Abstract
Cyclic mechanical stress (CMS) leadsQ1 to alterations of cellular functions in the trabecular meshwork (TM), including the up-regulation of transforming growth factor beta 1 (TGFβ1), that can potentially contribute to the pathogenesis of glaucoma. Although microRNAs (miRNAs) are known to play important roles in many biological functions, little is known about their potential involvement in the cellular responses elicited by mechanical stress. Here we analyzed changes in miRNA expression induced by CMS, and examined the possible role of miR-24 in the response of human TM cells to CMS. CMS induced the expression of miR-24 that led to the down regulation of the subtilisin-like proprotein convertase FURIN, which is known to play a major role in the processing of TGFβ1. FURIN was confirmed as a novel target of miR-24 by 3′ UTR luciferase assay and western blot. Overexpression of miR-24 resulted in a significant decrease in activated TGFβ1. This effect was mimicked by down regulation of FURIN by siRNA. Conversely, inhibition of miR-24 expression with a specific antagomir led to a small but significant increase in TGFβ1. Furthermore, the increase in active TGFβ1 induced by CMS in HTM cells was prevented by miR-24. Altogether, our results suggest that miRNAs might contribute to the regulation of responses to CMS in TM cells. Specifically, miR-24 might play an important role in modulating the induction of TGFβ1 mediated by CMS through direct targeting of FURIN.
Introduction
The trabecular meshwork (TM) and Schlemm’s canal form the major route by which the aqueous humor exits the anterior chamber of the human eye and constitute the site of the abnormal increase in outflow resistance that leads to elevated intraocular pressure (IOP) in glaucoma (Fautsch and Johnson, 2006; Johnson, 2006; Overby et al., 2009; Tektas and Lutjen-Drecoll, 2009). There is abundant evidence that the TM is deformed when IOP increases and the anatomy is restored as the IOP decreases (Johnstone, 1979; Johnstone, 2004). Thus, it is likely that transient IOP oscillations such as those resulting from the ocular pulse lead to constant cycles of stretching and then relaxation of the TM cells. Previous studies have demonstrated that TM cells are sensitive to mechanical forces (Borras, 2003; WuDunn, 2009) and that cyclic mechanical stress (CMS) induces changes in cell morphology and gene expression that can potentially exert important effects on the physiology of the TM (Luna et al., 2009a, b; Ramos et al., 2009). Some of the responses elicited by CMS in TM cells, including the increased expression of MMP3 (Luna et al., 2009a) and release of PGE2 (Luna et al., 2009b), have been hypothesized to constitute homeostatic mechanisms aimed at increasing aqueous humor outflow in response to mechanical stress resulting from elevations in IOP (Luna et al., 2009a, b). In contrast, other responses mediated by CMS, such as increased expression of TGFβ1 (Liton et al., 2005), BMP2 (Luna et al., 2009a), CTGF (Chudgar et al., 2006), or increased cell contractility (Ramos et al., 2009) may contribute to pathological alterations of the TM and then secondarily further increase IOP. In spite of the physiologic relevance of the effects of CMS on the TM, the molecular mechanisms involved in the alterations of gene expression induced by CMS in TM cells are still poorly understood.
MicroRNAs (miRNAs) are known to play an important role in the regulation of many cellular functions by either repressing translation or inducing mRNA degradation of multiple specific gene targets (Ying et al., 2006; Bushati and Cohen, 2007). MiRNAs can participate in the regulation of gene expression changes induced by several types of stress (Marsit et al., 2006; Martin et al., 2010Q2), including hypoxic stress (Crosby et al., 2009), cold stress (Dresios et al., 2005), and oxidative stress (Cheng et al., 2009; Lin et al., 2009; Luna et al., 2009c). However, only a few studies have analyzed the potential involvement of miRNAs on the responses induced by mechanical forces in general (Qin et al. 2010a; Weber et al., 2010), and nothing is known about their role in the responses induced by CMS. Therefore, in the present study, we examined whether CMS could lead to changes in miRNA expression in HTM cells, and investigated the mechanisms by which some of these changes might influence the responses induced by CMS in HTM cells.
Material and Methods
Cell culture and treatments
Human trabecular meshwork (HTM) cell cultures were generated from cadaver eyes, with no history of eye disease, within 48 h post mortem as previously reported (Stamer et al., 1995). All procedures involving human tissue were conducted in accordance with the tenets of the Declaration of Helsinski. Cell cultures were maintained at 37ºC in 5% CO2 in media (low glucose Dulbecco’s Modified Eagle Medium with L-glutamine, 110 mg/ml sodium pyruvate, 10% fetal bovine serum, 100 μM non-essential amino-acids, 100 units/ml penicillin, 100 μg/ml streptomycin sulfate, and 0.25 μg/ml amphotericin B). All the reagents were obtained from Invitrogen Corporation (Carlsbad, CA). SB431542 (Tocris Bioscience, Ellisville, MO) a TGFβ1 inhibitor was added at 10 μM concentration in serum free media during 30 min after which cells were washed and TGFβ1 (Sigma, St Louis, MO) was added at a concentration of 1 ng/ml in serum free media, and cells were analyzed after 24 h.
Cyclic mechanical stress (CMS)
HTM cell cultures (passages 3–5) were plated on type I collagen-coated flexible silicone bottom plates (Flexcell, Hillsborough, NC). For miRNA analysis cells were grown to confluence, and for transfections with miRNAs, cells were plated at 50–70% confluency, transfected 24 h later and subjected to CMS 72 h after transfection. Medium was switched to serum-free DMEM 2 h before CMS and cells were stretched for 3 or 16 h (20% stretching, 1 cycle per second), using the computer-controlled, vacuum-operated FX-3000 Flexercell Strain Unit (Flexcell, Hillsborough, NC). Frequency of 1 cycle per second was selected to mimic cardiac frequency. Control cells were cultured under the same conditions but no mechanical force was applied.
Transfections
HTM cells were transfected with hsa-miR-24 mimic or control mimic (scramble) (20–40 pmolar) (Dharmacon, Chicago, IL) using an endothelial nucleofactor Kit (Lonza, Basel, Switzerland) or lipofectamine 2000 (Invitrogen), following manufacturer’s instructions. Co-transfections of 293A cells with luciferase 3′UTR constructs (0.3 μg), miR-24 mimic or control mimic (20 pmolar) were accomplished using Effectene (Qiagen, Valencia, CA) following manufacturer’s instructions.
RNA isolation, Quantitative PCR (Q-PCR), and miRNA PCR arrays
Total RNA was isolated using a RNeasy kit (Qiagen Inc.) or Trizol (Invitrogen) according to the manufacturer’s instructions. RNA yields were measured using RiboGreen fluorescent dye (Invitrogen). First strand cDNA was synthesized from total RNA (1 μg) by reverse transcription using oligodT and Superscript II reverse transcriptase (Invitrogen) according to manufacturer’s instructions. Q-PCR reactions were performed in 20 μl mixture containing 1 μl of the cDNA preparation, 1X iQ SYBR Green Supermix (Biorad, Hercules, CA), using the following PCR parameters: 95ºC for 5 min followed by 50 cycles of 95ºC for 15 sec, 65ºC for 15 sec, and 72ºC for 15 sec. β-Actin was used as an internal standard of mRNA expression. The absence of non-specific products was confirmed by both the analysis of the melt curves and by electrophoresis in 3% Super acryl-Agarose gels. The primers used for Q-PCR amplification are shown in Table 1. MicroRNAs were extracted from total RNA using RT2 qPCR-Grade miRNA isolation kit (SABiosciences, Frederick, MD). For PCR Arrays, miRNAs cDNA (100 ng) were amplified using RT2 miRNA First Strand Kit and RT2 miRNA PCR Array (MAH-001A) following manufacturer’s instructions (SABiosciences, Frederick, MD). For miR-24 amplification we used TaqMan microRNA reverse transcription kit, specific primers for hsa-miR-24 and U6B, as a standard, and 25 ng of enriched small microRNAs. Q-PCR products were amplified using TaqMan® Universal PCR Master Mix, following manufacturer’s instructions (Applied Biosystems, Foster City, CA). The fluorescence threshold value (Ct) was calculated using the iCycle system software. The results were expressed as mean value ± SD in three independent experiments.
TABLE 1.
Primers used for Quantitative PCR
| Gene symbol | Forward 5′-3′ | Reverse 5′-3′ |
|---|---|---|
| CNDP2 | GCACAGCCACAAGAAAGACA | ATGGAGAACTTGCCAACCAC |
| F2RL1 | GATGTGGTCCAAACCCTCTG | TTTTGCCACTTAGAATAGCATTTG |
| KIF3B | CCAGGGAGCTGAAACTCAAG | TCTGGACGAATCATCATGGA |
| RASF2 | GGTCTTCCTGCACTTGAAGC | CACATAGGTGGCTGCTCAGA |
| CXCL6 | TTGAAGAGTGTGGGGGAAAG | CCACAGCCTTTTCGGTAAGA |
| SLC11A2 | ATGTCCTTCCTGGACTGTGG | CACACTGGCTCTGATGGCTA |
| NTPX1 | TGTACGCCTTCACTGTCTGC | TGATGACAAAAGGCAGCTTG |
| PDXK | GTCTCCGTGTTTGTCCCTGT | TTTTCACAAAGCACGACTGG |
| ENTDP6 | TTGCCTCTTCCTTGGGTATG | ACGTCACTCAAGCAGCACAG |
| STC2 | TCTGCACCTCGGCCATCCAG | TCAGAATACTCAGACTGTTC |
| IL8 | AGGACAAGAGCCAGGAAGAA | ACTGCACCTTCACACAGAGC |
| FURIN | ACAACTATGGGACGCTGACC | TGGACACAGCTCTTCTGGTG |
| ACTB | CCTCGCCTTTGCCGATCCG | GCCGGAGCCGTTGTCGACG |
Gene microarray analysis
Gene array analysis was conducted in three independent sets of transfections with either miR-24 mimic or mimic control of the same HTM cell line. Total RNA was extracted 3 days post-transfection using RNeasy kit (Qiagen), amplified (1 round amplification) using One cycle target labeling and control reagents (Affymetrix, Santa Clara, CA) and hybridized to Human Genome U133A2 Arrays (Affymetrix) at the Duke University Microarray facility. Raw data were normalized using quantile normalization and analyzed using GeneSpring 10 (Silicon Genetics). ANOVA test was performed (P-values ≤ 0.05) for genes differentially expressed using the Benjamin and Hochberg False Discovery Rate correction test. The list of genes was compared to three databases that predict targets for microRNAs: Microcosm (http://www.ebi.ac.uk/enright-srv/microcosm/htdocs/targets/v5/), TargetScan (http://www.targetscan.org), and PicTar-Vert (http://pictar.mdc-berlin.de/). To study the potential biological significance of the changes observed in the arrays, we performed network analysis of differentially expressed genes using Metacore pathway analysis (GeneGo, St. Joseph, MI).
Interaction between miR-24 and FURIN 3′-untranslated region (3′-UTR)
The entire 3′UTR from FURIN was amplified using the following primers FURIN 3′-F-ggCTCGAGgcaagaggggtggagactgc and FURIN 3′-R-gggGCGGCCGCctgtgcaccaacccagcatc, respectively, which carried XhoI and NotI restriction sites in the forward or the reverse position. PCR amplifications from 3′UTR and the complementary sequences were confirmed by sequencing and cloned into XhoI and NotI sites downstream of Renilla luciferase in the psiCheck2 vector (Promega, Madison, WI). For analysis of luciferase activity, 293A cells were seeded in 12 well plates, 24 h before transfection, transfected with psicheck 3′UTR or the complementary sequence from FURIN (0.3 μg), and miRNAs for miR-24 mimic or control mimic (20 pmolar). Luciferase was measured using the Dual Luciferase Kit (Promega, Madison, WI) following manufacturer’s instructions and read in a TD-20/20 luminometer (Turner Designs, Sunnyvale, CA).
Protein extraction and Western blotting
For protein extraction, cells were harvested 72 h after transfection, washed in PBS and lysated in 1X cold RIPA buffer, and protein concentration was determined using Micro BCA Protein Assay Kit (Pierce, Rockford, IL). Equal loading was run in 8% SDS–PAGE and transferred to PVDF membranes. Membranes were incubated overnight at 4ºC, with antibodies against FURIN or tubulin (Santa Cruz Biotechnology, Santa Cruz, CA). Blots were developed using a chemiluminescence detection system (ECL-Plus from Amersham, Buckinghamshire, UK).
TGFβ1 and TGFβ2 measurements
TGFβ1 and TGFβ2 were measured 72 h after transfection using Quantikine Human TGFβ1 and TGFβ2 (R&D systems, Minneapolis, MN) following manufacturer’s instructions. These are “sandwich” enzyme linked immunoassays that measure activated TGFβs. For CMS, supernatants were collected immediately after stretching (3 or 16 h). For other TGFβ measurements, supernatant was collected after 48 h, all media was serum free.
Results
Cyclic mechanical stress induced changes in miRNAs
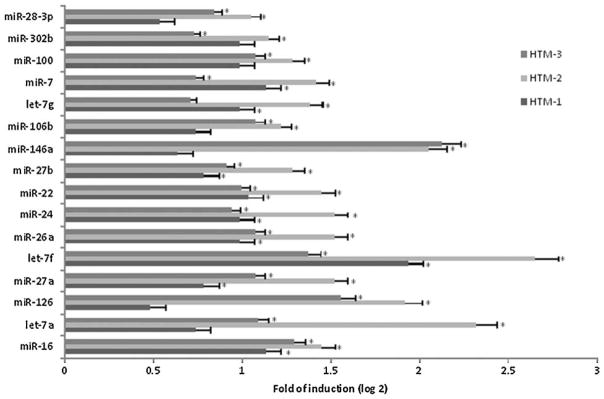
Three HTM cell lines were seeded by triplicates in collagen bioflex plates and subjected to CMS for 3 h at 20% stretching, 1 cycle per second. Non-mechanical force was applied to the controls. MicroRNAs were extracted from stressed and control cells and analyzed using miRNA PCR Arrays (Supplemental Material). Seven miRNAs were consistently up-regulated in all cell lines (miR-16, miR-27a, miR-27b, miR-7, let-7f, miR-26a, and miR-24) and another nine were significantly up-regulated in two cell lines (Fig. 1).
Fig. 1.
Changes in miRNA expressionQ3 induced by CMS in HTM cells. Three primary HTM cell lines were subjected to CMS (20% stretching, 1 cycle per second) for 3 h. Changes in microRNA expression were analyzed using miRNA PCR Arrays (MAH-001A). Control cells were incubated under the same conditions but no mechanical force was applied. The figure represents the logarithm of fold change in expression induced by CMS. Bars represent standard deviation from three different experiments; one asterisks means P ≤ 0.05.
Changes in gene expression induced by miR-24
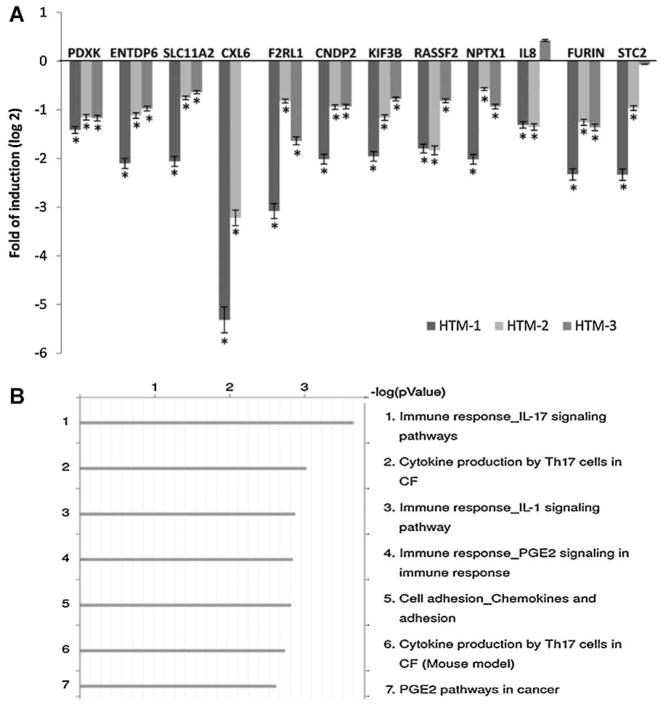
Since miR-24 is believed to be involved in the regulation of TGFβ1 signaling, which has been associated with multiple pathologic conditions associated with mechanical stress, we investigated the differences in gene expression induced by miR-24. For this purpose, one HTM cell line was transfected by triplicate with miR-24 or control mimic and the expression profile analyzed using Affymetrix U133A2 chips. Ninety-four genes were significantly (P ≤ 0.05) up- or down-regulated by miR-24 more than 1.5-fold. Twenty-one of these genes were predicted as putative targets of miR-24 by computational analysis (Table 2 shows genes up- or down- regulated by more than 1.8-fold). To validate Affymetrix microarray data, changes in expression of 12 genes were analyzed by Q-PCR in three independent HTM cell lines (Fig. 2A). To identify pathways, the genes significantly up- or down-regulated by more than 1.5-fold were analyzed using Metacore pathway analysis. The four canonical pathways more significantly affected by miR-24 were Immune Response IL-17 signaling pathway (P-value 5.5 × 10−4), Cytokine production by Th17 cells in cystic fibrosis (8.5 × 10−3), Immune response IL-1 signaling pathway (7.3 × 10−3), and Immune response PGE2 signaling pathway (7 × 10−3) (Fig. 2B).
TABLE 2.
Genes up- or down-regulated by 1.8-fold in human trabecular meshwork cells after over expression of miR-24 mimic
| Gene symbol | Fold change | Regulation | P-value | Database | Gene title |
|---|---|---|---|---|---|
| PTHLH | 3.0565975 | Down | 3.97E-04 | M,T,P | Parathyroid hormone-like hormone |
| CNDP2 | 2.4332333 | Down | 4.35E-05 | CNDP dipeptidase 2 | |
| IL8 | 2.2841344 | Down | 1.86E-04 | Interleukin 8 | |
| F2RL1 | 2.2676232 | Down | 4.22E-05 | T | Coagulation factor II (thrombin) receptor-like 1 |
| IL8 | 2.2576025 | Down | 4.99E-04 | P | Interleukin 8 |
| KIF3B | 2.2168906 | Down | 2.55E-05 | Kinesin family member 3B | |
| GALNT12 | 2.1888528 | Down | 0.0029251 | M | UDP-N-acetyl-alpha-D-galactosamine |
| RASSF2 | 2.15163 | Down | 2.69E-06 | Ras associationdomain family member 2 | |
| CXCL6 | 2.13901 | Down | 0.0017257 | Chemokine (C-X-C motif) ligand 6 | |
| SLC11A2 | 2.0708907 | Down | 1.35E-04 | Solute carrier family 11, member 2 | |
| C14orf2 | 2.0584471 | Down | 2.15E-05 | Chromosome 14 open reading frame 2 | |
| ANXA10 | 2.0433328 | Down | 3.64E-04 | Annexin A10 | |
| NPTX1 | 2.027866 | Down | 0.0187071 | Neuronal pentraxin I | |
| PDXK | 2.0209773 | Down | 1.01E-05 | T,P | Pyridoxal kinase |
| ENTPD6 | 2.0142882 | Down | 1.31E-04 | Ectonucleoside triphosphate diphosphohydrolase 6 | |
| KCNK3 | 1.9923251 | Down | 1.82E-04 | T,P | Potassium channel, subfamily K, member 3 |
| STC2 | 1.9881191 | Down | 1.43E-05 | T | Stanniocalcin 2 |
| HHLA3 | 1.9669507 | Down | 0.0013616 | HERV-H LTR-associating 3 | |
| CXCL1 | 1.9485825 | Down | 2.07E-04 | Chemokine (C-X-C motif) ligand 1 | |
| ACVR1B | 1.9390614 | Down | 2.84E-04 | M,T | Activin A receptor, type IB |
| MED16 | 1.9286143 | Down | 6.86E-04 | Mediator complex subunit 16 | |
| STC2 | 1.9134991 | Down | 5.14E-05 | Stanniocalcin 2 | |
| SYT1 | 1.9131047 | Up | 0.0040418 | Synaptotagmin I | |
| MICB | 1.8921673 | Down | 6.76E-05 | MHC class I polypeptide-related sequence B | |
| AVL9 | 1.8742172 | Down | 2.70E-06 | AVL9 homolog (S. cerevisiase) | |
| PTGS2 | 1.8690909 | Down | 6.22E-05 | Prostaglandin-endoperoxide synthase 2 | |
| GRINA | 1.8647411 | Down | 0.0017328 | Glutamate receptor, ionotropic | |
| BTN3A2/BTN3A3 | 1.857413 | Down | 1.95E-04 | Butyrophilin, subfamily 3, member A2//A3 | |
| FURIN | 1.8437475 | Down | 9.03E-04 | Furin (paired basic amino acid cleaving enzyme) | |
| STC1 | 1.8405004 | Down | 2.87E-05 | Stanniocalcin 1 | |
| RAP2C | 1.839736 | Down | 4.87E-04 | RAP2C, member of RAS oncogene family | |
| ADAM12 | 1.8122648 | Down | 6.18E-04 | ADAM metallopeptidase domain 12 | |
| GREM1 | 1.8109983 | Down | 5.69E-04 | Gremlin 1, cysteine knot superfamily, homolog | |
| GREM1 | 1.8081648 | Down | 1.07E-04 | M | Gremlin 1, cysteine knot superfamily, homolog |
M, T, P means Microcosm, TargetScan, and PicTar-Vert miRNA target prediction databases.
Fig. 2.
Validation of Affymetrix microarray data and analysis of canonical pathways affected by miR-24. Panel A represents the logarithm of the fold change on gene expression of HTM cells transfected with miR-24 mimic compared to cells transfected with scramble control in three primary cell lines. The expression of 12 genes significantly down-regulated on the array (Gene symbol: PDXK, ENTDP6, SLC11A2, CXXL6, F2RL1, CNDP2, KIF3B, RASF2, NPTX1, IL8, FURIN, and STC2) were analyzed by Q-PCR. Bars represent standard deviation from three different experiments; one asteriskmeans P ≤ 0.05. Panel B represents the seven canonical pathways most significantly affected by miR-24 mimic compared to controls and was generated using Metacore pathway analysis and the genes significantly up- or down-regulated by more than 1.5-fold (P ≤ 0.05) on the Affymetrix U133A2 arrays (CF = cystic fibrosis).
Targeting of FURIN by miR-24
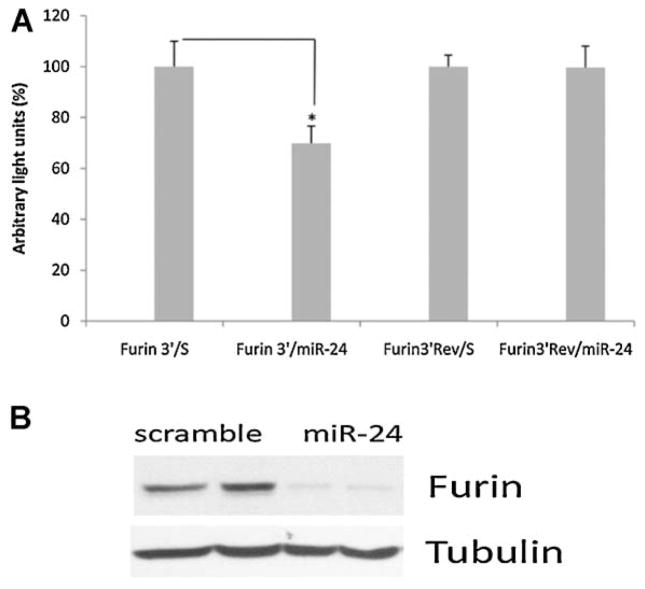
FURIN was among the genes significantly down-regulated by miR-24 and computational predictions indicated that miR-24 shares complementarity with the 3′UTR of FURIN. We confirmed that the 3′UTR of FURIN interacts with miR-24 using the psiCheck2 luciferase assay system. MiR-24 reduced the luciferase activity in cells co-transfected with FURIN 3′UTR compared to control (scrambled). This effect was prevented when the 3′UTR complementary sequence of FURIN was used. FURIN down-regulation was confirmed at protein level after transfection with miR-24 or scrambled (Fig. 3A,B).
Fig. 3.
FURIN is a novel target of miR-24. Panel A represents the luciferase activity (%) in 293 cells co-transfected with psicheckvectors, containing the 3′UTR or complementary sequence (R) from FURIN, and miR-24, compared to cells co-transfected with the same vectors and scramble control. Bars represent standard deviation from three different experiments; two asterisks means P ≤ 0.01. In panel B HTM cells were transfected with miR-24 mimic or scramble control and the expression of FURIN and tubulin were assayed by Western blot 3 days after transfection.
Regulation of TGFβ1 by miR-24
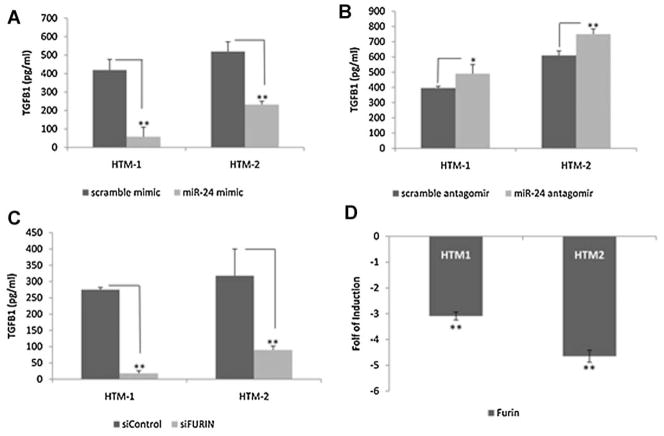
FURIN is known to play an important role in TGFβ1 processing. To evaluate whether miR-24 could affect the expression of active TGFβ1 we transfected two HTM cell lines with miR-24 or scramble control and measured the activated TGFβ1 and TGFβ2 by ELISA. TGFβ1 was significantly down-regulated by miR-24 by 83 and 54%, and miR-24 antagomir increased activated TGFβ1 by 21 and 22% (Fig. 4A,B). TGFβ2 was not significantly affected by miR-24 in the same experiments (data not showed).
Fig. 4.
Effects of changes in expression of miR-24 and FURIN on the production of active TGFβ1 in HTM cells. Activated TGFβ1 (pg/ml) was measured by ELISA on cell culture supernatant from two HTM cell lines transfected with miR-24 mimic, scramble control, siFURIN or siControl. Panel A represents TGFβ1 expression on cells transfected with miR-24 mimic or scramble control. Panel B represents a similar experiment in cell lines transfected with miR-24 antagomir or scramble antagomir. Panel C represents cell lines transfected with RNA interference against FURIN (siFURIN)or RNA interference control (siControl). Panel D shows the down-regulation of FURIN mRNA in the same experiments, measured by Q-PCR. Bars represent standard deviation from three different experiments. One, two, and three asterisks means P ≤ 0.05, 0.01, and 0.005.
RNA interference against FURIN down-regulated TGFβ1 expression
To evaluate whether specific down-regulation of FURIN was enough to inhibit the expression of active TGFβ1 in HTM cells, we transfected two cell lines with small interference RNA against FURIN or scramble RNA and measured the activated TGFβ1 by ELISA. siFURIN down-regulated significantly (88 and 69%) the amount of activated TGFβ1 compared to the control (Fig. 4C). The knockdown of FURIN was confirmed by Q-PCR (Fig. 4D).
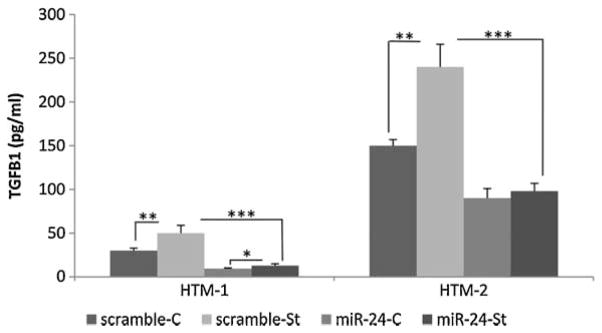
MiR-24 prevented the up-regulation of active TGFβ1 induced by CMS
Two HTM cell lines transfected with scramble or miR-24 were subjected to CMS, during 16 h and the amount of activated TGFβ1 was measured by ELISA. TGFβ1 increased significantly in cells subjected to CMS compared to non-stressed cells by 64 and 47%. Cells transfected with miR-24 and subjected to CMS increased TGFβ1 by 35 and 5%, respectively (Fig. 5).
Fig. 5.
Increase in active TGFβ1 induced by CMS was prevented by miR-24. Figure represents the amount of TGFβ1 (pg/ml) in two HTM cell lines transfected with miR-24 or scramble control and subjected to CMS for 16 h (St) or no mechanical force was applied in the control (C). Bars represent standard deviation from three different experiments. One, two, and three asterisks means P ≤ 0.05, 0.01, and 0.001.
TGFβ1 up-regulated miR-24 expression
In order to know if a feedback exists between miR-24 and TGFβ1, we treated two HTM cell lines with 1 ng per ml of TGFβ1 or TGFβ1 plus a TGFβ1 inhibitor (SB431542, 10 μM) and analyzed the expression of miR24 by Q-PCR. TGFβ1 increased significantly the expression of miR-24 and SB431542 abolished this increase (Fig. 6).
Fig. 6.
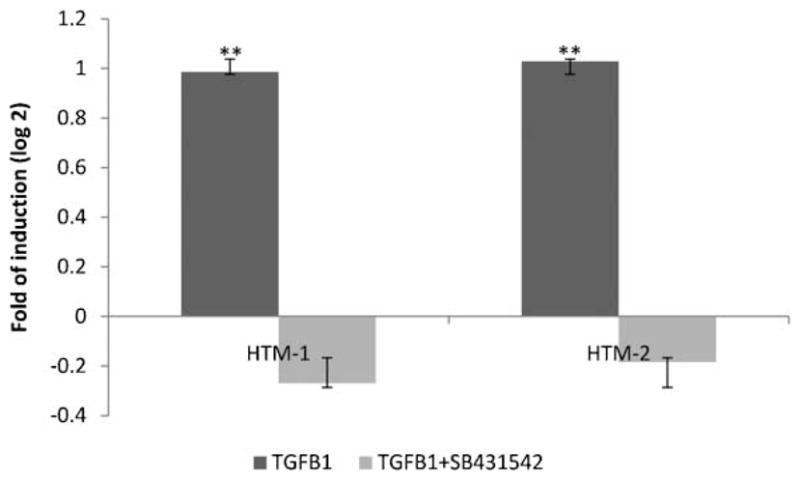
TGFβ1 increased expression of miR-24. Figure represents the logarithm of the fold change of miR-24 after 24 h of treatment with TGFβ1 (1 ng/ml) or TGFβ1 plus SB431542 (10 μM), a TGFβ1 inhibitor. Bars represent standard deviation from three different experiments in two HTM cell lines; two asterisks means P ≤ 0.01.
Discussion
Cellular responses to CMS are believed to play an important role in the physiology of the outflow pathway. Our results showed that CMS induced the expression of several miRNAs in HTM cells and that one of them, miR-24, can contribute to the regulation of the levels of TGFβ1 induced by CMS in HTM cells.
To our knowledge the only two previous studies addressing the effects of mechanical stress on miRNA expression in mammalian cells have been conducted in vascular endothelial cells using shear stress as a model (Qin et al., 2010a; Weber et al., 2010). Although the cell types and mode of mechanical stress reported in these publications were very different from those used in our study, an interesting common effect of shear stress in vascular endothelial cells and CMS in HTM cells was the up-regulation of members of the miR-23a/24-2/27a and miR-23b/24-1/27b paralog clusters. Therefore, up-regulation of miRNAs from these two clusters might constitute a common response to different forms of mechanical stress in several cell types.
The MicroRNA-23b/24-1/27b cluster has been demonstrated to regulate transforming growth factor-beta (TGFβ)/bone morphogenetic protein signaling and liver stem cell differentiation by different mechanisms including the direct targeting of Smads (Rogler et al., 2009). Specifically, miR-24, which is expressed from both miR-23a/24-2/27a and miR-23b/24-1/27b clusters, has been shown to antagonize with TGFβ signaling through post-transcriptional regulation of Tribbles-like protein-3 (Trb3) (Chan et al., 2009Q4). Trb3 mediates degradation of the SMAD specific E3 ubiquitin protein ligase 1, Smurf1 (Chan et al., 2007), which is involved in degradation of Smads and facilitates the antagonistic action of Smad7 by targeting Smad7 at the plasma membrane (Suzuki et al., 2002). Another example of the antagonistic activity of miR-24 with the TGFβ superfamily has been reported in hematopoietic progenitor cells where miR-24 targets the activin type I receptor ALK4 (ACVR1B) that interferes with activin-induced Smad 2 phosphorylation thus delaying activin-induced maturation of hematopoietic progenitor cells in cell cultures (Wang et al., 2008).
Induction of TGFβ is a common response to mechanical stress in several cell types (Yasuda et al., 1996; Li et al., 1998; Skutek et al., 2001; Sakata et al., 2004; Mohamed and Boriek, 2010) including TM (Liton et al., 2005). Such induction of TGFβ contributes to the pathogenesis of multiple conditions such as cystic fibrosis, liver disease, lung fibrosis, tubulointerstitial fibrosis in the kidney, myocardial fibrosis, glomerular sclerosis, diabetic nephropathy, and asthma (Yasuda et al., 1996; Hirakata et al., 1997; Ihn, 2002a, b; Kelly et al., 2003; Lee et al., 2006; Wolf and Ziyadeh, 2007; Kassiri et al., 2009; Mohamed and Boriek, 2010; Rohatgi and Flores, 2010). TGFβ1 is believed to play an important role in both the normal physiology of the TM and the pathogenesis of this tissue in glaucoma (Zhao et al., 2004; Tan et al., 2006; Acott and Kelley, 2008; Fatma et al., 2009; Fuchshofer and Tamm, 2009; Agarwal and Agarwal, 2010). For example, over-expression of TGFβ1 in rat eyes changed the morphology of the anterior segment and increased IOP, and elevation of TGFβ1 has been associated with pseudoexfoliative and neovascular glaucomas (Yu et al., 2007; Robertson et al., 2010; Schlotzer-Schrehardt et al., 2001; Zenkel et al., 2010).
Given the potential involvement of miR-24 in the response to mechanical stress in different cell types and the relevance of TGFβ in the pathogenic responses induced by mechanical stress, we analyzed the influence of this miRNA on gene expression in HTM cells. Our results showed that miR-24 induced significant changes in expression of several genes involved in immune response and cytokine production, including the previously reported target ACVR1B (ALK4, fold −1.94). Our results also showed a significant decrease in expression of the Subtilisin-like proprotein convertase FURIN (fold −1.84), which was confirmed as a novel bona fide target of this microRNA. Targeting of FURIN by miR-24 can be particularly relevant to the understanding of the antagonistic effects of this microRNA with the TGFβ pathway since FURIN is known to play a major role in the processing of TGFβ1 (Dubois et al., 2001; Kusakabe et al., 2008).
TGFβs are secreted as pro-proteins and cleaved to mature TGFβs and their latency associated peptides (LAPs) by the convertase family of endoproteases. After cleavage TGFβs and LAP remain non-covalently attached as latent complexes and secreted to the pericellular space associated with the ECM (Koli et al., 2001; Annes et al., 2003; ten Dijke and Arthur, 2007) (Dubois et al., 2001; Kusakabe et al., 2008; Hynes, 2009). Consistent with the role of FURIN on the processing of TGFβ1, miR-24 was able to decrease the levels of active TGFβ1 in HTM cell cultures, and this effect was mimicked by the inhibition of FURIN expression with a specific siRNA. The observed lack of effects of miR-24 on the presence of active TGFβ2 was also consistent with the known insensitivity of latent TGFβ2 to FURIN that results from the differences between the tertiary structure of the LAP regions of TGFβ1 and TGFβ2 (Kusakabe et al., 2008).
Our results also indicated that induction of miR-24 might contribute to regulate the amount of TGFβ1 activated by CMS in HTM cells since miR-24 mimic decreased and miR-24 antagomir increased the levels of active TGFβ1 produced by HTM. Furthermore, the observation that miR-24 was up-regulated by TGFβ1 in HTM cells suggests the presence of a negative feedback loop between TGFβ1 and miR-24 in which miR-24 might be up-regulated as a homeostatic response to increased levels of active TGFβ1. Thus, the up-regulation of miR-24 during CMS could potentially serve to prevent excessive activation of TGFβ1 and limit some of the pathogenic effects of this cytokine in the outflow pathway. The up-regulation of miR-24 by TGFβ1 observed in HTM cells may be cell-type or context dependent, since TGFβ1 has been reported to both, repress miR-24 transcription during skeletal muscle differentiation (Konishi et al., 1991), and induce the expression of the miR-23a/27a/24 cluster in Huh-7 cells (Huang et al., 2008).
In addition to its role in regulating the TGFβ superfamily, miR-24 is believed to act as a tumor suppressor by regulating cell cycle progression, apoptosis, and DNA damage responses through several validated targets (Lal et al., 2009a, b; Mishra et al., 2009; Qin et al., 2010b; Takagi et al., 2010; Zaidi et al., 2009). The observed targeting of FURIN by miR-24 could also be particularly relevant to the tumor suppressor effects mediated by miR-24. FURIN is known to play an important role in the acquisition of malignant phenotype and metastatic potential in tumor cells by processing growth factors (Basak et al., 2010), by enable tumors to evade the antiangiogenic effects of sema3B (Varshavsky et al., 2008), and also inhibition of FURIN has been shown to significantly inhibit invasion and migration of human cancer cells (Bassi et al., 2001; Zhou et al., 2009).
In conclusion, CMS induced significant alterations in the expression of several microRNAs that could contribute to the regulation of some of the responses to mechanical stress in HTM cells. Specifically, up-regulation of miR-24 and the subsequent down-regulation of its target FURIN may serve as a homeostatic mechanism to limit the amount of TGFβ1 activated by CMS and to prevent some potentially pathogenic effects of this cytokine in the outflow pathway.
Acknowledgments
This work was supported by NEI EY01894, NEI EY016228, NEI EY05722, and Research to Prevent Blindness.
Literature Cited
- Acott TS, Kelley MJ. Extracellular matrix in the trabecular meshwork. Exp Eye Res. 2008;86:543–561. doi: 10.1016/j.exer.2008.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agarwal R, Agarwal P. Future target molecules in antiglaucoma therapy: tgf-Beta may have a role to play. Ophthalmic Res. 2010;43:1–10. doi: 10.1159/000246571. [DOI] [PubMed] [Google Scholar]
- Annes JP, Munger JS, Rifkin DB. Making sense of latent TGFbeta activation. J Cell Sci. 2003;116:217–224. doi: 10.1242/jcs.00229. [DOI] [PubMed] [Google Scholar]
- BasakQ5 A, Chen A, Scamuffa N, Mohottalage D, Basak S, Khatib AM. Blockade of furin activity and furin-induced tumor cells malignant phenotypes by the chemically synthesized human furin prodomain. Curr Med Chem. 2010 doi: 10.2174/092986710791331040. [DOI] [PubMed] [Google Scholar]
- Bassi DE, Lopez De Cicco R, Mahloogi H, Zucker S, Thomas G, Klein-Szanto AJ. Furin inhibition results in absent or decreased invasiveness and tumorigenicity of human cancer cells. Proc Natl Acad Sci U S A. 2001;98:10326–10331. doi: 10.1073/pnas.191199198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borras T. Gene expression in the trabecular meshwork and the influence of intraocular pressure. Prog Retin Eye Res. 2003;22:435–463. doi: 10.1016/s1350-9462(03)00018-1. [DOI] [PubMed] [Google Scholar]
- Bushati N, Cohen SM. microRNA functions. Annu Rev Cell Dev Biol. 2007;23:175–205. doi: 10.1146/annurev.cellbio.23.090506.123406. [DOI] [PubMed] [Google Scholar]
- Chan MC, Hilyard AC, Wu C, Davis BN, Hill NS, Lal A, Lieberman J, Lagna G, Hata A. Molecular basis for antagonism between PDGF and the TGFbeta family of signalling pathways by control of miR-24 expression. EMBO J. 2009;29:559–573. doi: 10.1038/emboj.2009.370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan MC, Nguyen PH, Davis BN, Ohoka N, Hayashi H, Du K, Lagna G, Hata A. A novel regulatory mechanism of the bone morphogenetic protein (BMP) signaling pathway involving the carboxyl-terminal tail domain of BMP type II receptor. Mol Cell Biol. 2007;27:5776–5789. doi: 10.1128/MCB.00218-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y, Liu X, Zhang S, Lin Y, Yang J, Zhang C. MicroRNA-21 protects against the H(2)O(2)-induced injury on cardiac myocytes via its target gene PDCD4. J Mol Cell Cardiol. 2009;47:5–14. doi: 10.1016/j.yjmcc.2009.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chudgar SM, Deng P, Maddala R, Epstein DL, Rao PV. Regulation of connective tissue growth factor expression in the aqueous humor outflow pathway. Mol Vis. 2006;12:1117–1126. [PubMed] [Google Scholar]
- Crosby ME, Kulshreshtha R, Ivan M, Glazer PM. MicroRNA regulation of DNA repair gene expression in hypoxic stress. Cancer Res. 2009;69:1221–1229. doi: 10.1158/0008-5472.CAN-08-2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dresios J, Aschrafi A, Owens GC, Vanderklish PW, Edelman GM, Mauro VP. Cold stress-induced protein Rbm3 binds 60S ribosomal subunits, alters microRNA levels, and enhances global protein synthesis. Proc Natl Acad Sci U S A. 2005;102:1865–1870. doi: 10.1073/pnas.0409764102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubois CM, Blanchette F, Laprise MH, Leduc R, Grondin F, Seidah NG. Evidence that furin is an authentic transforming growth factor-beta1-converting enzyme. Am J Pathol. 2001;158:305–316. doi: 10.1016/s0002-9440(10)63970-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FatmaQ6 N, Kubo E, Toris CB, Stamer WD, Camras CB, Singh DP. PRDX6 attenuates oxidative stress- and TGFbeta-induced abnormalities of human trabecular meshwork cells. Free Radic Res. 2009:1–13. doi: 10.1080/10715760903062887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fautsch MP, Johnson DH. Aqueous humor outflow: what do we know? Where will it lead us? Invest Ophthalmol Vis Sci. 2006;47:4181–4187. doi: 10.1167/iovs.06-0830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchshofer R, Tamm ER. Modulation of extracellular matrix turnover in the trabecular meshwork. Exp Eye Res. 2009;88:683–688. doi: 10.1016/j.exer.2009.01.005. [DOI] [PubMed] [Google Scholar]
- Hirakata M, Kaname S, Chung UG, Joki N, Hori Y, Noda M, Takuwa Y, Okazaki T, Fujita T, Katoh T, Kurokawa K. Tyrosine kinase dependent expression of TGF-beta induced by stretch in mesangial cells. Kidney Int. 1997;51:1028–1036. doi: 10.1038/ki.1997.144. [DOI] [PubMed] [Google Scholar]
- Huang S, He X, Ding J, Liang L, Zhao Y, Zhang Z, Yao X, Pan Z, Zhang P, Li J, Wan D, Gu J. Upregulation of miR-23a approximately 27a approximately 24 decreases transforming growth factor-beta-induced tumor-suppressive activities in human hepatocellular carcinoma cells. Int J Cancer. 2008;123:972–978. doi: 10.1002/ijc.23580. [DOI] [PubMed] [Google Scholar]
- Hynes RO. The extracellular matrix: not just pretty fibrils. Science. 2009;326:1216–1219. doi: 10.1126/science.1176009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ihn H. Pathogenesis of fibrosis: role of TGF-beta and CTGF. Curr Opin Rheumatol. 2002a;14:681–685. doi: 10.1097/00002281-200211000-00009. [DOI] [PubMed] [Google Scholar]
- Ihn H. The role of TGF-beta signaling in the pathogenesis of fibrosis in scleroderma. Arch Immunol Ther Exp (Warsz) 2002b;50:325–331. [PubMed] [Google Scholar]
- Johnson M. What controls aqueous humour outflow resistance? Exp Eye Res. 2006;82:545–557. doi: 10.1016/j.exer.2005.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnstone MA. Pressure-dependent changes in nuclei and the process origins of the endothelial cells lining Schlemm’s canal. Invest Ophthalmol Vis Sci. 1979;18:44–51. [PubMed] [Google Scholar]
- Johnstone MA. The aqueous outflow system as a mechanical pump: evidence from examination of tissue and aqueous movement in human and non-human primates. J Glaucoma. 2004;13:421–438. doi: 10.1097/01.ijg.0000131757.63542.24. [DOI] [PubMed] [Google Scholar]
- Kassiri Z, Defamie V, Hariri M, Oudit GY, Anthwal S, Dawood F, Liu P, Khokha R. Simultaneous transforming growth factor beta-tumor necrosis factor activation and crosstalk cause aberrant remodeling response and myocardial fibrosis in Timp3-deficient heart. J Biol Chem. 2009;284:29893–29904. doi: 10.1074/jbc.M109.028449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly M, Kolb M, Bonniaud P, Gauldie J. Re-evaluation of fibrogenic cytokines in lung fibrosis. Curr Pharm Des. 2003;9:39–49. doi: 10.2174/1381612033392341. [DOI] [PubMed] [Google Scholar]
- Koli K, Saharinen J, Hyytiainen M, Penttinen C, Keski-Oja J. Latency, activation, and binding proteins of TGF-beta. Microsc Res Tech. 2001;52:354–362. doi: 10.1002/1097-0029(20010215)52:4<354::AID-JEMT1020>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- Konishi T, Takehara T, Tsuji T, Ohsato K, Matsumoto K, Nakamura T. Scatter factor from human embryonic lung fibroblasts is probably identical to hepatocyte growth factor. Biochem Biophys Res Commun. 1991;180:765–773. doi: 10.1016/s0006-291x(05)81131-3. [DOI] [PubMed] [Google Scholar]
- Kusakabe M, Cheong PL, Nikfar R, McLennan IS, Koishi K. The structure of the TGF-beta latency associated peptide region determines the ability of the proprotein convertase furin to cleave TGF-betas. J Cell Biochem. 2008;103:311–320. doi: 10.1002/jcb.21407. [DOI] [PubMed] [Google Scholar]
- Lal A, Navarro F, Maher CA, Maliszewski LE, Yan N, O’Day E, Chowdhury D, Dykxhoorn DM, Tsai P, Hofmann O, Becker KG, Gorospe M, Hide W, Lieberman J. miR-24 Inhibits cell proliferation by targeting E2F2, MYC, and other cell-cycle genes via binding to “seedless” 3′UTR microRNA recognition elements. Mol Cell. 2009a;35:610–625. doi: 10.1016/j.molcel.2009.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lal A, Pan Y, Navarro F, Dykxhoorn DM, Moreau L, Meire E, Bentwich Z, Lieberman J, Chowdhury D. miR-24-mediated downregulation of H2AX suppresses DNA repair in terminally differentiated blood cells. Nat Struct Mol Biol. 2009b;16:492–498. doi: 10.1038/nsmb.1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CG, Kang HR, Homer RJ, Chupp G, Elias JA. Transgenic modeling of transforming growth factor-beta(1): role of apoptosis in fibrosis and alveolar remodeling. Proc Am Thorac Soc. 2006;3:418–423. doi: 10.1513/pats.200602-017AW. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Muragaki Y, Hatamura I, Ueno H, Ooshima A. Stretch-induced collagen synthesis in cultured smooth muscle cells from rabbit aortic media and a possible involvement of angiotensin II and transforming growth factor-beta. J Vasc Res. 1998;35:93–103. doi: 10.1159/000025570. [DOI] [PubMed] [Google Scholar]
- Lin Y, Liu X, Cheng Y, Yang J, Huo Y, Zhang C. Involvement of MicroRNAs in hydrogen peroxide-mediated gene regulation and cellular injury response in vascular smooth muscle cells. J Biol Chem. 2009;284:7903–7913. doi: 10.1074/jbc.M806920200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liton PB, Liu X, Challa P, Epstein DL, Gonzalez P. Induction of TGF-beta1 in the trabecular meshwork under cyclic mechanical stress. J Cell Physiol. 2005;205:364–371. doi: 10.1002/jcp.20404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luna C, Li G, Liton PB, Epstein DL, Gonzalez P. Alterations in gene expression induced by cyclic mechanical stress in trabecular meshwork cells. Mol Vis. 2009a;15:534–544. [PMC free article] [PubMed] [Google Scholar]
- Luna C, Li G, Qiu J, Challa P, Epstein DL, Gonzalez P. Extracellular release of ATP mediated by cyclic mechanical stress leads to mobilization of AA in trabecular meshwork cells. Invest Ophthalmol Vis Sci. 2009b;50:5805–5810. doi: 10.1167/iovs.09-3796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luna C, Li G, Qiu J, Epstein DL, Gonzalez P. Role of miR-29b on the regulation of the extracellular matrix in human trabecular meshwork cells under chronic oxidative stress. Mol Vis. 2009c;15:2488–2497. [PMC free article] [PubMed] [Google Scholar]
- Marsit CJ, Eddy K, Kelsey KT. MicroRNA responses to cellular stress. Cancer Res. 2006;66:10843–10848. doi: 10.1158/0008-5472.CAN-06-1894. [DOI] [PubMed] [Google Scholar]
- Martin RC, Liu PP, Goloviznina NA, Nonogaki H. microRNA, seeds, and Darwin?: diverse function of miRNA in seed biology and plant responses to stress. J Exp Bot. 2010;61:2229–2234. doi: 10.1093/jxb/erq063. [DOI] [PubMed] [Google Scholar]
- Mishra PJ, Song B, Wang Y, Humeniuk R, Banerjee D, Merlino G, Ju J, Bertino JR. MiR-24 tumor suppressor activity is regulated independent of p53 and through a target site polymorphism. PLoS One. 2009;4:e8445. doi: 10.1371/journal.pone.0008445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MohamedQ7 JS, Boriek AM. Stretch augments TGF-{beta}1 Expression Through RhoA/ROCK1/2, PTK and PI3K in Airway Smooth Muscle Cells. Am J Physiol Lung Cell Mol Physiol. 2010 doi: 10.1152/ajplung.90628.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overby DR, Stamer WD, Johnson M. The changing paradigm of outflow resistance generation: towards synergistic models of the JCT and inner wall endothelium. Exp Eye Res. 2009;88:656–670. doi: 10.1016/j.exer.2008.11.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin W, Shi Y, Zhao B, Yao C, Jin L, Ma J, Jin Y. miR-24 regulates apoptosis by targeting the open reading frame (ORF) region of FAF1 in cancer cells. PLoS One. 2010b;5:e9429. doi: 10.1371/journal.pone.0009429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin X, Wang X, Wang Y, Tang Z, Cui Q, Xi J, Li YS, Chien S, Wang N. MicroRNA-19a mediates the suppressive effect of laminar flow on cyclin D1 expression in human umbilical vein endothelial cells. Proc Natl Acad Sci U S A. 2010a;107:3240–3244. doi: 10.1073/pnas.0914882107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos RF, Sumida GM, Stamer WD. Cyclic mechanical stress and trabecular meshwork cell contractility. Invest Ophthalmol Vis Sci. 2009;50:3826–3832. doi: 10.1167/iovs.08-2694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson JV, Golesic E, Gauldie J, West-Mays JA. Ocular gene transfer of active TGF-beta induces changes in anterior segment morphology and elevated IOP in rats. Invest Ophthalmol Vis Sci. 2010;51:308–318. doi: 10.1167/iovs.09-3380. [DOI] [PubMed] [Google Scholar]
- Rogler CE, Levoci L, Ader T, Massimi A, Tchaikovskaya T, Norel R, Rogler LE. MicroRNA-23b cluster microRNAs regulate transforming growth factor-beta/bone morphogenetic protein signaling and liver stem cell differentiation by targeting Smads. Hepatology. 2009;50:575–584. doi: 10.1002/hep.22982. [DOI] [PubMed] [Google Scholar]
- Rohatgi R, Flores D. Intratubular hydrodynamic forces influence tubulointerstitial fibrosis in the kidney. Curr Opin Nephrol Hypertens. 2010;19:65–71. doi: 10.1097/MNH.0b013e32833327f3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakata R, Ueno T, Nakamura T, Ueno H, Sata M. Mechanical stretch induces TGF-beta synthesis in hepatic stellate cells. Eur J Clin Invest. 2004;34:129–136. doi: 10.1111/j.1365-2362.2004.01302.x. [DOI] [PubMed] [Google Scholar]
- Schlotzer-Schrehardt U, Zenkel M, Kuchle M, Sakai LY, Naumann GO. Role of transforming growth factor-beta1 and its latent form binding protein in pseudoexfoliation syndrome. Exp Eye Res. 2001;73:765–780. doi: 10.1006/exer.2001.1084. [DOI] [PubMed] [Google Scholar]
- Skutek M, van Griensven M, Zeichen J, Brauer N, Bosch U. Cyclic mechanical stretching modulates secretion pattern of growth factors in human tendon fibroblasts. Eur J Appl Physiol. 2001;86:48–52. doi: 10.1007/s004210100502. [DOI] [PubMed] [Google Scholar]
- Stamer WD, Seftor RE, Williams SK, Samaha HA, Snyder RW. Isolation and culture of human trabecular meshwork cells by extracellular matrix digestion. Curr Eye Res. 1995;14:611–617. doi: 10.3109/02713689508998409. [DOI] [PubMed] [Google Scholar]
- Suzuki C, Murakami G, Fukuchi M, Shimanuki T, Shikauchi Y, Imamura T, Miyazono K. Smurf1 regulates the inhibitory activity of Smad7 by targeting Smad7 to the plasma membrane. J Biol Chem. 2002;277:39919–39925. doi: 10.1074/jbc.M201901200. [DOI] [PubMed] [Google Scholar]
- Takagi S, Nakajima M, Kida K, Yamaura Y, Fukami T, Yokoi T. MicroRNAs regulate human hepatocyte nuclear factor 4alpha, modulating the expression of metabolic enzymes and cell cycle. J Biol Chem. 2010;285:4415–4422. doi: 10.1074/jbc.M109.085431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan JC, Peters DM, Kaufman PL. Recent developments in understanding the pathophysiology of elevated intraocular pressure. Curr Opin Ophthalmol. 2006;17:168–174. doi: 10.1097/01.icu.0000193079.55240.18. [DOI] [PubMed] [Google Scholar]
- Tektas OY, Lutjen-Drecoll E. Structural changes of the trabecular meshwork in different kinds of glaucoma. Exp Eye Res. 2009;88:769–775. doi: 10.1016/j.exer.2008.11.025. [DOI] [PubMed] [Google Scholar]
- ten Dijke P, Arthur HM. Extracellular control of TGFbeta signalling in vascular development and disease. Nat Rev Mol Cell Biol. 2007;8:857–869. doi: 10.1038/nrm2262. [DOI] [PubMed] [Google Scholar]
- Varshavsky A, Kessler O, Abramovitch S, Kigel B, Zaffryar S, Akiri G, Neufeld G. Semaphorin-3B is an angiogenesis inhibitor that is inactivated by furin-like pro-protein convertases. Cancer Res. 2008;68:6922–6931. doi: 10.1158/0008-5472.CAN-07-5408. [DOI] [PubMed] [Google Scholar]
- Wang Q, Huang Z, Xue H, Jin C, Ju XL, Han JD, Chen YG. MicroRNA miR-24 inhibits erythropoiesis by targeting activin type I receptor ALK4. Blood. 2008;111:588–595. doi: 10.1182/blood-2007-05-092718. [DOI] [PubMed] [Google Scholar]
- Weber M, Baker MB, Moore JP, Searles CD. MiR-21 is induced in endothelial cells by shear stress and modulates apoptosis and eNOS activity. Biochem Biophys Res Commun. 2010;393:643–648. doi: 10.1016/j.bbrc.2010.02.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf G, Ziyadeh FN. Cellular and molecular mechanisms of proteinuria in diabetic nephropathy. Nephron Physiol. 2007;106:26–31. doi: 10.1159/000101797. [DOI] [PubMed] [Google Scholar]
- WuDunn D. Mechanobiology of trabecular meshwork cells. Exp Eye Res. 2009;88:718–723. doi: 10.1016/j.exer.2008.11.008. [DOI] [PubMed] [Google Scholar]
- Yasuda T, Kondo S, Homma T, Harris RC. Regulation of extracellular matrix by mechanical stress in rat glomerular mesangial cells. J Clin Invest. 1996;98:1991–2000. doi: 10.1172/JCI119003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying SY, Chang DC, Miller JD, Lin SL. The microRNA: overview of the RNA gene that modulates gene functions. Methods Mol Biol. 2006;342:1–18. doi: 10.1385/1-59745-123-1:1. [DOI] [PubMed] [Google Scholar]
- Yu XB, Sun XH, Dahan E, Guo WY, Qian SH, Meng FR, Song YL, Simon GJ. Increased levels of transforming growth factor-betal and -beta2 in the aqueous humor of patients with neovascular glaucoma. Ophthalmic Surg Lasers Imaging. 2007;38:6–14. doi: 10.3928/15428877-20070101-01. [DOI] [PubMed] [Google Scholar]
- Zaidi SK, Dowdy CR, van Wijnen AJ, Lian JB, Raza A, Stein JL, Croce CM, Stein GS. Altered Runx1 subnuclear targeting enhances myeloid cell proliferation and blocks differentiation by activating a miR-24/MKP-7/MAPK network. Cancer Res. 2009;69:8249–8255. doi: 10.1158/0008-5472.CAN-09-1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zenkel M, Lewczuk P, Junemann A, Kruse FE, Naumann GO, Schlotzer-Schrehardt U. Proinflammatory cytokines are involved in the initiation of the abnormal matrix process in pseudoexfoliation syndrome/glaucoma. Am J Pathol. 2010;176:2868–2879. doi: 10.2353/ajpath.2010.090914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Ramsey KE, Stephan DA, Russell P. Gene and protein expression changes in human trabecular meshwork cells treated with transforming growth factor-beta. Invest Ophthalmol Vis Sci. 2004;45:4023–4034. doi: 10.1167/iovs.04-0535. [DOI] [PubMed] [Google Scholar]
- Zhou Z, Shen T, Zhang BH, Lv XY, Lin HY, Zhu C, Xue LQ, Wang H. The proprotein convertase furin in human trophoblast: possible role in promoting trophoblast cell migration and invasion. Placenta. 2009;30:929–938. doi: 10.1016/j.placenta.2009.09.003. [DOI] [PubMed] [Google Scholar]