the release of cytochrome c from mitochondria (66, 71). The prooxidative activities of p53 also include the inhibition of the expression of antioxidant genes, leading to increased cellular oxidative stresses to induce apoptosis. For example, p53 could repress the expression of SOD2 and Nrf2, resulting in sensitivity to oxidative stress or inducing apoptosis (28, 34, 91). Interestingly, p53-induced upregulation of MnSOD and GPX, but not catalase, increases oxidative stress and apoptosis (54), suggesting that the balance of antioxidant enzyme and oxidative stress is important for cell survival. In summary, p53 plays important but context-dependent roles in regulating cellular oxidative stresses, and the levels of oxidative-stress damage dictate whether the p53 behavior is that of a protector or a killer (100).
p53 Interacts with Other Pathways Involved in Oxidative Stress and Aging
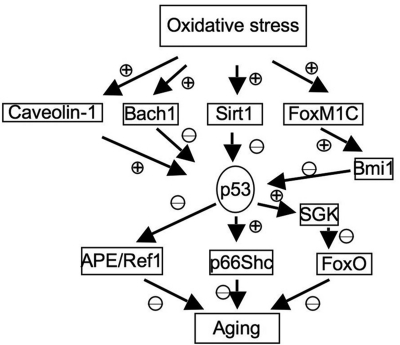
In addition to its direct regulation of genes involved in oxidative stresses, p53 also interact with other pathways that are involved in aging and oxidative stresses, which are summarized here (Fig. 5).
FIG. 5.
Functional interaction between p53 and other pathways important for oxidative stress response and aging.
Sirt1
The Sirt1 gene encodes the NAD-dependent histone deacetylase, which is important for the longevity in yeast and mammalian species by calorie restriction (42, 60, 64, 122). Sirt1 can deacetylate and inactivate p53, leading to impaired cell growth arrest and apoptosis in response to oxidative stresses (101). In addition, the expression of a dominant-negative version of Sirt1 increases the cellular sensitivity to oxidative stress, further indicating its antioxidant roles in cellular responses to oxidative stresses. However, the roles of Sirt1 in suppressing p53 in response to oxidative stresses remain to be fully established. In contrast to the prediction that Sirt1 deficiency would increase p53 activity, recent studies indicate that deficiency of Sirt1 extends the replicative capacity of mouse embryonic fibroblasts (MEFs) under the conditions of chronic oxidative stress due to the inefficient activation of p53 (21). However, the physiological relevance of replicative senescence in aging is not clear, because it primarily reflects a cell-culture phenomenon in the presence of nonphysiologically high levels of oxygen. Because Sirt1 is an NAD-dependent deacetylase, and NAD levels are regulated by cellular metabolism and levels of ROS, these findings implicate a complex functional interaction of p53, Sirt1, oxidative stresses, and aging.
p66Shc
p66Shc, a downstream target of p53, is indispensable for p53-dependent elevation of intracellular oxidative stresses and apoptosis (118). p66Shc is a splice variant of p52Shc/p46Shc, a cytoplasmic signal transducer involved in the transmission of mitogenic signal from activated receptors to Ras (93). However, p66Shc is not involved in regulating Ras signal but instead is involved in inducing apoptosis in response to oxidative stresses (80). The important role of p66Shc in oxidative stresses and aging is indicated by the findings that ablation of p66Shc enhances cellular resistance to apoptosis induced by oxidative stresses and extends the life span of p66Shc-deficient mice (79). In this context, cytochrome c release after oxidative signals is impaired in p66Shc-deficient cells (90). Therefore, p66Shc functionally links p53 to oxidative stress response and aging.
FoxO
Forkhead box O (FoxO) transcription factors are important mediators of the PI3K/Akt signaling pathway and regulate the cellular responses to oxidative stresses and the life span (56, 105). p53 negatively regulates the activities of FoxO by inducing the expression of serum- and glucocorticoid-inducible kinase (SGK), a negative regulator of FoxO and PTEN (37). In addition, Sirt1 can deacetylate FoxO3 and FoxO4, thus attenuating FoxO-induced apoptosis and cell-cycle arrest (41). Therefore, the balance of the functional interaction among Sirt1, FoxO, and p53 might play important roles in regulating oxidative stresses and aging.
APE/Ref1
The expression of APE/Ref1 is decreased in senescent human bone marrow–derived mesenchymal stem cells (hBMSCs) with increased endogenous ROS levels. Overexpression of APE1/Ref-1 suppresses superoxide production and decreases senescence in hBMSCs (48). In addition, aging mice have an impaired induction of APE in response to oxidative damage (15). The activities of APE/Ref1 are negatively regulated by p53 (131), implicating another pathway for p53 to modulate oxidative stresses and aging.
Caveolin-1
Expression of Caveolin-1 is induced in fibroblasts undergoing oxidative stress–induced senescence, and the antioxidant prevents the senescence and upregulation of Caveolin-1 (36, 126). Overexpression of Caveolin-1 in MEF induces the premature senescence through a p53-p21–dependent pathway, suggesting that Caveolin-1 could activate p53-dependent premature senescence after oxidative stresses (36). In this context, Caveolin-1 binds to Mdm2 and disrupts the binding of Mdm2 to p53, leading to the activation of p53 in response to oxidative stresses. The activation of p53 and induction of premature senescence are compromised in the Caveolin-1–null MEFs, confirming that Caveolin-1 is an upstream activator of p53 in response to oxidative stresses (7).
FoxM1C-Bmi1 pathway
Bmi1 is a negative regulator of the Ink4a/Arf and p53; FoxM1C induces the expression of Bmi1 to prevent the oxidative stress–induced cellular senescence by inhibiting the expression of p53 (13, 18, 33, 89). Bmi1 is important to repress the prooxidant activities of p53 in neurons and to suppress oxidative stress–induced apoptosis and premature aging-like phenotypes (18). In addition, targeted depletion of Bmi1 sensitizes tumor cells to p53-mediated apoptosis in response to radiation therapy (2).
Bach1
For transcription factors, the recruitment of co-activators or co-repressors to p53 target promoters is critical for p53-dependent transcription. Bach1 is induced by oxidative stresses and forms a complex with p53, histone deacetylase 1, and nuclear co-repressor N-coR, promoting histone deacetylation and suppression of certain p53 target genes (26). In this context, Bach1 inhibits oxidative stress–induced cellular senescence by disrupting p53-dependent gene expression (26).
Conclusion
The accumulation of oxidative stress and oxidative damage is a major inducer of aging. Many pathways involved in cellular responses to oxidative stresses regulate the aging process and the life spans of various organisms. p53 plays important but context-dependent roles in cellular responses to low or high levels of oxidative stresses. In response to low levels of oxidative stresses, p53 exhibits antioxidant activities and promotes cellular survival; in response to high levels of oxidative stresses, p53 exhibits prooxidative activities to induce cellular apoptosis. Both functions of p53 can prevent the accumulation of oxidative damage in cells and thus maintain genomic stability. p53 accomplishes these functions by direct transcriptional regulation of genes involved in oxidative-stress responses or modulating other pathways important in oxidative-stress responses.
Consistent with its context-specific roles in oxidative-stress responses, the roles of p53 in aging appear to be complex as well. In this context, increased p53 activities can accelerate aging in some transgenic mouse models but not in others (72, 74, 78, 119). In addition, the increase of the gene dosage of ARF and p53 does not promote aging but increases the life span of transgenic mice (74). Therefore, the roles of p53 in aging could also be context dependent. The accumulation of oxidative stresses in old mice could turn on the apoptotic or senescent roles of p53, thus promoting the aging process. However, increased dosages of p53 and ARF could ensure more efficient elimination of oxidative stress and thus prevent the accumulation of oxidative stresses to high levels in old mice. In support of this notion, a significant reduction of DNA damage occurs in old transgenic mice with additional copies of p53 and ARF (74). p53 primarily plays a protective role to increase the life span in these transgenic mice. Therefore, further elucidation of the mechanism that governs the context-dependent roles of p53 in oxidative-stress responses and the functional outcomes of the interaction between p53 and other pathways involved in cellular responses to oxidative stresses will shed light on its role in aging.
Abbreviations Used
- ALDH
aldehyde dehydrogenase
- APE/Ref1
apurinic/apyrimidinic endonuclease/redox factor-1
- BAI1
brain-specific angiogenesis inhibitor 1
- Dmp53
Drosophila melanogaster p53
- ES cell
embryonic stem cell
- FoxO
forkhead box O
- GPX
glutathione peroxidase
- GST
glutathione S-transferase
- hBMSCs
human bone marrow–derived mesenchymal stem cells
- MEF
mouse embryonic fibroblast
- NQO1
NAD(P)H dehydrogenase [quinone] 1
- Nrf1
NF-E2-related factor 2
- PGM
phosphoglycerate mutase
- PIG3
p53-inducible gene 3
- Puma
p53 upregulated modulator of apoptosis
- ORS
reactive oxygen species
- SGK
serum- and glucocorticoid-inducible kinase
- SOD
superoxide dismutase
- TIGAR
TP53-induced glycolysis and apoptosis regulator
- TP53INP1
tumor protein 53–induced nuclear protein 1
- TSP1
thrombospondin-1
Acknowledgment
This work is supported by an NIH grant to YX (R01 CA94254).
References
- 1.Airede AK. Copper, zinc and superoxide dismutase activities in premature infants: a review. East Afr Med J. 1993;70:441–444. [PubMed] [Google Scholar]
- 2.Alajez NM. Shi W. Hui AB. Yue S. Ng R. Lo KW. Bastianutto C. O'Sullivan B. Gullane P. Liu FF. Targeted depletion of BMI1 sensitizes tumor cells to P53-mediated apoptosis in response to radiation therapy. Cell Death Differ. 2009;16:1469–1479. doi: 10.1038/cdd.2009.85. [DOI] [PubMed] [Google Scholar]
- 3.Arking R. Buck S. Berrios A. Dwyer S. Baker GT., 3rd Elevated paraquat resistance can be used as a bioassay for longevity in a genetically based long-lived strain of Drosophila. Dev Genet. 1991;12:362–370. doi: 10.1002/dvg.1020120505. [DOI] [PubMed] [Google Scholar]
- 4.Arum O. Johnson TE. Reduced expression of the Caenorhabditis elegans p53 ortholog cep-1 results in increased longevity. J Gerontol A Biol Sci Med Sci. 2007;62:951–959. doi: 10.1093/gerona/62.9.951. [DOI] [PubMed] [Google Scholar]
- 5.Bannister JV. Bannister WH. Rotilio G. Aspects of the structure, function, and applications of superoxide dismutase. CRC Crit Rev Biochem. 1987;22:111–180. doi: 10.3109/10409238709083738. [DOI] [PubMed] [Google Scholar]
- 6.Barger JL. Kayo T. Vann JM. Arias EB. Wang J. Hacker TA. Wang Y. Raederstorff D. Morrow JD. Leeuwenburgh C. Allison DB. Saupe KW. Cartee GD. Weindruch R. Prolla TA. A low dose of dietary resveratrol partially mimics caloric restriction and retards aging parameters in mice. PLoS One. 2008;3:e2264. doi: 10.1371/journal.pone.0002264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bartholomew JN. Volonte D. Galbiati F. Caveolin-1 regulates the antagonistic pleiotropic properties of cellular senescence through a novel Mdm2/p53-mediated pathway. Cancer Res. 2009;69:2878–2886. doi: 10.1158/0008-5472.CAN-08-2857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bauer JH. Poon PC. Glatt-Deeley H. Abrams JM. Helfand SL. Neuronal expression of p53 dominant-negative proteins in adult Drosophila melanogaster extends life span. Curr Biol. 2005;15:2063–2068. doi: 10.1016/j.cub.2005.10.051. [DOI] [PubMed] [Google Scholar]
- 9.Bensaad K. Tsuruta A. Selak MA. Vidal MN. Nakano K. Bartrons R. Gottlieb E. Vousden KH. TIGAR, a p53-inducible regulator of glycolysis and apoptosis. Cell. 2006;126:107–120. doi: 10.1016/j.cell.2006.05.036. [DOI] [PubMed] [Google Scholar]
- 10.Bensaad K. Vousden KH. p53: new roles in metabolism. Trends Cell Biol. 2007;17:286–291. doi: 10.1016/j.tcb.2007.04.004. [DOI] [PubMed] [Google Scholar]
- 11.Berdichevsky A. Viswanathan M. Horvitz HR. Guarente L. C. elegans SIR-2.1 interacts with 14-3-3 proteins to activate DAF-16 and extend life span. Cell. 2006;125:1165–1177. doi: 10.1016/j.cell.2006.04.036. [DOI] [PubMed] [Google Scholar]
- 12.Bergamini E. Bizzarri R. Cavallini G. Cerbai B. Chiellini E. Donati A. Gori Z. Manfrini A. Parentini I. Signori F. Tamburini I. Ageing and oxidative stress: a role for dolichol in the antioxidant machinery of cell membranes? J Alzheimers Dis. 2004;6:129–135. doi: 10.3233/jad-2004-6204. [DOI] [PubMed] [Google Scholar]
- 13.Bruggeman SW. Valk-Lingbeek ME. van der Stoop PP. Jacobs JJ. Kieboom K. Tanger E. Hulsman D. Leung C. Arsenijevic Y. Marino S. van Lohuizen M. Ink4a and Arf differentially affect cell proliferation and neural stem cell self-renewal in Bmi1-deficient mice. Genes Dev. 2005;19:1438–1443. doi: 10.1101/gad.1299305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Budanov AV. Sablina AA. Feinstein E. Koonin EV. Chumakov PM. Regeneration of peroxiredoxins by p53-regulated sestrins, homologs of bacterial AhpD. Science. 2004;304:596–600. doi: 10.1126/science.1095569. [DOI] [PubMed] [Google Scholar]
- 15.Cabelof DC. Raffoul JJ. Ge Y. Van Remmen H. Matherly LH. Heydari AR. Age-related loss of the DNA repair response following exposure to oxidative stress. J Gerontol A Biol Sci Med Sci. 2006;61:427–434. doi: 10.1093/gerona/61.5.427. [DOI] [PubMed] [Google Scholar]
- 16.Cano CE. Gommeaux J. Pietri S. Culcasi M. Garcia S. Seux M. Barelier S. Vasseur S. Spoto RP. Pebusque MJ. Dusetti NJ. Iovanna JL. Carrier A. Tumor protein 53-induced nuclear protein 1 is a major mediator of p53 antioxidant function. Cancer Res. 2009;69:219–226. doi: 10.1158/0008-5472.CAN-08-2320. [DOI] [PubMed] [Google Scholar]
- 17.Chao C. Hergenhahn M. Kaeser MD. Wu Z. Saito S. Iggo R. Hollstein M. Appella E. anmd Xu Y. Cell type- and promoter-specific roles of Ser18 phosphorylation in regulating p53 responses. J Biol Chem. 2003;278:41028–41033. doi: 10.1074/jbc.M306938200. [DOI] [PubMed] [Google Scholar]
- 18.Chatoo W. Abdouh M. David J. Champagne MP. Ferreira J. Rodier F. Bernier G. The polycomb group gene Bmi1 regulates antioxidant defenses in neurons by repressing p53 pro-oxidant activity. J Neurosci. 2009;29:529–542. doi: 10.1523/JNEUROSCI.5303-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chelikani P. Fita I. Loewen PC. Diversity of structures and properties among catalases. Cell Mol Life Sci. 2004;61:192–208. doi: 10.1007/s00018-003-3206-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen JH. Hales CN. Ozanne SE. DNA damage, cellular senescence and organismal ageing: causal or correlative? Nucleic Acids Res. 2007;35:7417–7428. doi: 10.1093/nar/gkm681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chua KF. Mostoslavsky R. Lombard DB. Pang WW. Saito S. Franco S. Kaushal D. Cheng HL. Fischer MR. Stokes N. Murphy MM. Appella E. Alt FW. Mammalian SIRT1 limits replicative life span in response to chronic genotoxic stress. Cell Metab. 2005;2:67–76. doi: 10.1016/j.cmet.2005.06.007. [DOI] [PubMed] [Google Scholar]
- 22.Collins AR. Lyon CJ. Xia X. Liu JZ. Tangirala RK. Yin F. Boyadjian R. Bikineyeva A. Pratico D. Harrison DG. Hsueh WA. Age-accelerated atherosclerosis correlates with failure to upregulate antioxidant genes. Circ Res. 2009;104:e42–e54. doi: 10.1161/CIRCRESAHA.108.188771. [DOI] [PubMed] [Google Scholar]
- 23.Davies KJ. Oxidative stress: the paradox of aerobic life. Biochem Soc Symp. 1995;61:131. doi: 10.1042/bss0610001. [DOI] [PubMed] [Google Scholar]
- 24.Davis CD. Feng Y. Dietary copper, manganese and iron affect the formation of aberrant crypts in colon of rats administered 3,2′-dimethyl-4-aminobiphenyl. J Nutr. 1999;129:1060–1067. doi: 10.1093/jn/129.5.1060. [DOI] [PubMed] [Google Scholar]
- 25.De Bont R. van Larebeke N. Endogenous DNA damage in humans: a review of quantitative data. Mutagenesis. 2004;19:169–185. doi: 10.1093/mutage/geh025. [DOI] [PubMed] [Google Scholar]
- 26.Dohi Y. Ikura T. Hoshikawa Y. Katoh Y. Ota K. Nakanome A. Muto A. Omura S. Ohta T. Ito A. Yoshida M. Noda T. Igarashi K. Bach1 inhibits oxidative stress-induced cellular senescence by impeding p53 function on chromatin. Nat Struct Mol Biol. 2008;15:1246–1254. doi: 10.1038/nsmb.1516. [DOI] [PubMed] [Google Scholar]
- 27.Donald SP. Sun XY. Hu CA. Yu J. Mei JM. Valle D. Phang JM. Proline oxidase, encoded by p53-induced gene-6, catalyzes the generation of proline-dependent reactive oxygen species. Cancer Res. 2001;61:1810–1815. [PubMed] [Google Scholar]
- 28.Drane P. Bravard A. Bouvard V. May E. Reciprocal down-regulation of p53 and SOD2 gene expression-implication in p53 mediated apoptosis. Oncogene. 2001;20:430–439. doi: 10.1038/sj.onc.1204101. [DOI] [PubMed] [Google Scholar]
- 29.Droge W. Oxidative stress and ageing: is ageing a cysteine deficiency syndrome? Phil Trans R Soc Lond B Biol Sci. 2005;360:2355–2372. doi: 10.1098/rstb.2005.1770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.el-Deiry WS. Harper JW. O'Connor PM. Velculescu VE. Canman CE. Jackman J. Pietenpol JA. Burrell M. Hill DE. Wang Y, et al. WAF1/CIP1 is induced in p53-mediated G1 arrest and apoptosis. Cancer Res. 1994;54:1169–1174. [PubMed] [Google Scholar]
- 31.el-Deiry WS. Tokino T. Velculescu VE. Levy DB. Parsons R. Trent JM. Lin D. Mercer WE. Kinzler KW. Vogelstein B. WAF1, a potential mediator of p53 tumor suppression. Cell. 1993;75:817–825. doi: 10.1016/0092-8674(93)90500-p. [DOI] [PubMed] [Google Scholar]
- 32.Ethen CM. Reilly C. Feng X. Olsen TW. Ferrington DA. Age-related macular degeneration and retinal protein modification by 4-hydroxy-2-nonenal. Invest Ophthalmol Vis Sci. 2007;48:3469–3479. doi: 10.1167/iovs.06-1058. [DOI] [PubMed] [Google Scholar]
- 33.Fan C. He L. Kapoor A. Gillis A. Rybak AP. Cutz JC. Tang D. Bmi1 promotes prostate tumorigenesis via inhibiting p16(INK4A) and p14(ARF) expression. Biochim Biophys Acta. 2008;1782:642–648. doi: 10.1016/j.bbadis.2008.08.009. [DOI] [PubMed] [Google Scholar]
- 34.Faraonio R. Vergara P. Di Marzo D. Pierantoni MG. Napolitano M. Russo T. Cimino F. p53 suppresses the Nrf2-dependent transcription of antioxidant response genes. J Biol Chem. 2006;281:39776–39784. doi: 10.1074/jbc.M605707200. [DOI] [PubMed] [Google Scholar]
- 35.Freeman J. Schmidt S. Scharer E. Iggo R. Mutation of conserved domain II alters the sequence specificity of DNA binding by the p53 protein. EMBO J. 1994;13:5393–5400. doi: 10.1002/j.1460-2075.1994.tb06874.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Galbiati F. Volonte D. Liu J. Capozza F. Frank PG. Zhu L. Pestell RG. Lisanti MP. Caveolin-1 expression negatively regulates cell cycle progression by inducing G(0)/G(1) arrest via a p53/p21(WAF1/Cip1)-dependent mechanism. Mol Biol Cell. 2001;12:2229–2244. doi: 10.1091/mbc.12.8.2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Garinis GA. van der Horst GTJ. Vijg JHJ. Hoeijmakers J. DNA damage and ageing: new-age ideas for an age-old problem. Nat Cell Biol. 2008;10:1241–1247. doi: 10.1038/ncb1108-1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gaziano JM. Vitamin E and cardiovascular disease: observational studies. Ann N Y Acad Sci. 2004;1031:280–291. doi: 10.1196/annals.1331.028. [DOI] [PubMed] [Google Scholar]
- 39.Gems D. Partridge L. Insulin/IGF signalling and ageing: seeing the bigger picture. Curr Opin Genet Dev. 2001;11:287–292. doi: 10.1016/s0959-437x(00)00192-1. [DOI] [PubMed] [Google Scholar]
- 40.Gentry A. Venkatachalam S. Complicating the role of p53 in aging. Aging Cell. 2005;4:157–160. doi: 10.1111/j.1474-9726.2005.00154.x. [DOI] [PubMed] [Google Scholar]
- 41.Giannakou ME. Partridge L. The interaction between FOXO and SIRT1: tipping the balance towards survival. Trends Cell Biol. 2004;14:408–412. doi: 10.1016/j.tcb.2004.07.006. [DOI] [PubMed] [Google Scholar]
- 42.Guarente L. Sir2 links chromatin silencing, metabolism, and aging. Genes Dev. 2000;14:1021–1026. [PubMed] [Google Scholar]
- 43.Gudkov A. Microarray analysis of p53-mediated transcription: multi-thousand piece puzzle or invitation to collective thinking. Cancer Biol Ther. 2003;2:444–445. doi: 10.4161/cbt.2.4.480. [DOI] [PubMed] [Google Scholar]
- 44.Gupta A. Hasan M. Chander R. Kapoor NK. Age-related elevation of lipid peroxidation products: diminution of superoxide dismutase activity in the central nervous system of rats. Gerontology. 1991;37:305–309. doi: 10.1159/000213277. [DOI] [PubMed] [Google Scholar]
- 45.Hamilton ML. Van Remmen H. Drake JA. Yang H. Guo ZM. Kewitt K. Walter CA. Richardson A. Does oxidative damage to DNA increase with age? Proc Natl Acad Sci U S A. 2001;98:10469–10474. doi: 10.1073/pnas.171202698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Harman D. Aging: a theory based on free radical and radiation chemistry. J Gerontol. 1956;11:298–300. doi: 10.1093/geronj/11.3.298. [DOI] [PubMed] [Google Scholar]
- 47.Haupt Y. Maya R. Kazaz A. Oren M. Mdm2 promotes the rapid degradation of p53. Nature. 1997;387:296–299. doi: 10.1038/387296a0. [DOI] [PubMed] [Google Scholar]
- 48.Heo JY. Jing K. Song KS. Seo KS. Park JH. Kim JS. Jung YJ. Hur GM. Jo DY. Kweon GR. Yoon WH. Lim K. Hwang BD. Jeon BH. Park JI. Downregulation of APE1/Ref-1 is involved in the senescence of mesenchymal stem cells. Stem Cells. 2009;27:1455–1462. doi: 10.1002/stem.54. [DOI] [PubMed] [Google Scholar]
- 49.Hercberg S. Galan P. Preziosi P. Bertrais S. Mennen L. Malvy D. Roussel AM. Favier A. Briancon S. The SU.VI.MAX Study: a randomized, placebo-controlled trial of the health effects of antioxidant vitamins and minerals. Arch Intern Med. 2004;164:2335–2342. doi: 10.1001/archinte.164.21.2335. [DOI] [PubMed] [Google Scholar]
- 50.Hermeking H. Lengauer C. Polyak K. He TC. Zhang L. Thiagalingam S. Kinzler KW. Vogelstein B. 14-3-3 sigma is a p53-regulated inhibitor of G2/M progression. Mol Cell. 1997;1:3–11. doi: 10.1016/s1097-2765(00)80002-7. [DOI] [PubMed] [Google Scholar]
- 51.Honda R. Tanaka H. Yasuda H. Oncoprotein MDM2 is a ubiquitin ligase E3 for tumor suppressor p53. FEBS Lett. 1997;420:25–27. doi: 10.1016/s0014-5793(97)01480-4. [DOI] [PubMed] [Google Scholar]
- 52.Honda Y. Honda S. Oxidative stress and life span determination in the nematode Caenorhabditis elegans. Ann N Y Acad Sci. 2002;959:466–474. doi: 10.1111/j.1749-6632.2002.tb02117.x. [DOI] [PubMed] [Google Scholar]
- 53.Huang J. Logsdon N. Schmieg FI. Simmons DT. p53-mediated transcription induces resistance of DNA to UV inactivation. Oncogene. 1998;17:401–411. doi: 10.1038/sj.onc.1201951. [DOI] [PubMed] [Google Scholar]
- 54.Hussain SP. Amstad P. He P. Robles A. Lupold S. Kaneko I. Ichimiya M. Sengupta S. Mechanic L. Okamura S. Hofseth LJ. Moake M. Nagashima M. Forrester KS. Harris CC. p53-induced up-regulation of MnSOD and GPx but not catalase increases oxidative stress and apoptosis. Cancer Res. 2004;64:2350–2356. doi: 10.1158/0008-5472.can-2287-2. [DOI] [PubMed] [Google Scholar]
- 55.Jeffers JR. Parganas E. Lee Y. Yang C. Wang J. Brennan J. MacLean KH. Han J. Chittenden T. Ihle JN. McKinnon PJ. Cleveland JL. Zambetti GP. Puma is an essential mediator of p53-dependent and -independent apoptotic pathways. Cancer Cell. 2003;4:321–328. doi: 10.1016/s1535-6108(03)00244-7. [DOI] [PubMed] [Google Scholar]
- 56.Katic M. Kahn CR. The role of insulin and IGF-1 signaling in longevity. Cell Mol Life Sci. 2005;62:320–343. doi: 10.1007/s00018-004-4297-y. [DOI] [PubMed] [Google Scholar]
- 57.Kim J. Takahashi M. Shimizu T. Shirasawa T. Kajita M. Kanayama A. Miyamoto Y. Effects of a potent antioxidant, platinum nanoparticle, on the lifespan of Caenorhabditis elegans. Mech Ageing Dev. 2008;129:322–331. doi: 10.1016/j.mad.2008.02.011. [DOI] [PubMed] [Google Scholar]
- 58.Kondoh H. Lleonart ME. Gil J. Wang J. Degan P. Peters G. Martinez D. Carnero A. Beach D. Glycolytic enzymes can modulate cellular life span. Cancer Res. 2005;65:177–185. [PubMed] [Google Scholar]
- 59.Lamb DJ. Tickner ML. Hourani SM. Ferns GA. Dietary copper supplements modulate aortic superoxide dismutase, nitric oxide and atherosclerosis. Int J Exp Pathol. 2005;86:247–255. doi: 10.1111/j.0959-9673.2005.00432.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Langley E. Pearson M. Faretta M. Bauer UM. Frye RA. Minucci S. Pelicci PG. Kouzarides T. Human SIR2 deacetylates p53 and antagonizes PML/p53-induced cellular senescence. EMBO J. 2002;21:2383–2396. doi: 10.1093/emboj/21.10.2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lemon JA. Rollo CD. Boreham DR. Elevated DNA damage in a mouse model of oxidative stress: impacts of ionizing radiation and a protective dietary supplement. Mutagenesis. 2008;23:473–482. doi: 10.1093/mutage/gen036. [DOI] [PubMed] [Google Scholar]
- 62.Lenaz G. Role of mitochondria in oxidative stress and ageing. Biochim Biophys Acta. 1998;1366:53–67. doi: 10.1016/s0005-2728(98)00120-0. [DOI] [PubMed] [Google Scholar]
- 63.Lenaz G. Bovina C. Formiggini G. Parenti Castelli G. Mitochondria, oxidative stress, and antioxidant defences. Acta Biochim Pol. 1999;46:1–21. [PubMed] [Google Scholar]
- 64.Liu DP. Ou L. Clemenson GD., Jr Chao C. Lutske ME. Zambetti GP. Gage FH. Xu Y. Puma is required for p53-induced depletion of adult stem cells. Nat Cell Biol. 2010;12:993–998. doi: 10.1038/ncb2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Linnane AW. Marzuki S. Ozawa T. Tanaka M. Mitochondrial DNA mutations as an important contributor to ageing and degenerative diseases. Lancet. 1989;1:642–645. doi: 10.1016/s0140-6736(89)92145-4. [DOI] [PubMed] [Google Scholar]
- 66.Liu Z. Lu H. Shi H. Du Y. Yu J. Gu S. Chen X. Liu KJ. Hu CA. PUMA overexpression induces reactive oxygen species generation and proteasome-mediated stathmin degradation in colorectal cancer cells. Cancer Res. 2005;65:1647–1654. doi: 10.1158/0008-5472.CAN-04-1754. [DOI] [PubMed] [Google Scholar]
- 67.Lopez-Torres M. Barja G. Calorie restriction, oxidative stress and longevity. Rev Esp Geriatr Gerontol. 2008;43:252–260. doi: 10.1016/s0211-139x(08)71190-9. [DOI] [PubMed] [Google Scholar]
- 68.Lu H. Levine AJ. Human TAFII31 protein is a transcriptional coactivator of the p53 protein. Proc Natl Acad Sci U S A. 1995;92:5154–5158. doi: 10.1073/pnas.92.11.5154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lu H. Lin J. Chen J. Levine AJ. The regulation of p53-mediated transcription and the roles of hTAFII31 and mdm-2. Harvey Lect. 1994;90:81–93. [PubMed] [Google Scholar]
- 70.Machado A. Ayala A. Gordillo E. Revilla E. Santa Maria C. Relationship between enzymatic activity loss and post-translational protein modification in aging. Arch Gerontol Geriatr. 1991;12:187–197. doi: 10.1016/0167-4943(91)90027-n. [DOI] [PubMed] [Google Scholar]
- 71.Macip S. Igarashi M. Berggren P. Yu J. Lee SW. Aaronson SA. Influence of induced reactive oxygen species in p53-mediated cell fate decisions. Mol Cell Biol. 2003;23:8576–8585. doi: 10.1128/MCB.23.23.8576-8585.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Maier B. Gluba W. Bernier B. Turner T. Mohammad K. Guise T. Sutherland A. Thorner M. Scrable H. Modulation of mammalian life span by the short isoform of p53. Genes Dev. 2004;18:306–319. doi: 10.1101/gad.1162404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mansur CP. The regulation and function of the p53 tumor suppressor. Adv Dermatol. 1997;13:121–166. [PubMed] [Google Scholar]
- 74.Matheu A. Maraver A. Klatt P. Flores I. Garcia-Cao I. Borras C. Flores JM. Vina J. Blasco MA. Serrano M. Delayed ageing through damage protection by the Arf/p53 pathway. Nature. 2007;448:375–379. doi: 10.1038/nature05949. [DOI] [PubMed] [Google Scholar]
- 75.Mattson MP. Maudsley S. Martin B. A neural signaling triumvirate that influences ageing and age-related disease: insulin/IGF-1, BDNF and serotonin. Ageing Res Rev. 2004;3:445–464. doi: 10.1016/j.arr.2004.08.001. [DOI] [PubMed] [Google Scholar]
- 76.Meissner C. Mutations of mitochondrial DNA: cause or consequence of the ageing process? Z Gerontol Geriatr. 2007;40:325–333. doi: 10.1007/s00391-007-0481-z. [DOI] [PubMed] [Google Scholar]
- 77.Melov S. Schneider JA. Day BJ. Hinerfeld D. Coskun P. Mirra SS. Crapo JD. Wallace DC. A novel neurological phenotype in mice lacking mitochondrial manganese superoxide dismutase. Nat Genet. 1998;18:159–163. doi: 10.1038/ng0298-159. [DOI] [PubMed] [Google Scholar]
- 78.Mendrysa SM. O'Leary KA. McElwee MK. Michalowski J. Eisenman RN. Powell DA. Perry ME. Tumor suppression and normal aging in mice with constitutively high p53 activity. Genes Dev. 2006;20:16–21. doi: 10.1101/gad.1378506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Migliaccio E. Giorgio M. Mele S. Pelicci G. Reboldi P. Pandolfi PP. Lanfrancone L. Pelicci PG. The p66shc adaptor protein controls oxidative stress response and life span in mammals. Nature. 1999;402:309–313. doi: 10.1038/46311. [DOI] [PubMed] [Google Scholar]
- 80.Migliaccio E. Mele S. Salcini AE. Pelicci G. Lai KM. Superti-Furga G. Pawson T. Di Fiore PP. Lanfrancone L. Pelicci PG. Opposite effects of the p52shc/p46shc and p66shc splicing isoforms on the EGF receptor-MAP kinase-fos signalling pathway. EMBO J. 1997;16:706–716. doi: 10.1093/emboj/16.4.706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Mocchegiani E. Malavolta M. Muti E. Costarelli L. Cipriano C. Piacenza F. Tesei S. Giacconi R. Lattanzio F. Zinc, metallothioneins and longevity: interrelationships with niacin and selenium. Curr Pharm Des. 2008;14:2719–2732. doi: 10.2174/138161208786264188. [DOI] [PubMed] [Google Scholar]
- 82.Montaner B. O'Donovan P. Reelfs O. Perrett CM. Zhang X. Xu YZ. Ren X. Macpherson P. Frith D. Karran P. Reactive oxygen-mediated damage to a human DNA replication and repair protein. EMBO Rep. 2007;8:1074–1079. doi: 10.1038/sj.embor.7401084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Montine TJ. Neely MD. Quinn JF. Beal MF. Markesbery WR. Roberts LJ. Morrow JD. Lipid peroxidation in aging brain and Alzheimer's disease. Free Radic Biol Med. 2002;33:620–626. doi: 10.1016/s0891-5849(02)00807-9. [DOI] [PubMed] [Google Scholar]
- 84.Nakamizo A. Amano T. Zhang W. Zhang XQ. Ramdas L. Liu TJ. Bekele BN. Shono T. Sasaki T. Benedict WF. Sawaya R. Lang FF. Phosphorylation of Thr18 and Ser20 of p53 in Ad-p53-induced apoptosis. Neurol Oncol. 2008;10:275–291. doi: 10.1215/15228517-2008-015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Neilsen PM. Cheney KM. Li CW. Chen JD. Cawrse JE. Schulz RB. Powell JA. Kumar R. Callen DF. Identification of ANKRD11 as a p53 coactivator. J Cell Sci. 2008;121:3541–3552. doi: 10.1242/jcs.026351. [DOI] [PubMed] [Google Scholar]
- 86.Neumann CA. Krause DS. Carman CV. Das S. Dubey DP. Abraham JL. Bronson RT. Fujiwara Y. Orkin SH. Van Etten RA. Essential role for the peroxiredoxin Prdx1 in erythrocyte antioxidant defence and tumour suppression. Nature. 2003;424:561–565. doi: 10.1038/nature01819. [DOI] [PubMed] [Google Scholar]
- 87.Nowak M. Swietochowska E. Wielkoszynski T. Marek B. Karpe J. Gorski J. Glogowska-Szelag J. Kos-Kudla B. Ostrowska Z. Changes in blood antioxidants and several lipid peroxidation products in women with age-related macular degeneration. Eur J Ophthalmol. 2003;13:281–286. doi: 10.1177/112067210301300307. [DOI] [PubMed] [Google Scholar]
- 88.Ogata M. Acatalasemia. Hum Genet. 1991;86:331–340. doi: 10.1007/BF00201829. [DOI] [PubMed] [Google Scholar]
- 89.Oguro H. Iwama A. Morita Y. Kamijo T. van Lohuizen M. Nakauchi H. Differential impact of Ink4a and Arf on hematopoietic stem cells and their bone marrow microenvironment in Bmi1-deficient mice. J Exp Med. 2006;203:2247–2253. doi: 10.1084/jem.20052477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Orsini F. Migliaccio E. Moroni M. Contursi C. Raker VA. Piccini D. Martin-Padura I. Pelliccia G. Trinei M. Bono M. Puri C. Tacchetti C. Ferrini M. Mannucci R. Nicoletti I. Lanfrancone L. Giorgio M. Pelicci PG. The life span determinant p66Shc localizes to mitochondria where it associates with mitochondrial heat shock protein 70 and regulates trans-membrane potential. J Biol Chem. 2004;279:25689–25695. doi: 10.1074/jbc.M401844200. [DOI] [PubMed] [Google Scholar]
- 91.Pani G. Bedogni B. Anzevino R. Colavitti R. Palazzotti B. Borrello S. Galeotti T. Deregulated manganese superoxide dismutase expression and resistance to oxidative injury in p53-deficient cells. Cancer Res. 2000;60:4654–4660. [PubMed] [Google Scholar]
- 92.Pavletich NP. Chambers KA. Pabo CO. The DNA-binding domain of p53 contains the four conserved regions and the major mutation hot spots. Genes Dev. 1993;7:2556–2564. doi: 10.1101/gad.7.12b.2556. [DOI] [PubMed] [Google Scholar]
- 93.Pelicci G. Lanfrancone L. Grignani F. McGlade J. Cavallo F. Forni G. Nicoletti I. Grignani F. Pawson T. Pelicci PG. A novel transforming protein (SHC) with an SH2 domain is implicated in mitogenic signal transduction. Cell. 1992;70:93–104. doi: 10.1016/0092-8674(92)90536-l. [DOI] [PubMed] [Google Scholar]
- 94.Pinkston JM. Garigan D. Hansen M. Kenyon C. Mutations that increase the life span of C. elegans inhibit tumor growth. Science. 2006;313:971–975. doi: 10.1126/science.1121908. [DOI] [PubMed] [Google Scholar]
- 95.Polyak K. Xia Y. Zweier JL. Kinzler KW. Vogelstein B. A model for p53-induced apoptosis. Nature. 1997;389:300–305. doi: 10.1038/38525. [DOI] [PubMed] [Google Scholar]
- 96.Poulsen HE. Oxidative DNA modifications. Exp Toxicol Pathol. 2005;57(suppl 1):161–169. doi: 10.1016/j.etp.2005.05.015. [DOI] [PubMed] [Google Scholar]
- 97.Pratico D. Lipid peroxidation and the aging process. Sci Aging Knowledge Environ. 2002;50:1–4. doi: 10.1126/sageke.2002.50.re5. [DOI] [PubMed] [Google Scholar]
- 98.Reaume AG. Elliott JL. Hoffman EK. Kowall NW. Ferrante RJ. Siwek DF. Wilcox HM. Flood DG. Beal MF. Brown RH Jr. Scott RW. Snider WD. Motor neurons in Cu/Zn superoxide dismutase-deficient mice develop normally but exhibit enhanced cell death after axonal injury. Nat Genet. 1996;13:43–47. doi: 10.1038/ng0596-43. [DOI] [PubMed] [Google Scholar]
- 99.Rhee SG. Chae HZ. Kim K. Peroxiredoxins: a historical overview and speculative preview of novel mechanisms and emerging concepts in cell signaling. Free Radic Biol Med. 2005;38:1543–1552. doi: 10.1016/j.freeradbiomed.2005.02.026. [DOI] [PubMed] [Google Scholar]
- 100.Sablina AA. Budanov AV. Ilyinskaya GV. Agapova LS. Kravchenko JE. Chumakov PM. The antioxidant function of the p53 tumor suppressor. Nat Med. 2005;11:1306–1313. doi: 10.1038/nm1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Sakaguchi K. Herrera JE. Saito S. Miki T. Bustin M. Vassilev A. Anderson CW. Appella E. DNA damage activates p53 through a phosphorylation-acetylation cascade. Genes Dev. 1998;12:2831–2841. doi: 10.1101/gad.12.18.2831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Sandor J. Ambrus T. Ember I. The function of the p53 gene suppressor in carcinogenesis. Orv Hetil. 1995;136:1875–1883. [PubMed] [Google Scholar]
- 103.Schoneich C. Protein modification in aging: an update. Exp Gerontol. 2006;41:807–812. doi: 10.1016/j.exger.2006.07.002. [DOI] [PubMed] [Google Scholar]
- 104.Scolnick DM. Chehab NH. Stavridi ES. Lien MC. Caruso L. Moran E. Berger SL. Halazonetis TD. CREB-binding protein and p300/CBP-associated factor are transcriptional coactivators of the p53 tumor suppressor protein. Cancer Res. 1997;57:3693–3696. [PubMed] [Google Scholar]
- 105.Sedding DG. FoxO transcription factors in oxidative stress response and ageing: a new fork on the way to longevity? Biol Chem. 2008;389:279–283. doi: 10.1515/BC.2008.033. [DOI] [PubMed] [Google Scholar]
- 106.Selenius M. Rundlof AK. Olm E. Fernandes AP. Bjornstedt M. Selenium and the selenoprotein thioredoxin reductase in the prevention, treatment and diagnostics of cancer. Antioxid Redox Signal. 2009;12:867–880. doi: 10.1089/ars.2009.2884. [DOI] [PubMed] [Google Scholar]
- 107.Sesso HD. Buring JE. Christen WG. Kurth T. Belanger C. MacFadyen J. Bubes V. Manson JE. Glynn RJ. Gaziano JM. Vitamins E and C in the prevention of cardiovascular disease in men: the Physicians' Health Study II randomized controlled trial. JAMA. 2008;300:2123–2133. doi: 10.1001/jama.2008.600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Shinohara R. Mano T. Nagasaka A. Hayashi R. Uchimura K. Nakano I. Watanabe F. Tsugawa T. Makino M. Kakizawa H. Nagata M. Iwase K. Ishizuki Y. Itoh M. Lipid peroxidation levels in rat cardiac muscle are affected by age and thyroid status. J Endocrinol. 2000;164:97–102. doi: 10.1677/joe.0.1640097. [DOI] [PubMed] [Google Scholar]
- 109.Siedlak SL. Casadesus G. Webber KM. Pappolla MA. Atwood CS. Smith MA. Perry G. Chronic antioxidant therapy reduces oxidative stress in a mouse model of Alzheimer's disease. Free Radic Res. 2009;43:156–164. doi: 10.1080/10715760802644694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Sonntag WE. Lynch C. Thornton P. Khan A. Bennett S. Ingram R. The effects of growth hormone and IGF-1 deficiency on cerebrovascular and brain ageing. J Anat. 2000;197:575–585. doi: 10.1046/j.1469-7580.2000.19740575.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Soskic V. Groebe K. Schrattenholz A. Nonenzymatic posttranslational protein modifications in ageing. Exp Gerontol. 2008;43:247–257. doi: 10.1016/j.exger.2007.12.001. [DOI] [PubMed] [Google Scholar]
- 112.Stadtman ER. Protein modification in aging. J Gerontol. 1988;43:B112–B120. doi: 10.1093/geronj/43.5.b112. [DOI] [PubMed] [Google Scholar]
- 113.Stewart RR. Bewley JD. Lipid peroxidation associated with accelerated aging of soybean axes. Plant Physiol. 1980;65:245–248. doi: 10.1104/pp.65.2.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Takubo K. Ohmura M. Azuma M. Nagamatsu G. Yamada W. Arai F. Hirao A. Suda T. Stem cell defects in ATM-deficient undifferentiated spermatogonia through DNA damage-induced cell-cycle arrest. Cell Stem Cell. 2008;2:170–182. doi: 10.1016/j.stem.2007.10.023. [DOI] [PubMed] [Google Scholar]
- 115.Tan M. Li S. Swaroop M. Guan K. Oberley LW. Sun Y. Transcriptional activation of the human glutathione peroxidase promoter by p53. J Biol Chem. 1999;274:12061–12066. doi: 10.1074/jbc.274.17.12061. [DOI] [PubMed] [Google Scholar]
- 116.Terzioglu M. Larsson NG. Mitochondrial dysfunction in mammalian ageing. Novartis Found Symp. 2007;287:197–208. doi: 10.1002/9780470725207.ch14. [DOI] [PubMed] [Google Scholar]
- 117.Toescu EC. Myronova N. Verkhratsky A. Age-related structural and functional changes of brain mitochondria. Cell Calcium. 2000;28:329–338. doi: 10.1054/ceca.2000.0167. [DOI] [PubMed] [Google Scholar]
- 118.Trinei M. Giorgio M. Cicalese A. Barozzi S. Ventura A. Migliaccio E. Milia E. Padura IM. Raker VA. Maccarana M. Petronilli V. Minucci S. Bernardi P. Lanfrancone L. Pelicci PG. A p53-p66Shc signalling pathway controls intracellular redox status, levels of oxidation-damaged DNA and oxidative stress-induced apoptosis. Oncogene. 2002;21:3872–3878. doi: 10.1038/sj.onc.1205513. [DOI] [PubMed] [Google Scholar]
- 119.Tyner SD. Venkatachalam S. Choi J. Jones S. Ghebranious N. Igelmann H. Lu X. Soron G. Cooper B. Brayton C. Hee Park S. Thompson T. Karsenty G. Bradley A. Donehower LA. p53 mutant mice that display early ageing-associated phenotypes. Nature. 2002;415:45–53. doi: 10.1038/415045a. [DOI] [PubMed] [Google Scholar]
- 120.van Heemst D. Mooijaart SP. Beekman M. Schreuder J. de Craen AJ. Brandt BW. Slagboom PE. Westendorp RG. Variation in the human TP53 gene affects old age survival and cancer mortality. Exp Gerontol. 2005;40:11–15. doi: 10.1016/j.exger.2004.10.001. [DOI] [PubMed] [Google Scholar]
- 121.Vanfleteren JR. Braeckman BP. Mechanisms of life span determination in Caenorhabditis elegans. Neurobiol Aging. 1999;20:487–502. doi: 10.1016/s0197-4580(99)00087-1. [DOI] [PubMed] [Google Scholar]
- 122.Vaziri H. Dessain SK. Ng Eaton E. Imai SI. Frye RA. Pandita TK. Guarente L. Weinberg RA. hSIR2(SIRT1) functions as an NAD-dependent p53 deacetylase. Cell. 2001;107:149–159. doi: 10.1016/s0092-8674(01)00527-x. [DOI] [PubMed] [Google Scholar]
- 123.Videla LA. Fernandez V. Valenzuela A. Age-dependent changes in rat liver lipid peroxidation and glutathione content induced by acute ethanol ingestion. Cell Biochem Funct. 1987;5:273–280. doi: 10.1002/cbf.290050406. [DOI] [PubMed] [Google Scholar]
- 124.Viner RI. Ferrington DA. Williams TD. Bigelow DJ. Schoneich C. Protein modification during biological aging: selective tyrosine nitration of the SERCA2a isoform of the sarcoplasmic reticulum Ca2+-ATPase in skeletal muscle. Biochem J. 1999;340:657–669. [PMC free article] [PubMed] [Google Scholar]
- 125.Vivekananthan DP. Penn MS. Sapp SK. Hsu A. Topol EJ. Use of antioxidant vitamins for the prevention of cardiovascular disease: meta-analysis of randomised trials. Lancet. 2003;361:2017–2023. doi: 10.1016/S0140-6736(03)13637-9. [DOI] [PubMed] [Google Scholar]
- 126.Volonte D. Zhang K. Lisanti MP. Galbiati F. Expression of caveolin-1 induces premature cellular senescence in primary cultures of murine fibroblasts. Mol Biol Cell. 2002;13:2502–2517. doi: 10.1091/mbc.01-11-0529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Walker DR. Bond JP. Tarone RE. Harris CC. Makalowski W. Boguski MS. Greenblatt MS. Evolutionary conservation and somatic mutation hotspot maps of p53: correlation with p53 protein structural and functional features. Oncogene. 1999;18:211–218. doi: 10.1038/sj.onc.1202298. [DOI] [PubMed] [Google Scholar]
- 128.Wood ZA. Schroder E. Robin Harris J. Poole LB. Structure, mechanism and regulation of peroxiredoxins. Trends Biochem Sci. 2003;28:32–40. doi: 10.1016/s0968-0004(02)00003-8. [DOI] [PubMed] [Google Scholar]
- 129.Xu Y. Regulation of p53 responses by post-translational modifications. Cell Death Differ. 2003;10:400–403. doi: 10.1038/sj.cdd.4401182. [DOI] [PubMed] [Google Scholar]
- 130.Yoon KA. Nakamura Y. Arakawa H. Identification of ALDH4 as a p53-inducible gene and its protective role in cellular stresses. J Hum Genet. 2004;49:134–140. doi: 10.1007/s10038-003-0122-3. [DOI] [PubMed] [Google Scholar]
- 131.Zaky A. Busso C. Izumi T. Chattopadhyay R. Bassiouny A. Mitra S. Bhakat KK. Regulation of the human AP-endonuclease (APE1/Ref-1) expression by the tumor suppressor p53 in response to DNA damage. Nucleic Acids Res. 2008;36:1555–1566. doi: 10.1093/nar/gkm1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Zamocky M. Koller F. Understanding the structure and function of catalases: clues from molecular evolution and in vitro mutagenesis. Prog Biophys Mol Biol. 1999;72:19–66. doi: 10.1016/s0079-6107(98)00058-3. [DOI] [PubMed] [Google Scholar]
- 133.Zhu D. Wu J. Spee C. Ryan SJ. Hinton DR. BMP4 mediates oxidative stress-induced retinal pigment epithelial cell senescence and is overexpressed in age-related macular degeneration. J Biol Chem. 2009;284:9529–9539. doi: 10.1074/jbc.M809393200. [DOI] [PMC free article] [PubMed] [Google Scholar]