Abstract
Genomic instability is related to a wide-range of human diseases. Here, we show that mitochondrial iron–sulfur cluster biosynthesis is important for the maintenance of nuclear genome stability in Saccharomyces cerevisiae. Cells lacking the mitochondrial chaperone Zim17 (Tim15/Hep1), a component of the iron–sulfur biosynthesis machinery, have limited respiration activity, mimic the metabolic response to iron starvation and suffer a dramatic increase in nuclear genome recombination. Increased oxidative damage or deficient DNA repair do not account for the observed genomic hyperrecombination. Impaired cell-cycle progression and genetic interactions of ZIM17 with components of the RFC-like complex involved in mitotic checkpoints indicate that replicative stress causes hyperrecombination in zim17Δ mutants. Furthermore, nuclear accumulation of pre-ribosomal particles in zim17Δ mutants reinforces the importance of iron–sulfur clusters in normal ribosome biosynthesis. We propose that compromised ribosome biosynthesis and cell-cycle progression are interconnected, together contributing to replicative stress and nuclear genome instability in zim17Δ mutants.
INTRODUCTION
A wide-range of pathological disorders in humans are the consequence of genomic instability (1). To ensure genome stability, cells have developed an intricate network of mechanisms to control their cell cycle and to repair spontaneous and inducible DNA damage. Genetic alterations can be caused by radiation, exogenous chemical agents or endogenous DNA-dependent processes such as transcription and replication. Reactive oxygen species (ROS), produced in mitochondria, provide another source of genomic instability. ROS result in well-studied DNA modifications, such as 8-oxo-2′deoxyguanosine (2). Nuclear and mitochondrial DNA (mtDNA), as well as RNA, proteins and lipids are susceptible to ROS-induced damage (3). Most cells tolerate the toxic byproducts of aerobic metabolism because respiratory energy production is vital for most eukaryotic organisms. However, some organisms are able to thrive in anaerobic conditions, such as Saccharomyces cerevisiae that is able to obtain energy by fermentation (4).
Mitochondrial iron–sulfur cluster (mtISC) synthesis is essential for eukaryotic cell survival, most likely because ISCs serve as protein co-factors needed in ribosome biogenesis and other essential cellular processes (5). In human, the frataxin gene (Yfh1 in S. cerevisiae) (6) is involved in ISC biosynthesis (7) and its dysfunction has been associated to Friedreich's ataxia, an inherited disease that causes progressive damage to the nervous system (8). All proteins involved in mtISC synthesis are encoded by nuclear genes, and ISCs (or molecules dependent on mitochondrial ISC biosynthesis) are exported from the mitochondrial matrix to the cytosol prior to their assembly into ISC-containing proteins via the cytosolic iron assembly (CIA) machinery (5). In S. cerevisiae, components involved in mtISC biosynthesis (Yah1), transport (Atm1), as well as CIA machinery (Nar1) are essential for growth. Most known ISC-containing proteins are located within the mitochondria, although in recent years, increasing numbers of cytosolic and nuclear ISC-containing proteins have also been identified. In yeast, ISCs are important for cytosolic enzymes with biosynthetic functions in amino acid metabolism (9), as well as for nuclear proteins involved in DNA metabolism, rRNA processing and translation initiation (5). Recent work has elegantly connected defective ISC transport (reduction in mitochondrial membrane potential), as well as cytosolic ISC protein assembly (Nar1 depletion), to nuclear genomic instability (10).
Yeast Zim17 (Tim15) is a nuclear-encoded, mitochondrially localized protein, with orthologs in numerous eukaryotic organisms. It is initially synthesized as a cytosolic ∼23 kDa protein with two conserved CXXC zinc finger motifs, which is then directed to the mitochondria where it is processed to a ∼15–17 kDa protein (11). The zim17Δ mutants exhibit a strong growth defect and only propagate at low temperature, underscoring the importance of Zim17 for cell viability. Some evidence suggest that Zim17 may function as a heat-shock protein and chaperone, maintaining the activity of other chaperones such as the mtHsp70 homologs, Ssc1 and Ssq1 (12–14). Ssc1 and Ssq1 function in mitochondrial precursor protein import and mtISC biosynthesis, respectively (15,16). Recently, a human Zim17 ortholog, named Hep (human Hsp70 escort protein), has been discovered. Hep possesses the unique ability to stimulate the ATPase activity of human mtHsp70, as well as being able to prevent the aggregation of unfolded target proteins (17).
Genomic instability can comprise a number of features including, an increase in DNA double-strand breaks (DSBs), recombination, mutation and chromosome instability (1). In a genome-wide screen for S. cerevisiae mutants that display increased nuclear genome instability, we found that the deletion of ZIM17 results in an extreme hyperrecombinant phenotype. This phenotype is not caused by increased ROS formation or impaired DNA repair. Genomic instability of zim17Δ mutants further extends to an increase in mutation rates and DSB formation, and a decrease in plasmid stability. Cell-cycle progression in zim17Δ mutants is delayed leading to a G1 arrest and/or entry into S-phase. Cells that enter S-phase appear to be challenged by replicative stress because cell survival in the presence of hydroxyurea (HU) depends on components of the replication factor C (RFC)-like clamp loader complex (18). In addition, zim17Δ mutants display a metabolic response to iron starvation and are defective in the export of pre-ribosome particles, phenotypes that have been associated with impaired mtISC biosynthesis (19–21). Consequently, the increased recombination in ssq1Δ and npa1-1 mutants, affected in either ISC or ribosome biosynthesis (16,22), additionally support the idea that metabolic failures precede genome instability. Together, our findings provide evidence that impaired mtISC biosynthesis is linked to alterations in ribosome biogenesis and replicative stress challenging genome stability.
MATERIALS AND METHODS
Strains and plasmids
Yeast strains and plasmids are described in Supplementary Table S1. In order to maintain plasmids, cells were grown in SC media [synthetic minimal medium (Difco) supplemented with 2% glucose, and amino acids at the final concentration described by Sherman et al. (23)], lacking tryptophan (SC-T), leucine (SC-L) or uracil (SC-U), depending on the selectable marker used. The leu2-k::URA3-ADE2::leu2-k recombination system was introduced in the indicated strains by crossing with strain WRK5-3C (24) and the ribosomal rDNA::URA3-ADE2::rDNA recombination system by crossing with strain KM84 (25).
Induction of respiration-deficient strains
Spontaneously arising respiration-deficient strains (rho−) or rho° colonies (26) were isolated from zim17Δ mutants and compared to rho− control cells (WFNL-5A) generated by ethidium bromide (EtBr) treatment (26). Lack of respiration capacity of rho− strains was confirmed by growth inhibition on non-fermentable carbon sources.
Tetrad analysis
After inducing sporulation of diploid cells in SPO plates (1% potassium acetate, 0.1% yeast extract, 0.005% glucose, supplemented with half the amount of each amino acid as used for SC media), spores were micromanipulated on YEPD medium (1% yeast extract, 2% peptone, 2% glucose supplemented with 20 mg/l of adenine). Genetic markers of the spores were inferred by replica plating onto selective medium.
Recombination and mutation frequencies
Yeast strains were grown on solid SC media containing all amino acids for seven days at 26°C. Cells were taken from single colonies, resuspended in water and serial dilutions plated onto SC-5-FOA (500 mg/ml of 5-Fluorotic acid, 5-FOA) or SC-T-L (SC media lacking tryptophan and leucine), and compared to total number of colonies obtained on SC (leu2-k system) or SC-T media (L system). Mutation frequencies were obtained as described above, comparing the number of colonies growing on SC-Arg plates (SC media lacking arginine) containing 60mg/ml canavanine to the total number of colonies obtained on SC. Each recombination or mutation value was obtained by at least three different fluctuation tests and each fluctuation test represents the median value of six independent colonies.
Determination of plasmid loss and drug sensitivity
Yeast cells transformed with the pRS315 plasmid were grown overnight in SC-L selective liquid media. Cells were re-inoculated into YEPD non-selective media and grown for 12 h. Cells were then plated onto SC-T, to determine the number of plasmid-containing cells, and on YEPD, to obtain the number of total cells. The rate of plasmid loss per generation was calculated by taking the doubling time of each strain into consideration. For drug sensitivity analysis, serial 1:10 dilutions were spotted on YEPD in the absence or presence of drugs as indicated. 4-Nitroquinoline 1-oxide (4-NQO; 0.2 mg/ml stock solution in 99.9% acetone, Sigma) was added to a final concentration of 0.1 µg/ml and plates were kept in the dark. Methyl methane sulfonate (MMS; 250 M stock solution, Fluka) was added to a final concentration of 3, 6, 100 or 150 mM. Hydroxyurea (HU; USBiological) was added to a final concentration of 100 or 150 mM.
Quantification of Rad52-YFP foci
Rad52-YFP foci formation was analyzed in cells transformed with the Rad52-YFP expressing plasmid, pWJ1344. Transformants were grown at 26°C in SC-L liquid cultures and diluted to an OD600 = 0.1. After continuous growth for 3 h, Rad52-YFP foci formation was determined by wide-field microscopy (DM-6000B, Leica). YFP fluorescence was detected using 480 nm (excitation) and 527 nm (emission) wavelengths. A total of 200 cells were counted, with the number of budded (S/G2-phase) and unbudded cells determined by visual inspection. Means and standard deviations were derived from at least four independent experiments. P-values were obtained from a Student's t-test comparing values for the WT and the zim17Δ mutants (n ≥ 4 in all cases). Differences were considered statistically significant for P-values < 0.05.
Microarray analysis
Gene expression profiles were determined using the Affymetrix 3′-Expression Microarray facility of the CABIMER Genomics Unit (Seville, Spain). Strains were grown in 50 ml YEPD medium to an OD600 = 0.5 and total RNA was isolated using the RNeasy® Midi kit (Qiagen) and RNA quality was confirmed with the Bioanalyzer® 2100 (Agilent technology). Synthesis, labeling and hybridization were performed with RNA from three independent cultures according to the Affymetrix protocols (http://www.affymetrix.com/index.affx). Probe signal intensities were captured and processed with GeneChip® Operating Software 1.4.0.036 (Affymetrix), and the resulting CEL files were reprocessed using Robust Multi-array Average (RMA) normalization (27). Fold change (log2) values and their P-values were calculated with LIMMA [Linear Models for Microarray Analysis (28) using the affylmGUI interface (29)]. Limma uses an empirical Bayes method to moderate the standard errors of the estimated log-fold changes. Gene ontology terms and statistical significances were obtained using the GO Term Finder (version 0.83) freely available from the SGD-database (http://www.yeastgenome.org/).
Protein carbonylation and ROS detection
The amount of oxidized protein was inferred from the fraction of carbonylated proteins present in crude yeast cell extracts. Protein carbonylation was assayed by immunodetection of peptides bound to 2,4-dinitrophenylhydrazones (DNPs) (30). Oxidative damage in control cells was induced by tertiary-butylhydroperoxide (t-BOOH). The amount of cellular superoxide radicals was determined by dihydroethidium in order to deduce intracellular ROS levels (31).
Cell-cycle analysis
Flow cytometry was performed following standard procedures. For G1 and G2 synchronization, cells were incubated for 210 min in 1 µg/ml α-factor or 90 min in 15 µg/ml nocodazole, respectively. Cells were released from α-factor and nocodazole treatment by washing three times in pre-warmed, fresh media.
Detection of GFP-tagged ribosomal protein Rps2 and Rpl25
The zim17Δ and WT strains were transformed with plasmids pRS316-RPS2-eGFP or pRS316-RPL25-eGFP and grown in SC-U at 26°C. Images were obtained by creating maximum projections of a focal plane z-series derived from wide-field fluorescence microscopy (DM-6000B, Leica) at ×100 magnification. Fluorescence was detected using standard filters for DAPI (360 nm excitation/470 nm emission) and GFP (480 nm excitation/527 nm emission). For each experiment a total of 400 cells were analyzed. P-values were obtained from three independent experiments by the Student's t-test comparing the values for the WT and the zim17Δ mutants (n ≥ 4 in all cases). Differences were considered statistically significant for P < 0.05.
RESULTS
The absence of the mitochondrial protein Zim17/Tim15 leads to nuclear genome hyperrecombination
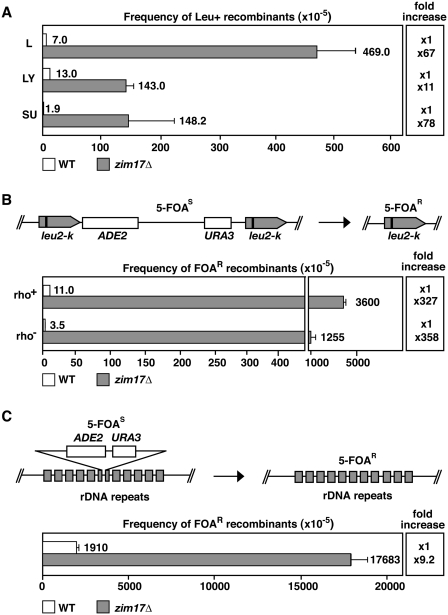
To identify novel genes with a role in genome stability, we performed a partial genome-wide screen of S. cerevisiae deletion strains for hyperrecombinant mutations. We analyzed a total of 610 of the 1000 deletion strains constructed by the EUROFAN consortium. All strains were transformed with pRS314 centromeric plasmids carrying three different recombination systems, L, LY and SU, as described previously (32). These systems are based on either direct (L and LY) or inverted (SU) repeats of a 0.6 kb internal fragment of the LEU2 ORF generated with two truncated copies of the LEU2 gene (leu2Δ3′ and leu2Δ5′) containing different DNA sequences in between. Deletions (L and LY systems) and inversions (SU) were scored as Leu+ events via fluctuation tests performed with six independent transformant colonies obtained for each system and each mutant strain. From all the strains analyzed, we found that the strain deleted for YNL310c, now termed ZIM17 (11), showed the strongest hyperrecombination phenotype and was hyperrecombinant for the three recombination systems analyzed (Figure 1A). Recombination frequencies in zim17Δ strains were 67, 11 and 78 times the WT levels for the L, LY and SU systems, respectively. The observation that different kinds of recombination events are increased in zim17Δ, suggests that zim17Δ-dependent DNA damage is not channeled into a specific repair system.
Figure 1.
ZIM17 deletion confers nuclear genome instability. (A) The zim17Δ leads to hyperrecombination in plasmidic recombination systems (L: direct repeats, LY: direct repeats disrupted by a transcribed sequence and SU: inverted repeats). (B) Schematic representation of the leu2-k::URA3-ADE2::leu2-k recombination system (top). Recombination between the direct leu2-k repeats results in the loss of URA3 and ADE2 markers and resistance against FOA (5-FOAR). Recombination frequencies in WT (white) and zim17Δ (gray) of respiratory competent (rho+) and deficient (rho−) strains are indicated (bottom). Recombinants were selected by plating cells onto 5-FOA-containing SC medium. The relative fold increase in recombination with respect to the adequate WT strain is represented (right). (C) Schematic representation of the rDNA::URA3-ADE2::rDNA recombination system (top). Recombination frequencies are shown (bottom). Description as in (B).
Zim17 has been shown to be a mitochondrially localized Zn-finger protein, essential for mitochondrial protein import and appears to be indispensable for growth at elevated temperatures (11,14). Based on its mitochondrial localization, we asked whether the genetic instability of zim17Δ mutants depends on the respiratory capacity of mitochondria. We therefore compared recombination frequencies in respiration-capable strains (rho+) and rho− (see ‘Materials and Methods’ section). Recombination frequencies in these cells were determined by the chromosomal leu2-k::ADE2-URA3::leu2-k (leu2-k) recombination system (33) in which recombinogenic events are scored by the appearance of 5-FOA resistant (5-FOAR) colonies (Figure 1B). Comparing WT rho− to zim17Δ rho− and WT rho+ to zim17Δ rho+, recombination in zim17Δ mutants was increased by >300-fold. Interestingly, in both WT and zim17Δ mutants, the relative recombination frequency was always higher in rho+ cells. To assess whether recombination was also stimulated in other compartments of the nucleus, recombination frequencies in the nucleolar, ribosomal DNA repeats (rDNA) were determined (Figure 1C). To this end, the loss of a functional copy of the URA3 gene inserted into the rDNA repeats (25) was measured by 5-FOAR colony formation. In rho+ zim17Δ mutants, rDNA recombination increased ∼9-fold with respect to WT cells confirming the general nuclear hyperrecombinant phenotype occurring in the absence of a functional Zim17 protein.
Hypermutation and high levels of DNA damage in zim17Δ strains
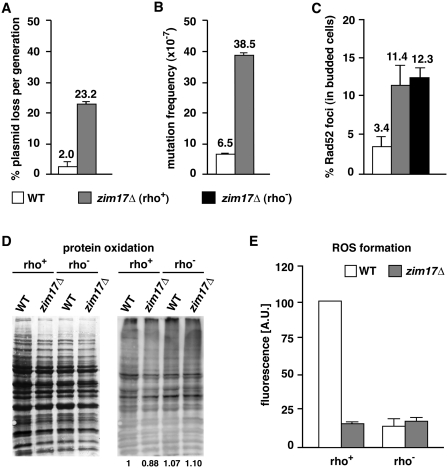
Increased mutation frequencies and diminished plasmid stability have been associated with genomic instability (1). To confirm the importance of Zim17 in the maintenance of genetic stability, we first determined the rate of plasmid loss in zim17Δ mutants transformed with the replicative plasmid pRS315. We found that, per generation, ∼2% of WT and 23% of zim17Δ cells spontaneously lost the plasmid (Figure 2A; corresponding to an 11-fold increase in plasmid loss in zim17Δ mutants). Mutation frequencies in zim17Δ mutants were detected by the spontaneous appearance of colonies in a medium supplemented with the toxic amino acid analog canavanine (Figure 2B). In this way, we identified a hypermutator phenotype in zim17Δ cells, since, the frequency of canavanine resistant colonies increased 5.9-fold in zim17Δ mutants as compared to the WT. Thus, increased plasmid loss and mutation frequencies confirm the importance of Zim17 in nuclear genome maintenance.
Figure 2.
The zim17Δ mutants exhibit higher levels of plasmid loss, mutation frequency and Rad52-YFP foci and reduced levels of protein oxidation and respiration. (A) Plasmid loss as determined by the percentage of pRS315-retaining cells after overnight growth on non-selective medium. (B) Mutation frequencies were calculated by fluctuation tests of canavanine resistant colony formation. (C) Rad52-YFP foci formation in S/G2 phase of rho+ (gray bar) and rho− (black bar) strains. (D) Total amount of protein determined by Coomassie staining (left) and protein carbonylation (right). Quantification is indicated (bottom). (E) Intracellular ROS levels determined by dihydroethidium.
DNA DSB formation could be a major component of the recombinogenic damage occurring in zim17Δ and contribute significantly to the hyperrecombination phenotype. To follow the formation of DSB, we took advantage of a Rad52-YFP fusion protein (34). Rad52 is a core component of homologous recombination-based DSB repair [reviewed in (35)], and is recruited to the DSB and assembled along the single-stranded DNA intermediate that represents the first step of the recombination process (36,37). Thus, Rad52-YFP fluorescent foci can be used to monitor the frequency of DSB repair events within the nucleus. Interestingly, the frequency of Rad52-YFP foci in both rho+ and rho− zim17Δ mutants was ∼3.5-fold higher than WT levels (Figure 2C). These results provide evidence that the zim17Δ hyperrecombination phenotype is general phenomena, rather than being restricted to the recombination systems analyzed, and also, that this phenotype is likely to be due to an increase in spontaneous DSB formation.
Iron can act as a powerful catalytic for the generation of toxic and highly reactive hydroxyl radicals via the so-called Fenton's reaction (38). Increased mitochondrial iron levels could therefore contribute to ROS formation and protein oxidation. To test if this is the case, protein oxidation and ROS formation were analyzed in WT and zim17Δ mutants in both rho+ and rho− strains. The level of protein oxidation was determined by measuring protein carbonyl content (Figure 2D), while intracellular ROS levels were quantified by following dihydroethidium fluorescence emission, primarily a measure of superoxide formation (Figure 2E). Protein oxidation levels remained the same and upon peroxide treatment, zim17Δ cells displayed the same levels of protein hypercarbonylation as wild-type cells (Supplementary Figure S1A). As expected, the background levels of ROS were reduced in rho− WT cells relative to respiration competent rho+ WT cells but notably, a reduction in ROS was observed in both rho− and rho+ zim17Δ cells. Decreased ROS formation and the downregulation of genes involved in respiration correlate with basal levels of respiration in zim17Δ mutants, excluding an increase in ROS formation as the cause of genetic instability.
RFC- and resolvase-like activities are important to protect zim17Δ cells from replicative stress and inappropriate S-phase entry
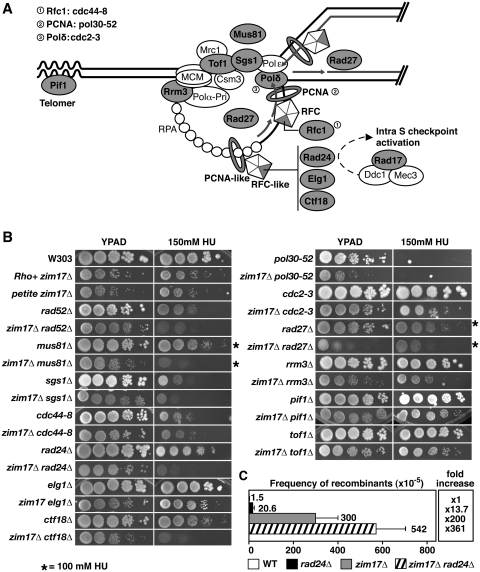
Impaired DNA repair and replication have been shown to be a major source of genetic instability (1). To search for genetic interactions, zim17Δ strains were crossed with a selection of 14 mutants covering representative steps in DNA replication and/or DNA repair (Figure 3A, shadowed in gray). Except for zim17Δ rad17Δ (Supplementary Figure S1B), all double mutants generated were viable and growth was assessed under conditions of replicative stress or DNA damage. To generate replicative stress by nucleotide depletion, cells were grown on HU-containing plates (Figure 3B). Interestingly, combinations of zim17Δ with mutants affected in cell-cycle checkpoints and replication fork maintenance were more HU sensitive compared to the respective single mutants. Deletions of proteins that confer hypersensitivity to HU in zim17Δ include the Rfc1-like proteins Rad24 and Ctf18, and the endonuclease Mus81. Rad24 and Ctf18 may have overlapping functions in the DNA-damage-replication checkpoint, where Rad24 acts in the Mec1-dependent phosphorylation of Rad53 (39), and Mus81 is involved in the resolution of cruciform structures and replication restart (40). The HU hypersensitivity of zim17Δ mus81Δ, zim17Δ ctf18Δ and zim17Δ rad24Δ double mutants suggests that the S-phase checkpoint is needed for the G1/S transition and/or to protect replication forks in zim17Δ mutants.
Figure 3.
The zim17Δ mutants are sensitive to HU-mediated replicative stress in the absence of Rad24, Ctf18 and Mus81. (A) Schematic representation of the replicon. Allelic mutants are indicated by circles. (B) Drop-test analysis of serial dilutions was performed in YEPD plates in the absence or presence of 100 or 150 mM HU (asterisks). (C) Recombination rates of zim17Δ and rad24Δ zim17Δ mutants determined by the leu2-k system. For details see Figure 1A.
To assess whether DSB repair is impeded in zim17Δ, sensitivity to DNA damage induced by the alkylating agent MMS, UV-light and the oxidative and UV-damage mimicking agent 4-NQO was assayed by serial dilution analysis. The rate of WT and zim17Δ mutant colony formation was similar (see Supplementary Figure S1C), indicating that the hyperrecombination phenotype of zim17Δ is not linked to impaired DNA repair. Interestingly, zim17Δ rad27Δ mutants exhibited a mild-synthetic-growth defect. Since zim17Δ and rad27Δ cause hypermutator phenotypes alone [(see Figure 2B (41)], the growth defect of zim17Δ rad27Δ double mutants might be mediated by a synergistic increase in the mutation rate. However, given the role of the Rad27 FLAP endonuclease in Okazaki fragment maturation, this synthetic growth defect could also be the result of the accumulation of spontaneous DNA damage in zim17Δ mutants that interferes with DNA replication.
The HU hypersensitivity of the zim17Δ mus81Δ, zim17Δ ctf18Δ and zim17Δ rad24Δ double mutants prompted us to assay whether HU hypersensitivity and hyperrecombination are linked. We determined the recombination levels of the zim17Δ rad24Δ double mutant as a way of assaying whether hyperrecombination could be the result of replication impairment. To achieve this, we generated a zim17Δ rad24Δ double mutant with an integrated chromosomal leu2-k recombination system (Figure 3C). Whilst recombination was 13-fold increased in rad24Δ mutants and 200-fold in zim17Δ mutants, a 360-fold increase in recombination was detected in zim17Δ rad24Δ double mutants with respect to the WT. It is important to note that in contrast to zim17Δ and zim17Δ rad24Δ (P = 0.14), the difference in recombination rates between WT and rad24Δ appears to be statistically significant (P = 0.0162). Since incompletely replicated or damaged chromosomes rely on Rad24-dependent cell-cycle checkpoint functions (18), it is conceivable that replicative stress contributes to zim17Δ-mediated genome instability.
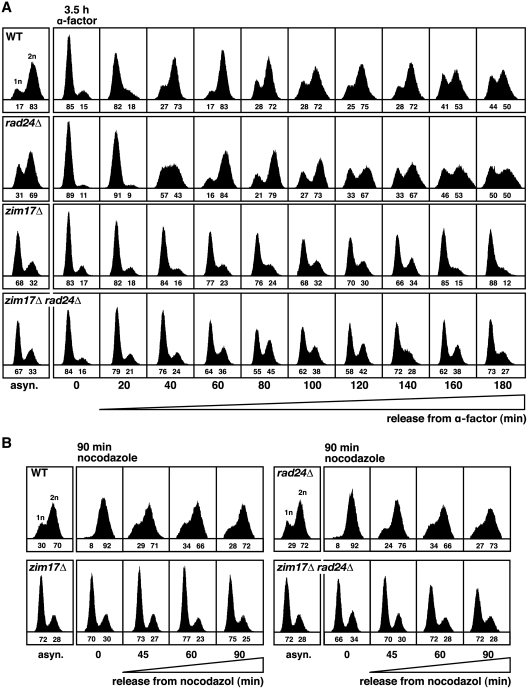
Delayed S-phase progression is a typical feature of replication and cell-cycle mutants. In an effort to further define the cellular functions of Zim17, we analyzed cycle progression in zim17Δ, rad24Δ and zim17Δ rad24Δ mutants by flow cytometry. Interestingly, asynchronous zim17Δ and zim17Δ rad24Δ cultures were characterized by an accumulation of cells with an 1n DNA content (Figure 4A). To determine the kinetics of S-phase progression, cells were blocked at G1/S with α-factor. In contrast to WT cells, after α-factor release, a high percentage of zim17Δ and zim17Δ rad24Δ cells did not enter into the S-phase. However, the fraction of cells progressing through S-phase was higher in zim17Δ rad24Δ (45% 2 n cells after 80 min) versus zim17Δ mutants (24% 2 n cells after 80 min). This observation could be explained by a reduced ability of zim17Δ cells to enter S-phase due to a G1 progression defect or to a G1/S-phase transition defect, however it is also possible that these phenotypes reflect an abnormal response to α-phactor treatment in zim17Δ cells. As an alternative approach, we attempted to synchronize cells in the G2 phase using nocodazole treatment. After 90 min of nocodazole treatment, WT as well as the rad24Δ mutants were completely synchronized in G2 (Figure 4B). In contrast, we were unable to synchronize zim17Δ cells in G2. These data confirm that zim17Δ cells are not able to progress adequately through the cell cycle leading to a high percentage of non-replicating cells.
Figure 4.
Cell-cycle progression is affected in zim17Δ and rad24Δ zim17Δ mutants. FACS analysis of S and G2 phase progression for (A) cells synchronized in G1/S for 3.5 h in the presence of α-factor or (B) treated for 90 min with nocodazole prior to release into YEPD medium. The fractions of unreplicated (1 n) or replicated (2 n) cells are indicated below (1 n + 2 n = 100).
The zim17Δ induces the transcriptional signature of iron starvation and respiration repression
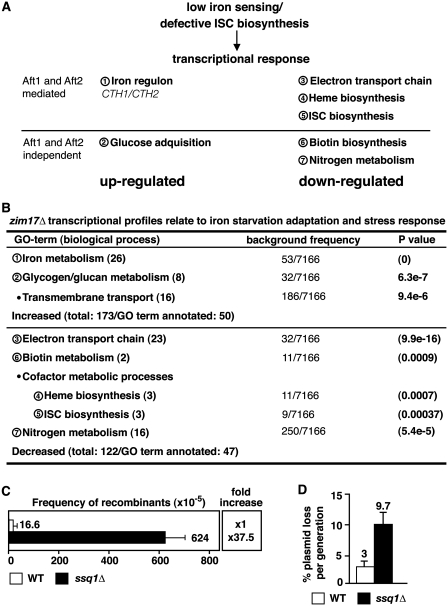
ISC biogenesis-defective yeast have been shown to adapt their metabolism if subjected to iron deprivation conditions (42,43). The iron starvation response includes alterations in the expression patterns of genes involved in iron transport and metabolism, as well as in metabolic processes in which ISC-containing proteins play a relevant role, such as respiration and biotin biosynthesis [see Figure 5A; (44)]. The transcriptional activators Aft1 and Aft2 (45) upregulate transcription of genes belonging to the iron regulon, as well as of genes coding for the mRNA-binding proteins Cth1 and Cth2 (also named Tis11). Cth1 and Cth2 decrease the stability of mRNAs coding for proteins involved in the major iron-consuming processes (mitochondrial electron transport, heme-biosynthesis and ISC biogenesis) and thereby mediate a rapid downregulation of mitochondrial respiration (46,47). To compensate for the downregulation of mitochondrial respiration, genes involved in glucose acquisition are induced. To evaluate the role of these kinds of metabolic adaptations in zim17Δ cells, we decided to determine the genome-wide transcriptional profile of these mutants. Therefore, we performed microarray analysis to compare the level of transcription of zim17Δ with WT strains. Interestingly, we found that a significant number of genes (81/295) whose transcription was significantly increased or decreased (≥2-fold; P < 0.05) in zim17Δ mutants were related to iron starvation adaptation (Figure 5B). The genome ontology (GO) terms of upregulated genes was related to iron metabolism, e.g. FIT1, HMX1, ARN1 (44) or glucose acquisition, e.g. GPH1, GYS2, PGM2 (47). Remarkably, CTH2 was 41.7-fold upregulated and accordingly, most genes whose mRNA levels were downregulated, are targets of Cth2 (electron transport chain, heme and ISC cluster biogenesis). Transcription of BIO2 and BIO4, two genes important for biotin biosynthesis but poorly expressed under iron deprivation conditions (44) and impaired ISC biogenesis (48), were also downregulated.
Figure 5.
The zim17Δ mutants show the transcriptional signature of iron starvation and deletion mutants of its interacting chaperone Ssq1 display nuclear genome instability. (A) Schematic representation of GO-terms (biological processes; encircled numbers) previously related to iron starvation (based on (5,21,43,44,46–48). See text for details. (B) Distribution of genes in which transcription is up- or downregulated in zim17Δ mutants (≥2 fold; P < 0.05). See Supplementary Table S2 for detail. The number of genes that cluster within predictable GO-terms versus all S. cerevisiae ORFs (7166) are shown. For comparison see Figure 5A. Recombination rates determined by the leu2-k system (C) and plasmid loss (D) of ssq1Δ mutants. For details, see Figures 1A, B and 2B, respectively.
The transcriptional profile of iron starvation raises the possibility that genomic instability in zim17Δ mutants is connected to impaired ISC biosynthesis. Zim17 acts as a chaperone for the non-essential Hsp70-type chaperone, Ssq1, that is required for the assembly of iron–sulfur clusters into proteins after cluster synthesis, and for the maturation of Yfh1 (16). In contrast to Ssc1, Ssq1 has been clearly shown to be part of the ISC biosynthesis machinery rather than that involved in mitochondrial protein import (16). We therefore tested if the deletion of SSQ1 would, in part, resemble the genomic instability phenotype obtained in zim17Δ mutants. This was indeed the case, as ssq1Δ mutants displayed a 37.5-fold increase in recombination and a 3-fold increase in plasmid loss, respectively (Figures 5C and D). From this result, we conclude that Zim17 contributes to ISC biosynthesis and that functional ISC biosynthesis appears to be important for the maintenance of genome stability.
Ribosome biogenesis defects are a hallmark of zim17Δ mutants and potentially contribute to genome instability
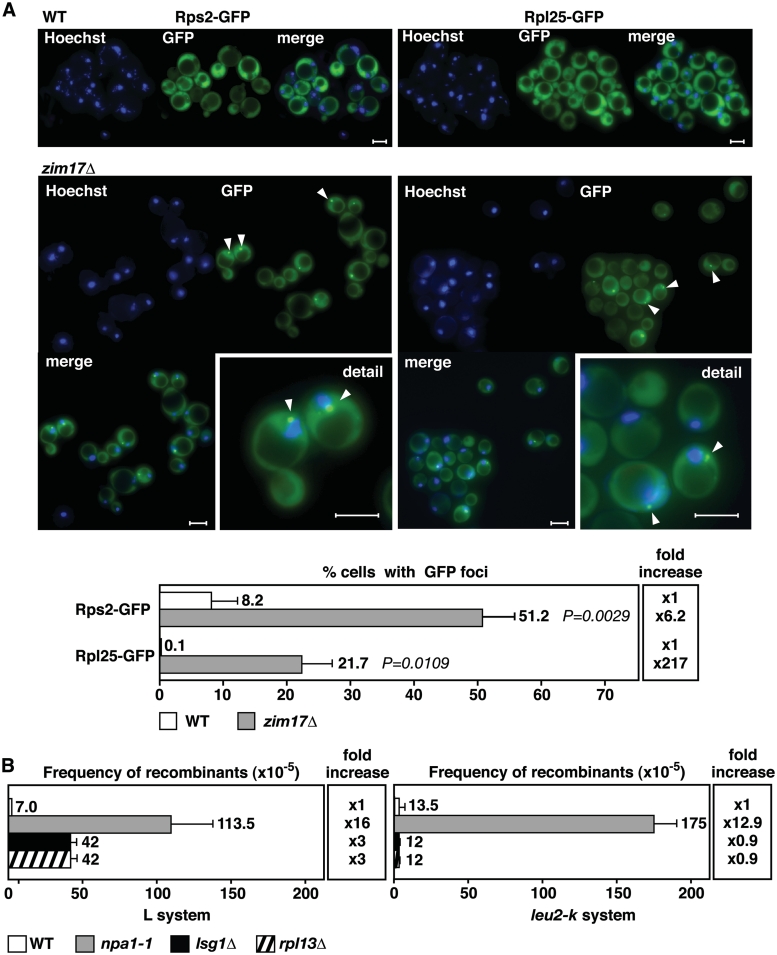
The inactivation of cytosolic CIA machinery leads to defects in ribosome export from the nucleolus to the cytosol, as has been shown for mutants lacking the ISC-containing protein Rli1 (19). Such defects in ribosome export can be detected by the nucleolar accumulation of ribosomal subunits. Because Zim17 has been predicted to have a role in mtISC biosynthesis (14), we wanted to determine if zim17Δ has a detrimental impact on the export of ribosomal subunits from the nucleus. Ribosomal export was assayed by following the localization of fluorescent-tagged ribosomal proteins, Rps2 and Rpl25, associated with the 40 S and 60 S ribosomal subunits, respectively. In contrast to WT cells, an accumulation of GFP-foci within a crescent shaped, nuclear region immediately adjacent to Hoechst33342-stained nuclear DNA was observed for both ribosomal subunits in the zim17Δ mutant (Figure 6A). Such a crescent-shaped nuclear signal is indicative of a perinuclear or nucleolar accumulation of the GFP-tagged proteins (49,50). Rps2-GFP and Rpl25-GFP foci formation was limited to 8.2% and 0.1% foci-containing cells in the WT, but were present in 51.2% and 21.7% of zim17Δ cells. Correspondingly, Rps2-GFP and Rpl25-GFP foci formation was 6.2- and 217-fold increased in zim17Δ mutants. These data indicate that zim17Δ mutants are weakly defective in 40 S and strongly defective in 60 S ribosome subunits export and therefore, ribosome biogenesis. To test for a putative stimulation of recombination by defective ribosome biogenesis, we assessed recombination in npa1-1 (22), lsg1Δ (51) and rpl13Δ (52) mutants, all of which have been shown to be impaired in 60 S ribosome maturation (Figure 6B). Indeed, depending on the mutant and recombination system, a no to 16-fold stimulation of recombination was observed in these mutants, thus linking impaired ribosome biogenesis to genome instability. However, we cannot exclude the possibility that impaired ribosome biogenesis negatively effects ISC cluster biosynthesis and it is unlikely that all mutants affected in ribosome biogenesis display genetic instability. Only in the npa1-1 mutant, recombination was >10-fold increased in both recombination systems. Moreover, tetrad analysis revealed that zim17Δ npa1-1 double mutants were not viable, supporting a vital role of Zim17 in ribosome biogenesis (Supplementary Figure S1D). Work in Schizosaccharomyces pombe has shown that a UV damage-induced G1 cell-cycle delay is mediated by the inhibition of protein translation (53). It is likely, that in zim17Δ mutants, impaired ribosome biogenesis and protein translation might delay G1- and S-phases of the cell cycle and that the shortage of proteins needed for faithful DNA replication might contribute to genome instability.
Figure 6.
ZIM17 deletion leads to a perinuclear or nucleolar accumulation of 40 S and 60 S pre-ribosomal particles. The combination of zim17Δ with mutants for genes involved in ribosome biogenesis. npa1 and emg1, leads to synthetic lethality. Recombination levels are increased in the npa1-1 mutant. (A) Rps2-GFP (40 S particles) and Rpl25-GFP (60 S particles) foci accumulation surrounding the nuclear periphery were detected by projecting of a series of focal plane images derived from wide-field fluorescence microscopy. Nuclear DNA was stained with Hoechst33342 (blue). White bars represent 5 µm. Quantification of Rps2-GFP and Rpl25-GFP foci formation (bottom). P-values are shown. (B) Recombination rates of the npa1-1, lsg1Δ and rpl13Δ mutants determined by the L and leu2-k systems. For details, see Figure 1A and B.
DISCUSSION
This work reveals a novel role for the mitochondrial Zim17 protein in the maintenance of nuclear DNA stability. Zim17 has been shown to be important for mitochondrial morphology, protein import and ISC biosynthesis (14). Various observations suggest that Zim17 is a chaperone of two structurally similar mtHsp70 homologs, namely Ssc1 and Ssq1 (12–14). Ssq1 has a role in mtISC biosynthesis, iron-homeostasis and frataxin maturation (16), and ssq1Δ mutants are viable, which suggest redundant functions in mtISC biosynthesis. In contrast, ssc1Δ mutants are inviable, since Ssc1 is essential for the mitochondrial protein import, and thus mtISC biosynthesis (15). Both Ssq1 and Ssc1 depend on Zim17 to prevent protein self-aggregation and concomitant loss of function. Consequently, Ssq1 and Ssc1 self-aggregation at elevated temperatures could explain the thermo-sensitivity of zim17Δ mutants. Mitochondrial ISC export, as well as cytosolic ISC-protein assembly, have recently been related to loss of heterozygosity (LOH) in S. cerevisiae (10). LOH was increased by a sudden loss of mitochondrial membrane potential, by the transient abrogation of mtISC export by ethidium bromide or inactivation of the CIA component Nar1. Being part of the mtISC biosynthesis pathway, Zim17 and Ssq1 act upstream of the ISC export and cytosolic ISC–protein assembly. Although to different extents, both zim17Δ and ssq1Δ mutants are prone to hyperrecombination, suggesting that failures in any step of ISC biogenesis could trigger nuclear genome instability.
As mitochondrial respiration is a major source of ROS (54), we examined whether or not ROS contributes to the genetic instability of zim17Δ. Although we cannot completely rule out this possibility, ROS does not appear to be a major cause of nuclear DNA hyperrecombination in zim17Δ mutants. zim17Δ cells are impaired in respiration and only display low levels of protein oxidation. Accordingly, numerous genes coding for components of the respiratory chain are transcriptionally repressed in the zim17Δ mutant. Indeed, recombination is generally decreased in respiration-deficient (rho−) zim17Δ and WT cells (Figure 1B), but impaired respiration did not suppress the nuclear hyperrecombination phenotype of zim17Δ. The zim17Δ mutants suffer from a high rate of rho− colony formation in line with the transcriptional downregulation of genes important for proper respiration. Importantly, zim17Δ transcription profiles show similarities to those of the iron starvation response (Figure 5A and B). Interestingly, iron sensing by Aft1/Aft2 is not linked to the cytosolic/nuclear ISC–protein assembly, but disruption of the mitochondrial ISC-biosynthesis (Nfs1, Yfh1 or Grx5 depletion) or transport (Atm1 depletion) result in the constitutive expression of genes in the iron regulon (21,55). Thus, it is conceivable that the sensing of cellular iron content depends on the amount of ISCs generated in, and exported from, the mitochondria. Mutants of genes coding for enzymes that are central to DNA replication and DNA damage response are sensitive to the reduction of intracellular iron levels in aft1Δ cells (56). These genetic interactions reinforce the idea that a reduced availability of ISCs lowers the activity of ISC-requiring enzymes that act in DNA replication and the damage response pathways that are needed to preserve genome stability.
Given the pleiotrophic effects of impaired mtISC biosynthesis on cellular metabolism, one possibility is that genetic instability in zim17Δ mutants results from a combination of cellular incapacities. Thus, the function of ISC-containing DNA-repair enzymes such as Ntg2 and Rad3 (5) could be challenged in zim17Δ mutants. However, zim17Δ mutants are not hypersensitive to DNA damage caused by MMS, 4NQO or UV light (Supplementary Figure S1C). Therefore, zim17Δ cells appear to be DNA repair proficient. Interestingly, our search for genetic interactions that confer increased sensitivity to HU in combination with zim17Δ, provides a link between genomic instability and replicative stress. Increased HU-sensitivity was observed in mutants lacking of ZIM17 and genes coding for the Rfc1-like proteins Rad24 and Ctf18 or the endonuclease, Mus81 (Figure 3B). Mus81 associates with Mms4 to form a DNA endonuclease complex that is required for the tolerance of arrested or broken replication forks (57). In response to replicative stress, the alternative clamp loaders Rad24 and Ctf18 play key roles in checkpoint and sister chromatid cohesion establishment, respectively (58,59). Rad24 is involved in Mec1 kinase-dependent activation of the Rad53-mediated S-phase checkpoint, and G1/S and G2/M checkpoint delays have been shown to be absent in rad24Δ mutants upon UV damage (18). The Rad24-RFC functions to load the PCNA-like clamp Rad17/Mec3/Ddc1 (the equivalent of the human 9-1-1 complex) onto DNA (60), and we have shown that zim17Δ is synthetic lethal in a rad17Δ strain background. Apparently, zim17Δ cells require the activation of the alternative RFC and PCNA like complexes to overcome alterations in the DNA metabolism. Only a small fraction of zim17Δ cells adequately enter and proceed through S-phase, but the relative number of S-phase entering cells is increased in zim17Δ rad24Δ double mutants (Figure 4A). We observed that replicating zim17Δ cells are more prone to DSB formation, raising the possibility that DNA damage is present early in S-phase and, is sensed by Rad24 at this stage. In a recent study, a relationship between Rad24 S-phase checkpoint-related functions and hyperrecombination has been established for hpr1Δ mutants of the THO complex (61). In contrast to other hyperrecombination mutants, such as rad3-102 (62), there is no evidence that zim17Δ leads to extensive replication fork breakage and thus, might explain why zim17Δ is not synthetic lethal with deletions of RAD52 or the MRX complex, or with other DSB repair functions in the presence of HU. Together, our results imply that replication constraints cause the hyperrecombination phenotype of zim17Δ mutants and that the Rad24 S-phase checkpoint acts in the avoidance or resolution of such constraints.
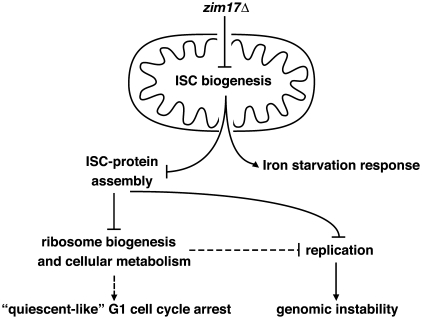
Several interesting questions remain: why do most zim17Δ cells arrest in a non-replicative G1 state, and whether this phenomenon could be related to genomic instability. The presence of an adequate number of functional ribosomes is important to produce a sufficient amount of proteins for cell division and DNA replication. Protein synthesis and cell-cycle regulation are intimately associated, with the coordination of cell growth and S-phase entry depending on ribosome-coupled translation (63). Studies in yeast suggest than aberrant ribosome biogenesis affects cell-cycle progression such that ribosome biogenesis is sensed by the START cell-cycle checkpoint (64). We observed that pre-ribosome particle export is hindered in zim17Δ cells and this phenotype might well be a consequence of impaired mtISC biosynthesis. Importantly, the ISC-containing ABC family-like ATPase Rli1 has been shown to be essential for ribosome biogenesis and translation initiation (65). Similar to zim17Δ, inactivation of Rli1, e.g. due to mutations that prevent ISC–Rli1 assembly, lead to defects in the transport of pre-ribosomal particles to the cytosol (19). Suboptimal ribosome biogenesis might contribute to both, the cell-cycle defect and the susceptibility of zim17Δ to genetic instability (Figure 7). It is conceivable that impaired ribosome biosynthesis during replication leads to a transient drop in factors that are essential for proper replication. These factors could include components of the replication machinery, as well as factors related to chromatin structure. Further evidence for such a link is provided by the nuclear genome stability observed in npa1-1, lsg1Δ and rpl13Δ mutants. Like zim17Δ, these mutants are impaired in the biogenesis of the 60 S ribosomal subunit (22,51,52) pointing to the possibility that other ribosome biosynthesis mutants are also prone to genetic instability.
Figure 7.
Model to connect zim17Δ mutant phenotypes and nuclear genome instability. See text for details.
Impairment of mitochondrial function has been shown to be the cause of human neurodegenerative and muscular diseases, and accounts for accelerated aging. Apart from Friedrich Ataxia (7), over the past years an increasing number of human diseases have been related to deficient ISC biogenesis and processing (5). Zim17 orthologs have been found in many eukaryotic organisms (11). Although direct evidence relating Hep, the mammalian Zim17 ortholog, to human disease is currently lacking, the observation that Hep1 is a chaperone of human mtHsp70, and that mtHsp70 dysfunction is connected to a human myelodysplastic syndrome, suggests this may be a interesting area of future study (17).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
European Union (EUROFAN II to A.A.); Spanish Ministry of Science and Innovation (BIO2003-07172 and BIO2006-08051 to R.E.W. and Consolider Ingenio 2010, CSD2007-0015); Junta de Andalucía (P08-CTS-04297 to R.E.W.); Pre-doctoral FPI fellowship from the Spanish Ministry of Science and Innovation (to M.D.L.). Funding for open access charge: Proyecto de investigación de excelencia (P08-CTS-04297) of the Junta de Andalucía.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank C. Brachmann, J. de la Cruz, D. Durocher, J. Haber, T. Lithgow, S. Roeder and R. Rothstein for reagents; J. García-Gómez, H. Gaillard and R. Stuckey for critical reading of the article; P. Domínguez, E. Andujar and M. Pérez from CABIMER Microscopy and Genomic Units, for assistance in microscopy and microarray analysis
REFERENCES
- 1.Aguilera A, Gomez-Gonzalez B. Genome instability: a mechanistic view of its causes and consequences. Nature Rev. Genetics. 2008;9:204–217. doi: 10.1038/nrg2268. [DOI] [PubMed] [Google Scholar]
- 2.Cooke MS, Evans MD, Dizdaroglu M, Lunec J. Oxidative DNA damage: mechanisms, mutation, and disease. FASEB J. 2003;17:1195–1214. doi: 10.1096/fj.02-0752rev. [DOI] [PubMed] [Google Scholar]
- 3.Fang YZ, Yang S, Wu G. Free radicals, antioxidants, and nutrition. Nutrition. 2002;18:872–879. doi: 10.1016/s0899-9007(02)00916-4. [DOI] [PubMed] [Google Scholar]
- 4.Stuckey R, Díaz de la Loza MC, Wellinger RE. Mitochondria: Structure, Functions and Dysfunctions. New York, NY: Nova Science Publishers Haupaugge; 2010. Cellular responses to mitochondrial DNA damage in yeast; pp. 709–729. [Google Scholar]
- 5.Lill R, Muhlenhoff U. Maturation of iron-sulfur proteins in eukaryotes: mechanisms, connected processes, and diseases. Ann. Rev. Bioch. 2008;77:669–700. doi: 10.1146/annurev.biochem.76.052705.162653. [DOI] [PubMed] [Google Scholar]
- 6.Babcock M, de Silva D, Oaks R, Davis-Kaplan S, Jiralerspong S, Montermini L, Pandolfo M, Kaplan J. Regulation of mitochondrial iron accumulation by Yfh1p, a putative homolog of frataxin. Science. 1997;276:1709–1712. doi: 10.1126/science.276.5319.1709. [DOI] [PubMed] [Google Scholar]
- 7.Campuzano V, Montermini L, Lutz Y, Cova L, Hindelang C, Jiralerspong S, Trottier Y, Kish SJ, Faucheux B, Trouillas P, et al. Frataxin is reduced in Friedreich ataxia patients and is associated with mitochondrial membranes. Hum. Mol. Genet. 1997;6:1771–1780. doi: 10.1093/hmg/6.11.1771. [DOI] [PubMed] [Google Scholar]
- 8.Friedreich N. Virchows Archiv für pathologische Anatomie und Physiologie und für klinische Medizin. Vol. 26. Berlin: 1863. Ueber degenerative Atrophie der spinalen Hirnstraenge; pp. 391–419. [Google Scholar]
- 9.Kispal G, Csere P, Prohl C, Lill R. The mitochondrial proteins Atm1p and Nfs1p are essential for biogenesis of cytosolic Fe/S proteins. EMBO J. 1999;18:3981–3989. doi: 10.1093/emboj/18.14.3981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Veatch JR, McMurray MA, Nelson ZW, Gottschling DE. Mitochondrial dysfunction leads to nuclear genome instability via an iron-sulfur cluster defect. Cell. 2009;137:1247–1258. doi: 10.1016/j.cell.2009.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Burri L, Vascotto K, Fredersdorf S, Tiedt R, Hall MN, Lithgow T. Zim17, a novel zinc finger protein essential for protein import into mitochondria. J. Biol. Chem. 2004;279:50243–50249. doi: 10.1074/jbc.M409194200. [DOI] [PubMed] [Google Scholar]
- 12.Sichting M, Mokranjac D, Azem A, Neupert W, Hell K. Maintenance of structure and function of mitochondrial Hsp70 chaperones requires the chaperone Hep1. EMBO J. 2005;24:1046–1056. doi: 10.1038/sj.emboj.7600580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yamamoto H, Momose T, Yatsukawa Y, Ohshima C, Ishikawa D, Sato T, Tamura Y, Ohwa Y, Endo T. Identification of a novel member of yeast mitochondrial Hsp70-associated motor and chaperone proteins that facilitates protein translocation across the inner membrane. FEBS Lett. 2005;579:507–511. doi: 10.1016/j.febslet.2004.12.018. [DOI] [PubMed] [Google Scholar]
- 14.Sanjuan Szklarz LK, Guiard B, Rissler M, Wiedemann N, Kozjak V, van der Laan M, Lohaus C, Marcus K, Meyer HE, Chacinska A, et al. Inactivation of the mitochondrial heat shock protein zim17 leads to aggregation of matrix hsp70s followed by pleiotropic effects on morphology and protein biogenesis. J. Mol. Biol. 2005;351:206–218. doi: 10.1016/j.jmb.2005.05.068. [DOI] [PubMed] [Google Scholar]
- 15.Bukau B, Horwich AL. The Hsp70 and Hsp60 chaperone machines. Cell. 1998;92:351–366. doi: 10.1016/s0092-8674(00)80928-9. [DOI] [PubMed] [Google Scholar]
- 16.Knight SA, Sepuri NB, Pain D, Dancis A. Mt-Hsp70 homolog, Ssc2p, required for maturation of yeast frataxin and mitochondrial iron homeostasis. J. Biol. Chem. 1998;273:18389–18393. doi: 10.1074/jbc.273.29.18389. [DOI] [PubMed] [Google Scholar]
- 17.Goswami AV, Chittoor B, D'Silva P. Understanding the functional interplay between mammalian mitochondrial Hsp70 chaperone machine components. J. Biol. Chem. 2010;18:19472–19482. doi: 10.1074/jbc.M110.105957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.de la Torre-Ruiz MA, Green CM, Lowndes NF. RAD9 and RAD24 define two additive, interacting branches of the DNA damage checkpoint pathway in budding yeast normally required for Rad53 modification and activation. EMBO J. 1998;17:2687–2698. doi: 10.1093/emboj/17.9.2687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kispal G, Sipos K, Lange H, Fekete Z, Bedekovics T, Janaky T, Bassler J, Aguilar Netz DJ, Balk J, Rotte C, et al. Biogenesis of cytosolic ribosomes requires the essential iron-sulphur protein Rli1p and mitochondria. EMBO J. 2005;24:589–598. doi: 10.1038/sj.emboj.7600541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yarunin A, Panse VG, Petfalski E, Dez C, Tollervey D, Hurt EC. Functional link between ribosome formation and biogenesis of iron-sulfur proteins. EMBO J. 2005;24:580–588. doi: 10.1038/sj.emboj.7600540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rutherford JC, Ojeda L, Balk J, Muhlenhoff U, Lill R, Winge DR. Activation of the iron regulon by the yeast Aft1/Aft2 transcription factors depends on mitochondrial but not cytosolic iron-sulfur protein biogenesis. J. Biol. Chem. 2005;280:10135–10140. doi: 10.1074/jbc.M413731200. [DOI] [PubMed] [Google Scholar]
- 22.Rosado IV, de la Cruz J. Npa1p is an essential trans-acting factor required for an early step in the assembly of 60S ribosomal subunits in Saccharomyces cerevisiae. RNA. 2004;10:1073–1083. doi: 10.1261/rna.7340404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sherman F, Fink GR, Hicks JB. New York: Cold Spring Harbor Laboratory; 1986. Laboratory Course Manual for Methods in Yeast Genetics. [Google Scholar]
- 24.Piruat JI, Aguilera A. A novel yeast gene, THO2, is involved in RNA pol II transcription and provides new evidence for transcriptional elongation-associated recombination. EMBO J. 1998;17:4859–4872. doi: 10.1093/emboj/17.16.4859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Keil RL, Roeder GS. Cis-acting, recombination-stimulating activity in a fragment of the ribosomal DNA of S. cerevisiae. Cell. 1984;39:377–386. doi: 10.1016/0092-8674(84)90016-3. [DOI] [PubMed] [Google Scholar]
- 26.Goldring ES, Grossman LI, Krupnick D, Cryer DR, Marmur J. The petite mutation in yeast. Loss of mitochondrial deoxyribonucleic acid during induction of petites with ethidium bromide. J. Mol. Biol. 1970;52:323–335. doi: 10.1016/0022-2836(70)90033-1. [DOI] [PubMed] [Google Scholar]
- 27.Irizarry RA, Ooi SL, Wu Z, Boeke JD. Use of mixture models in a microarray-based screening procedure for detecting differentially represented yeast mutants. Stat. Appl. Genet. Mol. Biol. 2003;2 doi: 10.2202/1544-6115.1002. Article 1. [DOI] [PubMed] [Google Scholar]
- 28.Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 2004;3 doi: 10.2202/1544-6115.1027. Article 3. [DOI] [PubMed] [Google Scholar]
- 29.Wettenhall JM, Simpson KM, Satterley K, Smyth GK. affylmGUI: a graphical user interface for linear modeling of single channel microarray data. Bioinformatics. 2006;22:897–899. doi: 10.1093/bioinformatics/btl025. [DOI] [PubMed] [Google Scholar]
- 30.Cabiscol E, Piulats E, Echave P, Herrero E, Ros J. Oxidative stress promotes specific protein damage in Saccharomyces cerevisiae. J. Biol. Chem. 2000;275:27393–27398. doi: 10.1074/jbc.M003140200. [DOI] [PubMed] [Google Scholar]
- 31.Aguilaniu H, Gustafsson L, Rigoulet M, Nystrom T. Asymmetric inheritance of oxidatively damaged proteins during cytokinesis. Science. 2003;299:1751–1753. doi: 10.1126/science.1080418. [DOI] [PubMed] [Google Scholar]
- 32.Prado F, Aguilera A. Role of reciprocal exchange, one-ended invasion crossover and single-strand annealing on inverted and direct repeat recombination in yeast: different requirements for the RAD1, RAD10, and RAD52 genes. Genetics. 1995;139:109–123. doi: 10.1093/genetics/139.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Piruat JI, Aguilera A. Mutations in the yeast SRB2 general transcription factor suppress hpr1-induced recombination and show defects in DNA repair. Genetics. 1996;143:1533–1542. doi: 10.1093/genetics/143.4.1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lisby M, Rothstein R, Mortensen UH. Rad52 forms DNA repair and recombination centers during S phase. Proc. Natl Acad. Sci. USA. 2001;98:8276–8282. doi: 10.1073/pnas.121006298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mortensen UH, Lisby M, Rothstein R. Rad52. Curr. Biol. 2009;19:R676–R677. doi: 10.1016/j.cub.2009.06.001. [DOI] [PubMed] [Google Scholar]
- 36.Mortensen UH, Bendixen C, Sunjevaric I, Rothstein R. DNA strand annealing is promoted by the yeast Rad52 protein. Proc. Natl Acad. Sci. USA. 1996;93:10729–10734. doi: 10.1073/pnas.93.20.10729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Van Dyck E, Stasiak AZ, Stasiak A, West SC. Binding of double-strand breaks in DNA by human Rad52 protein. Nature. 1999;398:728–731. doi: 10.1038/19560. [DOI] [PubMed] [Google Scholar]
- 38.Meneghini R. Iron homeostasis, oxidative stress, and DNA damage. Free Rad. Biol. Med. 1997;23:783–792. doi: 10.1016/s0891-5849(97)00016-6. [DOI] [PubMed] [Google Scholar]
- 39.Majka J, Niedziela-Majka A, Burgers PM. The checkpoint clamp activates Mec1 kinase during initiation of the DNA damage checkpoint. Mol. Cell. 2006;24:891–901. doi: 10.1016/j.molcel.2006.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hanada K, Budzowska M, Davies SL, van Drunen E, Onizawa H, Beverloo HB, Maas A, Essers J, Hickson ID, Kanaar R. The structure-specific endonuclease Mus81 contributes to replication restart by generating double-strand DNA breaks. Nature Struc. Mol. Biol. 2007;14:1096–1104. doi: 10.1038/nsmb1313. [DOI] [PubMed] [Google Scholar]
- 41.Reagan MS, Pittenger C, Siede W, Friedberg EC. Characterization of a mutant strain of Saccharomyces cerevisiae with a deletion of the RAD27 gene, a structural homolog of the RAD2 nucleotide excision repair gene. J. Bact. 1995;177:364–371. doi: 10.1128/jb.177.2.364-371.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ojeda L, Keller G, Muhlenhoff U, Rutherford JC, Lill R, Winge DR. Role of glutaredoxin-3 and glutaredoxin-4 in the iron regulation of the Aft1 transcriptional activator in Saccharomyces cerevisiae. J. Biol. Chem. 2006;281:17661–17669. doi: 10.1074/jbc.M602165200. [DOI] [PubMed] [Google Scholar]
- 43.Hausmann A, Samans B, Lill R, Muhlenhoff U. Cellular and mitochondrial remodeling upon defects in iron-sulfur protein biogenesis. J. Biol. Chem. 2008;283:8318–8330. doi: 10.1074/jbc.M705570200. [DOI] [PubMed] [Google Scholar]
- 44.Shakoury-Elizeh M, Tiedeman J, Rashford J, Ferea T, Demeter J, Garcia E, Rolfes R, Brown PO, Botstein D, Philpott CC. Transcriptional remodeling in response to iron deprivation in Saccharomyces cerevisiae. Mol. Biol. Cell. 2004;15:1233–1243. doi: 10.1091/mbc.E03-09-0642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rutherford JC, Jaron S, Winge DR. Aft1p and Aft2p mediate iron-responsive gene expression in yeast through related promoter elements. J. Biol. Chem. 2003;278:27636–27643. doi: 10.1074/jbc.M300076200. [DOI] [PubMed] [Google Scholar]
- 46.Puig S, Askeland E, Thiele DJ. Coordinated remodeling of cellular metabolism during iron deficiency through targeted mRNA degradation. Cell. 2005;120:99–110. doi: 10.1016/j.cell.2004.11.032. [DOI] [PubMed] [Google Scholar]
- 47.Puig S, Vergara SV, Thiele DJ. Cooperation of two mRNA-binding proteins drives metabolic adaptation to iron deficiency. Cell Metab. 2008;7:555–564. doi: 10.1016/j.cmet.2008.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Muhlenhoff U, Gerl MJ, Flauger B, Pirner HM, Balser S, Richhardt N, Lill R, Stolz J. The ISC [corrected] proteins Isa1 and Isa2 are required for the function but not for the de novo synthesis of the Fe/S clusters of biotin synthase in Saccharomyces cerevisiae. Eukaryotic Cell. 2007;6:495–504. doi: 10.1128/EC.00191-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gadal O, Strauss D, Kessl J, Trumpower B, Tollervey D, Hurt E. Nuclear export of 60s ribosomal subunits depends on Xpo1p and requires a nuclear export sequence-containing factor, Nmd3p, that associates with the large subunit protein Rpl10p. Mol. Cell. Biol. 2001;21:3405–3415. doi: 10.1128/MCB.21.10.3405-3415.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Milkereit P, Strauss D, Bassler J, Gadal O, Kuhn H, Schutz S, Gas N, Lechner J, Hurt E, Tschochner H. A Noc complex specifically involved in the formation and nuclear export of ribosomal 40 S subunits. J. Biol. Chem. 2003;278:4072–4081. doi: 10.1074/jbc.M208898200. [DOI] [PubMed] [Google Scholar]
- 51.Kallstrom G, Hedges J, Johnson A. The putative GTPases Nog1p and Lsg1p are required for 60S ribosomal subunit biogenesis and are localized to the nucleus and cytoplasm, respectively. Mol. Cell. Biol. 2003;23:4344–4355. doi: 10.1128/MCB.23.12.4344-4355.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Moritz M, Paulovich AG, Tsay YF, Woolford JL., Jr Depletion of yeast ribosomal proteins L16 or rp59 disrupts ribosome assembly. J. Cell Biol. 1990;111:2261–2274. doi: 10.1083/jcb.111.6.2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tvegard T, Soltani H, Skjolberg HC, Krohn M, Nilssen EA, Kearsey SE, Grallert B, Boye E. A novel checkpoint mechanism regulating the G1/S transition. Genes Dev. 2007;21:649–654. doi: 10.1101/gad.421807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Turrens JF. Mitochondrial formation of reactive oxygen species. J. Physiol. 2003;552:335–344. doi: 10.1113/jphysiol.2003.049478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chen OS, Crisp RJ, Valachovic M, Bard M, Winge DR, Kaplan J. Transcription of the yeast iron regulon does not respond directly to iron but rather to iron-sulfur cluster biosynthesis. J. Biol. Chem. 2004;279:29513–29518. doi: 10.1074/jbc.M403209200. [DOI] [PubMed] [Google Scholar]
- 56.Berthelet S, Usher J, Shulist K, Hamza A, Maltez N, Johnston A, Fong Y, Harris LJ, Baetz K. Functional genomics analysis of the Saccharomyces cerevisiae iron responsive transcription factor Aft1 reveals iron-independent functions. Genetics. 185:1111–1128. doi: 10.1534/genetics.110.117531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Boddy MN, Gaillard PH, McDonald WH, Shanahan P, Yates JR, 3rd, Russell P. Mus81-Eme1 are essential components of a Holliday junction resolvase. Cell. 2001;107:537–548. doi: 10.1016/s0092-8674(01)00536-0. [DOI] [PubMed] [Google Scholar]
- 58.Green CM, Erdjument-Bromage H, Tempst P, Lowndes NF. A novel Rad24 checkpoint protein complex closely related to replication factor C. Curr. Biol. 2000;10:39–42. doi: 10.1016/s0960-9822(99)00263-8. [DOI] [PubMed] [Google Scholar]
- 59.Mayer ML, Gygi SP, Aebersold R, Hieter P. Identification of RFC(Ctf18p, Ctf8p, Dcc1p): an alternative RFC complex required for sister chromatid cohesion in S. cerevisiae. Mol. Cell. 2001;7:959–970. doi: 10.1016/s1097-2765(01)00254-4. [DOI] [PubMed] [Google Scholar]
- 60.Majka J, Burgers PM. Yeast Rad17/Mec3/Ddc1: a sliding clamp for the DNA damage checkpoint. Proc. Natl Acad. Sci. USA. 2003;100:2249–2254. doi: 10.1073/pnas.0437148100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gomez-Gonzalez B, Felipe-Abrio I, Aguilera A. The S-phase checkpoint is required to respond to R-loops accumulated in THO mutants. Mol. Cell Biol. 2009;29:5203–5213. doi: 10.1128/MCB.00402-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Moriel-Carretero M, Aguilera A. A postincision-deficient TFIIH causes replication fork breakage and uncovers alternative Rad51- or Pol32-mediated restart mechanisms. Mol. Cell. 37:690–701. doi: 10.1016/j.molcel.2010.02.008. [DOI] [PubMed] [Google Scholar]
- 63.Deisenroth C, Zhang Y. Ribosome biogenesis surveillance: probing the ribosomal protein-Mdm2-p53 pathway. Oncogene. 2010;29:4253–60. doi: 10.1038/onc.2010.189. [DOI] [PubMed] [Google Scholar]
- 64.Bernstein KA, Bleichert F, Bean JM, Cross FR, Baserga SJ. Ribosome biogenesis is sensed at the Start cell cycle checkpoint. Mol. Biol. Cell. 2007;18:953–964. doi: 10.1091/mbc.E06-06-0512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Dong J, Lai R, Nielsen K, Fekete CA, Qiu H, Hinnebusch AG. The essential ATP-binding cassette protein RLI1 functions in translation by promoting preinitiation complex assembly. J. Biol. Chem. 2004;279:42157–42168. doi: 10.1074/jbc.M404502200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.