Abstract
Meiotic homologous recombination serves three principal roles. First, recombination reassorts the linkages between newly-arising alleles to provide genetic diversity upon which natural selection can act. Second, recombination is used to repair certain types of DNA damage to provide a mechanism of genomic homeostasis. Third, with few exceptions homologous recombination is required for the appropriate segregation of homologous chromosomes during meiosis. Recombination rates are elevated near DNA sites called “recombination hotspots.” These sites influence the distribution of recombination along chromosomes and the timing of recombination during the life cycle. Recent advances have revealed biochemical steps of hotspot activation and have suggested that hotspots may regulate when and where recombination occurs. Two models for hotspot activation, one in which hotspots act early in the recombination pathway and one in which hotspots act late in the recombination pathway, are presented. The latter model can account for changes at hypervariable minisatellite DNA in metazoan genomes by invoking resolution of Holliday junctions at minisatellite DNA repeats.
I. Introduction
Meiosis and homologous recombination have vital roles in the evolution of eukaryotes. Meiosis and subsequent joining of haploid gametes generates new genotypes by shuffling linkage groups (chromosomes) and recombination generates new genotypes by shuffling alleles of genes within individual chromosomes. Both processes supply genetic diversity upon which natural selection may act. Thus, meiotic reassortment and recombination provide eukaryotes a mechanism for sampling different combinations of newly-arising genetic variants. Recombination also has an apparent role in chromosome dynamics during meiosis: In the absence of recombination, homologous chromosomes fail to segregate appropriately, suggesting that recombination is required for proper chromosomal disjunction. Meiotic recombination rates are induced up to 1000-fold relative to mitotic rates, presumably to maximize the amount of genetic variability in the meiotic products or to insure homolog disjunction or both. Recombination rates are higher than average in the vicinity of DNA sites called “recombination hotspots.”
Recombination hotspots may contribute a significant fraction of all recombination that occurs in the genome (Wahls et al., 1990a; Smith, 1991; Wahls and Smith, 1994; Wu and Lichten, 1994; Lichten and Goldman, 1995; Kon et al., submitted). While the possibility that these sites regulate recombination is speculative, it is clear that hotspots must enhance rate-limiting steps in the pathway of recombination. They thus provide a larger window through which to view the molecular mechanisms of recombination.
Although eukaryotic recombination has been studied genetically for most of this century, the study of the biochemistry of eukaryotic recombination is in its infancy. The problem is one of scale. The products of recombination, whole chromosomes, are huge and can differ by as little as broken and reformed phosphodiester bonds. Thus, much of what we know about the machinery of recombination is inferred from genetic studies. The identification of recombination hotspots was first made in genetically-tractable organisms, such as bacteriophages and fungi, but it is now apparent that hotspots are ubiquitous and active in all organisms. Given that recombination occurs at a higher rate near hotspots, it is not surprising that recent advances in understanding the mechanisms of eukaryotic recombination have come from the study of hotspots.
This review outlines the identification of eukaryotic recombination hotspots and presents select examples of hotspots in greater detail. It is written for a general audience. For those who want an introductory recombination text, I recommend highly the new textbook by David Leach (1996). For those looking for a more in-depth treatment of specific subject matter, primary references and recent review articles are cited. The aim is to provide a historical framework within which current understanding of hotspot activity is considered. Although recombination hotspots have been studied principally in fungi, there are a few studies of hotspots in multicellular eukaryotes that have progressed beyond the descriptive stage. These are presented. And finally, two models for hotspot activation, one in which hotspots act early in the recombination pathway and one in which hotspots act late in the recombination pathway, are presented.
II. General Features of Chromosome Dynamics During Meiosis
During meiosis, chromosomes undergo one round of DNA replication, homologous chromosomes somehow find each other to pair and undergo recombination, and two rounds of chromosome segregation result in haploid meiotic products. The cytogenetic behavior of chromosomes during meiosis is fairly well characterized. Studies by Barbara McClintock using maize (McClintock, 1929) and Neuorspora (McClintock, 1945) firmly established general features of pairing and segregation of chromosomes during meiosis. McClintock also provided the original evidence that reciprocal recombination between genes on chromosomes during meiosis is accompanied by a physical exchange of chromosome parts (Creighton and McClintock, 1931). With a few notable exceptions, homologous recombination also correlates with the appropriate segregation of meiotic chromosomes (see Hassold, Chapter _____). While it is generally accepted that there is a relationship between the initial homologous alignment of chromosomes, formation of tightly-associated pairs held together by the synaptonemal complex (SC) (see Moens, Chapter _____), homologous recombination, and appropriate segregation of chromosomes, the nature of that relationship is not clear. It is within the macromolecular structure of the SC that homologous recombination occurs.
Genetic analyses of homologous recombination, conducted principally in various fungi, have led to theoretical models for how recombination occurs (Holliday, 1964; Meselson and Radding, 1975; Szostak et al., 1983). The models share common steps: (a) some sort of homology search and pairing of chromosomes; (b) a double-strand DNA (dsDNA) or single-strand DNA (ssDNA) break that initiates recombination; (c) processing of those DNA breaks in some fashion; (d) invasion of one homolog with a DNA strand from the other homolog; (e) repair synthesis and DNA ligation to create one or more a Holliday junctions, points where two homologous chromosomes are covalently linked by crossed DNA strands; (f) branch migration of the Holliday junction; (g) cleavage of the crossed strands in the Holliday junction to separate the homologous chromosomes; (h) ligation of the resulting nicks to produce recombinant chromosomes; and (i) mismatch repair of heteroduplex DNA . Physical and biochemical analyses have confirmed some of these theoretical steps in the pathway. For example, processed dsDNA breaks (steps “b” and “c”) have been detected (Sun et al., 1989; Cao et al., 1990), a nuclease that produces those breaks has been identified (Bergerat et al., 1997; Keeney et al., 1997), enzymes likely involved in strand exchange (step “d”) have been characterized (Bishop et al., 1992; Shinohara et al., 1992; Benson et al., 1994; Bishop, 1994) and an exonuclease with a role in mismatch correction (step “i”) has been purified (Szankasi and Smith, 1992; Szankasi and Smith, 1995). Many of these advances have been made by studying recombination hotspots.
III. Genetic Identification of Recombination Hotspots
A. Disparity between genetic and physical maps
The first indication that rates of recombination are not a uniform function of physical distance along the DNA came from mapping experiments. Mosig (1966) first pointed out such discrepancies from studies of the bacteriophage T4. While in most areas of the T4 genome the measured distances in DNA were proportional to genetic map distances determined from recombinant frequencies, the region including genes 34 and 35 were especially prone to recombination: Mosig states, “The discrepancies suggest that genetic recombination frequency in this region is increased by local factors other than distances between markers.”
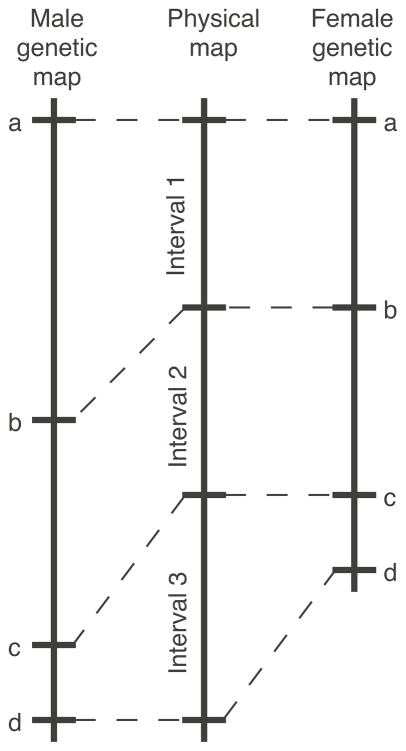
As contiguous physical maps of chromosomes become available, invariably there are expansions and contractions of the genetic maps relative to the physical maps (Fig. 1). These differences, which can be quite pronounced (Andersson et al., 1988; Zimmerer and Passmore, 1991; Bryda et al., 1992; Shiroishi et al., 1993), reflect variable frequencies of recombination in different regions of chromosomes. In addition to regional variability, the strain (Soper et al., 1988; Zimmerer and Passmore, 1991; Shiroishi et al., 1993) and sex (Shiroishi et al., 1990; Shiroishi et al., 1991) in which meiosis is occurring can markedly influence regional recombination frequencies. Both cis-acting and trans-acting factors are implicated. It has become clear that regional differences in reciprocal recombination between any two genetic markers are common. These recombination hotspots are inferred to contain some feature that promotes nearby recombination.
Figure 1. Schematic diagram of differences between physical and meiotic genetic maps.
Expansion and contraction of genetic intervals relative to physical intervals identifies regions of the genome with rates of recombination higher (interval 1) and lower (interval 3) than average, respectively. In this idealized example recombination is higher in interval 1 during male meiosis relative to female meiosis, revealing sex-specific differences of recombination rates. Cis-acting elements within or nearby the intervals are inferred to influence their rates of recombination.
B. Marker effects and polarity of recombination
The second indication that rates of recombination are not a uniform function of physical distance along the DNA came from the analysis of how individual genetic markers behave during recombination. After premeiotic DNA replication there are four chromatids (eight ssDNA strands) between which recombination may occur (Fig. 2A). Two rounds of chromosome segregation separate the chromatids into the four meiotic products. In some organisms, such as Saccharomyces cerevisiae, the four products of individual meioses are spores held together in an ascus, collectively called a tetrad. In other organisms, such as Neurospora crassa or Ascobolus immersus, a single round of mitosis occurs after meiosis and eight spores are formed in the ascus, called an octad. It is possible to dissect individual asci, isolate all the spores, and grow individual spore colonies. This permits phenotypic analysis of all four (or eight) products of a single meiosis.
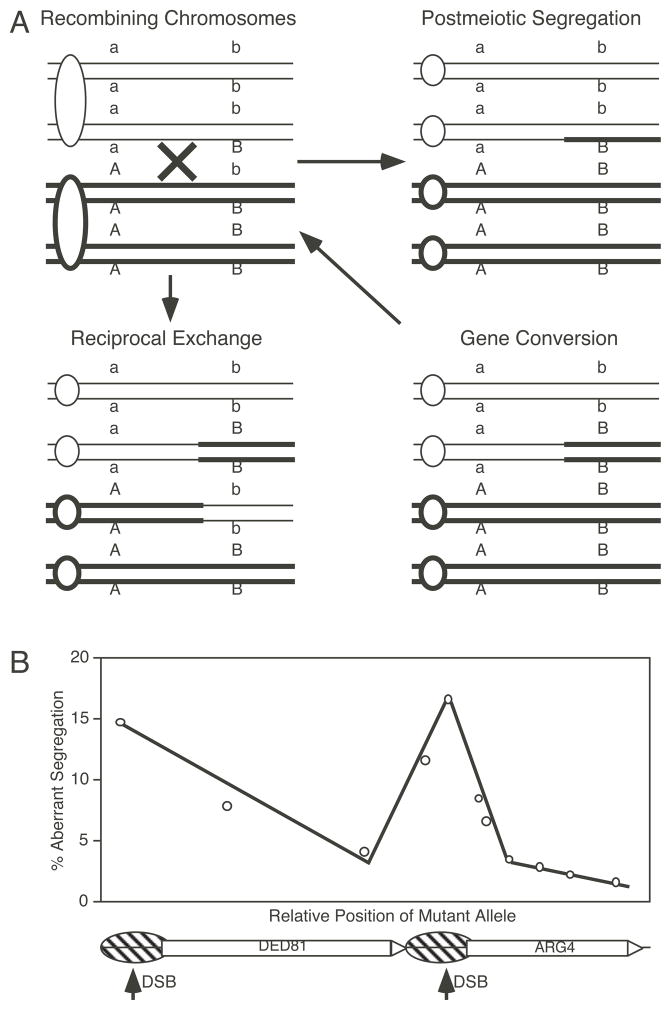
Figure 2. Polarity of recombination.
(A) Reciprocal exchange is conservative and switches the linkage between markers flanking the point of recombination. Gene conversion and postmeiotic segregation (PMS) are nonconservative (aberrant) recombination events that result from the transfer of the genetic information from one allele to the other. Other types of aberrant segregation (not shown) may also occur Aberrant segregation events in unselected tetrads can be used to gauge recombination rates at individual genetic markers. (B) An example of polarity resulting from different frequencies of aberrant segregation for multiple markers at the DED81 and ARG4 loci in S. cerevisiae. Regions of increased accessibility of DNA within meiotic chromatin (striped balloons) and meiosis-specific dsDNA breaks (DSB arrows) map to the high-frequency end of polarity gradients, suggesting that recombination initiates at those sites. Data are from (Fogel et al., 1981; White et al., 1985; Nicolas et al., 1989; Sun et al., 1989; Cao et al., 1990; Schultes and Szostak, 1990; Lichten, 1995).
By studying the products of a single meiosis, it is possible to determine whether recombination is conservative or nonconservative (Fig. 2A). Reciprocal exchange events are conservative; genetic information is neither lost nor gained and the alleles are inherited in a Mendelian 2:2 ratio. In contrast, during gene conversion the genetic identity of one allele is lost and is replaced by that of the opposite allele, leading to nonMendelian inheritance with 1:3 or 3:1 ratios (also called 2:6 or 6:2 ratios for 8-spored fungi). A second type of nonMendelian inheritance occurs when 2 spores of a tetrad yield colonies have phenotype A, 1 spore colony has phenotype a and one spore colony has the sectored phenotype A/a. These sectored colonies provide evidence for heteroduplex DNA that contains one DNA strand from each of two parental chromatids. During the first mitotic division those two strands separate and each other and each of the two daughter cells receives a different allele. This is called “post meiotic segregation” (PMS). Tetrads exhibiting PMS are classified as 5:3 or 3:5 tetrads (using nomenclature of 8-spored fungi). Some tetrads, aberrant 4:4 tetrads, have two spore colonies with PMS, indicating the presence of symmetrical heteroduplex DNA in two of the four chromatids. In general, the frequency of gene conversion is much higher than that of PMS for most markers, suggesting that the heteroduplex DNA is efficiently repaired. This repair leads to either gene conversion or to restoration of the original allele. Thus, a combination of heteroduplex formation and mismatch correction can lead to gene conversion events. The relative proportions of PMS, gene conversion, and symmetrical or asymmetrical heteroduplex DNA in different organisms is accounted for by several related models of recombination (Holliday, 1964; Meselson and Radding, 1975; Szostak et al., 1983). For a comparison of recombination models, see the review by Petes et al. (1991). Since all models propose that reciprocal exchange and gene conversion are alternative resolutions of a common intermediate in the pathway of recombination, it is possible to infer overall recombination rates at individual markers within any given locus by determining their frequencies of gene conversion.
The frequency of meiotic gene conversion varies depending upon the locus and alleles being studied. In S. cerevisiae, about 1% to 50% of tetrads will have gene conversion event at any given locus (Fogel et al., 1981; Petes et al., 1991). When multiple different alleles within a gene are studied there is usually a gradient of conversion frequencies across the gene (Fig. 2B). This is known as polarity. Polarity was discovered simultaneously in two fungi, Ascobolus immersus (Lissouba and Rizet, 1960) and Neurospora crassa (Murray, 1960). Murray also showed that polarity was conferred by elements close to the gene; chromosomal inversions also inverted the polarity gradient (Murray, 1968). Polarity has been demonstrated for several genes in S. cerevisiae including ARG4 (Fogel et al., 1981; Nicolas et al., 1989), CYS3 (Chaudhuri and Messing, 1992), DED81 (Schultes and Szostak, 1990), HIS1 (Fogel et al., 1967; Savage and Hastings, 1981), HIS2 (Malone et al., 1992), and HIS4 (Detloff et al., 1992). Polarity gradients generally have a high frequency at one end of the gene, a low frequency at the other end, and a relatively linear range in between, but there are some exceptions: Genes lacking polarity (Kitani and Olive, 1967) or having polarity gradients with the low frequency in the middle of the gene (Pees, 1966) have been described. The enhancement across yeast polarity gradients ranges from about a 3-fold (at HIS2) to about 10-fold (at ARG4). In cases of a uniform polarity gradient, the high-frequency conversion end of the gene usually corresponds to the 5′ end, but in the HIS2 gene the polarity gradient slopes up towards the 3′ end.
The shape of polarity gradients suggests that there are discrete sites at which recombination initiates. In this view, recombination events initiate preferentially near the high-frequency peak and propagate outwards. The slope and extent of the gradient reflects the processivity of recombination events initiating at each hotspot. dsDNA breaks have been found at these hotspots (Nicolas et al., 1989; Sun et al., 1989; Cao et al., 1990), suggesting that the hotspots within polarity gradients are sites where recombination initiates. These initiating lesions are discussed in more detail below (Section IV). On the other hand, one must consider the alternative explanation for the polarity, namely that a repressor of recombination acts on the low-frequency end of the gradient and polarity results from a distance-dependent propagation of repression. For example, several trans-acting gene products repress recombination in the mat2-mat3 interval of the fission yeast, Schizosaccharomyces pombe (Klar and Bonaduce, 1991; Thon and Klar, 1993; Thon et al., 1994), thereby making a recombination coldspot. However, this may reflect the special need to reduce recombination at the mat loci to achieve the appropriate regulation of mating-type switching. In some cases, polarity may be largely due to the extent of heteroduplex DNA that is formed to the right, left, or both sides of an initiating event. In other cases, the extent of the mismatch correction tract likely has a significant role in the shape of the polarity gradient. For a more comprehensive discussion of polarity a recent review is available (Nicolas and Petes, 1994).
C. Recombination hotspots of the mouse MHC
The best examples of recombination hotspots in multicellular eukaryotes are those found in the mouse major histocompatibility complex (MHC). The MHC is a large genomic region that encodes multiple cell surface proteins of the immune system involved in self, non-self recognition. Due to its importance in immune function, the MHC has been extensively studied. The MHC is highly polymorphic and different haplotypes (the particular combination of genes within the MHC) can be determined serologically, thus providing a large number of allelic markers for loci throughout the region. Those markers can be used to study recombination. Intriguingly, crosses between some mouse strains lead to an increase in recombination within the MHC relative to crosses between other strains (Shiroishi et al., 1982; Kobori et al., 1984). A strain-specific recombination enhancer (recombination hotspot) is inferred.
The generation of contiguous DNA clones spanning large intervals of the MHC (e.g., (Steinmetz et al., 1986)) made possible a comparison of the physical and genetic maps. By using RFLP analysis and DNA sequencing, it is possible to narrow down the intervals within which the reciprocal exchange events occurred. The data reveal that most recombination within the MHC is clustered within recombination hotspots. One hotspot is located in the Eβ second intron (Kobori et al., 1984; Kobori et al., 1986; Lafuse and David, 1986; Steinmetz et al., 1986), one is within the Pb-Ob intergenic interval (Lafuse et al., 1990), another is found in the Int3:Tnx interval (Yoshino et al., 1994a) and so on. In each case, as fine physical mapping was done, it was discovered that most of the exchange events occurred within small physical intervals of a few thousand base pairs or less. These points of exchange may be even more tightly clustered, however resolution is limited by the proximity of the nearest heterologies flanking the points of exchange. Some of these MHC hotspots can be really hot: Genetic crosses between Asian wild mice with the wm7 haplotype and laboratory strains generate about a 2% recombinant frequency within a physical interval of about 1000 base pairs at the Pb-Ob hotspot (Shiroishi et al., 1993). This reflects a meiotic recombination rate that is >1000-fold higher than the genomic average. As with the mouse MHC, hotspots have been identified in the human (van Endert et al., 1992; McAdam et al., 1994; Martin et al., 1995), cow (Andersson et al., 1988), and chicken (Hepkema et al., 1993) MHC regions, suggesting that hotspots in the MHC are evolutionarily conserved.
DNA sequence comparisons failed to reveal any major features common to all the MHC hotspots. However, the Eβ and the Lmp-2 hotspots do have three motifs in common. They each contain an isolated, ancestral retroviral LTR element, a middle repetitive element of the MT family, and tandem repeats of a tetrameric sequence resembling hypervariable minisatellite DNA. Two of these three elements (LTR and hypervariable minisatellite) are recombinogenic in plasmid-based or in vitro recombination assays (Edelmann et al., 1989; Wahls et al., 1990a). Activity of the Lmp2 hotspot requires significant homology between the recombining chromosomes, with the number of copies of the minisatellite-like sequences being most important (Yoshino et al., 1995). Furthermore, the HLA-DQ hotspot in humans has similar DNA sequence elements: Hypervariable minisatellite DNA and the repeating dinucleotide d(TG)n, which are hotspots in human cells (Wahls et al., 1990a; Wahls et al., 1990b), are located close to (or at) the point of exchange. These repeated DNA elements may be partially responsible for hotspot activity. DNAseI hypersensitive sites have been found in the vicinity of these elements, suggesting that open chromatin may play a role in hotspot activation (Shenkar et al., 1991; Mizuno et al., 1996). Roles for hypervariable minisatellite DNA and open chromatin are discussed below (Sections VII and V, respectively).
The control of hotspot function seems to be complex. The presence of two MHC hotspots located about 100 kilobase pairs apart does not have an additive effect upon recombinant frequencies (Yoshino et al., 1994b). (A similar phenomenon occurs for two pairs of hotspots located near to each other in fission yeast (Zahn-Zabal et al., 1995) and in budding yeast (Fan et al., 1997)). Presumably there is either competition for limiting recombination factors or direct regulation involved. Perhaps the most exciting findings have been for the Pb-Ob hotspot. Shiroishi and colleagues (1990) have shown that elements outside of the primary sequence in the hotspot region are required for activity. Two distinct components are inferred: An element centromere proximal to the hotspot is required for the hotspot to function at all, and an element that is distal to the hotspot represses hotspot activity in male meiosis (Shiroishi et al., 1991; Shiroishi et al., 1995). (Trans-acting repression of hotspot activity also occurs at the cog hotspot of N. crassa (Catcheside and Angel, 1974; Catcheside, 1977)). These findings reveal that hotspot activity is not simply a function of some local, cis-acting feature. At minimum, two other factors or DNA sequences located away from the Pb-Ob hotspot must be required for function. It may be possible to identify the factors that control hotspot activity from a distance.
D. Mammalian recombination hotspots reveled in plasmid recombination assays
Genetic studies of hotspot activity in mammalian meioses are time-consuming, expensive and restricted to well-characterized regions such as the MHC. In somatic cells, however, recombination between mutant DNA viruses (such as SV40) can be studied (Dubbs et al., 1974). The development of shuttle vectors derived from these DNA viruses and carrying a dominant, selectable marker gene (e.g., neomycin phosphotransferase) permits analysis of recombination in many cell types (Southern and Berg, 1982; Subramani and Southern, 1983). The general approach is to measure recombination between two similar plasmids, each of which has a different mutation in the marker gene. Recombination between the two defective genes restores a functional, wild-type gene that can be expressed in either bacterial or mammalian cells. Thus, it is possible to select directly for recombinants within mammalian cells that have been cotransfected with the two plasmids. Alternatively, one can select for recombinant plasmids within bacteria after incubating the plasmid recombination substrates in cell-free extracts of mammalian cells. The utility of this approach and a discussion of somatic recombination can be found in a review by Subramani and Seaton (1988). The general view is that plasmid-based recombination uses the same enzymes and pathways as chromosomal recombination. Thus, DNA sequences that are hotspots in the plasmid recombination system might also be active within chromosomes.
1. Z-DNA
Certain DNA sequences, such as the repeating dinucleotides d(CG)n and d(TG)n, can undergo a structural transition from the familiar right-handed B-DNA form to a left-handed Z-DNA form. Extensive stretches of these repeats are not detectable in the genomes of eubacteria, archaebacteria or mitochondria (Gross and Garrard, 1986). On the other hand, there are ≥ 105 copies of these dinucleotide repeats scattered throughout the mammalian genome. The discovery that Z-DNA can form in supercoiled DNA molecules under physiological conditions (Klysik et al., 1981; Singleton et al., 1982) and evidence that Z-DNA might exist in vivo within chromosomes (Lancillotti et al., 1987) suggested a biological role for Z-DNA.
Hotspot activity of the Z-DNA motif d(TG)n was initially seen as an increase in intramolecular recombination between two tracts of d(TG)n in SV40 viruses replicating in monkey cells (Stringer, 1985). Recombination between tandem copies of the Z-DNA motif is about 8-fold higher than recombination between other tandem repeats. At about the same time it was shown that a single tract of the Z-DNA motif can enhance intramolecular recombination between adjacent tandem DNA sequences in replicating SV40 virus (Bullock et al., 1986). Subsequently, the effect of the Z-DNA motif upon intermolecular recombination between non-replicating plasmids introduced into human cells in culture was tested (Wahls et al., 1990b). A (dTG)30 sequence enhances recombination about 20-fold and exerts its effect at a distance. The hotspot promotes both reciprocal exchange and gene conversion to an equal extent, and during gene conversion the substrate containing (dTG)30 is preferentially converted to wild-type. Two models for hotspot activity can account for these observations (Wahls et al., 1990b). Furthermore, binding of SV40 T antigen adjacent to the (dTG)30 sequence abolishes hotspot activity, suggesting that T antigen either prevents the (dTG)30 sequence from acquiring a recombinogenic configuration (such as left-handed Z-DNA), or it stearically blocks the interaction of recombination proteins with the hotspot (Wahls and Moore, 1990). A protein that may have a role in recombination, HPP-1, was partially purified by Z-DNA affinity chromatography from the extracts of mammalian cells (Fishel et al., 1988; Moore and Fishel, 1990; Moore et al., 1991). Whether HPP-1 is a factor that binds to the d(TG)n sequence to activate the hotspot remains to be seen.
Other results also implicate the Z-DNA motif as being recombinogenic. Purified Ustilago maydis rec1 protein, like RecA of E. coli, is a recombination enzyme that promotes synapsis and strand exchange between homologous DNA molecules (Kmiec and Holloman, 1983). Z-DNA forms during the initial synapsis of homologous DNA promoted by rec1 (Kmiec and Holloman, 1984) and the rec1 protein can increase the pairing of two DNA molecules that contain Z-DNA (Kmiec and Holloman, 1986). Addition of anti-Z-DNA antibodies or competitor Z-DNA, or altering the superhelical density of the plasmids inhibits the pairing reaction. This suggests that Z-DNA may be required for rec1-mediated pairing of duplex DNA molecules during recombination. Similarly, the RecA protein of E. coli binds more stabley to Z-DNA than B-DNA (Blaho and Wells, 1987), again suggesting that recombination enzymes may have a preference for these recombinogenic sequences. Since the biochemistry of eukaryotic meiotic recombination now being revealed in yeast (reviewed by Petes et al., 1991), it is worthwhile to consider one more example of d(TG)n hotspot activity. A short fragment of DNA from the human β-globin gene cluster, containing d(TG)40, enhances meiotic recombination during meiosis in yeast (Treco and Arnheim, 1986). Thus, further study of how this hotspot is activated during yeast meiosis might be productive.
2. Hypervariable minisatellite DNA
These highly polymorphic DNA sequences were first identified in a random clone from human chromosome 14 (Wyman and White, 1980). Related sequences were discovered subsequently at a variety of loci (Higgs et al., 1981; Bell et al., 1982; Capon et al., 1983; Jeffreys et al., 1985a). The common feature of hypervariable minisatellites is a short consensus core sequence, which may or may not have variable-length flanking sequences, that is repeated in tandem head-to-tail arrays at ≥15,000 loci in the human genome (Melmer, 1994). Allelic variation is due to differences in the number of repeats at each locus and typical loci have overall lengths from 0.1 to 50 kilobase pairs. Locus-specific and multilocus probes can be used to study multiple polymorphic loci by Southern blotting, giving rise to individual-specific DNA “fingerprints” (Jeffreys et al., 1985b) that can be used to establish genetic relationships.
The importance of DNA fingerprinting has fueled study of how the polymorphism arises and is maintained. The hypervariable repeat length at individual loci was initially thought to result from unequal reciprocal exchange between alleles, suggesting a high rate of meiotic recombination (Jeffreys et al., 1985a). But, analysis of markers flanking newly-arising minisatellite alleles revealed a lack of expected reciprocal exchange (Wolff et al., 1989; Kasperczyk et al., 1990; Kelly et al., 1991; Buard and Vergnaud, 1994). This led many to conclude that minisatellite DNA variability cannot be related to recombination. In fact, it only shows that reciprocal exchange within or near to minisatellite repeats seldom occurs when new length alleles are generated. The core sequences within individual repeats at any given locus and allele can vary slightly, which permitted further analysis of how new length variants arise. Surprisingly, alleles often exhibit polarized variability (Armour et al., 1993a; Armour et al., 1993b; Buard and Vergnaud, 1994; Ellsworth et al., 1995). New variants have one parental type across most of the locus and at a discrete point, usually within a few tens of repeats from one end of the array, they become mosaic for both parental types. This implies that minisatellite change begins or ends at a discrete position within each array and extends in a directional fashion towards one end. Furthermore, the mosaic pattern suggests either complex mutational events or, as proposed below (Section VII. B.), several short patches mismatch correction within a length of heteroduplex DNA extending to a discrete point of exchange.
Considerable circumstantial evidence suggests that minisatellites are recombinogenic. New alleles can arise at a remarkably high rate, more than 10% for some loci, during meiosis (Buard and Vergnaud, 1994) and very rarely during mitotic growth (Kelly et al., 1989). The pairing of homologous chromosomes and the high induction of recombination during meiosis are thus good candidates for a meiotic mechanism of minisatellite change. The frequency of change at minisatellite loci can be markedly different in female and male meioses (Buard and Vergnaud, 1994; Henke and Henke, 1995), again analogous to sex-specific patterns of meiotic recombination. In addition, hypervariable minisatellite DNA has homology to the Chi recombination hotspot of Escherichia coli (reviewed by Kowalczykowski et al., 1994), however, that homology may simply be coincidental. As discussed previously (Section III. C.), minisatellite DNA motifs are found within recombination hotspots of the MHC. In addition, these sequences are sometimes found in the vicinity of translocation breakpoints (Krowczynska et al., 1990; Jaeger et al., 1994), suggesting that they have a role in formation of DNA breaks that occasionally lead to aberrant recombination. Similarly, independent insertion events in the hamster aprt locus are accompanied by loss of a sequence resembling the minisatellite core (Meuth et al., 1987). A recent study identified minisatellite DNA among DNA that was preferentially undergoing repair in meiosis (Ramachandra and Rao, 1994), suggesting that these sites may enhance repair as well as recombination. On a larger scale, cytogenetic analyses have found hypervariable minisatellite DNA at chiasmata (Chandley and Mitchell, 1988), sites within chromosomes where recombination is thought to occur.
Direct evidence for hotspot activity was obtained with a plasmid-based assay. Six copies of a hypervariable minisatellite DNA core sequence enhanced intermolecular recombination up to 20-fold between plasmids introduced into human cells (Wahls et al., 1990a). The hotspot increases recombination within a distant test interval and exhibits three characteristics different from those of most other hotspots. First, there is parity during gene conversion; the hotspot-containing substrate functions equally well as the donor or as the recipient of genetic information. If the minisatellite DNA promoted dsDNA break formation, then disparity of conversion would be expected (Szostak et al., 1983). Second, the effects of the hotspot and a distal dsDNA break are additive. If most recombination initiates at dsDNA breaks, then this provides further evidence that the minisatellite hotspot does not function in the initiation of recombination. Third, the hotspot enhances reciprocal exchange events much more than gene conversion events. This provides compelling evidence that the minisatellite acts late in the pathway of recombination. Reciprocal exchange and gene conversion are alternative resolutions of an intermediate in the pathway of recombination, the Holliday junction (Holliday, 1964; Meselson and Radding, 1975; Szostak et al., 1983). I propose that hypervariable minisatellite DNA functions as a hotspot for recombination by acting as a resolution point for Holliday junctions (Section VII. B.)
A number of hypervariable minisatellite DNA binding proteins have been detected (Wahls, 1989; Collick et al., 1991; Shinder et al., 1994) and purified (Wahls et al., 1991) from nuclear extracts of mammalian cells. However, little is known about the biochemistry of these factors nor have any sequence data been reported. Intriguingly, several of these proteins seem indistinguishable from transcription factors of the rel/NF-kappa B family (Trepicchio and Krontiris, 1992) and the minisatellite DNA repeat can influence transcription of reporter genes (Green and Krontiris, 1993). Binding of these proteins to minisatellite DNA also seems to be required for hotspot activation (W.P. Wahls and P.D. Moore, unpublished). However, until further mechanistic data are available, the link between these proteins and hotspot activity or transcription must remain anecdotal.
3. Mouse retroviral LTR element
As mentioned above, an LTR element is located near two hotspots in the MHC. In an in vitro assay, recombination between LTR elements is about 13-fold higher than between two control DNA sequences (Edelmann et al., 1989). This hotspot activity seems to be meiosis-specific; enhancement occurs within extracts of mouse testes cells but not within extracts of ascites cells. Deletion analyses revealed that some feature within a 37 bp region is essential for hotspot activity and two different nuclear proteins bind in a sequence-specific fashion. Intriguingly, one of the proteins was absent from extracts of testes cells, raising the possibility that it represses hotspot activity. Recent purification of two proteins that interact with the hotspot (Goller et al., 1994) may facilitate further investigations.
IV. Double-Strand DNA Breaks and Open Chromatin
A. Double-strand DNA breaks activate recombination hotspots
Broken DNA can be deadly or recombinogenic. Based upon observations with irradiated S. cerevisiae cells, Resnick (1976) concluded that a single unrepaired dsDNA break may be lethal. This led to a model in which recombination is used to repair dsDNA breaks (Resnick, 1976; Resnick et al., 1992). Subsequent studies of yeast transformation showed that an artificially-introduced dsDNA break can markedly increase the frequency of transformation by both non-reciprocal (gene conversion) and reciprocal recombination mechanisms (Orr-Weaver et al., 1981; Orr-Weaver et al., 1983), giving rise to the dsDNA break repair model of recombination (Szostak et al., 1983). Meiotically-induced, dsDNA breaks were first detected within the ARG4 hotspot (Nicolas et al., 1989; Sun et al., 1989) and dsDNA breaks are present at other hotspots surveyed to date, as well as throughout the genome at sites not yet characterized genetically (Cao et al., 1990; Game, 1992; Zenvirth et al., 1992; Nag and Petes, 1993; Wu and Lichten, 1994; Fan et al., 1995; Wu and Lichten, 1995). At any given locus he amount of cleaved DNA is roughly proportional to recombination rates. Furthermore, the frequency of dsDNA breaks varies coordinately with hotspot activity for cis- and trans-acting mutations affecting recombination (Leem and Ogawa, 1992; Fan et al., 1995; Rockmill et al., 1995; Wu and Lichten, 1995; Xu and Kleckner, 1995; Fan and Petes, 1996), except in some instances such as when there is apparent competition between adjacent hotspots (Fan et al., 1997). These data suggest that most meiotic recombination in yeast initiates at dsDNA breaks.
In wild-type cells meiotic dsDNA breaks are transient and they are heterogeneous, resulting from resection of about 600 nucleotides of the 5′ terminated stand on each side of the break point (Sun et al., 1989; Cao et al., 1990; Sun et al., 1991). At least nine genes involved in the formation or processing of dsDNA breaks have been identified. The most widely-studied is a non-null mutation of RAD50 (rad50s) (Alani et al., 1990) that causes mutants to lack both the resection and subsequent repair of dsDNA breaks. This mutant has been particularly useful for gauging the overall frequency of dsDNA breaks at various hotspots. Other genes whose products are required for the formation or processing of dsDNA breaks include SPO11 (Cao et al., 1990), MEI4 (Menees et al., 1992; Scott Keeney, pers. comm.), MER2 (Rockmill et al., 1995), MRE11 (Ajimura et al., 1993; Johzuka and Ogawa, 1995), REC102, REC104 and REC114 (Bullard et al., 1996) and XRS2 (Ivanov et al., 1992). Intriguingly, in rad50s mutants the unprocessed dsDNA breaks accumulate with a protein covalently-bound to the 5′ ends of the DNA (de Massy et al., 1995; Keeney and Kleckner, 1995; Liu et al., 1995). A brute force purification (Keeney et al., 1997) and a genetic approach (Bergerat et al., 1997) revealed that Spo11 is the bound protein. The Spo11 protein has blocks of homology with type II topoisomerases (Bergerat et al., 1997; Keeney et al., 1997) and mutation of a conserved tyrosine residue abolishes dsDNA break formation (Bergerat et al., 1997). These data suggest that the biochemical function of Spo11 is the same as that of topoisomerase II, at least for the strand scission step. Genes homologous to SPO11 have been identified in archeabacteria, fission yeast, and nematodes, suggesting conservation of activity across Kingdoms. The fission yeast homolog, rec12, is a meiotically-induced gene required for meiotic recombination (DeVeaux et al., 1992; Lin and Smith, 1994). While dsDNA breaks have not been reported for hotspots of any organism other than budding yeast, conservation of SPO11 suggests that dsDNA break-promoted recombination may be evolutionarily conserved.
B. DNA accessibility within chromatin
One factor that dictates where Spo11 protein introduces dsDNA breaks may be the accessibility of DNA. Increased accessibility of DNA within chromatin (as gauged by increased sensitivity to enzymes such as micrococcal nuclease or DNAseI) was first observed in the vicinity of the Eβ recombination hotspot of the mouse MHC (Shenkar et al., 1991). This increased sensitivity, called “open chromatin,” is found in the vicinity of all natural and created dsDNA break sites that have been examined (Ohta et al., 1994; Wu and Lichten, 1994; Liu et al., 1995; Wu and Lichten, 1995; Fan and Petes, 1996), as well as at recombination hotspots where dsDNA breaks have not yet been reported (Shenkar et al., 1991; Mizuno et al., 1996; Mizuno et al., 1997). The association of recombination hotspot activity with open chromatin and dsDNA breaks in promoter regions is very strong (Lichten and Goldman, 1995). However, there are exceptions. Some hotspots are not in promoter regions (Malone et al., 1992) and hotspot activation can be uncoupled from levels of transcription (Grimm et al., 1991; Schultes and Szostak, 1991; White et al., 1992). The position of open chromatin does not always correlate precisely with the patterns of dsDNA break formation (Liu et al., 1995; Wu and Lichten, 1995; Fan and Petes, 1996). Furthermore, some regions of increased DNA accessibility within chromatin do not undergo high frequency dsDNA break formation (Wu and Lichten, 1995; Fan and Petes, 1996). It seems clear that accessibility of DNA within chromatin is not sufficient for hotspot activation. Intriguingly, the magnitude of hotspot activity can depend more upon binding of certain transcription factors than upon the relative abundance of open chromatin. Replacement of Bas1, Bas2, Gcn4 and Rap1 transcription factor binding sites in the HIS4 promoter region with two Rap1 binding sites results in very strong dsDNA break formation and hotspot activity (Fan et al., 1995), but no additional increase in DNA accessibility to nucleases. Three possible explanations were considered:. First, open chromatin may be necessary and sufficient to confer hotspot activity, but the reagents used to assay DNA accessibility within chromatin do not accurately reveal the chromatin structure. Second, open chromatin structure might be necessary, but not sufficient, and some other factor (such as a protein bound nearby) is required. Third, hotspots might not require open chromatin for action, but only a factor that is usually associated with open chromatin. Activation of the ade6-M26 hotspot by binding of Mts1/Mts2 transcription factor supports the latter two possibilities (Kon et al., submitted), particularly since hotspot-dependent, meiotically-induced open chromatin requires presence of the Mts1/Mts2 heterodimer (K. Ohta and W.P. Wahls, unpublished observations).
V. Roles of Protein-DNA Binding in Hotspot Activation
Presumably, as for cis-acting transcriptional regulatory elements, there are proteins that interact with hotspots to mediate their biological activity. The paradigm is found in Escherichia coli: the RecBCD enzyme interacts with Chi sites to enhance recombination (reviewed by Kowalczykowski et al., 1994). The cog hotspot near the his3 locus in Neurospora crassa is repressed by unlinked, dominant loci (Catcheside and Angel, 1974; Catcheside, 1977;), suggesting that trans-acting factors can negatively regulate eukaryotic hotspots. Hotspots in the MHC require distal elements that positively and negatively regulate activity (Shiroishi et al., 1990; Shiroishi et al., 1991; Shiroishi et al., 1993), perhaps due to trans-acting factors that recognize yet-unidentified cis-acting sites. In addition, proteins that interact with the hypervariable minisatellite DNA hotspot have been purified (Wahls et al., 1991) and circumstantial evidences suggests that protein binding is required for activity (W.P. Wahls and P.D. Moore, unpublished). An uncomplicated model would propose that discrete DNA-binding proteins are responsible for hotspot activation.
Attempts to identify discrete DNA elements that activate hotspots in S. cerevisiae have been largely unsuccessful, suggesting that hotspot activity is not conferred by a single protein binding to a discrete DNA site (Rocco et al., 1992; de Massy and Nicolas, 1993; Goyon and Lichten, 1993). Similarly, comparison of DNA sequences in the vicinity of dsDNA breaks failed to reveal any clear consensus sequences at the site of the breaks (de Massy et al., 1995). The prevailing view is that the dsDNA breaks at recombination hotspots are dictated by some feature of the overall chromatin structure (reviewed by Lichten and Goldman, 1995). It is not clear why some regions of open chromatin contain hotspots and others do not, but the answer is likely to be found within the individual protein components of chromatin. For example, proteins binding near yeast hotspots can influence hotspot activity. Full activity of the HIS4 hotspot in S. cerevisiae requires binding of Bas1, Bas2 and Rap1 transcription factors to the promoter region, but none of the individual factors seems responsible for complete hotspot activation (White et al., 1991; White et al., 1993). Deletion of all the binding sites for all these transcription factors abolishes hotspot activity and replacement with two Rap1 binding sites restores high hotspot activity, suggesting that Rap1 protein binding might be sufficient to activate the hotspot (Fan et al., 1995). The production of dsDNA breaks might be due to direct interaction of a meiosis-specific nuclease with one or a few transcription factors that are bound within regions of open chromatin (Fan and Petes, 1996; Kon et al., submitted). Alternatively, the topoisomerase II-like nuclease that makes the dsDNA break may have a loose consensus site that must be present within open chromatin for cleavage. Thus, other regions of open chromatin that lack the requisite subset of transcription factors or consensus sequence for the nuclease would not be recombinogenic. The lack of clear consensus sequences at dsDNA break locations (de Massy et al., 1995) favors the first possibility.
One particularly well-studied eukaryotic recombination hotspot is the M26 hotspot of the fission yeast, Schizosaccharomyces pombe. M26 was first identified as an ade6 mutant that exhibited a high frequency of meiotic recombination within the ade6 gene (Gutz, 1971). The M26 mutation is a single base pair substitution (Ponticelli et al., 1988; Szankasi et al., 1988) that increases meiotic recombination within ade6 about 15-fold relative to the nearby control allele, M375 (Gutz, 1971; Schuchert and Kohli, 1988). The hotspot functions during meiosis but not during mitosis (Gutz, 1971; Ponticelli et al., 1988; Schuchert and Kohli, 1988) and exerts its effect in the vicinity of ade6 , as well as in nearby intervals (Grimm et al., 1990), but not at other more-distant or unlinked loci (Ponticelli et al., 1988). M26 stimulates both gene conversion (Gutz, 1971) and reciprocal exchange events (Grimm et al., 1994) to an approximately equal extent, but during gene conversion there is strong disparity with the M26 allele being converted to wild-type about 10-fold higher than vice versa (i.e., M26 acts as a recipient of information during gene conversion events) (Gutz, 1971). Gene conversion events at M26 often coconvert alleles to the left, right, or both directions and the average conversion tract length is about 700 base pairs, consistent with the notion that M26 is an initiator of recombination (Gutz, 1971; Grimm et al., 1994). Preferential transfer of the transcribed strand occurs (Schar and Kohli, 1994), suggesting that the mechanism of hotspot activation does not involve extensive resection of both DNA strands from a dsDNA break. These data are consistent with M26 being the site of a ssDNA or dsDNA break that initiates recombination. Other data suggest that recombination may not initiate precisely at M26. M26-enhanced recombination events do not always include conversion of the M26 site (Grimm et al., 1990) and an M26-independent recombination initiation site near the 5′ end of the ade6 gene has been inferred (Grimm et al., 1994), suggesting that M26 may enhance recombination that initiates near M26 but not necessarily within the site itself.
Kohli and colleagues used in vitro mutagenesis to alter individual base pairs in the vicinity of the M26 site, they introduced the mutations into the ade6 gene and they then tested the resulting mutants for hotspot activity in vivo (Schuchert et al., 1991). This revealed that a specific 7 base pair DNA sequence, 5′-ATGACGT-3′, is absolutely required for activity of the M26 hotspot, suggesting that the M26 mutation might create a binding site for a protein involved in hotspot activation. (Such single base pair changes may be the best way to identify cis-acting sites involved in recombination at other hotspots because more extensive alterations, such as deletions, could perturb a local chromatin conformation that is necessary but not sufficient for hotspot activity. It may therefore be worthwhile to use a similar point-mutation approach to search for cis-acting sites near other hotspots.) A heterodimeric protein that binds to the M26 site has been identified and purified and the binding activity in vitro correlates with hotspot activity in vivo (Wahls and Smith, 1994). Binding of the heterodimer is essential for hotspot activation but not for basal recombination levels (Kon et al., submitted). Intriguingly, the Mts1/Mts2 heterodimer is also a putative transcription factor that has roles in sexual development and stress response (Takeda et al., 1995; Shiozaki and Russell, 1996; Watanabe and Yamamoto, 1996; Wilkinson et al., 1996). However, hotspot activity is not directly related to transcription because there is no detectable difference in the level of ade6 mRNA during meiosis in strains exhibiting normal versus hotspot recombination (Grimm et al., 1991). (Similarly, for hotspots in S. cerevisiae some promoter mutations that alter gene expression do not alter hotspot activity (Schultes and Szostak, 1991; White et al., 1992)). Mts1/Mts2 protein therefore has multiple biological roles: one in hotspot activation, one in regulating genes involved in sexual development, and one in regulating genes during stress conditions. It is proposed that the Mts1/Mts2 heterodimer has evolved separate roles in transcriptional regulation and hotspot activation (Kon et al., submitted). In each case, the protein is constitutively present (Wahls and Smith, 1994) but apparently inactive during normal mitotic growth, permitting rapid induction of activity in times of crisis (such as nitrogen starvation which leads to induction of meiotic differentiation). Activation of the protein involves signal transduction via MAP kinase and cAMP-dependent kinase pathways, and perhaps others (Takeda et al., 1995; Shiozaki and Russell, 1996; Watanabe and Yamamoto, 1996; Wilkinson et al., 1996) (N. Kon and W.P. Wahls, unpublished).
While DNA breaks at the M26 hotspot have not been reported, M26 site-dependent, meiotically-induced open chromatin is present at the M26 site and in the promoter region (Mizuno et al., 1997). A promoter deletion in cis to the M26 hotspot, ending 112 base pairs upstream of the M26 site, abolishes hotspot activity, while a promoter deletion in trans to M26 has no effect (Zahn-Zabal et al., 1995). Thus, in addition to Mts1/Mts2 binding to the M26 site, some other feature removed by the promoter deletion is required for hotspot activation. Remodeling of meiotic chromatin at M26 requires Mts1/Mts2 protein (K. Ohta and W.P. Wahls, unpublished observations), suggesting that Mts1/Mts2 protein binding, possibly interacting with a factor bound in the promoter region, is responsible for the open chromatin structure and hotspot activation. It remains to be seen whether Mts1/Mts2 recruits recombination enzymes via protein-protein interactions, or whether recombination enzymes assemble at chromatin conformationally altered by Mts1/Mts2 binding to M26 DNA. In either case, binding of Mts1/Mts2 to M26 sites increases recombination above basal levels, suggesting that the hotspot with its binding protein helps regulate where and when recombination occurs within the genome.
The M26 heptamer is about five-fold underrepresented in the S. pombe genome. If each naturally-occurring M26 site is, on average, as active in promoting recombination as the M26 site in the ade6 gene (Gutz, 1971; Schuchert and Kohli, 1988), then about 50% of all meiotic recombination events could be due to Mts1/Mts2 heterodimer binding those sites (Wahls and Smith, 1994). This proposal, while speculative, is testable. So far no study has reported whether or not any other, naturally-occurring M26 sites exhibit hotspot activity. However, intergenic recombination is reduced in mts mutants (N. Kon and W.P. Wahls, unpublished) and the M26 heptamer has hotspot activity when created at any of five locations in ade6 and ura4 (M.E. Fox, J.B. Virgin, J. Metzger, G.R. Smith, pers. comm.). These findings support the view that natural M26 sites are active in meiosis and contribute to elevated meiotic recombination levels throughout the genome.
VI. Control of Recombination in Cis and in Trans, Near and Far
The control of recombination likely involves a complex network of structural and enzymatic machinery. Hotspots act locally to enhance recombination events within a few tens of thousands of base pairs of their location. As discussed above, hotspots can be regulated positively and negatively by both cis- and trans-acting factors. On a wider scale, the timing and distribution of recombination is also regulated. Two examples of this control are provided here.
On the chromosome-wide scale, the distribution of recombination events is non-random. It has long been recognized that chiasmata (the cytological structures thought to represent points of reciprocal exchange between chromosomes) are not randomly distributed (reviewed by Maguire, 1995). Individual chromosomes have some means of insuring that at least 1, but not many, chiasmata occur on each chromosome arm. That regulation is likely conferred (at least in part) by structural proteins of the synaptonemal complex (Sym and Roeder, 1994; Kleckner, 1996). Presence of chiasmata usually correlates with appropriate segregation of chromosomes (see Hawley, Chapter _____). This correlation is supported by genetic observations in yeast where smaller chromosomes have higher rates of recombination than larger chromosomes, when adjusted for the physical size of the chromosomes (Kaback et al., 1992). Thus, recombination rates and distributions are controlled relative to individual chromosomes.
In the fission yeast S. pombe 18 complementation groups of mutants affecting meiotic recombination have been identified (Schmidt et al., 1987; Ponticelli and Smith, 1989; DeVeaux et al., 1992; Tavassoli et al., 1995). Three of the mutants, rec8, rec10 and rec11, profoundly reduce recombination over all tested intervals on chromosome III, but they leave recombination essentially normal on the other 2 chromosomes (DeVeaux and Smith, 1994). Since these genes are recessive and are encoded on 3 different chromosomes, they must encode trans-activators of recombination that recognize specific cis-acting determinants of chromosome III. The gene products are likely involved in either the initial pairing of chromosome III, which is required for meiotic recombination, or they are involved in chromosome III-specific recombination, perhaps by recognizing DNA sequences such as recombination hotspots that are preferentially located on chromosome III.
Additional examples of genome-wide regulation of recombination may be found in a recent review article (Lichten and Goldman, 1995)
VII. Hotspots as Initiators or Resolvers of Recombination: Two Models
A. Recombination hotspots enhancing an early step in recombination: convergence of genetics and biochemistry
The dsDNA break model of recombination (Resnick, 1976; Szostak et al., 1983) can explain most recombination in budding yeast and this pathway is likely conserved in other eukaryotes (Bergerat et al., 1997; Keeney et al., 1997). The modified model in Fig. 3 incorporates resection of the 5′ terminated strands at dsDNA breaks (Sun et al., 1989; Cao et al., 1990; Sun et al., 1991), a role for DNA accessibility within chromatin (Shenkar et al., 1991; Ohta et al., 1994; Wu and Lichten, 1994; Wu and Lichten, 1995), a requirement for specific transcription factors but not for transcription (Fan and Petes, 1996; Kon et al., submitted), and the recently-discovered biochemical activity of Spo11 protein as the nuclease that forms dsDNA breaks (Bergerat et al., 1997; Keeney et al., 1997).
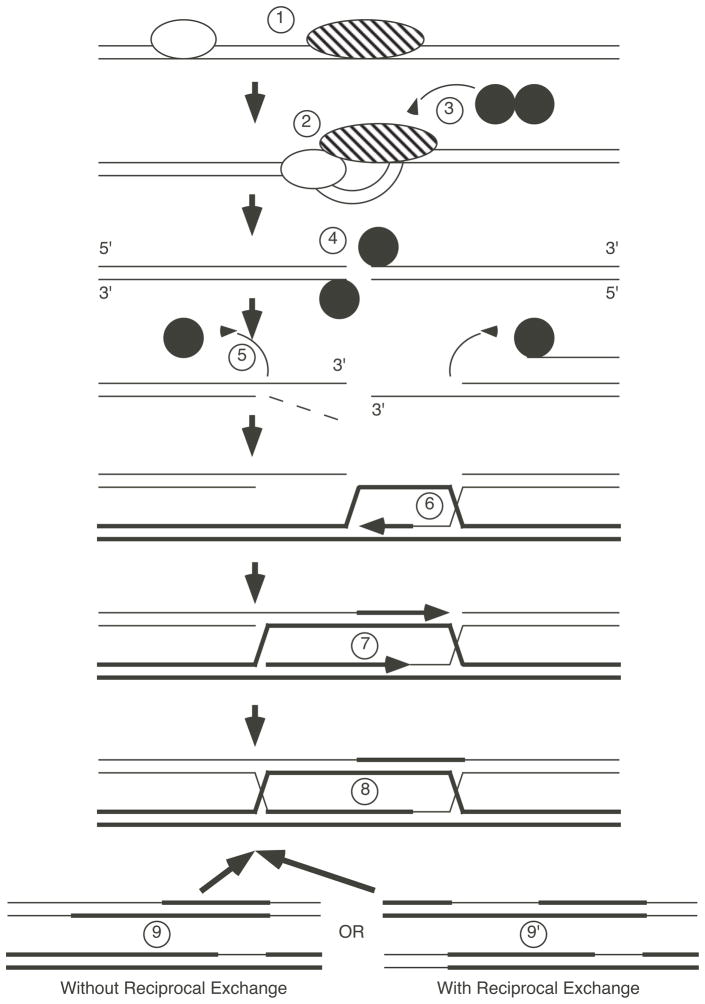
Figure 3. A dsDNA break repair model of recombination.
1. Mitotic chromosomes with associated DNA binding proteins (balloons). 2. During meiosis, the binding proteins are altered by post-translational modification or by interaction with meiotic factors to generate chromatin in which the DNA becomes more accessible. 3. Spo11 protein (filled circle) assembles at the site, either via protein-protein interactions or by recognizing an altered chromatin conformation at the hotspot. 4. A topoisomerase II-like, dsDNA cleavage reaction by Spo11 protein produces covalent protein-DNA intermediates with phosphodiester linkages between active site tyrosines and the 5′ terminal phosphate groups of the broken strands. 5. Release of protein is achieved by hydrolysis of the phosphotyrosine linkage or by nucleolytic cleavage of the covalently-bound strand (step 5′). Exonucleolytic resection (dotted line) generates free, 3′ ssDNA tails which can invade the homologous dsDNA to produce heteroduplex DNA. 6. The invading strand is extended by DNA synthesis (arrow) to produce a displacement loop. 7. Annealing of the remaining free 3′ strand to the displacement loop and further DNA synthesis fills the existing gaps. 8. Ligation of DNA strands results in chromosomes attached by two Holliday junctions. Branch migration of the junctions may occur (not shown). The junctions are resolved by cleavage of two DNA strands. Depending upon which strands are cleaved, the resulting chromosomes either lack reciprocal exchange of flanking markers (step 9) or have reciprocal exchange (step 9′). Additional details and references are provided in the text.
In step “1” chromosomes have associated DNA binding proteins involved in gene expression and a characteristic mitotic chromatin configuration. For example, the Mts1/Mts2 hotspot-activating protein of fission yeast is uniformly expressed and has equivalent DNA binding activity in mitotic and meiotic cells (Wahls and Smith, 1994), presumably acting as a transcription factor (Takeda et al., 1995; Watanabe and Yamamoto, 1996), but the M26 hotspot functions only during meiosis (Gutz, 1971; Schuchert and Kohli, 1988; Ponticelli and Smith, 1989). Similarly, multifunctional, constitutively-expressed RAP1 protein of budding yeast activates a meiotic hotspot (White et al., 1991; White et al., 1993). These observations suggest that the hotspot-activating proteins are always interacting with their DNA sites, but during meiosis some change in these proteins (or their environment) triggers recombinogenic activity.
During meiosis, as in step “2,” post-translational modification of the DNA binding proteins occurs. It is proposed that the machinery of eukaryotic recombination has evolved to use some transcription factors for induction of meiotic recombination at hotspots (Kon et al., submitted). Two transcription factors are known to fully activate meiotic recombination hotspots (White et al., 1993; Wahls and Smith, 1994; Kon et al., submitted), however, recombination-enhancing and transcription-regulating activities are separable (Grimm et al., 1991; Schultes and Szostak, 1991; White et al., 1992). The protein changes involve signal transduction via MAP kinase and cAMP-dependent kinase pathways, and perhaps others (Shiozaki and Russell, 1996; Wilkinson et al., 1996). Alteration of the DNA binding proteins either directly or indirectly results in a conformational change in the surrounding chromatin, making the DNA more accessible to nuclease digestion (Shenkar et al., 1991; Ohta et al., 1994; Wu and Lichten, 1994; Wu and Lichten, 1995; Mizuno et al., 1997). Current evidence is consistent with open chromatin being either the cause or the effect of hotspot activation.
In step “3” the meiosis-specific, topoisomerase II homolog Spo11 arrives at the hotspot and introduces a dsDNA break (Bergerat et al., 1997; Keeney et al., 1997). It remains to be seen whether Spo11 is recruited directly via protein-protein interactions with the transcription factors bound at the hotspot, or whether Spo11 assembles at chromatin that is conformationally altered by the hotspot binding proteins. In either case, in step “4” two subunits of Spo11 cleave the DNA by transesterase reactions in which active site tyrosines make closely-spaced ssDNA nicks and concomitantly form phosphodiester bonds with the 5′ termini of the broken DNA (Bergerat et al., 1997; Keeney et al., 1997). Because the energy of strand scission is retained in the phospho-tyrosine linkage, the dsDNA break could be religated during protein release. (Genetic evidence suggests this may occur: A high frequency of dsDNA breaks created by insertion of a RAP1 binding site within HIS4 does not have genetic hotspot activity when cis-linked to a natural hotspot located nearby, presumably because of competition between the two sites (Fan et al., 1997)). Alternatively, the covalent intermediate is further processed in the recombination pathway.
Release of the covalently-bound protein likely occurs by one of two mechanisms: Hydrolysis of the phosphodiester bond (step “5”) releases intact Spo11 monomer and leaves behind a dsDNA break that is subsequently processed by exonucleolytic resection of the 5′ terminated strand (Sun et al., 1989; Cao et al., 1990; Sun et al., 1991). Alternatively, Spo11 is released by endonucleolytic cleavage of one DNA strand, in cis to the bound protein, that produces a variable length ssDNA fragment that remains covalently-attached to the released Spo11 monomer (step “5 prime”). Genetic and biochemical studies suggest that these reactions are catalyzed by some combination of the Rad50, Mre11 and Xrs2 proteins. A more detailed discussion of this possibility is available (Keeney et al., 1997).
The remaining steps of this model are as proposed (Szostak et al., 1983) and are only briefly outlined here. For the sake of clarity, only two of the four chromatids are shown. One of the free, 3′-terminated, ssDNA ends at the dsDNA break invades the homologous chromosome to form heteroduplex DNA (step “6”). DNA synthesis extrudes a displacement loop that can serve as a template for DNA synthesis primed by the remaining 3′ ssDNA tail (step “7”). Ligation of DNA nicks (step “8”) and cleavage of the resulting Holliday junctions results in recombinant chromosomes without (“9”) or with (“9 prime”) reciprocal exchange of the flanking markers.
B. Recombination hotspots enhancing a late step in recombination: hypervariable minisatellite DNA as a site of resolution of Holliday junctions
Recombination hotspots enhance some rate-limiting step in the biochemical pathway of recombination. The prevailing view, as exemplified for recombination via dsDNA break repair in S. cerevisiae, is that the rate-limiting step is early, presumably acting as the initiating step. However, as with other biochemical pathways, the rate-limiting step need not be early. I propose that some eukaryotic recombination hotspots act late, as resolvers of recombination intermediates. This model can be summarized by the following hypothesis: Hypervariable minisatellite DNA is a site of resolution of Holliday junctions. I initially proposed this model at the 11th International Chromosome Conference (Edinburgh, 1992) to account for the observation of polarized changes at minisatellite DNA, presented at the meeting (Armour et al., 1993b), and the observation that minisatellites enhance recombination and have an additive influence when dsDNA breaks are introduced into the substrates (Wahls et al., 1990a). A number of findings since then further support the model shown in Fig. 4.
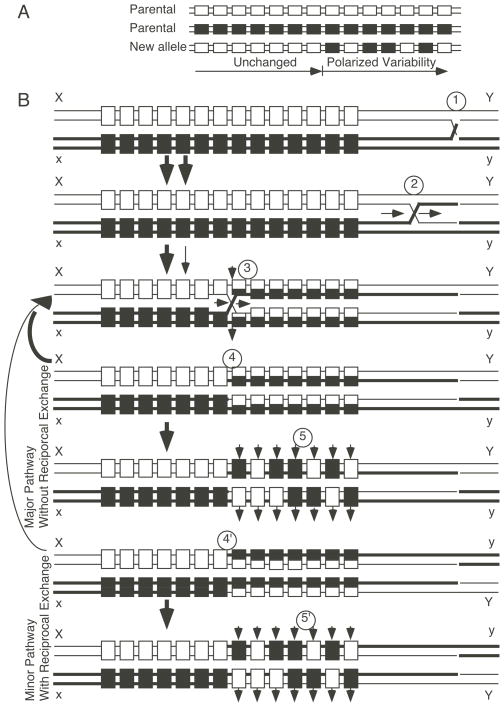
Figure 4. A model for recombination hotspot activity and formation of polarized variability at hypervariable minisatellite DNA.
(A) Schematic representation of polarized variability. Individual minisatellite repeats (boxes) can be sometimes be distinguished by minor sequence differences within each repeat (black or white). Arising alleles are uniform across most of the locus and then become mosaic towards one end of the locus. (B) Model for recombination hotspot activity and generation of polarized variability. 1. Initiating event leads to strand exchange and generation of a Holliday junction. 2. Branch migration of the Holliday junction. 3. An element within each minisatellite DNA repeat provides the signal to resolve the Holliday junction. Resolution can occur by the major pathway (nick strands at horizontal arrows) or by the minor pathway (nick strands at vertical arrows). 4. Resolution by the major pathway yields recombinants with heteroduplex DNA and no reciprocal exchange of flanking markers. 5. Short patch mismatch repair (up and down arrows) acts independently on each repeat to generate polarized variability. Loci with incomplete mismatch repair will exhibit postmeiotic segregation (not shown). 4′. Resolution by the minor pathway yields recombinants with heteroduplex DNA and reciprocal exchange of flanking markers. 5′. Short patch mismatch repair of the minor product also generates polarized variability. Additional details and references are provided in the text.
New minisatellite alleles can arise at some loci in as many as 10% of meioses (Buard and Vergnaud, 1994) and alleles often exhibit polarized variability (Armour et al., 1993a; Armour et al., 1993b; Buard and Vergnaud, 1994; Ellsworth et al., 1995). The polarized variability is shown schematically in Fig. 4A. Since reciprocal exchange of markers flanking minisatellite loci is seldom observed (Wolff et al., 1989; Kasperczyk et al., 1990; Kelly et al., 1991; Buard and Vergnaud, 1994), most models for minisatellite change have discounted recombination in favor of other, more complex mechanisms. The model proposed in Fig. 4B can account for all of the change via meiotic recombination and associated heteroduplex DNA repair.
In step “1” recombination initiates at some point distal to the minisatellite array but proximal to the nearest flanking marker, the “Y” locus. For the sake of clarity, only two of the four chromatids are shown. Minisatellite enhanced recombination can have at least one endpoint in a distant interval (Wahls et al., 1990a) and allelic changes at minisatellite arrays can be strongly influenced by cis-acting sites outside of the array (Monckton et al., 1994; Andreassen et al., 1996). The initiating lesion may be promoted by minisatellite DNA binding proteins (Wahls et al., 1991) or it may involve other cis- and trans-acting factors such as those inferred for hotspots in the mouse MHC (Shiroishi et al., 1991; Shiroishi et al., 1995). Open chromatin has been found near the minisatellite DNA in the Eβ hotspot in the mouse MHC (Shenkar et al., 1991), suggesting that the initiating event may is analogous to those at hotspots in yeast. Symmetrical ssDNA nicks are shown as per the Holliday model of recombination (Holliday, 1964), however, other types of initiating events (Meselson and Radding, 1975; Szostak et al., 1983) will work equally well as long as they lead to formation of at least one Holliday junction between the recombining chromosomes.
In step “2” the recombining chromosomes have exchanged single strands to form a Holliday junction and branch migration has moved the junction half way to the minisatellite array. On naked DNA a random walk can result in the junction moving in either direction. In E. coli the RuvAB heteromultimer catalyzes unidirectional migration of the junction (reviewed by Muller and West, 1994) and presumably eukaryotic counterparts carry out the same process. The extent of branch migration is dictated by the processivity of the enzyme. Branch migration might be reversed by dissociation and reassociation of the machinery in the opposite orientation, which would lead to “aborted” recombination events. Thus, in the absence of resolution , the Holliday junction is removed by reverse migration or it is pushed farther along the recombining chromosomes until it encounters a signal for resolution, the minisatellite DNA.
In step “3” branch migration has generated heteroduplex DNA that extends from the flanking region into the minisatellite array. Some component within each minisatellite DNA repeat acts as a signal for resolution of the Holliday junction. The signal is quite strong but not absolute, because polarized variability can occur within a few tens of repeats from the end of the minisatellite array (Armour et al., 1993a). I calculate that there is a probability of between 5% and 10% that a Holliday junction that is migrating past a minisatellite repeat unit will be resolved at that site. Thus, about 50% of resolutions will occur within the first ten repeats, about 75% within the first twenty repeats, and so on, reflecting the distribution of polarized changes that has been described. This proposal is considerably strengthened by recent findings of how the E. coli Holliday junction resolving enzyme, RuvC, functions. RuvC preferentially resolves Holliday junctions at a consensus sequence in an orientation-dependent fashion leading to preferential cleavage in one of two possible directions (Shah et al., 1994). That also supports the next step of the model, which is that resolution of the Holliday junction in the “patch” configuration (strand nicks at horizontal arrows) occurs with higher frequency than resolution in the “splice” configuration (strand nicks at vertical arrows).
In step “4,” the major pathway by which the junction is resolved, cleavage of two strands in the horizontal orientation allows separation of the recombining chromatids. There is no reciprocal exchange of the flanking markers distal to the minisatellite locus. A “patch” of heteroduplex DNA extends from the point of recombination initiation to the point of resolution within the minisatellite array. When enhancing recombination between plasmids, minisatellite DNA preferentially increases recombination events accompanied by reciprocal exchange (Wahls et al., 1990a). In the model, preferential increase of non-reciprocal resolution is proposed. The factors that dictate resolution in this orientation are unknown, but they may be contained within the minisatellite repeat itself or they may be conferred by the higher-order structure of the recombining chromosomes. In yeast, proteins that are central components of the synaptonemal complex, such as Zip1, clearly influence the outcome of recombination, but not necessarily its frequency (Sym et al., 1993). Yeast with the zip1 mutation have nearly-normal recombination frequencies, but they no longer exhibit crossover interference (Sym and Roeder, 1994). A recent model proposes that the physical constraints of the synaptonemal complex, specifically the tension created by initial steps of recombination within the semi-rigid structure, leads to reciprocal exchanges which alleviate the built up tension (Kleckner, 1996). Similar forces could be operating during recombination at the minisatellite loci. Resolution of the Holliday junction, signaled by the minisatellite DNA and catalyzed by interacting proteins, might be influenced by constraints of the surrounding chromatin.
In step “5” the resulting heteroduplex DNA is corrected by mismatch correction. The best described mismatch repair systems in mammalian cells have very short (a few nucleotides) excision tracts, but there is evidence for long patch repair (reviewed by Friedberg et al., 1995). Heteroduplex DNA that contains mismatches within the minisatellite loci is repaired by the short patch mismatch repair machinery. Each minisatellite repeat is corrected individually to excise mismatched nucleotides on the upper strand (upward-pointing arrows) or on the lower strand (downward-pointing arrows). This would generate the polarized variability. The observation that hypervariable minisatellite DNA undergoes a high rate of repair in mammalian meiosis (Ramachandra and Rao, 1994) supports the model. Furthermore, there is genetic evidence for heteroduplex DNA at minisatellites during meiosis: The rate of meiotic change of minisatellites is high (Buard and Vergnaud, 1994) but the rate of change in mitosis is very low. Intriguingly, “somatic mutation during early development” has been reported (Kelly et al., 1989; Gibbs et al., 1993). The Hm-2 locus experiences a high rate of allelic change per meiosis and, surprisingly, about 20% of mice with new variants of the Hm-2 locus are mosaic. Based upon the pattern of mosaicism, it was concluded that somatic mutant alleles arise within the first two cell divisions after fertilization (Kelly et al., 1989; Gibbs et al., 1993). I suggest that the mosaicism is actually due to postmeiotic segregation (PMS) of uncorrected heteroduplex DNA that is formed during meiotic recombination. Homotypic variants arise from recombination events with complete mismatch correction of heteroduplex DNA. The remaining new variants undergo partial repair of heteroduplex DNA and produce PMS that, upon fertilization, generates mosaic conceptuses, further supporting a role for short patch mismatch correction.
In step “4 prime” the minor pathway by which the junction is resolved, cleavage of two strands in the vertical orientation allows separation of the recombining chromatids. This results in a recombination event in which there is reciprocal exchange of the markers flanking the minisatellite locus.
In step “5 prime” the processing of the heteroduplex DNA is exactly as for step “4.” Tracts of short patch mismatch repair can independently correct each minisatellite repeat, with a random probability that either the top or the bottom strand will be excised and replaced at each repeat. As with the non-reciprocal recombinants, the reciprocal recombinants will have polarized variability.
The model in Figure 4 is drawn with an equal number of repeats in each allele for the sake of clarity. Individual alleles often have different numbers of repeats, so what is the consequence of recombination between two alleles of different lengths? If the flanking sequences to the right of the minisatellite array are identical for both alleles (as in recombination between sister chromatids), then the Holliday junction would encounter minisatellite DNA simultaneously on both strands. Recombination would proceed as in Figure 4 and the recombinant products would exhibit polarized variability for any internal allelic differences in the parental molecules, but no length change. These recombination events would be invisible to typical experimental approaches that score minisatellite change based upon the appearance of new length alleles.
If the allele lengths are different or the neighboring sequences are different (e.g., one allele extends farther into the adjacent DNA than the other), then strand exchange would likely bypass the heterology and branch migration would continue into the minisatellite array before encountering the signal to resolve the Holliday junction. The resulting heteroduplex DNA would be the same as shown in Figure 4 except the right end would have additional minisatellite DNA repeats in one strand opposite a strand of non-minisatellite DNA (a loop opposite a shorter opposing strand could also form). Mismatch repair could replace either strand. Thus, individual DNA strands within the heteroduplex DNA at the minisatellite array may become longer, shorter, or stay the same length, depending upon the initial minisatellite lengths and the direction of the mismatch correction. This mechanism can therefore generate new length alleles as well as account for polarized variability. However, it depends upon pre-existing length polymorphism within the population. How is the length polymorphism generated?
Most models for minisatellite length change invoke a combination of duplications and deletions involving both inter- and intra-chromosomal interactions (Buard and Vergnaud, 1994) and additional independent mutations leading to polarized variability (Ellsworth et al., 1995). Length changes are sometimes attributed to DNA polymerase slippage on the template strand during DNA replication followed by failure of mismatch repair, as is the case for expansion and contraction of dinucleotide repeats (Strand et al., 1993; Farber et al., 1994; Heale and Petes, 1995; Kolodner, 1995). This seems unlikely for minisatellites because they are relatively stable mitotically and they undergo a high rate of length change during meiosis. There is no evidence for reduced fidelity of replicative DNA polymerases during meiosis, nor any reason to believe that mismatch repair would be compromised. I propose that some length changes are due to DNA polymerase slippage on the template strand, but these events do not use the replicative DNA polymerases. Rather, the strand slippage probably occurs when repair-specific DNA polymerases help correct heteroduplex DNA present after resolution of recombination within minisatellite loci. The high rate of DNA repair at minisatellite DNA in mammalian meiosis, which seems to use DNA polymerase β (Ramachandra and Rao, 1994), supports this model.
In conclusion, most of the complex changes that occur at hypervariable minisatellite DNA may be due to minisatellites acting as sites where Holliday junctions are resolved. The polarity of change, the variability of change, the changes previously attributed to somatic mutation during early development, and allele expansion and contraction can all be accounted for by a model invoking meiotic homologous recombination followed by heteroduplex DNA repair. This suggests that rate-limiting steps can occur late in pathway of recombination as well as early. While primarily a meiotic event, the process of minisatellite recombination might also occur between sister chromatids or homologous chromosomes during mitotic growth. This would provide a mechanism for recombinational repair of DNA damage that preferentially uses gene conversion. If so, it would also provide a mechanism for some aberrant recombination events. For example, several independent translocation breakpoints at c-myc and bcl2 occurred precisely at minisatellite consensus sequences (Krowczynska et al., 1990), strongly suggesting that the aberrant recombination was resolved (or initiated) at the minisatellite DNA.
VIII. Summary
Recombination is increased up to 1000-fold during meiosis in order to promote accurate segregation of homologous chromosomes and to enhance genetic diversity. Part of this induction is due to recombination hotspots and their interacting factors. For at least one eukaryotic hotspot a discrete DNA sequence and interacting protein is required for activity (Schuchert et al., 1991; Kon et al., submitted) and for another hotspot a specific protein (presumably binding a discrete DNA site) is required (White et al., 1993; Fan et al., 1995). For other hotspots no single, discrete activating factors have been identified, but this could be due to redundancy of sites clustered near hotspots or limitations of the experimental approaches used. It remains to be seen whether hotspot activation is regulated specifically and independently, or whether it is a consequence of other biological processes such as transcription.
Yeast are genetically parsimonious eukaryotes that have evolved to the point of having about a 50% gene density. Meiotic recombination initiates at dsDNA breaks that appear in regions of increased accessibility of DNA within chromatin. While hotspots are usually found in the vicinity of transcriptional promoters, the emerging view is that recombination is not directly linked to transcription. Rather, it seems likely that some component of the transcription machinery, most likely one or a subset of transcription factors, has evolved dual roles in transcription and recombination (Fan and Petes, 1996; Kon et al., submitted). It remains to be seen whether hotspot binding proteins directly recruit recombination enzymes via protein-protein interactions, or whether recombination enzymes assemble at conformational changes in chromatin conferred by hotspot binding proteins. The recent discovery of a topoisomerase II-like nuclease that makes the dsDNA break (Bergerat et al., 1997; Keeney et al., 1997), and conservation of this protein between distant species, suggests that the initiation of recombination via dsDNA breaks is evolutionarily conserved. Identification of the factors that direct this nuclease to recombination hotspots and elucidation of the biochemical steps following strand scission are reasonable near-term objectives. In the longer term, study of yeast hotspots will reveal other biochemical activities in the pathway of recombination and yield information about whether recombination throughout the genome is regulated predominantly by hotspots.
Mammals, in contrast to experimental organisms such as yeast, have a very low gene density, an abundance of non-coding DNA, and a life cycle that makes genetic experimentation time-consuming and expensive. Despite the challenges of conducting such experiments, compelling evidence suggests cis- and trans-acting regulation of recombination at discrete sites in the mammalian genome, recombination hotspots. Furthermore, it seems that the fundamental mechanism of meiotic recombination in all metazoans is similar to that in yeast. Recombination probably initiates at dsDNA breaks and the basic molecular mechanisms are likely conserved. It is proposed that one type of non-coding, repetitive DNA found in metazoans, hypervariable minisatellite DNA, acts late in the pathway of meiotic recombination as a signal to resolve Holliday junctions. It is tempting to speculate that other types of noncoding DNA in metazoan genomes have additional roles in regulating the pairing, recombination and segregation of meiotic chromosomes.
Acknowledgments
I thank Bernard de Massy, Scott Keeney and Nancy Kleckner, Mary Fox and Gerry Smith and Tom Petes for sharing results prior to publication. I also thank my readers and colleagues for constructive comments and Mary Ann Handel for her hard work and patience in putting together this volume. This work was supported by the Vanderbilt University Department of Biochemistry and a grant from the National Institutes of Health. W.P.W. is a Leukemia Society of America Special Fellow.
IX. References
- Ajimura M, Leem SH, Ogawa H. Identification of new genes required for meiotic recombination in Saccharomyces cerevisiae. Genetics. 1993;133:51–66. doi: 10.1093/genetics/133.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alani E, Padmore R, Kleckner N. Analysis of wild-type and rad50 mutants of yeast suggests an intimate relationship between meiotic chromosome synapsis and recombination. Cell. 1990;61:419–36. doi: 10.1016/0092-8674(90)90524-i. [DOI] [PubMed] [Google Scholar]
- Andersson L, Lunden A, Sigurdardottir S, Davies CJ, Rask L. Linkage relationships in the bovine MHC region. High recombination frequency between class II subregions. Immunogenetics. 1988;27:273–80. doi: 10.1007/BF00376122. [DOI] [PubMed] [Google Scholar]
- Andreassen R, Egeland T, Olaisen B. Mutation rate in the hypervariable VNTR g3 (D7S22) is affected by allele length and a flanking DNA sequence polymorphism near the repeat array. Am J Human Genet. 1996;59:360–7. [PMC free article] [PubMed] [Google Scholar]
- Armour JA, Harris PC, Jeffreys AJ. Allelic diversity at minisatellite MS205 (D16S309): evidence for polarized variability. Human Mol Genet. 1993a;2:1137–45. doi: 10.1093/hmg/2.8.1137. [DOI] [PubMed] [Google Scholar]
- Armour JAL, Moncton DG, Neil DL, Crosier M, Tamaka K, Macleod A, Jeffreys AJ. Minisatellite mutation and recombination. In: Sumner AT, Chandley AC, editors. Chromosomes Today. Chapman & Hall; London: 1993b. pp. 337–350. [Google Scholar]
- Bell GI, Selby MJ, Rutter WJ. The highly polymorphic region near the human insulin gene is composed of simple tandemly repeating sequences. Nature. 1982;295:31–5. doi: 10.1038/295031a0. [DOI] [PubMed] [Google Scholar]
- Benson FE, Stasiak A, West SC. Purification and characterization of the human Rad51 protein, an analogue of E. coli RecA. EMBO Journal. 1994;13:5764–71. doi: 10.1002/j.1460-2075.1994.tb06914.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergerat A, de Massy B, Gadelle D, Varoutas P-C, Nicolas A, Forterre P. An atypical topoisomerase II from archaea with implication for meiotic recombination. Nature. 1997 doi: 10.1038/386414a0. In press. [DOI] [PubMed] [Google Scholar]
- Bishop DK. RecA homologs Dmc1 and Rad51 interact to form multiple nuclear complexes prior to meiotic chromosome synapsis. Cell. 1994;79:1081–92. doi: 10.1016/0092-8674(94)90038-8. [DOI] [PubMed] [Google Scholar]
- Bishop DK, Park D, Xu L, Kleckner N. DMC1: a meiosis-specific yeast homolog of E. coli recA required for recombination, synaptonemal complex formation, and cell cycle progression. Cell. 1992;69:439–56. doi: 10.1016/0092-8674(92)90446-j. [DOI] [PubMed] [Google Scholar]
- Blaho JA, Wells RD. Left-handed Z-DNA binding by the recA protein of Escherichia coli [published erratum appears in J. Biol Chem 1988 263, 11015] J Biol Chem. 1987;262:6082–8. [PubMed] [Google Scholar]
- Bryda EC, DePari JA, Sant’Angelo DB, Murphy DB, Passmore HC. Multiple sites of crossing over within the Eβ recombinational hotspot in the mouse. Mammalian Genome. 1992;2:123–9. doi: 10.1007/BF00353860. [DOI] [PubMed] [Google Scholar]
- Buard J, Vergnaud G. Complex recombination events at the hypermutable minisatellite CEB1 (D2S90) EMBO Journal. 1994;13:3203–10. doi: 10.1002/j.1460-2075.1994.tb06619.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullard SA, Kim S, Galbraith AM, Malone RE. Double strand breaks at the HIS2 recombination hot spot in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1996;93:13054–13059. doi: 10.1073/pnas.93.23.13054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullock P, Miller J, Botchan M. Effects of poly[d(pGpT). d(pApC)] and poly[d(pCpG).d(pCpG)] repeats on homologous recombination in somatic cells. Mol Cell Biol. 1986;6:3948–53. doi: 10.1128/mcb.6.11.3948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao L, Alani E, Kleckner N. A pathway for generation and processing of double-strand breaks during meiotic recombination in S. cerevisiae. Cell. 1990;61:1089–101. doi: 10.1016/0092-8674(90)90072-m. [DOI] [PubMed] [Google Scholar]
- Capon DJ, Chen EY, Levinson AD, Seeburg PH, Goeddel DV. Complete nucleotide sequences of the T24 human bladder carcinoma oncogene and its normal homologue. Nature. 1983;302:33–7. doi: 10.1038/302033a0. [DOI] [PubMed] [Google Scholar]
- Catcheside DG. The Genetics of Recombination. University Park Press; Baltimore, MD: 1977. [Google Scholar]
- Catcheside DG, Angel T. A histidine-3 mutant, in Neurospora crassa, due to an interchange. Aust J Biol Sci. 1974;27:219–229. doi: 10.1071/bi9740219. [DOI] [PubMed] [Google Scholar]
- Chandley AC, Mitchell AR. Hypervariable minisatellite regions are sites for crossing-over at meiosis in man. Cytogenet & Cell Genet. 1988;48:152–5. doi: 10.1159/000132613. [DOI] [PubMed] [Google Scholar]
- Chaudhuri S, Messing J. Genetic analysis of a new mutation conferring cysteine auxotrophy in Saccharomyces cerevisiae: updating of the sulfer metabolism pathway. Genetics. 1992;130:51–58. doi: 10.1093/genetics/130.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collick A, Dunn MG, Jeffreys AJ. Minisatellite binding protein Msbp-1 is a sequence-specific single-stranded DNA-binding protein. Nucleic Acids Res. 1991;19:6399–404. doi: 10.1093/nar/19.23.6399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creighton HB, McClintock B. A correlation of cytologicla and genetical crossing-over in Zea mays. Proc Natl Acad Sci USA. 1931;17 doi: 10.1073/pnas.17.8.492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Massy B, Nicolas A. The control in cis of the position and the amount of the ARG4 meiotic double-strand break of Saccharomyces cerevisiae. EMBO J. 1993;12:1459–66. doi: 10.1002/j.1460-2075.1993.tb05789.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Massy B, Rocco V, Nicolas A. The nucleotide mapping of DNA double-strand breaks at the CYS3 initiation site of meiotic recombination in Saccharomyces cerevisiae. EMBO J. 1995;14:4589–98. doi: 10.1002/j.1460-2075.1995.tb00138.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Detloff P, White MA, Petes TD. Analysis of a gene conversion gradient at the HIS4 locus in Saccharomyces cerevisiae. Genetics. 1992;132:113–23. doi: 10.1093/genetics/132.1.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeVeaux LC, Hoagland NA, Smith GR. Seventeen complementation groups of mutations decreasing meiotic recombination in Schizosaccharomyces pombe. Genetics. 1992;130:251–62. doi: 10.1093/genetics/130.2.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeVeaux LC, Smith GR. Region-specific activators of meiotic recombination in Schizosaccharomyces pombe. Genes & Dev. 1994;8:203–10. doi: 10.1101/gad.8.2.203. [DOI] [PubMed] [Google Scholar]
- Dubbs DR, Rachmeler M, Kit S. Recombination between temperature-sensitive mutants of simian virus 40. Virology. 1974;57:161–174. doi: 10.1016/0042-6822(74)90117-2. [DOI] [PubMed] [Google Scholar]
- Edelmann W, Kroger B, Goller M, Horak I. A recombination hotspot in the LTR of a mouse retrotransposon identified in an in vitro system. Cell. 1989;57:937–46. doi: 10.1016/0092-8674(89)90332-2. [DOI] [PubMed] [Google Scholar]
- Ellsworth DL, Shriver MD, Boerwinkle E. Nucleotide sequence analysis of the apolipoprotein B 3′ VNTR. Human Mol Genet. 1995;4:937–44. doi: 10.1093/hmg/4.5.937. [DOI] [PubMed] [Google Scholar]
- Fan Q, Xu F, Petes TD. Meiosis-specific double-strand DNA breaks at the HIS4 recombination hot spot in the yeast Saccharomyces cerevisiae: control in cis and trans. Mol Cell Biol. 1995;15:1679–88. doi: 10.1128/mcb.15.3.1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Q-Q, Xu F, White MA, Petes TD. Competition between adjacent meiotic recombination hotspots in the yeastSaccharomyces cerevisiae. Genetics. 1997 doi: 10.1093/genetics/145.3.661. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan QQ, Petes TD. Relationship between nuclease-hypersensitive sites and meiotic recombination hot spot activity at the HIS4 locus of Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:2037–43. doi: 10.1128/mcb.16.5.2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farber RA, Petes TD, Dominska M, Hudgens SS, Liskay RM. Instability of simple sequence repeats in a mammalian cell line. Human Mol Genet. 1994;3:253–6. doi: 10.1093/hmg/3.2.253. [DOI] [PubMed] [Google Scholar]
- Fishel RA, Detmer K, Rich A. Identification of homologous pairing and strand-exchange activity from a human tumor cell line based on Z-DNA affinity chromatography. Proc Natl Acad Sci USA. 1988;85:36–40. doi: 10.1073/pnas.85.1.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fogel S, Mortimer RK, Lusnak K. Meiotic gene conversion in yeast tetrrads and the theory of recombination. Genetics. 1967;57:455–481. doi: 10.1093/genetics/57.2.455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fogel S, Mortimer RK, Lusnak K. Mechanisms of meiotic gene conversion, or "wanderings on a foreign strand". In: Broach JR, Jones EW, Strathern JN, editors. The Molecular Biology of the Yeast Saccharomyces. Cold Spring Harbor Laboratory; Cold Spring Harbor: 1981. pp. 289–339. [Google Scholar]
- Friedberg EC, Walker GC, Siede W. DNA Repair and Mutagenesis. American Society for Microbiology; Washington, D.C: 1995. p. 698. [Google Scholar]
- Game JC. Pulse-field gel analysis of the pattern of DNA double-strand breaks in Saccharomyces genome during meiosis. Dev Genet. 1992;13:485–497. doi: 10.1002/dvg.1020130610. [DOI] [PubMed] [Google Scholar]
- Gibbs M, Collick A, Kelly RG, Jeffreys AJ. A tetranucleotide repeat mouse minisatellite displaying substantial somatic instability during early preimplantation development. Genomics. 1993;17:121–8. doi: 10.1006/geno.1993.1292. [DOI] [PubMed] [Google Scholar]
- Goller M, Funke B, Gehe-Becker C, Kroger B, Lottspeich F, Horak I. Murine protein which binds preferentially to oligo-C-rich single-stranded nucleic acids. Nucleic Acids Res. 1994;22:1885–9. doi: 10.1093/nar/22.10.1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goyon C, Lichten M. Timing of molecular events in meiosis in Saccharomyces cerevisiae: stable heteroduplex DNA is formed late in meiotic prophase. Mol Cell Biol. 1993;13:373–82. doi: 10.1128/mcb.13.1.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green M, Krontiris TG. Allelic variation of reporter gene activation by the HRAS1 minisatellite. Genomics. 1993;17:429–34. doi: 10.1006/geno.1993.1343. [DOI] [PubMed] [Google Scholar]
- Grimm C, Bahler J, Kohli J. M26 recombinational hotspot and physical conversion tract analysis in the ade6 gene of Schizosaccharomyces pombe. Genetics. 1994;136:41–51. doi: 10.1093/genetics/136.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimm C, Munz P, Kohli J. The recombinational hot spot mutation ade6-M26 of Schizosaccharomyces pombe stimulates recombination at sites in a nearby interval. Curr Genet. 1990;18:193–7. doi: 10.1007/BF00318379. [DOI] [PubMed] [Google Scholar]
- Grimm C, Schaer P, Munz P, Kohli J. The strong ADH1 promoter stimulates mitotic and meiotic recombination at the ADE6 gene of Schizosaccharomyces pombe. Mol Cell Biol. 1991;11:289–98. doi: 10.1128/mcb.11.1.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross DS, Garrard WT. The ubiquitous potential Z-forming sequence of eukaryoties, (dT-dG)n-(dC-dA)n, is not detectable in the genomes of eubacteria, archaebacteria, or mitochondria. Mol Cell Biol. 1986;6:3010–3013. doi: 10.1128/mcb.6.8.3010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutz H. Site specific induction of gene conversion in Schizosaccharomyces pombe. Genetics. 1971;69:331–337. doi: 10.1093/genetics/69.3.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heale SM, Petes TD. The stabilization of repetitive tracts of DNA by variant repeats requires a functional DNA mismatch repair system. Cell. 1995;83:539–45. doi: 10.1016/0092-8674(95)90093-4. [DOI] [PubMed] [Google Scholar]
- Henke J, Henke L. Recent observations in human DNA-minisatellite mutations. Int J Legal Med. 1995;107:204–8. doi: 10.1007/BF01428407. [DOI] [PubMed] [Google Scholar]
- Hepkema BG, Tilanus MG, Blankert HJ, Albers GA, Grosfeld-Stulemeyer MC, Hensen EJ. A high recombination frequency within the chicken major histocompatibility (B) complex. Animal Genet. 1993;24:389–91. doi: 10.1111/j.1365-2052.1993.tb00346.x. [DOI] [PubMed] [Google Scholar]
- Higgs DR, Goodbourn SE, Wainscoat JS, Clegg JB, Weatherall DJ. Highly variable regions of DNA flank the human alpha globin genes. Nucleic Acids Res. 1981;9:4213–24. doi: 10.1093/nar/9.17.4213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holliday R. A mechanism for gene conversion in fungi. Genet Res. 1964;78:273–287. doi: 10.1017/S0016672308009476. [DOI] [PubMed] [Google Scholar]
- Ivanov EL, Korolev VG, Fabre F. XRS2, a DNA repair gene of Saccharomyces cerevisiae, is needed for meiotic recombination. Genetics. 1992;132:651–64. doi: 10.1093/genetics/132.3.651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaeger U, Karth GD, Knapp S, Friedl J, Laczika K, Kusec R. Molecular mechanism of the t(14;18)--a model for lymphoid-specific chromosomal translocations. Leukemia & Lymphoma. 1994;14:197–202. doi: 10.3109/10428199409049669. [DOI] [PubMed] [Google Scholar]
- Jeffreys AJ, Wilson V, Thein SL. Hypervariable ‘minisatellite’ regions in human DNA. Nature. 1985a;314:67–73. doi: 10.1038/314067a0. [DOI] [PubMed] [Google Scholar]
- Jeffreys AJ, Wilson V, Thein SL. Individual-specific ‘fingerprints’ of human DNA. Nature. 1985b;316:76–9. doi: 10.1038/316076a0. [DOI] [PubMed] [Google Scholar]
- Johzuka K, Ogawa H. Interaction of Mre11 and Rad50: two proteins required for DNA repair and meiosis-specific double-strand break formation in Saccharomyces cerevisiae. Genetics. 1995;139:1521–32. doi: 10.1093/genetics/139.4.1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaback DB, Guacci V, Barber D, Mahon JW. Chromosome size-dependent control of meiotic recombination. Science. 1992;256:228–32. doi: 10.1126/science.1566070. [DOI] [PubMed] [Google Scholar]
- Kasperczyk A, DiMartino NA, Krontiris TG. Minisatellite allele diversification: the origin of rare alleles at the HRAS1 locus. Am J Human Genet. 1990;47:854–9. [PMC free article] [PubMed] [Google Scholar]
- Keeney S, Giroux CN, Kleckner N. Meiosis-specific DNA double-strand breaks are catalyzed by Spo11, a member of a widely conserved protein family. Cell. 1997;88:375–384. doi: 10.1016/s0092-8674(00)81876-0. [DOI] [PubMed] [Google Scholar]
- Keeney S, Kleckner N. Covalent protein-DNA complexes at the 5′ strand termini of meiosis-specific double-strand breaks in yeast. Proc Natl Acad Sci USA. 1995;92:11274–8. doi: 10.1073/pnas.92.24.11274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly R, Bulfield G, Collick A, Gibbs M, Jeffreys AJ. Characterization of a highly unstable mouse minisatellite locus: evidence for somatic mutation during early development. Genomics. 1989;5:844–56. doi: 10.1016/0888-7543(89)90126-2. [DOI] [PubMed] [Google Scholar]
- Kelly R, Gibbs M, Collick A, Jeffreys AJ. Spontaneous mutation at the hypervariable mouse minisatellite locus Ms6-hm: flanking DNA sequence and analysis of germline and early somatic mutation events. Proc Royal Soc of London Series B: Biological Sciences. 1991;245:235–45. doi: 10.1098/rspb.1991.0115. [DOI] [PubMed] [Google Scholar]
- Kitani Y, Olive LS. Genetics of Sordaria fimicolaVI Gene conversion at the g locus in mutant x wild-type crosses. Genetics. 1967;57:767–782. doi: 10.1093/genetics/57.4.767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klar AJ, Bonaduce MJ. swi6, a gene required for mating-type switching, prohibits meiotic recombination in the mat2-mat3 "cold spot" of fission yeast. Genetics. 1991;129:1033–42. doi: 10.1093/genetics/129.4.1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleckner N. Meiosis: How could it work? Proc Natl Acad Sci USA. 1996;93:8167–8174. doi: 10.1073/pnas.93.16.8167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klysik J, Stirdivant SM, Larson JE, Hart PA, Wells RD. Left-handed DNA in restriction fragments and a recombinant plasmid. Nature. 1981;290:672–7. doi: 10.1038/290672a0. [DOI] [PubMed] [Google Scholar]
- Kmiec EB, Holloman WK. Heteroduplex formation and polarity during strand transfere promoted by Ustilago rec1 protein. Cell. 1983;33:857–864. doi: 10.1016/0092-8674(83)90028-4. [DOI] [PubMed] [Google Scholar]
- Kmiec EB, Holloman WK. Synapsis promoted by Ustilago rec1 protein. Cell. 1984;36:593–598. doi: 10.1016/0092-8674(84)90338-6. [DOI] [PubMed] [Google Scholar]
- Kmiec EB, Holloman WK. Homologous pairing of DNA molecules by Ustilago rec1 protein is promoted by sequences of Z-DNA. Cell. 1986;44:545–54. doi: 10.1016/0092-8674(86)90264-3. [DOI] [PubMed] [Google Scholar]
- Kobori JA, Strauss E, Minard K, Hood L. Molecular analysis of the hotspot of recombination in the murine major histocompatibility complex. Science. 1986;234:173–9. doi: 10.1126/science.3018929. [DOI] [PubMed] [Google Scholar]
- Kobori JA, Winoto A, McNicholas J, Hood L. Molecular characterization of the recombination region of six murine major histocompatibility complex (MHC) I-region recombinants. J Mol Cell Immunol. 1984;1:125–35. [PubMed] [Google Scholar]
- Kolodner RD. Mismatch repair: mechanisms and relationship to cancer susceptibility. Trends in Biochem Sci. 1995;20:397–401. doi: 10.1016/s0968-0004(00)89087-8. [DOI] [PubMed] [Google Scholar]
- Kon N, Krawchuk MD, Warren BG, Smith GR, Wahls WP. Transcription factor Mts1/Mts2 (Atf1/Pcr1) activates the M26 meiotic recombination hotspot in S. pombe. doi: 10.1073/pnas.94.25.13765. submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalczykowski SC, Dixon DA, Eggleston AK, Lauder SD, Rehrauer WM. Biochemistry of homologous recombination in Escherichia coli. Microbiol Rev. 1994;58:401–65. doi: 10.1128/mr.58.3.401-465.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krowczynska AM, Rudders RA, Krontiris TG. The human minisatellite consensus at breakpoints of oncogene translocations. Nucleic Acids Res. 1990;18:1121–7. doi: 10.1093/nar/18.5.1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafuse WP, David CS. Recombination hot spots within the I region of the mouse H-2 complex map to the E beta and E alpha genes. Immunogenetics. 1986;24:352–60. doi: 10.1007/BF00377952. [DOI] [PubMed] [Google Scholar]
- Lafuse WP, Lee ST, David CS. Molecular analysis of two recombinant mouse strains with crossovers in the E alpha recombination hot spot. J Immunogenetics. 1990;17:169–76. doi: 10.1111/j.1744-313x.1990.tb00869.x. [DOI] [PubMed] [Google Scholar]
- Lancillotti F, Lopez MC, Arias P, Alonso C. Z-DNA in transcriptionally active chromosomes. Proc Natl Acad Sci USA. 1987;84:1560–1564. doi: 10.1073/pnas.84.6.1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leach DRF. Genetic Recombination. Blackwell Science Ltd; Oxford: 1996. p. 192. [Google Scholar]
- Leem SH, Ogawa H. The MRE4 gene encodes a novel protein kinase homologue required for meiotic recombination in Saccharomyces cerevisiae. Nucleic Acids Res. 1992;20:449–57. doi: 10.1093/nar/20.3.449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichten M, Goldman ASH. Meiotic recombination hotspts. Annu Rev Genet. 1995;29:423–444. doi: 10.1146/annurev.ge.29.120195.002231. [DOI] [PubMed] [Google Scholar]
- Lin Y, Smith GR. Transient, meiosis-induced expression of the rec6 and rec12 genes of Schizosaccharomyces pombe. Genetics. 1994;136:769–79. doi: 10.1093/genetics/136.3.769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lissouba P, Rizet G. Sur l’existaence d’une unite génétique polarisée ne subissant que des échanges non réciproques. CR Acad Sci, Paris. 1960;250:3408–3410. [PubMed] [Google Scholar]
- Liu J, Wu TC, Lichten M. The location and structure of double-strand DNA breaks induced during yeast meiosis: evidence for a covalently linked DNA-protein intermediate. EMBO J. 1995;14:4599–608. doi: 10.1002/j.1460-2075.1995.tb00139.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maguire MP. Is the synaptonemal complex a disjunction machine? J Heredity. 1995;86:330–40. doi: 10.1093/oxfordjournals.jhered.a111600. [DOI] [PubMed] [Google Scholar]
- Malone RE, Bullard S, Lundquist S, Kim S, Tarkowski T. A meiotic gene conversion gradient opposite to the direction of transcription. Nature. 1992;359:154–5. doi: 10.1038/359154a0. [DOI] [PubMed] [Google Scholar]
- Martin M, Mann D, Carrington M. Recombination rates across the HLA complex: use of microsatellites as a rapid screen for recombinant chromosomes. Human Mol Genet. 1995;4:423–8. doi: 10.1093/hmg/4.3.423. [DOI] [PubMed] [Google Scholar]
- McAdam SN, Boyson JE, Liu X, Garber TL, Hughes AL, Bontrop RE, Watkins DI. A uniquely high level of recombination at the HLA-B locus. Proc Natl Acad Sci USA. 1994;91:5893–7. doi: 10.1073/pnas.91.13.5893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClintock B. A cytological and genetical study of triploid maize. Genetics. 1929;14:180–122. doi: 10.1093/genetics/14.2.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClintock B. Neurospora. I Preliminary observations on the chromosomes of Neurospora crassa . Am J Bot. 1945;32:671–678. [Google Scholar]
- Melmer G. Hybridization of the variable number of tandem repeat core oligonucleotide GNNGTGGG to 350 cosmids from chromosome 7. Electrophoresis. 1994;15:175–6. doi: 10.1002/elps.1150150130. [DOI] [PubMed] [Google Scholar]
- Menees TM, Ross-MacDonald PB, Roeder GS. MEI4, a meiosis-specific yeast gene required for chromosome synapsis. Mol Cell Biol. 1992;12:1340–51. doi: 10.1128/mcb.12.3.1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meselson MS, Radding CM. A general model for genetic recombination. Proc Natl Acad Sci USA. 1975;72:358–61. doi: 10.1073/pnas.72.1.358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meuth M, Nalban-Toflu J, Phear D, Miles C. Banbury Report 28: Mammalian Cell Mutagenesis. Cold Spring Harbor Laboratory; Cold Spring Harbor: 1987. Molecular basis of genome rearrangements at the hamster aprt locus; pp. 183–191. [Google Scholar]
- Mizuno K, Koide T, Sagai T, Moriwaki K, Shiroishi T. Molecular analysis of a recombinational hotspot adjacent to Lmp2 gene in the mouse MHC: fine location and chromatin structure. Mammalian Genome. 1996;7:490–6. doi: 10.1007/s003359900149. [DOI] [PubMed] [Google Scholar]
- Mizuno K-I, Emura Y, Baur M, Kohli J, Ohta K, Shibata T. Remodeling of chromatin structure around a single nucleotide mutation in ade6-M26 that creates a meiotic recombination hotspot in fission yeast. Genes & Dev. 1997 doi: 10.1101/gad.11.7.876. in press. [DOI] [PubMed] [Google Scholar]
- Monckton DG, Neumann R, Guram T, Fretwell N, Tamaki K, MacLeod A, Jeffreys AJ. Minisatellite mutation rate variation associated with a flanking DNA sequence polymorphism. Nature Genetics. 1994;8:162–70. doi: 10.1038/ng1094-162. [DOI] [PubMed] [Google Scholar]
- Moore SP, Erdile L, Kelly T, Fishel R. The human homologous pairing protein HPP-1 is specifically stimulated by the cognate single-stranded binding protein hRP-A. Proc Natl Acad Sci USA. 1991;88:9067–71. doi: 10.1073/pnas.88.20.9067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore SP, Fishel R. Purification and characterization of a protein from human cells which promotes homologous pairing of DNA. J Biol Chem. 1990;265:11108–17. [PubMed] [Google Scholar]
- Mosig G. Distances separating genetic markers in T4 DNA. Proc Natl Acad Sci, USA. 1966;56:1177–1183. doi: 10.1073/pnas.56.4.1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller B, West SC. Processing of Holliday junctions by the Escherichia coli RuvA, RuvB, RuvC and RecG proteins. Experientia. 1994;50:216–22. doi: 10.1007/BF01924004. [DOI] [PubMed] [Google Scholar]
- Murray NE. Complementation and recombination between methionine-1 alleles in Neurospora crassa. Genetics. 1960;48:1163–1183. doi: 10.1093/genetics/48.9.1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray NE. Polarized intragenic recombination in chromosome rearrangements in Neurospora. Genetics. 1968;58:181–191. doi: 10.1093/genetics/58.2.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nag DK, Petes TD. Physical detection of heteroduplexes during meiotic recombination in the yeast Saccharomyces cerevisiae. Mol Cell Biol. 1993;13:2324–31. doi: 10.1128/mcb.13.4.2324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolas A, Petes TD. Polarity of meiotic gene conversion in fungi: contrasting views. Experientia. 1994;50:242–52. doi: 10.1007/BF01924007. [DOI] [PubMed] [Google Scholar]
- Nicolas A, Treco D, Schultes NP, Szostak JW. An initiation site for meiotic gene conversion in the yeast Saccharomyces cerevisiae. Nature. 1989;338:35–9. doi: 10.1038/338035a0. [DOI] [PubMed] [Google Scholar]
- Ohta K, Shibata T, Nicolas A. Changes in chromatin structure at recombination initiation sites during yeast meiosis. EMBO J. 1994;13:5754–63. doi: 10.1002/j.1460-2075.1994.tb06913.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr-Weaver TL, Szostak JW, Rothstein RJ. Yeast transformation: a model system for the study of recombination. Proc Natl Acad Sci USA. 1981;78:6354–8. doi: 10.1073/pnas.78.10.6354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr-Weaver TL, Szostak JW, Rothstein RJ. Genetic applications of yeast transformation with linear and gapped plasmids. Methods in Enzymology. 1983;101:228–45. doi: 10.1016/0076-6879(83)01017-4. [DOI] [PubMed] [Google Scholar]
- Pees E. Genetic fine structure and polarized negative interference at the lys-51 (FL) locus of Aspergillus nidulans. Genetica. 1966;38:275–304. [PubMed] [Google Scholar]
- Petes TD, Malone RE, Symington LS. Recombination in yeast. In: Broach JR, Jones EW, Pringle JR, editors. The Molecular and Cellular Biology of the Yeast Saccharomyces. Cold Spring Harbor Laboratory; Cold Spring Harbor: 1991. pp. 407–521. [Google Scholar]
- Ponticelli AS, Sena EP, Smith GR. Genetic and physical analysis of the M26 recombination hotspot of Schizosaccharomyces pombe. Genetics. 1988;119:491–7. doi: 10.1093/genetics/119.3.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponticelli AS, Smith GR. Meiotic recombination-deficient mutants of Schizosaccharomyces pombe. Genetics. 1989;123:45–54. doi: 10.1093/genetics/123.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramachandra L, Rao MR. Identification and sequence characterization of a 1.3 Kb EcoRI repeat fragment that harbors a DNA repair site of rat pachytene spermatocytes. Chromosoma. 1994;103:486–501. doi: 10.1007/BF00337387. [DOI] [PubMed] [Google Scholar]
- Resnick MA. The repair of double-strand breaks in DNA; a model involving recombination. J Theor Biol. 1976;59:97–106. doi: 10.1016/s0022-5193(76)80025-2. [DOI] [PubMed] [Google Scholar]
- Resnick MA, Zgaga Z, Hieter P, Westmoreland J, Fogel S, Nilsson-Tillgren T. Recombinant repair of diverged DNAs: a study of homoeologous chromosomes and mammalian YACs in yeast. Mol Gen Genet. 1992;234:65–73. doi: 10.1007/BF00272346. [DOI] [PubMed] [Google Scholar]
- Rocco V, de Massy B, Nicolas A. The Saccharomyces cerevisiae ARG4 initiator of meiotic gene conversion and its associated double-strand DNA breaks can be inhibited by transcriptional interference. Proc Natl Acad Sci USA. 1992;89:12068–72. doi: 10.1073/pnas.89.24.12068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockmill B, Engebrecht JA, Scherthan H, Loidl J, Roeder GS. The yeast MER2 gene is required for chromosome synapsis and the initiation of meiotic recombination. Genetics. 1995;141:49–59. doi: 10.1093/genetics/141.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savage EA, Hastings PJ. Marker effects and the nature of the recombination event at the HIS1 locus of Saccharomyces cerevisiae. Curr Genet. 1981;3:37. doi: 10.1007/BF00419579. [DOI] [PubMed] [Google Scholar]
- Schar P, Kohli J. Preferential strand transfer and hybrid DNA formation at the recombination hotspot ade6-M26 of Schizosaccharomyces pombe. EMBO J. 1994;13:5212–9. doi: 10.1002/j.1460-2075.1994.tb06852.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt H, Kapitza P, Gutz H. Switching genes in Schizosaccharomyces pombe: their influence on cell viability and recombination. Curr Genet. 1987;11:303–308. [Google Scholar]
- Schuchert P, Kohli J. The ade6-M26 mutation of Schizosaccharomyces pombe increases the frequency of crossing over. Genetic. 1988;119:507–515. doi: 10.1093/genetics/119.3.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuchert P, Langsford M, Kaslin E, Kohli J. A specific DNA sequence is required for high frequency of recombination in the ade6 gene of fission yeast. EMBO J. 1991;10:2157–63. doi: 10.1002/j.1460-2075.1991.tb07750.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultes NP, Szostak JW. Decreasing gradients of gene conversion on both sides of the initiation site for meiotic recombination at the ARG4 locus in yeast. Genetics. 1990;126:813–822. doi: 10.1093/genetics/126.4.813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultes NP, Szostak JW. A poly(dAdT) tract is a component of the recombination initiation site at the ARG4 locus in Saccharomyces cerevisiae. Mol Cell Biol. 1991;11:322–328. doi: 10.1128/mcb.11.1.322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah R, Bennett RJ, West SC. Genetic recombination in E. coli: RuvC protein cleaves Holliday junctions at resolution hotspots in vitro. Cell. 1994;79:853–64. doi: 10.1016/0092-8674(94)90074-4. [DOI] [PubMed] [Google Scholar]
- Shenkar R, Shen MH, Arnheim N. DNase I-hypersensitive sites and transcription factor-binding motifs within the mouse E beta meiotic recombination hot spot. Mol Cell Biol. 1991;11:1813–9. doi: 10.1128/mcb.11.4.1813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinder GA, Manam S, Ledwith BJ, Nichols WW. Minisatellite DNA-binding proteins in mouse brain, liver, and kidney. Exp Cell Res. 1994;213:107–12. doi: 10.1006/excr.1994.1179. [DOI] [PubMed] [Google Scholar]
- Shinohara A, Ogawa H, Ogawa T. Rad51 protein involved in repair and recombination in S. cerevisiae is a RecA-like protein [published erratum appears in Cell 1992 71, following 180] Cell. 1992;69:457–70. doi: 10.1016/0092-8674(92)90447-k. [DOI] [PubMed] [Google Scholar]
- Shiozaki K, Russell P. Conjugation, meiosis, and the osmotic stress response are regulated by Spc1 kinase through Atf1 transcription factor in fission yeast. Genes & Dev. 1996;10:2276–2228. doi: 10.1101/gad.10.18.2276. [DOI] [PubMed] [Google Scholar]
- Shiroishi T, Hanzawa N, Sagai T, Ishiura M, Gojobori T, Steinmetz M, Moriwaki K. Recombinational hotspot specific to female meiosis in the mouse major histocompatibility complex. Immunogenetics. 1990;31:79–88. doi: 10.1007/BF00661217. [DOI] [PubMed] [Google Scholar]
- Shiroishi T, Koide T, Yoshino M, Sagai T, Moriwaki K. Hotspots of homologous recombination in mouse meiosis. Adv Biophys. 1995;31:119–32. doi: 10.1016/0065-227x(95)99387-5. [DOI] [PubMed] [Google Scholar]
- Shiroishi T, Sagai T, Hanzawa N, Gotoh H, Moriwaki K. Genetic control of sex-dependent meiotic recombination in the major histocompatibility complex of the mouse. EMBO J. 1991;10:681–6. doi: 10.1002/j.1460-2075.1991.tb07997.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiroishi T, Sagai T, Moriwaki K. A new wild-derived H-2 haplotype enhancing K-IA recombination. Nature. 1982;300:370–2. doi: 10.1038/300370a0. [DOI] [PubMed] [Google Scholar]
- Shiroishi T, Sagai T, Moriwaki K. Hotspots of meiotic recombination in the mouse major histocompatibility complex. Genetica. 1993;88:187–96. doi: 10.1007/BF02424475. [DOI] [PubMed] [Google Scholar]
- Singleton CK, Klysik J, Stirdivant SM, Wells RD. Left-handed Z-DNA is induced by supercoiling in physiological ionic conditions. Nature. 1982;299:312–6. doi: 10.1038/299312a0. [DOI] [PubMed] [Google Scholar]
- Smith GR. Conjugational recombination in E. coli: myths and mechanisms. Cell. 1991;64:19–27. doi: 10.1016/0092-8674(91)90205-d. [DOI] [PubMed] [Google Scholar]
- Soper LA, Stein MB, Passmore HC. The E beta hot spot of recombination in wild-derived natural recombinant MHC haplotypes. Cross-over site mapping and the identification of a 1.0-kb E beta deletion in the p and w14 haplotypes. J Immunol. 1988;140:984–90. [PubMed] [Google Scholar]
- Southern PJ, Berg P. Transformation of mammalian cells to anitbiotic resistance with a bacterial gene under control of SV40 early region promoter. J Mol Appl Genet. 1982;1:327–341. [PubMed] [Google Scholar]
- Steinmetz M, Stephan D, Fischer Lindahl K. Gene organization and recombinational hotspots in the murine major histocompatibility complex. Cell. 1986;44:895–904. doi: 10.1016/0092-8674(86)90012-7. [DOI] [PubMed] [Google Scholar]
- Strand M, Prolla TA, Liskay RM, Petes TD. Destabilization of tracts of simple repetitive DNA in yeast by mutations affecting DNA mismatch repair [published erratum appears in Nature 1994 368, 569] Nature. 1993;365:274–6. doi: 10.1038/365274a0. [DOI] [PubMed] [Google Scholar]
- Stringer JR. Recombination between poly[d(GT). d(CA)] sequences in simian virus 40-infected cultured cells. Mol Cell Biol. 1985;5:1247–59. doi: 10.1128/mcb.5.6.1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramani S, Seaton BL. Homologous recombination in mitotically dividing mammalian cells. In: Kucherlapati R, Smith GR, editors. Genetic Recombination. American Society for Microbiology; Washington, D.C: 1988. pp. 549–573. [Google Scholar]
- Subramani S, Southern PJ. Analysis of gene expression using simian virus 40 vectors. Anal Biochem. 1983;135:1–15. doi: 10.1016/0003-2697(83)90723-6. [DOI] [PubMed] [Google Scholar]
- Sun H, Treco D, Schultes NP, Szostak JW. Double-strand breaks at an initiation site for meiotic gene conversion. Nature. 1989;338:87–90. doi: 10.1038/338087a0. [DOI] [PubMed] [Google Scholar]
- Sun H, Treco D, Szostak JW. Extensive 3′-overhanging, single-stranded DNA associated with the meiosis-specific double-strand breaks at the ARG4 recombination initiation site. Cell. 1991;64:1155–61. doi: 10.1016/0092-8674(91)90270-9. [DOI] [PubMed] [Google Scholar]
- Sym M, Engebrecht JA, Roeder GS. ZIP1 is a synaptonemal complex protein required for meiotic chromosome synapsis. Cell. 1993;72:365–78. doi: 10.1016/0092-8674(93)90114-6. [DOI] [PubMed] [Google Scholar]
- Sym M, Roeder GS. Crossover interference is abolished in the absence of a synaptonemal complex protein. Cell. 1994;79:283–92. doi: 10.1016/0092-8674(94)90197-x. [DOI] [PubMed] [Google Scholar]
- Szankasi P, Heyer WD, Schuchert P, Kohli J. DNA sequence analysis of the ade6 gene of Schizosaccharomyces pombeWild-type and mutant alleles including the recombination host spot allele ade6-M26. J Mol Biol. 1988;204:917–25. doi: 10.1016/0022-2836(88)90051-4. [DOI] [PubMed] [Google Scholar]
- Szankasi P, Smith GR. A DNA exonuclease induced during meiosis of Schizosaccharomyces pombe. J Biol Chem. 1992;267:3014–23. [PubMed] [Google Scholar]
- Szankasi P, Smith GR. A role for exonuclease I from S. pombe in mutation avoidance and mismatch correction. Science. 1995;267:1166–9. doi: 10.1126/science.7855597. [DOI] [PubMed] [Google Scholar]
- Szostak JW, Orr-Weaver TL, Rothstein RJ, Stahl FW. The double-strand-break repair model for recombination. Cell. 1983;33:25–35. doi: 10.1016/0092-8674(83)90331-8. [DOI] [PubMed] [Google Scholar]
- Takeda T, Toda T, Kominami K, Kohnosu A, Yanagida M, Jones N. Schizosaccharomyces pombe atf1+ encodes a transcription factor required for sexual development and entry into stationary phase. EMBO J. 1995;14:6193–208. doi: 10.1002/j.1460-2075.1995.tb00310.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavassoli M, Shayeghi M, Nasim A, Watts FZ. Cloning and characterisation of the Schizosaccharomyces pombe rad32 gene: a gene required for repair of double strand breaks and recombination. Nucleic Acids Res. 1995;23:383–8. doi: 10.1093/nar/23.3.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thon G, Cohen A, Klar AJ. Three additional linkage groups that repress transcription and meiotic recombination in the mating-type region of Schizosaccharomyces pombe. Genetics. 1994;138:29–38. doi: 10.1093/genetics/138.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thon G, Klar AJ. Directionality of fission yeast mating-type interconversion is controlled by the location of the donor loci. Genetics. 1993;134:1045–54. doi: 10.1093/genetics/134.4.1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treco D, Arnheim The evolutionarily conserved repetitive sequence d(TG. AC)n promotes reciprocal exchange and generates unusual recombinant tetrads during yeast meiosis. Mol Cell Biol. 1986;6:3934–3947. doi: 10.1128/mcb.6.11.3934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trepicchio WL, Krontiris TG. Members of the rel/NF-kappa B family of transcriptional regulatory proteins bind the HRAS1 minisatellite DNA sequence. Nucleic Acids Res. 1992;20:2427–34. doi: 10.1093/nar/20.10.2427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Endert PM, Lopez MT, Patel SD, Monaco JJ, McDevitt HO. Genomic polymorphism, recombination, and linkage disequilibrium in human major histocompatibility complex-encoded antigen-processing genes. Proc Natl Acad Sci USA. 1992;89:11594–7. doi: 10.1073/pnas.89.23.11594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahls WP. Department of Microbiology and Immunology. University of Illinois at Chicago; Chicago: 1989. DNA sequences that stimulate homologous recombination in mammalian cells; p. 197. [Google Scholar]
- Wahls WP, Moore PD. Homologous recombination enhancement conferred by the Z-DNA motif d(TG)30 is abrogated by simian virus 40 T antigen binding to adjacent DNA sequences. Mol Cell Biol. 1990;10:794–800. doi: 10.1128/mcb.10.2.794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahls WP, Smith GR. A heteromeric protein that binds to a meiotic homologous recombination hot spot: correlation of binding and hot spot activity. Genes & Dev. 1994;8:1693–702. doi: 10.1101/gad.8.14.1693. [DOI] [PubMed] [Google Scholar]
- Wahls WP, Swenson G, Moore PD. Two hypervariable minisatellite DNA binding proteins. Nucleic Acids Res. 1991;19:3269–74. doi: 10.1093/nar/19.12.3269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahls WP, Wallace LJ, Moore PD. Hypervariable minisatellite DNA is a hotspot for homologous recombination in human cells. Cell. 1990a;60:95–103. doi: 10.1016/0092-8674(90)90719-u. [DOI] [PubMed] [Google Scholar]
- Wahls WP, Wallace LJ, Moore PD. The Z-DNA motif d(TG)30 promotes reception of information during gene conversion events while stimulating homologous recombination in human cells in culture. Mol Cell Biol. 1990b;10:785–93. doi: 10.1128/mcb.10.2.785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe Y, Yamamoto M. Schizosaccharomyces pombe pcr1+ encodes a CREB/ATF protein involved in regulation of gene expression for sexual development. Mol Cell Biol. 1996;16:704–711. doi: 10.1128/mcb.16.2.704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White JH, Lusnak K, Fogel S. Missmatch-specific post-meiotic segregation frequency in yeast suggests a heteroduplex recombination intermediate. Nature. 1985;315:350–352. doi: 10.1038/315350a0. [DOI] [PubMed] [Google Scholar]
- White MA, Detloff P, Strand M, Petes TD. A promoter deletion reduces the rate of mitotic, but not meiotic, recombination at the HIS4 locus in yeast. Curr Genet. 1992;21:109–16. doi: 10.1007/BF00318468. [DOI] [PubMed] [Google Scholar]
- White MA, Dominska M, Petes TD. Transcription factors are required for the meiotic recombination hotspot at the HIS4 locus in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1993;90:6621–5. doi: 10.1073/pnas.90.14.6621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White MA, Wierdl M, Detloff P, Petes TD. DNA-binding protein RAP1 stimulates meiotic recombination at the HIS4 locus in yeast. Proc Natl Acad Sci USA. 1991;88:9755–9. doi: 10.1073/pnas.88.21.9755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson MG, Samuels M, Takeda T, Toone WM, Shieh JC, Toda T, Millar JBA, Jones N. The Atf1 transcription factor is a target for the Sty1 stress-activted MAP kinase pathway in fission yeast. Genes & Dev. 1996;10:2289–2301. doi: 10.1101/gad.10.18.2289. [DOI] [PubMed] [Google Scholar]
- Wolff RK, Plaetke R, Jeffreys AJ, White R. Unequal crossingover between homologous chromosomes is not the major mechanism involved in the generation of new alleles at VNTR loci. Genomics. 1989;5:382–4. doi: 10.1016/0888-7543(89)90076-1. [DOI] [PubMed] [Google Scholar]
- Wu TC, Lichten M. Meiosis-induced double-strand break sites determined by yeast chromatin structure. Science. 1994;263:515–8. doi: 10.1126/science.8290959. [DOI] [PubMed] [Google Scholar]
- Wu TC, Lichten M. Factors that affect the location and frequency of meiosis-induced double-strand breaks in Saccharomyces cerevisiae. Genetics. 1995;140:55–66. doi: 10.1093/genetics/140.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyman A, White R. A highly polymorphic locus in human DNA. Proc Natl Acad Sci USA. 1980;77:6754–6758. doi: 10.1073/pnas.77.11.6754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L, Kleckner N. Sequence non-specific double-strand breaks and interhomolog interactions prior to double-strand break formation at a meiotic recombination hot spot in yeast. EMBO J. 1995;14:5115–28. doi: 10.1002/j.1460-2075.1995.tb00194.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshino M, Sagai T, Lindahl KF, Toyoda Y, Moriwaki K, Shiroishi T. Allele-dependent recombination frequency: homology requirement in meiotic recombination at the hot spot in the mouse major histocompatibility complex. Genomics. 1995;27:298–305. doi: 10.1006/geno.1995.1046. [DOI] [PubMed] [Google Scholar]
- Yoshino M, Sagai T, Lindahl KF, Toyoda Y, Shirayoshi Y, Matsumoto K, Sugaya K, Ikemura T, Moriwaki K, Shiroishi T. Recombination in the class III region of the mouse major histocompatibility complex. Immunogenetics. 1994a;40:280–6. doi: 10.1007/BF00189973. [DOI] [PubMed] [Google Scholar]
- Yoshino M, Sagai T, Lindahl KF, Toyoda Y, Shiroishi T, Moriwaki K. No dosage effect of recombinational hotspots in the mouse major histocompatibility complex. Immunogenetics. 1994b;39:381–9. doi: 10.1007/BF00176154. [DOI] [PubMed] [Google Scholar]
- Zahn-Zabal M, Lehmann E, Kohli J. Hot spots of recombination in fission yeast: inactivation of the M26 hot spot by deletion of the ade6 promoter and the novel hotspot ura4-aim. Genetics. 1995;140:469–78. doi: 10.1093/genetics/140.2.469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zenvirth D, Arbel T, Sherman A, Goldway M, Klein S, Simchen G. Multiple sites for double-strand breaks in whole meiotic chromosomes of Saccharomyces cerevisiae. EMBO J. 1992;11:3441–3447. doi: 10.1002/j.1460-2075.1992.tb05423.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerer EJ, Passmore HC. Structural and genetic properties of the Eb recombinational hotspot in the mouse. Immunogenetics. 1991;33:132–40. doi: 10.1007/BF00210827. [DOI] [PubMed] [Google Scholar]