Abstract
Growth of poly(2-hydroxyethyl methacrylate) brushes on magnetic nanoparticles and subsequent brush functionalization with nitrilotriacetate-Ni2+ yield magnetic beads that selectively capture polyhistidine-tagged (His-tagged) protein directly from cell extracts. Transmission electron microscopy, FT-IR spectroscopy, thermogravimetric analysis, and magnetization measurements confirm and quantify the formation of the brushes on magnetic particles, and multilayer protein adsorption to these brushes results in binding capacities (220 mg BSA/g of beads and 245 mg His-tagged Ubiquitin/g of beads) that are an order of magnitude greater than those of commercial magnetic beads. Moreover, the functionalized beads selectively capture His-tagged protein within 5 min. The high binding capacity and protein purity along with efficient protein capture in a short incubation time make brush-modified particles attractive for purification of recombinant proteins.
Keywords: polymer brush, magnetic beads, His-tagged proteins
Introduction
Many protein-purification methods employ a separation step based on specific interactions between immobilized ligands and affinity tags on the protein,1-3 and the most common affinity tag is polyhistidine, which binds to immobilized Ni2+ or Co2+ complexes. Isolation of His-tagged protein typically occurs in columns containing resins modified with Ni2+ complexes, often Ni2+-nitrilotriacetate (Ni2+-NTA), and these methods are attractive for their high protein loading, mild elution conditions, and easy regeneration of the metal complexes on the resin.4-6 However, slow intra-particle biomolecule diffusion to binding sites in porous micron-sized beads can lead to relatively long processing times7 that can be problematic when purifying unstable proteins.8
Functionalized magnetic particles are attractive for small-scale purification and analysis because they can be readily isolated from solutions using a strong magnetic field, in some cases with commercially available magnetic separators9 or automated magnetic bead processors.10, 11 Specific modification of the particles allows isolation of protein or DNA,12 as well as magnetic cell separation.9, 13 However, most commercial magnetic beads are micron sized and thus have a limited surface area and binding capacity (typically <25 mg protein/g of beads).12, 14 The use of highly porous beads can enhance capacity, but as mentioned above, slow diffusion in pores will limit binding rates. In principle, addition of more beads to a sample can overcome low binding capacities, but the greater volume of the particle slurry will dilute the sample and perhaps lead to increased non-specific adsorption or sample loss. In some instances, beads with high capacities may increase the binding efficiencies for low-abundance protein.25
Several studies examined modified magnetic nanoparticles for protein binding because these small particles have a large surface area to volume ratio and hence a high binding capacity.15-25 Xu and coworkers reported preliminary capacities of 2-3 g of protein per g of bead when using extremely small (diameters less than 3 nm) FePt nanoparticles modified with nitrilotriacetate (NTA),22 but such particles may be difficult to collect.23 A recent paper also suggested capacities as high as 2 g protein/ per g of beads for ~30 nm NTA-Au/Fe3O4 beads, although some nonspecific protein adsorption occurred on the base Au/Fe3O4 particle.25 However, in all cases the binding capacity did not exceed a monolayer of protein on the particle surface.
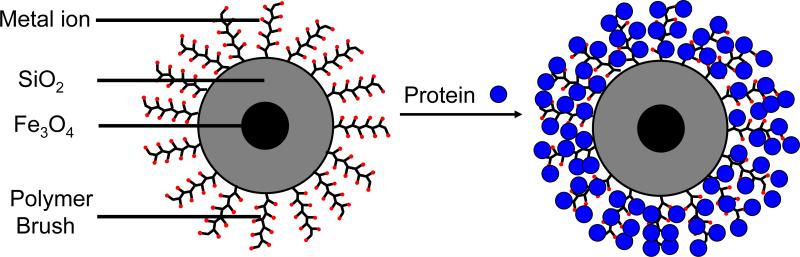
This work reports the modification of magnetic nanoparticles (MNPs) through growth of polymer brushes by atom transfer radical polymerization (ATRP) from immobilized initiators. Recent studies show that such brushes can increase protein-binding capacities in porous membranes,26, 27 and prior research on ATRP from silica demonstrates that this polymerization technique allows controlled brush growth without particle aggregation.28-34 Control over brush thickness is vital for negotiating the tradeoff between enhancing binding capacity and decreasing the magnetization of MNPs. ATRP of glycidyl methacrylate from magnetic beads permits covalent immobilization of proteins, but the highest reported binding capacity of such systems is 50 mg protein per g of beads.35, 36 This work shows that MNPs with ~50 nm-thick brushes can be collected with a permanent magnet and that multilayer binding of tagged protein (see Figure 1) leads to a binding capacity of 220-245 mg protein/g of beads. Moreover, protein isolation can occur directly from cell extracts in a few minutes.
Figure 1.
Schematic diagram of brush-modified beads and protein binding to these beads.
Experimental section
Materials
N,N-dimethylformamide (DMF, anhydrous, 99.8%), CuCl (99.999%), CuBr2 (99%), 2,2’-bipyridyl (bpy, 99%), 1-[3-(dimethylamino)propyl]-3-ethylcarbodiimide hydrochloride (EDC), N-hydroxysuccinimide (NHS), 4-dimethylaminopyridine (DMAP), imidazole (99%), 2-hydroxyethyl methacrylate (HEMA, 97%), TWEEN-20 surfactant, His-tagged ubiquitin (HisU), and bovine serum albumin (BSA) were obtained from Sigma Aldrich. Dynabeads® were purchased from Invitrogen. HEMA was passed through a column of activated basic alumina (Aldrich) prior to use. NiSO4·5H2O (Columbus Chemical), CuSO4·5H2O (CCI), NaH2PO4 (CCI), Na2HPO4 (Aldrich), tetraethyl orthosilicate (TEOS, Fluka), Nα,Nα-bis(carboxymethyl)-l-lysine hydrate (aminobutyl NTA, Fluka), succinic anhydride (SA, Matheson Coleman & Bell), Coomassie protein assay reagent (Pierce), and all other chemicals were of analytical grade, and used as received.
Synthesis of silica-coated magnetic nanoparticles
EMG 308 (1 mL, Ferrotec), a solution of Fe3O4 nanoparticles, was diluted with water (13.6 mL) prior to addition of isopropanol (80 mL) to precipitate the magnetite nanoparticles and remove excess surfactant. The supernatant was decanted, and the magnetically collected nanoparticles were then dispersed in 200 mL of 80% (v:v) ethanol (EtOH) in water. After sonication of this solution for 15 min, concentrated ammonia (5 mL) and TEOS (2 mL) were added to the solution,37, 38 and the mixture was mechanically stirred for 3 h. HCl (1 mL, 2M) was added to stop the reaction, and the silica-coated MNPs were magnetically collected and washed twice with 50 mL of EtOH (the same volume of solvent was used for all the washing steps), twice with water, and again with EtOH before drying under vacuum for 6 h.
Initiator attachment
SiO2-MNPs were dried at 120 °C overnight and then dispersed in toluene through sonication (0.2 g beads in 150 mL). Tricholorosilane initiator, (11-(2-bromo-2-methyl) propionyloxy)undecyltrichlorosilane39 was added (0.2 mL), and the mixture was stirred at 65 °C overnight while bubbling with N2. The initiator-modified beads were washed twice with 50 mL of toluene and twice with 50 mL of THF prior to drying in vacuum for 4 h.
Growth of poly(2-hydroxyethyl methacrylate) (PHEMA) on beads
Initiator-modified beads (0.1 g) were dispersed by sonication in purified HEMA (10 mL), and the mixture was degassed via 3 freeze-pump-thaw cycles. In another Schlenk flask, HEMA (10 mL) was mixed with water (10 mL) and bpy (366 mg), and after 3 degassing cycles CuCl (82.5 mg) and CuBr2 (54 mg) were added and another 2 degassing cycles were performed. In a glove bag, the two degassed HEMA mixtures were combined and magnetically stirred for 1 h. Subsequently, air was bubbled through the polymerization solution until its color turned from dark brown to dark green, and the beads were then washed sequentially with 20 mL of DMF (the same volume of solvent was used for all the washing steps), EtOH (twice), water, and EtOH, and dried in vacuum.
Derivatization
40 PHEMA-MNPs (200 mg) were dispersed in 10 mL DMF by sonication and mixed with 10 mL DMF containing SA (200 mg) and DMAP (200 mg) prior to stirring at 55 °C overnight. The SA-MNPs were washed with 20 mL of DMF (the same volume of solvent was used for all the washing steps), EtOH (twice) and then water (twice). Subsequently, an aqueous solution (20 mL) containing EDC (0.1 M) and NHS (0.1 M) was mixed with the SA-MNPs for 30 min followed by washing with EtOH three times. Then, the EDC/NHS-activated beads were exposed to a pH 10, aminobutyl-NTA (0.1 M) aqueous solution for 1 h with magnetic stirring and washed with water three times. Finally, the NTA-SA-PHEMA-MNPs were immersed in CuSO4 (0.05 M) for 1 h, washed with water three times and EtOH twice and dried under vacuum for 5 h.
Characterization methods
Thermogravimetric analysis (TGA) was run under air flow with a 3-step program: heating from 30 °C to 120 °C at a rate of 10 °C/min, holding at 120 °C for 30 min, and finally ramping the temperature to 800 °C at 10 °C/min. Transmission electron microscopy (TEM) images were obtained on a JEOL 100CX microscope. All the samples were prepared by a drop-dry method on carbon-coated copper grids. For dynamic light scattering (DLS), particles were dispersed in water, and the size distribution was determined on a ZetaPALS instrument (Brookhaven, Holtsville, NY) with a 35 mW 660 nm excitation.
Results and discussion
Preparation and derivatization of magnetic nanoparticles
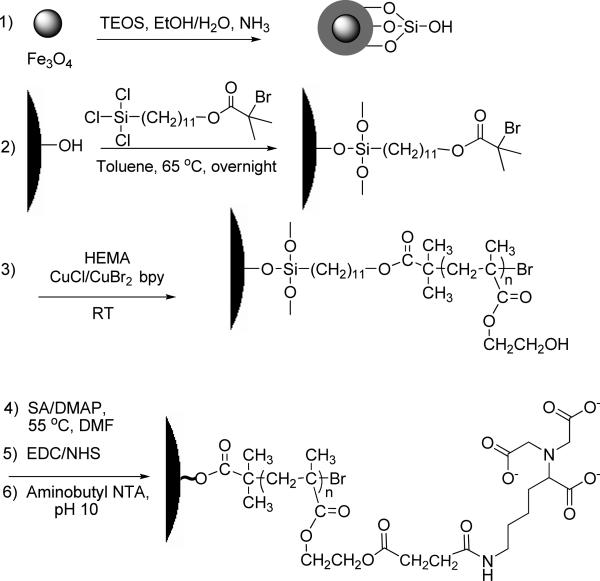
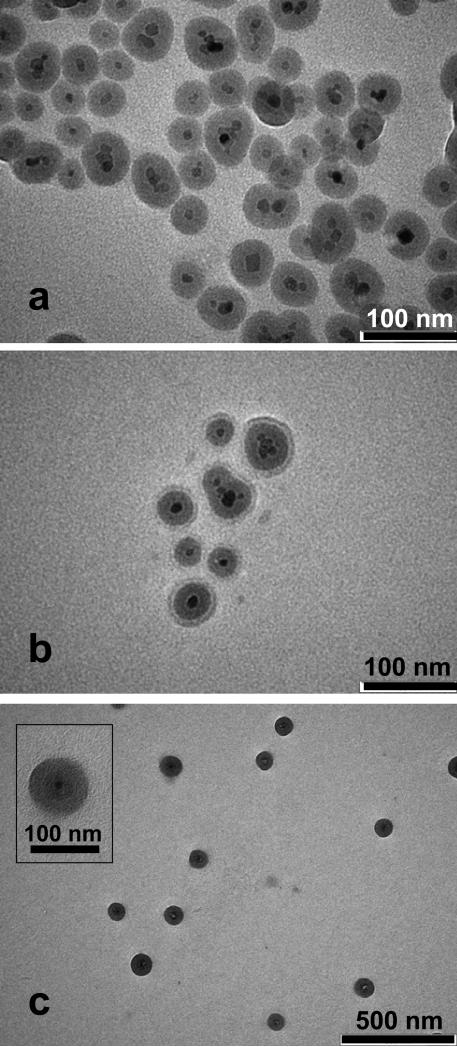
Scheme 1 shows the procedure for creating magnetic particles coated with functional polymer brushes that bind proteins. The first step is the formation of a silica coating on the magnetic nanoparticles to prevent oxidation and dissolution of the Fe3O4. The high density of –OH groups on the silica also allows further modification of the particles through silanization. To coat Fe3O4 nanoparticles with silica, we employ standard sol gel methods with some important modifications.37, 38 Among these modifications, precipitation of the commercial Fe3O4 nanoparticles in an isopropanol/water mixture (prior to their resuspension in an ethanol/water solution) presumably removes excess surfactant to minimize particle aggregation during deposition of silica through hydrolysis of TEOS and condensation on the Fe3O4 seeds. After formation of the encapsulated Fe3O4, rapid neutralization of the silica-coating solution by addition of HCl is critical to avoid the formation of large clusters during collection of the particles with a magnet. TEM images show unaggregated particles with a dark Fe3O4 core, which may consist of several Fe3O4 nanoparticles, inside a silica shell (Figure 2a).
Scheme 1.
Synthesis and derivatization of polymer brush-modified magnetic nanoparticles.
Figure 2.
TEM images of (a) SiO2-Fe3O4, (b) PHEMA-SiO2-Fe3O4, and (c) NTA-SA-PHEMA-SiO2-Fe3O4 nanoparticles. The inset in image (c) shows an NTA-SA-PHEMA-SiO2-Fe3O4 particle at higher magnification.
Growth of polymer brushes from silica-coated nanoparticles requires immobilization of polymerization initiators on the surface via silanization (Scheme 1, step 2). However, this reaction releases HCl, which may dissolve the Fe3O4 core even after coating with silica because of the high reaction temperature (65 °C) and relatively long time (~15 h). In initial studies, TEM images revealed white spots in the center of some of the nanoparticles after silanization as well as an absence of dark cores, suggesting the dissolution of the Fe3O4 (see Figure S1a, Figure numbers beginning with S refer to the supporting information). To overcome this problem, we sparge the silane solution with N2 during initiator attachment to both remove HCl and prevent the introduction of water vapor. With this procedure, all the initiator-modified nanoparticles in TEM images show dark Fe3O4 cores (Figure S1b).
Subsequent polymerization of HEMA from the immobilized initiators gives MNPs modified with polymer brushes, and quenching the polymerization with air prior to collecting the brush-modified nanoparticles helps to avoid particle aggregation. The ultimate goal of this work is to create readily collectible nanoparticles that exhibit high protein-binding capacities, and binding capacity should increase with the length of polymer chains. However, long brushes also decrease the magnetization of the MNPs and make their collection difficult, so we limit the polymerization time to 1 h. ATRP is attractive for modifying MNPs because it affords control over the brush molecular weight and film thickness through variation of polymerization time. Additionally, surface-initiated ATRP minimizes solution polymerization that could result in physisorbed polymers that cause particle aggregation.
Derivatization of the polymer brushes includes reaction with SA, activation of the resulting – COOH groups with EDC and NHS, and subsequent reaction with aminobutyl NTA (Scheme 1). Visible changes in the reaction suspensions qualitatively demonstrate the success of the derivatization procedure. For example, SA-modified MNPs typically disperse well in water because the side chains on the polymer brushes end in carboxylic acids, but after activation with EDC/NHS mixtures, the reacted particles tend to aggregate in water because they are more hydrophobic. In addition, the aggregation makes beads more collectable.
Characterization of particle size and the extent of polymerization and derivatization
TEM images confirm nanoparticle modification and provide estimates of particle diameters. Bare Fe3O4 particles have diameters of 5-15 nm, and after encasing these seeds in silica, some particles contain multiple Fe3O4 nuclei because of iron oxide aggregation prior to coating (Figure 2a).38 The size distribution for 290 silica-coated particles in TEM images (Figure S2) shows an average particle diameter of 43 ± 8 nm, and dynamic light scattering data (Figure S3, top) suggest that 90% of the particles have diameters between 43 and 57 nm. However, the light scattering data also show that about 10% of the particles are between 151 nm and 216 nm in diameter, which could be due to some particle aggregation.
After growth of PHEMA from the silica-coated nanoparticles, TEM images such as that in Figure 2b show the formation of a 10 nm-thick polymer layer on the ~45 nm-diameter SiO2-Fe3O4 particles. In contrast, light scattering measurements show that 90% of the PHEMA-modified particles have diameters between 134 and 170 nm, whereas the other 10% of particles are between 289 nm and 365 nm in diameter (Figure S3, bottom). The ~100 nm increase in diameter after polymerization suggests that the polymer layer is ~50 nm thick Light-scattering measurements occur in water, which likely swells the film to increase thickness. Such swelling of polymer brushes is vital for binding of multilayers of protein to modified beads. The particle diameter in TEM images is ~100 nm after modification with NTA, indicating a ~30 nm thick polymer layer on the SiO2-Fe3O4 core (Figure 2c). In the inset of Figure 2c, the contrast between the polymer and the underlying nanoparticle is relatively low, but the image suggests a ~30 nm film on the outside of the SiO2-Fe3O4 particle. Derivatization of the polymer with NTA more than triples (3.6 times) the molecular weight of the polymer repeat unit, so the large increase in thickness is not surprising. Notably, the particles are not aggregated after the derivatization process, presumably because of the high negative charge of NTA.
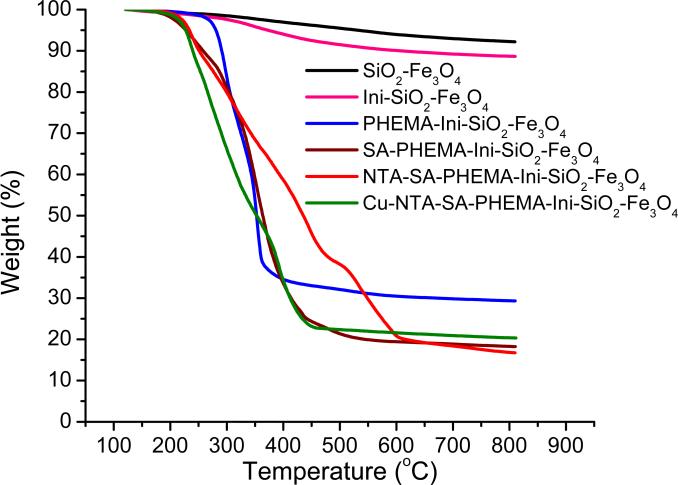
TGA plots of weight loss versus temperature (Figure 3) provide an estimate of the quantity of polymer grown from the nanoparticles. The coated particles consist of both thermally stable compounds that remain in the residue (silica, magnetite, and metal ions bound to the polymer) and decomposable polymer brushes and initiators that contribute to weight loss. Even with just silica-coated Fe3O4, however, a small amount of water and possibly residual ethoxy groups from the TEOS give rise to an 8% total weight loss. After growth of PHEMA from the particle, TGA shows a total weight loss of 71%, which corresponds to a PHEMA thickness of ~14 nm (see the supporting information for details of the thickness calculations from TGA data). This thickness value agrees reasonably well with the TEM thickness of ~10 nm. Reaction of PHEMA with SA leads to a total weight loss of 81%, which is consistent with essentially complete reaction between SA and PHEMA (see supporting information). After derivatization of SA-PHEMA with NTA and NTA-Cu2+, the percent mass loss of the particles during TGA does not increase significantly, probably because sodium and copper remain in the residue.
Figure 3.
Weight losses obtained from TGA of SiO2-Fe3O4 (black), Initiator-SiO2-Fe3O4 (pink), PHEMA-Initiator-SiO2-Fe3O4 (blue), SA-PHEMA-Initiator-SiO2-Fe3O4 (brown), NTA-SA-PHEMA-Initiator-SiO2-Fe3O4 (red), and Cu2+-NTA-SA-PHEMA-Initiator-SiO2-Fe3O4 (green) particles.
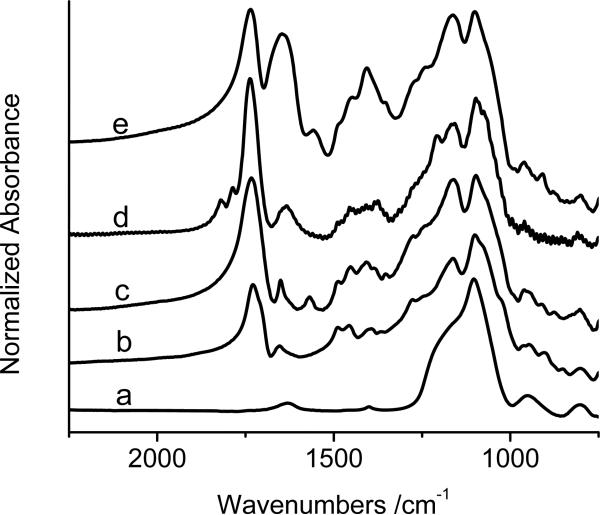
The FTIR spectra in Figure 4 also confirm the polymerization and derivatization reactions in Scheme 1. After growth of PHEMA on the SiO2-Fe3O4 particles, an absorption maximum due to carbonyl groups in the polymer appears at ~1740 cm-1 (spectrum 4b). Reaction of the PHEMA with SA results in a greatly increased absorbance at ~1740 cm-1 (spectrum 4c) because of the conversion of -OH groups to esters and the presence of carboxylic acid groups. Peaks due to formation of the succinimide ester (1817 and 1786 cm-1, spectrum 4d)41 appear upon activation of carboxylic acids with EDC/NHS, and these peaks disappear after reaction with NTA (spectrum 4e). A strong absorbance at 1680 cm-1 in spectrum 4e also suggests NTA immobilization and is probably due to the carboxylate groups from NTA as well as the amide bond that immobilizes the NTA.40
Figure 4.
KBr FTIR spectra of (a) SiO2-Fe3O4, (b) PHEMA-SiO2-Fe3O4, (c) SA-PHEMA-SiO2-Fe3O4, (d) NHS-SA-PHEMA-SiO2-Fe3O4, and (e) NTA-SA-PHEMA-SiO2-Fe3O4. The spectra are normalized with respect to the absorbance at 1099 cm-1, where most of the absorption should be due to silica.
Nanoparticle Magnetization
Brush-coated nanoparticles must be highly magnetic for collection with a magnet after protein binding. Figure S4 in the supporting information shows magnetization curves obtained with a vibrating sample magnetometer for Fe3O4 particles before and after different modification steps. As expected, the saturation magnetism per g of these nanoparticles decreases after silica coating (from 61.3 emu/g to 7.1 emu/g) and then decreases further to 2.5 emu/g after growth of polymer brushes. Derivatization with NTA and loading of Cu2+ reduces the magnetization to 0.94 emu/g because of the additional increase in mass of the particles. However, even after coating with Cu2+-NTA-SA-PHEMA, a rare earth NdFeB magnet (2” x 2” x 0.5”) can collect particles suspended in an Eppendorf tube in 5 min, which simplifies modification and washing steps. Optimization of the collection geometry should further decrease the time required for magnetic collection of the particles. (Centrifugation is not effective for collecting SiO2-Fe3O4 particles, but it is possible to accumulate modified particles using a bench-top centrifuge, presumably because of the larger size and increased tendency toward aggregation with the modified particles.)
Protein-binding capacities of magnetic beads modified with Cu2+-NTA-SA-PHEMA or Ni2+-NTA-SA-PHEMA
In our initial examination of protein binding to brush-modified MNPs, we mixed 0.45 mg of Cu2+-NTA-SA-PHEMA-modified nanoparticles in 50 μL of water with a 0.25-mL solution containing 0.3 mg of BSA in 20 mM pH 7.2 phosphate buffer. In this case, the protein binding likely occurs through interaction of BSA histidine residues with NTA-Cu2+ complexes. After a 30-min incubation with shaking, we isolated the beads with a magnet and determined the protein concentration in the supernatant with a Bradford assay. Based on a 33% decrease in the amount of protein in solution after mixing with the beads, the binding capacity is 220 ± 10 mg BSA per g of beads, which corresponds to approximately 1.8 × 10-16 g BSA per modified SiO2-Fe3O4 particle (a Cu2+-NTA-SA-PHEMA-modified particle has a mass of 8.2 × 10-16 g, see supporting information). This mass of BSA is the equivalent of ~8 monolayers on the bare SiO2-Fe3O4 nanoparticle, which has an average diameter of 43 nm, and the presence of the BSA alone would increase the diameter of the base SiO2-Fe3O4 particle to ~75 nm. The total particle size after protein adsorption is of course much larger than this because of the polymer. (See the supporting information for details of the calculation.) The mass of adsorbed BSA is about 25% of the mass of the Cu2+-NTA-SA-PHEMA brush on the beads. Consistent with BSA binding to NTA-Cu2+ complexes, a control experiment with NTA-SA-PHEMA-modified MNPs without Cu2+ showed no detectable binding of BSA.
Ni2+-NTA-SA-PHEMA-MNPs are especially attractive for purification of proteins tagged with polyhistidine. To measure the binding capacity of a His-tagged protein on Ni2+-NTA-SA-PHEMA-MNPs, we first mixed 25 μL of a solution containing 40 mg of modified beads/mL with 400 μL of 1 mg/mL HisU in 20 mM pH 7.2 phosphate buffer. After incubation for 30 min, washing with 20 mM pH 7.2 phosphate buffer containing 0.1% TWEEN-20 three times, and finally eluting with 250 μL elution buffer (20 mM pH 7.2 phosphate buffer with 0.5 M NaCl and 0.5 M imidazole), the concentration of HisU in the eluate was 0.98 mg/mL as determined by a Bradford assay. This amount of HisU in the eluate corresponds to a binding capacity of 245 mg HisU/g of beads, which is similar to the binding capacity for BSA on Cu2+-NTA-SA-PHEMA-MNPs.
Purification of His-tagged protein from cell lysate
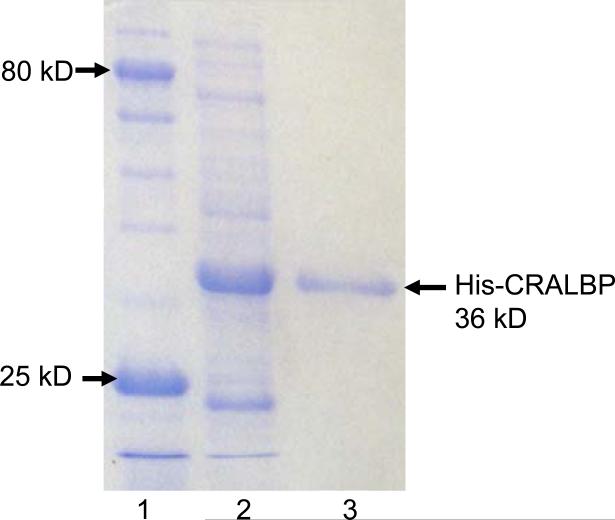
To demonstrate the utility of these beads for protein purification, we isolated over-expressed, His-tagged cellular retinaldehyde-binding protein (CRALBP) from a cell lysate obtained as described previously.27 The procedure simply involves mixing 100 μL of 20 mg/mL Ni2+-NTA-SA-PHEMA-modified beads with 400 μL of crude cell lysate, incubating for 30 min on a shaker table, collecting the beads with a magnet, washing them three times with 150 μL washing buffer (pH 7.2, 20 mM phosphate buffer containing 45 mM imidazole and 0.15 M NaCl), and eluting the protein by exposing the beads for 15 min (with shaking) to 40 μL of elution buffer (pH 7.2, 20 mM phosphate buffer containing 0.5 M imidazole and 0. 5 M NaCl). Figure 5 shows the gel electropherograms of both the cell lysate and the bead eluate and demonstrates that the Ni2+-NTA-SA-PHEMA nanoparticles are highly selective. Remarkably, the eluate electropherogram shows no impurity bands.
Figure 5.
SDS-PAGE analysis (Coomassie staining) of a cell lysate containing overexpressed His-tagged CRALBP before (lane 2) and after (lane 3) purification using Ni2+-NTA-SA-PHEMA-SiO2-Fe3O4 beads. Lane 1 shows a standard protein ladder.
One potential advantage of brush-modified nanoparticles over traditional porous microbeads is the elimination of mass-transport limitations due to diffusion within the bead. The short processing time is critical for successful purification of unstable proteins.8 To examine capture rates, we mixed 300 μL of an aqueous suspension containing 20 mg of Ni2+-NTA-modified beads per mL with 1.2 mL of ice-cold cell lysate and collected 350-μL aliquots of the mixture after 2, 5, 15, and 30 min. (The mixture was stirred with a vortex mixer for ~1 min at the beginning of the experiment, immediately before collection of an aliquot, and every 3-4 minutes during the incubation.) We centrifuged each aliquot, washed them four times, eluted the protein, and estimated protein concentration using a Bradford assay. (Collection of the modified beads via centrifugation facilitates kinetics experiments because it is more rapid (1 min) than magnetic collection, which takes ~5 min.) Importantly, the amount of eluted protein did not vary significantly with incubation times of 5 min, 15 min or 30 min, and beads reached 80-90% of their saturation capacities within 2 min of incubation (Figure S5). Thus, 5 min is sufficient time for the beads to bind protein. Gel electropherograms (Figure S6) confirm that the amount of protein binding does not vary greatly with incubation time. The fast capture presumably occurs because the swollen polymer brushes are only ~50 nm thick, so diffusion into the polymer is rapid. Typical procedures for purifications using agarose beads suggest either 30-min incubation times42-44 or only 2-5 min for incubation with beads of very limited capacity (1-5 mg protein/ mL beads).45, 46
To test the reusability of Ni2+-NTA-SA-PHEMA-modified beads, after elution of His-tagged CRALBP we extracted the Ni2+ from the beads with an ethylenediaminetetraacetic acid solution, reloaded the Ni2+, and again purified His-tagged CRALBP from a sample of cell lysate. Gel electropherograms (compare Figure 5 and Figure S7) are similar for the first and second purifications.
Qualitative assessment of protein recovery and comparison with commercial magnetic beads
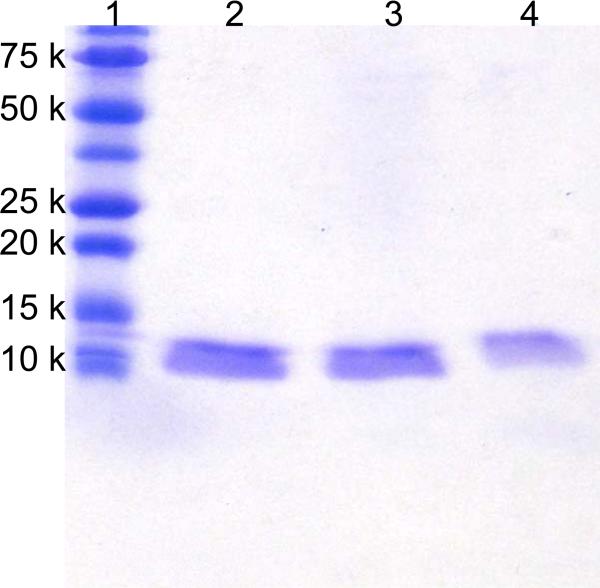
Along with selectivity, high recoveries are also vital in protein purification. Using gel electrophoresis, we qualitatively compared the recoveries of HisU in purifications with Ni2+-NTA-SA-PHEMA-MNPs and commercial Dynabeads® to determine if the high capacity of Ni2+-NTA-SA-PHEMA-MNPs leads to more efficient protein capture. Although Dynabeads® exhibit one of the highest protein-binding capacities among commercial magnetic beads (40 mg/g, see Table 1), their capacity is still <20% of that for Ni2+-NTA-SA-PHEMA-MNPs. In these experiments, we combined a 50 μL mixture containing 40 mg/mL beads (either Ni2+-NTA-SA-PHEMA-MNPs or commercial Dynabeads®) with 400 μL of 5 μg/mL HisU solution in 20 mM pH 7.2 phosphate buffer. After incubation for 30 min and washing with washing buffer, the bound protein was eluted with 15 μL elution buffer (see the protein-binding capacities section for buffer details) and analyzed qualitatively by SDS-PAGE by loading 2 μg of protein (assuming 100% recovery) in each lane. The Gel electropherogram in Figure 6 shows that the intensity of the band from HisU (10.7 kDa) enriched with Ni2+-NTA-SA-PHEMA-MNPs (lane 3) is similar to the intensity of the band resulting from directly loading of 2 μg of pure HisU (lane 2). In contrast the band of the eluate from commercially available beads (lane 4) is less intense, suggesting that the Ni2+-NTA-SA-PHEMA-MNPs have a higher recovery than the commercially available beads.
Table 1.
Properties of commercially available beads that bind His-tagged proteins.
| Beads | Company | Size | Incubation time (on ice) | Capacity | Matrix |
|---|---|---|---|---|---|
| PopCulture® His•Mag™ Purification Kit45 | Novagen | 3 μm | 5 min | 5 mg/mL | Agarose |
| MagneHis™ Ni-Particles46 | Promega | 2 min | 1 mg/mL | ||
| Ni-NTA Magnetic Agarose Beads42 | QIAGEN | 20–70 μm | 30 min | 0.25–1 mg/mL | Agarose |
| Dynabeads® TALON™47 | Dynal Biotech | 1.1 μm | 10 min | 40 mg/g | |
| HIS-Select® Nickel Magnetic Beads44 | Sigma | 20–75 μm | 30 min | ≥10 mg/mL | Agarose |
| μMACS His Isolation Kit43 | Miltenyi Biotec | 50 nm | 30 min | ||
| Ni2+-NTA-SA-PHEMA-Initiator-SiO2-Fe3O4 (this work) | N/A | ~100 nm | 5 min | 220-245 mg/mL | Polymer |
Figure 6.
SDS-PAGE analysis (Coomassie staining) of 2 μg HisU directly applied (lane 2) or enriched from 400 μL of a 5 μg/mL protein solution using 2 mg of Ni2+-NTA-SA-PHEMA-SiO2-Fe3O4 beads (lane 3) or 2 mg of Dynabeads® (lane 4). Lane 1 shows a standard protein ladder.
In this experiment, the amount of protein in the binding solution (2 μg) is considerably less than the fully loaded binding capacity of the beads (80 μg for the Dynabeads® and 490 μg for Ni2+-NTA-SA-PHEMA-MNPs). However, at such low concentrations (5 μg/mL), the equilibrium fraction of binding sites occupied by protein may be very low, and the equilibrium binding capacity of the Dynabeads® may still be significantly less than the amount of protein in the solution. At a similar or even slightly lower fractional filling of binding sites, the higher capacity of the Ni2+-NTA-SA-PHEMA-MNPs should allow binding of a larger fraction of the protein in solution. The advantages of high capacity may be particularly apparent at low protein concentrations.25
Table 1 summarizes some of the properties of commercially available magnetic beads used for isolation of His-tagged proteins. Most beads are micron sized and have binding capacities that are a factor of 5.5-200 lower than the 220-245 mg protein/g of brush-modified beads. In 2004, Xu and coworkers15 suggested that modified particles with diameters of ~2 nm bind His-tagged green fluorescent protein with a capacity of 2-3 g per g of beads, but as mentioned in the introduction such particles may be difficult to collect. Very recently, Xie et al. reported Ni2+-NTA-Au/Fe3O4 particles with capacity of 2 mg protein/mg beads, however, there was some non specific protein binding to the base Au/Fe3O4 particles.25 Such particles with high capacity may also prove useful in the collection of proteins at low concentrations.
Conclusions
Growth of polymer brushes on silica-coated Fe3O4 yields stable MNPs that bind an order of magnitude more protein than typical commercial magnetic microparticles. These particles can rapidly isolate multilayers of His-tagged protein directly from a cell extract with high purity, and the ~50 nm thickness of the polymer brushes reduces diffusion limitations to allow protein capture in as little as 5 min. Moreover, the brush-modified particles afford high protein recoveries and can be reused if desired.
Supplementary Material
Acknowledgement
We are grateful to the US National Institutes of Health (GM080511) and the US National Science Foundation (CHE-0616795) for supporting this research. We also thank Dr. Jinhua Dai for helpful ideas and discussions and Prof. Volodymyr Tarabara for use of the light-scattering instrument.
Footnotes
Supporting Information Available.
TEM images of initiator-modified SiO2-Fe3O4 particles prepared with and without nitrogen sparging; size distribution of particles determined by TEM and DLS; calculations for estimation of the extent of reaction of PHEMA with SA and the PHEMA thickness; calculation of the mass of fully modified beads and of the protein in a BSA monolayer on a SiO2-Fe3O4 particle; magnetization curves of the modified nanoparticles; SDS-PAGE and Bradford assay analysis of the His-CRALBP eluted from modified beads after incubation of the beads in a cell lysate for various times, and an electropherogram of CRALBP purified using beads with reloaded Ni2+. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Steen J, Uhlen M, Hober S, Ottosson J. Protein Expression Purif. 2006;46:173–178. doi: 10.1016/j.pep.2005.12.010. [DOI] [PubMed] [Google Scholar]
- 2.Roque ACA, Silva CSO, Taipa MA. J. Chromatogr., A. 2007;1160:44–55. doi: 10.1016/j.chroma.2007.05.109. [DOI] [PubMed] [Google Scholar]
- 3.McCarthy P, Chattopadhyay M, Millhauser GL, Tsarevsky NV, Bombalski L, Matyjaszewski K, Shimmin D, Avdalovic N, Pohl C. Anal. Biochem. 2007;366:1–8. doi: 10.1016/j.ab.2007.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gutierrez R, Martin del Valle EM, Galan MA. Sep. Purif. Rev. 2007;36:71–111. [Google Scholar]
- 5.Kaur-Atwal G, Weston DJ, Bonner PLR, Crosland S, Green PS, Creaser CS. Curr. Anal. Chem. 2008;4:127–135. [Google Scholar]
- 6.Gautrot JE, Huck WTS, Welch M, Ramstedt M. ACS Appl. Mater. Interfaces. 2009;2:193–202. doi: 10.1021/am9006484. [DOI] [PubMed] [Google Scholar]
- 7.Ghosh R. J. Chromatogr., A. 2002;952:13–27. doi: 10.1016/s0021-9673(02)00057-2. [DOI] [PubMed] [Google Scholar]
- 8.Kaufmann M. J. Chromatogr., B: Anal. Technol. Biomed. Life Sci. 1997;699:347–369. doi: 10.1016/s0378-4347(96)00512-9. [DOI] [PubMed] [Google Scholar]
- 9.Safarik I, Safarikova M. J. Chromatogr., B: Anal. Technol. Biomed. Life Sci. 1999;722:33–53. [PubMed] [Google Scholar]
- 10.Ficarro SB, Adelmant G, Tomar MN, Zhang Y, Cheng VJ, Marto JA. Anal. Chem. 2009;81:4566–4575. doi: 10.1021/ac9004452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Meng J, Walter JG, Kokpinar O, Stahl F, Scheper T. Chem. Eng. Technol. 2008;31:463–468. [Google Scholar]
- 12.Zhang L, Chen L, Wan Q-H. Chem. Mater. 2008;20:3345–3353. [Google Scholar]
- 13.Di Corato R, Piacenza P, Musaro M, Buonsanti R, Cozzoli PD, Zambianchi M, Barbarella G, Cingolani R, Manna L, Pellegrino T. Macromol. Biosci. 2009;9:952–958. doi: 10.1002/mabi.200900154. [DOI] [PubMed] [Google Scholar]
- 14.Felinto MCFC, Parra DF, Lugao AB, Batista MP, Higa OZ, Yamaura M, Camilo RL, Ribela MTC,P, Sampaio LC. Nucl. Instrum. Methods Phys. Res., Sect. B. 2005;236:495–500. [Google Scholar]
- 15.Li Y, Xu X, Yan B, Deng C, Yu W, Yang P, Zhang X. J. Proteome Res. 2007;6:2367–2375. doi: 10.1021/pr060558r. [DOI] [PubMed] [Google Scholar]
- 16.Ma Z, Guan Y, Liu H. J. Magn. Magn. Mater. 2006;301:469–477. [Google Scholar]
- 17.Xu C, Xu K, Gu H, Zheng R, Liu H, Zhang X, Guo Z, Xu B. J. Am. Chem. Soc. 2004;126:9938–9939. doi: 10.1021/ja0464802. [DOI] [PubMed] [Google Scholar]
- 18.Shieh D-B, Su C-H, Chang F-Y, Wu Y-N, Su W-C, Hwu JR, Chen J-H, Yeh C-S. Nanotechnology. 2006:4174. doi: 10.1088/0957-4484/17/16/030. [DOI] [PubMed] [Google Scholar]
- 19.Li Y-C, Lin Y-S, Tsai P-J, Chen C-T, Chen W-Y, Chen Y-C. Anal. Chem. 2007;79:7519–7525. doi: 10.1021/ac0711440. [DOI] [PubMed] [Google Scholar]
- 20.Liao Y, Cheng Y, Li Q. J. Chromatogr. A. 2007;1143:65–71. doi: 10.1016/j.chroma.2006.12.063. [DOI] [PubMed] [Google Scholar]
- 21.Tural B, Kaya M, Ozkan N, Volkan M. J. Nanosci. Nanotechnol. 2008;8:695–701. doi: 10.1166/jnn.2008.b270. [DOI] [PubMed] [Google Scholar]
- 22.Xu C, Xu K, Gu H, Zhong X, Guo Z, Zheng R, Zhang X, Xu B. J. Am. Chem. Soc. 2004;126:3392–3393. doi: 10.1021/ja031776d. [DOI] [PubMed] [Google Scholar]
- 23.Kim J, Piao Y, Lee N, Park YI, Lee I-H, Lee J-H, Paik SR, Hyeon T. Adv. Mater. 2010;22:57–60. doi: 10.1002/adma.200901858. [DOI] [PubMed] [Google Scholar]
- 24.Feng GD, Hu DD, Yang L, Cui Y, Cui XA, Li H. Sep. Purif. Technol. 2010;74:253–260. [Google Scholar]
- 25.Xie H-Y, Zhen R, Wang B, Feng Y-J, Chen P, Hao J. J. Phys. Chem. C. 2010;114:4825–4830. [Google Scholar]
- 26.Singh N, Wang J, Ulbricht M, Wickramasinghe SR, Husson SM. J. Membr. Sci. 2008;309:64–72. [Google Scholar]
- 27.Jain P, Vyas MK, Geiger JH, Baker GL, Bruening ML. Biomacromolecules. 2010;11:1019–1026. doi: 10.1021/bm9014792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang X, Wirth MJ. Anal. Chem. 1997;69:4577–4580. [Google Scholar]
- 29.Lattuada M, Hatton TA. Langmuir. 2007;23:2158–2168. doi: 10.1021/la062092x. [DOI] [PubMed] [Google Scholar]
- 30.Seino M, Yokomachi K, Hayakawa T, Kikuchi R, Kakimoto M, Horiuchi S. Polymer. 2006;47:1946–1952. [Google Scholar]
- 31.Bell NS, Piech M. Langmuir. 2006;22:1420–1427. doi: 10.1021/la0516375. [DOI] [PubMed] [Google Scholar]
- 32.Tsyalkovsky V, Klep V, Ramaratnam K, Lupitskyy R, Minko S, Luzinov I. Chem. Mater. 2007;20:317–325. [Google Scholar]
- 33.Chen X, Armes SP. Adv. Mater. 2003;15:1558–1562. [Google Scholar]
- 34.Perruchot C, Khan MA, Kamitsi A, Armes SP, von Werne T, Patten TE. Langmuir. 2001;17:4479–4481. [Google Scholar]
- 35.Huang JS, Han BZ, Yue W, Yan H. J. Mater. Chem. 2007;17:3812–3818. [Google Scholar]
- 36.Wang W, Xu Y, Wang DIC, Li Z. J. Am. Chem. Soc. 2009;131:12892–12893. doi: 10.1021/ja905477j. [DOI] [PubMed] [Google Scholar]
- 37.Deng Y, Wang C, Hu J, Yang W, Fu S. Colloids Surf., A. 2005;262:87–93. [Google Scholar]
- 38.Lu Y, Yin Y, Mayers BT, Xia Y. Nano Lett. 2002;2:183–186. [Google Scholar]
- 39.Matyjaszewski K, Miller PJ, Shukla N, Immaraporn B, Gelman A, Luokala BB, Siclovan TM, Kickelbick G, Vallant T, Hoffmann H, Pakula T. Macromolecules. 1999;32:8716–8724. [Google Scholar]
- 40.Dotzauer DM, Dai J, Sun L, Bruening ML. Nano Lett. 2006;6:2268–2272. doi: 10.1021/nl061700q. [DOI] [PubMed] [Google Scholar]
- 41.Lahiri J, Isaacs L, Tien J, Whitesides GM. Anal. Chem. 1999;71:777–790. doi: 10.1021/ac980959t. [DOI] [PubMed] [Google Scholar]
- 42. [02/16/2010]. http://www1.qiagen.com/literature/render.aspx?id=2047.
- 43. [02/16/2010]. http://www.miltenyibiotec.com/download/flyer_en/735/Protein_isolation_flyer.pdf.
- 44. [02/16/2010]. http://www.sigmaaldrich.com/etc/medialib/docs/Sigma/Bulletin/1/h9914bul.Par.0001.File.tmp/h9914bul.pdf.
- 45. [02/16/2010]. http://www.merck-chemicals.com/chemdat/en_US/Merck-International-Site/USD/ViewProductDocuments-File?ProductSKU=EMD_BIO-71002&DocumentType=USP&DocumentId=%2Femd%2Fbiosciences%2Fuserprotocols%2Fen-US%2FTB054.pdf&DocumentSource=GDS.
- 46. [02/16/2010]. http://www.promega.com/tbs/tm060/tm060.pdf.
- 47. [02/16/2010]. http://tools.invitrogen.com/content/sfs/manuals/101%2003D_04D_05D_Rev000.pdf.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.