Abstract
Microorganisms can degrade saturated hydrocarbons (alkanes) not only under oxic but also under anoxic conditions. Three denitrifying isolates (strains HxN1, OcN1, HdN1) able to grow under anoxic conditions by coupling alkane oxidation to CO2 with NO3− reduction to N2 were compared with respect to their alkane metabolism. Strains HxN1 and OcN1, which are both Betaproteobacteria, utilized n-alkanes from C6 to C8 and C8 to C12 respectively. Both activate alkanes anaerobically in a fumarate-dependent reaction yielding alkylsuccinates, as suggested by present and previous metabolite and gene analyses. However, strain HdN1 was unique in several respects. It belongs to the Gammaproteobacteria and was more versatile towards alkanes, utilizing the range from C6 to C30. Neither analysis of metabolites nor analysis of genes in the complete genome sequence of strain HdN1 hinted at fumarate-dependent alkane activation. Moreover, whereas strains HxN1 and OcN1 grew with alkanes and NO3−, NO2− or N2O added to the medium, strain HdN1 oxidized alkanes only with NO3− or NO2− but not with added N2O; but N2O was readily used for growth with long-chain alcohols or fatty acids. Results suggest that NO2− or a subsequently formed nitrogen compound other than N2O is needed for alkane activation in strain HdN1. From an energetic point of view, nitrogen–oxygen species are generally rather strong oxidants. They may enable enzymatic mechanisms that are not possible under conditions of sulfate reduction or methanogenesis and thus allow a special mode of alkane activation.
Introduction
Saturated hydrocarbons (alkanes) as major constituents of petroleum (Tissot and Welte, 1984) enter the environment via natural seeps or accidental spills, or due to the use of refined petroleum products. Furthermore, alkanes are widespread products of living organisms (Birch and Bachofen, 1988). Aerobic alkane biodegradation, in particular the initial O2-dependent activation by monooxygenases, has been studied since many decades (Rojo, 2009). In recent years, alkanes were also shown to be degraded anaerobically with nitrate (Ehrenreich et al., 2000; Bonin et al., 2004; Grossi et al., 2008; Callaghan et al., 2009) or sulfate (Aeckersberg et al., 1991; 1998; So and Young, 1999; Cravo-Laureau et al., 2004; Davidova et al., 2006; Kniemeyer et al., 2007; Higashioka et al., 2009) as electron acceptor, or under conditions of methanogenesis (Zengler et al., 1999; Anderson and Lovley, 2000; Jones et al., 2008). The only established mechanism for anaerobic activation of alkanes to date is the radical-catalysed addition to fumarate yielding alkylsuccinates (Kropp et al., 2000; Rabus et al., 2001; Heider, 2007). Genes [designated mas, for (1-methylalkyl)succinate synthase; or ass, for alkylsuccinate synthase] encoding the putative enzyme have been detected in a nitrate-reducing (Grundmann et al., 2008) and a sulfate-reducing (Callaghan et al., 2008) strain. Still, an alternative possibility for anaerobic alkane activation has been suggested on the basis of cell fatty acid and isotope labelling analysis (Aeckersberg et al., 1998; So et al., 2003; Callaghan et al., 2006).
Of three denitrifying strains, HxN1, OcN1 and HdN1, that were isolated with n-hexane, n-octane andn-hexadecane, respectively (Ehrenreich et al., 2000), only the first one has been formerly studied with respect to its alkane metabolism (Rabus et al., 2001; Wilkes et al., 2002; Grundmann et al., 2008). A subsequent comparative study including the two other strains revealed that also strain OcN1 formed alkylsuccinates during growth with alkanes and harboured a gene apparently encoding the responsible enzyme. In contrast, alkylsuccinates were not detectable in strain HdN1, and its complete genome sequence did not reveal any gene likely to encode (1-methylalkyl)succinate or alkylsuccinate synthase. A unique physiological characteristic of strain HdN1 was that it did not grow with alkanes if N2O was added instead of NO3−, whereas growth with alcohols and fatty acids readily occurred with N2O. In contrast, strains HxN1 and OcN1 grew well with N2O and alkanes. These findings suggest that alkane activation in strain HdN1 differs principally from alkane activation in strains HxN1 and OcN1 and requires an NO3−-derived compound other than N2O.
Results and discussion
Cultivation, phylogenetic relationships, morphology and purity control
The focus of this study is on strain HdN1 and its apparently unusual physiology with respect to n-alkane utilization. The isolation of strain HdN1 along with that of strains OcN1 and HxN1, a few substrate tests, and the capacity for complete alkane oxidation in anoxic medium with NO3− have been documented previously (Ehrenreich et al., 2000). Unless indicated otherwise, the strains were grown in conventional HCO3−/CO2-buffered defined medium (Rabus and Widdel, 1995) with alkanes as the only organic substrates.
The study of anaerobic microbial hydrocarbon utilization requires the strict exclusion of any traces of O2 from air which through monooxygenases could lead to hydroxyl compounds (which can be further degraded anaerobically). Hence, in addition to physical exclusion of air (Widdel and Bak, 1992), the presence of a reductant (‘redox buffer’) is advisable. Unlike sulfate-reducing bacteria that form a chemical reducing agent, sulfide, nitrate-reducing bacteria do not produce a reductant. Addition of sulfide (or other reducing sulfur compounds) is inappropriate because it is easily oxidized in by-reactions of the ‘high-potential’ nitrate reduction pathway, or because it can inhibit denitrifiers (F. Widdel, unpubl. results). We therefore added ascorbate (4 mM) as a mild reductant (Rabus and Widdel, 1995; Ehrenreich et al., 2000; Widdel, 2009). Ascorbate did not serve as substrate for growth and nitrate reduction, as revealed in control incubations with ascorbate alone. We also verified that ascorbate in our medium did not scavenge nitrite, the intermediate of nitrate reduction, by chemical reaction. If sterile medium with ascorbate (pH 7.2) and NaNO2 (2 mM) was incubated for 10 days and analysed by ion chromatography (Rabus and Widdel, 1995), there was no noticeable decrease of the nitrite concentration. At low pH, reduction of the protonated form (HNO2) by ascorbic acid to yield nitric oxide can be significant (Yamasaki, 2000). Furthermore, tests were carried out to exclude an adverse physiological effect of ascorbate. Strain HdN1 was grown with n-tetradecane and NO3− or NO2− in ascorbate-containing medium as well as in ascorbate-free medium deoxygenated by vigorous sparging with N2. The cultures with and without ascorbate grew equally well.
Strain HdN1 affiliates with the Gammaproteobacteria, whereas strain OcN1 and HxN1 are members of the Betaproteobacteria (Fig. 1). Most nitrate-reducing bacteria enriched and isolated with various aromatic or saturated petroleum hydrocarbons are Betaproteobacteria (Widdel et al., 2009). Some denitrifying strains that degrade petroleum hydrocarbons are Gammaproteobacteria; these also include alkane degraders affiliating with Marinobacter sp. (Bonin et al., 2004) and Pseudomonas balearica (Grossi et al., 2008).
Fig. 1.
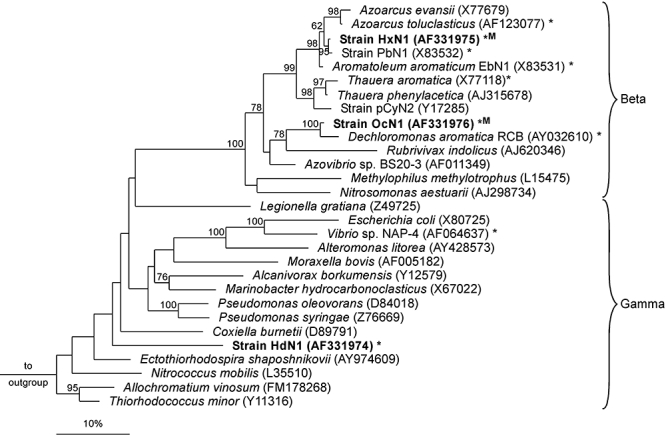
Phylogenetic (16S rRNA-based) affiliation of strain HdN1 with selected Beta- and Gammaproteobacteria including other strains able to degrade aromatic or saturated petroleum hydrocarbons with nitrate (*). Strains able to degrade n-alkanes anaerobically are highlighted in bold; occurrence of (1-methylalkyl)succinate formation for alkane activation is also indicated (M). Bootstrap values (%; only > 60% shown) were obtained after 1000 resamplings. Scale bar, 10% estimated sequence divergence.
The cell shape of strain HdN1 was unusually variable and significantly influenced by the organic growth substrate (Ehrenreich et al., 2000). In particular long-chain alkanes caused swelling of a large fraction of the cells. In such cells, spacious inclusions resembling storage compounds could be seen at high magnification (Fig. 2A). However, polyhydroxyalkanoates were not detectable (A. Steinbüchel, pers. comm.) by gas chromatography following acidic hydrolysis and methylation of freeze-dried cells (Steinbüchel and Wiese, 1992). Cells in alkane cultures tended to grow in close contact with the overlying insoluble hydrocarbon phase. The bulk of alkane-grown cells was buoyant, possibly due to association with or storage of alkane droplets. Alkane storage and buoyancy is a phenomenon known from aerobic alkane degraders (Scott and Finnerty, 1976). This behaviour rendered harvesting by centrifugation difficult. A minor fraction of the cells was motile.
Fig. 2.
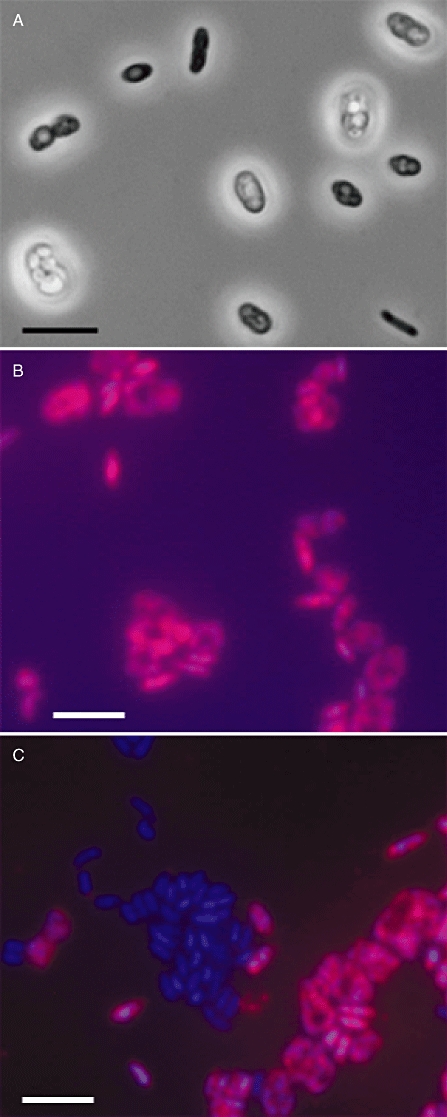
Microscopic images of strain HdN1.
A. Highly variable cell forms of strain HdN1 grown anaerobically with hexadecane and nitrate. Phase-contrast micrographs of viable cells. Bar, 5 µm.
B. Cells from a pure culture of strain HdN1 hybridized with a specific 16S rRNA-targeted oligonucleotide probe and stained with DAPI. The image represents an overlay of the probe and the DAPI signal. Bar, 5 µm.
C. Mixed cells of strains HdN1 and OcN1 hybridized, stained and visualized as in (B). Bar, 5 µm.
Thorough purity tests excluded that cell shape heterogeneity in cultures of strain HdN1 was due to accompanying microorganisms. First, repeated aerobic and anaerobic (with NO3−) liquid dilution series (according to the most probable number technique) were carried out separately with n-tetradecane or n-valerate (n-pentanoate). All cultures derived from the highest positive dilution tubes were microscopically indistinguishable and always able to use both, tetradecane and valerate. Second, cultures were streaked on agar plates containing valerate and yeast extract and incubated in a jar under air with 3% CO2. All well-separated valerate-grown colonies transferred to anoxic liquid media grew again with tetradecane, and cultures had the microscopic appearance as before. Third, strain HdN1 was mixed with strain OcN1, and a specific 16S rRNA-targeting fluorescent oligonucleotide probe (Appendix S1) was applied. Whereas in the pure culture all cells exhibited the specific hybridization signal (Fig. 2B), the mixed culture contained in addition the expected non-hybridizing cells that exhibited only the general fluorescent stain (Fig. 2C). Hence, strain HdN1 is in principle distinguishable from contaminants by specific probing.
Anaerobic growth tests with alkanes and alkanoates
The capability of strain HdN1 for complete hexadecane oxidation with nitrate according to 5 C16H34 + 98 NO3− + 18 H+→ 80 HCO3− + 49 N2 + 54 H2O has been verified formerly with small, precisely quantifiable amounts of alkane (Ehrenreich et al., 2000). In all subsequent experiments, significantly higher amounts of alkanes were added than could be oxidized by the electron acceptor (10 mM NO3−). In this way, a large contact area between the insoluble hydrocarbon and the aqueous phase was provided which favoured growth (Widdel, 2009). In further growth tests, alkanes with carbon chains ≤ C10 were provided as solutions in 2,2,4,4,6,8,8-heptamethylnonane (HMN) as an inert carrier phase to avoid toxic effects (Appendix S1). Tests revealed that strain HdN1 utilized n-alkanes from C6 (n-hexane) to C30 (n-triacontane) as carbon sources and electron donors (C6 to C20, C24, C26, C28, C30, C36 and C40 tested). Fastest growth was observed in the range from C14 (tetradecane) to C18 (octadecane). With an inoculum size of 1% (v/v), full growth and complete NO3− consumption occurred within 7 days. A doubling time of 11–13 h during early growth was estimated from an analysis of the nitrate consumption curve (see also Ehrenreich et al., 2000). (Inhomogeneous growth and alkane droplets prevented measurement of the optical density as a growth parameter.) Growth with alkanes of shorter or longer chains was slower (two- to threefold time required for full growth and NO3− consumption). The other strains, HxN1 and OcN1, utilized a significantly narrower range of alkanes, which was from C6 to C8 (n-octane) and C8 to C12 (n-dodecane) respectively. Also alkane-utilizing sulfate-reducing bacteria utilized a narrower range (Rueter et al., 1994; Aeckersberg et al., 1998).
Strain HdN1 utilized monocarboxylic acids (sodium salts; method of preparation and addition given by Widdel and Bak, 1992; see also Appendix S1) from acetate to stearate (C2–C18; higher fatty acids not tested), with best growth (roughly twice as fast as with alkanes) with valerate (C5) and with fatty acids from n-decanoate (C10) to stearate. Some primary linear alcohols were also tested (C8 and C10 provided as solutions in HMN; C14 and C16 added as solid compounds). Strain HdN1 grew well with 1-decanol, 1-tetradecanol and 1-hexadecanol; growth with 1-octanol was poor, and no growth occurred with ethanol.
Growth tests with different electron acceptors
All three strains grew also aerobically with alkanes. Examination of strain HdN1 in more detail revealed that almost the same range of n-alkanes (and fatty acids) was oxidized with O2 as in anaerobic cultures with NO3−. Only n-hexane was not utilized so far with O2. Another slight difference between aerobic and anaerobic alkane utilization was observed if cultures grown with hexadecane were transferred to medium with tridecane (C13) or dodecane (C12). Whereas aerobic cultures grew immediately with the lighter alkanes, anaerobic cultures exhibited a lag-phase of > 10 days.
The transient formation by strain HdN1 of NO2− (≤ 1.5 mM; not shown) and N2O (Fig. 3A and B, lower curves) at low concentration during NO3− reduction and the detection of N2 in all cultures grown under an argon atmosphere indicated the common denitrification pathway. To further examine the capability for efficient use of NO2− and N2O, these electron acceptors were tested individually in the absence of NO3−.
Fig. 3.
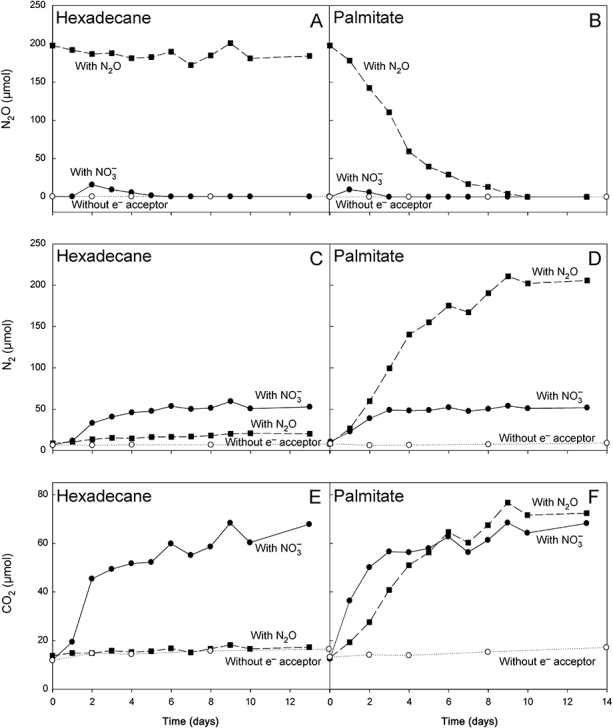
Time-courses of the formation of N2O (A and B), N2 (C and D) and CO2 (E and F) in anaerobic cultures of strain HdN1 with n-hexadecane (A, C and E) or palmitate (B, D and F). The electron acceptors were added in stoichiometrically limiting amounts (100 µmol of NO3−; c. 250 µmol of N2O) relative to the electron donor (171 µmol of hexadecane, advantage of large excess explained in text; 10 µmol of palmitate). Results show that alkane oxidation to CO2 was not possible with N2O, but readily occurred with NO3−. The functionalized compound, palmitate, was oxidized with N2O. Duplicates yielded the same results (not shown). Culture volumes of 10 ml (phosphate-buffered medium, pH ≈ 7.1, without addition of NaHCO3; Appendix S1) were incubated in 165 ml serum bottles under an argon headspace. N2O was injected as pure O2-free gas. Cultures were very gently shaken for a few minutes per day. Vigorous shaking had to be avoided because it impeded growth. Samples from the headspace were analysed with a gas chromatograph employing argon as carrier gas and a thermal conductivity detector. The calculated dissolved amounts of gases were added so as to obtain the total amounts in the bottles. Calculation was based on literature data (Wilhelm et al., 1977; Stumm and Morgan, 1995), assuming equilibrium (which may not have been fully reached due to limited agitation) and considering pH in the case of CO2.
Growth with alkanes also occurred with added NO2− (instead of NO3−), but was slightly slowed down if more than 5 mM NO2− was added. Furthermore, a lag-phase of c. 2 days was sometimes observed after inoculation of new medium with NO2−. Hence, several mM may be somewhat inhibitory.
Surprisingly, strain HdN1 did not grow with alkanes in the growth tests with N2O. In accordance with the lack of growth, N2O was not consumed (Fig. 3A, upper curve), and N2 (Fig. 3C) or CO2 (Fig. 3E) were not formed. In contrast, growth with 1-tetradecanol, 1-hexadecanol or fatty acids was possible with added N2O, and consumption of N2O (Fig. 3B) as well as formation of N2 (Fig. 3D) and CO2 (Fig. 3F) was obvious. A minor formation of N2 from N2O during incubation with hexadecane can be explained by reduction with an endogenous electron source in the inoculum. The formation of N2 from N2O requires only 2 e−, whereas formation of N2 from NO3− requires 10 e− from an electron donor. The lack of alkane utilization with N2O was not due to specific inhibition. The same amount of N2O added to a culture with hexadecane and NO3− did not inhibit growth. For physiological comparison, strains HxN1 and OcN1 were also incubated with N2O as the only electron acceptor and utilizable alkanes (n-hexane and n-octane respectively). These strains were able to grow with N2O and alkanes. Results are summarized in Fig. 4. The inability for coupling alkane utilization to N2O reduction is apparently unique for strain HdN1.
Fig. 4.
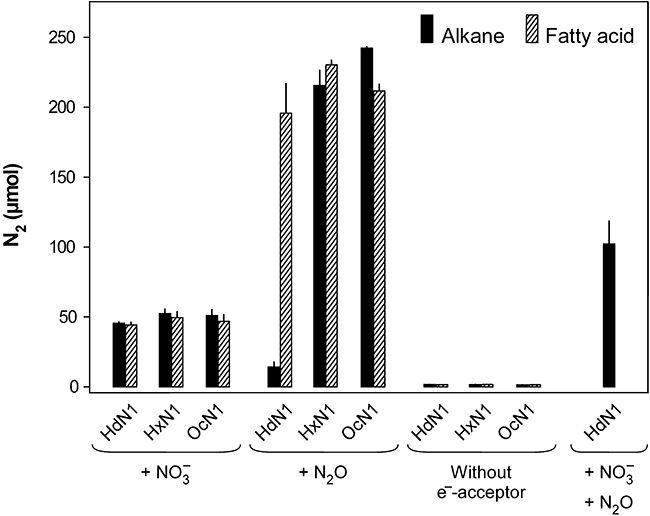
N2 formed in anaerobic cultures of strains HdN1, HxN1 and OcN1 with alkanes (black bars) or fatty acids (striated bars) and either NO3− (100 µmol) or N2O (250 µmol). A control experiment with strain HdN1 for excluding N2O toxicity received both, NO3− and N2O. Here, more N2 was formed than with NO3− alone. This indicated that not only NO3− but also N2O was used in the anaerobic respiratory chain if alkane degradation was enabled by NO3−. Data show that strain HdN1 could not use N2O alone for alkane degradation, in contrast to the other strains. Culture volumes of 10 ml were incubated in 20 ml butyl-rubber sealed tubes. Strain HdN1 received 171 µmol of pure n-hexadecane, or 10 µmol of palmitate. Strain HxN1 received 38 µmol of n-hexane (in 100 µl of heptamethylnonane as carrier), or 30 µmol of caproate. Strain OcN1 received 31 µmol of n-octane (in 100 µl of heptamethylnonane), or 30 µmol of caproate. Tubes were incubated nearly horizontally while contact of the hydrocarbon phase with the stopper was avoided (Widdel, 2009) as far as possible. Gas samples were withdrawn 11 days after inoculation and analysed (triplicates) as indicated in Fig. 3.
Other electron acceptors tested (concentrations in mM) but not utilized were sulfate (15), thiosulfate (5), sulfur (added as slurry), fumarate (10) and perchlorate (10). Toxic effects were excluded in controls containing in addition NO3−. In contrast, chlorate was toxic.
Search for metabolites and genes involved in alkane degradation
Investigation of metabolites and genes involved in alkane degradation via addition to fumarate have been reported for strain HxN1 (Rabus et al., 2001; Wilkes et al., 2002; Grundmann et al., 2008). The presently performed metabolite analysis of strain OcN1 upon growth with n-octane and NO3− revealed (1-methylheptyl)succinate (extraction, methylation and analysis as in Rabus et al., 2001; Wilkes et al., 2003), again indicating an activation via addition to fumarate. In contrast, alkyl-substituted succinates were never detectable in cultures and cells of strain HdN1. Another product searched for [by gas chromatography-mass spectrometry of extracts silylated with N,O-bis(trimethylsilyl)acetamide; Appendix S1] in anaerobic n-hexadecane cultures of strain HdN1 was 1-hexadecanol. If air was strictly excluded and if the culture was inactivated by heat (85°C; Rabus et al., 2001) before extraction, 1-hexadecanol was not detectable. In contrast, 1-hexadecanol was detected if the anaerobically grown culture was exposed to air for 20–30 min (data not shown). Such 1-alkanol formation is a long-known indicator of alkane monooxygenase activity (Britton, 1984). Metabolite analysis in anaerobic alkane degraders with facultative aerobic metabolism thus requires careful avoidance of artefacts due to reaction with O2 from air.
The gene possibly encoding the alkane-activating enzyme in strain OcN1 was retrieved via polymerase chain reaction with degenerate primers for mas and ass genes, generation of a probe and screening of a genomic library, similar as described for strain HxN1 (Grundmann et al., 2008). The derived amino acid sequence (Accession No. FN675935) revealed close relationships (Fig. S1) to the orthologue from strain HxN1 (Grundmann et al., 2008) and a sulfate-reducing bacterium (Callaghan et al., 2008). Attempts to amplify in an analogous manner mas- or ass-like genes from strain HdN1 failed. Therefore, a shotgun genomic library of strain HdN1 was established. This allowed assemblage of the complete genome sequence (4 587 455 bp; 3762 coding sequences; Accession No. FP929140; for some more details see Table S1). But neither this revealed mas- or ass-like genes (Table S2).
These findings suggested that the mechanism for alkane activation in strain HdN1, which has to involve the cleavage of a strong, apolar C−H bond, differs basically from the mechanism with fumarate as co-substrate in the two other strains.
Linkage of alkane activation in strain HdN1 to the nitrate reduction pathway?
The distinctive results of the incubation experiments with either alkanes or functionalized (O-group-containing) substrates and N2O may offer a clue as to how strain HdN1 could initiate alkane degradation under anoxic conditions. The electron acceptor tests with functionalized electron donors as well as identified genes (Table S3) indicate that strain HdN1 employs the common reduction sequence (NO3−→ NO2−→ NO → N2O → N2), viz. is in principle able to readily reduce N2O. Also during growth with alkanes as organic substrates and NO3− or NO2− as electron acceptors, N2O must have been a regular intermediate because N2 rather than N2O was the end-product. However, N2O added alone did not allow growth with alkanes. An early reaction during alkane utilization must thus depend on a nitrogen–oxygen (N–O) species other than N2O. The early reaction could be the biochemically crucial activation of the alkane. The required N–O species cannot be NO3−, because growth with alkanes was also possible if NO2− was added instead of NO3−. Hence, NO2− or NO (or a so far unknown product from NO2− reduction) may be essential for alkane activation. The basic hypothesis is depicted in Fig. 5. In further experiments, added NO (prepared from acidified NaNO2 and KI; Schreiber et al., 2008) turned out to be very toxic so that application of amounts theoretically sufficient to achieve measurable growth was not possible; NO at a partial pressure of c. 75 Pa [0.075% (v/v) in gas mixture of ambient pressure] in the headspace completely inhibited growth with NO3−. At a partial pressure of 50 Pa (0.05%) NO did not completely inhibit growth with NO3−, albeit growth was retarded. To test whether such still tolerated NO concentration is sufficient to initiate alkane degradation and in this way allow growth, 50 Pa NO was provided together with N2O (17 mmol l−1), the latter serving as main electron acceptor for anaerobic respiration. However, growth was not observed unless NO3− was added. It thus remains elusive whether NO2− or NO (or an unknown NO2−-derived species) is actually required to initiate alkane degradation.
Fig. 5.
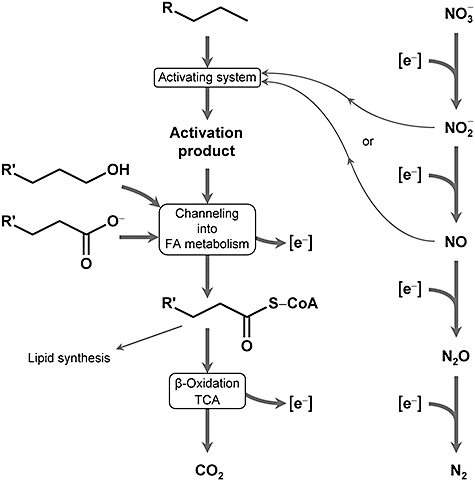
Hypothetical involvement of denitrification intermediates in alkane activation. The scheme offers an explanation for the inability of strain HdN1 to utilize n-alkanes with N2O alone (see Figs 3 and 4). It is assumed that a small proportion of NO2− or NO is deviated from the respiratory chain for alkane activation. They may be used for activation indirectly (by yielding O2 that is used by alkane monooxygenase; or by giving rise to another reactive factor or enzyme centre) or directly (as co-reactants introducing a polar group). The alkyl residue R′ may or may not be identical with the original residue R (depending on the activation mechanism and alkane C-atom being attacked). FA, fatty acid; TCA, tricarboxylic acid cycle.
From a thermodynamic point of view, an involvement of N–O species in alkane activation under anoxic conditions is an appealing hypothesis. N–O species other than NO3− (Fig. 6A) are all metastable (Garrels and Christ, 1965; Thauer et al., 1977) and represent or can provide strong potential oxidants; this property may be enzymatically exploited to achieve alkane activation. An indirect use to form another reactive compound as well as a direct use of an N–O species can be envisaged.
Fig. 6.
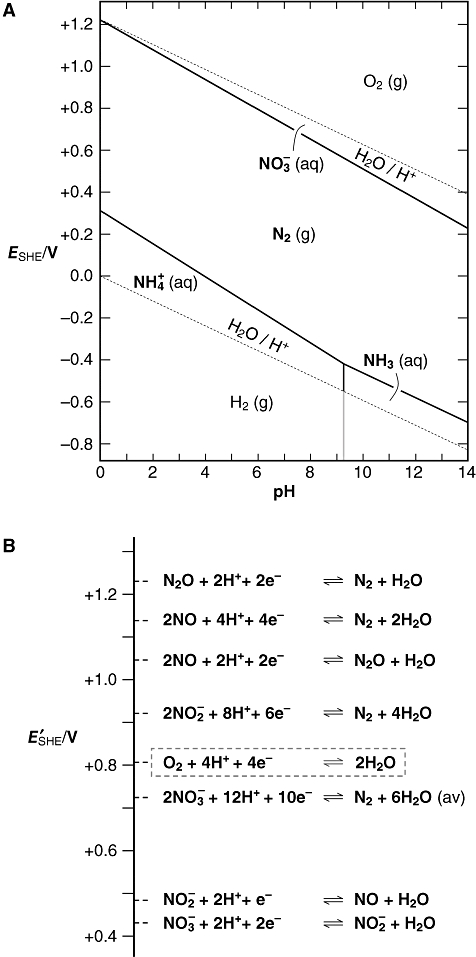
Some energetic aspects of N–O (and N–H) species. Graphs are for the following activities or fugacities: {NO3−}, {NO2−}, {NH3 (g)}, {NH4+} = 10−2; {N2 (g)} = 10−0.1 (78% in air); {O2 (g)} = 10−0.7 (21% in air); {H2} = 1; {NO (g)}, {N2O (g)} = 10−4.3 (approximately corresponding to dissolved concentrations monitored under natural conditions; Schreiber et al., 2009).A. E–pH (stability, Pourbaix) diagram of the system H–N–O. Only N2 and the lowest and highest oxidation states, NH4+, NH3 and NO3−, are thermodynamically stable. Other metabolically formed inorganic N-compounds are metastable (endergonic; e.g. Eqs 1 and 2) and can, in principle, spontaneously decompose (dismutate) into the species (including O2) depicted in the diagram. Endergonic N-compounds can be metabolically formed because they appear as co-products besides H2O (from reductive O elimination).B. Electrochemical half-reactions (including hypothetical ones) of N–O species and O2. If an endergonic N-compound does not react via dismutation (e.g. Eqs 1 and 2) but in an electrochemical half-reaction (yielding the same product as dismutation), this half-reaction has a higher redox potential than that of O2/2H2O (E°′ = +0.815 V). Again, this does not contradict the fact that NO3− and NO2− originate from a microbial oxidation process with O2 (see under A; also, the reaction sequence in nitrification is different: NH4+/NH3→ NH2OH → NO2−). Generally, the redox potentials (Ei) of subsequent reduction steps (i = 1, 2, …, m) of an overall reduction with free intermediates are linked to an average redox potential (Eav) according to (n1E1 + n2E2 + … + nmEm)/ntot = Eav; ni is the number of electrons involved in an individual step and ntot the total number of electrons. Eav is connected to the total free energy change, ΔGtot, of the overall reduction with an electron-donating reaction of the redox potential Edon according to Eav = −ΔGtot/(ntotF) + Edon, with F = 96 485 C mol−1 (explanation in Appendix S2). Eav of 2NO3−/N2 marks the borderline between the stability regions of NO3− and N2 in the E–pH diagram (A). In a real metabolic process, a strong oxidant formed in a reduction sequence can only appear as free intermediate if its further reduction is enzymatically controlled and if unspecific reactions with reductants are slower or do not take place. Also, overall irreversibility is required, but this is naturally given (in an equilibrium system, redox pairs with different redox potential, e.g. NO3−/ NO2− and NO2−/NO, cannot coexist).Calculations are based on standard free energy data (Garrels and Christ, 1965; Thauer et al., 1977). Derived standard redox potentials at pH = 7 (E°′/V): NO3−/NO2−, +0.431; 2NO3−/N2, +0.747 (av); NO2−/NO, +0.347; 2NO2−/N2, +0.958; 2NO/N2O, +1.172; 2NO/N2, +1.264; N2O/N2; +1.355.
One mode of indirect use of NO2− and NO could be their dismutation (formally an ‘internal’ reduction of N and oxidation of O) leading to O2, according to the following equations:
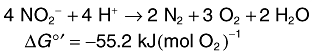 |
(1) |
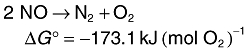 |
(2) |
O2 could then be used for an alkane monooxygenase reaction (alkane hydroxylation). Also N2O can in principle lead to O2[2 N2O → 2 N2 + O2; ΔG° = −208.4 kJ (mol O2)−1], but the present results exclude its use for an initiation of alkane degradation. There is indeed evidence for O2 formation at very low concentration during NO2− reduction in a methane-utilizing enrichment culture dominated by ‘Candidatus Methylomirabilis oxyfera’. The enrichment grew under exclusion of air and depended on NO2− addition (Ettwig et al., 2010). 18O2 formation from N18O2− (indirectly labelled through H218O) became detectable upon specific inhibition of methane monooxygenase. NO dismutation (Eq. 2) was suggested as the underlying mechanism. Neither was NO3− or N2O reduced, nor did the genome of the dominant bacterium harbour typical N2O-reductase genes. Results therefore suggested that NO dismutation was a main reaction during NO2− reduction in ‘Candidatus M. oxyfera’. An earlier example of a metastable inorganic oxo-compound enabling biodegradative reactions through O2 formation is chlorite, an intermediate of microbial chlorate reduction (ClO2−→ Cl− + O2; ΔG°′ = −148.4 kJ mol−1; Ginkel et al., 1996; Chakraborty and Coates, 2004; Tan et al., 2006; Weelink et al., 2008; Mehboob et al., 2009a,b;). The presently investigated alkane-degrading strain HdN1 differs metabolically from ‘M. oxyfera’ in several respects. Strain HdN1 does not utilize methane, grows with NO3− and obviously involves the conventional reduction sequence via N2O to N2. If strain HdN1 would employ NO2−- or NO-derived O2, the demand per hydrocarbon molecule utilized would be much lower than in ‘Candidatus M. oxyfera’. Long-chain alkane activation would require only a minor withdrawal of NO2− or NO from the respiratory path that mainly leads to N2 through N2O. For instance, n-hexadecanol resulting from oxygenation of n-hexadecane (C15H31CH3 + O2 + 2 [H]→ C15H31CH2OH + H2O) yields as many as 96 [H] (C15H31CH2OH + 31 H2O → 16 CO2 + 96 [H]) per substrate molecule. With 2 [H] consumed for activation, each oxygenation event thus leaves 94 [H] per C16H34 for respiratory energy conservation. In contrast, each oxygenation event in methane utilization provides only 4 [H] per CH4 for respiration. According to genomic data, strain HdN1 may form a di-iron monooxygenase, a P450-type monooxygenase and possibly a third type of monooxgenase (Table S4). Multiple monooxygenases are not uncommon in aerobic alkane degraders (Rojo, 2009).
O2 formation could not be detected so far in strain HdN1. We mixed a culture of strain HdN1 with a culture of luminous bacteria (isolated from herring using glycerol-peptone medium; Farmer and Hickman-Brenner, 2006) as sensitive O2 indicators (Chance et al., 1978); both cultures had been adapted to brackish water (180 mM NaCl and 20 mM MgSO4) medium. After extinction of luminescence due to oxygen consumption, neither addition of NO3− nor of NO2− or NO-saturated water caused the luminous reaction to resume (whereas air did immediately). Neither was oxygen detectable by means of an O2-microelectrode (lower detection limit, 1 µM; Revsbech, 1989) in cultures supplied with NO2− or NO. Nevertheless, results do not rule out O2 as an intermediate. A very low production rate and effective scavenging by alkane monooxygenase and competing respiratory enzymes (if present under anoxic conditions) such as high-affinity cbb3-type oxidases (Pitcher and Watmough, 2004; predicted for strain HdN1; Table S5) could maintain the O2 concentration below detection level. Also, the produced alcohol may be consumed effectively by the subsequent reaction. Only upon sudden exposure to air, the anaerobically grown cells accumulated detectable n-hexadecanol (see above).
The slight differences between the growth tests under oxic and anoxic conditions with alkanes of various chain lengths (see growth tests with different electron acceptors) do not necessarily contradict the hypothesis that monooxygenases are used under oxic as well as under anoxic cultivation conditions for alkane activation. There might be slight differences with respect to chain length specificity between the monooxygenase(s) formed in aerobic and denitrifying cultures.
Still, also other modes of an indirect use of N–O species for alkane activation can be envisaged. For instance, they may serve as high-potential (strongly oxidizing) electron acceptors (half reactions in Fig. 6B) to generate by electron withdrawal an oxidized, active (reactive) state of a factor or an enzyme site involved in alkane activation.
Another hypothesis would be the direct involvement of an N–O species in the activation reaction of the alkane. Such direct biochemical linkage of the disintegration of an N–O species to C−H bond cleavage and formation of a functionalized product from an alkane would probably require an intricate mechanism.
Concluding remarks
In conclusion, results suggest a mechanistic alternative to the fumarate-dependent reaction for anaerobic alkane activation. Also a sulfate-reducing bacterium, strain Hxd3, metabolized long-chain n-alkanes obviously via an initial reaction different from that in other anaerobic alkane degraders (Aeckersberg et al., 1998; So et al., 2003; Callaghan et al., 2006). This raises the question whether nitrate-reducing strain HdN1 and sulfate-reducing strain Hxd3 employ basically the same reaction or different reactions to initiate alkane degradation. If they would employ essentially the same fumarate-independent activation reaction, strain HdN1 cannot employ an N–O species or derived O2 directly in the mechanism because they are excluded in strain Hxd3; the sulfate reducer was grown without nitrate. Also other ways to generate O2 are essentially excluded in sulfate reducers; sulfate and its metabolites are all thermodynamically very stable and represent very weak oxidants. Hence, alkane activation by the same basic mechanism in strains HdN1 and Hxd3 would imply that the denitrifier uses an N–O species or O2 only indirectly to generate an alkane-activating factor, whereas the sulfate reducer would generate the same type of factor in a different manner. If strains HdN1 and Hxd3 use different mechanisms for alkane activation, which is more appealing to assume, the reaction in strain HdN1 would represent a third type of alkane activation under anoxic conditions, besides the fumarate-dependent mechanism and the speculative mechanism in strain Hxd3. More refined physiological experiments (preceded by an improved method for harvesting the buoyant cells associated with alkane) are needed to provide further hints as to the alkane activation mechanism in strain HdN1, with consideration of its apparently diverse monooxygenases.
Finally, the present results as well as the oxidation of methane with NO2− (Ettwig et al., 2010) indicate that NO3− or NO2− (either from NO3− reduction or directly from NH4+ oxidation) should not be considered merely as electron acceptors for anaerobic respiration. An intermediate formed during NO3− or NO2− reduction may provide or function as a co-reactant for the biochemical activation of various hydrocarbons or even of other chemically unreactive compounds. NO3− or NO2− in anoxic habitats could, in principle, promote or enable the degradation of certain organic fractions which tend to be refractory under conditions of sulfate reduction or methanogenesis.
Acknowledgments
We are indebted to Daniela Lange for experimental support, to Andreas Schummer for help with substrate tests, and to Alexander Steinbüchel for analysis of polyhydroxyalkanoic acids. This work was supported by the Max-Planck-Gesellschaft.
Supporting information
Additional Supporting Information may be found in the online version of this article:
Fig. S1. Relationship of the assumed catalytic (large) subunit (MasD) of the n-alkane-activating enzyme in strain OcN1 to other enzymes activating hydrocarbons by addition to fumarate.
Table S1. General genome features of strain HdN1.
Table S2. Genome-based search for genes (bssA and homologues) that may encode fumarate-dependent glycyl radical enzymes for anaerobic alkane activation.
Table S3. Genome-based prediction of enzymes for the denitrification pathway in strain HdN1.
Table S4. Genome-based prediction of alkane monooxygenases (alkane hydroxylases) in strain HdN1.
Table S5. Genome-based prediction of cbb3-type oxidases (oxidases with high O2 affinity) in strain HdN1.
Appendix S1. Cultivation of strains, physiological experiments.
Appendix S2. Formal derivation of the average redox potential.
Please note: Wiley-Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
References
- Aeckersberg F, Bak F, Widdel F. Anaerobic oxidation of saturated hydrocarbons to CO2 by a new type of sulfate-reducing bacterium. Arch Microbiol. 1991;156:5–14. [Google Scholar]
- Aeckersberg F, Rainey FA, Widdel F. Growth, natural relationships, cell fatty acids and metabolic adaptation of sulfate-reducing bacteria that utilize long-chain alkanes under anoxic conditions. Arch Microbiol. 1998;170:361–369. doi: 10.1007/s002030050654. [DOI] [PubMed] [Google Scholar]
- Anderson RT, Lovley DR. Hexadecane decay by methanogenesis. Nature. 2000;404:722–723. doi: 10.1038/35008145. [DOI] [PubMed] [Google Scholar]
- Birch LD, Bachofen R. Microbial production of hydrocarbons. In: Rehm HJ, Reed G, editors. Biotechnology. Vol 6b. Special Microbial Processes. Weinheim, Germany: VCH Verlagsgesellschaft; 1988. pp. 71–99. [Google Scholar]
- Bonin PC, Cravo-Laureau C, Michotey V, Hirschler-Réa A. The anaerobic hydrocarbon biodegrading bacteria: an overview. Ophelia. 2004;58:243–254. [Google Scholar]
- Britton LN. Microbial degradation of aliphatic hydrocarbons. In: Gibson TD, editor. Microbial Degradation of Organic Compounds. New York, USA: Marcel Dekker; 1984. pp. 89–129. [Google Scholar]
- Callaghan AV, Gieg LM, Kropp KG, Suflita JM, Young LY. Comparison of mechanisms of alkane metabolism under sulfate-reducing conditions among two bacterial isolates and a bacterial consortium. Appl Environ Microbiol. 2006;72:4274–4282. doi: 10.1128/AEM.02896-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callaghan AV, Wawrik B, Ní Chadhain SM, Young LY, Zylstra GJ. Anaerobic alkane-degrading strain AK-01 contains two alkylsuccinate synthase genes. Biochem Biophys Res Commun. 2008;366:142–148. doi: 10.1016/j.bbrc.2007.11.094. [DOI] [PubMed] [Google Scholar]
- Callaghan AV, Tierney M, Phelps CD, Young LY. Anaerobic biodegradation of n-hexadecane by a nitrate-reducing consortium. Appl Environ Microbiol. 2009;75:1339–1344. doi: 10.1128/AEM.02491-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty R, Coates JD. Anaerobic degradation of monoaromatic hydrocarbons. Appl Microbiol Biotechnol. 2004;64:437–446. doi: 10.1007/s00253-003-1526-x. [DOI] [PubMed] [Google Scholar]
- Chance B, Oshino R, Oshino N. Sensitive oxygen assay method by luminous bacteria. Methods Enzymol. 1978;54:499–505. doi: 10.1016/s0076-6879(78)54030-5. [DOI] [PubMed] [Google Scholar]
- Cravo-Laureau C, Matheron R, Cayol J-L, Joulian C, Hirschler-Réa A. Desulfatibacillum aliphaticivorans gen. nov., sp. nov., an n-alkane- and n-alkene-degrading, sulfate-reducing bacterium. Int J Syst Evol Microbiol. 2004;54:77–83. doi: 10.1099/ijs.0.02717-0. [DOI] [PubMed] [Google Scholar]
- Davidova IA, Duncan KE, Choi OK, Suflita JM. Desulfoglaeba alkanexedens gen. nov., sp. nov., an n-alkane-degrading, sulfate-reducing bacterium. Int J Syst Evol Microbiol. 2006;56:2737–2742. doi: 10.1099/ijs.0.64398-0. [DOI] [PubMed] [Google Scholar]
- Ehrenreich P, Behrends A, Harder J, Widdel F. Anaerobic oxidation of alkanes by newly isolated denitrifying bacteria. Arch Microbiol. 2000;173:58–64. doi: 10.1007/s002030050008. Erratum: Arch Microbiol173: 232. [DOI] [PubMed] [Google Scholar]
- Ettwig KF, Butler MK, Le Paslier D, Pelletier E, Mangenot S, Kuypers MMM, et al. Nitrite-driven anaerobic methane oxidation by oxygenic bacteria. Nature. 2010;464:543–548. doi: 10.1038/nature08883. [DOI] [PubMed] [Google Scholar]
- Farmer JJ, III, Hickman-Brenner FW. The genera Vibrio and Photobacterium. In: Dworkin M, Falkow S, Rosenberg E, Schleifer K-H, Stackebrandt E, editors. The Prokaryotes. Vol. 6. New York, USA: Springer; 2006. pp. 508–563. [Google Scholar]
- Garrels RM, Christ CL. Solutions, Minerals, and Equilibria. New York, USA: Harper & Row; 1965. [Google Scholar]
- Ginkel CG, Rikken GB, Kroon AGM, Kengen SWM. Purification and characterization of chlorite dismutase: a novel oxygen-generating enzyme. Arch Microbiol. 1996;166:321–326. doi: 10.1007/s002030050390. [DOI] [PubMed] [Google Scholar]
- Grossi V, Cravo-Laureau C, Guyoneaud R, Ranchou-Peyruse A, Hirschler-Rea A. Metabolism of n-alkanes and n-alkenes by anaerobic bacteria: a summary. Org Geochem. 2008;39:1197–1203. [Google Scholar]
- Grundmann O, Behrends A, Rabus R, Amann J, Halder T, Heider J, Widdel F. Genes encoding the candidate enzyme for anaerobic oxidation of n-alkanes in the denitrifying bacterium, strain HxN1. Environ Microbiol. 2008;10:376–385. doi: 10.1111/j.1462-2920.2007.01458.x. [DOI] [PubMed] [Google Scholar]
- Heider J. Adding handles to unhandy substrates: anaerobic hydrocarbon activation mechanisms. Curr Opin Chem Biol. 2007;11:188–194. doi: 10.1016/j.cbpa.2007.02.027. [DOI] [PubMed] [Google Scholar]
- Higashioka Y, Kojima H, Nakagawa T, Sato S, Fukui M. A novel n-alkane-degrading bacterium as a minor member of p-xylene-degrading sulfate-reducing consortium. Biodegradation. 2009;20:383–390. doi: 10.1007/s10532-008-9229-8. [DOI] [PubMed] [Google Scholar]
- Jones DM, Head IM, Gray ND, Adams JJ, Rowan AK, Aitken CM, et al. Crude-oil biodegradation via methanogenesis in subsurface petroleum reservoirs. Nature. 2008;451:176–180. doi: 10.1038/nature06484. [DOI] [PubMed] [Google Scholar]
- Kniemeyer O, Musat F, Sievert SM, Knittel K, Wilkes H, Blumenberg M, et al. Anaerobic oxidation of short-chain hydrocarbons by marine sulphate-reducing bacteria. Nature. 2007;449:898–901. doi: 10.1038/nature06200. [DOI] [PubMed] [Google Scholar]
- Kropp KG, Davidova IA, Suflita JM. Anaerobic oxidation of n-dodecane by an addition reaction in a sulfate-reducing bacterial enrichment culture. Appl Environ Microbiol. 2000;66:5393–5398. doi: 10.1128/aem.66.12.5393-5398.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehboob F, Junca H, Schraa G, Stams AJM. Growth of Pseudomonas chloritidismutans AW-1T on n-alkanes with chlorate as electron acceptor. Appl Microbiol Biotechnol. 2009a;83:739–747. doi: 10.1007/s00253-009-1985-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehboob F, Wolterink AF, Vermeulen AJ, Jiang B, Hagedoorn PL, Stams AJ, Kengen SW. Purification and characterization of a chlorite dismutase from Pseudomonas chloritidismutans. FEMS Microbiol Lett. 2009b;293:115–121. doi: 10.1111/j.1574-6968.2009.01517.x. [DOI] [PubMed] [Google Scholar]
- Pitcher RS, Watmough NJ. The bacterial cytochrome cbb3 oxidases. Biochim Biophys Acta. 2004;1655:388–399. doi: 10.1016/j.bbabio.2003.09.017. [DOI] [PubMed] [Google Scholar]
- Rabus R, Widdel F. Anaerobic degradation of ethylbenzene and other aromatic-hydrocarbons by new denitrifying bacteria. Arch Microbiol. 1995;163:96–103. doi: 10.1007/BF00381782. [DOI] [PubMed] [Google Scholar]
- Rabus R, Wilkes H, Behrends A, Armstroff A, Fischer T, Pierik AJ, Widdel F. Anaerobic initial reaction of n-alkanes: evidence for (1-methylpentyl)succinate as initial product and for involvement of an organic radical in the metabolism of n-hexane in a denitrifying bacterium. J Bacteriol. 2001;183:1707–1715. doi: 10.1128/JB.183.5.1707-1715.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revsbech NP. An oxygen microsensor with a guard cathode. Limnol Oceanogr. 1989;34:474–478. [Google Scholar]
- Rojo F. Enzymes for aerobic degradation of alkanes. In: Timmis KN, editor. Handbook of Hydrocarbon and Lipid Microbiology. Vol. 2. Heidelberg, Germany: Springer; 2009. pp. 781–797. [Google Scholar]
- Rueter P, Rabus R, Wilkes H, Aeckersberg F, Rainey FA, Jannasch HW, Widdel F. Anaerobic oxidation of hydrocarbons in crude-oil by new types of sulfate-reducing bacteria. Nature. 1994;372:455–458. doi: 10.1038/372455a0. [DOI] [PubMed] [Google Scholar]
- Schreiber F, Polerecky L, de Beer D. Nitric oxide microsensor for high spatial resolution measurements in biofilms and sediments. Anal Chem. 2008;80:1152–1158. doi: 10.1021/ac071563x. [DOI] [PubMed] [Google Scholar]
- Schreiber F, Loeffler B, Polerecky L, Kuypers MM, de Beer D. Mechanisms of transient nitric oxide and nitrous oxide production in a complex biofilm. ISME J. 2009;3:1301–1313. doi: 10.1038/ismej.2009.55. [DOI] [PubMed] [Google Scholar]
- Scott CCL, Finnerty WR. Comparative analysis of ultrastructure of hydrocarbon-oxidizing microorganisms. J Gen Microbiol. 1976;94:342–350. doi: 10.1099/00221287-94-2-342. [DOI] [PubMed] [Google Scholar]
- So CM, Young LY. Isolation and characterization of a sulfate-reducing bacterium that anaerobically degrades alkanes. Appl Environ Microbiol. 1999;65:2969–2976. doi: 10.1128/aem.65.7.2969-2976.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- So CM, Phelps CD, Young LY. Anaerobic transformation of alkanes to fatty acids by a sulfate-reducing bacterium, strain Hxd3. Appl Environ Microbiol. 2003;69:3892–3900. doi: 10.1128/AEM.69.7.3892-3900.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinbüchel A, Wiese S. A Pseudomonas strain accumulating polyesters of 3-hydroxybutyric acid and medium-chain-length 3-hydroxyalkanoic acids. Appl Microbiol Biotechnol. 1992;37:691–697. [Google Scholar]
- Stumm W, Morgan JJ. Aquatic Chemistry. New York, USA: John Wiley & Sons; 1995. [Google Scholar]
- Tan NC, van Doesburg W, Langenhoff AA, Stams AJ. Benzene degradation coupled with chlorate reduction in a soil column study. Biodegradation. 2006;17:113–119. doi: 10.1007/s10532-005-5335-z. [DOI] [PubMed] [Google Scholar]
- Thauer RK, Jungermann K, Decker K. Energy conservation in chemotrophic anaerobic bacteria. Bacteriol Rev. 1977;41:100–180. doi: 10.1128/br.41.1.100-180.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tissot BP, Welte DH. Petroleum Formation and Occurrence. Berlin, Germany: Springer; 1984. [Google Scholar]
- Weelink SA, Tan NC, ten Broeke H, van den Kieboom C, van Doesburg W, Langenhoff AA, et al. Isolation and characterization of Alicycliphilus denitrificans strain BC, which grows on benzene with chlorate as the electron acceptor. Appl Environ Microbiol. 2008;74:6672–6681. doi: 10.1128/AEM.00835-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widdel F. Cultivation of anaerobic microorganisms with hydrocarbons as growth substrates. In: Timmis KN, editor. Handbook of Hydrocarbon and Lipid Microbiology. Vol. 2. Heidelberg, Germany: Springer; 2009. pp. 3787–3798. [Google Scholar]
- Widdel F, Bak F. Gram-negative mesophilic sulfate-reducing bacteria. In: Balows A, Trüper HG, Dworkin M, Harder W, Schleifer K-H, editors. The Prokaryotes. New York, USA: Springer; 1992. pp. 3352–3378. [Google Scholar]
- Widdel F, Knittel K, Galushko A. Anaerobic hydrocarbon-degrading microorganisms – an overview. In: Timmis KN, editor. Handbook of Hydrocarbon and Lipid Microbiology. Vol. 2. Heidelberg, Germany: Springer; 2009. pp. 1997–2021. [Google Scholar]
- Wilhelm E, Battino R, Wilcock RJ. Low-pressure solubility of gases in water. Chem Rev. 1977;77:219–262. [Google Scholar]
- Wilkes H, Rabus R, Fischer T, Armstroff A, Behrends A, Widdel F. Anaerobic degradation of n-hexane in a denitrifying bacterium: further degradation of the initial intermediate (1-methylpentyl)succinate via C-skeleton rearrangement. Arch Microbiol. 2002;177:235–243. doi: 10.1007/s00203-001-0381-3. [DOI] [PubMed] [Google Scholar]
- Wilkes H, Kühner S, Bolm C, Fischer T, Classen A, Widdel F, Rabus R. Formation of n-alkane- and cycloalkane-derived organic acids during anaerobic growth of a denitrifying bacterium with crude oil. Org Geochem. 2003;34:1313–1323. [Google Scholar]
- Yamasaki H. Nitrite-dependent nitric oxide production pathway: implications for involvement of active nitrogen species in photoinhibition in vivo. Philos Trans R Soc Lond B Biol Sci. 2000;355:1477–1488. doi: 10.1098/rstb.2000.0708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zengler K, Richnow HH, Roselló-Mora R, Michaelis W, Widdel F. Methane formation from long-chain alkanes by anaerobic microorganisms. Nature. 1999;401:266–269. doi: 10.1038/45777. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


