Abstract
Membrane proteins play an important role in many biological functions. Solid-state NMR spectroscopy is uniquely suited for studying structure and dynamics of membrane proteins in a membranous environment. The major challenge to obtain high quality solid-state NMR spectra of membrane proteins is sensitivity, due to limited quantities of labeled high-molecular-weight proteins. Here we demonstrate the incorporation of paramagnetic metal (Cu2+) ions, through either EDTA or a chelator lipid, into membrane protein samples for rapid data collection under fast magic-angle spinning (MAS) and low power 1H decoupling. Spectral sensitivity of DsbB (20 kDa), an integral membrane protein, more than doubles in the same experimental time due to 1H T1 relaxation enhancement by Cu2+ ions, with DsbB native fold and active site intact. This technique can be implemented to acquire multidimensional solid-state NMR spectra for chemical shift assignments and structure elucidation of large membrane proteins with small sample quantities.
Keywords: paramagnetic relaxation enhancement, fast magic-angle spinning, solid-state NMR, large membrane protein, spectral sensitivity, chelator lipid
Solid-state NMR spectroscopy (SSNMR) is a powerful and versatile tool for studying structural biology and biophysics of membrane proteins in a membranous environment.1–14 A variety of structural and dynamic information, such as internuclear distances, dihedral angles, tensor orientations and order parameters, can be extracted by rational design of spin labeling and pulse sequences. The rich information can then be used to elucidate the functional mechanisms of membrane proteins.15 However, a major hurdle preventing SSNMR from fast acquirement of structural information is the low sensitivity of membrane proteins, because the molar quantity of isotopically labeled protein that can be packed into magic-angle spinning rotors is limited by the high molecular weight and the presence of required amount of lipids to mimic the membranous environment. New methods under development in recent years have great potential to significantly improve the sensitivity of SSNMR samples. For example, paramagnetic relaxation enhancement (PRE) effect of paramagnetic metal ions can reduce 1H spin-lattice relaxation times (T1), which govern experimental times of most SSNMR measurements.16 Previous demonstrations have shown SSNMR samples with paramagnetic metal ions to speed up the data collection, such as Cu(II)-EDTA for fibrillar protein,16 a Co(II) replaced metal center for metalloprotein17 and covalently bound paramagnetic tags for nanocrystalline protein mutants.18 All those experiments were carried out under fast (> 35 kHz) magic-angle spinning (MAS) to take advantage of low power 1H decoupling to minimize the radio-frequency induced sample heating, which allows for significant reduction of the recycle delay. The fast MAS SSNMR rotors also require smaller amounts of labeled protein, which are especially beneficial for the low molar quantities of large membrane proteins.
Recently, a novel copper chelated lipid was demonstrated to shorten 1H T1 of an antimicrobial membrane peptide in magnetically aligned bicelles with enhanced signals for static SSNMR experiments.19 The usage of paramagnetic metal ions in large membrane protein samples will significantly facilitate SSNMR data collection. Nevertheless, the effort of incorporating paramagnetic metal ions in large membrane protein samples is still nontrivial, and the effects of the metal ions and fast MAS on spectral characteristics and structures of membrane proteins remain unexplored.
Here we present the sensitivity enhancement of an integral membrane protein DsbB (20 kDa) under fast MAS conditions by incorporating Cu2+ ions through either EDTA or a chelator lipid. DsbB belongs to a disulfide bond generating enzymatic system in E. coli, including periplasmic protein DsbA, membrane protein DsbB and cofactor ubiquinone, which is responsible for forming disulfide bonds in substrate proteins.20 The combination of short 1H T1 and low power 1H decoupling under fast MAS resulted in five-fold reduction of experimental times of two-dimensional 13C-13C correlation spectra. The spectra show the same correlations as the spectra collected on the DsbB samples that do not contain copper. These results demonstrate that the PRE effect of paramagnetic metal ions can be applied to large membrane proteins, without disrupting the native fold and the active site, to enhance the sensitivity of fast MAS SSNMR experiments for chemical shift assignments and distance measurements, and thus accelerate structure determination of membrane proteins.
Three samples of uniformly labeled DsbB were prepared to investigate the PRE effect on membrane proteins: (1) DsbB without copper as a control, (2) DsbB with Cu2+ in the chelator lipid – 14:0 PE-DTPA (DsbB/CuDMPE-DTPA), (3) DsbB with Cu2+ in EDTA (DsbB/CuEDTA). The 1H T1 of the three samples were measured under the same condition with 36 kHz MAS rate and the sample temperature ~ −15 °C to achieve the optimal resolution and sensitivity as determined in the previous study.21 Table 1 summarizes the 1H T1 values of DsbB, DsbB/CuDMPE-DTPA and DsbB/CuEDTA. Clearly, the Cu2+ ions shorten the 1H T1 significantly by 10 and 6 times for CuDMPE-DTPA and CuEDTA, respectively. In principle, the recycle delay of the cross-polarization (CP) experiments from proton can be as short as three times the 1H T1, i.e. 0.15 s. However, in practice, the recycle delays of 0.3~0.5 s were used considering the high-power radio-frequency irradiation during CP and recoupling periods to keep the duty cycle under 3% in combination with fast MAS and low power decoupling during acquisitions. Even with this limitation the experimental time was shortened by up to 10 times with Cu2+ doping compared to the conventional experiments with the recycle delays of 1.5~3 s.
Table 1.
1H T1 values of DsbB, DsbB/CuDMPE-DTPA and DsbB/CuEDTA under 36 kHz MAS.
| Samples | DsbB | DsbB CuDMPEDTPA | DsbB CuEDTA |
|---|---|---|---|
| 1H T1/ms | 490 | 50 | 80 |
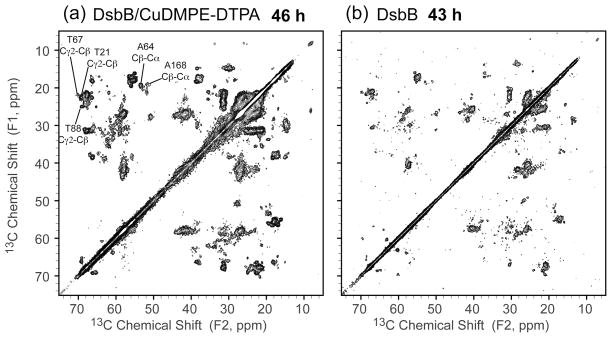
The samples of DsbB with and without Cu2+ ions were characterized by SSNMR two-dimensional (2D) 13C-13C correlation spectra, from which one can assess the PRE effects of Cu2+ ions on the resolution, sensitivity and secondary structure distribution of DsbB samples. Figure 1 shows the 2D 13C-13C correlation spectra of DsbB/CuDMPE-DTPA and DsbB with the similar experimental times (~40 h) and similar amounts of labeled proteins (~3 mg in 1.6 mm rotors). The difference between the two experiments lies in the recycle delays. The spectrum collected of DsbB with CuDMPE-DTPA was collected with a five-fold reduction in recycle delay (from 1.5 s to 0.3 s). Between these two samples, the observed chemical shift resonances remain the same, as shown in Table 2, indicating that the chelator lipids (DMPE-DTPA) did not disturb the secondary structure of DsbB in the membrane.
Figure 1.
2D 13C-13C correlation spectra of (a) DsbB/CuDMPE-DTPA (recycle delay 0.3 s, total time 46 h) and (b) DsbB (recycle delay 1.5 s, total time 43 h) under 36 kHz MAS with RFDR 7.1 ms mixing (B0 = 17.6 T, Tsample = −15 °C, maximum t1 = 15.36 ms, maximum t2 = 20.48 ms). The crosspeaks of residues that are resolved in both spectra are marked. Both data were processed with 30 and 30 Hz net line broadening in each dimension and plotted at the same contour levels, with the lowest contour at 5σ.
Table 2.
Comparison of 13C chemical shifts of resolved resonances in 2D 13C-13C correlation spectra of DsbB, DsbB/CuDMPE-DTPA and DsbB/CuEDTA.
| Chemical shift (ppm) | DsbB | DsbB CuDMPE-DTPA | DsbB CuEDTA |
|---|---|---|---|
| G11 Cα | 43.8 | 43.3 | 43.4 |
| G11 C | 172.4 | 172.3 | 172.4 |
| T21 Cγ2 | 22.6 | 22.3 | 22.4 |
| T21 Cβ | 69.0 | 69.0 | 68.9 |
| G61 Cα | 45.7 | 45.3 | 45.3 |
| G61 C | 173.6 | 173.6 | 173.6 |
| A64 Cβ | 19.2 | 18.9 | 19.0 |
| A64 Cα | 53.3 | 53.1 | 53.1 |
| T67 Cγ2 | 21.9 | 21.6 | 21.5 |
| T67 Cβ | 69.9 | 69.8 | 70.0 |
| T88 Cγ2 | 24.8 | 24.8 | 24.7 |
| T88 Cβ | 68.8 | 68.8 | 68.7 |
| G148 Cα | 47.9 | 47.6 | 47.6 |
| G148 C | 174.5 | 174.6 | 174.8 |
| A168 Cβ | 18.9 | 18.9 | 18.8 |
| A168 Cα | 51.2 | 51.1 | 51.1 |
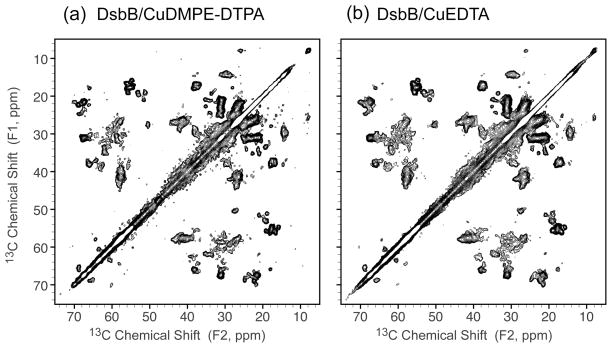
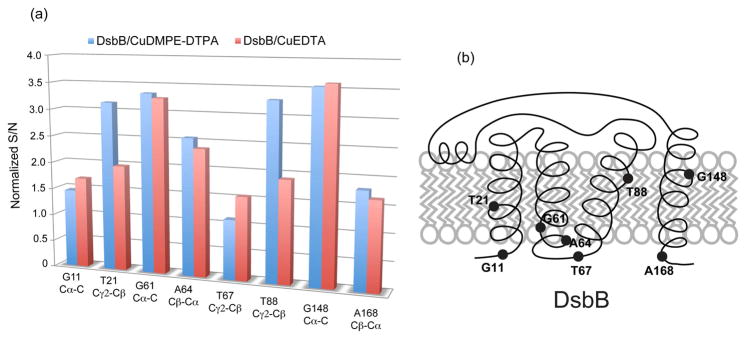
We further evaluated the various ways of introducing Cu2+ ions into membrane protein samples: (1) CuDMPE-DTPA in the membrane; (2) CuEDTA in solution. The concentrations of Cu2+ ions in DsbB/CuDMPE-DTPA and DsbB/CuEDTA are ~0.6 mM and 2.5 mM, respectively. Figure 2 shows the comparison of 2D 13C-13C correlation spectra of DsbB/CuDMPE-DTPA and DsbB/CuEDTA. Both spectra exhibit the same chemical shift resonances of DsbB as shown in Table 2, indicating that CuEDTA does not affect DsbB structure either. The resolved crosspeak signal-to-noise ratios (S/N) of the residues (G11, T21, G61, A64, T67, T88, G148, A168) in DsbB/CuDMPE-DTPA and DsbB/CuEDTA samples are summarized in Figure 3. The sensitivity of DsbB/CuDMPE-DTPA and DsbB/CuEDTA are consistently enhanced (1.1 ~ 3.6) compared to DsbB without copper due to PRE effect and reduced recycle delays. Interestingly, G11, T67 and A168 in the loop regions of DsbB have less sensitivity enhancement compared to the residues in the transmembrane helices. This is probably because the residues in the loops or termini are more mobile than the transmembrane residues, and thus have slower 1H spin diffusion rate, which is the mechanism for 1H magnetization transfer through the samples.16 Hence, the transmembrane residues are more rigid and spin-diffuse faster to obtain the 1H magnetization by PRE from Cu2+ ions. Another explanation would be that the residues in the loops or termini might be close to Cu2+ ions to have shorter T1ρs than transmembrane residues, and thus have less cross-polarization intensities.
Figure 2.
2D 13C-13C correlation spectra of (a) DsbB/CuDMPE-DTPA (recycle delay 0.3 s) and (b) DsbB/CuEDTA (recycle delay 0.5 s) under 36 kHz MAS with RFDR 3.6 ms mixing (B0 = 11.7 T, Tsample = −15 °C, maximum t1 = 15.36 ms, maximum t2 = 20.48 ms). The crosspeak pattern is the same between the two samples, indicating both CuDMPE-DTPA and CuEDTA are effective in shortening 1H T1 of membrane protein DsbB. Both data were processed with 40 and 40 Hz net line broadening in each dimension and plotted at the same contour levels, with the lowest contour at 5σ.
Figure 3.
(a) Relative signal to noise ratios (S/N) of DsbB/CuDMPE-DTPA (blue) and DsbB/CuEDTA (red) in the selected residues that are clearly resolved in all three 2D 13C-13C spectra. All S/N of corresponding crosspeaks are normalized to those in 2D 13C-13C spectrum of DsbB without copper, considering the same sample quantities and experimental times. (b) Schematic respresentation of DsbB topology in the membrane. Residues selected in (a) are marked to indicate their locations relative to the membrane. T21, G61, A64, T88 and G148 are in the transmembrane helices, while G11, T67 and A168 are in the loops or termini.
All these 2D 13C-13C correlation experiments used the radio-frequency field-driven dipolar recoupling22–24 (RFDR) for 13C-13C mixing. This mixing scheme was chosen because the commonly utilized dipolar recoupling schemes involving protons, like proton-driven spin diffusion (PDSD),25 dipolar-assisted rotational resonance (DARR)26 and radio-frequency-assisted diffusion (RAD),27 do not work well under fast MAS conditions. Additionally, RFDR takes advantage of the high power 13C B1 field without requiring high power 1H heteronuclear decoupling during the mixing to yield the efficient 13C-13C homonuclear recoupling of DsbB with rapid data collection, similar to the experiments performed on microcrystalline and fibrillar protein samples that were reported before.16, 28
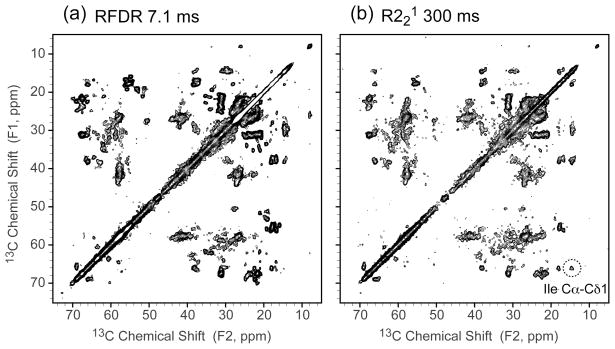
Recently, a few amplitude- or phase-modulated spin diffusion techniques have been developed for fast MAS, such as mixed rotational and rotary-resonance (MIRROR),29 phase-alternated recoupling irradiation scheme (PARIS)30 and rotor-synchronized symmetry-based recoupling (R221).31 We also utilized the R221 scheme to collect 2D 13C-13C correlation spectra to compare with RFDR. As shown in Figure 4, the 2D 13C-13C correlation spectra of DsbB/CuDMPE-DTPA with 7.1 ms of RFDR mixing and 300 ms of R221 mixing have similar correlations. In RFDR mixing, only one-bond or two-bond correlations were observed, while in R221 mixing there were additional three-bond correlations present, such as Ile Cα-Cδ1 and Leu Cα-Cδ. This is most likely due to R221 suffering less from dipolar truncation than RFDR.23–24, 31 However, R221 requires much longer mixing time than RFDR (300 ms vs. 7.1 ms) to yield similar crosspeak intensities due to its low power irradiation and reduced recoupling efficiency at fast MAS, which greatly increases the experimental time to collect the equal-quality spectra. Further, paramagnetic Cu2+ ions might reduce 13C T1 as well, causing lower sensitivity for prolonged 13C-13C longitudinal mixing time for R221. Due to short mixing time requirement, easy experimental setup and sequence robustness, RFDR is advantageous for fast MAS and rapid data collection.
Figure 4.
2D 13C-13C correlation spectra of DsbB/CuDMPE-DTPA (a) with RFDR 7.1 ms mixing and (b) with R221 300 ms mixing under 36 kHz MAS (B0 = 11.7 T, Tsample = −15 °C, recycle delay 0.3 s, maximum t1 = 15.36 ms, maximum t2 = 20.48 ms). The dashed circle indicates Ile Cα-Cδ1 correlations that are more prominent in R221 300 ms mixing. Both data were processed with 30 and 30 Hz net line broadening in each dimension and plotted at the same contour levels, with the lowest contour at 5σ.
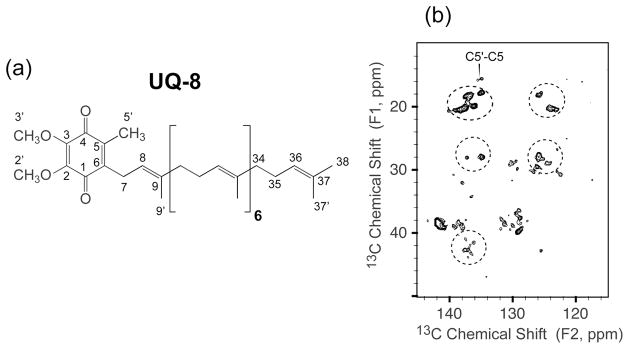
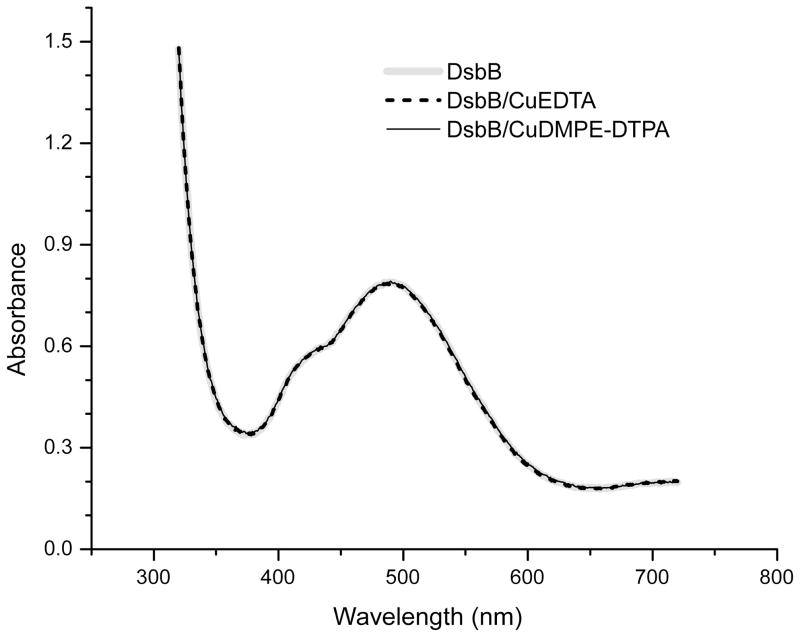
In addition, the sensitivity enhancement from rapid data collection and the broadband recoupling from RFDR provide good crosspeak intensities for the cofactor ubiquinone-8 (UQ-8) (Figure 5) for ~ 40 h, which required long time signal averaging (~ 70 h) to observe previously at moderate MAS frequencies (10–20 kHz) with the twice amount of labeled protein.21 The chemical shifts obtained from DsbB/CuDMPE-DTPA are the same as those assigned from the previous study,21 as listed in Table 3, indicating that Cu2+ ions in chelated forms do not perturb the active site of charge-transfer complex between DsbB and UQ-8.20 This is also supported by the identical UV-Vis spectra of three DsbB samples with and without copper (Figure 6) showing the signature 500 nm absorbance corresponding to the charge-transfer complex.
Figure 5.
(a) UQ-8 structure with IUPAC nomenclature. (b) Expansion of UQ-8 region of 2D 13C-13C correlation spectra of DsbB/CuDMPE-DTPA under 36 kHz MAS with RFDR 7.1 ms mixing (B0 = 11.7 T, Tsample = −15 °C, recycle delay 0.3 s, maximum t1 = 15.36 ms, maximum t2 = 20.48 ms). The UQ-8 C5′-C5 peak was marked and dashed circles highlighted the UQ-8 correlations. The data were processed with 30 and 30 Hz net line broadening in each dimension.
Table 3.
Comparison of 13C chemical shifts of UQ-8 in DsbB/CuDMPE-DTPA and DsbB. Chemical shifts of UQ-8 in DsbB are from the previous study.21
| Chemical Shift (ppm) | UQ-8 in DsbB CuDMPE-DTPA | UQ-8 in DsbB21 |
|---|---|---|
| CH3 (5′) | 15.6 | 15.9 |
| CH3 (9′~37′) | 20.5~17.8 | 20.7~17.9 |
| CH3 (38) | 28.0 | 28.2 |
| CH2 (11~35) | 30.4~27.5 | 30.3~27.3 |
| CH2 (10~34) | 43.8~41.5 | 43.4~41.6 |
| CH3O (2′, 3′) | 63.2, 63.7 | 63.6, 64.0 |
| C=C (12~36) | 126.0~123.0 | 126.6~123.6 |
| C=C (13~37) | 138.6~135.0 | 139.0~135.2 |
| C=C (9) | 140.2 | 140.2 |
| C=O (1, 4) | 154.5 | 154.7 |
Figure 6.
UV-Vis spectra of DsbB, DsbB/CuEDTA and DsbB/CuDMPE-DTPA.
In summary, we have demonstrated the sensitivity enhancement of membrane protein samples under fast MAS through the incorporation of paramagnetic metal ions (CuDMPE-DTPA and CuEDTA). The combination of fast T1 relaxation enhanced by Cu2+ ions and low power 1H decoupling enabled by fast MAS facilitated rapid data collection for a large membrane protein (20 kDa). The experimental times are reduced by more than five times. 2D 13C-13C spectra of DsbB with CuDMPE-DTPA or CuEDTA resulted in the same correlations as the normal DsbB sample without copper, indicating that the copper dopants did not affect the structure of DsbB, especially for the cofactor UQ-8 in the active site. The 13C-13C mixing scheme of RFDR has been found to be optimal for chemical shift assignments of intraresidual correlations considering the recoupling efficiency and the economy of overall experimental time. In principle, other mixing schemes for interresidue correlations can be implemented to obtain long-range distances, such as proton assisted recoupling (PAR)32 and CHHC.33 The 1H-detected experiments34–35 of perdeuterated membrane proteins can also benefit significantly from the sensitivity enhancement of Cu2+ ions given the low yield of perdeuterated membrane protein expression. Therefore, this widely applicable technique of sensitivity enhancement will greatly facilitate chemical shift assignments and structure determinations of membrane proteins.
EXPERIMENTAL SECTION
Sample preparation
Uniformly-labeled 13C, 15N DsbB C41S was expressed in E. coli C43 (DE3) using modified Studier Medium P, containing 10 ml/L 13C, 15N Bioexpress (Cambridge Isotope Laboratories, Andover, MA) as described previously.21 Cell membranes were isolated, and the solubilized DsbB was purified on a Talon cobalt column.21 Fractions containing DsbB were pooled, concentrated, and dialyzed against 25 mM HEPES, pH 7.8. The His-tag was removed by incubation with Factor Xa overnight at 23 °C. DsbB was then centrifuged to remove precipitated contaminants and further concentrated. The concentration of dodecylmaltoside (DDM) was determined to be 2% using a colorimetric assay.36 CuEDTA was added to 2.5 mM to half of the purified DsbB. To the other aliquot of DsbB, CuDMPE-DTPA (Avanti Polar Lipids, Inc., Alabaster, AL) in 5% DDM was added in an approximate ratio of 3 CuDMPE-DPTA:DsbB, resulting in a final concentration of 0.6 mM CuDMPE-DPTA. This solution was stirred 24 h at 23 °C to facilitate exchange of the CuDMPE-DPTA with the endogenous lipids bound to the solubilized DsbB. For both samples, addition of the chelated Cu2+ ions had no effect on the visible absorbance bands of DsbB C41S (Figure 6). The samples were then precipitated by removal of DDM over several days by addition of methyl-β-cyclodextrin and dialysis,21 and packed into 1.6 mm rotors for SSNMR. The UV-Vis spectra of DsbB(C41S) were acquired on an Evolution 600 Spectrophotometer (Thermo Scientific, Waltham, MA).
NMR spectroscopy
SSNMR experiments were performed on a 750 MHz Varian VNMRS spectrometer and a 500 MHz Varian VNMRS spectrometer (1H frequency). All spectrometers were equipped with 1.6 mm fast MAS probes. Typical π/2 pulse widths were 1.5~1.7 μs for 1H and 1.6 μs for 13C. The spinning speed was 36 kHz. All experiments utilized tangent ramped cross polarization (CP)37 with low power 1H SPINAL38 decoupling (~15 kHz) applied during acquisition and evolution periods. 1H T1s were measured by 1H 1D inversion recovery experiments. The 13C B1 field for RFDR was 140 kHz and no 1H decoupling was applied during RFDR. The 1H RF field for R221 was 18 kHz as half of the spinning speed. Detailed parameters are given in the figure captions. Sample temperatures were determined by ethylene glycol calibration.39 Chemical shifts were referenced to DSS, using adamantane as a secondary standard.40 Spectra were processed with NMRPipe41 and peak picking and assignments were performed in Sparky (T.D. Goddard and D.G. Kneller, University of California, San Francisco). Back linear prediction and polynomial baseline correction were applied to the direct dimension. Zero filling and Lorentzian-to-Gaussian apodization were used for each dimensions before Fourier transformation.
Acknowledgments
The authors thank the National Institutes of Health (R01GM075937 to C.M.R.) for funding, the School of Chemical Sciences NMR Facility at the University of Illinois at Urbana-Champaign for assistance with data acquisition, Lars K. Rikardsen and Dr. Anna E. Nesbitt for the help with sample preparation, and Dr. Lindsay J. Sperling for insightful discussions.
References
- 1.Cady SD, Schmidt-Rohr K, Wang J, Soto CS, Degrado WF, Hong M. Structure of the Amantadine Binding Site of Influenza M2 Proton Channels in Lipid Bilayers. Nature. 2010;463:689–692. doi: 10.1038/nature08722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hu F, Luo W, Hong M. Mechanisms of Proton Conduction and Gating in Influenza M2 Proton Channels from Solid-State NMR. Science. 2010;330:505–508. doi: 10.1126/science.1191714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ader C, Schneider R, Hornig S, Velisetty P, Wilson EM, Lange A, Giller K, Ohmert I, Martin-Eauclaire MF, Trauner D, et al. A Structural Link between Inactivation and Block of a K+ Channel. Nat Struct Mol Biol. 2008;15:605–612. doi: 10.1038/nsmb.1430. [DOI] [PubMed] [Google Scholar]
- 4.Qiang W, Bodner ML, Weliky DP. Solid-State NMR Spectroscopy of Human Immunodeficiency Virus Fusion Peptides Associated with Host-Cell-Like Membranes: 2D Correlation Spectra and Distance Measurements Support a Fully Extended Conformation and Models for Specific Antiparallel Strand Registries. J Am Chem Soc. 2008;130:5459–5471. doi: 10.1021/ja077302m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.De Angelis AA, Howell SC, Nevzorov AA, Opella SJ. Structure Determination of a Membrane Protein with Two Trans-Membrane Helices in Aligned Phospholipid Bicelles by Solid-State NMR Spectroscopy. J Am Chem Soc. 2006;128:12256–12267. doi: 10.1021/ja063640w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xu J, Durr UH, Im SC, Gan Z, Waskell L, Ramamoorthy A. Bicelle-Enabled Structural Studies on a Membrane-Associated Cytochrome B5 by Solid-State MAS NMR Spectroscopy. Angew Chem Int Ed Engl. 2008;47:7864–7867. doi: 10.1002/anie.200801338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bajaj VS, Mak-Jurkauskas ML, Belenky M, Herzfeld J, Griffin RG. Functional and Shunt States of Bacteriorhodopsin Resolved by 250 GHz Dynamic Nuclear Polarization-Enhanced Solid-State NMR. Proc Natl Acad Sci U S A. 2009;106:9244–9249. doi: 10.1073/pnas.0900908106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Traaseth NJ, Verardi R, Torgersen KD, Karim CB, Thomas DD, Veglia G. Spectroscopic Validation of the Pentameric Structure of Phospholarnban. Proc Natl Acad Sci U S A. 2007;104:14676–14681. doi: 10.1073/pnas.0701016104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sharma M, Yi M, Dong H, Qin H, Peterson E, Busath DD, Zhou HX, Cross TA. Insight into the Mechanism of the Influenza a Proton Channel from a Structure in a Lipid Bilayer. Science. 2010;330:509–512. doi: 10.1126/science.1191750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goncalves JA, South K, Ahuja S, Zaitseva E, Opefi CA, Eilers M, Vogel R, Reeves PJ, Smith SO. Highly Conserved Tyrosine Stabilizes the Active State of Rhodopsin. Proc Natl Acad Sci U S A. 2010;107:19861–19866. doi: 10.1073/pnas.1009405107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shi L, Ahmed MA, Zhang W, Whited G, Brown LS, Ladizhansky V. Three-Dimensional Solid-State NMR Study of a Seven-Helical Integral Membrane Proton Pump--Structural Insights. J Mol Biol. 2009;386:1078–1093. doi: 10.1016/j.jmb.2009.01.011. [DOI] [PubMed] [Google Scholar]
- 12.Li Y, Berthold DA, Gennis RB, Rienstra CM. Chemical Shift Assignment of the Transmembrane Helices of DsbB, a 20-kDa Integral Membrane Enzyme, by 3D Magic-Angle Spinning NMR Spectroscopy. Protein Sci. 2008;17:199–204. doi: 10.1110/ps.073225008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Struts AV, Salgado GF, Tanaka K, Krane S, Nakanishi K, Brown MF. Structural Analysis and Dynamics of Retinal Chromophore in Dark and Meta I States of Rhodopsin from 2H NMR of Aligned Membranes. J Mol Biol. 2007;372:50–66. doi: 10.1016/j.jmb.2007.03.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lange V, Becker-Baldus J, Kunert B, van Rossum BJ, Casagrande F, Engel A, Roske Y, Scheffel FM, Schneider E, Oschkinat H. A MAS NMR Study of the Bacterial ABC Transporter ArtMP. Chem Bio Chem. 2010;11:547–555. doi: 10.1002/cbic.200900472. [DOI] [PubMed] [Google Scholar]
- 15.McDermott A. Structure and Dynamics of Membrane Proteins by Magic Angle Spinning Solid-State NMR. Annu Rev Biophys. 2009;38:385–403. doi: 10.1146/annurev.biophys.050708.133719. [DOI] [PubMed] [Google Scholar]
- 16.Wickramasinghe NP, Parthasarathy S, Jones CR, Bhardwaj C, Long F, Kotecha M, Mehboob S, Fung LWM, Past J, Samoson A, et al. Nanomole-Scale Protein Solid-State NMR by Breaking Intrinsic 1H T1 Boundaries. Nat Methods. 2009;6:215–218. doi: 10.1038/nmeth.1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bertini I, Emsley L, Lelli M, Luchinat C, Mao J, Pintacuda G. Ultrafast MAS Solid-State NMR Permits Extensive 13C and 1H Detection in Paramagnetic Metalloproteins. J Am Chem Soc. 2010;132:5558–5559. doi: 10.1021/ja100398q. [DOI] [PubMed] [Google Scholar]
- 18.Nadaud PS, Helmus JJ, Sengupta I, Jaroniec CP. Rapid Acquisition of Multidimensional Solid-State NMR Spectra of Proteins Facilitated by Covalently Bound Paramagnetic Tags. J Am Chem Soc. 2010;132:9561–9563. doi: 10.1021/ja103545e. [DOI] [PubMed] [Google Scholar]
- 19.Yamamoto K, Xu J, Kawulka KE, Vederas JC, Ramamoorthy A. Use of a Copper-Chelated Lipid Speeds up NMR Measurements from Membrane Proteins. J Am Chem Soc. 2010;132:6929–6931. doi: 10.1021/ja102103n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Inaba K, Ito K. Structure and Mechanisms of the DsbB-DsbA Disulfide Bond Generation Machine. Biochim Biophys Acta, Mol Cell Res. 2008;1783:520–529. doi: 10.1016/j.bbamcr.2007.11.006. [DOI] [PubMed] [Google Scholar]
- 21.Tang M, Sperling LJ, Berthold DA, Nesbitt AE, Gennis RB, Rienstra CM. Solid-State NMR Study of the Charge-Transfer Complex between Ubiquinone-8 and Disulfide Bond Generating Membrane Protein DsbB. J Am Chem Soc. 2011;133:4359–4366. doi: 10.1021/ja107775w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bennett AE, Ok JH, Griffin RG, Vega S. Chemical Shift Correlation Spectroscopy in Rotating Solids: Radio-Frequency Dipolar Recoupling and Longitudinal Exchange. J Chem Phys. 1992;96:8624–8627. [Google Scholar]
- 23.Ishii Y, Balbach JJ, Tycko R. Measurement of Dipole-Coupled Lineshapes in a Many-Spin System by Constant-Time Two-Dimensional Solid State NMR with High-Speed Magic-Angle Spinning. Chem Phys. 2001;266:231–236. [Google Scholar]
- 24.Ishii Y. 13C-13C Dipolar Recoupling under Very Fast Magic Angle Spinning in Solid-State Nuclear Magnetic Resonance: Applications to Distance Measurements, Spectral Assignments, and High-Throughput Secondary-Structure Determination. J Chem Phys. 2001;114:8473–8483. [Google Scholar]
- 25.Bloembergen N. On the Interaction of Nuclear Spins in a Crystalline Lattice. Physica. 1949;15:386–426. [Google Scholar]
- 26.Takegoshi K, Nakamura S, Terao T. 13C-1H Dipolar-Driven 13C-13C Recoupling without 13C Rf Irradiation in Nuclear Magnetic Resonance of Rotating Solids. J Chem Phys. 2003;118:2325–2341. [Google Scholar]
- 27.Morcombe CR, Gaponenko V, Byrd RA, Zilm KW. Diluting Abundant Spins by Isotope Edited Radio Frequency Field Assisted Diffusion. J Am Chem Soc. 2004;126:7196–7197. doi: 10.1021/ja047919t. [DOI] [PubMed] [Google Scholar]
- 28.Wickramasinghe NP, Kotecha M, Samoson A, Past J, Ishii Y. Sensitivity Enhancement in 13C Solid-State NMR of Protein Microcrystals by Use of Paramagnetic Metal Ions for Optimizing 1H T1 Relaxation. J Magn Reson. 2007;184:350–356. doi: 10.1016/j.jmr.2006.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Scholz I, Huber M, Manolikas T, Meier BH, Ernst M. MIRROR Recoupling and Its Application to Spin Diffusion under Fast Magic-Angle Spinning. Chem Phys Lett. 2008;460:278–283. [Google Scholar]
- 30.Weingarth M, Demco DE, Bodenhausen G, Tekely P. Improved Magnetization Transfer in Solid-State NMR with Fast Magic Angle Spinning. Chem Phys Lett. 2009;469:342–348. [Google Scholar]
- 31.Hou G, Yan S, Sun S, Han Y, Byeon IJ, Ahn J, Concel J, Samoson A, Gronenborn AM, Polenova T. Spin Diffusion Driven by R-Symmetry Sequences: Applications to Homonuclear Correlation Spectroscopy in MAS NMR of Biological and Organic Solids. J Am Chem Soc. 2011;133:3943–3953. doi: 10.1021/ja108650x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lewandowski JR, De Paepe G, Eddy MT, Struppe J, Maas W, Griffin RG. Proton Assisted Recoupling at High Spinning Frequencies. J Phys Chem B. 2009;113:9062–9069. doi: 10.1021/jp810280t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lange A, Luca S, Baldus M. Structural Constraints from Proton-Mediated Rare-Spin Correlation Spectroscopy in Rotating Solids. J Am Chem Soc. 2002;124:9704–9705. doi: 10.1021/ja026691b. [DOI] [PubMed] [Google Scholar]
- 34.Zhou DH, Shea JJ, Nieuwkoop AJ, Franks WT, Wylie BJ, Mullen C, Sandoz D, Rienstra CM. Solid-State Protein-Structure Determination with Proton-Detected Triple-Resonance 3D Magic-Angle Spinning NMR Spectroscopy. Angew Chem Int Ed. 2007;46:8380–8383. doi: 10.1002/anie.200702905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Linser R, Fink U, Reif B. Proton-Detected Scalar Coupling Based Assignment Strategies in MAS Solid-State NMR Spectroscopy Applied to Perdeuterated Proteins. J Magn Reson. 2008;193:89–93. doi: 10.1016/j.jmr.2008.04.021. [DOI] [PubMed] [Google Scholar]
- 36.Lau FW, Bowie JU. A Method for Assessing the Stability of a Membrane Protein. Biochemistry. 1997;36:5884–5892. doi: 10.1021/bi963095j. [DOI] [PubMed] [Google Scholar]
- 37.Hediger S, Meier BH, Kurur ND, Bodenhausen G, Ernst RR. NMR Cross-Polarization by Adiabatic Passage through the Hartmann-Hahn Condition (APHH) Chem Phys Lett. 1994;223:283–288. [Google Scholar]
- 38.Fung BM, Khitrin AK, Ermolaev K. An Improved Broadband Decoupling Sequence for Liquid Crystals and Solids. J Magn Reson. 2000;142:97–101. doi: 10.1006/jmre.1999.1896. [DOI] [PubMed] [Google Scholar]
- 39.Raiford DS, Fisk CL, Becker ED. Calibration of Methanol and Ethylene Glycol Nuclear Magnetic Resonance Thermometers. Anal Chem. 1979;51:2050–2051. [Google Scholar]
- 40.Morcombe CR, Zilm KW. Chemical Shift Referencing in MAS Solid State NMR. J Magn Reson. 2003;162:479–486. doi: 10.1016/s1090-7807(03)00082-x. [DOI] [PubMed] [Google Scholar]
- 41.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. Nmrpipe - a Multidimensional Spectral Processing System Based on Unix Pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]