Abstract
The third intracellular loop (IL3) of G protein-coupled receptors (GPCRs) is an important contact domain between GPCRs and their G proteins. Previously, the IL3 of Ste2p, a Saccharomyces cerevisiae GPCR, was suggested to undergo a conformational change upon activation as detected by differential protease susceptibility in the presence and absence of ligand. In this study using disulfide crosslinking experiments we show that the Ste2p cytoplasmic ends of helix 5 (TM5) and helix 6 (TM6) that flank the amino and carboxyl sides of IL3 undergo conformational changes upon ligand binding whereas the center of the IL3 loop does not. Single Cys substitution of residues in the middle of IL3 led to receptors that formed high levels of crosslinked Ste2p, whereas Cys substitution at the interface of IL3 and the contiguous cytoplasmic ends of TM5 and TM6 resulted in minimal disulfide mediated crosslinked receptor. The alternating pattern of residues involved in crosslinking suggested the presence of a 310 helix in the middle of IL3. Agonist (WHWLQLKPGQPNleY) induced Ste2p activation reduced crosslinking mediated by Cys substitutions at the cytoplasmic ends of TM5 and TM6 but not by residues in the middle of IL3. Thus the cytoplasmic ends of TM5 and TM6 undergo conformational change upon ligand binding. An α-factor antagonist (des-Trp, des-His-alpha-factor) did not influence disulfide mediated Ste2p crosslinking suggesting that the interaction of the N-terminus of α-factor with Ste2p is critical for inducing conformational changes at TM5 and TM6. We propose that the changes in conformation revealed for residues at the ends of TM5 and TM6 are affected by the presence of G protein but not G protein activation. This study provides new information about role of specific residues of a GPCR in signal transduction and how peptide ligand binding activates the receptor.
G protein-coupled receptors (GPCRs) are integral membrane proteins that are known to play important roles in cell communication by activating intracellular events through both G protein-dependent and -independent processes (1–3). These receptors are encoded by the largest gene family in mammals, constitute the main target of prescribed drugs, and represent promising targets for new drug development (1, 3–6). They are composed of seven transmembrane (TM) helical segments connected by intracellular and extracellular loops. The core region of the receptor containing the seven TMs has been found to be generally responsible for binding of small ligands whereas peptide ligands often bind to the extracellular portions as well as the core of GPCRs (7–8). In the classical model of ligand activation of a GPCR signal transduction pathway, the binding of an agonist to the receptor induces conformational changes in the TMs propagated to the cytoplasmic ends of a GPCR for G protein activation and transduction to various intracellular metabolic events. (1, 9).
The cytoplasmic surfaces of GPCRs contain multiple contact regions responsible for receptor coupling to signal transduction complexes. The most prominent domains in GPCRs for coupling are the second and third (IL3) intracellular loops as well as the C-terminal tail and TM boundaries (1–3). Comparing the crystal structure of opsin and rhodopsin shows that the cytoplasmic end of TM6 is shifted outwards from the center of the bundle relative to its position in the inactive state (rhodopsin) and is closer to TM5 in the active state (opsin) (10). Such movements have also been observed in other receptor such as M3 and adrenergic receptors using disulfide cross-linking of Cys mutant receptors. In the M3 receptor, agonist binding increased the proximity between the cytoplasmic ends of TM5 and TM6. Possible TM6 rotational movements were also observed in the M3 receptor (11).
Several studies have shown that peptide ligands bind GPCRs and occupy a pocket defined by side chains from extracellular loops and helices. The binding of the ligand induces changes at the extracellular surface of the receptor resulting in conformational changes at the cytoplasmic surfaces. These phenomena are believed to be critical for GPCR activation (2). An explosion of recent crystal structures of ligand-bound (activated) β1-adrenergic (12), β2-adrenergic (13), and A2a (14), receptors indicated major changes in the cytoplasmic ends of TM5 and TM6 as a result of receptor activation.
Ste2p, the yeast Saccharomyces cerevisiae receptor for the pheromone α-factor, is believed to have a structure similar to that of mammalian GPCRs. The third intracellular loop of Ste2p has been shown to play an important role in signal transduction and is involved in Gpa1p (the S. cerevisiae Gα protein) activation (15–18). Analysis of the Ste2p IL3 demonstrated that IL3 becomes hypersensitive to proteolytic digestion with trypsin in response to ligand binding indicating that IL3 undergoes a conformational change that is likely to be important for G protein activation (19), although specific residues that are involved in these conformational changes were not identified. The TM5 and TM6 that flank the IL3 have also been shown to interact with each other and have been suggested to play a critical role in signaling (20). It is important to note that the Gα protein of the heteromeric G protein complex need not dissociate from Ste2p during receptor activation as indicated by the full activity of a Ste2p-Gpa1p fusion chimera (21).
In this study, we have investigated IL3 and flanking TM5 and TM6 conformational changes by determining the propensity for disulfide cross-linking between receptors carrying Cys substitutions. The IL3-Cys mutant receptors were observed to form dimers with the pattern of dimer formation suggesting the presence of a 310 helix in the middle of the IL3 loop. Addition of α-factor affected the levels of disulfide formation in Cys-substituted receptors in portions of TM5 and TM6 contiguous with IL3 implying that activation of Ste2p involved ligand-induced conformational changes in the cytoplasmic ends of TM5 and TM6. In contrast disulfide formation involving residues in the middle of IL3 was not sensitive to ligand addition indicating that residues in the IL3 loop do not change conformation or availability during receptor activation. The presence of an α-factor antagonist or a non hydrolyzeable GTP analog did not influence the levels of disulfide mediated dimerization suggesting that receptor activation but not Gα activation was necessary for the observed conformational changes.
EXPERIMENTAL PROCEDURES
Media, Reagents, and Strains and Transformation
Saccharomyces cerevisiae strain LM102 [MATa, bar1, leu2, trp1, ura3, FUS1-lacZ::URA3, ste2Δ (22)] was used for Ste2p growth arrest and FUS1-LacZ assays. The protease deficient strain BJS21 [MATa, prc1-407 prb1-1122 pep4-3 leu2 trp1 ura3-52 ste2::KanR (23)] and TM5117 [MATa, bar1, leu2, his3, ura3, FUS1-lacZ::URA3, ste2Δ, gpa1Δ (22)] were used for protein isolation and immunoblot analysis and disulfide cross-linking. Plasmids coding for the STE2 mutants and GPA1 mutants were transformed by the method previously described (22) into LM102, TM5117 and BJS21 cells. Transformants were selected by growth on minimal medium (24) lacking tryptophan (designated as MLT) or lacking both tryptophan and uracil (designated as MLTU) to maintain selection for the plasmids. All media components were obtained from Becton-Dickinson (Franklin Lakes, NJ).
Cysteine scanning mutagenesis
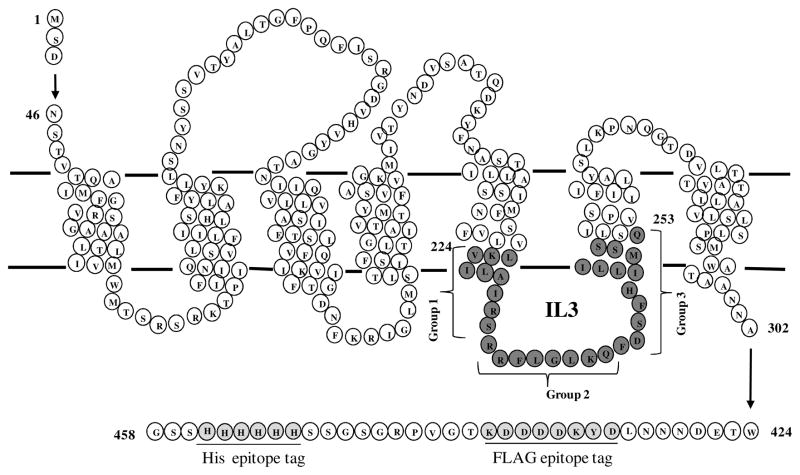
C-terminal FLAG and His tagged STE2 with the two native cysteine residues (Cys59 and Cys252) substituted with serine and cloned into the plasmid p424- GPD (high copy plasmid) to yield plasmid pBEC2 (22) was used as the backbone for mutagenesis of Ste2p. The plasmid pBEC2 was engineered by site-directed mutagenesis to individually replace 30 residues in Ste2p (V224-Q253, see Fig. 1) with cysteine as previously described (22).
Figure 1.
Schematic diagram of Ste2p. Single cysteine at positions 224 to 253 (solid circles) were introduced into the IL3 region of the STE2 gene. The FLAG and His tags are underlined in the C-terminal tail.
Membrane extraction and Immunoblots
BJS21 cells expressing STE2 and GPA1 constructs grown in their selective media were used to prepare total cell membranes isolated as previously described (25). Cells were harvested and lysed by agitation with glass beads in 700 μL of HEPES buffer (50 mM HEPES, 150 mM NaCl, pH 7.5) and the following concentrations of protease inhibitors: 1.0 mM leupeptin, 10 μM pepstatin A, and 5.0 mM phenylmethanesulfonyl fluoride. The lysate was cleared by centrifugation at 3000g for 2 minutes. The membrane fraction was harvested by centrifugation at 15000g for 30 minutes and was then resuspended in the HEPES buffer with 20% glycerol. Protein concentration was determined by BioRad (BioRad, Hercules, CA) protein assay as previously described(22). For immunoblot analyses the membrane extract was solubilized in SDS sample buffer (BioRad, Hercules, CA) and 5 μg were fractioned by 10% SDS-PAGE. Blots were probed with FLAG™ antibody (Sigma/Aldrich Chemical, St. Louis, MO) to detect Ste2p. The signals generated were analyzed using Quantity One software (Version 4.5.1) on a Chemi-Doc XRS photodocumentation system (BioRad, Hercules, CA). The student’s T-test was used to analyze differences in dimer formation (cross-linking) between pheromone treated and non-treated receptors. A probability of p≤0.01 was regarded as statistically significant. Statistical analyses were performed with Prism (GraphPad Software, San Diego, CA).
Growth Arrest Assays
LM102 cells expressing C-terminal FLAG and His tagged Ste2p were grown at 30 ºC in MLT, harvested by centrifugation, washed three times with water and resuspended at a final concentration of 5 × 106 cells/ml according to previously described procedures (23). Cells (1 ml) were combined with 3.5 ml agar noble (1.1 %) and poured as a top agar lawn onto MLT medium agar plates. Filter disks (Whatman #1 paper) impregnated with α-factor pheromone (0.125 – 2.0 μg) were placed on the top agar. The plates were incubated at 30 ºC for 18 hours and then observed for clear halos around the discs. The diameter of halos around the discs were measured, plotted as diameter versus pheromone amount, and analyzed by linear regression analysis using Prism software (GraphPad Software, San Diego, CA) to determine the amount of pheromone that yielded a 15 mm halo. The experiment was repeated at least three times and reported values represent the mean of these tests.
Fus1-LacZ Assays
LM102 cells expressing C-terminal FLAG and His tagged Ste2p Cys mutants were grown at 30 ºC in selective media, harvested, washed three times with fresh media and resuspended at a final concentration of 5 × 107 cells/ml. Cells (0.5 ml) were combined with α-factor pheromone (final concentration of 1 μM) and incubated at 30 ºC for 90 minutes. The cells were transferred to a 96-well flatbottom plate in triplicates, permeabilized with 0.5% Triton X-100 in 25 mM PIPES buffer and then β-galactosidase assays were carried out using fluorescein di-β-galactopyranoside (Molecular Probes, OR) as a substrate (26). The reaction mixtures were incubated at 37 °C for 60 minutes and 1.0 M Na2CO3 was added to stop the reaction. The fluorescence of the samples (excitation of 485nm and emission of 530 nM) was determined using a 96-well plate reader Synergy2 (BioTek Instruments, Inc., Winooski, VT). The data were analyzed using Prism software (GraphPad Software, San Diego, CA). The experiments were repeated at least three times and reported values represent the mean of these tests.
Saturation Binding Assay
Tritiated α-factor (9.3 Ci/mmol) was used in saturation binding assays on whole cells. LM102 cells expressing Ste2p Cys-less as the wild-type and mutants with the Cys-substitutions in IL3 (L228C, A229C, R233C, R234C, L248C and I249C) were harvested, washed three times with YM1 [yeast minimum medium with 0.5 M potassium phosphate (pH 6.24) containing 10 mM TAME, 10 mM sodium azide, 10 mM potassium fluoride,and 1% BSA] and adjusted to a final concentration of 4 × 107 cells/ml in binding medium YM1i (YM1 plus protease inhibitors). Cells (630 μl) were combined with 70 μl of 10X YM1i supplemented with [3H]α-factor and incubated at room temperature for 30 minutes. The final concentration of [3H]α-factor ranged from 0.4 to 50 × 10−9 M. Upon completion of the incubation interval, 200 μl aliquots of the cell-pheromone mixture were collected in triplicate and washed over glass fiber filter mats using the Standard Cell Harvester (Skatron Instruments, Sterling, VA). Retained radioactivity on the filter was counted by liquid scintillation spectroscopy. LM102 cells lacking Ste2p were used as a non-specific binding control for the assays. Specific binding for each mutant receptor was calculated by subtracting the non-specific values from those obtained for total binding. Specific binding data were analyzed by non-linear regression analysis for single-site binding using Prism software (GraphPad Software, San Diego, CA) to determine the Kd (nM) and Bmax values (receptors/cell) for each mutant receptor were calculated. The final values represent the binding constants from at least three independent experiments.
Binding Competition Assays
This assay was performed using LM102 cells expressing C-terminal FLAG and His-tagged Ste2p. Tritiated [3H]-α-factor (10.2 Ci/mmol, 12 μM) prepared as previously described (23, 27) was used in competition binding assays on whole cells. The cells were grown at 30 ºC in MLT, harvested, washed three times with YM1 [ 0.5 M potassium phosphate (pH 6.24) containing 10 mM TAME, 10 mM sodium azide, 10 mM potassium fluoride,and 1% BSA] and adjusted to a final concentration of 2 × 107 cells per ml in YM1 plus protease inhibitors [YM1i (25)]. For the competition binding studies, cells (600 μl) were combined with 150 μl of ice-cold 5X YM1i supplemented with 6nM [3H]α-factor in the presence or absence α-factor or α-factor antagonist and incubated at room temperature for 30 min. The final concentrations of α-factor and α-factor antagonist varied from 0.5 × 10−10 to 1 × 10−6 M. After incubation, triplicate samples of 200 μl aliquots were filtered and washed over glass fiber filter mats using the Standard Cell Harvester (Skatron Instruments, Sterling, VA) and placed in scintillation vials. The radioactivity [3H] on the filter was counted by liquid scintillation spectroscopy. The binding data were analyzed by non-linear regression analysis for one-site competition binding using Prism software (GraphPad Software, San Diego, CA) (28). The final values represent the binding constants from at least three independent experiments
Disulfide cross-linking
Membrane proteins (in HEPES buffer, 20% glycerol) extracted from BJS21 cells expressing Ste2p mutants co-expressed with wild type Gpa1p were treated with CuP (1.0 μM Cu and 3.0 μM 1,10-phenanthroline) and incubated at room temperature for 30 minutes. The reaction was quenched by addition of EDTA (ethylenediamine-tetraacetic acid) and NEM (N-ethylmaleimide) to final concentration of 10 mM. The cross-linked samples were resuspended in non-reducing SDS sample buffer (BioRad, Hercules, CA) and resolved on 10% SDS-PAGE as described above. The disulfide reaction was reversed by reducing with 4% 2-mercaptoethanol (β-ME) in the SDS sample buffer before resolving it on SDS-PAGE. To investigate the effects of ligand or GTP-γ-S on cross-linking, membrane fractions were incubated in HEPES buffer containing 5μM α-factor (WHWLQLKPGQPNleY) or 5μM α-factor antagonist (desW,desH-α-factor = WLQLKPGQPNleY) in the presence or absence of 0.1 mM GTP-γ-S (1) before CuP treatment.
For in vivo (whole cell) cross-linking, cells were grown to mid-log phase, harvested and washed with water by centrifugation (4000g for 5 min). The cells were resuspended in 100 mM LiAc and incubated at 30 °C for 30 minutes with mixing. After centrifugation, the cells were resuspended in 25% PEG with 250 mM LiAc and incubated for 60 minutes to make them permeable to CuP. Cells were washed with HEPES buffer three times after LiAc treatment and then resuspended in YM1i buffer containing 5 μM α-factor or α-factor-antagonist and incubated at room temperature for 30 minutes. Disulfide cross-linking was induced by adding CuP to final concentrations of 0.5 mM Cu and 1.5 mM 1,10-phenanthroline as previously described for yeast whole cell cross-linking (29). The mixture was incubated at room temperature for 60 minutes with end-over-end mixing. EDTA and NEM) to final concentrations of 10 mM were added to stop the reaction. The cells were washed three times with HEPES buffer containing EDTA (10 mM) and NEM (10 mM). Membrane extraction was carried as described above with HEPES buffer containing 10 mM EDTA/NEM. Reducing conditions of disulfide bond were carried out by resuspending membrane samples in SDS-PAGE sample buffer containing 4% β-ME and incubating at room temperature for at least 15 min. To investigate the role of Gpa1p in the TM5-IL3-TM6 conformational changes, disulfide cross-linking of Cysless, L228C (TM5), L236 (IL3) and I249C (TM6) were carried out in a Gpa1p deleted strain, TM5117 as described above. For the time course experiments, the CuP mediated disulfide cross-linking in BJS21 was terminated at the indicated time points by addition of EDTA and NEM to final concentrations of 10 mM.
Prediction and modeling
All models of 3-D structures were generated by the Phyre Structural Bioinformatics Group prediction tools(30). The structures were viewed and labeled with the PyMOL pdb viewer software (DeLano Scientific LLC, Palo Alto, CA).
RESULTS
Phenotypes of Ste2p IL3 Cys substitution mutants
The signaling activities of the cysteine mutants were examined by α-factor induced growth arrest and FUS1-LacZ induction assays. The growth arrest assay is a sensitive test that measures the ability of cells expressing Ste2p to maintain pheromone-induced cell division arrest at the G1 phase over a 24–36 hour time frame, whereas the FUS1-lacZ induction assay measures an early response of the yeast cell to pheromone. The strains used in this study contain a reporter gene construct consisting of a fusion between FUS1 promoter and the lacZ gene encoding the enzyme β-galactosidase (26). This allows for rapid, sensitive detection of mating pathway activation by assessing β-galactosidase activity in response to mating pheromone.
The Cys-less Ste2p engineered as the background for the specific cysteine mutations has been shown to have biological activities identical to the wild type receptor (22). In this study we grouped the IL3 residues into three categories based on their position as follows: group 1 (TM5-IL3 boundary) residues, V224-S232; group 2 (middle IL3) residues, R233-Q240; and group 3 (IL3-TM6 boundary) residues, F241-Q253 (Fig. 1.). Substitution of single cysteine residues in intracellular loop 3 (IL3) of Cys-less Ste2p resulted in constructs that were expressed and had biological activities identical or similar to that of the Cys-less receptor, except for K225C which was biologically inactive (see Table 1 and supporting information Fig. S1). The decreased sensitivity in biological assays caused by the K225C mutation has been observed in previous studies (31).
Table 1.
Summary of phenotypes of Ste2p IL3 Cys substitution mutants. The expression levels as measured by western blots (Fig. 3 and Fig. 6) and biological activities all had standard deviations between ±2 and ±5 percent. All assays were done at least three times independently. NA: No activity was observed.
| Mutation | Protein Expression (%) | Growth arrest activities (%) | Fus1-LacZ induction (%) |
|---|---|---|---|
| Cys-less | 100 | 100 | 100 |
| V224C | 100 | 95 | 90 |
| K225C | 100 | NA | NA |
| L226C | 98 | 90 | 80 |
| I227C | 100 | 100 | 105 |
| L228C | 100 | 90 | 110 |
| A229C | 100 | 90 | 75 |
| I230C | 100 | 85 | 100 |
| R231C | 100 | 90 | 100 |
| S232C | 100 | 100 | 90 |
| R233C | 65 | 70 | 60 |
| R234C | 75 | 100 | 80 |
| F235C | 75 | 100 | 80 |
| L236C | 75 | 75 | 65 |
| G237C | 75 | 100 | 75 |
| L238C | 75 | 100 | 75 |
| K239C | 75 | 75 | 65 |
| Q240C | 75 | 100 | 80 |
| F241C | 100 | 100 | 80 |
| D242C | 85 | 85 | 85 |
| S243C | 100 | 95 | 100 |
| F244C | 100 | 90 | 95 |
| H245C | 90 | 90 | 70 |
| I246C | 100 | 90 | 70 |
| L247C | 100 | 90 | 75 |
| L248C | 100 | 85 | 95 |
| I249C | 100 | 85 | 100 |
| M250C | 100 | 95 | 80 |
| S251C | 100 | 100 | 110 |
| S252C | 100 | 95 | 90 |
| Q253C | 100 | 90 | 65 |
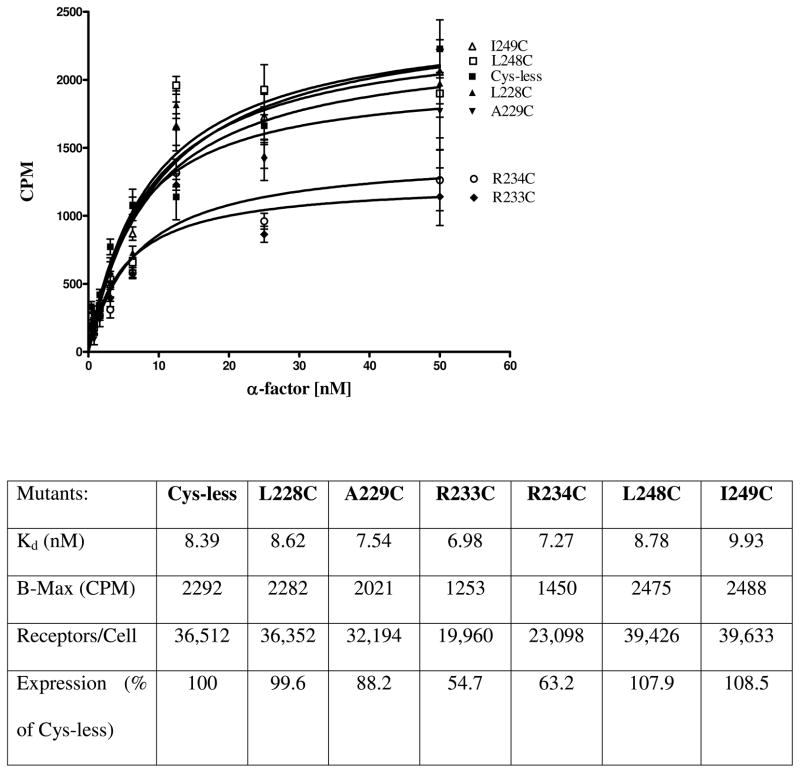
Receptor expression was measured by western blot signals that were quantitated using Quantity One software (Version 4.5.1) on a Chemi-Doc XRS photodocumentation system (BioRad, Hercules, CA). The Cys-less receptor signal was used as the measure of 100% expression. In addition to quantitation of the western blot signals, saturation binding assays were carried out on selected mutants (L228C and A229C) from the TM5-IL3 boundary, (R233C and R234C) from the middle of IL3, and (L248C and I249C) from the IL3-TM6 boundary (Fig. 2). The receptors showed almost equal Kd values for ligand binding and receptors L228C, A229C, L248C, and I249C were expressed at levels within 12 % of the Cys-less control, whereas R233C and R234C were expressed to about 55–63% of control, respectively, as judged by Bmax values.
Figure 2.
Whole cell saturation binding assay of [3H]α-factor to Cysless and IL3 Cys mutant receptors. A graph of CPM (count per minute of radioactivity of cells) versus the concentration of the [3H]α-factor is shown. The data represents specific binding to cells as determined by subtracting the binding to cells lacking the receptor from binding to cells containing the Ste2p Cys mutant receptors. Below the graph is a table containing a summary of the binding affinities (Kd) and surface expression of the receptors.
IL3 Cys-mutants form dimers
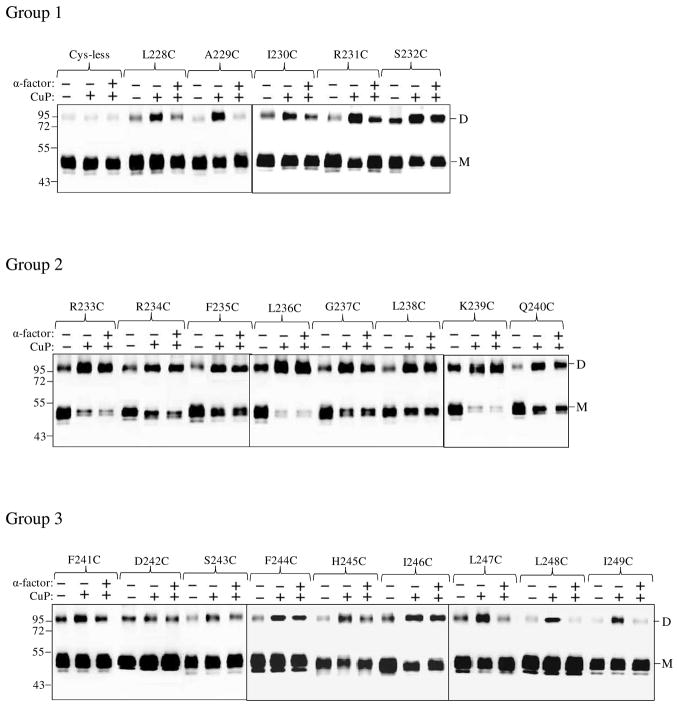
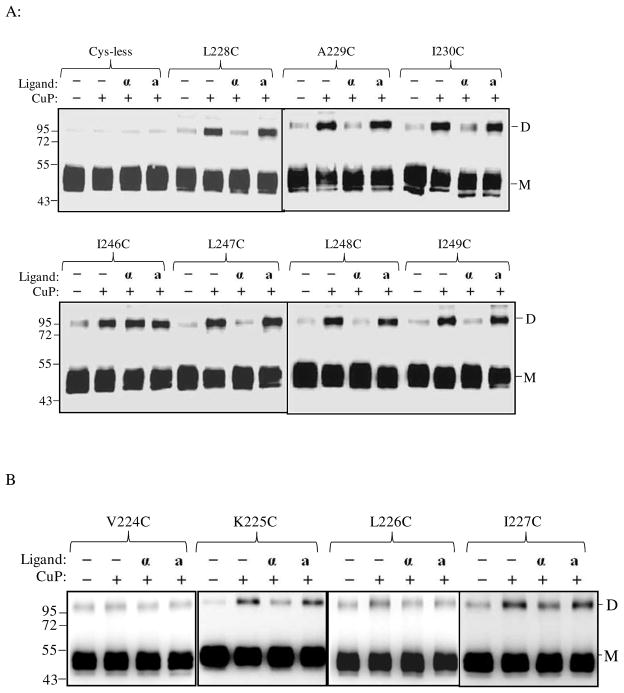
One of the most commonly used strategies to investigate GPCR agonist-induced conformational change is disulfide cross-linking involving cysteine-substituted mutant GPCRs. This approach involves determination of differences in disulfide formation between two receptor monomers containing Cys residues in the presence and absence of ligand (29, 32–33). Ste2p predominates in SDS-PAGE as a monomer of about 50 kDa as non-covalent interactions between receptors are disrupted by the conditions of the SDS-PAGE. As observed in many studies (8, 22) some SDS-resistant dimers persist in all lanes including that of the Cys-less receptor (Fig. 3). We observed strong dimer formation in some of the IL3 Cys mutant receptors when treated with CuP (Supplementary Fig. S2). Compared to the Cys-less receptor, which showed little or no increase in the dimer band at 100 kDa upon incubation of membranes with CuP, both the R233C and R234C receptors showed marked increases in the dimer band. This increase was completely (R234C) or almost completely (R233C) reversed by addition of β-ME indicating the involvement of disulfide bonds in stabilization of the dimer.
Figure 3.
Ste2p IL3 Cys mutants form dimers. Membranes from various mutants were incubated with or without CuP in the presence or absence of α-factor as described in the Methods. The membrane extracts were run on SDS-PAGE gels, the gels were blotted and probed with anti-FLAG antibody to detect the presence of Ste2p at either the monomer (M) or dimer (D) positions. Each preparation was untreated (lanes −, −) or treated with CuP (lanes −, +) or with α-factor and CuP (lanes +, +).
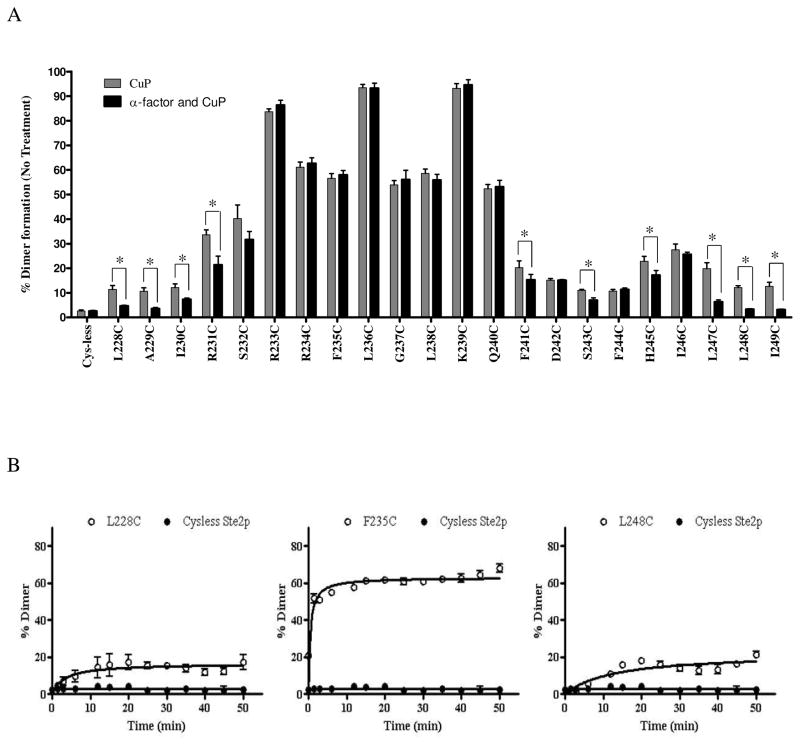
Given that the disulfide cross-linking was observed with Ste2p(R233C) and Ste2p(R234C) a comprehensive examination of the cross-linking of Cys mutants in the IL3 region of the receptor was conducted. These experiments were done in the presence and absence of ligand in order to determine whether crosslinking changed during activation of the receptor (Fig. 3). Examination of the ratio of dimer to monomer in the gels of all the IL3 Cys mutants showed that some mutants exhibited high levels of disulfide formation [e.g., Ste2p(L236C), Group 2; Fig. 3.)] whereas others exhibited only a small increase in dimer content with the monomer remaining the predominant species (e.g., L228C, Group 1; Fig.3). When treated with CuP in the absence of ligand, disulfide crosslinked receptor formation for the 22 single Cys mutants studied varied from 10% to 96% of the total Ste2p related bands (monomer plus dimer) (Fig. 3, compare lanes “− −”and “− +”). Group 1 (L228C-S232C) and group 3 (F241C-I249C) residues displayed about 10–45% increase in disulfide formation upon incubation with CuP in the absence of ligand compared to the untreated receptor. In contrast, upon treatment with CuP in the absence of ligand Group 2 (R233C-Q240C) mutants showed a greater increase (54–96%) in percent of crosslinked receptor.
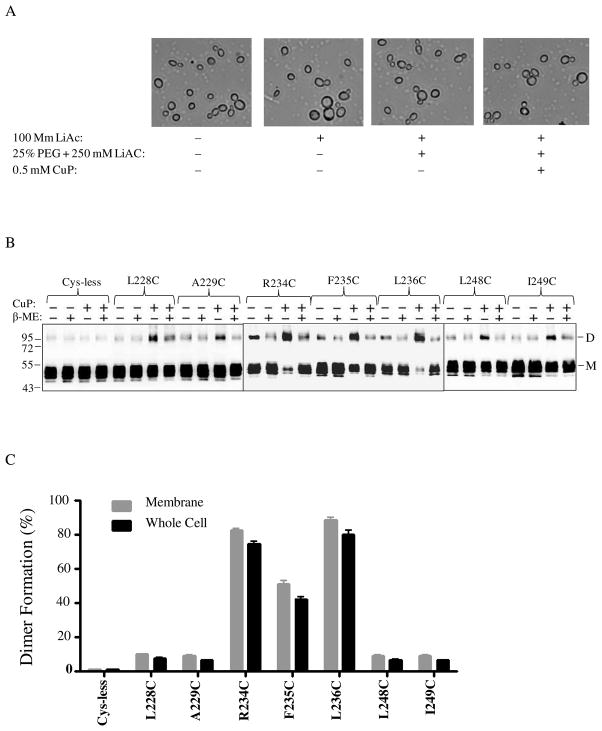
The high degree of disulfide crosslinking found in some of the IL3 Cys mutants suggested that IL3 loops in two receptor subunits were within close proximity. However, we were concerned that some of these protein-protein interactions may have been artifacts related to the use of membranes. To determine whether disulfide formation was the same or different in whole cells in comparison to membrane preparations, an in vivo CuP treatment assay was performed. For this experiment we chose seven IL3 Cys-mutants representing each group in IL3 and treated these receptors with CuP in live permeabilized cells (Fig. 4). Microscopic observation (Fig. 4A) showed that the cells were not morphologically affected by CuP treatment and these treated cells were fully viable upon plating on growth media (data not shown). As was true with membranes, the disulfide formation catalyzed by CuP was reversed to various extents in the presence of β-ME (Fig. 4B, +/+ lane). Comparison of the percent crosslinking in membranes and whole cells (Fig. 4B, immunoblots, and Fig. 4C, bar graph dimer/monomer ratio) showed that the trend for dimer formation in different IL3 mutants was similar in both preparations. A quantitative summary of the percentage of disulfide formation in all IL3 Cys mutants with and without Cu-P treatment or with ligand plus Cu-P treatment is listed in the supporting information (Table S1). Dimerization of some mutants was reduced or disappeared upon α-factor addition; this together with the variations in the amount of dimerization among the Ste2p-IL3 Cys mutants observed in at least three independent experiments allows us to conclude that the dimer formation was reproducible and specific.
Figure 4.
Analyses of Ste2p dimerization in whole yeast cells. Cells expressing various Ste2p IL3 cysteine mutants were permeabilized and exposed to CuP. A: Images of cells treated with various reagents used during the whole cell disulfide cross-linking CuP treatment. B: Blots of seven IL3 Cys mutants selected for whole cell cross-linking evaluation. The samples were probed with anti-FLAG antibody to detect the presence of Ste2p at either the monomer (M) or dimer (D) positions. Each preparation was untreated (lanes −, −) or treated with β-ME (lanes −+) or CuP (lanes +, −) or with CuP and β-ME (lanes +, +). C: A graph comparing the percentage of the Ste2p mutant receptors that formed crosslinked dimers in membrane preparations and whole cells. D. Whole cells expressing receptors L228C and I249C were untreated or incubated with ligand prior to Cu-P treatments. The blots were probed with anti-FLAG antibody to detect the presence of Ste2p at either the monomer or dimer positions, respectively. Each preparation was untreated (lanes −, −) or treated with Cu-P (lanes −, +) or with α-factor and Cu-P (lanes α, +) or with α-factor antagonist and Cu-P (lanes a, +).
Conformational changes in TM5 and TM6 upon α-factor binding
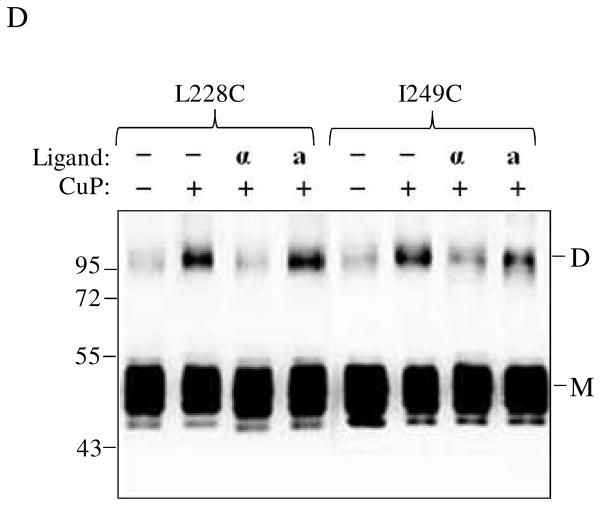
In previous studies it has been shown that binding of α-factor affected Ste2p dimer formation mediated through the TM regions suggesting that these regions undergo conformational changes (29, 34–35). We investigated whether incubation with α-factor would have any effect on CuP-mediated crosslinking of the IL3 mutants. While ligand binding had only a minor effect on disulfide formation in receptors carrying mutations in group 2 residues, (Fig. 3, R233C to Q240C, compare lanes “− +”and “+ +”), a reduction in crosslinking was observed for mutation near TM5 (I230C and R231C) and TM6 (F241C, S243C and H245C). Dimer formation was almost completely inhibited at boundaries between IL3 and TM5 (L228C and A229C) and TM6 (L247C, L248C and I249C). The results suggest that α-factor binding induces conformational changes at the TM5 and TM6 cytoplasmic ends of Ste2p that change the availability of the Cys residues for chemical crosslinking, whereas in most of the IL3 loop the reactivity of these residues is not affected by ligand binding. We observed similar inhibition of cross-linking by α-factor in experiments using intact whole cells (Fig. 4D).
A graphical representation showing the pattern of crosslink formation of IL3 Cys mutants of Ste2p in the inactive (ligand unbound) and active (ligand bound) states is presented in Fig. 5A. Analysis of the time course of disulfide (dimer) formation (Figure 5B) showed that the TM5 (e.g. A228C) and TM6 (e.g. L248C) Ste2p mutants formed dimers with a half maximal time of about 3 minutes whereas residues in the middle of IL3 loop (e.g. F235C) exhibited half maximal time of about 1.5 minutes. The slower dimer formation at the TM5 and TM6 extracellular ends may suggest that these residues are farther from each other compared to residues in the IL3 loop. It is clear from these time course studies that formation of dimer is complete by 30 minutes under these experimental conditions.
Figure 5.
Dimer formation in Ste2p IL3. A. Relative amount (%) of IL3-mediated dimer formation in the presence and absence of pheromone. The graph was plotted from three independent experimental values. *The differences between the mean values were statistically significant (P < 0.01). B. Time course of dimer formation after CuP treatment of Ste2p mutants L228C (TM5), F235C (IL3) and L248C (TM6). Preparations were treated for the indicated time and then analyzed on Western blots for dimer formation. Results represent the average of four to five independent experiments. Error bars indicate the standard deviation.
α-factor antagonist did not induce conformational changes in TM5 and TM6
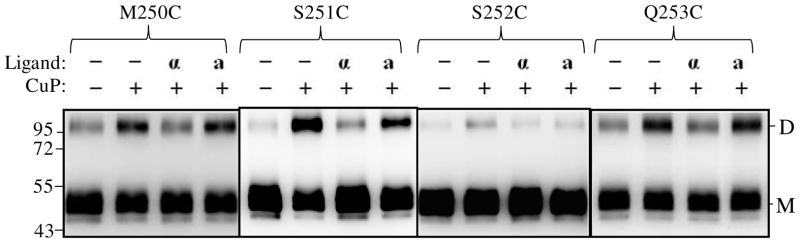
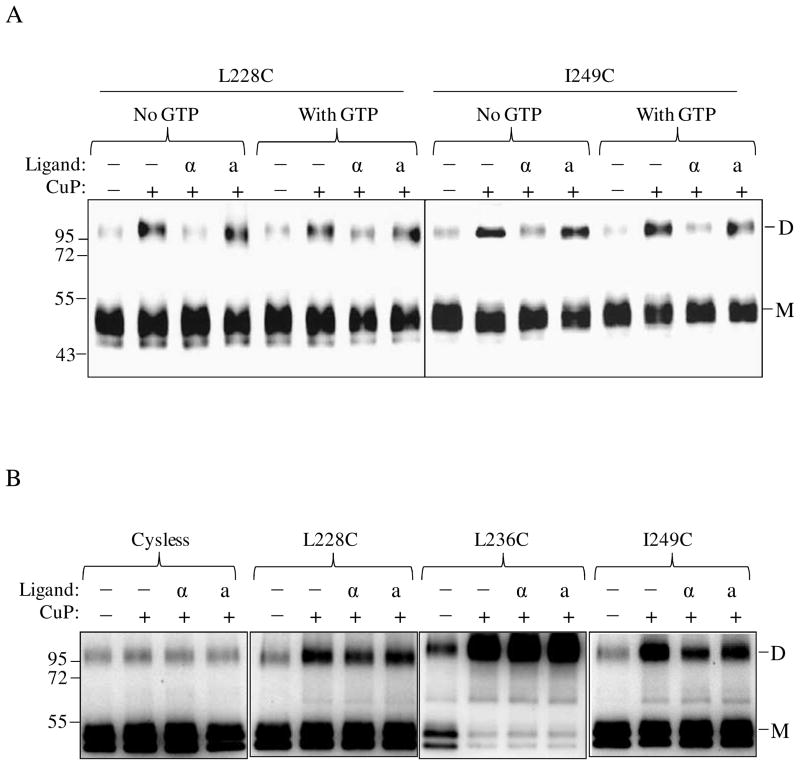
Biochemical and cross-linking studies in our lab have shown that the N-terminus of α-factor is necessary for activation but not binding to Ste2p (36–38); see also supplementary Fig. S3. Recently we showed that Trp1 of α-factor interacts with Lys269 (TM6) of Ste2p (8). The question of whether receptor activation involving the N-terminus of α-factor is important for the changes in conformation at the cytoplasmic ends of TM5 and TM6 was investigated. Membrane samples containing TM5 (L228C, A229C, I230C) and TM6 (I246C, L247C, L248C, I249C) receptors were incubated with α-factor (WHWLQLKPGQPNle12Y) or an α-factor antagonist (desW1,desH2-WLQLKPGQPNle12Y) prior to Cu-P treatment. The results (Fig. 6A, compare lanes label “α” and “a”) showed that in contrast to the native α-factor, treatment with antagonist did not significantly change the percent of crosslinked product relative to that found in the unliganded receptor. Specifically for the L228, A229, I230 L247, L248, I249 Cys mutant receptors the crosslink percentages for the unliganded and antagonist bound receptors were nearly identical whereas minimal crosslinking was found in the presence of α-factor. We also examined conformational changes in residues at positions 224–227 and 250–253 at the ends of the TM5 and TM6 helices, respectively. Dimer (disulfide bond) formation was observed in some residues (K225C, I227C, M250C, S251C and Q253C) as shown in Fig. 6B that were also affected by the presence of α-factor suggesting that the cytoplasmic ends of the TM5/TM5 and TM6/TM6 helices in a dimeric receptor are indeed in very close proximity and that they participate in the conformational change associated with ligand binding.
Figure 6.
Effects α-factor antagonist on dimerization of TM5 and TM6 Cys mutants. A: membranes from cells expressing Ste2p cysteine mutants at cytoplasmic ends of the helices and B: membranes from cells expressing Ste2p cysteine mutants in helices that are suggested to be buried in the membrane were treated were incubated with CuP in the presence or absence of α-factor or α-factor antagonist as described in the Methods. The membrane extracts were run on SDS-PAGE gels, The blots were probed with anti-FLAG antibody to detect the presence of Ste2p at either the monomer (M, 53–55 kDa) or dimer (D, 106–110 kDa) positions, respectively. Each preparation was untreated (lanes −, −) or treated with Cu-P (lanes −, +) or with α-factor and Cu-P (lanes α, +) or with α-factor antagonist and Cu-P (lanes a, +).
GTP did not affect dimer formation and conformational changes
The recent crystal structure of opsin with the 11 C-terminal residues of the Gα protein led the authors to conclude that the active conformation of this GPCR was stabilized by its interaction with Gα (10). Another study also reported GTP may affect the interaction of receptor with Gα (39). We therefore determined whether the presence or the absence of GTP would affect disulfide formation observed in this study. We co-expressed the IL3 Cys-mutants A228C and I249C with wild-type Gpa1p under the control of the same promoter in order to maintain equal level of the receptor and its Gα protein and repeated the Cu-P treatment in the presence of GTP-γ-S. The results (Fig. 7A) suggest that GTP-γ-S addition had no effect on the level of disulfide formation indicating that Gα protein activation is not required for the observed conformational changes. In contrast, when cross-linking was carried out in a Gpa1p deleted strain (Fig. 7B) the level of disulfide formation in the presence of ligand was not significantly reduced at the TM5 and TM6 ends in comparison to that observed in the Gpa1p background (Fig. 7A, Fig. 3 and Fig. 6A).
Figure 7.
Effects of G protein expression and activation on TM5 and TM6 ligand induced conformational changes. A. Whole cell membranes containing receptors, A228C and I249C were treated with or without ligand in the presence or absence of 0.1mM GTP-γ-S prior to Cu-P treatments. The blots were probed with anti-FLAG antibody to detect the presence of Ste2p at either the monomer (M, 53–55 kDa) or dimer (D, 106–110 kDa) positions, respectively. Each preparation was untreated (lanes −, −) or treated with Cu-P (lanes −, +) or with α-factor and Cu-P (lanes α, +) or with α-factor antagonist and Cu-P (lanes a, +). B. Analyses of Ste2p dimerization in yeast cells (TM5117) devoid of Gα protein. Whole cell membranes containing Cysless, A228C, L236C and I249C Ste2p, from cells lacking Gα were treated with or without ligand prior to Cu-P treatments. The blots were probed with anti-FLAG antibody to detect the presence of Ste2p at either the monomer or dimer positions, respectively.
DISCUSSION
We have used disulfide cross-linking analysis to probe conformational changes of residues in the third intracellular loop (IL3) and the ends of the contiguous fifth (TM5) and sixth (TM6) transmembrane domains of Ste2p during ligand-induced activation. An assumption of this approach is that the cysteine mutations may act as surrogates of the native residues in Ste2p. Therefore, it is critical that the mutations not affect the activity of the receptor, its conformation or its spatial relationships to other receptor molecules or interacting proteins. Several studies have reported that individual residue mutations in Ste2p IL3 did not significantly affect receptor activity (15–17). Consistent with previous studies (31), Ste2p with mutations in IL3 was observed to tolerate Cys substitutions in our experiments except for the K225C receptor. The tolerance for substitutions in IL3 suggests that the contact points revealed in the cross-linking analysis likely exist in the native receptor and that activation of G protein by Ste2p involves multiple intermolecular interactions as observed for mammalian GPCRs (40–41).
Given that IL3 has been suggested to be involved in receptor-Gα protein (Ste2p-Gpa1p) interactions (15–17) it is interesting that strong dimer formation occurred in the IL3 Cys mutants, This is consistent with the conclusion that IL3 in rhodopsin has been identified to play a role in receptor oligomerization (42–44). In addition, molecular dynamics simulations, correlated-mutation analysis, and evolutionary-trace analysis all predict the involvement of IL3 in GPCR dimerization (32). The dimer formations observed in our study were also detected in intact cells, indicating that the disulfide bond formation between the two monomers of Ste2p occurs in the native environment of the receptor. Substitution cysteine accessibility studies on Ste2p-IL3 residues suggested all the residues, L228-I249 were accessible, and even though residues I246C and L247C were about 2-fold more accessible than most IL3 (R233-Q240) residues (31), in our study when treated with CuP these residues (I246C, L247C) exhibited only 22–30% dimer formation compared to the 54–96% dimer formation found for residues R233C-Q240C. In addition, though K225C, I227C, M250C, S251C and Q253C at the cytoplasmic ends of the TM5 and TM6 helices have been shown to be buried in the membrane and not solvent accessible (31), we observed disulfide formation involving these residues that was reduced by the presence of α-factor suggesting that the cytoplasmic ends of the TM5/TM5 and TM6/TM6 helices in the inactive state of Ste2p are in closer proximity than when activated by ligand. The differences in the results of the cysteine accessibility and disulfide cross-linking studies of Ste2p IL3 cysteine mutants suggest that the cross-linking (dimer formation) that we observed is not due to the fact that these residues are accessible and randomly cross-linking. Rather we suggest that these results indicate that specific residues in the IL3 region are in positions and orientations that permit disulfide bond formation while others are not, irrespective of their accessibility.
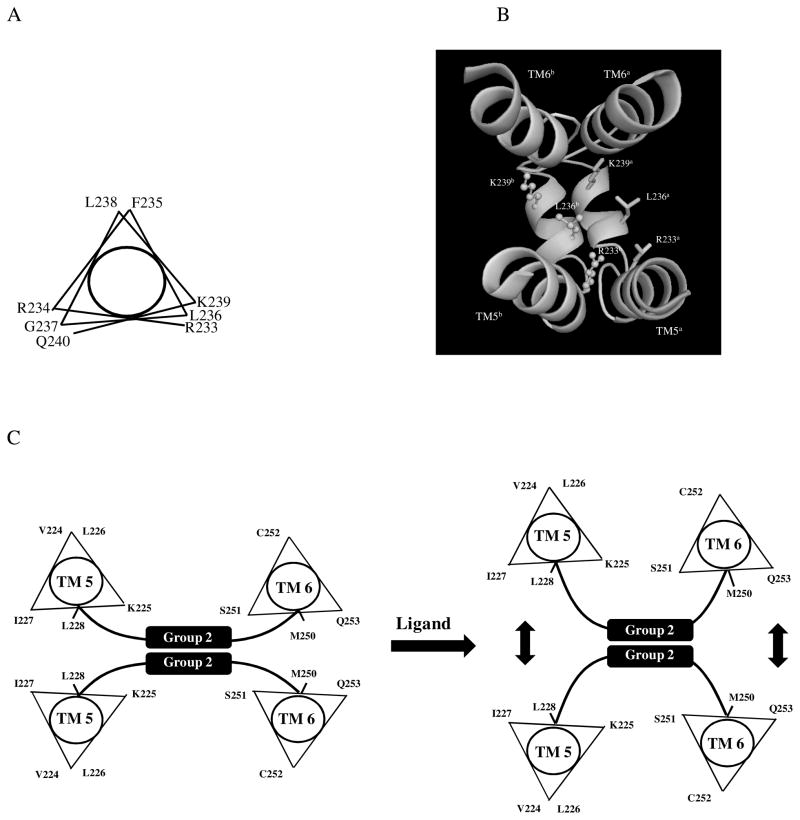
The IL3 of Ste2p has been suggested to have the potential to form an a-helix with a distinct amphipathic character (45). The pattern of disulfide formation in our study, with R233C, L236C and K239C mutant receptors having the highest percent of disulfide formation (Fig. 3 and 5) suggests that the group 2 residues in IL3 (R233-Q240) may form a 310 helix as shown in Fig. 8A. The Phyre structural bioinformatics group tools (30) also predicted a 310 helix in the 3-D structure of IL3 (Figure 8B) consistent with our cross-linking studies. The IL3 310 helix (RRFLGLKQ) is highly positively charged. It might be expected that in the wild type Ste2p dimer there should be charge repulsion between IL3 loop residues in the two Ste2p monomers. However, in the mutants studied herein the data strongly suggest that the substituted Cys residues are close enough to effectively form disulfide bonds. One might suggest that the Cys substitution alone changes the physical chemical characteristics of the loop leading to non-native interactions. However, the L236C mutant still contains all three positive residues (R233, R234 and K239) yet is capable of forming about 95% dimer, indicating that the IL3 residues in the dimeric receptor are indeed in very close proximity. We conclude that in native dimers of Ste2p the IL3 residues are close enough to make contacts that can be captured in disulfide crosslinking experiments. As pointed out by others the interpretation of such dimers in terms of receptor function must be tempered by the possible changes in the monomer-dimer equilibrium that are effected by chemical crosslinking (29, 35).
Figure 8.
Helical wheel projections and 3D model of IL3 residues involved in Ste2p-Ste2p interactions. A: The IL3 residues R233 to Q240 are suggested to form a 310 helix structure with a periodicity of 3.2 residues per turn in the helical wheel (58). B: The 3D structure of two Ste2p molecules showing regions TM5, TM6 and IL3 with a 310 helix in IL3 as predicted by the Phyre Structural Bioinformatics Group prediction tools (30). The IL3 310 helixes of two Ste2p subunits are suggested to be in close proximity that permit disulfide cross-linking. C: A schematic diagram showing possible shifts at the cytoplasmic ends of TM5 and TM6 upon ligand binding. The vertical arrows indicate a shift of the TMs away from each other.
A previous study used limited trypsin digestion of Ste2p to identify conformational changes induced by the binding of α-factor. The presence of α-factor caused the third intracellular loop of the receptor to become more accessible to trypsin suggesting that α-factor binding to Ste2p induced conformational changes of IL3 (19). However, specific residues or regions of the IL3 involved in such changes were not identified. As judged by their availability to form disulfide crosslinks our data suggest that the residues in the middle of IL3 do not undergo conformational change whereas the residues at the TM5-IL3 and IL3-TM6 boundaries do, leading to a change in their availability for disulfide formation.
The recent crystal structure of chemokine receptor, CXCR4 in the dimer form suggests that the monomers interact only at the extracellular side of helices TM5 and TM6 to play an important role in stabilizing the dimeric receptor (7). Our results suggest that Ste2p TM5/TM5 and TM6/TM6 interact at the cytoplasmic ends. We propose that the Ste2p residues in TM5 (K225, I227, L228, A229, I230), and in TM6 (L247, L248, I249, S251, and Q253) at the cytoplasmic ends may be involved in Ste2p-Ste2p interactions. Such interactions are disrupted upon ligand binding as shown by the disappearance or reduction of disulfide-mediated dimerization of the various Cys mutants in the presence of α-factor. Alpha-factor induced similar changes in the cross-linking of the various receptors in intact whole cells (Fig. 4D). Since in this latter experiment α-factor should only bind to the receptors at the cell surface (at the plasma membrane) the results suggest that the changes at the TM5 and TM6 cytoplasmic ends found in disulfide crosslinking experiments reflect the native state of these receptors. Disulfide crosslinking efficiencies reflect both the spatial proximity of the sulfhydryl groups and their mobility. Thus changes in conformation at the ends of TM5 and TM6 upon α-factor induced Ste2p activation can be inferred from the data. Another possibility is that α-factor activation changes the flexibility of these regions of Ste2p. A model consistent with our experimental data would have the TM5 and TM6 ends of a Ste2p dimer shift away from each other so that it is not possible to form a disulfide bond as shown in Fig. 8C. A similar reorientation of TM5 and TM6 has been proposed for activated forms of mammalian GPCRs (11, 46–48) and shown recently in a series of x-ray crystallographic studies (12–14, 49). Antagonist-bond dopamine receptor also showed reorientation of TM regions (49).
It is possible that the 100 kDa band observed in the gels represents interaction of Ste2p with some protein other than itself. The fact that all Ste2p Cys mutants we examined form this band makes this possibility highly unlikely. In addition, a number of papers (8, 22–23, 29, 35, 50–53) showed that Ste2p labeled by a variety of epitopes (FLAG, MYC, RHO) react with antibodies to these epitopes at positions in the gels that correspond to the molecular weight of a dimer of Ste2p. The antibodies react with no other bands except proteins at the Ste2p dimer or monomer position in these SDS-PAGE immunoblots. Similar results are obtained with an antibody generated against the N-terminus of Ste2p. Furthermore, purified Ste2p confirmed by MALDI-TOF and nanospray MS exhibits a 100 kDa at the same position we have attributed to the Ste2p dimer (54). Most importantly, the major conclusion of this paper is that the results indicate that the cytoplasmic ends of TM5 and TM6 undergo conformational change upon ligand binding. This conclusion would remain even if there were a second partner.
At the TM6 cytoplasmic end, residues L247, L248 I249, M250, S251, and Q253 all formed about 10–20% Ste2p-Ste2p dimers, however in the presence of ligand the percent of Ste2p-Ste2p dimer is greatly reduced, and residues L247C and I249C interact with Gpa1p (Umanah, unpublished). The switch of L247C and I249C from involvement in Ste2p-Ste2p interaction in the inactive state to Ste2p-Gpa1p heterodimer interactions in the active state would be consistent with a clock-wise rotation of this region of the receptor as observed in other mammalian GPCRs (11, 46–48). In the inactive state the TM5 cytoplasmic ends residues K225, I227, L228, A229 and I230 also formed about 10–15% Ste2p-Ste2p homodimers, and in the presence of ligand, these residues displayed reduced Ste2p-Ste2p homodimer formation and L228C formed a Ste2p-Gpa1p heterodimer (Umanah, unpublished). We therefore propose that the TM5 cytoplasmic end undergoes an anti-clockwise rotation as observed in other GPCRs (55).
Previously we showed that Ste2p Tyr266 (on the extracellular end of TM6) interacted with Asn205 (on the extracellular end of TM5) only in the active conformation of the receptor (56) suggesting that these extracellular regions of the Ste2p TM5 and TM6 undergo conformational changes upon α-factor binding. Thus our crosslinking results would support a model where binding of α-factor to Ste2p leads to conformational changes at the extracellular regions of TM5 and TM6 which are propagated to the cytoplasmic ends of these helices and result in exposure of L247C, L248 and I249C for activation of Gα.
The N-terminus of α-factor pheromone has been suggested to interact with Ste2p residues, S251-M294 which comprises part of TM6, the third extracellular loop, and part of TM7 (36–38). Recently, we show that Trp1 of α-factor interacts with K269 located at the extracellular boundary of TM6 (8). Previous studies showed that the α-factor antagonist was capable of competing out α-factor binding to Ste2p but did not exhibit measurable biological activities suggesting that the interactions of the first two amino acids of α-factor with residues at extracellular regions of Ste2p TM5/TM6 are critical for receptor activation. The experiments presented in this study suggest that the antagonist was not able to block or reduce disulfide formation compared to the native α-factor implying that antagonist binding to Ste2p does not induce the conformational changes or changes in flexibility at the TM5 and TM6 cytoplasmic ends.
It has been shown that the exchange of GDP for GTP in Gα proteins during receptor activation is induced by interactions of Gα with the receptor during activation and also that GTP binding to Gα may affect ligand affinity for its GPCR (1). The addition of GTP-γ-S to membrane fractions prior to CuP treatment to induce disulfide cross-linking did not have any observable effects on dimer formation though Ste2p and Gpa1p were expressed under the same promoter. Cross-linking carried out in a Gpa1p deleted strain was not affected by a large excess of agonist suggesting that Gpa1p-Ste2p interactions may influence conformational changes or changes in side chain flexibility at the cytoplasmic ends of TM5 and TM6. We conclude that the putative conformational changes at the cytoplasmic ends of TM5 and TM6 in Ste2p require interactions of the N-terminal amino residues of α-factor and are affected by the interactions with Gα, but not by Gα activation.
In conclusion, we show for the first time changes in receptor conformation or flexibility that influence the availability for disulfide formation of residues in the region of IL3 of Ste2p close to the contiguous TM5 and TM6 helices. The hydrophilic residues located in the middle of the IL3 loop (R231-Q240) do not change conformation/availability during receptor activation whereas many hydrophobic residues at the TM5 and TM6 cytoplasmic junctions do. These conformational changes require ligand-induced activation of Ste2p and appear to depend on Ste2p-Gα protein interactions. The pattern of disulfide formation observed is consistent with a 310 helical structure in the center of IL3 and suggests that the IL3 loop of two Ste2p subunits remain in close proximity both in the active and inactive states of the receptors. Since Ste2p has been shown to have structure-function relationships similar to the physiologically and pharmacologically important rhodopsin-like GPCR family (57), the role of IL3 observed in this study has implications not only for Ste2p but also other GPCR systems
Supplementary Material
Acknowledgments
This work was supported by GM-22087 from the National Institute of General Medical Sciences of the National Institutes of Health.
The abbreviations used are
- GPCR
G protein-coupled receptor
- G protein
heterotrimeric GTP-binding protein
- Gpa1p
Gα subunit of G protein encoded by the GPA1 gene
- IL3
the third intracellular loop of Ste2p
- Ste2p
α-factor receptor encoded by the STE2 gene
- TM
transmembrane domain
Footnotes
Supporting Information Available: Supporting information contains the results of the growth arrest and Fus1-LacZ assays of all IL3 cysteine mutants. The binding and growth arrest assays of the native and antagonist α-factor are also included. All supplemental materials may be accessed free of charge online at http://pubs.acs.org.
References
- 1.Ratnala VR, Kobilka B. Understanding the ligand-receptor-G protein ternary complex for GPCR drug discovery. Methods Mol Biol. 2009;552:67–77. doi: 10.1007/978-1-60327-317-6_5. [DOI] [PubMed] [Google Scholar]
- 2.Rosenbaum DM, Rasmussen SG, Kobilka BK. The structure and function of G-protein-coupled receptors. Nature. 2009;459:356–363. doi: 10.1038/nature08144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Williams C, Hill SJ. GPCR signaling: understanding the pathway to successful drug discovery. Methods Mol Biol. 2009;552:39–50. doi: 10.1007/978-1-60327-317-6_3. [DOI] [PubMed] [Google Scholar]
- 4.Panetta R, Greenwood MT. Physiological relevance of GPCR oligomerization and its impact on drug discovery. Drug Discov Today. 2008;13:1059–1066. doi: 10.1016/j.drudis.2008.09.002. [DOI] [PubMed] [Google Scholar]
- 5.De Amici M, Dallanoce C, Holzgrabe U, Trankle C, Mohr K. Allosteric ligands for G protein-coupled receptors: a novel strategy with attractive therapeutic opportunities. Med Res Rev. 2010;30:463–549. doi: 10.1002/med.20166. [DOI] [PubMed] [Google Scholar]
- 6.Lundstrom K. An overview on GPCRs and drug discovery: structure-based drug design and structural biology on GPCRs. Methods Mol Biol. 2009;552:51–66. doi: 10.1007/978-1-60327-317-6_4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wu B, Chien EY, Mol CD, Fenalti G, Liu W, Katritch V, Abagyan R, Brooun A, Wells P, Bi FC, Hamel DJ, Kuhn P, Handel TM, Cherezov V, Stevens RC. Structures of the CXCR4 chemokine GPCR with small-molecule and cyclic peptide antagonists. Science. 2010;330:1066–1071. doi: 10.1126/science.1194396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Umanah GK, Huang L, Ding FX, Arshava B, Farley AR, Link AJ, Naider F, Becker JM. Identification of residue-to-residue contact between a peptide ligand and its G protein-coupled receptor using periodate-mediated dihydroxyphenylalanine cross-linking and mass spectrometry. J Biol Chem. 2010;285:39425–39436. doi: 10.1074/jbc.M110.149500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cabrera-Vera TM, Vanhauwe J, Thomas TO, Medkova M, Preininger A, Mazzoni MR, Hamm HE. Insights into G protein structure, function, and regulation. Endocr Rev. 2003;24:765–781. doi: 10.1210/er.2000-0026. [DOI] [PubMed] [Google Scholar]
- 10.Scheerer P, Park JH, Hildebrand PW, Kim YJ, Krauss N, Choe HW, Hofmann KP, Ernst OP. Crystal structure of opsin in its G-protein-interacting conformation. Nature. 2008;455:497–502. doi: 10.1038/nature07330. [DOI] [PubMed] [Google Scholar]
- 11.Wess J, Han SJ, Kim SK, Jacobson KA, Li JH. Conformational changes involved in G-protein-coupled-receptor activation. Trends Pharmacol Sci. 2008;29:616–625. doi: 10.1016/j.tips.2008.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Warne T, Moukhametzianov R, Baker JG, Nehme R, Edwards PC, Leslie AG, Schertler GF, Tate CG. The structural basis for agonist and partial agonist action on a beta(1)-adrenergic receptor. Nature. 2011;469:241–244. doi: 10.1038/nature09746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rosenbaum DM, Zhang C, Lyons JA, Holl R, Aragao D, Arlow DH, Rasmussen SG, Choi HJ, Devree BT, Sunahara RK, Chae PS, Gellman SH, Dror RO, Shaw DE, Weis WI, Caffrey M, Gmeiner P, Kobilka BK. Structure and function of an irreversible agonist-beta(2) adrenoceptor complex. Nature. 2011;469:236–240. doi: 10.1038/nature09665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xu F, Wu H, Katritch V, Han GW, Jacobson KA, Gao ZG, Cherezov V, Stevens RC. Structure of an agonist-bound human A2A adenosine receptor. Science. 2011;332:322–327. doi: 10.1126/science.1202793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Clark CD, Palzkill T, Botstein D. Systematic mutagenesis of the yeast mating pheromone receptor third intracellular loop. J Biol Chem. 1994;269:8831–8841. [PubMed] [Google Scholar]
- 16.Stefan CJ, Blumer KJ. The third cytoplasmic loop of a yeast G-protein-coupled receptor controls pathway activation, ligand discrimination, and receptor internalization. Mol Cell Biol. 1994;14:3339–3349. doi: 10.1128/mcb.14.5.3339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Celic A, Martin NP, Son CD, Becker JM, Naider F, Dumont ME. Sequences in the intracellular loops of the yeast pheromone receptor Ste2p required for G protein activation. Biochemistry. 2003;42:3004–3017. doi: 10.1021/bi0269308. [DOI] [PubMed] [Google Scholar]
- 18.Gladue DP, Konopka JB. Scanning mutagenesis of regions in the Galpha protein Gpa1 that are predicted to interact with yeast mating pheromone receptors. FEMS Yeast Res. 2008;8:71–80. doi: 10.1111/j.1567-1364.2007.00311.x. [DOI] [PubMed] [Google Scholar]
- 19.Bukusoglu G, Jenness DD. Agonist-specific conformational changes in the yeast alpha-factor pheromone receptor. Mol Cell Biol. 1996;16:4818–4823. doi: 10.1128/mcb.16.9.4818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dube P, DeCostanzo A, Konopka JB. Interaction between transmembrane domains five and six of the alpha -factor receptor. J Biol Chem. 2000;275:26492–26499. doi: 10.1074/jbc.M002767200. [DOI] [PubMed] [Google Scholar]
- 21.Medici R, Bianchi E, Di Segni G, Tocchini-Valentini GP. Efficient signal transduction by a chimeric yeast-mammalian G protein alpha subunit Gpa1-Gsalpha covalently fused to the yeast receptor Ste2. EMBO J. 1997;16:7241–7249. doi: 10.1093/emboj/16.24.7241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hauser M, Kauffman S, Lee BK, Naider F, Becker JM. The first extracellular loop of the Saccharomyces cerevisiae G protein-coupled receptor Ste2p undergoes a conformational change upon ligand binding. J Biol Chem. 2007;282:10387–10397. doi: 10.1074/jbc.M608903200. [DOI] [PubMed] [Google Scholar]
- 23.Huang LY, Umanah G, Hauser M, Son C, Arshava B, Naider F, Becker JM. Unnatural amino acid replacement in a yeast G protein-coupled receptor in its native environment. Biochemistry. 2008;47:5638–5648. doi: 10.1021/bi701866e. [DOI] [PubMed] [Google Scholar]
- 24.Sherman F. Getting started with yeast. Methods Enzymol. 1991;194:3–21. doi: 10.1016/0076-6879(91)94004-v. [DOI] [PubMed] [Google Scholar]
- 25.David NE, Gee M, Andersen B, Naider F, Thorner J, Stevens RC. Expression and purification of the Saccharomyces cerevisiae alpha-factor receptor (Ste2p), a 7-transmembrane-segment G protein-coupled receptor. J Biol Chem. 1997;272:15553–15561. doi: 10.1074/jbc.272.24.15553. [DOI] [PubMed] [Google Scholar]
- 26.Trueheart J, Boeke JD, Fink GR. Two genes required for cell fusion during yeast conjugation: evidence for a pheromone-induced surface protein. Mol Cell Biol. 1987;7:2316–2328. doi: 10.1128/mcb.7.7.2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Raths SK, Naider F, Becker JM. Peptide analogues compete with the binding of alpha-factor to its receptor in Saccharomyces cerevisiae. J Biol Chem. 1988;263:17333–17341. [PubMed] [Google Scholar]
- 28.Lee BK, Khare S, Naider F, Becker JM. Identification of residues of the Saccharomyces cerevisiae G protein-coupled receptor contributing to alpha-factor pheromone binding. J Biol Chem. 2001;276:37950–37961. doi: 10.1074/jbc.M103579200. [DOI] [PubMed] [Google Scholar]
- 29.Wang HX, Konopka JB. Identification of amino acids at two dimer interface regions of the alpha-factor receptor (Ste2) Biochemistry. 2009;48:7132–7139. doi: 10.1021/bi900424h. [DOI] [PubMed] [Google Scholar]
- 30.Kelley LA, Sternberg MJ. Protein structure prediction on the Web: a case study using the Phyre server. Nat Protoc. 2009;4:363–371. doi: 10.1038/nprot.2009.2. [DOI] [PubMed] [Google Scholar]
- 31.Choi Y, Konopka JB. Accessibility of cysteine residues substituted into the cytoplasmic regions of the alpha-factor receptor identifies the intracellular residues that are available for G protein interaction. Biochemistry. 2006;45:15310–15317. doi: 10.1021/bi0614939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bouvier M. Oligomerization of G-protein-coupled transmitter receptors. Nat Rev Neurosci. 2001;2:274–286. doi: 10.1038/35067575. [DOI] [PubMed] [Google Scholar]
- 33.Park PS, Filipek S, Wells JW, Palczewski K. Oligomerization of G proteincoupled receptors: past, present, and future. Biochemistry. 2004;43:15643–15656. doi: 10.1021/bi047907k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Overton MC, Blumer KJ. The extracellular N-terminal domain and transmembrane domains 1 and 2 mediate oligomerization of a yeast G protein-coupled receptor. J Biol Chem. 2002;277:41463–41472. doi: 10.1074/jbc.M205368200. [DOI] [PubMed] [Google Scholar]
- 35.Kim H, Lee BK, Naider F, Becker JM. Identification of specific transmembrane residues and ligand-induced interface changes involved in homo-dimer formation of a yeast G protein-coupled receptor. Biochemistry. 2009;48:10976–10987. doi: 10.1021/bi901291c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Eriotou-Bargiota E, Xue CB, Naider F, Becker JM. Antagonistic and synergistic peptide analogues of the tridecapeptide mating pheromone of Saccharomyces cerevisiae. Biochemistry. 1992;31:551–557. doi: 10.1021/bi00117a036. [DOI] [PubMed] [Google Scholar]
- 37.Henry LK, Khare S, Son C, Babu VV, Naider F, Becker JM. Identification of a contact region between the tridecapeptide alpha-factor mating pheromone of Saccharomyces cerevisiae and its G protein-coupled receptor by photoaffinity labeling. Biochemistry. 2002;41:6128–6139. doi: 10.1021/bi015863z. [DOI] [PubMed] [Google Scholar]
- 38.Son CD, Sargsyan H, Hurst GB, Naider F, Becker JM. Analysis of ligand-receptor cross-linked fragments by mass spectrometry. J Pept Res. 2005;65:418–426. doi: 10.1111/j.1399-3011.2005.00248.x. [DOI] [PubMed] [Google Scholar]
- 39.Smith B, Hill C, Godfrey EL, Rand D, van den Berg H, Thornton S, Hodgkin M, Davey J, Ladds G. Dual positive and negative regulation of GPCR signaling by GTP hydrolysis. Cell Signal. 2009;21:1151–1160. doi: 10.1016/j.cellsig.2009.03.004. [DOI] [PubMed] [Google Scholar]
- 40.Slessareva JE, Ma H, Depree KM, Flood LA, Bae H, Cabrera-Vera TM, Hamm HE, Graber SG. Closely related G-protein-coupled receptors use multiple and distinct domains on G-protein alpha-subunits for selective coupling. J Biol Chem. 2003;278:50530–50536. doi: 10.1074/jbc.M304417200. [DOI] [PubMed] [Google Scholar]
- 41.Kristiansen K. Molecular mechanisms of ligand binding, signaling, and regulation within the superfamily of G-protein-coupled receptors: molecular modeling and mutagenesis approaches to receptor structure and function. Pharmacol Ther. 2004;103:21–80. doi: 10.1016/j.pharmthera.2004.05.002. [DOI] [PubMed] [Google Scholar]
- 42.Fotiadis D, Liang Y, Filipek S, Saperstein DA, Engel A, Palczewski K. Atomic-force microscopy: Rhodopsin dimers in native disc membranes. Nature. 2003;421:127–128. doi: 10.1038/421127a. [DOI] [PubMed] [Google Scholar]
- 43.Liang Y, Fotiadis D, Filipek S, Saperstein DA, Palczewski K, Engel A. Organization of the G protein-coupled receptors rhodopsin and opsin in native membranes. J Biol Chem. 2003;278:21655–21662. doi: 10.1074/jbc.M302536200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Filizola M, Wang SX, Weinstein H. Dynamic models of G-protein coupled receptor dimers: indications of asymmetry in the rhodopsin dimer from molecular dynamics simulations in a POPC bilayer. J Comput Aided Mol Des. 2006;20:405–416. doi: 10.1007/s10822-006-9053-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Blumer KJ, Thorner J. Receptor-G protein signaling in yeast. Annu Rev Physiol. 1991;53:37–57. doi: 10.1146/annurev.ph.53.030191.000345. [DOI] [PubMed] [Google Scholar]
- 46.Bhattacharya S, Hall SE, Vaidehi N. Agonist-induced conformational changes in bovine rhodopsin: insight into activation of G-protein-coupled receptors. J Mol Biol. 2008;382:539–555. doi: 10.1016/j.jmb.2008.06.084. [DOI] [PubMed] [Google Scholar]
- 47.Jaakola VP, Griffith MT, Hanson MA, Cherezov V, Chien EY, Lane JR, Ijzerman AP, Stevens RC. The 2.6 angstrom crystal structure of a human A2A adenosine receptor bound to an antagonist. Science. 2008;322:1211–1217. doi: 10.1126/science.1164772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ludeke S, Mahalingam M, Vogel R. Rhodopsin activation switches in a native membrane environment. Photochem Photobiol. 2009;85:437–441. doi: 10.1111/j.1751-1097.2008.00490.x. [DOI] [PubMed] [Google Scholar]
- 49.Chien EY, Liu W, Zhao Q, Katritch V, Han GW, Hanson MA, Shi L, Newman AH, Javitch JA, Cherezov V, Stevens RC. Structure of the human dopamine D3 receptor in complex with a D2/D3 selective antagonist. Science. 2010;330:1091–1095. doi: 10.1126/science.1197410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Blumer KJ, Reneke JE, Thorner J. The STE2 gene product is the ligand-binding component of the alpha-factor receptor of Saccharomyces cerevisiae. J Biol Chem. 1988:263. [PubMed] [Google Scholar]
- 51.Konopka JB, Jenness DD, Hartwell LH. The C-terminus of the S. cerevisiae alpha-pheromone receptor mediates an adaptive response to pheromone. Cell. 1988;54:609–620. doi: 10.1016/s0092-8674(88)80005-9. [DOI] [PubMed] [Google Scholar]
- 52.Yesilaltay A, Jenness DD. Homo-oligomeric complexes of the yeast alpha-factor pheromone receptor are functional units of endocytosis. Mol Biol Cell. 2000;11:2873–2884. doi: 10.1091/mbc.11.9.2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shi C, Shin YO, Hanson J, Cass B, Loewen MC, Durocher Y. Purification and characterization of a recombinant G-protein-coupled receptor, Saccharomyces cerevisiae Ste2p, transiently expressed in HEK293 EBNA1 cells. Biochemistry. 2005;44:15705–15714. doi: 10.1021/bi051292p. [DOI] [PubMed] [Google Scholar]
- 54.Lee BK, Jung KS, Son C, Kim H, VerBerkmoes NC, Arshava B, Naider F, Becker JM. Affinity purification and characterization of a G-protein coupled receptor, Saccharomyces cerevisiae Ste2p. Protein Expr Purif. 2007;56:62–71. doi: 10.1016/j.pep.2007.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Domazet I, Martin SS, Holleran BJ, Morin ME, Lacasse P, Lavigne P, Escher E, Leduc R, Guillemette G. The fifth transmembrane domain of angiotensin II Type 1 receptor participates in the formation of the ligand-binding pocket and undergoes a counterclockwise rotation upon receptor activation. J Biol Chem. 2009;284:31953–31961. doi: 10.1074/jbc.M109.051839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lee YH, Naider F, Becker JM. Interacting residues in an activated state of a G protein-coupled receptor. J Biol Chem. 2006;281:2263–2272. doi: 10.1074/jbc.M509987200. [DOI] [PubMed] [Google Scholar]
- 57.Eilers M, Hornak V, Smith SO, Konopka JB. Comparison of class A and D G protein-coupled receptors: common features in structure and activation. Biochemistry. 2005;44:8959–8975. doi: 10.1021/bi047316u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kim S, Cross TA. 2D solid state NMR spectral simulation of 3(10), alpha, and pi-helices. J Magn Reson. 2004;168:187–193. doi: 10.1016/j.jmr.2004.02.009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.