Editing of the human genome to correct disease-causing mutations is a promising approach for the treatment of genetic disorders. Genome editing improves on simple gene replacement strategies by effecting in situ correction of a mutant gene, leading to restoration of normal gene function under the control of endogenous regulatory elements, and reducing risks associated with random insertion into the genome. Gene-specific targeting has historically been limited to mouse embryonic stem (ES) cells. The development of zinc finger nucleases (ZFNs) has permitted efficient genome editing in transformed and primary cells previously thought intractable to such genetic manipulation1. In vitro, ZFNs have been shown to promote efficient genome editing via homology-directed repair (HDR) by inducing a site-specific double-strand break (DSB) at a target locus2–4, but it is unclear whether ZFNs can induce DSBs and stimulate genome editing at a clinically meaningful level in vivo. Here we show that ZFNs are able to induce DSBs efficiently when delivered directly to mouse liver, and when co-delivered with an appropriately designed gene targeting vector, stimulate gene replacement through both homology-directed and homology-independent targeted gene insertion at the ZFN-specified locus (referred to here as gene targeting). Importantly, the level of gene targeting achieved is sufficient to correct the prolonged clotting times in a mouse model of hemophilia B, and remained persistent following induced liver regeneration. Thus, ZFN-driven gene correction can be achieved in vivo raising the possibility of genome editing as a viable strategy for the treatment of genetic disease.
Viral vector-mediated transfer of the wild-type (w.t.) copy of a gene that is defective in disease (gene replacement therapy) has been successfully performed in a variety of animal models, and humans5–9. However, disadvantages of gene replacement include risks related to insertional mutagenesis10–12, and loss of endogenous regulatory signals that control gene expression. Gene-specific targeting in mouse induced pluripotent stem (iPS) cells has highlighted the potential to overcome these challenges through ex vivo correction of a disease-causing mutation13, but the majority of genetic diseases affect organ systems where ex vivo manipulation of target cells is not feasible. One such organ is the liver, the major site of plasma protein synthesis including the blood coagulation factors. A model genetic disease for liver gene therapy is hemophilia B, which is caused by deficiency of blood coagulation factor IX (F.IX), encoded by the F9 gene. Most affected individuals have circulating F.IX levels below 1% of normal (5,000 ng/mL), but restoration to ~5% activity (250 ng/mL) converts severe hemophilia B to a mild form14. The majority of mutations in the F9 gene are distributed across the coding sequences for exons 2–8 (Figure 1a)15. Thus, specific targeting of any single mutant allele would not allow complete coverage of the wide spectrum of mutations found in the human population. However, ZFN-mediated targeting of a promoterless therapeutic gene fragment 2,16 (i.e. a partial cDNA preceded by a splice acceptor site) into the first intron of F9 would allow for splicing of a w.t. coding sequence with exon 1, leading to expression of functionally active F.IX and rescue of the defect caused by most mutations. We thus sought to investigate whether ZFNs combined with a targeting vector carrying the w.t. F9 exons 2–8 could induce gene targeting in vivo and correct a mutated F9 gene in situ.
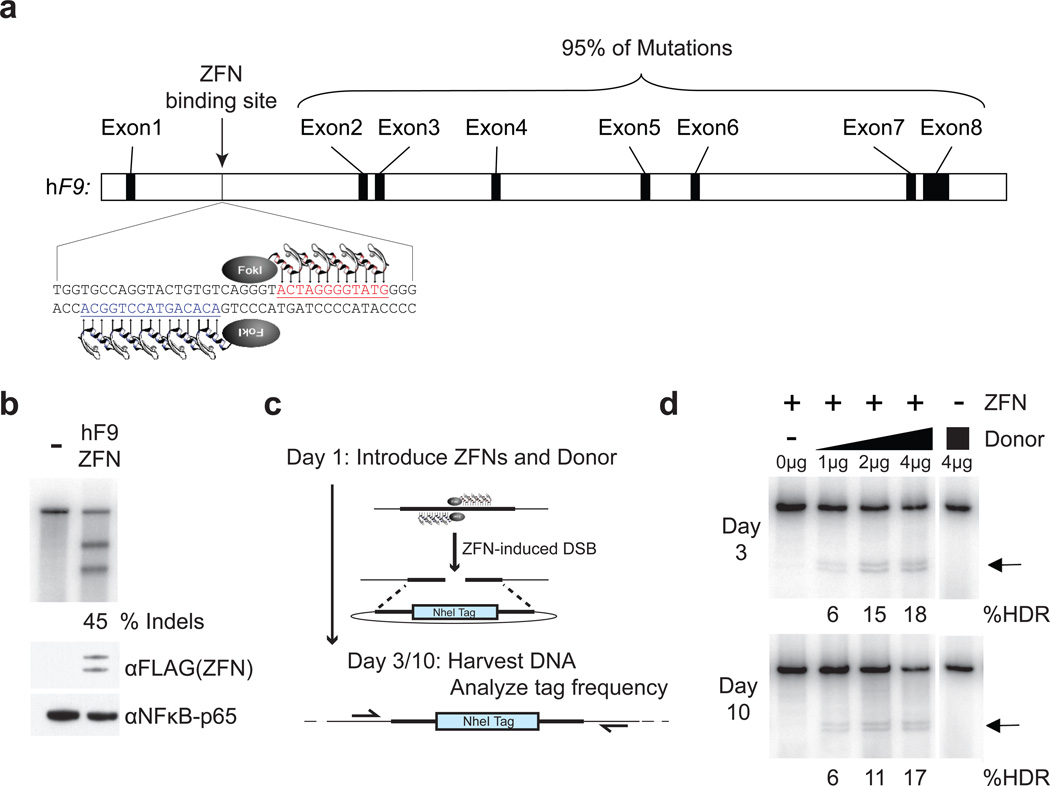
Figure 1. F9 ZFNs cleave human F9 intron 1 and induce homology-directed repair in vitro.
a, F9 ZFNs target intron 1 of the human F9 gene, allowing for homology-directed repair upstream of 95% of F9 mutations. b, ZFN expression constructs (400 ng) were transfected (right lane) or not (left lane) into K562 cells and genomic DNA was harvested 3 days post-transfection. The Cel-I assay was used to determine the frequency of ZFN-induced insertions and deletions (indels) in both samples, indicated as the % Indels at the base of the right lane. ZFN (FLAG-tagged) expression is confirmed by α-FLAG immunoblotting, and αNFκB-p65 serves as a loading control. c, Schematic of RFLP assay detailing ZFN-mediated targeting of a NheI restriction site tag to the human F9 gene. d, Co-transfection of 400 ng of ZFN expression plasmid with increasing amounts of NheI donor plasmid (0–4 µg) results in increasing levels of homology-directed repair (HDR) at days 3 and 10 post-transfection, whereas transfection of the NheI donor alone (4 µg) does not result in detectable HDR. Black arrows denote NheI-sensitive cleavage products resulting from HDR. PCR performed using 32P-labeled nucleotides, followed by PAGE and band intensity quantification by autoradiography. Lanes with no quantification had no detectable HDR.
We designed ZFNs (F9 ZFNs) targeting intron 1 of the hF9 gene (Supplementary Figure 1) and confirmed their capacity to introduce a DSB at the intended targeted site (Figure 1b) and stimulate genome editing by homology-directed repair (HDR) in human cells (Figure 1c, d). This ZFN pair was found to be highly active, driving up to 45% small insertions and/or deletions (indels) characteristic of DSB repair by non-homologous end-joining (NHEJ) and ~17–18% of alleles stably carrying the NheI restriction site diagnostic of repair by HDR using a homologous donor template designed to insert a novel restriction enzyme site in the F9 locus. Similar results were obtained in the Hep3B human hepatocyte line (Supplementary Figure 2). For in vivo evaluation we generated a humanized mouse model of hemophilia B (HB) since the F9 ZFNs target a site present in hF9 intron 1 absent from the murine gene. We constructed an hF9 mini-gene17 under control of a liver-specific enhancer and promoter18, which mimics a previously identified mutation (Y155stop)19 that results in the absence of circulating F.IX protein. We knocked in this mini-gene at the mouse ROSA26 locus20, confirmed its genotype (Figure 2a), and showed that the resulting transgenic mice had no detectable circulating hF.IX (Figure 2b). We then crossed these mice (hereafter referred to as hF9mut mice) with an existing mouse model that has a deletion of the murine F9 gene21 to generate hF9mut/HB mice to test ZFN-driven gene correction activity in vivo (Figure 3a).
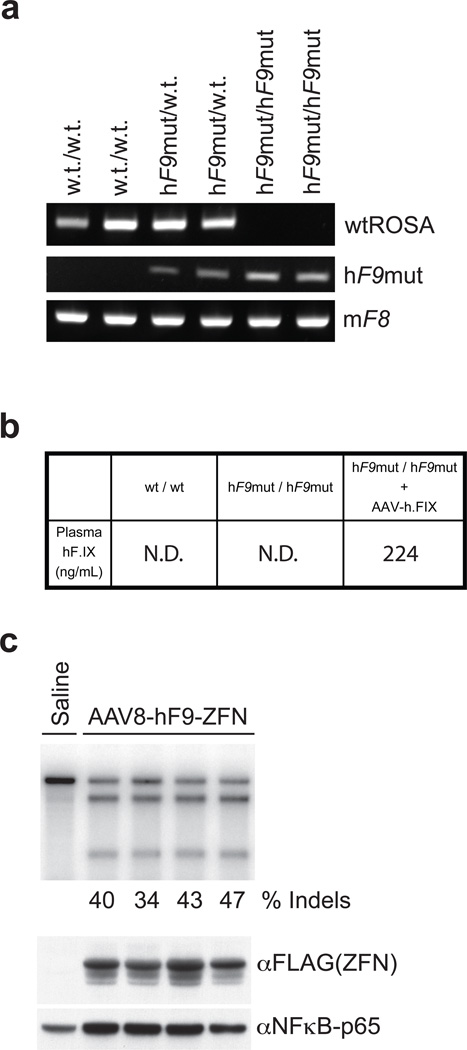
Figure 2. AAV8-mediated delivery of F9 ZFNs to hF9mut mouse liver results in cleavage of hF9mut intron 1 in vivo.
a, PCR genotyping of parental strain (w.t./w.t.), mouse heterozygous for human F9 mutant (hF9mut) construct knocked into the ROSA 26 locus (hF9mut/w.t.), and mouse homozygous for hF9mut knocked into the ROSA 26 locus (hF9mut/hF9mut). The murine Factor VIII (mF8) PCR product indicates no inhibition of PCR. b, Plasma hF.IX levels, assayed by hF.IX ELISA, in w.t. mice, homozygous hF9mut mice, and hF9mut mice injected with a viral vector expressing hF.IX (1e9 v.g. AAV-hF.IX20 injected via tail vein). N.D.= none detected. c, Tail vein injection of 1e11 v.g. AAV8-ZFN expression vector into hF9mut mice results in cleavage of intron 1. The Cel-I assay was performed on liver DNA isolated at day 7 post-injection to determine the frequency of ZFN-induced insertions and deletions (indels), indicated as the % Indels at the base of each lane, resulting from cleavage of the hF9mut intron. Lane with no quantification had no detectable cleavage products. Each lane represents an individual mouse. ZFN (FLAG-tagged) expression confirmed by α-FLAG immunoblotting of whole liver lysate.
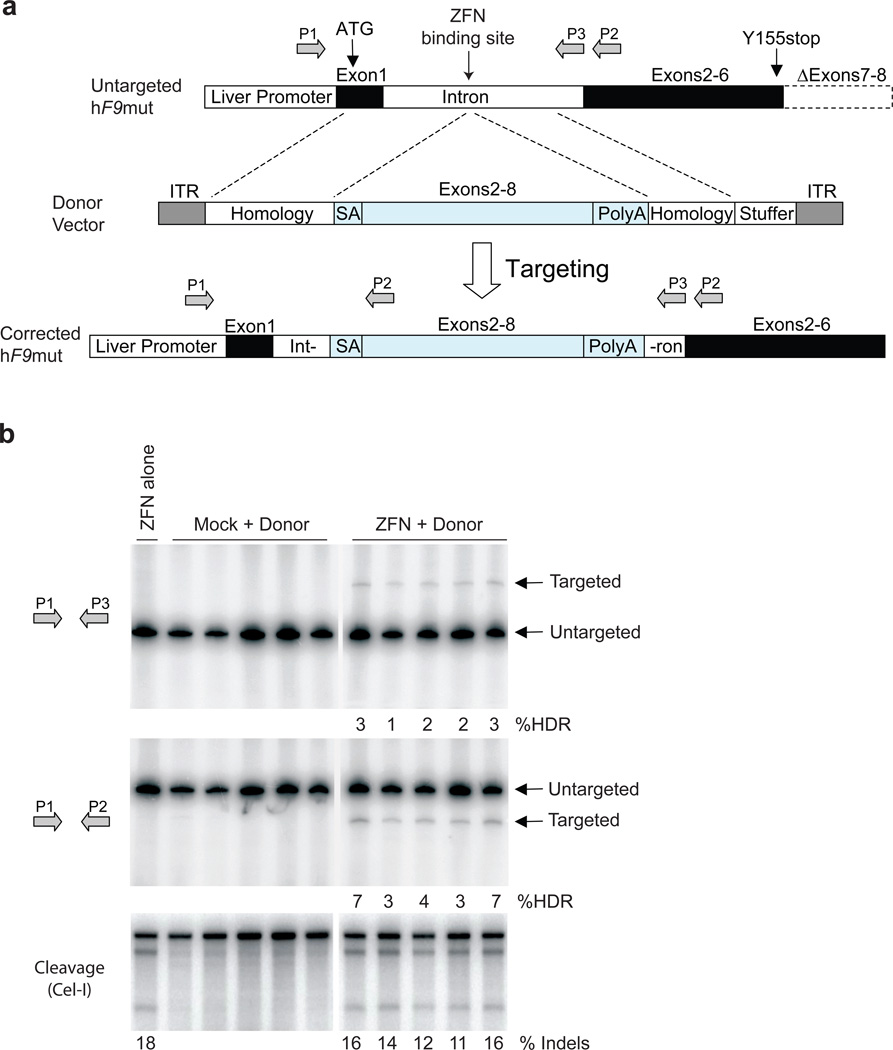
Figure 3. F9 ZFNs promote AAV-mediated targeting of wild-type F9 exons 2–8 to hF9mut intron 1 in vivo.
a, The hF9mut gene mutation (truncation of exons 7&8) can be bypassed by targeted integration of hF9 exons 2–8 into intron 1. Targeted and untargeted hF9mut alleles can be differentiated through PCR using primers P1, P2, and P3. Location of methionine start codon, and premature stop mutation indicated by arrows. The left arm of homology spans from the beginning of exon 1 to the ZFN target site. (Deletion of exon 1 from left homology arm does not alter results, Supplementary Figure 13). The right arm of homology spans intronic sequence 3’ of the ZFN target site. b, PCR analysis with primer pairs P1/P3 (upper panel) and P1/P2 (middle panel) demonstrating successful gene targeting by HDR upon I.P. co-injection of 5e10 v.g. AAV8-ZFN and 2.5e11 v.g. AAV8-Donor in hF9mut/HB mice at day 2 of life (n=5), but not with injection of 5e10 v.g. AAV8-ZFN alone (n=1), or co-injection of 5e10 v.g. AAV8-Mock and 2.5e11 v.g. AAV8-Donor (n=5). Mock vector replaces F9 ZFN coding sequences with Renilla Luciferase. PCR was performed using 32P-labeled nucleotides, followed by PAGE and product band intensity quantification by autoradiography to evaluate targeting frequency. Targeting frequencies are rounded down to the nearest whole number. Lower panel. IP injection of AAV8-ZFN expression vector into hF9mut mice results in cleavage of intron 1. The Cel-I assay was performed on liver DNA to determine the frequency of ZFN-induced insertions and deletions (indels), indicated as the % Indels at the base of each lane, resulting from cleavage of the hF9mut intron. Lanes with no quantification had no detectable HDR or indels. Each lane represents an individual mouse.
To deliver the F9 ZFNs to the liver we generated a hepatotropic adeno-associated virus vector, serotype 8 (AAV8-ZFN) expressing the F9 ZFNs from a liver-specific enhancer and promoter18. To test the cleavage activity of the F9 ZFNs in vivo we injected hF9mut mice with AAV8-ZFN through the tail vein and isolated liver DNA at day 7 post-injection. Cleavage activity was measured via the Surveyor Nuclease (Cel-I) assay22 which determines the frequency of the small insertions and deletions (indels) characteristic of DSB repair by NHEJ. We observed mutation frequencies ranging from 34 to 47%, demonstrating that coupling of the F9 ZFNs with AAV8-mediated delivery promotes highly efficient genome modification in mouse liver (Figure 2c), results which were confirmed by direct sequencing of the target locus (Supplementary Figure 3).
To correct the mutated hF9 gene in situ, we generated an AAV donor template vector (AAV8-Donor) for gene targeting, with arms of homology flanking a corrective, partial cDNA cassette (“SA – wild-type hF9 exons 2–8 – polyA”) (Figure 3a). Having first established that we could readily detect HDR in vitro (Supplementary Figure 4) we co-injected hF9mut/HB mice at day 2 of life by intra-peritoneal (I.P.) injection with AAV8-ZFN + AAV8-Donor, AAV8-Mock + AAV8-Donor, or AAV8-ZFN alone. (Note that I.P. injection in neonatal mice is less efficient than tail vein injection in adult mice [compare Cel-I results in Figure 3b to those in Figure 2c], but is used since it leads to higher survival rates). At week 10 of life we extracted liver DNA to assay for gene replacement at the hF9 locus via HDR. Using primers that hybridize to the chromosome outside of the donor homology arms, generating a larger amplicon for a targeted allele, (Figure 3a, primers P1/P3, Figure 3b, upper panel) we observed HDR only in mice receiving both the donor and F9 ZFNs with targeting efficiencies in the 1–3% range (Figure 3b, upper panel). We confirmed HDR using alternative primers that hybridize to sites outside the donor homology arms and within the inserted cassette, respectively (Figure 3a, primers P1/P2 and Figure 3b, middle panel). Thus co-delivery of ZFNs and a donor template using AAV vectors leads to HDR in vivo.
To determine if ZFN-mediated gene targeting results in production of circulating hF.IX, we injected hF9mut mice at day 2 of life I.P. with AAV8-ZFN alone, AAV8-Mock + AAV8-Donor, or AAV8-ZFN + AAV8-Donor. Plasma hF.IX levels for mice receiving ZFN alone or Mock+Donor averaged <15 ng/mL (the lower limit of detection of the assay), while mice receiving ZFN+Donor averaged 116–121 ng/mL (corresponding to 2–3% of normal) (Figure 4a), significantly greater than mice receiving ZFN alone and mice receiving Mock+Donor (p≤0.006 at all time points, 2-tailed T-test, Supplementary Figure 5). Importantly, in individual mice, the amount of circulating hF.IX directly correlated with the detected level of gene targeting via HDR (Supplementary Figure 6).
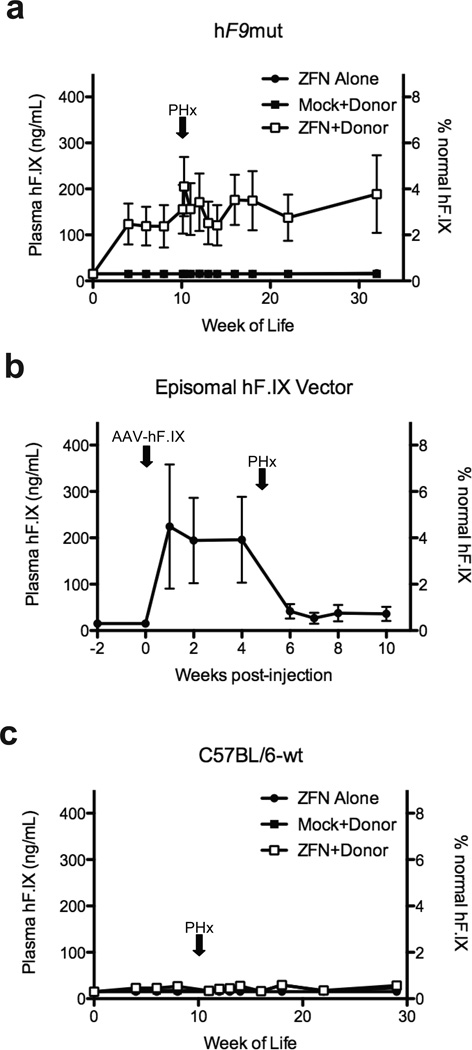
Figure 4. In vivo hF9mut gene correction results in stable circulating F.IX.
a, Plasma hF.IX levels in hF9mut mice following I.P. injection at day 2 of life with either 5e10 v.g. AAV8-ZFN alone (n=7), 5e10 v.g. AAV8-ZFN and 2.5e11 v.g. AAV8-Donor (n=7), or 5e10 v.g. AAV8-Mock and 2.5e11 v.g. AAV8-Donor (n=6). Partial hepatectomy (PHx) performed at time indicated by arrow. Plasma hF.IX assayed by ELISA. Error bars denote standard error. b, Plasma hF.IX levels in wild-type mice (n=3) following tail vein injection of 1e9 v.g. AAV-hF.IX (predominantly episomal) with subsequent PHx. Plasma hF.IX assayed by ELISA. Error bars denote standard error. c, Plasma hF.IX levels in w.t. C57BL/6J mice following I.P. injection at day 2 of life with either 5e10 v.g. AAV8-ZFN alone (n=8 pre-PHx, n=4 post-PHx), 5e10 v.g. AAV8-ZFN and 2.5e11 v.g. AAV8-Donor (n=9 pre-PHx, n=5 post-PHx), or 5e10 v.g. AAV8-Mock and 2.5e11 v.g. AAV8-Donor (n=6 pre-PHx, n=5 post-PHx). Plasma hF.IX assayed by ELISA. Error bars denote standard error.
To confirm stable genomic correction, we performed partial hepatectomies (PHx). Levels of hF.IX persist after hepatectomy following genome editing (Figure 4a), whereas an episomal AAV vector expressing hF.IX (AAV-hF.IX, Figure 4b) revealed markedly reduced hF.IX expression, since extra-chromosomal episomes are lost during liver regeneration23 (Figure 4b). Control mice receiving ZFN alone or Mock+Donor continued to average <15 ng/mL post-hepatectomy (Figure 4a) (p≤0.01 at all time points, 2-tailed T-test, Supplementary Figure 5).
To ensure hF.IX expression did not result from random donor integration into the genome, we injected wild-type mice (lacking the hF9mut mini-gene) at day 2 of life I.P. with AAV8-ZFN alone, AAV8-Mock + AAV8-Donor, or AAV8-ZFN + AAV8-Donor. Importantly, plasma hF.IX levels for mice in all groups averaged < 15, 30, and 30 ng/mL, respectively (Figure 4c), indicating the majority of hF.IX expression in ZFN+Donor treated hF9mut mice came from specific gene correction. PCR targeting assays in these w.t. control mice were negative, indicating amplicons used to quantify HDR were target gene specific (Supplementary Figure 7).
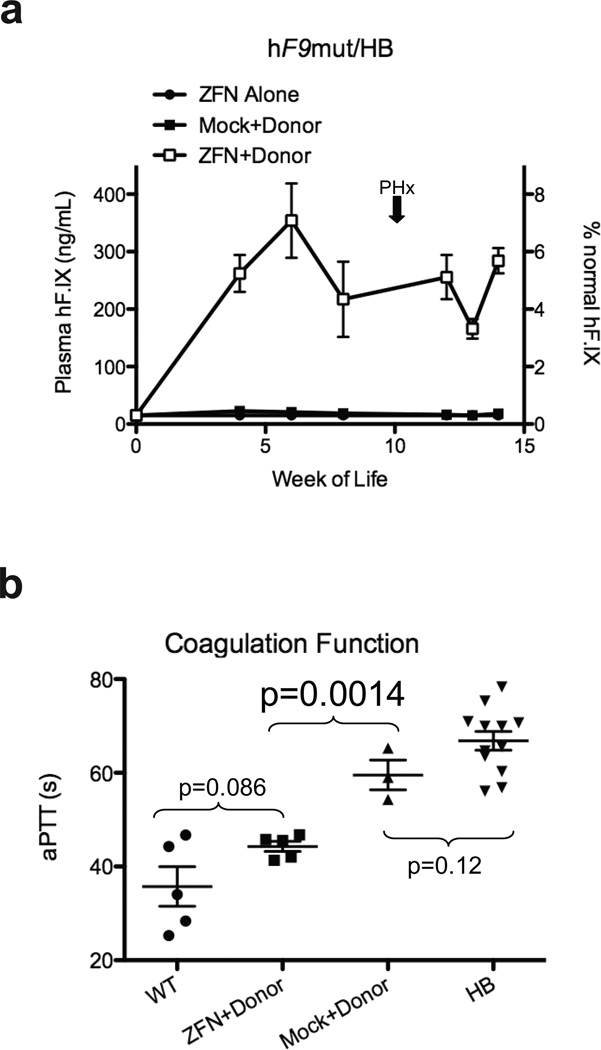
To determine if ZFN-mediated gene targeting would provide circulating hF.IX levels sufficient to correct the HB phenotype, we injected hF9mut/HB mice at day 2 of life I.P. with AAV8-ZFN alone, AAV8-Mock + AAV8-Donor, or AAV8- ZFN + AAV8-Donor. Plasma hF.IX levels for mice receiving ZFN alone again averaged < 15 ng/mL. Mice receiving Mock+Donor averaged < 25 ng/mL, and mice receiving ZFN+Donor had significantly higher hF.IX levels (p≤0.04 at all time points compared to Mock+Donor, 2-tailed T-test, Supplementary Figure 5), averaging 166–354 ng/mL (3–7% of normal circulating levels) (Figure 5a). A titration of AAV-Donor showed degree of correction was dependent on AAV-Donor dose (Supplementary Figure 8). To assay whether the HB phenotype was corrected, we assayed activated partial thromboplastin time (aPTT), a measure of clot formation kinetics that is markedly prolonged in hemophilia. The average aPTTs for wild-type mice (n=5) and HB mice (n=12) were 36 seconds and 67 seconds, respectively (Figure 5b). Mice receiving Mock+Donor (n=3) averaged 60 seconds, while mice receiving ZFN+Donor (n=5) had significantly shortened aPTTs, averaging 44 seconds (p=0.0014 compared to Mock+Donor, 2-tailed T-test). Clotting times for ZFN+Donor and wild-type mice were not significantly different (p=0.086, 2-tailed T-test) (Figure 5b). Together, these data demonstrate clinically significant correction of the coagulation defect in hemophilia B via direct in vivo delivery of ZFNs to mediate permanent correction of the genome in mouse hepatocytes.
Figure 5. Hepatic hF9mut gene correction results in phenotypic correction of hemophilia B.
a, Plasma hF.IX levels in hF9mut/HB mice following I.P. injection at day 2 of life with either 5e10 v.g. AAV8-ZFN alone (n=10 pre-PHx, n=1 post-PHx), 5e10 v.g. AAV8-ZFN and 2.5e11 v.g. AAV8-Donor (n=9 pre-PHx, n=5 post-PHx), or 5e10 v.g. AAV8-Mock and 2.5e11 v.g. AAV8-Donor (n=9 pre-PHx, n=3 post-PHx). Plasma hF.IX assayed by ELISA. Error bars denote standard error. b, Test of clot formation by aPTT at week 14 of life of mice receiving I.P. injection at day 2 of life with 5e10 v.g. AAV8-ZFN and 2.5e11 v.g. AAV8-Donor (n=5), or 5e10 v.g. AAV8-Mock and 2.5e11 v.g. AAV8-Donor (n=3). aPTTs of wild-type (WT, n=5) and hemophilia B (HB, n=12) mice are shown for comparison (p-values from 2-tailed Student’s T-test of WT vs. ZFN+Donor, ZFN+Donor vs. Mock+Donor, and Mock+Donor vs. HB). Error bars denote standard error.
To begin to evaluate the specificity of this approach, we used a SELEX-based method22 to identify the top 20 potential off-target sites for the F9 ZFNs within the mouse genome. Cel-I assays performed at each of these sites were unable to detect cleavage in 19 out of 20 (lower limit of detection 1%). At the 20th site, located in an intergenic region at mouse chromosome 9qE3.1, we detected cleavage at 1/10 the frequency seen at the F9 target site (Supplementary Figure 9). Thus the specificity of the hF9 ZFNs are comparable to the CCR5-specific ZFNs by this analysis22.
To further investigate the specificity of the ZFN approach we employed LM-PCR and 454 pyrosequencing to detect sites of AAV vector integration genome wide24. Comparison of ZFN+ Donor and Mock + Donor mice revealed similar distributions of AAV integration sites across the mouse genome (Supplementary Figure 10), which, consistent with previously reported data, favored genes24,25, but not oncogenes as integration sites. We next validated the prediction from in vitro studies26 that a ZFN-induced DSB would capture the AAV vector itself, by employing a direct PCR approach using primers that anneal to the hF9mut locus and the AAV8 ITR (Supplementary Figure 11). This assay confirmed AAV integration at the ZFN target site in ZFN + Donor mice but not in the Mock + Donor mice. Finally, pre-clinical evaluation of toxicity in injected and control mice showed no effects on growth or weight gain in either hF9mut or wild-type mice (n=43) over 8 months of observation (data not shown), and no changes in LFTs at 4, 29, and 32 weeks following injection (Supplementary Figure 12), suggesting that the treatment was well-tolerated.
Studies showing that ZFNs can efficiently mediate gene correction through the introduction of site-specific DSBs, and induce HDR in cultured cells have provided important proof-of-concept results for the clinical application of engineered nucleases for diseases affecting cells that can be removed and returned to the patient. However, the necessity to isolate and manipulate cells ex vivo limits application to a subset of genetic diseases. Our results demonstrate that AAV delivery of donor template and ZFNs in vivo induces gene targeting, resulting in measurable circulating F.IX levels. This therapeutic strategy is sufficient to restore hemostasis in a mouse model of hemophilia B, thus demonstrating genome editing in an animal model of a disease. Clinical translation of these results will require optimization of correction efficiency and a thorough analysis of off-target effects within the human genome, an issue that we have begun to monitor. Together these data demonstrate that AAV-mediated delivery of ZFNs and a donor template effect persistent and clinically meaningful levels of genome editing in vivo, and thus can be an effective strategy for targeted gene disruption or in situ correction of genetic disease in vivo.
METHODS SUMMARY
Zinc finger nucleases targeting the hF9 gene were designed and validated as described in Methods. ZFN expression, donor template, and AAV vector production plasmids were constructed using standard molecular biology techniques. AAV vectors were produced through triple transfection of HEK293 cells. K562, Hep3B, and HEK293T cells were cultured and transfected using standard techniques. hF9mut mice were created by targeted transgenesis as described in Methods. Mouse injections, plasma collection, and surgical procedures were approved by the Children’s Hospital of Philadelphia IACUC and performed as described in Methods. Cel-I assay, target site sequencing, RFLP knock-in assay, targeting assay, and hFIX RT-PCR were performed as described in Methods. hF.IX ELISA, aPTT, and LFTs were performed as described in Methods. SELEX, LM-PCR, and 454 sequencing were performed as described in Methods.
METHODS
ZFN reagents
ZFNs targeting the hF9 gene were designed by modular assembly using an archive of zinc finger proteins as previously described2. The full amino acid sequence of the F9 ZFN pair are disclosed in the supporting supplementary information (Supplementary Figure 1). The ZFN expression vector used in vitro was assembled as previously described27. The F9 ZFN AAV production plasmid was constructed by transferring the coding sequence into pRS115, a vector containing the AAV2 ITRs. ZFN expression was under the control of the ApoE enhancer and hα1AT promoter from the previously described pAAV-hFIX16 plasmid17.
Targeting vectors
The NheI RFLP donor plasmid was constructed by amplifying 1 kb regions flanking the ZFN cleavage site from K562 cell genomic DNA. A short sequence containing the NheI restriction site was subsequently introduced between the left and right arms of homology as described16. The NotI RFLP donor plasmid was constructed by amplifying the left (1kb) and right (0.6kb) arms of homology flanking the ZFN cleavage site from hF9mut mouse genomic DNA (see below) and cloned into the production plasmid pRS165 that contains the AAV2 ITRs. A short sequence containing the NotI restriction site was subsequently introduced between the left and right arms of homology as described16. The targeting vector used in vivo was built by cloning the “splice acceptor – exons 2–8 coding sequence – bovine growth hormone polyA signal” cassette from the pAAV-hFIX16 plasmid20 into the NotI RFLP donor plasmid.
Cell culture and transfection. K562 cells (ATCC) were maintained at 37°C under 5% CO2 in RPMI medium supplemented with 10% FBS and were transfected using the 96-well Nucleofector® Kit SF (Lonza) as per the manufacturer recommendations. Hep3B cells (ATCC) were maintained at 37°C under 5% CO2 in DMEM medium supplemented with 10% FBS and were transfected using the 96-well Nucleofector® Kit SE (Lonza). HEK293T cells (ATCC) were maintained at 37°C under 5% CO2 in DMEM medium supplemented with 10% FBS and were transfected using the 96-well Nucleofector® Kit SF (Lonza). Lentiviral vector for hF9mut mini-gene stable transduction into HEK293T cells was made using the ViraPower HiPerform Lentiviral Expression System (Invitrogen).
Surveyor Nuclease (Cel-I) assay and Target site seqencing. Genomic DNA from K562 and Hep3B cells, was extracted from cells using the QuickExtract™ DNA Extraction Solution (Epicentre Biotechnologies). ZFN target loci were amplified by PCR (30 cycles, 60°C annealing and 30 sec elongation at 68°C) using the hF9cel-1 For (TCGGTGAGTGATTTGCTGAG) and hF9cel1 Rev (AACCTCTCACCTGGCCTCAT) primers. Genomic DNA from mouse liver was isolated using the MasterPure Complete DNA Purification kit (Epicentre Biotechnologies). Primers for Cel-I of the hF9mut construct were hF9mut-cel1 For (CTAGTAGCTGACAGTACC) and hF9mut-cel1 Rev (GAAGAACAGAAGCCTAATTATG). The locus was amplified for 30 cycles (50°C annealing and 30 sec elongation at 68°C). The assays were carried out as described previously22. For target site sequencing, amplicons were cloned into the pCR-TOPO vector (Invitrogen) and sequenced using the primers M13forward (GTAAAACGACGGCCAGTG) and M13reverse (GGAAACAGCTATGACCATG). RFLP knock-in and targeting assays. Genomic DNA was extracted from K562 and Hep3B cells using the QuickExtract™ DNA Extraction Solution (Epicentre Biotechnologies). Genomic DNA from mouse liver was isolated using the MasterPure Complete DNA Purification kit (Epicentre Biotechnologies). The hF9 locus was amplified by 25 cycles of PCR (3 min extension time at 68°C and 30 sec annealing at 55°C) in the presence of radiolabeled dNTPs using the hF9-TI For (GGCCTTATTTACACAAAAAGTCTG) and hF9-TI Rev (TTTGCTCTAACTCCTGTTATCCATC) primers. The PCR products were then purified with G50 columns, digested with NheI, resolved by 5% PAGE, and autoradiographed. RFLP Assays in HEK293T cells transduced with the hF9mut mini-gene were genotyped as described above using the P1 (ACGGTATCGATAAGCTTGATATCGAATTCTAG) and P2 (CACTGATCTCCATCAACATACTGC) primers and the PCR products (25 cycles (63°C annealing and 2 min extension at 65°C) were digested with NotI. To quantify the targeting of the “splice acceptor – exons 2–8 coding sequence – bovine growth hormone polyA signal” cassette, gDNA was amplified using the P1 and P3 (GAATAATTCTTTAGTTTTAGCAA) or the P1 and P2 primer pairs by 25 cycles of PCR (4 min extension time at 65°C and 30 sec annealing at 48°C) in the presence of radiolabeled dNTPs. The PCR products were then purified with G50 columns, resolved by 5% PAGE, and autoradiographed. All PCR reactions were performed using Accuprime Taq HiFi (Invitrogen). To capture the NHEJ-mediated insertion of the AAV vector at the hF9 ZFN cut site, gDNA was amplified using P1 and P4 (AGG AAC CCC TAG TGA TGG AG) primer pairs by 25 cycles of PCR (1 min and 20 sec extension time at 65°C) in the presence of radiolabeled dNTPs. The PCR reactions were performed using the Phusion High-Fidelity DNA Polymerase (New England BioLabs) in conjunction with GC Buffer and 3% DMSO. The PCR products were then purified with G50 columns, resolved by 5% PAGE, and autoradiographed.
hF9mut mouse generation
The hF9mut construct (sequence provided in Supplementary Figure 14) was constructed by gene synthesis (Genscript) and ligated into the pUC57 plasmid. The hF9mut construct was then excised and ligated into a proprietary plasmid between FLP recombinase sites compatible for recombinase-mediated cassette exchange (RCME) (Taconic-Artemis) to create the hF9mut KI plasmid. The hF9mut KI plasmid and a FLP recombinase expression plasmid (Taconic-Artemis) were transfected into B6S6F1 ES cells containing FLP recombinase sites compatible for RCME at the ROSA26 locus20. Correctly targeted B6S6F1-hF9mut ES cell clones were identified by Southern blot and injected into B6D2F1 blastocysts. Pure ES cell derived B6S6F1-hF9mut mice (G0) were delivered by natural birth, and chimeric pups were back-crossed with C57BL/6J mice (Jackson Laboratories) for 5 generations (in vivo cleavage experiments) or 7–10 generations (in vivo gene targeting experiments). hF9mut mice were genotyped using primers hF9mut Oligo 1 (ACTGTCCTCTCATGCGTTGG), hF9mut Oligo 2 (GATGTTGGAGGTGGCATGG), wtROSA Oligo 1 (CATGTCTTTAATCTACCTCGATGG), wtROSA Oligo 2 (CTCCCTCGTGATCTGCAACTCC), mFVIII Oligo1 (GAGCAAATTCCTGTACTGAC), and mFVIII Oligo 2 (TGCAAGGCCTGGGCTTATTT). HB mice have been back-crossed with C57BL/6J mice (Jackson Laboratories) for >10 generations and were genotyped using previously described primers20. C57BL/6J mice (Jackson Laboratories) were used for hF9mut-negative gene targeting experiments.
AAV vector production
AAV serotype 8 vectors were produced by triple transfection methods into HEK293 cells and subsequent CsCl density gradient purification as previously described28.
Animal experiments
AAV vector was diluted to 200 µL with PBS prior to tail vein injection. AAV vector was diluted to 20 µL with PBS prior to neonatal I.P. injection. Plasma for hF.IX ELISA was obtained by retro-orbital bleeding into heparinized capillary tubes. Plasma for aPTT was obtained by tail bleeding 9:1 into 3.8% sodium citrate. Partial hepatectomies were performed as previously described29. Tissue for nucleic acid analysis was immediately frozen on dry ice after necropsy. All animal procedures were approved by the Institutional Animal Care and Use Committee of the Children’s Hospital of Philadelphia.
RT-PCR
RNA from frozen mouse tissue was isolated using the RNeasy kit (Qiagen) and the RNase-free DNase kit (Qiagen). cDNA synthesis was performed using the iSCRIPT reverse transcription kit (Bio-Rad). RT-PCR for hF.IX transcript was performed using primers hFIX Gen1 For (ACCAGCAGTGCCATTTCCA) and hFIX Gen1 Rev (GAATTGACCTGGTTTGGCATCT)
SELEX
In silico identification of potential off-target ZFN cleavage sites was performed by identifying homologous regions within the genome as previously described22.
LM-PCR and 454 sequencing
AAV-Donor integration junctions were cloned and sequenced as previously described24. In brief, genomic DNA from mouse liver was isolated using the MasterPure Complete DNA Purification kit (Epicentre Biotechnologies). 1 µg of DNA was digested with MseI (New England Biolabs) and 1 µg of DNA was digested with CviQ1 (New England Biolabs) for 16 hrs at 37°C. These 2 enzymes were chosen for their ideal proximity to the target site. Digested DNA was purified using a PCR purification kit (Qiagen), and a previously described double-stranded linker24 was ligated to digested DNA ends using T4 DNA Ligase (New England Biolabs) for 16 hrs at 16°C. Integration junctions were then PCR amplified using an adapter primer (GTAATACGACTCACTATAGGGC) and a stuffer primer (CTCCAACTCCTAATCTCAGGTGATCTACCC). PCR products were then diluted 1 in 200 in TE buffer and integration junctions were PCR amplified again using a second adapter primer (CGTATCGCCTCCCTCGCGCCATCAGnnnnnnnnnnAGGGCTCCGCTTAAGGGAC, where nnnnnnnnnn is a sample specific barcode) and a second stuffer primer (CTATGCGCCTTGCCAGCCCGCTCAGNNNNNNNNNNACCTTGGCCTCCCAAATTGCTGGG, where nnnnnnnnnn is a sample specific barcode). Amplified integration junctions were then sequenced using a Genome Sequencer FLX pyrosequencer (Roche/454).
Integration Site Analysis
Pyrosequencing reads were first decoded using DNA barcodes, separating sequence reads by mouse. Reads were then aligned against the linker and stuffer primers using the Crossmatch program (-minmatch 8 -penalty -2 -minscore 6). Reads matching one or the other primer were then aligned using BLAT against three target sequences: the stuffer, the AAV-ITR, and the hF9mut construct. BLAT parameters were optimized to find repetitive and/or short sequence hits against each target sequence (-stepSize=3 -tileSize=8 -repMatch=16384 -minScore=5 -minIdentity=50 -oneOff=1). Additionally, BLAT fastMap option was included for alignment against the stuffer and the hF9mut construct. BLAT hits originating from the ITR were processed as previously described24. BLAT hits originating from the stuffer and the hF9mut construct were identified by requiring a unique high scoring match requiring at least 90% sequence identity with <=5bp gap. All the BLAT hits from each of the three target sequences were consolidated and ordered by their location within each read. Reads having stuffer and/or linker with ITR but no hF9mut construct were segregated and aligned using BLAT against the mouse genome. BLAT hits in the mouse genome were scored using the same criteria as described above and were required to not overlap with hits originating from stuffer, ITR, and linker. A master table of all the reads and their respective target hits was constructed to manage the alignment data and associated metadata. All the subsequent 454 analysis was carried out using this master table. Sequence analysis and control of mispriming was carried out separately for reads originating from each primer (stuffer or linker). To remove reads originating from mispriming at the stuffer primer, we required that each read involving the stuffer primer extend through 30 bp of adjoining stuffer sequence and at least 13 bp of the flanking ITR. For reads originating on the linker side, we required that reads include at least 13 bp of ITR and at least 15 bases of the stuffer. Integration sites in the mouse genome were analyzed as previously described30.
hF.IX quantification and functional analysis
Quantification of plasma hF.IX was performed using an hF.IX ELISA kit (Affinity Biologicals), with a standard curve from pooled normal human plasma (Trinity Biotech). All readings below the last value of the standard curve (15 ng/mL) were arbitrarily given that value of 15 ng/mL, the limit of detection. aPTT was performed by mixing sample plasma 1:1:1 with pooled HB human plasma (George King Biomedical, Inc.) and aPTT reagent (Trinity Biotech). Clot formation was initiated by addition of 25 mM calcium chloride.
LFTs
Quantification of plasma ALT values was performed using an ALT(SGPT) Reagent set (Teco Diagnostics) colorimetric assay.
Statistics
Student’s T-test was used as described. Linear regressions were performed using Prism (Graphpad). In all tests, differences were considered significant at p<0.05.
Supplementary Material
ACKNOWLEDGEMENTS
This work was funded by the National Institutes of Health and the Howard Hughes Medical Institute.
Footnotes
AUTHOR CONTRIBUTIONS H.L., V.H., Y.D., T.L., S.L.M., P.D.G., M.C.H., and K.A.H. designed the experiments. H.L., V.H., Y.D., T.L., S.Y.W., A.S.B., N.M., X.M.A., R.S., L.I., S.L.M., J.D.F., F.R.K., S.Z., D.E.P., and E.J.R. generated reagents and performed the experiments. H.L., Y.D., F.D.B., P.D.G., M.C.H., and K.A.H. wrote and edited the manuscript.
AUTHOR INFORMATION H.L., V.H., A.S.B., N.M., X.M.A., R.S., L.I., S.L.M., J.D.F. F.R.K., S.Z., and F.D.B. have no competing financial interests to declare. Y.D., T.L., S.Y.W., D.E.P., E.J.R., P.D.G., and M.C.H. were all employees of Sangamo Biosciences when involved in this work. K.A.H. holds patents related to AAV vectors in the treatment of hemophilia.
REFERENCES
- 1.Urnov FD, Rebar EJ, Holmes MC, Zhang HS, Gregory PD. Genome editing with engineered zinc finger nucleases. Nat Rev Genet. 2010;11(9):636–646. doi: 10.1038/nrg2842. [DOI] [PubMed] [Google Scholar]
- 2.Urnov FD, et al. Highly efficient endogenous human gene correction using designed zinc-finger nucleases. Nature. 2005;435(7042):646–651. doi: 10.1038/nature03556. [DOI] [PubMed] [Google Scholar]
- 3.Porteus MH, Baltimore D. Chimeric nucleases stimulate gene targeting in human cells. Science. 2003;300(5620):763. doi: 10.1126/science.1078395. [DOI] [PubMed] [Google Scholar]
- 4.Bibikova M, et al. Stimulation of homologous recombination through targeted cleavage by chimeric nucleases. Mol Cell Biol. 2001;21(1):289–297. doi: 10.1128/MCB.21.1.289-297.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cartier N, et al. Hematopoietic stem cell gene therapy with a lentiviral vector in X-linked adrenoleukodystrophy. Science. 2009;326(5954):818–823. doi: 10.1126/science.1171242. [DOI] [PubMed] [Google Scholar]
- 6.Aiuti A, et al. Gene therapy for immunodeficiency due to adenosine deaminase deficiency. N Engl J Med. 2009;360(5):447–458. doi: 10.1056/NEJMoa0805817. [DOI] [PubMed] [Google Scholar]
- 7.Cideciyan AV, et al. Human gene therapy for RPE65 isomerase deficiency activates the retinoid cycle of vision but with slow rod kinetics. Proc Natl Acad Sci U S A. 2008;105(39):15112–15117. doi: 10.1073/pnas.0807027105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Maguire AM, et al. Safety and efficacy of gene transfer for Leber's congenital amaurosis. N Engl J Med. 2008;358(21):2240–2248. doi: 10.1056/NEJMoa0802315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bainbridge JW, et al. Effect of gene therapy on visual function in Leber's congenital amaurosis. N Engl J Med. 2008;358(21):2231–2239. doi: 10.1056/NEJMoa0802268. [DOI] [PubMed] [Google Scholar]
- 10.Cavazzana-Calvo M, et al. Transfusion independence and HMGA2 activation after gene therapy of human beta-thalassaemia. Nature. 2010;467(7313):318–322. doi: 10.1038/nature09328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Howe SJ, et al. Insertional mutagenesis combined with acquired somatic mutations causes leukemogenesis following gene therapy of SCID-X1 patients. J Clin Invest. 2008;118(9):3143–3150. doi: 10.1172/JCI35798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hacein-Bey-Abina S, et al. LMO2-associated clonal T cell proliferation in two patients after gene therapy for SCID-X1. Science. 2003;302(5644):415–419. doi: 10.1126/science.1088547. [DOI] [PubMed] [Google Scholar]
- 13.Hanna J, et al. Treatment of sickle cell anemia mouse model with iPS cells generated from autologous skin. Science. 2007;318(5858):1920–1923. doi: 10.1126/science.1152092. [DOI] [PubMed] [Google Scholar]
- 14.Scriver CR, Beaudet AL, Valle D, Sly WS, editors. The Metabolic and Molecular Bases of Inherited Disease. New York: McGraw-Hill; 2001. [Google Scholar]
- 15.Green P, Haemophilia B. Mutation Database. 2004 http://www.kcl.ac.uk/ip/petergreen/haemBdatabase.html.
- 16.Moehle EA, et al. Targeted gene addition into a specified location in the human genome using designed zinc finger nucleases. Proc Natl Acad Sci U S A. 2007;104(9):3055–3060. doi: 10.1073/pnas.0611478104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Manno CS, et al. Successful transduction of liver in hemophilia by AAV-Factor IX and limitations imposed by the host immune response. Nat Med. 2006;12(3):342–347. doi: 10.1038/nm1358. [DOI] [PubMed] [Google Scholar]
- 18.Miao CH, et al. Inclusion of the hepatic locus control region, an intron, and untranslated region increases and stabilizes hepatic factor IX gene expression in vivo but not in vitro. Mol Ther. 2000;1(6):522–532. doi: 10.1006/mthe.2000.0075. [DOI] [PubMed] [Google Scholar]
- 19.Thompson AR, Chen SH. Germ line origins of de novo mutations in hemophilia B families. Hum Genet. 1994;94(3):299–302. doi: 10.1007/BF00208288. [DOI] [PubMed] [Google Scholar]
- 20.Zambrowicz BP, et al. Disruption of overlapping transcripts in the ROSA beta geo 26 gene trap strain leads to widespread expression of beta-galactosidase in mouse embryos and hematopoietic cells. Proc Natl Acad Sci U S A. 1997;94(8):3789–3794. doi: 10.1073/pnas.94.8.3789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lin HF, Maeda N, Smithies O, Straight DL, Stafford DW. A coagulation factor IX-deficient mouse model for human hemophilia B. Blood. 1997;90(10):3962–3966. [PubMed] [Google Scholar]
- 22.Perez EE, et al. Establishment of HIV-1 resistance in CD4+ T cells by genome editing using zinc-finger nucleases. Nat Biotechnol. 2008;26(7):808–816. doi: 10.1038/nbt1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nakai H, et al. Extrachromosomal recombinant adeno-associated virus vector genomes are primarily responsible for stable liver transduction in vivo. J Virol. 2001;75(15):6969–6976. doi: 10.1128/JVI.75.15.6969-6976.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li H, et al. Assessing the potential for AAV vector genotoxicity in a murine model. Blood. 2011;117(12):3311–3319. doi: 10.1182/blood-2010-08-302729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nakai H, et al. AAV serotype 2 vectors preferentially integrate into active genes in mice. Nat Genet. 2003;34(3):297–302. doi: 10.1038/ng1179. [DOI] [PubMed] [Google Scholar]
- 26.Miller DG, Petek LM, Russell DW. Adeno-associated virus vectors integrate at chromosome breakage sites. Nat Genet. 2004;36(7):767–773. doi: 10.1038/ng1380. [DOI] [PubMed] [Google Scholar]
- 27.Doyon Y, et al. Heritable targeted gene disruption in zebrafish using designed zinc-finger nucleases. Nat Biotechnol. 2008;26(6):702–708. doi: 10.1038/nbt1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ayuso E, et al. High AAV vector purity results in serotype- and tissue-independent enhancement of transduction efficiency. Gene Ther. 2010;17(4):503–510. doi: 10.1038/gt.2009.157. [DOI] [PubMed] [Google Scholar]
- 29.Mitchell C, Willenbring H. A reproducible and well-tolerated method for 2/3 partial hepatectomy in mice. Nat Protoc. 2008;3(7):1167–1170. doi: 10.1038/nprot.2008.80. [DOI] [PubMed] [Google Scholar]
- 30.Berry C, Hannenhalli S, Leipzig J, Bushman FD. Selection of target sites for mobile DNA integration in the human genome. PLoS Comput Biol. 2006;2(11):e157. doi: 10.1371/journal.pcbi.0020157. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.