Abstract
Developing ontologies to account for the complexity of biological systems requires the time intensive collaboration of many participants with expertise in various fields. While each participant may contribute to construct a list of terms for ontology development, no objective methods have been developed to evaluate how relevant each of these terms is to the intended domain. We have developed a computational method based on a hypergeometric enrichment test to evaluate the relevance of such terms to the intended domain. The proposed method uses the PubMed literature database to evaluate whether each potential term for ontology development is overrepresented in the abstracts that discuss the particular domain. This evaluation provides an objective approach to assess terms and prioritize them for ontology development.
Keywords: Ontology development, hypergeometric test, PubMed, text mining
1 INTRODUCTION
High-quality ontologies such as the Gene Ontology (GO) [1] have been instrumental in analyzing data generated from microarray experiments [2–13]. However, developing such high-quality ontologies still poses significant challenges, as wide spectra of literatures and domain experts need to be involved. To aid ontology development, numerous methods have been developed to extract terms from literature automatically. Daille proposed combined techniques to extract terms automatically from corpora by combining linguistic filters and statistical methods [14]. Frantzi et al developed a C-value/NC-value method to extract multi-word terms automatically [15]. By taking advantage of semantic relations encoded between terms, Grabar and Zweigenbaum developed a two-step approach to collect semantically related terms and to align morphologically linked work forms for term extraction [16]. A “weirdness metric” was proposed by Ahmad and Rogers to evaluate terms overrepresented in the domain-specific corpus for ontology development [17]. Savova et al developed a data-driven approach to extract the “most specific term” for ontology development using an algorithm combining statistical and linguistic approaches [18]. Another tool developed to extract terms for ontology development was Text2Onto, which was built upon the Probabilistic Ontology Model [19]. In addition, Smith et al proposed a machine learning approach to retrieve definitional content for ontology development [20]. Concept maps have also been used by Castro et al in the ontology development process [21]. Alexopoulou et al developed two additional methods, one based on the relative frequency of a term in the corpus and the other using the document frequency derived from all phrases contained in PubMed abstract database, to extract terms for ontology development [22]. While these methods focuses on extracting terms from published literatures, two other studies also proposed to extract terms from web resources for concept and ontology development [23, 24]. Despite these efforts, it is still widely recognized that manual curation is the most reliable method for ontology development [25], and these automatic term extraction methods are rarely used as a mainstream approach in the current biomedical ontology development process.
In the manual curation process, curators read as much scientific literature as possible for a particular biological domain in order to identify corresponding ontology terms and to classify their relationships to the domain and to other terms within it (such as “is_a” and “has_a” relationships). One significant challenge during this process is to determine which term should be used as the basic building block from which to develop an ontology for a particular domain. This challenge is compounded by the fact that many curators with diverse backgrounds may be involved in developing the ontology for a given domain; diversity in their backgrounds can result in the selection of a wide variety of terms to be compiled within the ontology. This term selection process relies on the expertise of individual curators, without either a preliminary or confirmatory test using some objective method and measure. A quantitative approach to evaluate whether terms are appropriate to develop an ontology for a particular domain would provide this objective method and measure, improve the utility of the resulting ontology, and reduce the amount of work imposed on curators.
While the above mentioned term extraction algorithms or their underlying metrics have the potential to be used to evaluate terms assembled by experts during manual curation for ontology development, several limitations exist for such an application. First, most of these algorithms are developed to extract terms from a corpus of selected literatures already identified as relevant to a specific domain. Lacking such a pre-defined corpus, as is the case for many ontology development projects, these methods’ effectiveness is not clear. Second, the volume of existing literature databases like PubMed is significantly larger than a corpus related to a specific domain; this corpus size difference raises questions about the performance of these algorithms if applied to evaluate terms using PubMed abstracts in the absence of a domain specific corpus. Furthermore, these methods have not been widely tested against manually assembled ontology terms. New approaches capable of dealing with large databases and confirmed through comparison to manually curated ontologies need to be developed.
One objective criterion to evaluate a term’s suitability for incorporation in an ontology for a particular concept domain is to quantify the term’s relevance to the domain within published biomedical literature. If a term occurs at high frequency in PubMed abstracts relevant to the concept domain, then it should be more suitable for ontology development than other terms occurring at low frequencies. Because ontology development aims to describe particular biological domains, we hypothesized that the terms used within an already existing ontology for a particular domain would be overrepresented in the PubMed abstracts relevant to that domain (Domain PubMed Abstract, DPA). We further hypothesized that the degree of overrepresentation could be detected by employing a hypergeometric enrichment test.
Testing based on hypergeometric distribution has been applied in the analysis of GO overrepresentation on biologically-interesting gene sets (review see [26]). Hypergeometric testing can measure the association between a term and the domain by calculating the probability of observing the term within the DPA as long as both categories are sampled without replacement from a finite population. Such a probability can be used as a direct measure of how relevant a term is to the domain: the higher probability we observe a term in the DPA, the more overrepresented this term is in the DPA, and the more relevant this term is. The degree of overrepresentation of terms relevant to a domain in the DPA can indicate the usefulness of the terms for developing this domain’s ontology. Experts from diverse fields could use the information gleaned through such a hypergeometric evaluation to narrow candidate terms for a given ontology. The test could significantly reduce the manual effort involved in ontology development.
In this study, we first used GO [1] as a control to evaluate whether the proposed text-mining approach could detect the overrepresentation of ontology terms in the corresponding DPA. We demonstrated that the hypergeometric test could capture the relevant terms in the DPA and reflect their relative importance by their overrepresentation. We then demonstrated that this approach could be used to evaluate putative ontology terms generated by different experts for the development of a Clinical Trial Ontology/Ontology for Clinical Investigation [27]. Our results indicated that such a computational algorithm can provide an objective measure for the selection of putative ontology terms to facilitate ontology development.
2 METHODS
2.1 PubMed Database Preparation
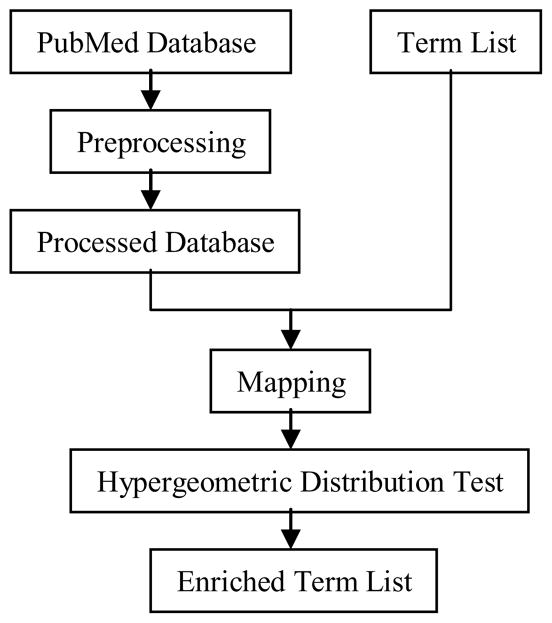
Figure 1 illustrates a condensed version of our process. The entire PubMed database (2007) in XML format was downloaded from NCBI. The database was processed to extract all abstracts. Necessary formatting, such as capitalizing all the abstracts and removing special characters, was performed (box ‘preprocessing’). All the software was implemented in C++. PubMed stopwords were also downloaded from NCBI.
Figure 1.
Workflow of enrichment test to evaluate terms for ontology development
2.2 Collection of GO and other Terms
Terms for evaluation by hypergeometric enrichment test
In order to test whether terms relevant to a domain were overrepresented in the corresponding DPA, we identified test sets where each set had a domain term and a list of terms known to be relevant to that domain. To generate these test sets, we took advantage of the hierarchical structure of GO. In an ontology such as GO, terms with specific meaning are children of terms that are more general, thus comprising an “is_a” relationship. We viewed a parent term as a domain and the child term as a term relevant to that domain. This approach can be extrapolated to multi-level hierarchies such that an ontology term at a high level of the hierarchy can be viewed as a domain term, and all of its child terms can be viewed as terms that are relevant to this domain.
GO was downloaded from the GO Consortium website (http://www.geneontology.org, June, 2007). Two terms were selected from GO as domain terms for our study. The criteria for selection were: 1) each term had more than 50 child terms under an “is_a” relationship to yield a significant number of child terms relevant to the domain; 2) the selected domain term was not a child term of another domain term. Two domain terms, Monosaccharide Metabolic Process (GO:0005996) and Protein Kinase Activity (GO:0004672), were selected for this purpose. Monosaccharide Metabolic Process is categorized as a biological process within GO, and Protein Kinase Activity falls within the molecular function category. Descendents of these two terms were also collected. Since we focus on the terms that describe biological systems, common words such as “activity” or “process” were removed from these descendent terms(henceforth, such terms are shown in brackets). This practice was limited to the GO terms used for validation purposes and was not applied to the putative ontology terms evaluated in subsequent analyses. The number of unique terms after removing these common words was 56 and 97 for Monosaccharide [Metabolic Process] and Protein Kinase [Activity], respectively. The degree of overrepresentation of these descendent terms in the DPA was tested as described below.
We also selected additional GO terms as controls. These terms were randomly chosen from a pool of GO terms that had no descendant-ancestor relationships with Monosaccharide [Metabolic Process] or Protein Kinase [Activity]. The control terms identified as overrepresented in abstracts that also included Monosaccharide [Metabolic Process] or Protein Kinase [Activity] were counted as false positives in constructing Precision-Recall curves.
Terms for evaluating putative ontology terms for Clinical Trial Ontology
Putative ontology terms were downloaded from the wiki for the Ontology for Clinical Investigations [27]. These 316 terms were assembled by experts with various experience and backgrounds in areas related to clinical trials. The domain of clinical trials focuses on clinical trial design, implementation and outcome analysis, while the domain of GO mainly characterizes gene functions. Therefore, we assumed that the overlap between these two domains would be small and not significant. This assumption is supported by the fact that the PubMed abstracts containing the word “protein kinase” comprise only 0.1% of those containing the keyword “clinical trial”, and the PubMed abstracts that contain the word “clinical trial” only comprise 0.4% of those with the keyword “protein kinase”. The low frequency of GO terms appearing in the PubMed abstracts that contain the keyword “clinical trial” also supports our assumption (data not shown). Therefore, randomly selected GO terms were used as negative controls: if any of these GO terms were overrepresented in the PubMed abstracts that discussed clinical trials, they were counted as false positives. The selected terms were mapped to the PubMed abstracts, and their occurrences in the abstracts relevant to the clinical trial domain were counted. For the clinical trial domain, we identified any PubMed abstracts that contained the term “Clinical Trial” or “Clinical-Trial” as relevant. The final test was run for all 316 putative ontology terms and identified terms that were overrepresented in the DPA for the clinical trial domain.
2.3 Mapping Terms to PubMed Abstracts
We implemented the efficient Boyer-Moore string search algorithm [28] in C++ to map terms to PubMed abstracts. The algorithm identifies single or multiword terms between white space in the abstracts that match query terms. Mapping of terms to PubMed abstracts was performed on a 76 node computer cluster. Each cluster node had two dual-core Intel Xeon 3GB processors. The PubMed database was split into 100 files, and mapping was performed on 100 cluster nodes for the selected GO terms, the putative ontology terms for Clinical Trial Ontology development, and the domain terms. The numbers of abstracts that contained individual terms were then summed. The results were assembled and analyzed by the hypergeometric test.
2.4. Performing the Hypergeometric Test and the Weirdness Metric Test
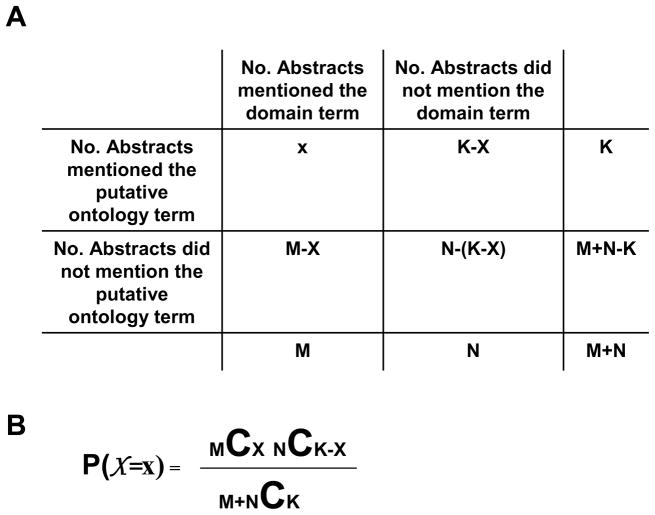
In this study, we used a one-sided hypergeometric test [26] to compute the overrepresentation of each ontology term in the corresponding DPA. To assess whether a putative ontology term is relevant to a domain, the PubMed abstracts can be divided into four different categories, using the clinical trial domain as an example: 1) PubMed abstracts that contain both the putative ontology term and the domain term “Clinical Trial” or “Clinical-Trial” (top left cell in Figure 2A, the number of these abstracts = X); 2) PubMed abstracts that contain only the putative ontology term but not the domain term (top middle cell in Figure 2A, the number of these abstracts = K − X); 3) PubMed abstracts that contain the domain term but not the putative ontology term (left middle cell in Figure 2A, the number of these abstracts = M-X); and 4) PubMed abstracts that contain neither the putative ontology term nor the domain term (middle cell in Figure 2A, the number of these abstracts = N-(K-X)). Therefore, the total number of PubMed abstracts that contains the domain term is M, the total number of PubMed abstracts that contains the putative ontology term is K, and the total number of PubMed abstracts is M+N. For a given putative ontology term, the probability of getting X number of annotated abstracts among M number of abstracts that contain the domain term, given that the total number of abstracts containing the putative ontology term is K , is shown in Figure 2B [29]. Due to the discreteness of the hypergeometric distribution, mid-P-values [30] were used in the test: pmid = P (X > x ) + 0.5P (X > x) , where P ( X > x) is the probability of observing more than x abstracts annotated with both the putative ontology and domain terms, and P ( X > x) is the probability of observing exactly x abstracts. We used the hypergeometric distribution functions (i.e. dhyper and phyper) implemented in the statistics package of R [31] to perform the one-sided test (for a discussion about one-sided and two-sided tests, see [26]) and to calculate the mid-p-value. The null and alternative hypotheses for the one-sided test are Ho : r1 ≤ r2 and H1 : r1 > r2 , respectively; where r1 is the probability of the ontology term being relevant to the domain, while r2 is the probability of the ontology term being relevant to other domains. The resulting p-value indicates the degree of overrepresentation of the term in the PubMed abstracts discussing the domain term “Clinical Trial” or “Clinical-Trial”. In order to perform this test in a high-throughput manner, we also implemented analysis software in C++ to perform the test using the functions (gsl_ran_hypergeometric_pdf and gsl_cdf_hypergeometric_P) in the GNU scientific library [32].
Figure 2.
Using PubMed abstract database and hypergeometric test to evaluate terms for ontology development. A) X is the number of abstracts that contains both a term for ontology development (putative ontology term to be evaluated) and the domain term; M and N are the abstracts with and without the term for the domain, respectively; K is the number of abstracts with the term. M+N is the total number of PubMed Abstracts examined. For example, to evaluate whether hexokinase activity is an appropriate term to develop an ontology for monosaccharide metabolism, X is the number of PubMed abstracts that contain both hexokinase activity and monosaccharide metabolism. B) The equation shows the probabil-a! ity of observing X by employing the hypergeometric formulation, where is the binomial coefficient.
We tested the overrepresentation of each term under the corresponding chosen domain (i.e. “Monosaccharide [Metabolic Process]”, “Protein Kinase [Activity]”, and “Clinical Trial”) against all the abstracts in PubMed. Recall and Precision were calculated for different p-value cut offs, where and . A Precision-Recall curve generated from ROCR [33] was used to plot each test and to evaluate the text mining method. To evaluate whether child terms were overrepresented for the domain terms Monosaccharide [Metabolic Process] or Protein Kinase [Activity], we randomly sampled 100 times the same number of control terms as the domain terms to evaluate our method. We also applied the hypergeometric test to assess whether the distribution of Clinical Data Interchange Standards Consortium (CDISC) [34] terms was overrepresented or not in the DPA containing the domain term “Clinical Trial” or “Clinical-Trial”.
We then compared our hypergeometric enrichment test-based approach to a similar approach based on the Weirdness Metric proposed by Ahmad and Rogers [17]. This approach counts the frequency of a term in the DPA and non-DPA and then takes the ratio of these two frequencies to generate the Weirdness Metric. We used this approach to test the overrepresentation of each term under the corresponding chosen domain (i.e. “Monosaccharide [Metabolic Process]” and “Protein Kinase [Activity]”) against all the abstracts in PubMed and compared the Precision-Recall curves of this approach with our hypergeo-metric enrichment test.
3 RESULTS
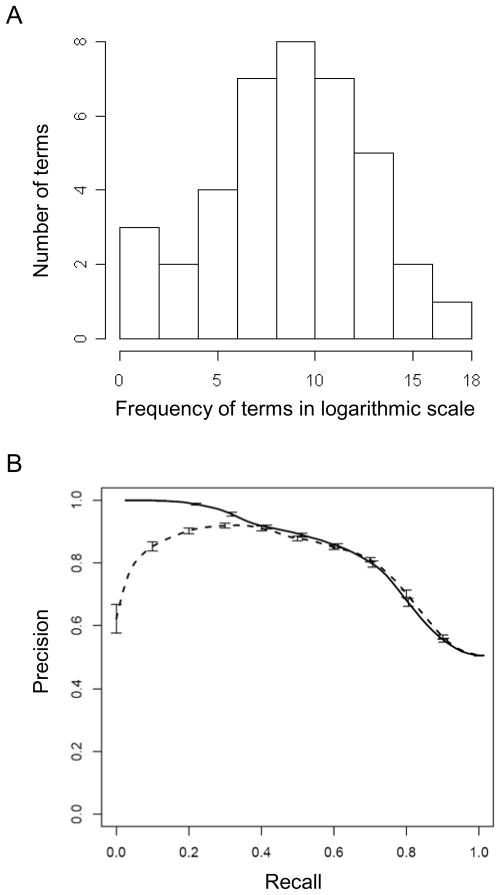
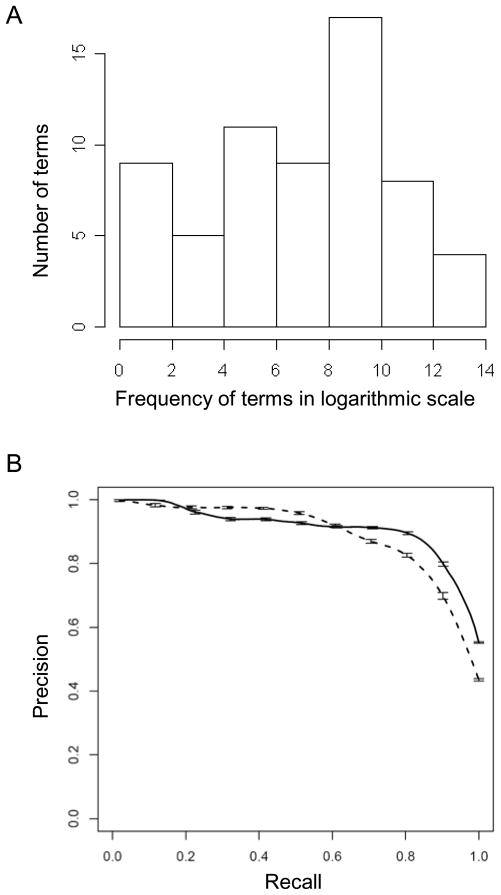
We examined 8355343 PubMed abstracts, and the number of abstracts that contained “Monosaccharide [Metabolic Process]” and “Protein Kinase [Activity]” were 4241 and 86531, respectively. Out of 56 “Monosaccharide [Metabolic Process]” children, 39 could be mapped to the PubMed abstracts, and 63 out of 97 “Protein Kinase [Activity]” children could be mapped. In addition, 588 randomly selected GO terms that had no parent-child relationship to “Monosaccharide [Metabolic Process]” and “Protein Kinase [Activity]” were used as control. The distribution of the occurrence of these terms in PubMed abstracts is shown in Figure 3A and 4A for “Monosaccharide [Metabolic Process]” and “Protein Kinase [Activity],” respectively.
Figure 3.
Hypergeometric test can be applied to identify children of a particular Biological Process ontology term in well-developed Gene Ontology. A) The occurrence of child terms of Monosaccharide Metabolic Process in PubMed Abstract. X axis is the frequency of each term in the entire PubMed abstracts in logarithmic scale. Y axis is the number of terms at a particular frequency. Terms are binned together based on their frequency. B) The Precision/Recall curve created for the overrepresentation of all the children of parent term “Monosaccharide Metabolic Process” in the abstracts that contain the parent term. This curve indicates that the hypergeometric test or Weirdness Metric indeed can identify the children (ontology terms) of the parent term (domain), and the result of this computational method is consistent with the Gene Ontology created by domain experts.
Figure 4.
Hypergeometric test can be applied to identify children of a particular Molecular Function ontology term in well-developed Gene Ontology. A) The occurrence of children of “Protein Kinase Activity” in PubMed Abstract. X axis is the frequency of each term in the entire PubMed abstracts in logarithmic scale. Y axis is the number of terms at a particular frequency. Terms are binned together based on their frequency. B) The Precision/Recall curve created for the overrepresentation of all the children of parent term “Protein Kinase Activity” in the abstracts that contain the parent term, indicating that the same enrichment test also applies to a second domain of a well-developed ontology. For all these terms, the common words “metabolic”, “catabolic”, “biosynthetic”, “process” and “activity” were removed.
The hypergeometric distribution test showed that a majority of the children of “Monosaccharide [Metabolic Process]” were overrepresented in the abstracts that contained the word “Monosaccharide [Metabolic Process]”. This also held true for the children of “Protein Kinase [Activity]”. The results from the GO analysis are shown in the Precision-Recall curves (Figure 3B and 4B, solid line). We achieved a Precision of 0.84 at the Recall level of 0.77 (F-measure=0.8) for Monosaccharide [Metabolic Process] and a Precision of 0.89 at the Recall level of 0.88 (F-measure=0.88) for Protein Kinase [Activity]. The corresponding Precision and Recall rates for the Weirdness Metric approach [17] were 0.83 and 0.77 (F-measure=0.8) for Monosaccharide Metabolic Process and 0.84, 0.88 (F-measure=0.86) for Protein Kinase [Activity], respectively, indicating our approach performed slightly better.
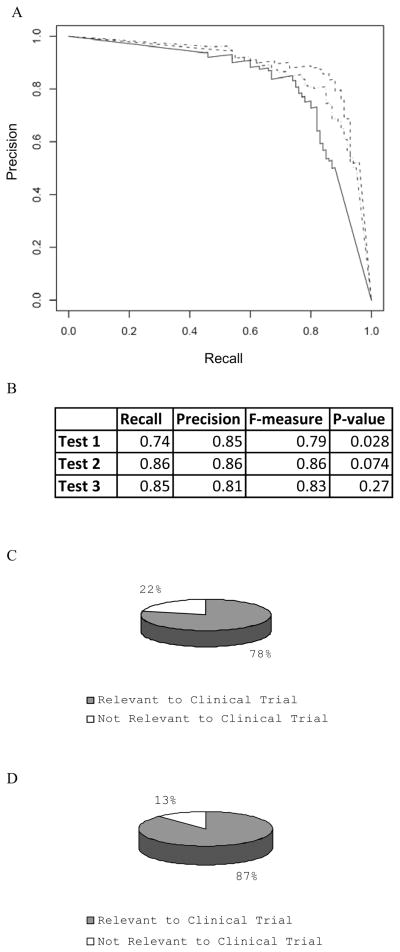
In order to apply the same test to evaluate the appropriateness of putative ontology terms for the clinical trial domain, we performed two tests. First, we tested whether selected GO terms could be separated from the putative ontology terms for the clinical trial domain. We picked 300 terms from the putative ontology term list and randomly assigned them into three groups, with each group having 100 terms. As a control, three groups of control GO terms with 100 terms each were generated. We used one group of putative ontology terms and one group of GO terms to test whether these terms were overrepresented in the DPA that contained the domain term “Clinical Trial” or “Clinical-Trial”. The test was performed for the other two pairs as well, and the Precision-Recall curve for the analysis is shown in Figure 5A. The (Precision, Recall, F-measure) for these three tests were (0.74, 0.85, 0.79), (0.86, 0.86, 0.86) and (0.85, 0.81, 0.83), respectively, indicating that the enrichment test indeed separated putative ontology terms for the clinical trial domain from control GO terms. We also selected the p-value 0.0744 that gave the highest F-measure (Figure 5B) as the threshold to evaluate all putative ontology terms downloaded from the CTO wiki. Note that this threshold functioned to separate putative ontology terms for clinical trial domain from GO terms. Using 85881 PubMed abstracts that contained the terms “Clinical Trial” or “Clinical-Trial,” we classified a term as overrepresented if its p-value from the hypergeometric test was less than 0.0744. By this standard, twenty two percent of the terms were not overrepresented while the rest, seventy eight percent, were (Figure 5C). Table 1 lists some selected representative terms and their enrichment p-values.
Figure 5.
Separation of terms for developing Clinical Trial ontology and identifying irrelevant ontology terms randomly selected from Gene Ontology. A) The Precision/Recall curves for three sets of data; each set contains the terms assembled by experts for developing the Clinical Trial ontology, as well as randomly selected ontology terms from GO. The overrepresentation of each data set was computed under the domain terms “Clinical Trial” or “Clinical-Trial”. B) p-value from three tests in A) where highest F-measures were obtained. C) Terms developed by experts are evaluated using the p-value=0.0744 as a threshold for enrichment test. The list of terms and their p-values are in the supplemental Table 1. About 22% of the terms are not overrepresented in the abstracts that have the term “Clinical Trial” or “Clinical-Trial”. D) Out of 316 terms developed by experts, there are 136 terms in the glossary of CDISC. For these 136 terms, 87% (119) are evaluated as overrepresented in the PubMed abstracts that contain “Clinical Trial” or “Clinical-Trial”. A hypergeometric test indicates that this distribution had a p-value of 0.0003, indicating our enrichment test on 316 terms indeed had a strong bias toward terms involved in clinical trials.
Table 1.
Selected putative ontology terms for clinical trial domain. Terms were assembled by domain experts and evaluated using a hypergeometric enrichment test. A threshold is determined using the p-value that generates the highest F-measure to separate clinical trial terms from irrelevant terms (Figure 5C). CDISC terms are also indicated. The full list of the terms is provided as Supplemental material.
| Terms | Included in CDISC | P-value One sided (ALL) | P-value < 0.074 | Included in CDISC with signficant p-value |
|---|---|---|---|---|
| ADVERSE EVENT | Yes | 0.00 | Yes | Yes |
| COHORT | Yes | 0.00 | Yes | Yes |
| EFFICACY | Yes | 0.00 | Yes | Yes |
| GROUP SEQUENTIAL | Yes | 0.00 | Yes | Yes |
| MISSING DATA | Yes | 0.00 | Yes | Yes |
| TRIAL-DESIGN | No | 0.01 | Yes | No |
| TREATMENT-ASSIGNMENT | No | 0.01 | Yes | No |
| INDEPENDENT REVIEW BOARD | No | 0.02 | Yes | No |
| ENROLLED POPULATION | Yes | 0.02 | Yes | Yes |
| EXCLUDED POPULATION | No | 0.02 | Yes | No |
| ASSESSMENT SCHEDULE | No | 0.03 | Yes | No |
| PERMUTED BLOCK RANDOMIZATION | No | 0.04 | Yes | No |
| DATA COLLECTION SCHEDULE | Yes | 0.05 | Yes | Yes |
| DIAGNOSTIC TRIAL | No | 0.06 | Yes | No |
| NUTRIENT | No | 0.12 | No | No |
| WITHDRAWAL CONSENT | Yes | 0.12 | No | No |
| ELECTRONIC SIGNATURE | Yes | 0.22 | No | No |
| VULNERABLE SUBJECT | Yes | 0.31 | No | No |
| DOCUMENT ROLE | Yes | 0.49 | No | No |
| SUPPLIER | Yes | 0.55 | No | No |
| POSITIVE CONTROL | No | 0.66 | No | No |
| SURVEY | Yes | 0.90 | No | No |
| NEGATIVE CONTROL | No | 1.00 | No | No |
| CASE HISTORY | Yes | 1.00 | No | No |
In order to further assess this evaluation, we took advantage of existing terms known to be relevant to the clinical trial domain. The CDISC glossary has been adopted by the FDA as the data standard for clinical trials. Therefore, terms from CDISC were used as positive controls, testing whether they were overrepresented in the DPA relevant to the clinical trial domain. We identified 136 terms in CDISC that had exact matches with the putative ontology terms downloaded from the CTO wiki. Eighty seven percent (n=119) of these 136 terms were classified as overrepresented in the DPA for clinical trial domain, where only thirteen percent (n=17) out of these 136 terms fall into the not overrepresented category (Figure 5D, and a set of selected terms are included in Table 1). A one-tailed hypergeometric test indicated that the probability for such an enrichment level to occur was 0.0003. Therefore, our approach demonstrated that terms’ overrepresentation in the DPA indicated relevance to the domain, and the method provided an objective and effective way to evaluate terms for ontology development.
4 DISCUSSION
Ontology development is typically a community effort involving many expert individuals. Their backgrounds and experience will and should affect ontology development. Despite their expertise, however, an initial term list developed by many people will contain many terms that may not ultimately be deemed relevant to the intended domain for ontology development. The algorithm we have developed provided an objective assessment of terms that could help to speed and consolidate term acquisition from a wide variety of sources. Unlike many other term extraction algorithms, the hypergeometric enrichment test used all PubMed abstracts and did not require an a priori corpus of literature for a specific domain. Our method performed slightly better than a similar approach using the Weirdness Metric [17]. In addition, since the Weirdness Metric is calculated only from the ratio of two frequencies of word occurrence, our approach provided statistical means to assess terms based on their overrepresentation as determined by a hypergeometric enrichment test.
One challenge of our approach is that some terms may not map to any PubMed abstract. While this circumstance may affect the method’s (as well as other methods’) ability to evaluate every single term important for ontology development, a significant percentage of terms indeed can be mapped to the PubMed abstracts at high or median frequencies, consistent with the observation by Verspoor et al [35]. In the relatively rare situation in which a domain term cannot be mapped to any PubMed abstract, ontology developers should consider identifying potential synonyms or semantic equivalents. We also noted that existing ontologies such as GO contain many artifacts in order to define their terms precisely. These artifacts may prevent the terms from being mapped to PubMed abstracts. However, compared to developed ontology terms, the putative terms that are hand-selected by experts contain fewer of these artifacts, are closer to the linguistic phrases used in the literature, and are suitable to be evaluated by our approach.
The diverse topics, synonyms, and semantic variants may also have significant impacts on mapping efficiency. After all, researchers often use different terms to describe research results. Despite the fact that researchers in the field of Natural Language Process and text mining have made progress in this mapping effort [14, 35–37], it remains difficult to identify all PubMed abstracts relevant to a particular term. Hypergeometric enrichment testing can provide certain degrees of tolerance to the inefficient mapping of terms to PubMed abstracts, especially since the ratio between the frequency of a term in domain specific and non-domain specific abstracts (x/(k-x) from Figure 2) is likely to be the same as the ratio for the synonyms/syntactic variants of the term. Under such a condition, the probability of observing a term in the DPA would remain at similar levels to its syntactic variants/synonyms, and the overrepresentation could still be detected.
Our approach surveys all PubMed abstracts to perform the hypergeometric test instead of focusing on a pre-determined corpus of abstracts for some specialized field. However, the hypergeometric test takes into consideration four different categories of PubMed abstracts (Figure 2). The combination of three of these categories (PubMed abstracts containing: 1) both ontology and the domain term; 2) ontology terms but not the domain term; 3) the domain term but not ontology terms) can be viewed as a corpus. Based on the distribution of ontology and domain terms among these categories, each abstract that has both the domain term and the child term increases our confidence that the two are relevant. The mapping of terms also helps us to separate commonly used terms that are widely used by the community from obscure terms that are rarely used. Terms that are present with high frequency in the abstracts may have a better chance to be adopted as ontology terms since these are common terms already widely accepted by the research community. Finally, we are aware that current mapping does not identify all relevant articles through PubMed abstracts (i.e. terms are not mentioned in the abstract but are used in the text). However, for those terms we are able to identify, our approach showed that the enrichment test can prioritize relevant terms for ontology development.
We noticed that a number of putative ontology terms were not overrepresented, and their enrichment p-values were very high. Judged by domain experts we consulted, these few terms were very general and their uses were not limited to clinical trials. As a result, these terms would likely have lower priority for ontology development in the clinical trial domain. A high p-value may also indicate the ambiguity of a term. The specificity or ambiguity of a term is typically associated with its use across many different contexts. Therefore, ambiguous terms are used for many different domains, and their degree of overrepresentation in a particular domain is less than terms with very specific meanings for that domain. For a term that is important to a domain (such as “Withdrawal Consent” and “Electronic Signature” for the clinical trial domain in Table 1), a high enrichment p-value may also suggest that a namespace would be needed to avoid ambiguity and to distinguish the use of such terms within the intended domain from its use in other domains.
Our approach may help to develop an automatic/semi-automatic approach for ontology development. Ontology development largely relies on scientific facts to depict the reality of an intended domain. Often, a set of reviewer articles and textbook chapters can capture these facts in great detail for a particular domain. Putative ontology terms can be extracted from this corpus of selected articles and evaluated against the domain using our hypergeometric enrichment test. In addition, the enrichment p-value can serve as a quantitative measure of the relevance of a putative term to the domain term. Specific (child) terms may be overrepresented to a greater degree than a general (parent) term in the DPA. Therefore, the enrichment p-value may be used as a measure to place terms within an ontology hierarchy. Such a scenario may help to improve the efficiency of the current ontology development process and alleviate burdens on ontology developers/curators.
5 CONCLUSION
We have developed a computational method that utilizes a text mining approach based on hypergeometric enrichment test to evaluate terms for ontology development. We analyzed the occurrence of terms in the PubMed database and evaluated their relevance for ontology development for a particular domain. Such an application can facilitate the current manual process for ontology development.
Supplementary Material
Acknowledgments
The work was inspired by the Ontology of Clinical Investigation project and benefited from many discussions with the group. We also would like to thank Dr. Jijun Tang to allow us to use the 152-CPU Linux cluster in the Department of Computer Science and Engineering, University of South Carolina, and Dr. Tom Smith for proof reading the manuscript. We are very grateful for the valuable suggestions by the two anonymous reviewers and the journal editors. This work is partly supported by grants IRG 97-219-08 from the American Cancer Society, a pilot project of Grant 5 P20 RR017696-05 and PhRMA Foundation Research Starter Grant to W. Jim Zheng. Ravi Patel was supported by Grant No. P20 RR016461 for the summer support from the NCRR to the SC INBRE Program. Lam C. Tsoi is supported by NLM training grant 5-T15-LM007438-02.
References
- 1.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25(1):25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Osier MV, Zhao H, Cheung KH. Handling multiple testing while interpreting microarrays with the Gene Ontology Database. BMC Bioinformatics. 2004;5(1):124. doi: 10.1186/1471-2105-5-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Al-Shahrour F, Diaz-Uriarte R, Dopazo J. FatiGO: a web tool for finding significant associations of Gene Ontology terms with groups of genes. Bioinformatics. 2004;20(4):578–580. doi: 10.1093/bioinformatics/btg455. [DOI] [PubMed] [Google Scholar]
- 4.Ahn WS, Kim KW, Bae SM, Yoon JH, Lee JM, Namkoong SE, Kim JH, Kim CK, Lee YJ, Kim YW. Targeted cellular process profiling approach for uterine leiomyoma using cDNA microarray, proteomics and gene ontology analysis. Int J Exp Pathol. 2003;84(6):267–279. doi: 10.1111/j.0959-9673.2003.00362.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Badea L. Functional discrimination of gene expression patterns in terms of the gene ontology. Pac Symp Biocomput. 2003:565–576. [PubMed] [Google Scholar]
- 6.Doniger SW, Salomonis N, Dahlquist KD, Vranizan K, Lawlor SC, Conklin BR. MAPPFinder: using Gene Ontology and GenMAPP to create a global gene-expression profile from microarray data. Genome Biol. 2003;4(1):R7. doi: 10.1186/gb-2003-4-1-r7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hvidsten TR, Laegreid A, Komorowski J. Learning rule-based models of biological process from gene expression time profiles using gene ontology. Bioinformatics. 2003;19(9):1116–1123. doi: 10.1093/bioinformatics/btg047. [DOI] [PubMed] [Google Scholar]
- 8.Troyanskaya OG, Dolinski K, Owen AB, Altman RB, Botstein D. A Bayesian framework for combining heterogeneous data sources for gene function prediction (in Saccharomyces cerevisiae) Proc Natl Acad Sci U S A. 2003;100(14):8348–8353. doi: 10.1073/pnas.0832373100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhong S, Li C, Wong WH. ChipInfo: Software for extracting gene annotation and gene ontology information for microarray analysis. Nucleic Acids Res. 2003;31(13):3483–3486. doi: 10.1093/nar/gkg598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Baehrecke EH, Dang N, Babaria K, Shneiderman B. Visualization and analysis of microarray and gene ontology data with treemaps. BMC Bioinformatics. 2004;5(1):84. doi: 10.1186/1471-2105-5-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boyle EI, Weng S, Gollub J, Jin H, Botstein D, Cherry JM, Sherlock G. GO::TermFinder--open source software for accessing Gene Ontology information and finding significantly enriched Gene Ontology terms associated with a list of genes. Bioinformat-ics. 2004;20(18):3710–3715. doi: 10.1093/bioinformatics/bth456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Khatri P, Bhavsar P, Bawa G, Draghici S. Onto-Tools: an ensemble of web-accessible, ontology-based tools for the functional design and interpretation of high-throughput gene expression experiments. Nucleic Acids Res. 2004;32(Web Server issue):W449–456. doi: 10.1093/nar/gkh409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pavlidis P, Qin J, Arango V, Mann JJ, Sibille E. Using the gene ontology for microarray data mining: a comparison of methods and application to age effects in human prefrontal cortex. Neurochem Res. 2004;29(6):1213–1222. doi: 10.1023/b:nere.0000023608.29741.45. [DOI] [PubMed] [Google Scholar]
- 14.Daille B. Study and Implementation of Combined Techniques for Automatic Extraction of Terminology. In: Resnik P, Klavans J, editors. The Balancing Act: Combining Symbolic and Statistical Approaches to Language. Cambridge, MA: MIT Press; 1996. pp. 49–66. [Google Scholar]
- 15.Frantzi K, Ananiadou S, Mima H. Automatic Recognition of Multi-Word Terms: the C-value/NC-value Method. International Journal on Digital Libraries. 2000;3(2):115–130. [Google Scholar]
- 16.Grabar N, Zweigenbaum P. A general method for sifting linguistic knowledge from structured terminologies. Proc AMIA Symp. 2000:310–314. [PMC free article] [PubMed] [Google Scholar]
- 17.Ahmad K, Rogers MA. Corpus Linguistics and Terminology Extraction. In: Wright S, Budin G, editors. Handbook of Terminology Management. Vol. 2. Amsterdam & Philadelphia: John Benjamins Publishing Co; 2001. pp. 725–760. [Google Scholar]
- 18.Savova GK, Harris M, Johnson T, Pakhomov SV, Chute CG. A data-driven approach for extracting "the most specific term" for ontology development. AMIA Annu Symp Proc. 2003:579–583. [PMC free article] [PubMed] [Google Scholar]
- 19.Cimiano P, Volker J. Text2Onto - A Framework for Ontology Learning and Data-driven Change Discovery. 10th International Conference on Applications of Natural Language to Information Systems (NLDB 2005); 2005; Alicante, Spain: Springer; 2005. pp. 227–238. [Google Scholar]
- 20.Smith L, Wilbur WJ. Retrieving definitional content for ontology development. Comput Biol Chem. 2004;28(5–6):387–391. doi: 10.1016/j.compbiolchem.2004.09.007. [DOI] [PubMed] [Google Scholar]
- 21.Castro AG, Rocca-Serra P, Stevens R, Taylor C, Nashar K, Ragan MA, Sansone SA. The use of concept maps during knowledge elicitation in ontology development processes--the nutrigenomics use case. BMC Bioinformatics. 2006;7:267. doi: 10.1186/1471-2105-7-267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Alexopoulou D, Wachter T, Pickersgill L, Eyre C, Schroeder M. Terminologies for text-mining; an experiment in the lipoprotein metabolism domain. BMC Bioinformatics. 2008;9 (Suppl 4):S2. doi: 10.1186/1471-2105-9-S4-S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Navigli R, Velardi P. Learning Domain Ontologies from Document Warehouses and Dedicated Web Sites. Computational Linguistics. 2004;30(2):151–179. [Google Scholar]
- 24.Li Q, Shilane P, Noy NF, Musen MA. Ontology acquisition from on-line knowledge sources. Proc AMIA Symp. 2000:497–501. [PMC free article] [PubMed] [Google Scholar]
- 25.Camon E, Magrane M, Barrell D, Binns D, Fleischmann W, Kersey P, Mulder N, Oinn T, Maslen J, Cox A, et al. The Gene Ontology Annotation (GOA) project: implementation of GO in SWISS-PROT, TrEMBL, and InterPro. Genome Res. 2003;13(4):662–672. doi: 10.1101/gr.461403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rivals I, Personnaz L, Taing L, Potier MC. Enrichment or depletion of a GO category within a class of genes: which test? Bioin-formatics. 2007;23(4):401–407. doi: 10.1093/bioinformatics/btl633. [DOI] [PubMed] [Google Scholar]
- 27.OCI. http://www.bioontology.org/wiki/index.php/OCI:Main_Page.
- 28.Boyer R, Moore J. A fast string searching algorithm. Communications of the ACM. 1977;20:762–772. [Google Scholar]
- 29.Lieberman GJ, Owen DB. Tables of the Hypergeometric probability distribution. Stanford, Calif: Stanford University Press; 1961. [Google Scholar]
- 30.Agresti A. Categorical Data Analysis. 2. Hoboken, New Jersey: John Wiley & Sons, Inc; 2002. [Google Scholar]
- 31.R Development Core Team. R: a language and environment for statistical computing. 2007 http://www.R-project.org.
- 32.GNU Scientific Library
- 33.Goswami R, Singh D, Phillips G, Kilkus J, Dawson G. Ceramide regulation of the tumor suppressor phosphatase PTEN in rafts isolated from neurotumor cell lines. J Neurosci Res. 2005;81(4):541–550. doi: 10.1002/jnr.20550. [DOI] [PubMed] [Google Scholar]
- 34.http://www.cdisc.org/index.html
- 35.Verspoor CM, Joslyn C, Papcun GJ. The Gene Ontology as a Source of Lexical Semantic Knowledge for a Biological Natural Language Processing Application. SIGIR'03 Workshop on Text Analysis and Search for Bioinformatics; August, 1st 2003; Toronto, CA. 2003. pp. 51–56. [Google Scholar]
- 36.Justeson JS, Katz SM. Principled disambiguation: discriminating adjective senses with modified nouns. Computational Linguistics. 1995;21(1):1–27. [Google Scholar]
- 37.Baumgartner WA, Jr, Cohen KB, Hunter L. An open-source framework for large-scale, flexible evaluation of biomedical text mining systems. J Biomed Discov Collab. 2008;3:1. doi: 10.1186/1747-5333-3-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.