Abstract
p53 regulates the cell cycle and deoxyribonucleic acid (DNA) repair pathways as part of its unequivocally important function to maintain genomic stability. Intriguingly, recent studies show that p53 can also transactivate genes involved in coordinating the two major pathways of energy generation to promote aerobic metabolism, but how this serves to maintain genomic stability is less clear. In an attempt to understand the biology, this review presents human epidemiologic data on the inverse relationship between aerobic capacity and cancer incidence that appears to be mirrored by the impact of p53 on aerobic capacity in mouse models. The review summarizes mechanisms by which p53 regulates mitochondrial respiration and proposes how this might contribute to maintaining genomic stability. Although disparate in nature, the data taken together suggest that the promotion of aerobic metabolism by p53 serves as an important tumor suppressor activity and may provide insights for cancer prevention strategies in the future. Antioxid. Redox Signal. 15, 1739–1748.
Introduction
The loss of p53 activity in the majority of malignant human cancers is a testament to its critical role in maintaining genomic stability and preventing tumorigenesis, hence the reference to p53 as “guardian of the genome” (47, 52). Major advances in our understanding of p53 in cancer biology have been made by investigating its activities in regulating the cell cycle, senescence, apoptosis, and genomic stability (108). The involvement of p53 in functions as diverse as fecundity and metabolism continue to be uncovered (109). p53 is a gene that evolved before the beginning of multicellular life (56); therefore, studying its function is likely to yield valuable information beyond that relevant for cancer biology.
The promotion of aerobic metabolism by 53 through the stimulation of mitochondrial respiration and the concurrent inhibition of glycolysis have attracted attention because the frequent inactivation of p53 in cancers would be predicted to contribute to the observed phenomenon of increased glycolysis (62, 110, 112, 122). Increased glycolysis, also known to be stimulated by a number of oncogenes, may be advantageous to proliferating cancer cells as it generates high energy reducing equivalents, tricarboxcylic acid cycle substrates, and other carbon-back bone intermediates needed for the biosynthesis of lipids, proteins, and nucleic acids (24, 43). Likely reflecting altered cellular demands, cancer cells show other changes in metabolism, such as a greater dependence on glutamine as a bioenergetic substrate, which p53 also appears to regulate (36, 99, 113).
Consistent with abundant biochemical and genetic data demonstrating mitochondrial dysfunction in cancer cells (18, 66, 111), p53, as a tumor suppressor, appears to counteract this and promotes respiration.
The impedance of electron transfer, caused by mutations in nuclear and mitochondrial genes encoding the respiratory chain complex genes, may result in increased reactive oxygen species (ROS) and genomic deoxyribonucleic acid (DNA) damage (111). A recent study has demonstrated the importance of the mitochondria even at later stages of cancer progression by transferring metastatic potential between cancer cell lines through the mitochondria (39). This review begins by summarizing epidemiologic data that show an inverse relationship between cancer and cardiorespiratory fitness (CRF), a phenotype that is strongly associated with mitochondrial biogenesis (114). In light of this human data, the marked decrease in exercise capacity of p53-deficient mice appears to take on additional significance (62, 76, 88), and the current state of knowledge on both the physiological and molecular aspects of p53 regulation of mitochondrial respiration is presented. The remarkable consistency with which p53 promotes aerobic metabolism suggests that this phenotype may be important for tumor suppression and that it may reveal insights useful for devising new preventive strategies.
Cancer and CRF
Clues to the biological significance of the relationship between p53 and aerobic metabolism may be gleaned from the many clinical studies that have established the benefits of physical activity against cancer. Exercise can have cancer preventive effects, both before and after disease diagnosis (72), and therefore may affect both tumor initiation and progression (86). A number of comprehensive reviews have summarized evidence that supports this link between cancer prevention and physical activity in epidemiologic studies (29) and experimental models (35). To quantify physical activity and exercise habits, researchers have measured CRF (also termed aerobic endurance or aerobic fitness) by maximal exercise treadmill testing and resting heart rate (8, 12).
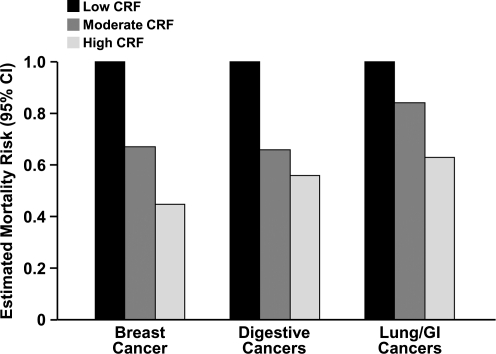
The strongest epidemiologic evidence for the protective effects of physical activity is seen in breast (67) and colon cancer (104). There is a dose-dependent relationship between CRF and estimated mortality risk from breast cancer (78), digestive cancer (79), and primarily lung and gastrointestinal cancers (49) (Fig. 1). In an evaluation of 29 case-controlled and 19 cohort epidemiologic studies, physical activity was associated with a 15%–20% decrease in the combined risk of pre- and postmenopausal breast cancer with stronger evidence for the latter. Conversely, 13%–14% of colon cancer incidence can be attributed to physical inactivity (34). Emerging evidence suggests that the risk of developing other types of cancers, such as that of the prostate (72), lung (49, 51), and gastrointestinal or digestive system (49, 79), may be decreased by physical activity as well.
FIG. 1.
The inverse relationship between cardiorespiratory fitness (CRF) and cancer mortality. Estimated mortality risks (95% confidence interval [95% CI]) were extrapolated from multivariate hazard ratio (breast and digestive cancers) and relative risk values (primarily lung and gastrointestinal [GI] cancers) in primary epidemiological studies: breast cancer (78), digestive cancer (79), and primarily lung and GI cancers (49).
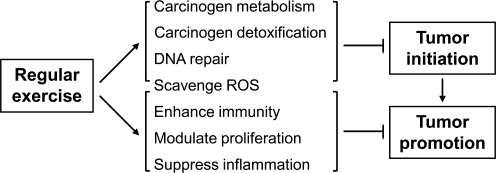
The cancer-type-specific benefits of exercise further increase the complexity of the potential mechanisms and indicate a need for improved understanding of this phenomenon (31, 70). The beneficial effects of exercise are often attributed to factors such as lowered risk of chronic diseases, including mental illnesses, decreased osteoporosis and sarcopenia, and decreased obesity. The specific mechanisms by which exercise may inhibit cancer formation can be summarized using the multistep model of carcinogenesis (Fig. 2) (86). Such a model provides a framework for outlining the complex network of cellular processes affected by exercise, some of which are better characterized than others.
FIG. 2.
Some of the proposed pathways by which exercise inhibits cancer are summarized using the multistep model of carcinogenesis (86). Such a model provides a useful framework for outlining the complex network of cellular processes affecting initiation and promotion/progression of cancer formation. The strength of evidence supporting the specific mechanisms is variable.
The first step in the multistep carcinogenesis model is initiation involving carcinogen metabolism and detoxification processes. Exercise has been shown to enhance cytochrome P450 and other enzymes such as NAD(P)H:quinone oxidoreductase 1 (NQO1) for carcinogen handling by the body (82, 107). Although acute, high-intensity exercise may increase mitochondrial ROS production (93), moderate long-term physical activity reduces oxidative damage by increasing antioxidant enzymes (26, 71, 83). Another direct mechanism by which moderate exercise could affect cancer may be through enhancing DNA damage repair enzymes such as human 8-hydroxyguanine DNA-glycosylase (hOGG1) (83). Notably, p53 has been shown to interact with, or regulate, many of the components discussed above (4, 73) and with both nuclear and mitochondrial genomic DNA repair pathways (1, 17).
Exercise affects a heterogeneous set of physiological processes to suppress tumorigenesis that include boosting innate immunity (69, 117) and decreasing chronic inflammation linked to cancer (21, 44, 86). Transcription factors important for mediating inflammatory signals, such as nuclear factor κ-light-chain-enhancer of activated B cells (NFKB) and activator protein-1 (AP-1), have been shown to be reduced by regular exercise (81). Besides the immune system, exercise modulates a host of other processes involved in cancer progression, for example, by suppressing ROS-induced mutagenesis even after initiation and balancing cell proliferation and apoptosis (86). The relative importance of the specific mechanisms by which exercise decreases cancer likely depends on multiple factors, including the type of mutagen, tissue, and species. Despite these complexities, the important fact is that increased CRF has a striking effect in preventing human cancers.
The Physiological Effects of p53 on CRF
Given the inverse relationship between CRF and cancer, the positive effect of p53 on exercise capacity is consistent with its role as a tumor suppressor (62). However, it was initially a surprising observation that mice deficient in p53 (p53−/−) had markedly lower exercise capacity (∼50%) in comparison to their wild-type (p53+/+) littermates using a forced swimming test (62). Further confirming this observation, p53−/− mice ran only a fraction of the distance of p53+/+ mice during an 8-week period of voluntary wheel running (88). Although maximum exercise testing is considered to be the gold standard for assessing human CRF (30, 68), its interpretation can be complicated in mice because forced exercise testing can activate stress responses in mice (3), while voluntary wheel running can be affected by circadian and neuro-behavioral factors. Although most animal exercise tests have their limitations, the combination of voluntary wheel running, forced swimming, and treadmill exercise data has provided convincing evidence for a defect in the exercise capacity of p53−/− mice (76, 88).
The decreased exercise capacity of p53−/− mice was further revealed biochemically after submaximum exercise (76). The blood lactate level was increased approximately threefold in p53−/− mice compared to wild-type, suggesting a lower aerobic metabolic capacity, and this difference was magnified after a 5-week period of treadmill training. The maximum work capacity of p53+/+ mice was significantly increased by training, whereas that of p53−/− mice was relatively unresponsive. There was further widening of the differences in aerobic capacity as monitored by both the lactate level and respiratory exchange ratio (RER, CO2 production/O2 consumption) during exercise, the latter being a parameter that is highly correlated to CRF in human subjects (85).
How p53 affects CRF warranted closer investigation, as the overt behavioral and morphometric features of p53+/+ and p53−/− mice are similar. CRF is determined by a number of important physiological parameters such as cardiac, pulmonary, and skeletal muscle function. Compensatory augmentation of cardiac output, in response to increased circulatory demand, is a major physiological determinant of exercise capacity. By cardiac magnetic resonance imaging, the baseline left ventricular function and inotropic reserve, as elicited by dobutamine stimulation, showed no significant dependence on p53 (76). In addition, there was no gross difference in the motor coordination or skeletal muscle strength of p53−/− mice (76, 88). Consistent with this finding, both genotypes demonstrated equivalent levels of high intensity sprinting in which glycolytic muscle fibers play an important role (76). Other factors that could affect exercise capacity such as hemoglobin concentration and skeletal muscle capillary density were also similar.
The lack of a significant difference in these important physiological parameters between p53−/− and p53+/+ mice indicated that the previous finding of p53-regulated mitochondrial respiration may contribute to the difference in aerobic exercise capacity (62). While muscle strength depends on short-term anaerobic energy generation, muscle endurance (or fatigue) is mainly determined by mitochondrial function and lactate disposition (25). p53−/− mice showed 50% greater fatigue than p53+/+ mice by an in situ muscle stimulation experiment, and p53−/− intermyofibrillar mitochondria had lower state 3 respiration and higher respiration-induced ROS production (88). p53−/− skeletal muscle also has decreased levels of the respiratory chain complex succinate dehydrogenase enzyme activity and lower mitochondrial DNA (mtDNA) content, further suggesting a decrease in functional mitochondria (76). In summary, experiments examining exercise metabolism and skeletal muscle revealed that p53 may contribute to CRF by regulating mitochondrial function.
p53 may also regulate other physiological parameters that determine cardiovascular function. In addition to the adaptive changes in skeletal muscle, prolonged exercise induces balanced cardiac hypertrophy in humans, which increases ventricular stroke volume (cardiac output), thus increasing exercise capacity (90). A recent study demonstrated that the downregulation of hypoxia-inducible factor 1, alpha subunit by p53 during sustained pressure overload of the heart impairs angiogenesis and may cause maladaptive cardiac hypertrophy in mice (89). In this model, p53 apparently serves to decrease cardiac function under pathological states, whereas under physiological exercise training conditions, p53 appears to promote aerobic exercise capacity. Apart from the cardiovascular system, p53 has been shown to inhibit insulin-like growth factor signaling (53) that is associated with glycolytic metabolism. Thus, through this pathway as well, p53 would be predicted to promote oxidative metabolism.
p53 Suppresses Anaerobic Metabolism
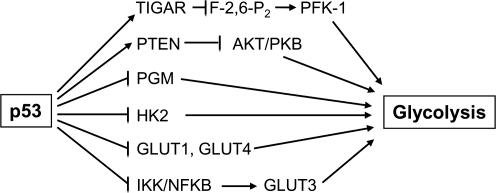
The phenotype of increased glycolysis observed in many cancer cells and the promotion of oxidative phosphorylation by p53 is intriguing as they represent the contrasting pathways of energy generation through anaerobic and aerobic metabolism, respectively. Even in primary human fibroblasts, the depletion of p53 increases glycolysis (15). Before addressing p53 and aerobic metabolism, the studies linking p53 to the regulation of anaerobic metabolism (glycolysis) are summarized (Fig. 3). The earliest studies showing that p53 can affect metabolism involved mutant p53 transactivating the hexokinase 2 (HK2) gene in hepatoma cells predictive of increased glycolysis in a p53 defective background (61). Subsequently, mutant p53 was shown to increase the activity of another glycolytic enzyme phosphoglycerate mutase (PGM) (45). In contrast, wild-type p53 was shown to interact with and transactivate PGM promoter constructs in vitro and in muscle cells, suggesting cell-type-specific effects of unclear significance (87). However, these initial metabolic studies collectively indicated that in the setting of defective p53 signaling, there is an increase in glycolytic metabolism.
FIG. 3.
The suppression of anaerobic metabolism by p53 is mediated through multiple pathways. Glycolysis is inhibited by decreasing metabolic substrate availability through downregulation of glucose transporters and by modulating key enzymes in the glycolytic pathway hexokinase 2 (HK2), 6-phosphofructo-1-kinase (PFK-1), and phosphoglycerate mutase (PGM).
More recently, the TP53-induced glycolysis and apoptosis regulator (TIGAR) gene has been shown to decrease the cellular levels of fructose-2,6-bisphosphate (F-2,6-P2), a potent allosteric activator of the enzyme 6-phosphofructo-1-kinase (PFK-1) that serves as a control point of the glycolytic pathway (11). The expression of TIGAR not only decreases glycolytic activity by dephosphorylating F-2,6-P2, but also reduces ROS levels and apoptosis by redirecting metabolites into the pentose phosphate shunt and promoting glutathione production, further supporting an antioxidant role for p53 (11, 73). p53 may also regulate glycolysis by downregulating the plasma membrane glucose transporters 1 and 4 (GLUT1 and GLUT4) (91). In addition, loss of p53 activates IKK, which through the NFKB pathway increases GLUT3 and glucose availability for glycolysis (41). Taken together, these various studies show that wild-type p53 activity suppresses anaerobic metabolism.
p53 Promotes Mitochondrial Function
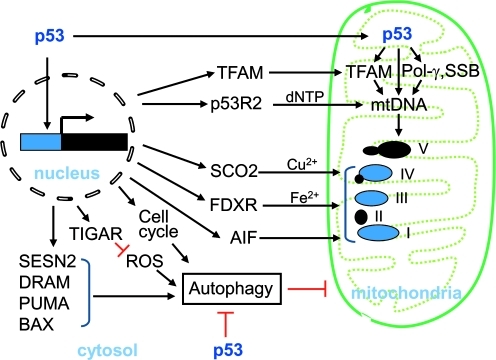
The importance of p53 in regulating normal mitochondrial function was first suggested by its apparent requirement for the transition from anaerobic to aerobic metabolism in mouse embryo development (38). In the absence of p53, there was a marked decrease in cytochrome c oxidase (COX) staining in association with lower ATP and mitochondrial 16S ribosomal RNA levels in day 8 mouse embryos (7–8 somite stage). Early studies focusing on p53-induced apoptosis clearly demonstrated that p53 regulates redox homeostasis, including the transactivation of a number of genes bearing homology to redox enzymes (40, 80). Since that time, significant progress has been made to advance our understanding of p53 redox biology (73) and emerging data show that p53 also regulates mitochondrial respiration through multiple pathways (Fig. 4).
FIG. 4.
The multiple pathways of mitochondrial regulation by p53. These are: (1) assembly of the oxidative phosphorylation complexes; (2) mitochondrial deoxyribonucleic acid (mtDNA) repair, replication, and transcription through DNA polymerase γ, p53-inducible ribonucleotide reductase 2 (p53R2), and TF1M; and (3) autophagy for normal mitochondrial homeostasis. (To see this illustration in color the reader is referred to the web version of this article at www.liebertonline.com/ars).
The mechanisms by which p53 regulates respiration are being elucidated, but the underlying biology is less clear. The recent creation of a nonrespiring human cell line, through somatic cell homologous recombination, has demonstrated the critical role of respiration in regulating both intracellular oxygen and redox homeostasis, which are both important for preventing oxygen-associated genomic DNA damage (98). This observation reinforces the original concept underlying the symbiotic theory of the mitochondrion. Protection from oxygen toxicity, rather than the bioenergetic benefits of oxidative phosphorylation, was proposed to be the evolutionary driving force for the incorporation of oxygen consuming proteobacteria into primordial anaerobic eukaryotes (59).
Assembly of the respiratory chain complexes
The identification of ferrodoxin reductase, encoded by the p53-transactivated gene FDXR, was an early clue that p53 may regulate respiration as this mitochondrial NADPH P450 reductase is required for iron–sulfur protein and heme biogenesis, both essential for the function of respiratory complexes (37, 54). COX enzymatic activity was shown to be decreased in the p53-deficient human colon cancer HCT116 cell line in parallel with reduced COX II subunit protein level (122). However, the genetic mechanism by which p53 affected COX II subunit expression remained unclear, as its transcript levels were reported to be unchanged (122).
Through gene expression analysis, the synthesis of cytochrome c oxidase 2 (SCO2) gene was identified as a p53 transcriptional target gene (37, 62). Given its critical role in COX assembly as a copper delivery chaperone, SCO2 provided a defined genetic mechanism by which p53 directly regulated the eukaryotic center of oxygen consumption (62, 75). Like SCO2, mitochondrial flavoprotein apoptosis-inducing factor (AIF) also appeared to contribute to mitochondrial complex assembly, as its depletion results in reduced content and activity of respiratory chain complex I and increased glycolysis (105). The AIF gene was later found to have a p53 binding element that is responsive to basal levels of p53, further supporting a physiologic function (95). Thus, p53 appears to transcriptionally regulate at least three proteins that are involved in the assembly of respiratory complexes.
Mitochondrial DNA
In analogy to its role in maintaining nuclear genomic integrity, p53 is also important for the mitochondrial genome. p53 can translocate into mitochondria and interact with DNA polymerase γ and single-stranded DNA-binding (SSB) protein to regulate mtDNA repair, replication, and recombination (2, 7, 116) as well as directly bind to mtDNA to regulate nucleoid remodeling (5). It can also interact with mitochondrial transcription factor A (TFAM), essential for regulating both mtDNA transcription and replication (48), and modulate its binding to mtDNA (120). A recent study has shown that the interaction of TFAM with both the N-terminal transactivation and C-terminal regulatory domains of p53 has implications for the repair of damaged mtDNA (115). Besides TFAM, cytoplasmic p53-inducible ribonucleotide reductase 2 (p53R2) is essential for maintaining mtDNA through regulating deoxyribonucleotide (dNTP) homeostasis (33). In the skeletal muscle of both mice and humans, p53R2 mutations cause severe mtDNA depletion (13, 14).
The observation of higher mtDNA content in the skeletal muscle of p53+/+ compared to p53−/− mice was consistent with the expected major role that skeletal muscle plays in determining exercise capacity. It was particularly notable that the relative difference in mtDNA content between the p53+/+ and p53−/− mice was greater in the oxidative (soleus, plantaris) versus glycolytic (tibialis anterior, extensor digitorum longus) skeletal muscles, underscoring the observation that p53 promotes aerobic metabolism (76). The lower mtDNA content in p53−/− muscle correlated with reduced respiratory activity (76, 88). The maintenance of mtDNA by p53 is not limited to skeletal muscle, as it is also reduced in p53−/− mouse embryonic fibroblasts and p53-depleted human fibroblasts in association with reductions in TFAM and/or p53R2 (46, 50). Although the modulation of TFAM by p53 appears complex and may involve post-transcriptional mechanisms (50), the mouse TFAM gene also has p53 responsive elements (76). This may explain the augmentation of TFAM expression by p53 in mouse skeletal muscle with exercise (76). Whether p53 is also involved in the regulation of exercise-induced TFAM expression in humans remains to be seen (9).
Mitochondrial autophagy
Autophagy is a membrane-trafficking process used to degrade cytosolic proteins and organelles and is important in regulating mitochondrial function. The selective degradation of dysfunctional mitochondria (mitophagy) is required for normal mitochondrial homeostasis as deletion of the critical autophagy gene ATG7 in muscle results in the accumulation of abnormal mitochondria (42, 60). Intriguingly, like the anti- and pro-oxidant effects of p53, the regulation of autophagy by p53 also displays a dual nature (101, 109). In response to genotoxic stress, nuclear p53 induces autophagy by transactivating damage-regulated autophagy modulator (DRAM), a gene essential for both p53-triggered autophagy and apoptosis (22). In contrast, basal levels of cytoplasmic p53 suppress autophagy by inhibiting 5′adenosine monophosphate-activated protein kinase and activating mammalian target of rapamycin whereas the loss of p53 induces autophagy in both normal and cancer cells (28, 102).
Compared with the established role of p53-induced apoptosis in tumor suppression, the dual nature of p53-regulated autophagy on cancer is less clear. DRAM is downregulated in several human cancer cell lines, suggesting a tumor suppressor role for p53-regulated autophagy (22). A recent study showed that another p53 target gene, Sestrin 2 (SESN2), induces autophagy in cells with wild-type p53, whereas depletion of SESN2 or DRAM was equally efficient in preventing autophagy (57). Autophagy also shares pathways that promote apoptosis, further linking it to tumor suppression. p53 transactivated apoptotic genes, p53 upregulated modulator of apoptosis (PUMA), and Bcl-2-associated X protein (BAX) can coordinately trigger autophagy that selectively degrades the mitochondria (119). Mitochondrial autophagy itself facilitates apoptosis-associated cytochrome c release and likewise the antiapoptotic protein Bcl-2 suppresses autophagy (121).
In contrast to its proautophagic function associated with apoptosis, p53 also suppresses autophagy and this may facilitate cell survival under conditions of physiologic stress. In addition to inhibiting glycolysis, p53-induced TIGAR decreases ROS production in response to nutrient starvation, thereby inhibiting autophagy (10). While SESN2 has been reported to promote autophagy, it remains to be seen whether other p53-regulated sestrin family members that are known to have antioxidant function have inhibitory activities on autophagy (73). Mitochondrial morphology and homeostasis are connected to the cell cycle and a “mitochondrial checkpoint” in the G1 phase has been proposed (65). Mitochondrial respiration is high in G1 and its dysfunction causes G1 arrest (58, 63). The depletion of p53 induces a periodic peak of autophagy at the G1 and S phases but not in the G2 or M phase (103). Thus, the antiautophagic function of p53 appears to be cell cycle specific and possibly related to variations in bioenergetic requirements during different phases of the cell cycle. In summary, p53-regulated autophagy may play an important role in modulating mitochondrial function to meet different cellular and physiologic requirements.
Redox Regulation of p53
p53 activity is sensitive to the cellular redox state, suggesting its role as a mediator of redox homeostasis. p53 regulates mitochondrial respiration, which has a profound effect on the cellular redox state (98), and other accumulating evidence supports the role of p53 as a redox sensor (Fig. 5) (55). The presence of cellular reducing agents are required for the binding of p53 to its target DNA in vitro (32), whereas increased cellular oxidants such as copper inhibit its DNA-binding activity in mammalian cells (106). Depleting yeast thioredoxin reductase (TRR) causes accumulation of an oxidized, inactive form of p53 (77), whereas knocking down mammalian TRR does not affect p53 disulfide status but instead stabilizes p53 and increases its activity (92). The redox regulation of p53 is specifically dependent on an intact thioredoxin system and genetically altering the ratio of oxidized to reduced glutathione does not affect p53 activity (64, 96). In mammalian cells, p53 also binds to apurinic (apyrimidinic) endonuclease/redox-factor1 (APE/REF-1). Depleting APE/REF-1 results in the degradation of p53 although its basal level does not change when both TRR and APE/REF-1 are depleted (92). Therefore, without apparently affecting its disulfide status, TRR and APE/REF-1 differentially regulate p53 basal and stress-induced levels, nuclear translocation, and DNA binding.
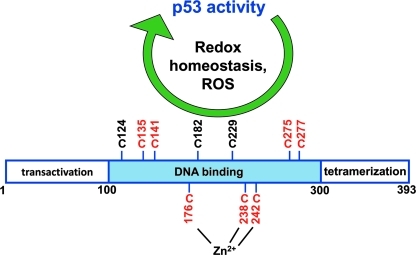
FIG. 5.
p53 as a sensor and regulator of cellular redox homeostasis. All 10 cysteine residues of human p53 are localized to the DNA binding domain. Cysteine residues numbered in red are those that have been shown to be important for regulating p53 activity and those coordinated with zinc have been shown to be protected from oxidation. (To see this illustration in color the reader is referred to the web version of this article at www.liebertonline.com/ars).
Human p53 contains 10 cysteine residues, all in the DNA-binding domain, and their redox state is critical for p53 function (Fig. 5). Three cysteine residues C176, C238, and C242 are tightly bound to zinc, which protects them from being oxidized, and their substitution by serine abolishes p53 activity (19, 96). Oxidation of the cysteine residues and formation of disulfide bonds inhibit p53 tetramerization and DNA binding activity (97). Other cysteine residues such as C135, C141, C275, and C277 have also been reported to be important for the selective modulation of p53 activity (16, 96). For example, intramolecular disulfide bond formation between C275 and C277 in response to DNA damage specifically prevents p53 binding to the promoter sequence of GADD45 but not that of p21 (16, 94). Similarly, using DNA-mediated charge transport to selectively oxidize DNA-bound proteins in vitro to simulate severe stress, oxidized p53 loses its ability to bind to the GADD45 but not the p21 promoter (6). The importance of these specific cysteine residues for p53 activity is further supported by the absence of the nonessential human p53 C229 in mouse and by the observation that the three additional cysteines in mouse p53 (C40, C293, and C308) are not required for activity (84). In summary, mitochondrial function is an important determinant of cellular redox state and, in turn, the sensitivity of p53 to cellular redox state points to a homeostatic relationship.
Conclusions and Perspectives
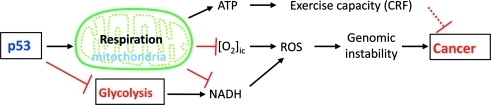
In the context of the reviewed data, the capacity of p53 to regulate mitochondrial respiration and thereby promote aerobic metabolism are proposed to have a tumor suppressor function. As a gene that is represented in the earliest of unicellular life, its ancestral function has been proposed to extend beyond protection against tumor formation in the multicellular animal (56). Given its early evolutionary origin, its promotion of aerobic metabolism likely pertains to more basic functions such as adaptations for aerobic life. In support of this notion, when mitochondrial respiration is ablated by the targeted disruption of both copies of the p53 target gene SCO2, the essential substrates for ROS generation (intracellular oxygen and reducing equivalents) are increased with resultant oxidative genomic DNA damage (Fig. 6) (98). Studying such primary biologic functions reveals at least one reason why p53 might promote aerobic metabolism, for genomic protection in an oxidative environment, and may provide more insights into cancer biology.
FIG. 6.
p53 promotion of mitochondrial functions improves CRF and decreases oxidative stress to prevent DNA damage. Respiration consumes intracellular oxygen ([O2]ic) and NADH, both of which contribute to reactive oxygen species (ROS) generation and genomic instability. Concurrent inhibition of glycolysis decreases the generation of reducing equivalents (NADH) that serve as electron source to molecular oxygen. The pleiotropic nature of the mechanism by which CRF may contribute to tumor suppression is indicated by a dashed line. (To see this illustration in color the reader is referred to the web version of this article at www.liebertonline.com/ars).
Recent work further underscores the important effect of metabolism on tumorigenesis. A specific mutation in the cytosolic enzyme isocitrate dehydrogenase 1 results in a gain of function that may contribute to cancer pathogenesis (23, 118) and the switching of pyruvate kinase enzyme isoforms improves cancer cell survival (20). In retrospect, the previous observations that mitochondrial enzymes succinate dehydrogenase and fumarate hydratase act as tumor suppressor genes (27) also presaged the finding that p53 promotes aerobic metabolism. The hope is that translating these basic principles to specific populations with genetic susceptibility to cancers, such as in Li-Fraumeni syndrome (74, 100), or to the general population may result in new strategies to prevent cancer and improve overall human health.
Abbreviations Used
- 95% CI
associated 95% confidence intervals
- AIF
apoptosis-inducing factor
- APE/REF-1
apurinic (apurimidinic) endonuclease/redox-factor 1
- BAX
Bcl-2-associated X protein
- COX
cytochrome c oxidase
- CRF
cardiorespiratory fitness
- DNA
deoxyribonucleic acid
- dNTP
deoxyribonucleotide
- DRAM
damage-regulated autophagy modulator
- F-2,6-P2
fructose-2,6-bisphosphate
- FDXR
ferrodoxin reductase
- GI
gastrointestinal
- GLUT
glucose transporter
- HK2
hexokinase 2
- IKK/NFKB
IκB kinase/nuclear factor κ-light-chain-enhancer of activated B cells
- mtDNA
mitochondrial DNA
- NADH
reduced nicotinamide adenine dinucleotide
- [O2]ic
intracellular oxygen concentration
- p53R2
p53-inducible ribonucleotide reductase 2
- PFK-1
6-phosphofructo-1-kinase
- PGM
phosphoglycerate mutase
- Pol-γ
DNA polymerase γ
- PTEN
phosphatase and tensin homolog
- PUMA
p53 upregulated modulator of apoptosis
- ROS
reactive oxygen species
- SCO2
synthesis of cytochrome c oxidase 2
- SESN2
sestrin 2
- SSB
single-stranded DNA binding protein
- TFAM
mitochondrial transcription factor A
- TIGAR
TP53-induced glycolysis and apoptosis regulator
- TRR
thioredoxin reductase
References
- 1.Achanta G. Huang P. Role of p53 in sensing oxidative DNA damage in response to reactive oxygen species-generating agents. Cancer Res. 2004;64:6233–6239. doi: 10.1158/0008-5472.CAN-04-0494. [DOI] [PubMed] [Google Scholar]
- 2.Achanta G. Sasaki R. Feng L. Carew JS. Lu W. Pelicano H. Keating MJ. Huang P. Novel role of p53 in maintaining mitochondrial genetic stability through interaction with DNA Pol gamma. EMBO J. 2005;24:3482–3492. doi: 10.1038/sj.emboj.7600819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arida RM. Scorza CA. da Silva AV. Scorza FA. Cavalheiro EA. Differential effects of spontaneous versus forced exercise in rats on the staining of parvalbumin-positive neurons in the hippocampal formation. Neurosci Lett. 2004;364:135–138. doi: 10.1016/j.neulet.2004.03.086. [DOI] [PubMed] [Google Scholar]
- 4.Asher G. Lotem J. Kama R. Sachs L. Shaul Y. NQO1 stabilizes p53 through a distinct pathway. Proc Natl Acad Sci U S A. 2002;99:3099–3104. doi: 10.1073/pnas.052706799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ashley N. Poulton J. Anticancer DNA intercalators cause p53-dependent mitochondrial DNA nucleoid re-modelling. Oncogene. 2009;28:3880–3891. doi: 10.1038/onc.2009.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Augustyn KE. Merino EJ. Barton JK. A role for DNA-mediated charge transport in regulating p53: oxidation of the DNA-bound protein from a distance. Proc Natl Acad Sci U S A. 2007;104:18907–18912. doi: 10.1073/pnas.0709326104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bakhanashvili M. Grinberg S. Bonda E. Simon AJ. Moshitch-Moshkovitz S. Rahav G. p53 in mitochondria enhances the accuracy of DNA synthesis. Cell Death Differ. 2008;15:1865–1874. doi: 10.1038/cdd.2008.122. [DOI] [PubMed] [Google Scholar]
- 8.Balke B. Ware RW. An experimental study of physical fitness of Air Force personnel. U S Armed Forces Med J. 1959;10:675–688. [PubMed] [Google Scholar]
- 9.Bengtsson J. Gustafsson T. Widegren U. Jansson E. Sundberg CJ. Mitochondrial transcription factor A and respiratory complex IV increase in response to exercise training in humans. Pflugers Arch. 2001;443:61–66. doi: 10.1007/s004240100628. [DOI] [PubMed] [Google Scholar]
- 10.Bensaad K. Cheung EC. Vousden KH. Modulation of intracellular ROS levels by TIGAR controls autophagy. EMBO J. 2009;28:3015–3026. doi: 10.1038/emboj.2009.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bensaad K. Tsuruta A. Selak MA. Vidal MN. Nakano K. Bartrons R. Gottlieb E. Vousden KH. TIGAR, a p53-inducible regulator of glycolysis and apoptosis. Cell. 2006;126:107–120. doi: 10.1016/j.cell.2006.05.036. [DOI] [PubMed] [Google Scholar]
- 12.Blair SN. Kohl HW., 3rd Paffenbarger RS., Jr. Clark DG. Cooper KH. Gibbons LW. Physical fitness and all-cause mortality. A prospective study of healthy men and women. JAMA. 1989;262:2395–2401. doi: 10.1001/jama.262.17.2395. [DOI] [PubMed] [Google Scholar]
- 13.Bornstein B. Area E. Flanigan KM. Ganesh J. Jayakar P. Swoboda KJ. Coku J. Naini A. Shanske S. Tanji K. Hirano M. DiMauro S. Mitochondrial DNA depletion syndrome due to mutations in the RRM2B gene. Neuromuscul Disord. 2008;18:453–459. doi: 10.1016/j.nmd.2008.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bourdon A. Minai L. Serre V. Jais JP. Sarzi E. Aubert S. Chretien D. de Lonlay P. Paquis-Flucklinger V. Arakawa H. Nakamura Y. Munnich A. Rotig A. Mutation of RRM2B, encoding p53-controlled ribonucleotide reductase (p53R2), causes severe mitochondrial DNA depletion. Nat Genet. 2007;39:776–780. doi: 10.1038/ng2040. [DOI] [PubMed] [Google Scholar]
- 15.Burns DM. Richter JD. CPEB regulation of human cellular senescence, energy metabolism, and p53 mRNA translation. Genes Dev. 2008;22:3449–3460. doi: 10.1101/gad.1697808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Buzek J. Latonen L. Kurki S. Peltonen K. Laiho M. Redox state of tumor suppressor p53 regulates its sequence-specific DNA binding in DNA-damaged cells by cysteine 277. Nucleic Acids Res. 2002;30:2340–2348. doi: 10.1093/nar/30.11.2340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chatterjee A. Mambo E. Osada M. Upadhyay S. Sidransky D. The effect of p53-RNAi and p53 knockout on human 8-oxoguanine DNA glycosylase (hOgg1) activity. FASEB J. 2006;20:112–114. doi: 10.1096/fj.04-3423fje. [DOI] [PubMed] [Google Scholar]
- 18.Chatterjee A. Mambo E. Sidransky D. Mitochondrial DNA mutations in human cancer. Oncogene. 2006;25:4663–4674. doi: 10.1038/sj.onc.1209604. [DOI] [PubMed] [Google Scholar]
- 19.Cho Y. Gorina S. Jeffrey PD. Pavletich NP. Crystal structure of a p53 tumor suppressor-DNA complex: understanding tumorigenic mutations. Science. 1994;265:346–355. doi: 10.1126/science.8023157. [DOI] [PubMed] [Google Scholar]
- 20.Christofk HR. Vander Heiden MG. Harris MH. Ramanathan A. Gerszten RE. Wei R. Fleming MD. Schreiber SL. Cantley LC. The M2 splice isoform of pyruvate kinase is important for cancer metabolism and tumour growth. Nature. 2008;452:230–233. doi: 10.1038/nature06734. [DOI] [PubMed] [Google Scholar]
- 21.Coussens LM. Werb Z. Inflammation and cancer. Nature. 2002;420:860–867. doi: 10.1038/nature01322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Crighton D. Wilkinson S. O'Prey J. Syed N. Smith P. Harrison PR. Gasco M. Garrone O. Crook T. Ryan KM. DRAM, a p53-induced modulator of autophagy, is critical for apoptosis. Cell. 2006;126:121–134. doi: 10.1016/j.cell.2006.05.034. [DOI] [PubMed] [Google Scholar]
- 23.Dang L. White DW. Gross S. Bennett BD. Bittinger MA. Driggers EM. Fantin VR. Jang HG. Jin S. Keenan MC. Marks KM. Prins RM. Ward PS. Yen KE. Liau LM. Rabinowitz JD. Cantley LC. Thompson CB. Vander Heiden MG. Su SM. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature. 2009;462:739–744. doi: 10.1038/nature08617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.DeBerardinis RJ. Lum JJ. Hatzivassiliou G. Thompson CB. The biology of cancer: metabolic reprogramming fuels cell growth and proliferation. Cell Metab. 2008;7:11–20. doi: 10.1016/j.cmet.2007.10.002. [DOI] [PubMed] [Google Scholar]
- 25.Dengler R. Wohlfarth K. Zierz S. Jobges M. Schubert M. Muscle fatigue, lactate, and pyruvate in mitochondrial myopathy with progressive external ophthalmoplegia. Muscle Nerve. 1996;19:456–462. doi: 10.1002/(SICI)1097-4598(199604)19:4<456::AID-MUS5>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 26.Duncan K. Harris S. Ardies CM. Running exercise may reduce risk for lung and liver cancer by inducing activity of antioxidant and phase II enzymes. Cancer Lett. 1997;116:151–158. doi: 10.1016/s0304-3835(97)00189-4. [DOI] [PubMed] [Google Scholar]
- 27.Eng C. Kiuru M. Fernandez MJ. Aaltonen LA. A role for mitochondrial enzymes in inherited neoplasia and beyond. Nat Rev Cancer. 2003;3:193–202. doi: 10.1038/nrc1013. [DOI] [PubMed] [Google Scholar]
- 28.Feng Z. Zhang H. Levine AJ. Jin S. The coordinate regulation of the p53 and mTOR pathways in cells. Proc Natl Acad Sci U S A. 2005;102:8204–8209. doi: 10.1073/pnas.0502857102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Friedenreich CM. Orenstein MR. Physical activity and cancer prevention: etiologic evidence and biological mechanisms. J Nutr. 2002;132:3456S–3464S. doi: 10.1093/jn/132.11.3456S. [DOI] [PubMed] [Google Scholar]
- 30.Gibbons LW. Mitchell TL. Wei M. Blair SN. Cooper KH. Maximal exercise test as a predictor of risk for mortality from coronary heart disease in asymptomatic men. Am J Cardiol. 2000;86:53–58. doi: 10.1016/s0002-9149(00)00827-4. [DOI] [PubMed] [Google Scholar]
- 31.Gill JM. Malkova D. Physical activity, fitness and cardiovascular disease risk in adults: interactions with insulin resistance and obesity. Clin Sci (Lond) 2006;110:409–425. doi: 10.1042/CS20050207. [DOI] [PubMed] [Google Scholar]
- 32.Hainaut P. Milner J. Redox modulation of p53 conformation and sequence-specific DNA binding in vitro. Cancer Res. 1993;53:4469–4473. [PubMed] [Google Scholar]
- 33.Hakansson P. Hofer A. Thelander L. Regulation of mammalian ribonucleotide reduction and dNTP pools after DNA damage and in resting cells. J Biol Chem. 2006;281:7834–7841. doi: 10.1074/jbc.M512894200. [DOI] [PubMed] [Google Scholar]
- 34.Harriss DJ. Cable NT. George K. Reilly T. Renehan AG. Haboubi N. Physical activity before and after diagnosis of colorectal cancer: disease risk, clinical outcomes, response pathways and biomarkers. Sports Med. 2007;37:947–960. doi: 10.2165/00007256-200737110-00003. [DOI] [PubMed] [Google Scholar]
- 35.Hoffman-Goetz L. Physical activity and cancer prevention: animal-tumor models. Med Sci Sports Exerc. 2003;35:1828–1833. doi: 10.1249/01.MSS.0000093621.09328.70. [DOI] [PubMed] [Google Scholar]
- 36.Hu W. Zhang C. Wu R. Sun Y. Levine A. Feng Z. Glutaminase 2, a novel p53 target gene regulating energy metabolism and antioxidant function. Proc Natl Acad Sci U S A. 2010;107:7455–7460. doi: 10.1073/pnas.1001006107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hwang PM. Bunz F. Yu J. Rago C. Chan TA. Murphy MP. Kelso GF. Smith RA. Kinzler KW. Vogelstein B. Ferredoxin reductase affects p53-dependent, 5-fluorouracil-induced apoptosis in colorectal cancer cells. Nat Med. 2001;7:1111–1117. doi: 10.1038/nm1001-1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ibrahim MM. Razmara M. Nguyen D. Donahue RJ. Wubah JA. Knudsen TB. Altered expression of mitochondrial 16S ribosomal RNA in p53-deficient mouse embryos revealed by differential display. Biochim Biophys Acta. 1998;1403:254–264. doi: 10.1016/s0167-4889(98)00066-4. [DOI] [PubMed] [Google Scholar]
- 39.Ishikawa K. Takenaga K. Akimoto M. Koshikawa N. Yamaguchi A. Imanishi H. Nakada K. Honma Y. Hayashi J. ROS-generating mitochondrial DNA mutations can regulate tumor cell metastasis. Science. 2008;320:661–664. doi: 10.1126/science.1156906. [DOI] [PubMed] [Google Scholar]
- 40.Johnson TM. Yu ZX. Ferrans VJ. Lowenstein RA. Finkel T. Reactive oxygen species are downstream mediators of p53-dependent apoptosis. Proc Natl Acad Sci U S A. 1996;93:11848–11852. doi: 10.1073/pnas.93.21.11848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kawauchi K. Araki K. Tobiume K. Tanaka N. p53 regulates glucose metabolism through an IKK-NF-kappaB pathway and inhibits cell transformation. Nat Cell Biol. 2008;10:611–618. doi: 10.1038/ncb1724. [DOI] [PubMed] [Google Scholar]
- 42.Kim I. Rodriguez-Enriquez S. Lemasters JJ. Selective degradation of mitochondria by mitophagy. Arch Biochem Biophys. 2007;462:245–253. doi: 10.1016/j.abb.2007.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim JW. Dang CV. Cancer's molecular sweet tooth and the Warburg effect. Cancer Res. 2006;66:8927–8930. doi: 10.1158/0008-5472.CAN-06-1501. [DOI] [PubMed] [Google Scholar]
- 44.King DE. Carek P. Mainous AG., 3rd Pearson WS. Inflammatory markers and exercise: differences related to exercise type. Med Sci Sports Exerc. 2003;35:575–581. doi: 10.1249/01.MSS.0000058440.28108.CC. [DOI] [PubMed] [Google Scholar]
- 45.Kondoh H. Lleonart ME. Gil J. Wang J. Degan P. Peters G. Martinez D. Carnero A. Beach D. Glycolytic enzymes can modulate cellular life span. Cancer Res. 2005;65:177–185. [PubMed] [Google Scholar]
- 46.Kulawiec M. Ayyasamy V. Singh KK. p53 regulates mtDNA copy number and mitocheckpoint pathway. J Carcinog. 2009;8:8. doi: 10.4103/1477-3163.50893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lane DP. Cancer. p53, guardian of the genome. Nature. 1992;358:15–16. doi: 10.1038/358015a0. [DOI] [PubMed] [Google Scholar]
- 48.Larsson NG. Wang J. Wilhelmsson H. Oldfors A. Rustin P. Lewandoski M. Barsh GS. Clayton DA. Mitochondrial transcription factor A is necessary for mtDNA maintenance and embryogenesis in mice. Nat Genet. 1998;18:231–236. doi: 10.1038/ng0398-231. [DOI] [PubMed] [Google Scholar]
- 49.Laukkanen JA. Pukkala E. Rauramaa R. Makikallio TH. Toriola AT. Kurl S. Cardiorespiratory fitness, lifestyle factors and cancer risk and mortality in Finnish men. Eur J Cancer. 2010;46:355–363. doi: 10.1016/j.ejca.2009.07.013. [DOI] [PubMed] [Google Scholar]
- 50.Lebedeva MA. Eaton JS. Shadel GS. Loss of p53 causes mitochondrial DNA depletion and altered mitochondrial reactive oxygen species homeostasis. Biochim Biophys Acta. 2009;1787:328–334. doi: 10.1016/j.bbabio.2009.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lee IM. Sesso HD. Paffenbarger RS., Jr Physical activity and risk of lung cancer. Int J Epidemiol. 1999;28:620–625. doi: 10.1093/ije/28.4.620. [DOI] [PubMed] [Google Scholar]
- 52.Levine AJ. p53, the cellular gatekeeper for growth and division. Cell. 1997;88:323–331. doi: 10.1016/s0092-8674(00)81871-1. [DOI] [PubMed] [Google Scholar]
- 53.Levine AJ. Feng Z. Mak TW. You H. Jin S. Coordination and communication between the p53 and IGF-1-AKT-TOR signal transduction pathways. Genes Dev. 2006;20:267–275. doi: 10.1101/gad.1363206. [DOI] [PubMed] [Google Scholar]
- 54.Lill R. Function and biogenesis of iron-sulphur proteins. Nature. 2009;460:831–838. doi: 10.1038/nature08301. [DOI] [PubMed] [Google Scholar]
- 55.Liu B. Chen Y. St. Clair DK. ROS and p53: a versatile partnership. Free Radic Biol Med. 2008;44:1529–1535. doi: 10.1016/j.freeradbiomed.2008.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lu WJ. Amatruda JF. Abrams JM. p53 ancestry: gazing through an evolutionary lens. Nat Rev Cancer. 2009;9:758–762. doi: 10.1038/nrc2732. [DOI] [PubMed] [Google Scholar]
- 57.Maiuri MC. Malik SA. Morselli E. Kepp O. Criollo A. Mouchel PL. Carnuccio R. Kroemer G. Stimulation of autophagy by the p53 target gene Sestrin2. Cell Cycle. 2009;8:1571–1576. doi: 10.4161/cc.8.10.8498. [DOI] [PubMed] [Google Scholar]
- 58.Mandal S. Guptan P. Owusu-Ansah E. Banerjee U. Mitochondrial regulation of cell cycle progression during development as revealed by the tenured mutation in Drosophila. Dev Cell. 2005;9:843–854. doi: 10.1016/j.devcel.2005.11.006. [DOI] [PubMed] [Google Scholar]
- 59.Margulis L. Symbiosis in Cell Evolution. New York: W.H. Freeman and Company; 1993. [Google Scholar]
- 60.Masiero E. Agatea L. Mammucari C. Blaauw B. Loro E. Komatsu M. Metzger D. Reggiani C. Schiaffino S. Sandri M. Autophagy is required to maintain muscle mass. Cell Metab. 2009;10:507–515. doi: 10.1016/j.cmet.2009.10.008. [DOI] [PubMed] [Google Scholar]
- 61.Mathupala SP. Heese C. Pedersen PL. Glucose catabolism in cancer cells. The type II hexokinase promoter contains functionally active response elements for the tumor suppressor p53. J Biol Chem. 1997;272:22776–22780. doi: 10.1074/jbc.272.36.22776. [DOI] [PubMed] [Google Scholar]
- 62.Matoba S. Kang JG. Patino WD. Wragg A. Boehm M. Gavrilova O. Hurley PJ. Bunz F. Hwang PM. p53 regulates mitochondrial respiration. Science. 2006;312:1650–1653. doi: 10.1126/science.1126863. [DOI] [PubMed] [Google Scholar]
- 63.Matsumoto T. Wang PY. Ma W. Sung HJ. Matoba S. Hwang PM. Polo-like kinases mediate cell survival in mitochondrial dysfunction. Proc Natl Acad Sci U S A. 2009;106:14542–14546. doi: 10.1073/pnas.0904229106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Merwin JR. Mustacich DJ. Muller EG. Pearson GD. Merrill GF. Reporter gene transactivation by human p53 is inhibited in thioredoxin reductase null yeast by a mechanism associated with thioredoxin oxidation and independent of changes in the redox state of glutathione. Carcinogenesis. 2002;23:1609–1615. doi: 10.1093/carcin/23.10.1609. [DOI] [PubMed] [Google Scholar]
- 65.Mitra K. Wunder C. Roysam B. Lin G. Lippincott-Schwartz J. A hyperfused mitochondrial state achieved at G1-S regulates cyclin E buildup and entry into S phase. Proc Natl Acad Sci U S A. 2009;106:11960–11965. doi: 10.1073/pnas.0904875106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Modica-Napolitano JS. Singh KK. Mitochondrial dysfunction in cancer. Mitochondrion. 2004;4:755–762. doi: 10.1016/j.mito.2004.07.027. [DOI] [PubMed] [Google Scholar]
- 67.Monninkhof EM. Elias SG. Vlems FA. van der Tweel I. Schuit AJ. Voskuil DW. van Leeuwen FE. Physical activity and breast cancer: a systematic review. Epidemiology. 2007;18:137–157. doi: 10.1097/01.ede.0000251167.75581.98. [DOI] [PubMed] [Google Scholar]
- 68.Mora S. Redberg RF. Cui Y. Whiteman MK. Flaws JA. Sharrett AR. Blumenthal RS. Ability of exercise testing to predict cardiovascular and all-cause death in asymptomatic women: a 20-year follow-up of the lipid research clinics prevalence study. JAMA. 2003;290:1600–1607. doi: 10.1001/jama.290.12.1600. [DOI] [PubMed] [Google Scholar]
- 69.Murphy EA. Davis JM. Brown AS. Carmichael MD. Mayer EP. Ghaffar A. Effects of moderate exercise and oat beta-glucan on lung tumor metastases and macrophage antitumor cytotoxicity. J Appl Physiol. 2004;97:955–959. doi: 10.1152/japplphysiol.00252.2004. [DOI] [PubMed] [Google Scholar]
- 70.Myers J. Prakash M. Froelicher V. Do D. Partington S. Atwood JE. Exercise capacity and mortality among men referred for exercise testing. N Engl J Med. 2002;346:793–801. doi: 10.1056/NEJMoa011858. [DOI] [PubMed] [Google Scholar]
- 71.Nakatani K. Komatsu M. Kato T. Yamanaka T. Takekura H. Wagatsuma A. Aoyama K. Xu B. Hirano T. Kasai H. Ando S. Takeuchi T. Habitual exercise induced resistance to oxidative stress. Free Radic Res. 2005;39:905–911. doi: 10.1080/10715760500183300. [DOI] [PubMed] [Google Scholar]
- 72.Newton RU. Galvao DA. Exercise in prevention and management of cancer. Curr Treat Options Oncol. 2008;9:135–146. doi: 10.1007/s11864-008-0065-1. [DOI] [PubMed] [Google Scholar]
- 73.Olovnikov IA. Kravchenko JE. Chumakov PM. Homeostatic functions of the p53 tumor suppressor: regulation of energy metabolism and antioxidant defense. Semin Cancer Biol. 2009;19:32–41. doi: 10.1016/j.semcancer.2008.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Palmero EI. Achatz MI. Ashton-Prolla P. Olivier M. Hainaut P. Tumor protein 53 mutations and inherited cancer: beyond Li-Fraumeni syndrome. Curr Opin Oncol. 2010;22:64–69. doi: 10.1097/CCO.0b013e328333bf00. [DOI] [PubMed] [Google Scholar]
- 75.Papadopoulou LC. Sue CM. Davidson MM. Tanji K. Nishino I. Sadlock JE. Krishna S. Walker W. Selby J. Glerum DM. Coster RV. Lyon G. Scalais E. Lebel R. Kaplan P. Shanske S. De Vivo DC. Bonilla E. Hirano M. DiMauro S. Schon EA. Fatal infantile cardioencephalomyopathy with COX deficiency and mutations in SCO2, a COX assembly gene. Nat Genet. 1999;23:333–337. doi: 10.1038/15513. [DOI] [PubMed] [Google Scholar]
- 76.Park JY. Wang PY. Matsumoto T. Sung HJ. Ma W. Choi JW. Anderson SA. Leary SC. Balaban RS. Kang JG. Hwang PM. p53 improves aerobic exercise capacity and augments skeletal muscle mitochondrial DNA content. Circ Res. 2009;105:705–712. doi: 10.1161/CIRCRESAHA.109.205310. 11 p following 712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Pearson GD. Merrill GF. Deletion of the Saccharomyces cerevisiae TRR1 gene encoding thioredoxin reductase inhibits p53-dependent reporter gene expression. J Biol Chem. 1998;273:5431–5434. doi: 10.1074/jbc.273.10.5431. [DOI] [PubMed] [Google Scholar]
- 78.Peel JB. Sui X. Adams SA. Hebert JR. Hardin JW. Blair SN. A prospective study of cardiorespiratory fitness and breast cancer mortality. Med Sci Sports Exerc. 2009;41:742–748. doi: 10.1249/MSS.0b013e31818edac7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Peel JB. Sui X. Matthews CE. Adams SA. Hebert JR. Hardin JW. Church TS. Blair SN. Cardiorespiratory fitness and digestive cancer mortality: findings from the aerobics center longitudinal study. Cancer Epidemiol Biomarkers Prev. 2009;18:1111–1117. doi: 10.1158/1055-9965.EPI-08-0846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Polyak K. Xia Y. Zweier JL. Kinzler KW. Vogelstein B. A model for p53-induced apoptosis. Nature. 1997;389:300–305. doi: 10.1038/38525. [DOI] [PubMed] [Google Scholar]
- 81.Radak Z. Chung HY. Naito H. Takahashi R. Jung KJ. Kim HJ. Goto S. Age-associated increase in oxidative stress and nuclear factor kappaB activation are attenuated in rat liver by regular exercise. FASEB J. 2004;18:749–750. doi: 10.1096/fj.03-0509fje. [DOI] [PubMed] [Google Scholar]
- 82.Radak Z. Kaneko T. Tahara S. Nakamoto H. Ohno H. Sasvari M. Nyakas C. Goto S. The effect of exercise training on oxidative damage of lipids, proteins, and DNA in rat skeletal muscle: evidence for beneficial outcomes. Free Radic Biol Med. 1999;27:69–74. doi: 10.1016/s0891-5849(99)00038-6. [DOI] [PubMed] [Google Scholar]
- 83.Radak Z. Naito H. Kaneko T. Tahara S. Nakamoto H. Takahashi R. Cardozo-Pelaez F. Goto S. Exercise training decreases DNA damage and increases DNA repair and resistance against oxidative stress of proteins in aged rat skeletal muscle. Pflugers Arch. 2002;445:273–278. doi: 10.1007/s00424-002-0918-6. [DOI] [PubMed] [Google Scholar]
- 84.Rainwater R. Parks D. Anderson ME. Tegtmeyer P. Mann K. Role of cysteine residues in regulation of p53 function. Mol Cell Biol. 1995;15:3892–3903. doi: 10.1128/mcb.15.7.3892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ramos-Jiménez A. Hernandez-Torres RP. Torres-Durán PV. Romero-Gonzalez J. Mascher M. Posadas-Romero C. Juárez-Oropeza MA. The respiratory exchange ratio is associated with fitness indicators both in trained and untrained men: a possible application for people with reduced exercise tolerance. Clin Med Circ Respir Pulm. 2008;2:1–9. doi: 10.4137/ccrpm.s449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Rogers CJ. Colbert LH. Greiner JW. Perkins SN. Hursting SD. Physical activity and cancer prevention: pathways and targets for intervention. Sports Med. 2008;38:271–296. doi: 10.2165/00007256-200838040-00002. [DOI] [PubMed] [Google Scholar]
- 87.Ruiz-Lozano P. Hixon ML. Wagner MW. Flores AI. Ikawa S. Baldwin AS., Jr. Chien KR. Gualberto A. p53 is a transcriptional activator of the muscle-specific phosphoglycerate mutase gene and contributes in vivo to the control of its cardiac expression. Cell Growth Differ. 1999;10:295–306. [PubMed] [Google Scholar]
- 88.Saleem A. Adhihetty PJ. Hood DA. Role of p53 in mitochondrial biogenesis and apoptosis in skeletal muscle. Physiol Genomics. 2009;37:58–66. doi: 10.1152/physiolgenomics.90346.2008. [DOI] [PubMed] [Google Scholar]
- 89.Sano M. Minamino T. Toko H. Miyauchi H. Orimo M. Qin Y. Akazawa H. Tateno K. Kayama Y. Harada M. Shimizu I. Asahara T. Hamada H. Tomita S. Molkentin JD. Zou Y. Komuro I. p53-induced inhibition of Hif-1 causes cardiac dysfunction during pressure overload. Nature. 2007;446:444–448. doi: 10.1038/nature05602. [DOI] [PubMed] [Google Scholar]
- 90.Scharhag J. Schneider G. Urhausen A. Rochette V. Kramann B. Kindermann W. Athlete's heart: right and left ventricular mass and function in male endurance athletes and untrained individuals determined by magnetic resonance imaging. J Am Coll Cardiol. 2002;40:1856–1863. doi: 10.1016/s0735-1097(02)02478-6. [DOI] [PubMed] [Google Scholar]
- 91.Schwartzenberg-Bar-Yoseph F. Armoni M. Karnieli E. The tumor suppressor p53 down-regulates glucose transporters GLUT1 and GLUT4 gene expression. Cancer Res. 2004;64:2627–2633. doi: 10.1158/0008-5472.can-03-0846. [DOI] [PubMed] [Google Scholar]
- 92.Seemann S. Hainaut P. Roles of thioredoxin reductase 1 and APE/Ref-1 in the control of basal p53 stability and activity. Oncogene. 2005;24:3853–3863. doi: 10.1038/sj.onc.1208549. [DOI] [PubMed] [Google Scholar]
- 93.Sen CK. Oxidants and antioxidants in exercise. J Appl Physiol. 1995;79:675–686. doi: 10.1152/jappl.1995.79.3.675. [DOI] [PubMed] [Google Scholar]
- 94.Seo YR. Kelley MR. Smith ML. Selenomethionine regulation of p53 by a ref1-dependent redox mechanism. Proc Natl Acad Sci U S A. 2002;99:14548–14553. doi: 10.1073/pnas.212319799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Stambolsky P. Weisz L. Shats I. Klein Y. Goldfinger N. Oren M. Rotter V. Regulation of AIF expression by p53. Cell Death Differ. 2006;13:2140–2149. doi: 10.1038/sj.cdd.4401965. [DOI] [PubMed] [Google Scholar]
- 96.Stoner CS. Pearson GD. Koc A. Merwin JR. Lopez NI. Merrill GF. Effect of thioredoxin deletion and p53 cysteine replacement on human p53 activity in wild-type and thioredoxin reductase null yeast. Biochemistry. 2009;48:9156–9169. doi: 10.1021/bi900757q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Sun XZ. Vinci C. Makmura L. Han S. Tran D. Nguyen J. Hamann M. Grazziani S. Sheppard S. Gutova M. Zhou F. Thomas J. Momand J. Formation of disulfide bond in p53 correlates with inhibition of DNA binding and tetramerization. Antioxid Redox Signal. 2003;5:655–665. doi: 10.1089/152308603770310338. [DOI] [PubMed] [Google Scholar]
- 98.Sung HJ. Ma W. Wang P-y. Hynes J. O'Riordan TC. Combs CA. McCoy JP. Bunz F. Kang J-G. Hwang PM. Mitochondrial respiration protects against oxygen-associated DNA damage. Nat Commun. 2010;1:1–8. doi: 10.1038/ncomms1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Suzuki S. Tanaka T. Poyurovsky MV. Nagano H. Mayama T. Ohkubo S. Lokshin M. Hosokawa H. Nakayama T. Suzuki Y. Sugano S. Sato E. Nagao T. Yokote K. Tatsuno I. Prives C. Phosphate-activated glutaminase (GLS2), a p53-inducible regulator of glutamine metabolism and reactive oxygen species. Proc Natl Acad Sci U S A. 2010;107:7461–7466. doi: 10.1073/pnas.1002459107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Tabori U. Malkin D. Risk stratification in cancer predisposition syndromes: lessons learned from novel molecular developments in Li-Fraumeni syndrome. Cancer Res. 2008;68:2053–2057. doi: 10.1158/0008-5472.CAN-07-2091. [DOI] [PubMed] [Google Scholar]
- 101.Tasdemir E. Chiara Maiuri M. Morselli E. Criollo A. D'Amelio M. Djavaheri-Mergny M. Cecconi F. Tavernarakis N. Kroemer G. A dual role of p53 in the control of autophagy. Autophagy. 2008;4:810–814. doi: 10.4161/auto.6486. [DOI] [PubMed] [Google Scholar]
- 102.Tasdemir E. Maiuri MC. Galluzzi L. Vitale I. Djavaheri-Mergny M. D'Amelio M. Criollo A. Morselli E. Zhu C. Harper F. Nannmark U. Samara C. Pinton P. Vicencio JM. Carnuccio R. Moll UM. Madeo F. Paterlini-Brechot P. Rizzuto R. Szabadkai G. Pierron G. Blomgren K. Tavernarakis N. Codogno P. Cecconi F. Kroemer G. Regulation of autophagy by cytoplasmic p53. Nat Cell Biol. 2008;10:676–687. doi: 10.1038/ncb1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Tasdemir E. Maiuri MC. Orhon I. Kepp O. Morselli E. Criollo A. Kroemer G. p53 represses autophagy in a cell cycle-dependent fashion. Cell Cycle. 2008;7:3006–3011. doi: 10.4161/cc.7.19.6702. [DOI] [PubMed] [Google Scholar]
- 104.Trojian TH. Mody K. Chain P. Exercise and colon cancer: primary and secondary prevention. Curr Sports Med Rep. 2007;6:120–124. doi: 10.1007/BF02941153. [DOI] [PubMed] [Google Scholar]
- 105.Vahsen N. Cande C. Briere JJ. Benit P. Joza N. Larochette N. Mastroberardino PG. Pequignot MO. Casares N. Lazar V. Feraud O. Debili N. Wissing S. Engelhardt S. Madeo F. Piacentini M. Penninger JM. Schagger H. Rustin P. Kroemer G. AIF deficiency compromises oxidative phosphorylation. EMBO J. 2004;23:4679–4689. doi: 10.1038/sj.emboj.7600461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Verhaegh GW. Richard MJ. Hainaut P. Regulation of p53 by metal ions and by antioxidants: dithiocarbamate down-regulates p53 DNA-binding activity by increasing the intracellular level of copper. Mol Cell Biol. 1997;17:5699–5706. doi: 10.1128/mcb.17.10.5699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Vistisen K. Loft S. Poulsen HE. Cytochrome P450 IA2 activity in man measured by caffeine metabolism: effect of smoking, broccoli and exercise. Adv Exp Med Biol. 1991;283:407–411. doi: 10.1007/978-1-4684-5877-0_55. [DOI] [PubMed] [Google Scholar]
- 108.Vogelstein B. Lane D. Levine AJ. Surfing the p53 network. Nature. 2000;408:307–310. doi: 10.1038/35042675. [DOI] [PubMed] [Google Scholar]
- 109.Vousden KH. Prives C. Blinded by the light: the growing complexity of p53. Cell. 2009;137:413–431. doi: 10.1016/j.cell.2009.04.037. [DOI] [PubMed] [Google Scholar]
- 110.Vousden KH. Ryan KM. p53 and metabolism. Nat Rev Cancer. 2009;9:691–700. doi: 10.1038/nrc2715. [DOI] [PubMed] [Google Scholar]
- 111.Wallace DC. A mitochondrial paradigm of metabolic and degenerative diseases, aging, and cancer: a dawn for evolutionary medicine. Annu Rev Genet. 2005;39:359–407. doi: 10.1146/annurev.genet.39.110304.095751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Warburg O. On the origin of cancer cells. Science. 1956;123:309–314. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- 113.Wise DR. DeBerardinis RJ. Mancuso A. Sayed N. Zhang XY. Pfeiffer HK. Nissim I. Daikhin E. Yudkoff M. McMahon SB. Thompson CB. Myc regulates a transcriptional program that stimulates mitochondrial glutaminolysis and leads to glutamine addiction. Proc Natl Acad Sci U S A. 2008;105:18782–18787. doi: 10.1073/pnas.0810199105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Wisloff U. Najjar SM. Ellingsen O. Haram PM. Swoap S. Al-Share Q. Fernstrom M. Rezaei K. Lee SJ. Koch LG. Britton SL. Cardiovascular risk factors emerge after artificial selection for low aerobic capacity. Science. 2005;307:418–420. doi: 10.1126/science.1108177. [DOI] [PubMed] [Google Scholar]
- 115.Wong TS. Rajagopalan S. Freund SM. Rutherford TJ. Andreeva A. Townsley FM. Petrovich M. Fersht AR. Biophysical characterizations of human mitochondrial transcription factor A and its binding to tumor suppressor p53. Nucleic Acids Res. 2009;37:6765–6783. doi: 10.1093/nar/gkp750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Wong TS. Rajagopalan S. Townsley FM. Freund SM. Petrovich M. Loakes D. Fersht AR. Physical and functional interactions between human mitochondrial single-stranded DNA-binding protein and tumour suppressor p53. Nucleic Acids Res. 2009;37:568–581. doi: 10.1093/nar/gkn974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Woods JA. Davis JM. Kohut ML. Ghaffar A. Mayer EP. Pate RR. Effects of exercise on the immune response to cancer. Med Sci Sports Exerc. 1994;26:1109–1115. [PubMed] [Google Scholar]
- 118.Yan H. Parsons DW. Jin G. McLendon R. Rasheed BA. Yuan W. Kos I. Batinic-Haberle I. Jones S. Riggins GJ. Friedman H. Friedman A. Reardon D. Herndon J. Kinzler KW. Velculescu VE. Vogelstein B. Bigner DD. IDH1 and IDH2 mutations in gliomas. N Engl J Med. 2009;360:765–773. doi: 10.1056/NEJMoa0808710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Yee KS. Wilkinson S. James J. Ryan KM. Vousden KH. PUMA- and Bax-induced autophagy contributes to apoptosis. Cell Death Differ. 2009;16:1135–1145. doi: 10.1038/cdd.2009.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Yoshida Y. Izumi H. Torigoe T. Ishiguchi H. Itoh H. Kang D. Kohno K. P53 physically interacts with mitochondrial transcription factor A and differentially regulates binding to damaged DNA. Cancer Res. 2003;63:3729–3734. [PubMed] [Google Scholar]
- 121.Zhang XD. Wang Y. Wu JC. Lin F. Han R. Han F. Fukunaga K. Qin ZH. Down-regulation of Bcl-2 enhances autophagy activation and cell death induced by mitochondrial dysfunction in rat striatum. J Neurosci Res. 2009;87:3600–3610. doi: 10.1002/jnr.22152. [DOI] [PubMed] [Google Scholar]
- 122.Zhou S. Kachhap S. Singh KK. Mitochondrial impairment in p53-deficient human cancer cells. Mutagenesis. 2003;18:287–292. doi: 10.1093/mutage/18.3.287. [DOI] [PubMed] [Google Scholar]