Abstract
Plants have several L-ascorbic acid (AsA) biosynthetic pathways, but the contribution of each one to the synthesis of AsA varyies between different species, organs, and developmental stages. Strawberry (Fragaria×ananassa) fruits are rich in AsA. The pathway that uses D-galacturonate as the initial substrate is functional in ripe fruits, but the contribution of other pathways to AsA biosynthesis has not been studied. The transcription of genes encoding biosynthetic enzymes such as D-galacturonate reductase (FaGalUR) and myo-inositol oxygenase (FaMIOX), and the AsA recycling enzyme monodehydroascorbate reductase (FaMDHAR) were positively correlated with the increase in AsA during fruit ripening. Fruit storage for 72 h in a cold room reduced the AsA content by 30%. Under an ozone atmosphere, this reduction was 15%. Ozone treatment increased the expression of the FaGalUR, FaMIOX, and L-galactose-1-phosphate phosphatase (FaGIPP) genes, and transcription of the L-galactono-1,4-lactone dehydrogenase (FaGLDH) and FAMDHAR genes was higher in the ozone-stored than in the air-stored fruits. Analysis of AsA content in a segregating population from two strawberry cultivars showed high variability, which did not correlate with the transcription of any of the genes studied. Study of GalUR protein in diverse cultivars of strawberry and different Fragaria species showed that a correlation between GalUR and AsA content was apparent in most cases, but it was not general. Three alleles were identified in strawberry, but any sequence effect on the AsA variability was eliminated by analysis of the allele-specific expression. Taken together, these results indicate that FaGalUR shares the control of AsA levels with other enzymes and regulatory elements in strawberry fruit.
Keywords: L-Ascorbic acid, D-galacturonic acid reductase, fruit ripening, gene expression, strawberry, oxidative stress
Introduction
L-Ascorbic acid (AsA) is the common name for the six-carbon sugar derivative L-threo-hex-2-enono-1,4-lactone. Humans, non-human primates, and a few other mammals cannot synthesize AsA because the gene encoding the enzyme catalysing the last step in the biosynthesis, L-gulono-1,4-lactone oxidase, is non-functional due to mutations (Chatterjee, 1973) (Fig. 1). This compound has important antioxidant and metabolic functions, making its incorporation into the human diet essential and is known as vitamin C. Plants, the main source of vitamin C for humans, also require this compound (Pastori et al., 2003). Strawberry fruits are particularly rich sources of vitamin C (Perkins-Veazie, 1995).
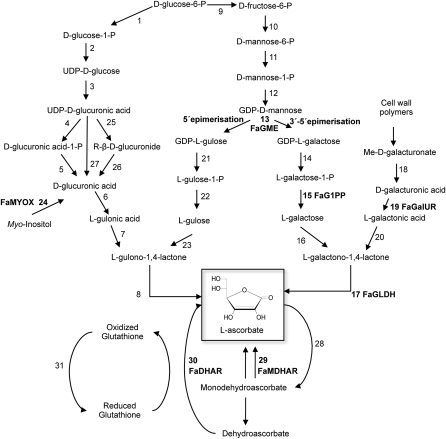
Fig. 1.
Proposed biosynthetic pathways of L-ascorbic acid in animals (reactions 1–8 and 25–27), plants (reactions 9–24) and the recycling pathway (reactions 28–31) are shown. The enzymes used in this work are in bold. The enzymes catalysing these reactions are the following: 1, phosphoglucomutase; 2, UDP-D-glucose pyrophosphorylase; 3, UDP-D-glucose dehydrogenase; 4, D-glucuronate-1-phosphate uridylyltransferase; 5, D-glucurono kinase; 6, D-glucuronate reductase; 7, aldono-lactonase; 8, L-gulono-1,4-lactone dehydrogenase; 9, D-glucose-6-phosphate isomerase; 10, D-mannose-6-phosphate isomerase; 11, phosphomannomutase; 12, GDP-mannose pyrophosphorylase (mannose-1-phosphate guanyltransferase); 13, GDP-mannose-3',5'-epimerase; 14, GDP-L-galactose phosphorylase; 15, L-galactose-1-phosphate phosphatase; 16, L-galactose dehydrogenase; 17, L-galactono-1,4-lactone dehydrogenase; 18, methylesterase; 19, D-galacturonate reductase; 20, aldono-lactonase; 21, phosphodiesterase; 22, sugar phosphatase; 23, L-gulose dehydrogenase; 24, myo-inositol oxygenase; 25, UDP-glucuronosyltransferase; 26, β-glucuronidase; 27, UDP-glucuronidase; 28, ascorbate peroxidase; 29, monodehydroascorbate reductase; 30, dehydroascorbate reductase; 31, glutathione reductase.
There are several known and proposed alternative pathways for AsA biosynthesis in plants (Fig. 1), but the prevalence of these pathways in different tissues and/or developmental stages is largely unknown (Davey et al., 2000; Jain and Nessler, 2000; Valpuesta and Botella, 2004). The first AsA biosynthetic pathway to be described in plants proceeds through GDP-D-mannose and L-galactose (Wheeler et al., 1998). In this pathway, D-glucose is converted to AsA through the following intermediate compounds: GDP-D-mannose, GDP- L-galactose, L-galactose-1-phosphate, L-galactose, and L-galactono-1,4-lactone (Fig. 1). All the genes involved in this pathway (Man/Gal pathway) have been characterized in plants, including those encoding GDP-D-mannose pyrophosphorylase (Conklin et al., 1999), GDP-D-mannose-3‘,5’-epimerase (Gibson et al., 2008; Wolucka and Van Montagu, 2003), GDP-L-galactose phosphorylase (Dowdle et al., 2007; Laing et al., 2007), L-galactose-1-phosphate phosphatase (Laing et al., 2004), L-galactose dehydrogenase (Gatzek et al., 2002), and L-galactono-1,4-lactone dehydrogenase (Imai et al., 1998; do Nascimento et al., 2005). Biochemical studies indicated that GDP-L-gulose was an alternative product of the enzyme GDP-D-mannose-3',5'-epimerase (Wolucka and Van Montagu, 2003). This reaction product established both a new branch of the pathway in plants and a link to the animal pathway (Fig. 1). The subsequent steps in the branch, however, have not yet been described in plants. In addition, a pathway initiating from D-galacturonate was proposed after the identification of a D-galacturonate reductase gene in strawberry (Agius et al., 2003) (Fig. 1). Finally, a myo-inositol-based pathway was proposed after the cloning of myo-inositol oxygenase from Arabidopsis (Lorence et al., 2004) (Fig. 1).
In plants, the AsA recycling pathway (Fig. 1) plays an important role in the response and adaptation to the stress (Stevens et al., 2008) and also contributes to the regulation of AsA levels jointly with the processes of synthesis, degradation, and transportation (Yin et al., 2010). Ascorbate peroxidase (APX) uses two AsA molecules to reduce H2O2 to water, with two molecules of monodehydroascorbate (MDHA) generated as by-products. MDHA is reduced back to AsA by monodehydroascorbate reductase (MDHAR) using NADH/NADPH as electron donors. In addition, because MDHA is an unstable radical, it can spontaneously disproportionate to AsA and dehydroascorbate (DHA). DHA is reduced to AsA by dehydroascorbate reductase (DHAR) using reduced glutathione (GSH) as a reducing compound. Therefore, DHAR and MDHAR are key components in the maintenance of the reduced pool of AsA and are of paramount importance in oxidative stress tolerance (Eltayeb et al., 2006)
In plants, AsA content has been shown to change with light (Tabata et al., 2002; Badejo et al., 2009; Li et al., 2009), hour of the day (Chen and Gallie, 2004; Tamaoki et al., 2003), age (Bartoli et al., 2000), plant tissue (Lorence et al., 2004), and cell compartment (Davey et al., 2000; Zechmann et al., 2011). A tightly regulated metabolic network of AsA biosynthesis, catabolism, and recycling might explain this fine modulation. Surprisingly, there have only been a few studies examining the various elements involved in this metabolic network (Linster and Clarke, 2008; Badejo et al., 2009; Cruz-Rus et al., 2010).
In strawberry, the nucleotide sequences for some of the genes encoding enzymes of the different AsA biosynthetic and recycling pathways are known. The AsA content of these fruits is known to vary and has also been reported to increase in ripe strawberry fruits after treatment with ozone (Pérez et al., 1999). For example, there is a continuous increase in the content of AsA during fruit development and ripening (Agius et al., 2003). Because of these observations, strawberry fruits were selected to study the correlation between the expression levels of the genes involved in the biosynthesis of AsA and the levels of AsA. In addition, the variability of AsA content in the ripe fruits of different cultivars of Fragaria×ananassa and species of Fragaria was evaluated to explore whether a relationship exists between AsA content and the expression and/or structure of D-galacturonate reductase.
Material and methods
Plant materials
Fragaria species and F.×ananassa varieties were sampled from the Fragaria Germplasm Collection (IFAPA, Churriana, Málaga, Spain). All plants were grown in a shade house under the typical prevailing field conditions of the area. In all cases, fruits were carefully selected at the same stage of ripeness.
The strawberry (F.×ananassa) mapping population (Y Zorrilla-Fontanesi et al., unpublished results) consisted of a full-sib family of 95 F1 progeny derived from an intraspecific cross between two IFAPA selection lines, 232 (‘Vilanova’×Sel. 4-43) and 1392 (‘Gaviota’בCamarosa’). These lines were chosen because they differ in important agronomic and fruit quality traits including production, time to flowering, fruit colour, acidity, and the content of sugars, anthocyanins, volatile compounds, and AsA.
The strawberry population was planted at the end of October in the strawberry-producing area of Moguer (Huelva, Spain). A total of six plants of each parental line and four plants of each genotype of the progeny were established on mulched raised beds that were 35 cm high and 50 cm wide in a double row in a zigzag formation. The population was grown under macro tunnels of polyethylene following conventional practices, with an inter-row distance of 30 cm and a distance of 25 cm between plants. Fruits for AsA determination were harvested at the same ripening stage for the whole population and immediately frozen in liquid nitrogen.
Ascorbic acid determination
The ascorbic acid content of strawberry fruits was determined by HPLC. Briefly, the fruits were harvested and pulverized in liquid nitrogen. Samples were suspended in 2.0% (w/v) metaphosphoric acid and 2 mM EDTA, chilled on ice for 20 min, and centrifuged at 13 000 g for 20 min at 4 °C, which corresponds to the protocol previously published (Davey et al., 1996) with minor modifications. The supernatants were filtered through a 0.45 μm syringe filter, and 15 μl of the sample was injected into the HPLC system. The flow rate was 0.6 ml min−1 with a mobile phase consisting of 0.1 M sodium phosphate monobasic, 0.2 mM EDTA (adjusted to pH 3.1 with orthophosphoric acid), and a Kromasil 100 C18 5 μm 25×0.46 column (Scharlab www.scharlab.com). The eluents were monitored with a UV detector at 254 nm (Harapanhalli et al., 1993). Three biological repetitions, with three technical replicas for each, were performed for every sample. The AsA content was calculated by comparison with the values obtained from a standard curve.
Ozone treatment
Strawberry (F.×ananassa Duch., cv. Camarosa) plants were grown in the south of Spain (Moguer, Huelva) under the same conditions previously described for the mapping population. Fruits were harvested, selected for uniformity of size and colour, and packed in polypropylene berry baskets (500 g each). Ozone was produced using an ozone generator (Laboratory Ozonizer 300.7) to a level of 35 ppm and applied to one set of fruit. Both sets, the treated and untreated fruit, were placed in the same cold room at 2–4 °C and 90% relative humidity. After 3 d in the cold room, the fruits were frozen at –80 °C until used for analysis.
RNA isolation and gene expression analysis
Total RNA was isolated from the Fragaria fruits according to the method described by Manning (1991). RT-PCR of the different genes was performed starting from 1 μg of total RNA using the iScript Select cDNA Synthesis Kit (Bio-Rad, http://www.bio-rad.com/). Expression was analysed by quantitative RT-PCR (qRT-PCR) using a SYBR® Premix Ex Taq™ sample (Takara, http://www.takara-bio.com/), a Rotor Gene detection system (Qiagen, http://www.qiagen.com) according to the manufacturer's instructions, and gene-specific primers (see Supplementary Table S1 at JXB online). At least two biological and three technical replicates were performed for every sample. The absence of primer dimers was confirmed by an examination of the dissociation curves. The results obtained were normalized against FaRib413 expression, which had been shown by Northern blot analysis to be constitutive in different strawberry plant tissues, including fruits at different stages of development (Casado-Díaz et al., 2006). The relative quantification of expression level was performed using the comparative Ct method (Pfaffl, 2001). Statistical analysis was performed using the StatGraphic Centurion program. Significant differences were determined by ANOVA. Differential allelic expression was determined using a standard PCR with specific primers for the open reading frame of FaGalUR. To differentiate between the distinct alleles, the PCR product was subsequently digested using the restriction enzymes MbiI and Eco52I (Fermentas, http://www.fermentas.com) according to the manufacturer's instructions, followed by the quantification of the bands using the Quantity One software (Bio-Rad, http://www.bio-rad.com).
Protein extraction and immunoblot analysis
The fruits were first pulverized in liquid nitrogen. Proteins were then extracted in 6× Laemmli buffer (Laemmli, 1970) and centrifuged for 5 min at 10 000 g. Protein separation and blotting were performed on 12% polyacrylamide gels and nitrocellulose membranes, respectively, as previously described by Cruz-Rus et al. (2010). The gels were stained with Coomassie blue to confirm that there was equal loading in all lanes. After being blocked in 5% milk, the membranes were incubated with polyclonal anti-FaGalUR antibodies (Agius et al., 2003) followed by anti-rabbit antibodies conjugated to horseradish peroxidase. The immunoblots were developed using the ECL detection kit from Amersham (Amersham Biosciences, http://www.gelifesciences.com/).
Results
The sequences of the strawberry genes encoding L-galactone-1,4-lactone dehydrogenase (FaGLDH) (do Nascimento et al., 2005) and L-galacturonic acid reductase (FaGalUR) (Agius et al., 2003) have previously been published. A tBLASTn search of the strawberry EST database (Bombarely et al., 2010) (http://fresa.uco.uma.es/srs71/), using the amino acid sequences previously described for enzymes of the AsA biosynthesis metabolic pathways in plants as queries, identified ESTs for GDP-mannose-3',5'-epimerase (FaGME), L-galactose-1-phosphate phosphatase (FaG1PP), and myo-inositol oxygenase (FaMYOX).
The sequence identity between the strawberry genes and the homologues found in other species was high (Table 1). Strawberry FaGME showed an 88% identity with its corresponding Arabidopsis enzyme, with similar levels of identity with proteins from other members of the Rosaceae family, such as Prunus persica (89%) and Malus domestica (85%). Strawberry FaG1PP had identities of 63% and 90% with its corresponding enzymes from Arabidopsis and Prunus persica, respectively. FaMYOX had a 56% identity with Arabidopsis MYOX4. As expected, the identity of each of the enzymes was highest when compared with the corresponding enzymes of other Rosaceae species.
Table 1.
Strawberry sequences corresponding to genes encoding the enzymes of the L-ascorbic acid biosynthetic and recycling pathwaysThe identity at the amino acid level between each of the enzymes and those of the reference species are shown.
| Accession number | Enzyme (EC) | Reference species | Identity (%) | |
| GW402740 | GDP-mannose-3',5'epimerase | Arabidopsis thaliana | 88 | |
| (EC 5.1.3.18) | (At5g28840) | |||
| GT150979 | L-Galactose-1-phosphate-phosphatase | Arabidopsis thaliana | 63 | |
| (EC 3.1.3 25) | (At3g02870) | |||
| AY102631 | L-Galactono-1,4-lactone dehydrogenase | Fragaria×ananassa | 100 | |
| (EC 1.3.2.3) | (cv. Campineiro) | |||
| AF039182 | D-Galacturonate reductase | Fragaria ×ananassa | 100 | |
| (EC 1.1.1.203) | (cv. Chandler) | |||
| GW402590 | myo-Inositol oxygenase | Arabidopsis thaliana | 56 | |
| (EC 1.13.99.1) | (At4g26260) | |||
| Cl248Cg1 | Monodehydroascorbate reductase | Arabidopsis thaliana | 76 | |
| (EC 1.6.5.4) | (At3g52880) | |||
| CO817488 | Dehydroascorbate reductase | Arabidopsis thaliana | 64 | |
| (EC 1.8.5.1) | (At1g19570) |
In addition, the sequences of monodehydroascorbate reductase (FaMDHAR) and a chloroplastic dehydroascorbate reductase (FaDHAR), two genes involved in AsA recycling, were identified in the strawberry EST database (Bombarely et al., 2010). FaMDHAR showed identities of 76% and 90% with the corresponding enzymes of Arabidopsis and Malus domestica, respectively. FaDHAR had identities of 64% and 81% with the corresponding enzymes from Arabidopsis and Malus domestica.
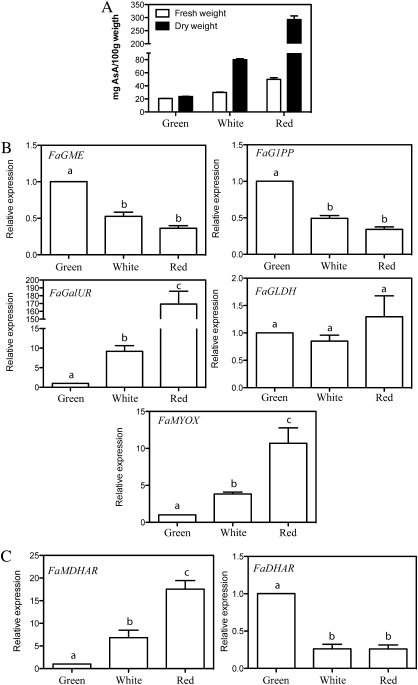
The content of AsA in the fruits of the strawberry cv. Camarosa increased over the course of development from the green immature stage to that of red ripe fruits, reaching a final concentration of 50 mg/100 g fresh weight (Fig. 2A). This level lies within the range of values previously reported for ripe fruits of other varieties of this species (Agius et al., 2003). Notably, the increase in AsA levels with ripening is much higher on a dry weight basis. Thus, the AsA content rose from 23.9 mg/100 g dry weight in green fruits to 292.5 mg/100 g dry weight in ripe fruits, a greater than 12.2-fold increase.
Fig. 2.
(A) Changes in AsA levels and (B, C) mRNA levels of genes involved in AsA biosynthesis (FaGME, FaG1PP, FaGalUR, FaGLDH, and FaMYOX) and recycling (FaMDHAR and FaDHAR) in fruits during three ripening stages. The values were normalized against the abundance of FaRib413, using green fruit value as the reference (value 1). The bars represent the mean of at least two independent biological samples ±SD. Different letters indicate a significant difference between samples according to the corresponding ANOVA (P <0.05).
In the same samples, the relative expression levels of the genes encoding the enzymes of the AsA biosynthetic pathways were evaluated by qRT-PCR (Fig. 2B). The expression pattern of both FaGalUR and FaMYOX was positively correlated with alterations in the levels of AsA. The increase associated with fruit development was higher for FaGalUR, changing more than 150-fold from the green to the red stage. By contrast, the expression of FaG1PP and FaGME was negatively associated with AsA levels, with the highest values in green fruits and the lowest in red fruit. The net changes were below 3-fold. With respect to the expression of FaGLDH, no significant change was observed among the three developmental stages.
The regulation of AsA levels in cells is tightly controlled, not only by its synthesis, but also by the recycling, degradation, and transport processes (Loewus, 1999; Pallanca and Smirnoff, 1999; Horemans et al., 2000). An examination of the expression patterns of the genes encoding monodehydroascorbate reductase (MDHAR) and dehydroascorbate reductase (DHAR) revealed contrasting associations. The FaMDHAR levels increased over 17-fold in the transition from green to ripe fruit (Fig. 2C), paralleling the increase in AsA levels. The levels of FaDHAR, however, were highest in green fruits, with decreasing levels in white and red fruits (Fig. 2C).
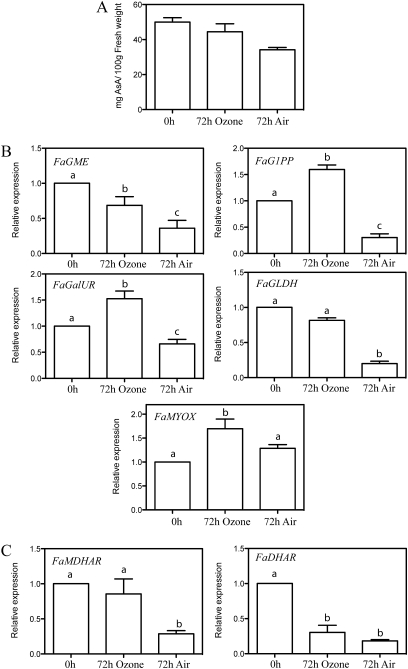
Strawberry is one of the most delicate and perishable fruits, being highly susceptible to mechanical injury, physiological deterioration, water loss, and fungal decay. Therefore, effective post-harvesting procedures are required to prevent deterioration during strawberry packing and transport. Installation of ozone generators in strawberry cooling rooms has been effectively used to increase the AsA content of the fruits (Pérez et al., 1999). The ozone treatment was successfully reproduced here, and the effectiveness of the ozone treatment was confirmed by the appearance of the fruits, which, when stored in air, were eventually naturally infected with fungi (Rhizopus sp.) while the ozone-treated fruits were not (results not shown). The AsA content was lower after 72 h in air than just after fruit detachment (0 h), but this reduction was significantly smaller in the ozone-treated fruits (Fig. 3A). Differences in expression between the air- and ozone-stored fruits were significant for each of the biosynthetic genes examined (Fig. 3B), ranging from 1.2-fold for FaMIOX to 5-fold for FaG1PP. With respect to the AsA recycling genes, the expression of FaMDHAR was 3-fold higher in fruits stored for 72 h in ozone compared with those stored in air, whereas this difference was not significant for the FaDHAR gene (Fig. 3C). In order to determine whether fungal infection influences plant responses, the same samples were analysed for the expression of PR5 and polygalacturonase-inhibiting protein (PGIP) using semi-quantitative RT-PCR, genes that have been reported to be induced by infection in strawberry fruits (Mehli et al., 2004; Osorio et al., 2008). There were no significant differences in the expression of these genes between fruits stored either in ozone or air (see Supplementary Fig. S1 at JXB online) suggesting that the changes in expression of AsA biosynthetic and recycling genes previously reported was mainly an effect caused by the ozone rather than a consequence of the infection.
Fig. 3.
(A) Changes in AsA levels and (B, C) mRNA levels of the genes involved in AsA biosynthesis (FaGME, FaG1PP, FaGalUR, FaGLDH, and FaMYOX) and recycling (FaMDHAR and FaDHAR) in fruits before (0 h) and after storage for 72 h in ozone or air atmosphere. The values were normalized against the abundance of FaRib413, using the 0 h value as the reference (value 1). The bars represent the mean of at least two independent biological samples ±SD. Different letters indicate a significant difference between samples according to the corresponding ANOVA (P <0.05).
The search for a possible relationship between gene expression and AsA was extended to a F.×ananassa segregating population, which provided highly variable levels of AsA in a limited genetic background. This population consisted of 95 lines derived from a cross between selections 1392 (fruits with high AsA content) and 232 (fruits with a low–medium AsA content). The AsA content was measured in the red fruits of all lines, and the mean values of fruit AsA were normally distributed and varied from 20–58 mg/100 g FW (see Supplementary Fig. S2 at JXB online). In addition to the parental lines, two progeny individuals with high AsA levels (93-30 and 93-34), one with a medium AsA level (93-29) and two with low AsA levels (93-67 and 93-72) (Fig. 4A), were selected for the analysis of the expression of genes encoding enzymes of the AsA biosynthetic pathways (Fig. 4B). No correlation was found between the expression of selected genes, including FaGalUR, and the levels of AsA. With respect to the genes involved in AsA recycling, the expression of FaMDHAR was generally higher in the high AsA content lines, although the differences were not significant (Fig. 4C).
Fig. 4.
(A) Changes in AsA levels and (B, C) mRNA levels of the genes involved in AsA biosynthesis (FaGME, FaG1PP, FaGalUR, FaGLDH, and FaMYOX) and recycling (FaMDHAR and FaDHAR) by QRT-PCR. (D) FaGalUR protein levels in the ripe fruits of the parentals (1392 and 232) as well as lines with high (93-30 and 93-34), medium (93-29), and low (93-67 and 93-72) AsA levels in the segregating population. The QRT-PCR values were normalized against the abundance of FaRib413, using the parental 1392 value as the reference (value 1). The bars represent the mean of at least two independent biological samples ±SD. Different letters indicate a significant difference between samples according to the corresponding ANOVA (P <0.05).
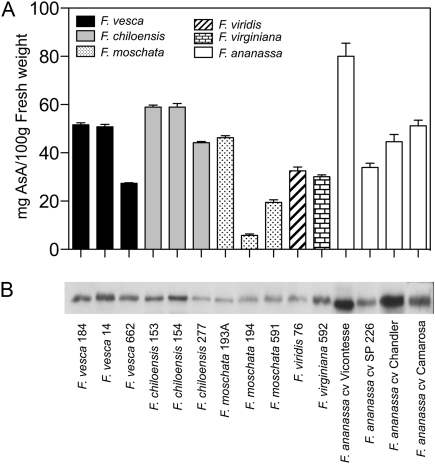
Transcription studies can be of limited value because discrepancies between transcripts and encoded proteins can occur. Therefore, the amounts of FaGalUR protein and AsA were analysed to determine if there was a correlation between the two. There were no significant differences in the amount of FaGalUR protein among the analysed individuals of the 232×1392 population (Fig. 4D) despite the different levels of AsA. This study was then extended to other genotypes belonging to different species of the Fragaria genus, which should provide a wider range of variability in the levels of AsA than in the full-sib population. Moreover, it had previously been found that a Fragaria moschata genotype with lower levels of AsA than F.×ananassa also had lower levels of the GalUR protein (Agius et al., 2003). The selected samples included three accessions of the wild diploid species Fragaria vesca, one accession of the diploid Fragaria viridis, three accessions of the hexaploid Fragaria moschata, three accessions of the octoploid Fragaria chiloensis, one accession of octoploid Fragaria virginiana, and four cultivars of octoploid F.×ananassa. As expected, the AsA content in the ripe fruits of these species was highly variable, not only between species, but also within them (Fig. 5A). The lowest values were found for F. moschata (nos 194, 591) and the highest for F.×ananassa cv. Vicontesse. Gene expression was evaluated at the protein level by Western blot analysis in the same fruit samples. Although there were some cases for which the levels of AsA and the GalUR protein were parallel, a tight correlation between these two parameters was not always found (Fig. 5B).
Fig. 5.
(A) Changes in AsA levels and (B) GaLUR levels by Western blot in the red fruits of different species from the genus Fragaria and different varieties of F ×ananassa.
A striking discrepancy between the levels of AsA and the GalUR protein was found between the two F.×ananassa cultivars, Camarosa and Chandler. Therefore, the possibility of differences in the primary structure of the enzyme was explored by the sequence analysis of 12 genomic clones of FaGalUR from each of these two cultivars. The genomic structure of this gene included three introns and four exons (Fig. 6A). A SSR in the first intron that was polymorphic in the two cultivars was identified (Fig. 6B), and it had been previously reported to also be polymorphic in other cultivars of this species (Zorrilla-Fontanesi et al., 2011). In addition, variations in a single nucleotide were found at four positions within the coding region (Fig. 6B). In summary, this analysis identified the same three alleles in both cultivars (see Supplementary Fig. S3 at JXB online). Only two of the substitutions in the coding region altered the amino acid sequence (Fig. 6B), with only the change in position 258, Gln/His, involving a non-conservative substitution. The location of this amino acid within the predicted tertiary structure indicated that it is not close to the active site but is located within to the C-terminal region, which is not highly conserved among the members of this protein family (Sanli et al., 2003). The possibility of a difference in the activity of the enzymes encoded by the different alleles, however, cannot be discarded. Therefore, differential expression of the alleles between cv. Camarosa and cv. Chandler could explain the discrepancy between the total FaGalUR protein and AsA levels (Fig. 5A). The changes in two of the four positions produced new restriction sites that could be used to identify which allele was expressed in each cultivar. At the first nucleotide change (Fig. 6B), Eco52I-digestion of alleles 1 and 2 in the cDNA generated two fragments of 554 bp and 406 bp. At the second nucleotide change (Fig. 6B), MbiI-digestion of alleles 1 and 2 in the cDNA generated two fragments of 651 bp and 309 bp (see Supplementary Table S2 at JXB online). Accordingly, the restriction study of the cDNAs would distinguish the joint expression of alleles 1 and 2 from that of allele 3. Using RNA extracted from the ripe fruits of cv. Camarosa and cv. Chandler and FaGalUR specific primers, RT-PCR followed by digestion with the appropriate restriction enzyme showed that in both cultivars, the fragment pattern was consistent with the expression of the three alleles (Fig. 6C). Using densitometry on the bands, the relative expression of each allele within each lane was estimated (Fig. 6B). Basically, there were no significant differences between the cultivars with respect to the joint expression of alleles 1 and 2 and that of allele 3.
Fig. 6.
(A) Schematic representation of the GalUR genomic DNA. The grey boxes represent exons, and the black lines represent introns. White arrows indicate synonymous mutations of a nucleotide, and grey arrows indicate non-synonymous mutations. The triangle marks the position of the SSR. (B) Changes in nucleotides and amino acids, highlighted in grey, as well as their positions in the different genomic clones of GalUR from strawberry cv. Chandler and cv. Camarosa are shown. The ‘Allele’ column indicates the abundance of the different alleles that were identified. The ‘Expression’ column indicates the expression of each allele in cv. Camarosa and cv. Chandler. The positions of nucleotide/amino acid correspond to Allele 3. (C) Digestion pattern of the open reading frame of FaGalUR from cv. Camarosa (1–3) and cv. Chandler (4–6) using specific primers and digested with Eco52I and MbiI. M, Marker; 1, FaGalUR from cv. Camarosa; 2, FaGalUR from cv. Camarosa digested with MbiI; 3, FaGalUR from cv. Camarosa digested with Eco52I; 4, FaGalUR from cv. Chandler; 5, FaGalUR from cv. Chandler digested with MbiI; 6, FaGalUR from cv. Chandler digested with Eco52I.
Discussion
Among those genes of the AsA biosynthetic pathways that were analysed, only two genes of the AsA biosynthetic pathways, FaGalUR and FaMIOX, showed an expression pattern that paralleled the increase of AsA in the strawberry fruit, and only one, FaGalUR, showed a striking increase (over 150-fold) during the development of the fruit from the green to the ripe stage. The AsA content in strawberry fruit from cv. Chandler increased from the green immature to the ripe stage, with a ≥12.2-fold increase on a dry weight basis. During this transition, the number of cells per fruit remains constant and the cell volume increases (Perkins-Veazie, 1995). The change in cell size is mainly due to an increased volume of vacuoles. Recent reports on AsA distribution in different cell compartments showed that the lowest levels were localized just in this compartment (Zechmann et al., 2011). If this distribution pattern is also seen in strawberry fruit cells, the increase in the AsA levels in compartments other than the vacuole during ripening would be dramatic.
Two other genes encoding enzymes of the mannose/galactose pathway, FaGME and FaG1PP (Wheeler et al., 1998), were down-regulated as fruit ripening proceeded from the green to the red stage. This observation suggests that this pathway is responsible for AsA biosynthesis in green fruits. The prevalence of one pathway over the others depending upon the developmental stage has previously been reported in the fruits of other plants such as grape (Cruz-Rus et al., 2010) and tomato (Ioannidi et al., 2009). This metabolic versatility toward AsA synthesis could be related to the different starting metabolites of each pathway, the availability of each of which is dependent on many environmental and/or developmental factors. In the mannose/galactose pathway, hexose phosphates are produced in the assimilatory biosynthetic pathways (Wheeler et al., 1998), whereas in the GalUR pathway, methyl-D-galacturonate results from the breakdown of pectin polymers (Agius et al., 2003). The first pathway is expected to be abundant in green fruits, whereas the second is expected to be abundant in the ripening fruits.
Interestingly, FaMDHAR expression was over 17-fold increased as the fruits ripened from green to red, whereas the expression of FaDHAR was higher in green fruits than in white and red fruits. These patterns indicate that FaMDHAR expression is associated with high AsA content in ripe fruit. The involvement of this enzyme in the reduction to AsA has been linked to the response of the plant to oxidative stress (Eltayeb et al., 2006; Stevens et al., 2008). Early transcriptional analysis during strawberry fruit ripening indicated that the strawberry ripening transcriptional programme is an oxidative, stress-induced process (Aharoni et al., 2002). Therefore, FaMDHAR might contribute, along with other enzymes of these biosynthetic pathways, to the maintenance of high AsA content in the ripe fruit of strawberries. In green fruit, however, AsA recycling appears to be performed by DHAR. These observations are consistent with previous studies on tomato (Ioannidi et al., 2009), kiwifruit (Bulley et al., 2009) and acerola (Eltelib et al., 2010). In these fruits, the two AsA recycling genes are differently regulated during ripening, depending on the plant species and the stage of ripening.
The ripe fruits stored under an ozone atmosphere suggest another scenario in which changes in the synthesis of AsA are expected. The most significant changes at the gene expression level were found in the FaG1PP, FaGalUR, and FaGLDH genes. This observation suggested the activation of both the mannose/galactose and the D-galacturonate pathways under this treatment. In fact, decreases in sucrose, glucose, and fructose have been reported in ozone-treated fruits (Pérez et al., 1999). The mannose/galactose pathway may consume these sugars. The analysis of the FaGalUR promoter revealed the presence of a G-box motif (Agius et al., 2005), which is present in the ozone-responsive promoter region of the grape stilbene synthase gene (Schubert et al., 1997). In addition, the 3-fold higher expression of FaMDHAR in the ozone-treated fruits compared with the air-stored fruits also supports a role of the corresponding enzyme in maintaining the AsA content in those fruits treated with ozone. In tomato fruits under stress conditions, a main role for MDHAR in maintaining the AsA levels has been reported (Stevens et al., 2008).
Information on the regulation of the AsA biosynthetic pathways is still limited. In the mannose/galactose pathway, substrate availability has been suggested to be a limiting factor (Wheeler et al., 1998). Other studies indicate that GME and/or GDP-L-galactose guanyltransferase (GGT) (Bulley et al., 2009; Gilbert et al., 2009) play key roles in the regulation of ascorbate biosynthesis in plants. During tomato fruit ripening, G1PP has been proposed to play a main role in the accumulation of AsA (Ioannidi et al., 2009). In the D-galacturonate pathway, a correlation between GalUR expression and AsA content was found (Agius et al., 2003). This study has extended the analysis of the AsA pathways to ripe fruits of a segregating population from a cross between two F. ananassa parents with a very close genetic background that corresponded to cultivars with a Californian pedigree (Rousseau-Gueutin et al., 2009). The lack of correlation between the transcriptional activity of the genes and the highly variable AsA content in the ripe fruits of this segregating population excluded the possibility of a single regulatory step and points to a co-ordinated action involving several of these genes.
Effectively, our results from the protein studies, both in different species of the Fragaria genus and in two closely related cultivars of F. ananassa, nullified the hypothesis that the enzyme GalUR had a unique regulatory role on AsA synthesis in ripe strawberry fruit. In plants, the AsA content in different tissues varies with environmental (Smirnoff and Wheeler, 2000; Tamaoki et al., 2003) and developmental (Pallanca and Smirnoff, 1999; Bartoli et al., 2000; Agius et al., 2003) conditions, which requires tight control of its biosynthesis, catabolism, and reductive recovery from the oxidized form. Therefore, a number of regulatory steps should be expected. In agreement with this theory, plants transformed with the genes from different pathways have shown only a limited increase in their AsA content (Ishikawa et al., 2006). This observation holds true for GalUR (Agius et al., 2003; Hemavathi et al., 2009), L-galactose dehydrogenase (Gatzek et al., 2002), GDP-L-galactose guanyltransferase (Bulley et al., 2009), GLDH (Tokunaga et al., 2005), and DHAR (Kwon et al., 2003). In addition, genetic studies also support this complexity because several quantitative trait loci contributing to AsA content have been identified in apple and tomato (Davey et al., 2006; Stevens et al., 2007).
Overall, our results indicate that the GalUR enzyme plays a prominent role in the regulation of the AsA biosynthetic pathway in ripe strawberry fruits. This regulatory role, however, is probably limited and shared with the enzymes of other pathways and even with other components of this AsA pathway, which might include the substrate availability. As in other plant tissues, the metabolic control of AsA levels is not executed by a single element, enzyme or substrate. Rather, this control is likely to be shared by several components in a complex and still unknown regulatory network that must be studied using species/tissue/stage-based approaches.
Supplementary data
Supplementary data can be found at JXB online
Supplementary Table S1. Sequence of the specific primers used for RT-PCR expression studies.
Supplementary Table S2. Table with the different restriction sites and the fragment length of the different cuts in the different alleles.
Supplementary Fig. S1. Changes in mRNA levels of PR5 and PG1P, two pathogen-induced genes (Osorio et al., 2008) in fruits before (0 h) and after storage for 72 h in ozone or air atmosphere by semi-quantitative RT-PCR.
Supplementary Fig. S2. Frequency distributions of AsA for the 95 individuals from the population segregation derived from the cross between ‘232’ and ‘1392’; arrows indicate the parental values.
Supplementary Fig. S3. Alignment of the three different alleles of FaGalUR
Acknowledgments
This work was supported by grants BIO2010-15630 (MICINN, Spain) and RTA2008-0029-00-00 (INIA, Spain). EC-R was awarded a ‘Formación de Doctores en Centros de Investigación y Universidades Andaluzas’ fellowship from the Conserjería de Innovación, Ciencia y Empresa de la Junta de Andalucía (Spain).
Glossary
Abbreviations
- AsA
L-ascorbic acid
- GME
GDP-mannose-3'-5'-epimerase
- G1PP
L-galactose-1-phosphate phosphatase
- GalUR
L-galacturonic acid reductase
- GLDH
L-galactone-1,4-lactone dehydrogenase
- MYOX
myo-inositol oxygenase
- MDHAR
monodehydroascorbate reductase
- DHAR
dehydroascorbate reductase
References
- Agius F, Amaya I, Botella MA, Valpuesta V. Functional analysis of homologous and heterologous promoters in strawberry fruits using transient expression. Journal of Experimental Botany. 2005;56:37–46. doi: 10.1093/jxb/eri004. [DOI] [PubMed] [Google Scholar]
- Agius F, González-Lamothe R, Caballero JL, Muñoz-Blanco J, Botella MA, Valpuesta V. Engineering increased vitamin C levels in plants by overexpression of a D-galacturonic acid reductase. Nature Biotechnology. 2003;21:177–181. doi: 10.1038/nbt777. [DOI] [PubMed] [Google Scholar]
- Aharoni A, Keizer LCP, Van Den Broeck HC, Blanco-Portales R, Muñoz-Blanco J, Bois G, Smit P, De Vos RCH, O'Connell AP. Novel insight into vascular, stress, and auxin-dependent and -independent gene expression programs in strawberry, a non-climacteric fruit. Plant Physiology. 2002;129:1019–1031. doi: 10.1104/pp.003558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badejo AA, Fujikawa Y, Esaka M. Gene expression of ascorbic acid biosynthesis related enzymes of the Smirnoff–Wheeler pathway in acerola (Malpighia glabra) Journal of Plant Physiology. 2009;166:652–660. doi: 10.1016/j.jplph.2008.09.004. [DOI] [PubMed] [Google Scholar]
- Bartoli CG, Pastori GM, Foyer CH. Ascorbate biosynthesis in mitochondria is linked to the electron transport chain between complexes III and IV. Plant Physiology. 2000;123:335–344. doi: 10.1104/pp.123.1.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bombarely A, Merchante C, Csukasi F, et al. Generation and analysis of ESTs from strawberry (Fragaria×ananassa) fruits and evaluation of their utility in genetic and molecular studies. BMC Genomics. 2010;11:503. doi: 10.1186/1471-2164-11-503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulley SM, Rassam M, Hoser D, Otto W, Schünemann N, Wright M, MacRae E, Gleave A, Laing W. Gene expression studies in kiwifruit and gene over-expression in Arabidopsis indicates that GDP-L-galactose guanyltransferase is a major control point of vitamin C biosynthesis. Journal of Experimental Botany. 2009;60:765–778. doi: 10.1093/jxb/ern327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casado-Díaz A, Encinas-Villarejo S, Santos Bdl, et al. Analysis of strawberry genes differentially expressed in response to Colletotrichum infection. Physiologia Plantarum. 2006;128:633–650. [Google Scholar]
- Chatterjee IB. Evolution and the biosynthesis of ascorbic acid. Science. 1973;182:1271–1272. doi: 10.1126/science.182.4118.1271. [DOI] [PubMed] [Google Scholar]
- Chen Z, Gallie DR. The ascorbic acid redox state controls guard cell signaling and stomatal movement. The Plant Cell. 2004;16:1143–1162. doi: 10.1105/tpc.021584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conklin PL, Norris SR, Wheeler GL, Williams EH, Smirnoff N, Last RL. Genetic evidence for the role of GDP-mannose in plant ascorbic acid (vitamin C) biosynthesis. Proceedings of the National Academy of Sciences, USA. 1999;96:4198–4203. doi: 10.1073/pnas.96.7.4198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz-Rus E, Botella MA, Valpuesta V, Gomez-Jimenez MC. Analysis of genes involved in L-ascorbic acid biosynthesis during growth and ripening of grape berries. Journal of Plant Physiology. 2010;167:739–748. doi: 10.1016/j.jplph.2009.12.017. [DOI] [PubMed] [Google Scholar]
- Davey MW, Van Montagu M, Inzé D, Sanmartin M, Kanellis A, Smirnoff N, Benzie IJJ, Strain JJ, Favell D, Fletcher J. Plant L-ascorbic acid: chemistry, function, metabolism, bioavailability and effects of processing. Journal of the Science of Food and Agriculture. 2000;80:825–860. [Google Scholar]
- Davey MW, Bauw G, Van Montagu M. Analysis of ascorbate in plant tissues by high-performance capillary zone electrophoresis. Analytical Biochemistry. 1996;239:8–19. doi: 10.1006/abio.1996.0284. [DOI] [PubMed] [Google Scholar]
- Davey MW, Kenis K, Keulemans J. Genetic control of fruit vitamin C contents. Plant Physiology. 2006;142:343–351. doi: 10.1104/pp.106.083279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- do Nascimento Jo, Higuchi B, Gómez M, Oshiro R, Lajolo F. L-Ascorbate biosynthesis in strawberries: L-galactono-1,4-lactone dehydrogenase expression during fruit development and ripening. Postharvest Biology and Technology. 2005;38:34–42. [Google Scholar]
- Dowdle J, Ishikawa T, Gatzek S, Rolinski S, Smirnoff N. Two genes in Arabidopsis thaliana encoding GDP-L-galactose phosphorylase are required for ascorbate biosynthesis and seedling viability. The Plant Journal. 2007;52:673–689. doi: 10.1111/j.1365-313X.2007.03266.x. [DOI] [PubMed] [Google Scholar]
- Eltayeb A, Kawano N, Badawi G, Kaminaka H, Sanekata T, Morishima I, Shibahara T, Inanaga S, Tanaka K. Enhanced tolerance to ozone and drought stresses in transgenic tobacco overexpressing dehydroascorbate reductase in cytosol. Physiologia Plantarum. 2006;127:57–65. [Google Scholar]
- Eltelib HA, Badejo AA, Fujikawa Y, Esaka M. Gene expression of monodehydroascorbate reductase and dehydroascorbate reductase during fruit ripening and in response to environmental stresses in acerola (Malpighia glabra) Journal of Plant Physiology. 2010;168:619–627. doi: 10.1016/j.jplph.2010.09.003. [DOI] [PubMed] [Google Scholar]
- Gatzek S, Wheeler GL, Smirnoff N. Antisense suppression of L-galactose dehydrogenase in Arabidopsis thaliana provides evidence for its role in ascorbate synthesis and reveals light modulated L-galactose synthesis. The Plant Journal. 2002;30:541–553. doi: 10.1046/j.1365-313x.2002.01315.x. [DOI] [PubMed] [Google Scholar]
- Gibson DG, Benders GA, Andrews-Pfannkoch C, et al. Complete chemical synthesis, assembly, and cloning of a Mycoplasma genitalium genome. Science. 2008;319:1215–1220. doi: 10.1126/science.1151721. [DOI] [PubMed] [Google Scholar]
- Gilbert L, Alhagdow M, Nunes-Nesi A, et al. GDP-D-mannose 3,5-epimerase (GME) plays a key role at the intersection of ascorbate and non-cellulosic cell-wall biosynthesis in tomato. The Plant Journal. 2009;60:499–508. doi: 10.1111/j.1365-313X.2009.03972.x. [DOI] [PubMed] [Google Scholar]
- Harapanhalli RS, Howell RW, Rao DV. Testicular and plasma ascorbic acid levels in mice following dietary intake: a high-performance liquid chromatographic analysis. Journal of Chromatography. 1993;614:233–243. doi: 10.1016/0378-4347(93)80314-t. [DOI] [PubMed] [Google Scholar]
- Hemavathi, Upadhyaya C, Young K, et al. Over-expression of strawberry D-galacturonic acid reductase in potato leads to accumulation of vitamin C with enhanced abiotic stress tolerance. Plant Science. 2009;177:659–667. [Google Scholar]
- Horemans N, Foyer CH, Asard H. Transport and action of ascorbate at the plant plasma membrane. Trends in Plant Science. 2000;5:263–267. doi: 10.1016/s1360-1385(00)01649-6. [DOI] [PubMed] [Google Scholar]
- Imai T, Karita S, Shiratori G, Hattori M, Nunome T, Oba K, Hirai M. L-galactono-γ-lactone dehydrogenase from sweet potato: purification and cDNA sequence analysis. Plant and Cell Physiology. 1998;39:1350–1358. doi: 10.1093/oxfordjournals.pcp.a029341. [DOI] [PubMed] [Google Scholar]
- Ioannidi E, Kalamaki MS, Engineer C, Pateraki I, Alexandrou D, Mellidou I, Giovannonni J, Kanellis AK. Expression profiling of ascorbic acid-related genes during tomato fruit development and ripening and in response to stress conditions. Journal of Experimental Botany. 2009;60:663–678. doi: 10.1093/jxb/ern322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa T, Dowdle J, Smirnoff N. Progress in manipulating ascorbic acid biosynthesis and accumulation in plants. Physiologia Plantarum. 2006;126:343–255. [Google Scholar]
- Jain A, Nessler C. Metabolic engineering of an alternative pathway for ascorbic acid biosynthesis in plants. Molecular Breeding. 2000;6:73–78. [Google Scholar]
- Kwon SY, Choi SM, Ahn YO, Lee HS, Lee HB, Park YM, Kwak SS. Enhanced stress-tolerance of transgenic tobacco plants expressing a human dehydroascorbate reductase gene. Journal of Plant Physiology. 2003;160:347–353. doi: 10.1078/0176-1617-00926. [DOI] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Laing WA, Bulley S, Wright M, Cooney J, Jensen D, Barraclough D, MacRae E. A highly specific L-galactose-1-phosphate phosphatase on the path to ascorbate biosynthesis. Proceedings of the National Academy of Sciences, USA. 2004;101:16976–16981. doi: 10.1073/pnas.0407453101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laing WA, Wright MA, Cooney J, Bulley SM. The missing step of the L-galactose pathway of ascorbate biosynthesis in plants, an L-galactose guanyltransferase, increases leaf ascorbate content. Proceedings of the National Academy of Sciences, USA. 2007;104:9534–9539. doi: 10.1073/pnas.0701625104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Ma F, Shang P, Zhang M, Hou C, Liang D. Influence of light on ascorbate formation and metabolism in apple fruits. Planta. 2009;230:39–51. doi: 10.1007/s00425-009-0925-3. [DOI] [PubMed] [Google Scholar]
- Linster C, Clarke S. L-Ascorbate biosynthesis in higher plants: the role of VTC2. Trends in Plant Science. 2008;13:567–573. doi: 10.1016/j.tplants.2008.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loewus F. Biosynthesis and metabolism of ascorbic acid in plants and of analogs of ascorbic acid in fungi. Phytochemistry. 1999;52:193–210. [Google Scholar]
- Lorence A, Chevone BI, Mendes P, Nessler CL. myo-inositol oxygenase offers a possible entry point into plant ascorbate biosynthesis. Plant Physiology. 2004;134:1200–1205. doi: 10.1104/pp.103.033936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning K. Isolation of nucleic acids from plants by differential solvent precipitation. Analytical Biochemistry. 1991;195:45–50. doi: 10.1016/0003-2697(91)90292-2. [DOI] [PubMed] [Google Scholar]
- Mehli L, Schaart J, Kjellsen T, Tran D, Salentijn E, Schouten H, Iversen T. A gene encoding a polygalacturonase-inhibiting protein (PGIP) shows developmental regulation and pathogen-induced expression in strawberry. New Phytologist. 2004;163:99–110. doi: 10.1111/j.1469-8137.2004.01088.x. [DOI] [PubMed] [Google Scholar]
- Osorio S, Castillejo C, Quesada MA, Medina-Escobar N, Brownsey GJ, Suau R, Heredia A, Botella MA, Valpuesta V. Partial demethylation of oligogalacturonides by pectin methyl esterase 1 is required for eliciting defence responses in wild strawberry (Fragaria vesca) The Plant Journal. 2008;54:43–55. doi: 10.1111/j.1365-313X.2007.03398.x. [DOI] [PubMed] [Google Scholar]
- Pallanca J, Smirnoff N. Ascorbic acid metabolism in pea seedlings. A comparison of D-glucosone, L-sorbosone, and L-galactono-1,4-lactone as ascorbate precursors. Plant Physiology. 1999;120:453–462. doi: 10.1104/pp.120.2.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastori GM, Kiddle G, Antoniw J, Bernard S, Veljovic-Jovanovic S, Verrier PJ, Noctor G, Foyer CH. Leaf vitamin C contents modulate plant defense transcripts and regulate genes that control development through hormone signaling. The Plant Cell. 2003;15:939–951. doi: 10.1105/tpc.010538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez AG, Sanz C, Ríos JJ, Olías R, Olías JM. Effects of ozone treatment on postharvest strawberry quality. Journal of Agricultural and Food Chemistry. 1999;47:1652–1656. doi: 10.1021/jf980829l. [DOI] [PubMed] [Google Scholar]
- Perkins-Veazie P. Growth and ripening of strawberry fruit. Horticultural Reviews. 1995;17:267–297. [Google Scholar]
- Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Research. 2001;29:2002–2007. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rousseau-Gueutin M, Gaston A, Aïnouche A, Aïnouche ML, Olbricht K, Staudt G, Richard L, Denoyes-Rothan B. Tracking the evolutionary history of polyploidy in Fragaria L. (strawberry): new insights from phylogenetic analyses of low-copy nuclear genes. Molecular Phylogenetics and Evolution. 2009;51:515–530. doi: 10.1016/j.ympev.2008.12.024. [DOI] [PubMed] [Google Scholar]
- Sanli G, Dudley JI, Blaber M. Structural biology of the aldo-keto reductase family of enzymes: catalysis and cofactor binding. Cell Biochemistry and Biophysics. 2003;38:79–101. doi: 10.1385/CBB:38:1:79. [DOI] [PubMed] [Google Scholar]
- Schubert R, Fischer R, Hain R, Schreier PH, Bahnweg G, Ernst D, Sandermann H. An ozone-responsive region of the grapevine resveratrol synthase promoter differs from the basal pathogen-responsive sequence. Plant Molecular Biology. 1997;34:417–426. doi: 10.1023/a:1005830714852. [DOI] [PubMed] [Google Scholar]
- Smirnoff N, Wheeler GL. Ascorbic acid in plants: biosynthesis and function. Critical Reviews in Biochemistry and Molecular Biology. 2000;35:291–314. doi: 10.1080/10409230008984166. [DOI] [PubMed] [Google Scholar]
- Stevens R, Buret M, Duffé P, Garchery C, Baldet P, Rothan C, Causse M. Candidate genes and quantitative trait loci affecting fruit ascorbic acid content in three tomato populations. Plant Physiology. 2007;143:1943–1953. doi: 10.1104/pp.106.091413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens R, Page D, Gouble B, Garchery C, Zamir D, Causse M. Tomato fruit ascorbic acid content is linked with monodehydroascorbate reductase activity and tolerance to chilling stress. Plant, Cell and Environment. 2008;31:1086–1096. doi: 10.1111/j.1365-3040.2008.01824.x. [DOI] [PubMed] [Google Scholar]
- Tabata K, Takaoka T, Esaka M. Gene expression of ascorbic acid-related enzymes in tobacco. Phytochemistry. 2002;61:631–635. doi: 10.1016/s0031-9422(02)00367-9. [DOI] [PubMed] [Google Scholar]
- Tamaoki M, Mukai F, Asai N, Nakajima N, Kubo A, Aono M, Saji H. Light-controlled expression of a gene encoding L-galactono-[gamma]-lactone dehydrogenase which affects ascorbate pool size in Arabidopsis thaliana. Plant Science. 2003;164:1111–1117. [Google Scholar]
- Tokunaga T, Miyahara K, Tabata K, Esaka M. Generation and properties of ascorbic acid-overproducing transgenic tobacco cells expressing sense RNA for L-galactono-1,4-lactone dehydrogenase. Planta. 2005;220:854–863. doi: 10.1007/s00425-004-1406-3. [DOI] [PubMed] [Google Scholar]
- Valpuesta V, Botella MA. Biosynthesis of L-ascorbic acid in plants: new pathways for an old antioxidant. Trends in Plant Science. 2004;9:573–577. doi: 10.1016/j.tplants.2004.10.002. [DOI] [PubMed] [Google Scholar]
- Wheeler GL, Jones MA, Smirnoff N. The biosynthetic pathway of vitamin C in higher plants. Nature. 1998;393:365–369. doi: 10.1038/30728. [DOI] [PubMed] [Google Scholar]
- Wolucka BA, Van Montagu M. GDP-mannose 3‘,5’-epimerase forms GDP-L-gulose, a putative intermediate for the de novo biosynthesis of vitamin C in plants. Journal of Biological Chemistry. 2003;278:47483–47490. doi: 10.1074/jbc.M309135200. [DOI] [PubMed] [Google Scholar]
- Yin L, Wang S, Eltayeb AE, Uddin MI, Yamamoto Y, Tsuji W, Takeuchi Y, Tanaka K. Overexpression of dehydroascorbate reductase, but not monodehydroascorbate reductase, confers tolerance to aluminum stress in transgenic tobacco. Planta. 2010;231:609–621. doi: 10.1007/s00425-009-1075-3. [DOI] [PubMed] [Google Scholar]
- Zechmann B, Stumpe M, Mauch F. Immunocytochemical determination of the subcellular distribution of ascorbate in plants. Planta. 2011;233:1–12. doi: 10.1007/s00425-010-1275-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zorrilla-Fontanesi Y, Cabeza A, Torres AM, Botella MA, Valpuesta V, Monfort A, Sanchez-Sevilla JF, Amaya I. Development and bin mapping of strawberry genic-SSRs in diploid Fragaria and their transferability across the Rosoideae subfamily. Molecular Breeding. 2011;27:137–156. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.