Abstract
Using reporter gene constructs, consisting of the bacterial uidA (GUS) coding region flanked by the 5′ and 3′ regions of the Chlamydomonas rbcL and psaB genes, respectively, we studied the degradation of mRNAs in the chloroplast of Chlamydomonas reinhardtii in vivo. Extending the 5′ terminus of transcripts of the reporter gene by more than 6 nucleotides triggered rapid degradation. Placing a poly(G) tract, known to pause exoribonucleases, in various positions downstream of the 5′ terminus blocked rapid degradation of the transcripts. In all these cases the 5′ ends of the accumulating GUS transcripts were found to be trimmed to the 5′ end of the poly(G) tracts indicating that a 5′→3′ exoribonuclease is involved in the degradation process. Several unstable variants of the GUS transcript could not be rescued from rapid degradation by a poly(G) tract showing that sequence/structure-dependent modes of mRNA breakdown exist in the Chlamydomonas chloroplast. Furthermore, degradation of poly(G)-stabilized transcripts that accumulated in cells maintained in the dark could be augmented by illuminating the cells, implying a photo-activated mode of mRNA degradation that is not blocked by a poly(G) tract. These results suggest sequence- and condition-dependent 5′→3′ mRNA-degrading pathways in the chloroplast of C. reinhardtii.
INTRODUCTION
Understanding the decay of messenger RNAs (mRNAs) in cells is essential for understanding mRNA turnover and overall regulation of gene expression. A number of mRNA-degrading pathways have been characterized to date in eukaryotes and prokaryotes but initiation and exact routes of mRNA breakdown are in many cases not clear. This is particularly true for mRNA degradation in plastids where the molecular machinery involved is not fully characterized to date (1–4).
The molecular biology of plastids is basically prokaryotic but important processes, including mRNA turnover, are known to be regulated by nucleus-encoded trans-acting factors (5–13). Compared to bacterial mRNAs, plastid mRNAs are relatively stable, most of them having half-lives of several hours. Longevity is conferred to plastid mRNAs by sequences in their 5′ and 3′ untranslated regions (UTRs) that often fold into specific RNA secondary structures (14–17). These cis-acting stabilizing elements are thought to be binding sites for nucleus-encoded proteins that are known to be required for stability of transcripts in plastids (3).
The plastid-located exo- and endoribonucleases that are thought to be central in plastid mRNA turnover (4) are in most cases encoded in the nuclear genome. Defining the roles of these ribonucleases in RNA decay has proven difficult because degradation intermediates do not accumulate to any extent under normal conditions in vivo suggesting that initiation is the rate-limiting step in breakdown of plastid mRNAs. In addition, most of the ribonucleases thought to be involved in mRNA breakdown also have a function in processing of transfer and ribosomal RNAs (18,19) leading to pleiotropic effects in ribonuclease mutants.
It is generally believed that homologs of bacterial endoribonucleases of the RNase E/G or RNase J type and the 3′ to 5′ exoribonucleases RNase II and polynucleotide phosphorylase (PNPase) are involved in mRNA decay in plastids (4,20–22). Indirect evidence also suggests that a 5′ to 3′ exoribonucleolytic activity plays a role in plastid mRNA degradation (23), although a protein with this activity has not yet been located in plastids to date.
To shed light on the pathways by which transcripts in the chloroplast of the unicellular green alga Chlamydomonas reinhardtii are degraded we studied in vivo the degradation of transcripts of a specific reporter gene consisting of the 5′ region of the Chlamydomonas chloroplast rbcL gene (from positions −70 to +157 relative to the transcription start site) fused 5′ to the coding region of the bacterial uidA (GUS) gene and terminated by the 3′ end of the Chlamydomonas chloroplast psaB gene. It was found that initiation of degradation of these transcripts starts at the 5′ end. Analyses of the stability of variants of the GUS transcript revealed sequence/structure-dependent and conditional mRNA degradation pathways that involve 5′ to 3′ exoribonucleolytic and, most likely, endoribonucleolytic activities.
MATERIALS AND METHODS
Growth of algae and chloroplast transformation
The photosynthesis-deficient mutant strain ac-uc-221 (CC373) of C. reinhardtii, originally obtained from the culture collection of the Chlamydomonas Genetics Center at Duke University, NC, USA, was maintained in low light (≈0.05 µmol s−1 m−2) on 1.5% agar plates in high salt (HS) medium supplemented with 2.5 g/l potassium acetate. Photosynthetic transformants of this strain were grown in HS medium on agar plates, in 250 ml Erlenmeyer flasks, or tubes in high light (≈50 µmol s−1 m−2). Tube cultures were grown at 32°C and continuously bubbled with 2% CO2-enriched air. When used for RNA isolation, cultures were grown in 12 h light/12 h dark cycles with daily dilutions to a cell density of ∼1 million cells/ml near the end of the dark period.
Chloroplast transformation was essentially as described (24,25). Transformants were screened for homoplasmicity by DNA slot and DNA gel (Southern) blots (25). Those transformants that did not accumulate GUS transcripts to high levels were selectively replated and screened until homoplasmic cell lines were found. Transformants that accumulated GUS transcripts to high levels were all estimated to be more than 50% homoplasmic but no effort was made to isolate 100% homoplasmic cell lines of those transformants.
GUS reporter gene constructs
Unless otherwise stated, standard DNA manipulation techniques were followed (26). SK+157, the plasmid from which all GUS reporter genes used in this study were derived, has been described (27). It consists of the 5′ region of the C. reinhardtii rbcL gene (extending from positions −70 to +157 relative to the transcription start site) fused 5′ to the coding region of the bacterial uidA (GUS) gene and cloned into the XhoI–XbaI sites of plasmid pBluescript SK+ (Stratagene, La Jolla, CA, USA). Restriction sites that were used for altering the sequence in the 5′ region are shown in Figure 1. For some of the constructs an Eco47III (AGCGCT) restriction site was introduced at position +95 using the QuikChange Site-Directed Mutagenesis kit (Stratagene, La Jolla, CA, USA). Sequences to be altered were synthesized as complementary oligonucleotides with ends compatible with the restriction sites. After cloning the oligonucleotides into their respective sites of Sk+157 the constructs were released from SK+157 by digestion with XhoI and XbaI and cloned into XhoI/XbaI-cut transformation vector pCrc32 (28) upstream of the 3′ region of the Chlamydomonas psaB gene (Figure 1). All constructs were sequenced prior to insertion into the Chlamydomonas chloroplast genome.
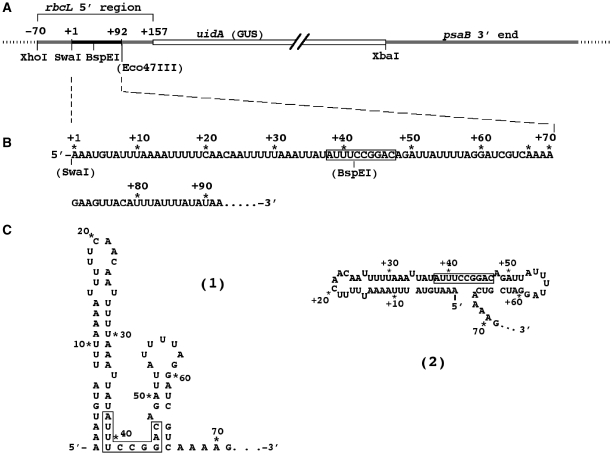
Figure 1.
GUS reporter gene construct used in this study. The construct has been described (27). (A) Map of the chimeric uidA (GUS) gene. The indicated restriction sites were used to introduce changes into the sequence by oligonucleotide replacement. The Eco47III restriction site (AGCGCT) was introduced at position +95 in order to be able to modify the sequence in the second half of the rbcL 5′ UTR in some constructs. Numbers above the map and the RNA sequence denote nucleotide positions relative to the transcription start site at +1. (B) Sequence of the rbcL 5′ UTR. The box in the RNA sequence marks a sequence element that is required for stability of the chimeric rbcL:GUS:psaB 3′ end transcripts (16). (C) Two schematic representations of the RNA secondary structure at the 5′ end of the rbcL 5′ UTR. The stem–loops shown were predicted by RNA folding programs and have been verified experimentally (16). A previously identified (16) stabilizing sequence element is boxed.
Analysis of transcript levels
Levels of transcripts of the GUS reporter gene were determined by RNA gel blot (northern) analysis as described (29). Briefly, total RNA was isolated from tube cultures growing in 12 h light/12 h dark cycles at the end of the dark period and/or 1 h into the light period. Four micrograms of total RNA were separated in a 1.3% formaldehyde agarose gel (26). Ribosomal RNA bands served as controls for loading of equal amounts of RNA into the lanes of the gel. After capillary transfer to a Zetaprobe nylon membrane (Bio-Rad Laboratories, Hercules, CA, USA), GUS transcripts were detected by hybridization to the random primer [32P]-labeled GUS sequence and exposure to an X-ray film (Kodak Biomax MS) as described (29).
Determining the 5′ end of transcripts
The 5′ ends of transcripts of the GUS reporter gene were determined by primer extension analyses using a 21-nt GUS primer (5′-CGCGCTTTCCCACCAACGCTG-3′) that binds to positions 94–115 of the coding region of the GUS transcript. The primer was 5′ end-labeled with γ-32P-ATP (3000 Ci/mmol) and T4 polynucleotide kinase using a standard end-labeling protocol (26). Primer extension was done with 20 µg total RNA using the RevertAid™ H Minus First Strand cDNA Synthesis Kit (Fermentas UAB, Vilnius, Lithuania) and subsequent purification of the primer extension products by phenol/chloroform extraction and standard Na-acetate/ethanol precipitation (26). Primer extension products were dissolved in 6 µl sterile water and 4 µl stop solution [95% formamide, 20 mM ethylenediaminetetraacetic acid (EDTA), 0.05% bromphenolblue, 0.05% xylene cyanol FF]. Two microlitres of the samples were separated in a TBE-buffered denaturing polyacrylamide (6.5%) gel (24) at 80 W constant power for about 6 h. The 33P-dATP-labeled sequence of plasmid SK+157, generated with the 21 nt GUS primer and the Sequenase Version 2.0 DNA Sequencing Kit (USB, Cleveland, OH, USA) served as a size ladder for determination of the length of the primer extension products. The dried gel was exposed to X-ray film (Hyperfilm MP, GE Healthcare, Pollards Wood, UK) with an intensifying screen at −80°C overnight.
RESULTS
Degradation of GUS transcripts is initiated at the 5′ end
The GUS reporter gene used in this study is schematically shown in Figure 1. When stably inserted into the Chlamydomonas chloroplast genome, transcripts of this construct have a half-life of 4–5 h in the dark (28) and accumulate to levels comparable to levels of transcripts of the endogenous rbcL gene (30). The 5′ terminus of these transcripts corresponds to a position in the DNA sequence that is just 4 nt downstream of the putative rbcL promoter (16) suggesting that the transcripts’ 5′ end, in contrast to the 5′ ends of most primary transcripts in the Chlamydomonas chloroplast (3), are not processed ribonucleolytically. Folding of the 5′ end into a terminal stem–loop structure (as shown in Figure 1C) is predicted by RNA folding programs (31) (http://www.bioinfo.rpi.edu/applications/hybrid/quikfold.php) and has been confirmed experimentally (16). Modifications in the rbcL 5′ UTR of this GUS reporter gene do not affect rates of transcription but can have an effect on transcript longevity (32). Therefore, the abundance of these transcripts in Chlamydomonas chloroplast transformants is an indirect measure of their relative stabilities in vivo.
To identify the pathways by which transcripts of the GUS reporter gene are degraded we first added extensions of up to 19 extra nucleotides to their 5′ end in order to trigger transcript degradation (Figure 2A). Most of the extra sequences were designed to produce single-stranded 5′ tails as predicted by RNA folding programs (31). Extensions of up to 6 extra nucleotides did not have a notable effect on transcript accumulation but transcripts with 5′ extensions of 8 or more nucleotides did not accumulate showing that they are unstable (Figure 2B).
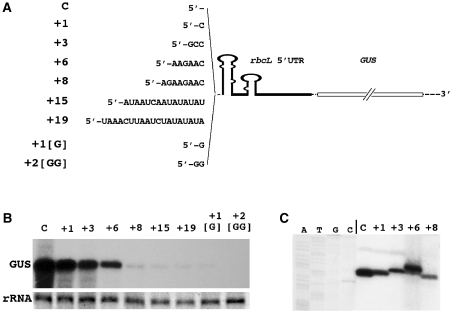
Figure 2.
Effect of extending the 5′ end of rbcL:GUS transcripts on accumulation of GUS transcripts in Chlamydomonas chloroplast transformants. (A) Extra nucleotide sequences added to the 5′ terminus of GUS transcripts. Except for extensions +1[G] and +2[GG], the sequences form single-stranded 5′ tails that do not base pair with sequences downstream. (B) RNA gel (northern) blot of total RNA isolated from Chlamydomonas chloroplast transformants harboring the constructs depicted in (A). GUS transcripts were detected by hybridization to a random primer-labeled GUS probe comprising the entire GUS coding region (25). (C) Primer extension analysis to determine the 5′ end of the GUS transcripts that accumulated in Chlamydomonas chloroplast transformants (see ‘Materials and Methods’ section for details). A Sanger dideoxynucleotide sequencing ladder, created with the same GUS primer (‘Materials and Methods’ section) used in the primer extension, was run to the left of the primer extension samples. Because of low GUS transcript levels, no primer extension analyses were done with RNA isolated from the +15, +19, +1[G], and +2[GG] transformants. C, control; rRNA, levels of 23 S ribosomal RNA (stained with ethidium bromide) used as control for loading of equal amounts of RNA on the agarose gel.
Primer extension analyses revealed that transcripts with 5′ extensions up to 6 nucleotides retained the extra nucleotides while accumulating in the chloroplast, whereas an extension of 8 nt was not maintained but seemed to be degraded transiently to the original 5′ terminus of the rbcL 5′ UTR (Figure 2C). These results show that a free single-stranded tail of 6 extra nucleotides at the rbcL 5′ terminus is largely inaccessible to ribonucleases. Transient degradation of the +8 nt tail to the +1 position shows that degradation triggered by an extension of more than 6 free extra nucleotides is initiated at the single-stranded RNA tail. Considering that the only difference between the single-stranded 5′ tail and any other single-stranded region of similar length within the transcript is the free 5′ phosphate group at the terminal nucleotide, it is very likely that the 5′ terminal nucleotide is recognized by a protein that initiates mRNA degradation.
In contrast to free single-stranded 5′ tails which require a length of more than 6 nt to destabilize the GUS transcripts, addition of just 1–2 guanine nucleotides that are predicted to base pair with the cytosine nucleotides in positions 42–43 (Figure 1C) suffices to trigger transcript degradation (+1[G] and +2[GG] in Figure 2). This confirms previous results (27) which showed that conformational changes at the base of the 5′ terminal stem–loop of the rbcL 5′ UTR destabilizes the reporter gene transcripts, presumably by altering the binding site for a protective RNA-binding protein (13,27).
A 5′ to 3′ exoribonucleolytic activity is involved in degradation of GUS transcripts
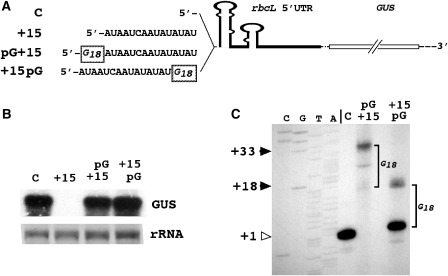
Previous studies indicate that 5′ to 3′ exoribonucleases normally participate in breakdown of transcripts in the Chlamydomonas chloroplast (23). To find out whether GUS transcripts that are destabilized by single-stranded 5′ tails of 8 or more nucleotides are degraded by 5′ to 3′ exoribonucleases, a tract of 18 guanine nucleotides was inserted upstream or downstream of the 15 nt 5′ extension (Figure 3A). Poly(G) tracts of this length, by way of their extremely stable secondary structure, are known to block movement of exoribonucleases (33). In both cases, transcripts that are normally rapidly degraded due to the free tail of 15 extra nucleotides at their 5′ ends were rescued by the poly(G) tracts (Figure 3B). The 5′ ends of the rescued transcripts corresponded to the 5′ ends of the poly(G) sequences (Figure 3C) showing for +15pG the participation of a 5′ to 3′ exoribonucleolytic activity in degradation. Dual bands in the pG+15 and +15pG lanes (Figure 3C, as well as in the primer extension analyses shown in Figures 4C and 5C) do not represent two GUS transcript populations of different lengths but two primer extension products from the same transcript population. They are caused by the RNA secondary structure of the poly(G) tract that at the temperature of the primer extension reaction (42°C) can block progress of the reverse transcriptase.
Figure 3.
Rescue of 5′ tail-destabilized GUS transcripts by a poly(G) tract. (A) The extension of 15 extra nucleotides and the two positions into which a tract of 18 extra guanine nucleotides (pG18) was inserted. (B) RNA gel (northern) blot of total RNA isolated from Chlamydomonas chloroplast transformants harboring the constructs depicted in (A). (C) Primer extension analysis to determine the 5′ ends of GUS transcripts accumulating in Chlamydomonas chloroplast transformants. Arrowheads to the left of the panel indicate the positions of the end of the longest primer extension product in each lane. The open arrowhead marks the 5′ end of the control transcript. The numbers relate to the nucleotide positions relative to the transcription start site as deduced from the sequencing ladder that was run alongside the samples. G18, 18 nt poly(G) tract. Further details as explained in the legend to Figure 2.
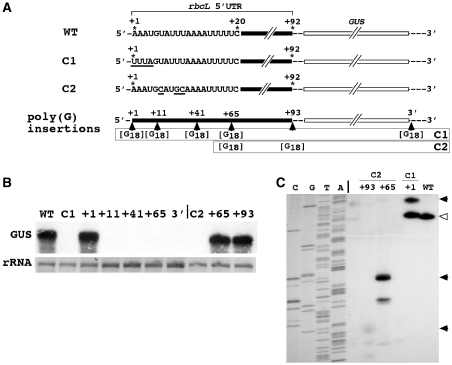
Figure 4.
The effect of poly(G) tracts inserted in various positions of the GUS transcript sequence. (A) Sequence differences in the transcripts analyzed and insertion points of poly(G) tracts in the transcript sequence. Differences in the nucleotide sequences between the original wild-type sequence and the sequences of the two unstable control transcripts (C1 and C2) are underlined. Poly(G) tracts were inserted into five positions of GUS transcripts with the C1 background and two positions with the C2 background. (B) RNA gel (northern) blot of total RNA isolated from Chlamydomonas chloroplast transformants harboring the constructs depicted in (A). (C) Primer extension analysis to determine the 5′ ends of GUS transcripts that accumulate in Chlamydomonas chloroplast transformants carrying the poly(G)-harboring constructs [as determined in (B)]. Arrowheads indicate the end of the longest primer extension product in each lane. The open arrowhead marks the 5′ end of the control (WT) transcript. WT, wild-type; G18, 18 nt poly(G) tract. Other details as explained in the legends to Figures 2 and 3.
Figure 5.
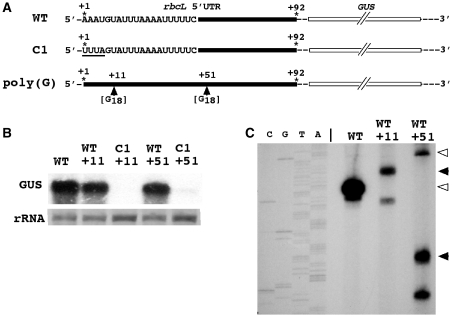
Comparison of the effect of poly(G) tracts on transcript degradation when inserted into the same position of wild-type and mutant (C1) GUS transcripts. (A) Sequence differences in the rbcL 5′ UTR of wild-type and C1 transcripts and identical points of insertion (+11 and +51) of poly(G) tracts into both transcripts. The sequence that differs in C1 from the wild-type sequence is underlined. (B) RNA gel (northern) blot of total RNA isolated from Chlamydomonas chloroplast transformants harboring the constructs depicted in (A). (C) Primer extension analysis to determine the 5′ ends of GUS transcripts that accumulate in Chlamydomonas chloroplast transformants carrying the poly(G) constructs [as determined in (B)]. Arrowheads indicate the ends of the primer extension products. Open arrowheads mark the native 5′ end of the rbcL 5′ UTR, filled arrowheads the ends of the poly(G) tracts. WT, wild-type; G18, 18 nt poly(G) tract. Other details as explained in the legends to Figures 2 and 3.
Degradation of GUS transcripts is sequence dependent
If the unstable GUS transcripts are degraded solely by exoribonucleases in 5′ to 3′ direction, without involvement of endoribonucleases, it should be possible to halt the degradation process at any position in the RNA sequence by insertion of a poly(G) tract. To test this notion, poly(G) tracts were inserted into various positions of the sequence of transcripts of the GUS reporter gene (Figure 4A). Two versions of unstable GUS transcripts that differed in the nucleotide sequence in the beginning of the rbcL 5′ UTR were chosen for these analyses (Figure 4A). One of the variants (named C1) starts with a 5′-UUUA … sequence instead of the normal 5′-AAAU … sequence, the second version of the transcript (C2) contains three altered nucleotides in the beginning of the 5′ UTR (Figure 4A). Both modifications render the GUS transcripts unstable (Figure 4B) presumably because their 5′ ends do not fold into the native 5′ stem–loop structure (Figure 1C) required for stability (27). Insertion of a poly(G) tract into the sequence of C1 transcripts in five different positions could not rescue them from rapid degradation, except when the poly(G) tract was inserted at the immediate 5′ terminus (Figure 4B). The inability of a poly(G) tract to pause degradation and trap decay intermediates of C1 transcripts suggests that these transcripts are not exclusively degraded by 5′ to 3′ exoribonucleases and that breakdown follows a mode that differs from degradation of the transcripts carrying a 5′ extension (Figure 3). Because a poly(G) tract inserted in the transcript’s 3′ region—between the GUS coding region and the 3′ UTR—did not rescue GUS transcripts from rapid degradation, it appears that 3′ to 5′ exoribonucleases do not play a major role in primary breakdown of GUS transcripts.
Rescue of C1 transcripts by a poly(G) tract at the transcripts’ 5′ terminus, whereas poly(G) tracts in any other position did not pause degradation, shows that a poly(G) tract at the immediate 5′ end affects a different part of the RNA decay pathway than in a position further downstream. Considering the results of the 5′ tail studies described above, the part impaired by the terminal poly(G) tract could be initiation of degradation at the 5′ terminus. Interference at initiation might also explain why the 5′ poly(G)-tailed C1 transcripts accumulate in the chloroplast to levels that are slightly higher than levels of unmodified control GUS transcripts (Figure 4B).
In contrast to C1 transcripts, intermediates of degradation of C2 transcripts could be trapped by poly(G) tracts at positions downstream of the 5′ terminus (Figure 4B) leading to accumulation of transcripts whose 5′ ends were found to be trimmed to the 5′ end of the poly(G) sequence (Figure 4C). Therefore, degradation of C1 and C2 transcript variants differs, either because of involvement of different ribonucleolytic activities or because of significant changes in the processivity of the same ribonucleases. It is noteworthy that C1 and C2 transcripts harboring a poly(G) tract in the same position (+65) are degraded differently (Figure 4B). Since these transcripts differ only by a few nucleotides at their 5′ ends (Figure 4A), these results indicate that modes of mRNA degradation vary in the Chlamydomonas chloroplast depending on sequence and/or RNA secondary structure.
Insertion of poly(G) tracts into the rbcL 5′ UTR sequence has most likely an effect on RNA secondary structure depending on the position of the insertion. It is therefore possible that differences in degradation pathways are caused by different positions of the poly(G) tracts and not by differences in the background mRNA sequence or structure. To eliminate this possibility and to further test the notion of diverse sequence/structure-dependent mRNA degradation modes in the Chlamydomonas chloroplast, we compared the effect of inserting poly(G) tracts into exactly the same positions (+11 and +51) of transcripts carrying the wild-type or the modified C1 rbcL 5′ UTR sequence (Figure 5A). The poly(G) tracts impeded degradation of transcripts when inserted into the wild type but not when inserted into the C1 UTR sequence (Figure 5B and C). These results substantiate the conclusion of sequence/structure-dependent modes of mRNA breakdown in the Chlamydomonas chloroplast. As seen in the primer extension analysis (Figure 5C), transcripts ending in the original +1 adenine nucleotide (Figure 5A) accumulated in WT+51 transformants carrying a GUS construct with a poly(G) tract in position +51, but not in WT+11 transformants carrying a construct with a poly(G) tract in position +11 (Figure 5C). This shows that insertion of the poly(G) tract into position +11, but not into position +51, of the rbcL 5′ UTR in itself destabilizes the transcripts supporting the notion of poly(G) insertion-dependent changes in RNA secondary structure. Lack of GUS transcripts ending in the original +1 nucleotide in WT+11 transformants also shows that integrity of the stem–loop structure at the 5′ end (Figure 1C) is crucial for stability of the reporter gene transcripts.
Photo-activated re-initiation of poly(G)-arrested degradation of GUS transcripts
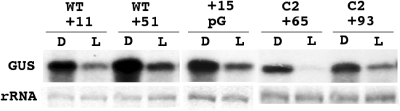
All analyses of transcript levels described above were done with RNA isolated from Chlamydomonas chloroplast transformants at the end of the dark period of a 12 h light/12 h dark growth regime. Additional support for diverse modes of RNA breakdown in the Chlamydomonas chloroplast comes from analyzing light/dark differences of RNA stability (Figure 6). It has been found previously (34) that levels of GUS transcripts in Chlamydomonas chloroplast transformants are significantly lower at 1 h into the light period than in the dark period of the light/dark cycle due to active degradation of these transcripts in the light. To find out to what extent the light-dependent active degradation of GUS transcripts is affected by poly(G) tracts, total RNA was isolated at the end of the dark period and at 1 h into the light period from transformants carrying GUS constructs WT+11, WT+51, +15pG, C2+65 and C2+93 (constructs shown in Figures 3–5). Transcripts of these constructs accumulated in the dark with a poly(G) sequence at their 5′ ends (Figures 3C, 4 C and 5 C). In all the cases analyzed, breakdown of 5′ poly(G)-ending transcripts is re-initiated by illuminating the cells (Figure 6). This shows that stalling of exoribonucleolytic RNA degradation by a poly(G) tract can be overcome by a photo-activated mechanism that either re-activates paused exoribonucleases or recruits additional proteins that actively promote mRNA breakdown.
Figure 6.
Photo-activated degradation of GUS transcripts that end 5′ with a poly(G) tract. Transformants carrying rbcL:GUS constructs with inserted poly(G) tracts (as indicated in Figures 3–5) were grown in 12 h light/12 h dark cycles and total RNA was isolated at 11 h in the dark (D) and 1 h in the light period (L). Note that the primer extension analyses shown in Figures 3–5 show that the GUS transcripts accumulating in the transformants in the dark all end 5′ with a poly(G) tract.
DISCUSSION
5′ initiation of RNA degradation in the Chlamydomonas chloroplast
The results of this study show that degradation of transcripts of the chimeric reporter gene constructs is initiated at the 5′ end, that a 5′ to 3′ exoribonucleolytic activity is involved early in the degradation process, and that different routes of mRNA breakdown can be followed depending on RNA sequence or structure and external cues, e.g. light/dark.
Initiation of degradation at the 5′ end can be concluded from the analyses of the stability of 5′-tailed transcripts (Figure 2). These analyses show that free single-stranded 5′ tails of more than 6 nt trigger transcript degradation. Assuming that the 5′ ends of chloroplast mRNAs are normally protected from degradation by RNA-binding proteins, there are two possible explanations for the effect of long 5′ tails on transcript stability. First, it is possible that a long 5′ tail interferes sterically with binding of a protein that normally protects the 5′ end of the transcripts from ribonucleolytic attack. In the case of the rbcL 5′ UTR the stabilizing sequence depicted in Figure 1B and C could be the binding site for such a protein. In a second possible explanation a 5′ tail of more than 6 nt does not interfere with binding of a 5′-protecting protein but protrudes from the protected region, thereby exposing the 5′ terminus of the RNA to the ribonucleolytic machinery in the chloroplast. In both cases, degradation would be initiated at the 5′ end of the transcripts. Initiation of degradation at the 5′ tail is supported by transient accumulation of RNAs whose ends are trimmed to the original +1 position (+8 in Figure 2C).
Therefore, we consider it very likely that an exposed 5′ terminal nucleotide with its unique 5′ phosphate group is the binding site of a protein that initiates mRNA breakdown in chloroplasts. The 5′ terminal phosphate is the only 5′ feature that is common to all mRNAs and is easy to protect from ribonucleolytic attack.
The situation is reminiscent of mRNA breakdown in Escherichia coli where a free 5′ end extension also enhances transcript degradation (35). In bacteria, accessibility of the 5′ end can be hindered by a 5′ RNA hairpin structure that presumably prevents proteins sterically from binding to the 5′ terminus (35). Recently, an RNA pyrophosphohydrolase (RppH) that converts the triphosphates at the mRNAs’ 5′ terminus into monophosphates has been identified in bacteria (36). It has been suggested that binding of this enzyme to the 5′ terminus of bacterial mRNAs may be the rate-limiting step in mRNA breakdown by the bacterial 5′ end-dependent degradation pathway (36) because monophosphorylated 5′ ends are the preferred substrates for RNase E/G and RNase J1/J2 (19,37), enzymes that are thought to be central in primary mRNA breakdown in E. coli or Bacillus subtilis.
Assuming that the mechanisms of mRNA breakdown in bacteria and the Chlamydomonas chloroplast are similar, very long half-lives of Chlamydomonas chloroplast mRNAs can be explained by protection of their 5′ and 3′ ends by proteins (4,7,13). The presence of Chlamydomonas nuclear mutants that lack specific chloroplast transcripts, while all other transcripts accumulate, suggest that 5′ and/or 3′ end-protecting proteins are transcript specific (3). For example, Chlamydomonas mutant mrl1 which fails to accumulate solely rbcL transcripts has been found to lack a conserved nucleus-encoded protein (MRL1) that binds to the rbcL 5′ UTR and stabilizes rbcL transcripts (11). To date, all Chlamydomonas mutants known to lack a specific transcript species in the chloroplast have been found to be deficient in 5′ UTR-binding proteins, suggesting that 5′ end protection is more variable and specific than protection of the 3′ ends in the Chlamydomonas chloroplast.
Different modes of ribonucleolytic degradation in the Chlamydomonas chloroplast
The results of this study provide indirect clues to the type of enzymes that degrade transcripts of the GUS reporter gene in the Chlamydomonas chloroplast. It has been shown previously (23) that a 5′ to 3′ exoribonuclease activity normally plays a role in mRNA breakdown in the Chlamydomonas chloroplast. The fact that degradation of an extension of 15 extra nucleotides at the 5′ end of GUS transcripts can be blocked by a poly(G) tract (Figure 3) confirms this finding and indicates that a 5′ to 3′ exoribonuclease activity is involved early in a 5′-initiated pathway. The finding that several unstable transcripts cannot be rescued by a poly(G) tract (Figure 4) suggests the involvement of additional ribonucleolytic activities that split GUS transcripts downstream of the poly(G) tract. Processive endoribonucleases, proposed to be involved in degradation of RNA downstream of processing signals in the 3′ end maturation pathway of atpB and, possibly, rbcL transcripts in the Chlamydomonas chloroplast (38,39), could be candidates for this type of ribonucleolytic activity.
Alternative to bypassing of poly(G) tracts by an endoribonucleolytic activity in 5′ to 3′ direction, complete loss of poly(G) harboring transcripts could also be due to degradation from their 3′ ends. While ribonucleases with 3′ to 5′ activities are present in chloroplasts and essential for complete breakdown of chloroplast mRNAs (4), it is not clear whether they play a major role in primary breakdown of the GUS reporter gene transcripts used in this study. A poly(G) tract inserted between the psaB 3′ end and the GUS coding sequence (Figure 1A) did not rescue GUS transcripts from rapid degradation (C1–3′ in Figure 4B) suggesting that a 3′ to 5′ exoribonuclease does not play a major role in primary breakdown of these transcripts. Initiation of degradation from the 3′ end triggered by sequence differences at the 5′ end would also require a hitherto unknown mechanism in chloroplasts to communicate 5′ structural and/or sequence features to the 3′ to 5′ degrading machinery.
It is also possible that poly(G) tracts in some of the very unstable GUS transcripts are rapidly degraded by a 5′ to 3′ exoribonuclease that changes processivity in a substrate-dependent manner. We consider this less likely because it is difficult to explain how such an exoribonuclease could distinguish between poly(G) tracts located in the same position of transcripts, e.g. WT+51 and C1+51 (Figure 5).
Finally, a protein with dual exo- and endoribonucleolytic activity may be involved in removing poly(G) tracts, switching its activity when encountering different RNA sequences and/or secondary structures, as proposed for RNase J1 of B. subtilis (40). Interestingly, an RNase J1 homolog appears to be present in Chlamydomonas (3).
Independent of the ribonucleases involved in 5′ to 3′ mRNA degradation, the sequences of the transcripts are important for routes of degradation (Figures 4 and 5). As mRNA degrading ribonucleases are unlikely to be sequence specific, sequence dependence is likely caused by differences in RNA secondary structures that may provide targets for different proteins involved in mRNA degradation. Alternatively, structural differences between the transcript variants could affect the relative activities of already bound ribonucleases leading to varying and shifting rates of ribonucleolytic activities.
Light/dark controlled RNA degradation
Routes of mRNA degradation appear to differ in cells kept in the dark and cells kept in light (Figure 6). It has been shown previously that photo-destabilization of GUS transcripts is mediated by redox changes in the Chlamydomonas chloroplast (41) and that sequences outside of the rbcL 5′ UTR are important for this pathway of RNA breakdown (42). As shown here (Figure 6), the light-dependent degradation pathway actively re-initiates degradation of GUS transcripts that accumulate in the dark. Because the transcripts analyzed all end 5′ with a poly(G) tract (Figures 3–5) photo-activation of mRNA degradation must involve a mechanism that degrades or ignores the poly(G) secondary structure. Whether the mechanism involves redox-mediated binding of new proteins to the mRNA or regulation of the activities of already bound proteins remains to be elucidated.
FUNDING
The Ministerio de Educación y Ciencia (BFU2006-07783/BMC to M.L.S.); Ministerio de Investigación, Ciencia e Innovación, Spain (BFU2009-11965/BMC to M.L.S.). Funding for open access charge: University of Valencia, University of Oslo.
Conflict of interest statement. None declared.
REFERENCES
- 1.Bollenbach TJ, Schuster G, Stern DB. Cooperation of endo- and exoribonucleases in chloroplast mRNA turnover. Prog. Nucleic Acid Res. Mol. Biol. 2004;78:305–337. doi: 10.1016/S0079-6603(04)78008-3. [DOI] [PubMed] [Google Scholar]
- 2.Bollenbach TJ, Schuster G, Portnoy V, Stern DB. Processing, degradation and polyadenylation of chloroplast transcripts. Topics Curr. Genet. 2008;19:175–211. [Google Scholar]
- 3.Herrin DL. Chloroplast RNA processing. In: Stern DB, editor. The Chlamydomonas Sourcebook. Vol. 2. Oxford: Academic Press; 2009. pp. 937–965. [Google Scholar]
- 4.Stern DB, Goldschmidt-Clermont M, Hanson M. Chloroplast RNA metabolism. Annu. Rev. Plant Biol. 2010;61:125–155. doi: 10.1146/annurev-arplant-042809-112242. [DOI] [PubMed] [Google Scholar]
- 5.Nickelsen J. Chloroplast RNA-binding proteins. Curr. Genet. 2003;43:329–399. doi: 10.1007/s00294-003-0425-0. [DOI] [PubMed] [Google Scholar]
- 6.Nakamura T, Schuster G, Sugiura M, Sugita M. Chloroplast RNA-binding and pentatricopeptide repeat proteins. Biochem. Soc. Trans. 2004;32:571–574. doi: 10.1042/BST0320571. [DOI] [PubMed] [Google Scholar]
- 7.Pfalz J, Bayraktar OA, Prikryl J, Barkan A. Site-specific binding of a PPR protein defines and stabilizes 5′ and 3′ mRNA termini in chloroplasts. EMBO J. 2009;28:2042–2052. doi: 10.1038/emboj.2009.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Saha D, Prasad AM, Srinivasan R. Pentatricopeptide repeat proteins and their emerging role in plants. Plant Physiol. Biochem. 2007;45:521–534. doi: 10.1016/j.plaphy.2007.03.026. [DOI] [PubMed] [Google Scholar]
- 9.Raynaud C, Loiselay C, Wostrikoff K, Kuras R, Girard-Bascou J, Wollman F-A, Choquet Y. Evidence for regulatory function of nucleus-encoded factors on mRNA stabilization and translation in the chloroplast. Proc. Natl Acad. Sci. USA. 2007;104:9093–9098. doi: 10.1073/pnas.0703162104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schwarz C, Elles I, Kortmann J, Piotrowski M, Nickelsen J. Synthesis of the D2 protein of photosystem II in Chlamydomonas is controlled by a high molecular mass complex containing the RNA stabilization factor Nac2 and the translational activator RBP40. Plant Cell. 2007;19:3627–3639. doi: 10.1105/tpc.107.051722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Johnson X, Wostrikoff K, Finazzi G, Kuras R, Schwarz C, Bujaldon S, Nickelsen J, Stern DB, Wollman F-A, Vallon O. MRL1, a conserved pentatricopeptide repeat protein, is required for stabilization of rbcL mRNA in Chlamydomonas and Arabidopsis. Plant Cell. 2010;22:234–248. doi: 10.1105/tpc.109.066266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jacobs J, Kück U. Function of chloroplast RNA-binding proteins. Cell. Mol. Life Sci. 2011;68:735–748. doi: 10.1007/s00018-010-0523-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Prikryl J, Rojas M, Schuster G, Barkan A. Mechanism of RNA stabilization and translational activation by a pentatricopeptide repeat protein. Proc. Natl Acad. Sci. USA. 2011;108:415–420. doi: 10.1073/pnas.1012076108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Higgs DC, Shapiro RS, Kindle KL, Stern DB. Small cis-acting sequences that specify secondary structures in a chloroplast mRNA are essential for RNA stability and translation. Mol. Cell Biol. 1999;19:8479–8491. doi: 10.1128/mcb.19.12.8479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nickelsen J, van Dillewijn J, Rahire M, Rochaix J-D. Determinants for stability of the chloroplast psbD RNA are located within its short leader region in Chlamydomonas reinhardtii. EMBO J. 1994;13:3182–3191. doi: 10.1002/j.1460-2075.1994.tb06617.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Anthonisen IL, Salvador ML, Klein U. Specific sequence elements in the 5′ untranslated regions of rbcL and atpB gene mRNAs stabilize transcripts in the chloroplast of Chlamydomonas reinhardtii. RNA. 2001;7:1024–1033. doi: 10.1017/s1355838201001479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zicker AA, Kadakia C, Herrin DL. Distinct roles for the 5′ and 3′ untranslated regions in the degradation and accumulation of chloroplast tufA mRNA: identification of an early intermediate in the in vivo degradation pathway. Plant Mol. Biol. 2007;63:689–702. doi: 10.1007/s11103-006-9117-8. [DOI] [PubMed] [Google Scholar]
- 18.Deutscher MP. Degradation of RNA in bacteria: comparison of mRNA and stable RNA. Nucleic Acids Res. 2006;34:659–666. doi: 10.1093/nar/gkj472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kime L, Jourdan SS, McDowall KJ. Identifying and characterizing substrates of the RNase E/G family of enzymes. Methods Enzymol. 2008;447:215–241. doi: 10.1016/S0076-6879(08)02212-X. [DOI] [PubMed] [Google Scholar]
- 20.Mudd EA, Sullivan S, Gisby MF, Mironov A, Kwon CS, Chung W-I, Day A. A 125 kDa RNase E/G-like protein is present in plastids and is essential for chloroplast development and autotrophic growth in Arabidopsis. J. Exp. Bot. 2008;5:2597–2610. doi: 10.1093/jxb/ern126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schein A, Sheffy-Levin S, Glaser F, Schuster G. The RNase E/G-type endoribonuclease of higher plants is located in the chloroplast and cleaves RNA similarly to the E. coli enzyme. RNA. 2008;14:1057–1068. doi: 10.1261/rna.907608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zimmer SL, Schein A, Zipor G, Stern DB, Schuster G. Polyadenylation in Arabidopsis and Chlamydomonas organelles: the input of nucleotidyltransferases, poly(A) polymerases and polynucleotide phosphorylase. Plant J. 2009;59:88–99. doi: 10.1111/j.1365-313X.2009.03853.x. [DOI] [PubMed] [Google Scholar]
- 23.Drager RG, Higgs DC, Kindle KL, Stern DB. 5′ to 3′ exoribonucleolytic activity is a normal component of chloroplast mRNA decay pathways. Plant J. 1999;19:521–531. doi: 10.1046/j.1365-313x.1999.00546.x. [DOI] [PubMed] [Google Scholar]
- 24.Boynton JE, Gillham NW, Harris EH, Hosler JP, Johnson AM, Jones AR, Randolph-Anderson BL, Robertson D, Klein TM, Shark KB, et al. Chloroplast transformation in Chlamydomonas with high velocity microprojectiles. Science. 1988;240:1534–1538. doi: 10.1126/science.2897716. [DOI] [PubMed] [Google Scholar]
- 25.Blowers AD, Bogorad L, Shark K, Sanford JC. Studies on Chlamydomonas chloroplast transformation: foreign DNA can be stably maintained in the chromosome. Plant Cell. 1989;1:123–132. doi: 10.1105/tpc.1.1.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sambrook J, Russell DW. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- 27.Suay L, Salvador ML, Abesha E, Klein U. Specific roles of 5′ RNA secondary structures in stabilizing transcripts in chloroplasts. Nucleic Acids Res. 2005;33:4754–4761. doi: 10.1093/nar/gki760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Blowers AD, Klein U, Ellmore GS, Bogorad L. Functional in vivo analyses of the 3′ flanking sequences of the Chlamydomonas chloroplast rbcL and psaB genes. Mol. Gen. Genet. 1993;238:339–349. doi: 10.1007/BF00291992. [DOI] [PubMed] [Google Scholar]
- 29.Salvador ML, Klein U, Bogorad L. Light-regulated and endogenous fluctuations of chloroplast transcript levels in Chlamydomonas: regulation by transcription and RNA degradation. Plant J. 1993;3:213–219. doi: 10.1046/j.1365-313x.1993.t01-13-00999.x. [DOI] [PubMed] [Google Scholar]
- 30.Klein U, Salvador ML, Bogorad L. Activity of the Chlamydomonas chloroplast rbcL gene promoter is enhanced by a remote sequence element. Proc. Natl Acad. Sci. USA. 1994;91:10819–10923. doi: 10.1073/pnas.91.23.10819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Markham NR, Zuker M. UNAFold: software for nucleic acid folding and hybriziation. In: Keith JM, editor. Methods in Molecular Biology, number 453. Bioinformatics. Vol. II. Totowa, NJ: Humana Press; 2008. pp. 3–31. [DOI] [PubMed] [Google Scholar]
- 32.Salvador ML, Suay L, Anthonisen IL, Klein U. Changes in the 5′ region of the rbcL gene accelerate transcript degradation more than 50-fold in the chloroplast of Chlamydomonas reinhardtii. Curr. Genet. 2004;45:176–182. doi: 10.1007/s00294-003-0470-8. [DOI] [PubMed] [Google Scholar]
- 33.Decker CJ, Parker R. A turnover pathway for both stable and unstable mRNAs in yeast: evidence for a requirement for deadenylation. Genes Dev. 1993;7:1632–1643. doi: 10.1101/gad.7.8.1632. [DOI] [PubMed] [Google Scholar]
- 34.Salvador ML, Klein U, Bogorad L. 5′ sequences are important positive and negative determinants of the longevity of Chlamydomonas chloroplast transcripts. Proc. Natl Acad. Sci. USA. 1993;90:1556–1560. doi: 10.1073/pnas.90.4.1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bouvet P, Belasco JG. Control of RNase E-mediated RNA degradation by 5′-terminal base pairing in E. coli. Nature. 1992;360:488–491. doi: 10.1038/360488a0. [DOI] [PubMed] [Google Scholar]
- 36.Deana A, Celesnik H, Belasco JG. The bacterial enzyme RppH triggers messenger RNA degradation by 5′ pyrophosphate removal. Nature. 2008;451:355–359. doi: 10.1038/nature06475. [DOI] [PubMed] [Google Scholar]
- 37.Even S, Pellegrini O, Zig L, Labas V, Vinh J, Bréchemmier-Baey D, Putzer H. Ribonucleases J1 and J2: two novel endoribonucleases in B. subtilis with functional homology to E. coli RNase E. Nucleic Acids Res. 2005;33:2141–2152. doi: 10.1093/nar/gki505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hicks A, Drager RG, Higgs DC, Stern DB. An mRNA 3′ processing site targets downstream sequences for rapid degradation in Chlamydomonas chloroplasts. J. Biol. Chem. 2002;277:3325–3333. doi: 10.1074/jbc.M108979200. [DOI] [PubMed] [Google Scholar]
- 39.Goldschmidt-Clermont M, Rahire M, Rochaix J-D. Redundant cis-acting determinants of 3′ processing and RNA stability in the chloroplast rbcL mRNA of Chlamydomonas. Plant J. 2008;53:566–577. doi: 10.1111/j.1365-313X.2007.03365.x. [DOI] [PubMed] [Google Scholar]
- 40.Mathy N, Bérnard L, Pellegrini O, Daou R, Wen T, Condon C. 5′-to-3′ exoribonuclease activity in bacteria: Role of RNase J1 in rRNA maturation and 5′ stability of mRNA. Cell. 2007;129:681–692. doi: 10.1016/j.cell.2007.02.051. [DOI] [PubMed] [Google Scholar]
- 41.Salvador ML, Klein U. The redox state regulates RNA degradation in the chloroplast of Chlamydomonas reinhardtii. Plant Physiol. 1999;121:1367–1374. doi: 10.1104/pp.121.4.1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Singh M, Boutanaev A, Zucchi P, Bogorad L. Gene elements that affect the longevity of rbcL sequence-containing transcripts in Chlamydomonas reinhardtii chloroplasts. Proc. Natl Acad. Sci. USA. 2001;98:2289–2294. doi: 10.1073/pnas.041609798. [DOI] [PMC free article] [PubMed] [Google Scholar]