Abstract
Splicing of RNA polymerase II (polII) transcripts is a crucial step in gene expression and a key generator of mRNA diversity. Splicing and transcription have been generally been studied in isolation, although in vivo pre-mRNA splicing occurs in concert with transcription. The two processes appear to be functionally connected because a number of variables that regulate transcription have been identified as also influencing splicing. However, the mechanisms that couple the two processes are largely unknown. In this review, I highlight the observations that implicate splicing as occurring during transcription and describe the evidence supporting functional interactions between the two processes. I discuss postulated models of how splicing couples to transcription and consider the potential impact that such coupling might have on exon recognition.
Pre-mRNA splicing is connected to transcription in vivo
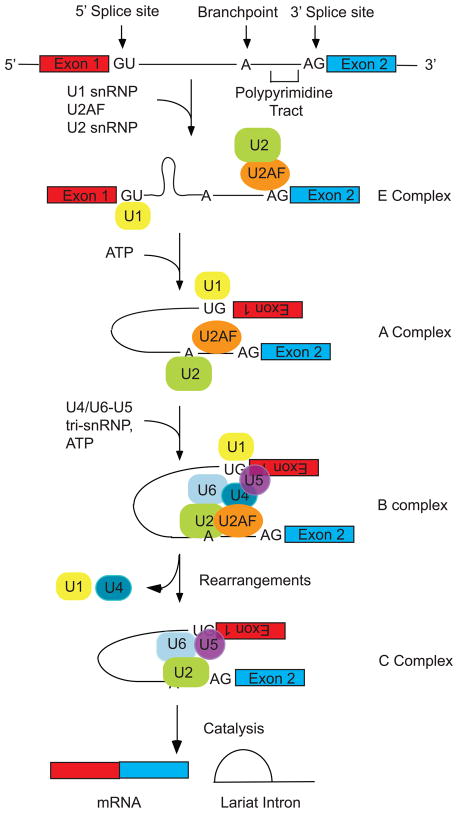
Precursor messenger RNA (pre-mRNA) splicing is the process by which intron sequences are identified and excised from pre-mRNA transcripts with concurrent ligation of the flanking exons. The splicing reaction is catalyzed by the spliceosome, a large ribonucleoprotein (RNP) complex that forms from the ordered assembly of five small nuclear ribonucleoprotein (snRNP) particles onto each intron (1, 2)(Figure 1). Four essential RNA elements direct spliceosome assembly (Figure 1). They are: the 5′ and 3′ splice sites that define the exon-intron and intron-exon junctions respectively, the polypyrimidine tract which aids in 3′ss recognition and the branchpoint sequence that provides the attacking 2′ hydroxyl in the first of the two transesterification reactions that comprise the chemical steps of splicing (for a complete review see (1)).
Figure 1. The spliceosome assembles in an ordered, step-wise manner in vitro.
In the E (early or commitment) complex the U1 snRNP binds the 5′ss via base pairing interactions, U2AF recognizes the PPT and additional factors bring the 5′ss and 3′ss into juxtaposition. This pairing is stabilized by addition of ATP, which allows binding of the U2 snRNP to the BP to form the A complex. The U4/U6-U5 tri-snRNP then joins as a single unit to form the B complex. Multiple ATP-dependent rearrangements result in the release of the U1 and U4 snRNPs and formation of the C complex, which catalyzes excision of the intron as a lariat and ligation of the exon sequences.
In humans, almost all intron-containing genes generate multiple mRNA isoforms through differential inclusion of alternative exons (3, 4). Splicing proteins regulate splice site choice by binding to RNA elements within exons or introns to promote or impede spliceosome assembly (5). Some splicing factors are developmentally or tissue-restricted in expression (5, 6), while others, such as those in the Serine/Arginine-rich (SR) family, are more ubiquitously expressed and exert more fundamental roles in general splicing (7, 8).
Mechanistic regulation of pre-mRNA splicing is generally studied independently of transcription, using pre-synthesized templates added to nuclear extracts (9–11)or injected into living cells (12). Experiments of this kind have established that active transcription, while necessary for RNA synthesis, is not required for spliceosome assembly or intron excision in vitro. Yet, in vivo splicing and transcription are intimately connected, and it is well accepted that spliceosome assembly initiates on pre-mRNAs that are still being synthesized, or that may be complete but remain tethered to the chromatin template (13–15).
A number of variables known to control transcription have recently been implicated in altering splicing outcomes. These include: RNA polymerase II (polII) and its Carboxy-Terminal Domain (CTD), promoter sequences, transcriptional activators, chromatin remodelers and short interfering RNAs (siRNAs)(16–22). These findings have generated considerable interest in understanding how splicing and transcription might influence one another. The two processes are presumed to functionally couple through mechanisms involving polII (20, 22), although testing this idea is not trivial because many of the variables described as affecting splicing are not easily reconstituted in vitro. This drawback has hindered efforts using tractable systems to understand the mechanism(s) of coupling. As a result, existing models for how these variables influence splicing remain speculative and elucidation of the regulatory mechanisms that connect splicing to transcription remains a major goal in the field. In this review, I focus primarily on work performed in mammalian systems, although much important research in this field is also being pursued in other organisms (23–26). I present the data that reveal nascent transcripts are spliceosomal substrates. I describe the variables that are known to influence splicing during transcription, discuss possible mechanisms for how coupling might be achieved, as well as consider the consequences of such coupling on exon recognition.
NASCENT pre-mRNAs ARE SUBSTRATES FOR THE SPLICEOSOME
The spliceosome assembles on nascent RNAs during transcription
The notion that splicing initiates during transcription was introduced 25 years ago when Osheim and Beyer published electron micrographs of Miller chromatin spreads from early D. melanogaster embryos (13). The micrographs depict protein particles assembled upon nascent RNAs still attached to the DNA template (13). Some particles appear to coalesce and in so doing, loop out the intervening sequence in a process reminiscent of splicing (27). Results showing that antibodies against snRNP, SR and hnRNP proteins localize to transcriptionally active regions of large insect polytene salivary chromosomes supported the presumption that these protein particles are spliceosomal complexes (28–30). In mammalian cells, transcriptional activation similarly results in the migration of splicing factors from nuclear speckles to sites of nascent RNA synthesis (31–33)and the accumulation of splicing factors at active gene loci(15). Interestingly, splicing factor deposition is not observed at single exon gene loci, bolstering the idea that splicing factors accumulate on intron-containing RNAs (15). Collectively, these data support a scenario in which spliceosomal components assemble on nascent pre-mRNAs still tethered to the chromatin template.
Quantifying the extent of intron removal in nascent pre-mRNAs
Intron excision is executed by the catalytic spliceosome that forms after multiple snRNP assembly steps and ATP-driven rearrangements(1). That spliceosomal components assemble on nascent RNAs suggests that splicing occurs co-transcriptionally, but does not confirm that introns are excised from pre-mRNAs during synthesis. However, quantifying the extent of intron removal in nascent pre-mRNAs is often limited by the difficulty of purifying such transient molecules.
An early approach to measuring co-transcriptional intron removal was though micro-dissection of chromatin puffs, regions of insect polytene chromosomes undergoing active transcription known as ‘Balbiani Rings’ (BR) (34). Analysis of nascent RNA isolated from contiguous segments of the BR1 gene found that intron 3 is completely excised prior to transcriptional termination while the fourth and terminal intron, which is positioned within 1kb of the polyadenylation site, is only excised in 5–10% of the same transcripts (35). Inspection of the BR3 transcript demonstrated that splicing generally proceeds in a 5′ to 3′ direction, although neighboring introns are not always excised in this order, nor are all introns completely removed during synthesis (36).
Similar results have been documented in mammals. Dystrophin (DMD) is one of the longest known human genes (2.3Mb) and is predicted to take over 16 hours to transcribe (37). DMD is active in myoblasts as they differentiate into myotubes. RT-PCR analysis of myoblast RNA shows detectable levels of exon 3-containing transcripts after 8 hours of differentiation. The exon 2 – 3 spliced product is detectable 12 hours prior to that of exon 69, showing that excision of 5′ proximal introns is achieved prior to complete synthesis of the DMD transcript. Further investigation has determined that splicing of the DMD pre-mRNA progresses in a general 5′ to 3′ direction (37). This result is corroborated by quantitative measurements of intron removal along the c-Src and Fibronectin (FN) transcripts in isolates of chromatin-associated RNA (14, 38). In this study, intron flanking alternative exons within the c-Src and FN transcripts were also excised prior to release of the transcript from the chromatin template. Interestingly, the order of removal for introns neighboring regulated exons does not proceed in an exclusive 5′ to 3′ direction, as illustrated by the alternative FN exons 25 and 33 in which the downstream introns are removed prior to the upstream ones under steady state conditions (14, 39). This pattern of intron removal is also found in some neural-specific alternative splicing events (40).
Measuring the rate of splicing relative to transcription provides information on how quickly introns are excised after being transcribed. One approach to doing this involves clearing nascent RNAs from the chromatin by treating cells with 5,6-dichlorobenzimidazole 1-beta-D-ribofuranoside (DRB). DRB inhibits the Positive Transcriptional Elongation Factor b (P-TEFb) (41). Phosphorylation of polII by p-TEFb facilitates the transition from transcriptional initiation to elongation and as such, DRB is thought to prevent transcriptional initiation complexes entering active elongation but does not appear to impede actively elongating molecules from completing transcription of a locus (41). Time course measurements by RT-PCR of multiple pre-RNA species synthesized after reversing 3 hours of DRB inhibition have shown that intron excision occurs within 5 – 10 minutes of synthesis of the downstream neighboring exon, irrespective of the length of the intron, the length of the gene or whether the intron is excised by the major or minor spliceosome (42).
An additional approach to inferring splicing kinetics harnesses the power of Fluorescence Recovery After Photobleaching (FRAP). FRAP measurements in mammalian cells reveal that the diffusion rate of GFP-tagged spliceosomal factors is up to 17 fold lower when measured at a transcriptionally active locus compared to the average rate of diffusion of the protein in the nucleoplasm (43). The decrease in diffusion rate has been interpreted as retention of the snRNPs at sites of transcription though binding of nascent RNAs. The time spent at the active loci can also be computed from the diffusion rate. The U2 and U5 snRNPs are present at the active loci for between 15 and 30seconds, while the U1, U4 and U6 snRNPs are found in residence for much shorter periods, suggesting that spliceosome assembly proceeds rapidly on nascent RNA and that intron excision might occur much faster than previously estimated (42, 43).
Collectively, these data confirm that nascent pre-mRNAs are splicing substrates and indicate that at least a majority of introns are recognized rapidly during transcription. Moreover, a large proportion of introns flanking both constitutive and alternative exons are seemingly excised prior to release of the RNA from the chromatin template.
TRANSCRIPTIONAL REGULATORY VARIABLES THAT IMPACT SPLICING DURING TRANSCRIPTION
The Carboxy-Terminal Domain (CTD) of RNA polymerase II
In mammals, the CTD comprises over half of the largest polII subunit, Rpb1 (POLR2A). It is composed of 364 amino acids grouped into 52 heptad repeats that form a consensus sequence of Tyr1-Ser2-Pro3-Thr4-Ser5-Pro6-Ser7 (44, 45)and is subject to extensive post-translational modification (46–50).
Initial evidence for involvement of polII in splicing regulation emerged from the observation that cells expressing a version of polII that lacks the CTD but retains transcriptional capabilities, do not support splicing or 3′ end cleavage of a reporter substrate (20). These cells also exhibit altered localization of splicing factors and reduced mRNA expression from endogenous genes (51, 52). In vitro, splicing of a reporter substrate is improved by addition of purified polII and this effect is dependent upon hyperphosphorylation of the CTD (53). These data implicate polII, and the CTD in particular, in splicing regulation.
Experiments testing the ability of the CTD to support splicing in an exogenous setting found that splicing of a reporter substrate transcribed by T7 RNA Polymerase (T7 RNAP) fused to the polII CTD is not enhanced when compared to transcription by wild type T7 RNAP (54). A similar result is obtained when tested in the context of RNA polymerase III (54), demonstrating that the CTD is only effective at coordinating splicing in the context of the polII holoenzyme.
Rate of transcription by RNA Polymerase II
In early studies examining how the order of exon synthesis enforced the linear order of exon ligation, Aebi and Weissmann noted that a set of 5′ss mutations in intron 2 of a 3 exon construct did not inhibit intron 2 excision, but instead resulted in skipping of exon 2 and ligation of exon 1 to exon 3 (55). While it is now recognized that exon skipping is the most common result of a splice site mutation (56), Aebi and Weissmann explained their findings by proposing a ‘first come, first served’ (FCFS) model, in which splicing commitment complexes would assemble during transcription to unite each cognate pair of splice sites as they were synthesized (55). They hypothesized that this would remove committed upstream junctions from competition with splice sites downstream, thereby preventing missplicing of multi-exon transcripts.
Experiments specifically examining the regulation of an alternative exon during transcription also support variations of the FCFS model. A splicing minigene carrying the FN alternative exon 33 exhibits increased use of the upstream alternative splice site when transcribed with a mutant ‘slow’ polymerase (22). While this effect is not observed with all test exons, a subset of regulated exons are clearly affected by modulations in transcriptional rate(22).
Promoter-related effects
Regulated exon inclusion in mRNAs produced from minigene reporter constructs is affected by variables involved in transcriptional initiation. For instance, inclusion of the FN alternative exon 33 varies substantially in mRNAs transcribed from minigene constructs driven by different promoter sequences (19). This change is not dependent on the overall expression from the promoter, since inclusion of exon 33 does not change upon induction of a CMV promoter with cyclic AMP, or an MMTV promoter with dexamethasone (19). One explanation for these observations is that different promoters might recruit transcriptional complexes of varying composition that can alter splicing outcomes.
TheU2AF-related CAPERα and β (co-activator of activating protein-1 and estrogen receptors) proteins bind and stimulate transcription from steroid-responsive promoters (18). When co-transfected into hormone-treated cells with a splicing sensitive minigene driven by a steroid responsive promoter, CAPERα and β also alter alternative exon inclusion (18). Although their mechanism(s) of action are not entirely clear, CAPERs encode RS and RNA recognition motif (RRM) domains that may permit them to interact with the RNA or other spliceosomal proteins (18).
The control of transcriptional regulatory factors on splicing is not limited to regulated events. Splicing of transcripts with a single splicing pattern is enhanced when activated by the strong chimeric Gal4-VP16 fusion rather than the weaker Gal4-SW6 protein (57). The polypyrimidine tract binding protein-associated splicing factor (PSF) and related p54nrb/NonO protein preferentially bind the VP16 over the SW6 activation domain, demonstrating that transcriptional activators can indeed interact with auxiliary splicing components (58). PSF further enhances splicing of a Gal4-VP16 dependent splicing substrate, indicating that the VP16-PSF interaction is functional (58). These data show that promoters and the proteins that bind to them can influence splicing outcomes.
Chromatin-related effects
SWI/SNF is a multi-subunit enzyme that remodels nucleosomes around promoter regions to allow binding of transcriptional regulatory proteins (59). Transient transfection of cells with the Brahma (Brm) subunit of mammalian SWI/SNF alters inclusion of a set of alternative exons (17). Notably, the affected transcripts are encoded by genes dependent on SWI/SNF remodeling for activation (17). Brm interacts with the U1 and U5, but not the U3 snRNAs, as detected by RT-PCR of RNA from Brm immunoprecipitates. Brm’s effect on splicing does not require its ATPase domain, suggesting that this function is independent of its remodeling ability (17). Thus, not unlike the transcriptional activators that affect splicing, it may be that Brm influences alternative exon inclusion by binding to splicing factors to interfere with spliceosome assembly rather than through altering chromatin structure.
The notion that chromatin packing can alter splicing outcomes garners support from an epigenetic process known as transcriptional gene silencing (TGS) (60–62). TGS is induced by short interfering RNAs (siRNA) targeted to promoter regions that shut off gene expression (63). The siRNAs induce methylation of cytosines within the DNA and H3K9 methylation at the promoter, via a mechanism that is dependent on the argonaute-1 (ago-1) and Heterochromatin Protein 1 (HP1) proteins (64–66). SiRNAs targeted to the intron downstream of FN exon 33 produce a two-fold increase in inclusion of endogenous FN exon 33 (16). Much like TGS, this effect is dependent on polII elongation, ago-1 and -2 and HP1a. The siRNAs also increase H3K9 di-methylation and H3K27 tri-methylation across proximal downstream regions from the siRNA target site in intron 33.
In contrast, depolarization of neuronal N2A cells with KCl reduces inclusion levels of exon 18 from Neuronal Cell Adhesion Molecule (NCAM) mRNA and increases H3K9 acetylation and H3K36 methylation around the exon 18 region (67). These chromatin modifications are associated with relaxed chromatin as determined by DNA sensitivity to nuclease digestion. The pathway from depolarizing stimulus to the DNA is not yet clear, but it appears to be independent of CamKIV, which was also shown to mediate a depolarization-induced signalling cascade that represses exon inclusion (68).
With the advent of high throughput sequencing technology, additional interesting connections between chromatin structure and splicing have been described. Whole genome chromatin immunoprecipitation and sequencing (ChIP-seq) from human T-cells have provided the first high-resolution map of nucleosome distribution along chromosomes (69). Multiple bioinformatic analyses of ChIP-seq data found that exon sequences, but not introns, have a approximately 1.5 fold enrichment of nucleosomes above background (69–76), which appears to be dependent on the biased nucleotide composition of exons (70). After controlling for nucleosome density, the same studies discovered higher than average levels of di-and tri-methylated H3K36 and mono-methylated H2BK5 in nucleosomes residing within exons. Exons also exhibited lower than average tri-methylation of H3K9 (70). Additional work has further correlated distinct sets of histone modifications with specific intragenic locations. For example, H3K4me3 is strongly associated with transcriptional start sites. H3K79me2 is found downstream of promoter sequences and displays a reciprocal relationship with H3K36me3, which is most abundant across regions encoding internal exons (77).
These data have led to the enticing idea that nucleosomes and certain histone modifications around exons are signals that aid in the recognition of exons. There are reports of chromatin modifications affecting splicing factor association (78, 79), although at this point, nearly all the data is correlative. It will be very important to develop experimental approaches to examine how chromatin affects exon recognition by the spliceosome.
HOW MIGHT SPLICING COUPLE TO TRANSCRIPTION?
The data described above clearly indicate that a number of variables known to regulate transcription also affect alternative splicing events. However, mechanistic understanding of how these variables exert their influence on splicing is elusive. This can be attributed in part to the fact that faithfully reconstituting coupled transcription-splicing conditions in vitro is extremely challenging. Current systems use short, naked DNA templates transcribed by polII from strong viral promoters (80–82). These setups can interrogate certain aspects of how transcription might influence splicing, but they fall short of replicating the endogenous environment. More significantly, it is difficult to demonstrate that a system is truly coupled. Splicing and transcription that occur simultaneously in the same reaction is not sufficient evidence for coupling, because they can proceed independently of one another (83). Furthermore, that a transcription regulatory factor can alter splicing suggests a connection between the two processes, but because the same protein can perform independent functions in similar settings, it does not confirm that coupling exists (84). Biochemically coupled reactions share a common intermediate and have an overall free energy of reaction equal to, or less than, zero (85). Pre-mRNA fulfills the common intermediate requirement because it is the product of transcription and the substrate for splicing. However, there is little evidence to support energetic coupling between the two processes. Thus, it is important to remember that the evidence in support of coupling between splicing and transcription is largely functional rather than biochemical.
A couple of models have been proposed to account for how some of the variables might exert their influence on splicing regulation and are discussed below.
The recruitment hypothesis
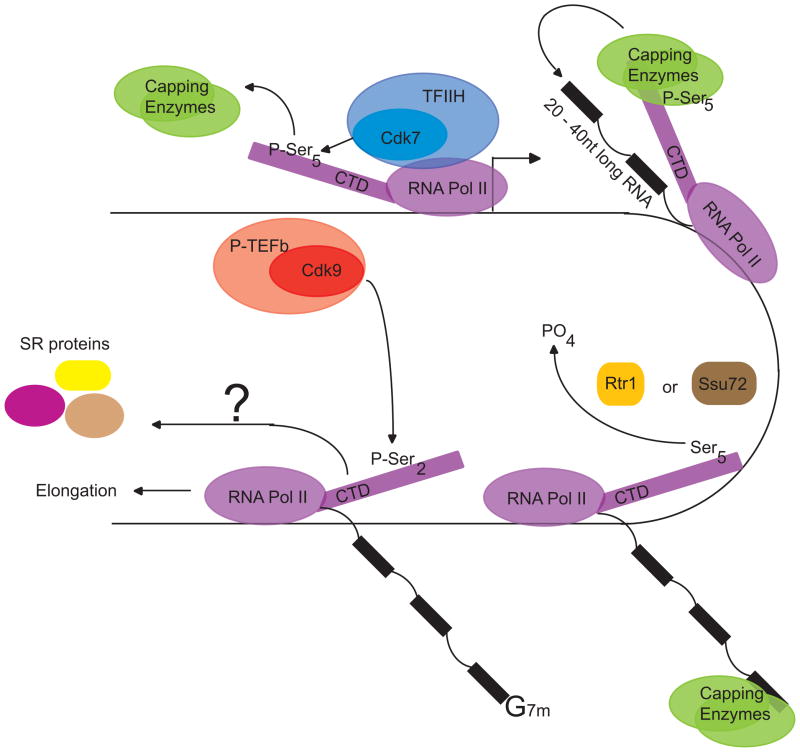
The finding that a version of polII lacking the CTD does not support splicing or 3′end cleavage of a reporter substrate (20)has lead to much work focused on the role of the CTD in mRNA maturation. How the CTD might function in this capacity is best understood in the context of pre-mRNA capping (Figure 2). The heptad repeats of the CTD each contain three serines that are targets for post-translational phosphorylation. Phospho-Ser2 and Ser5 and marks are associated with synthesis of protein-coding genes (48). Phospho-Ser7 is detected on polII molecules transcribing both protein-coding and snRNA genes (50, 86). The cdk7 (kin28 protein in yeast) subunit of transcription factor IIH (TFIIH) phosphorylates Ser5 residue(s) on polII molecules located within promoter regions of target genes (48, 87). The capping enzymes, which add the 7-methylguanosine cap to the 5′end of pre-mRNAs, are recruited to the promoter by binding to the CTD in a Kin28-dependent manner (88–91). In this instance, phosphorylation of the CTD enables recruitment of capping enzymes and thereby co-ordinates the 5′ processing of nascent RNAs with transcriptional initiation.
Figure 2. CTD phosphorylation recruits mRNA capping enzymes.
The cdk9 subunit of TFIIH phosphorylates Ser5 residues on the CTD of promoter proximal RNA polII molecules. The pre-mRNA capping enzymes bind to the CTD in a phospho-Ser5 dependent manner and catalyzes cap addition on the 5′ end of the pre-mRNA. Ser5 residues are dephosphorylated sometime after transcriptional initiation, probably by the Rtr1 or SSu72 phosphatase(s). Phospho-Ser2 marks are detected on elongating polymerases located within genes. The cdk9 subunit of p-TEFb is the Ser2 kinase. P-TEFb binds to promoter sequences and stimulates transcriptional elongation, but it is not known whether it has to be promoter bound to phosphorylate the CTD. RNA polymerase II can interact with SR proteins through recruitment mechanism(s) that are unclear, as indicated by a question mark.
The ability of the CTD to bind capping enzymes at a specific stage of the transcriptional process has provoked speculation that a similar mechanism might function to regulate splicing. This has led to a hypothesis, known as the recruitment model, which posits that the multiple modifications and conformations adopted by the CTD form a physical code that directs the dynamic engagement of RNA processing factors (20).
Support for the recruitment model as it pertains to splicing regulation comes from a few main observations. The first is that transcriptional induction causes splicing factors to localize to, and accumulate at, active gene loci (15, 31, 32). Re-localization is disrupted when transcription is executed by polII lacking the CTD. This implicates the CTD in mediating re-localization of splicing factors upon transcriptional induction(51).
Additionally, multiple studies have reported interactions between polII and RS-domain containing proteins. For instance, polII is found in immunoprecipitates of the SR family of proteins (mAb 104), and sm proteins (mAb Y12) from Hela nuclear extract (92). In another study, a transcriptionally active polII-complex purified over an SII column from Hela cell extract contains the U1, U2 and U4 snRNPs, U2AF65 and well as many SR proteins (93). In addition, some RS-domain containing proteins have been shown to bind to the CTD specifically(94, 95).
Lastly, the polII CTD displays phosphorylation patterns that appear restricted to protein-coding regions of genes. The phospho-Ser2 mark is found predominantly on polII molecules located downstream of the promoter (48). Phosphorylation at this residue is catalyzed by p-TEFb, which is required for the transition from transcriptional initiation to elongation (41, 96). The findings that hyperphosphorylated polII enhances splicing when added to an in vitro splicing reaction (53), and phospho-Ser2 residues are associated with the elongation phase of transcription (48), has lead to the idea that this moiety might mediate recruitment of splicing factors to the RNA during elongation, analogously to the function of phospho-Ser5 residues in facilitating pre-mRNA capping during transcriptional initiation.
While it is clear that the CTD is involved in splicing regulation, and it is tempting to adopt the hypothesis that the recruitment model explains aspects of splicing regulation during transcription, there are some outstanding issues that must first be addressed.
It has been shown that RS-domain containing proteins can interact with polII (92, 93, 97)and splicing factors like SRSF3 can regulate splicing in a CTD dependent manner (98). Yet, there is scant evidence for legitimate contacts between defined splicing factors and the polII CTD. Interactions that have been described between polII and U2AF65 or GFP fusions of multiple SR proteins are RNase sensitive and suggest that spliceosomal components interact with polII via an RNA tether rather than through direct protein-protein contacts (15, 99, 100). If it can be shown that splicing factors do indeed bind the CTD, it will also be important to determine whether recruitment is dependent upon phosphorylation of Ser2.
To date, much of the evidence for the recruitment model is based upon immunological detection methods. One caveat of this technique is that, without cross-linking, proteins may re-associate with one another after cell lysis and that protein-protein interactions detected via co-immunoprecipitation may not occur in vivo (101). Moreover, antibodies raised against different epitopes within the same protein will likely display disparate binding characteristics and when combined with varied incubation, buffer and wash conditions, it is perhaps not surprising that independent studies have each identified different sets of polII-interacting proteins (93, 97, 100). As a result, it is still unclear which, if any, splicing factors interact with polII and whether splicing factor accumulation on nascent RNA is specifically coordinated by polII or is simply a function of their high nuclear concentration (102). The increase in splicing efficiency observed with polII transcripts synthesized in vitro can also be attributed to the fact that polII transcripts display superior stability over those made by T7 RNAP (81).
A great deal of emphasis has been placed on the role of polII in connecting splicing to transcription, yet some reports suggest that SR proteins are equally necessary. For example, splicing of an adenoviral transcript in a seemingly coupled system is inhibited if transcription occurs in extracts depleted of SR proteins (100). This result is not unexpected, since SR proteins are required for splicing in vitro (103–105)and in vivo (106). What is striking is that add back of SR proteins rescues splicing only if addition occurs before, but not after, the onset of transcription. This results implies that that communication between the splicing and transcriptional machineries begins prior to transcriptional initiation and that SR proteins play an integral role in connecting the two processes (100). Additionally, in vivo accumulation of nascent RNA is lower in cells deleted of SRSF2, as detected by H3 labeling (107). ChIP-DNA selection and ligation (DSL) technology (108)detected higher polII density within the coding regions of some genes from the SRSF2 knock out cell lines, suggesting that the decrease in nascent RNA is due to less transcription rather than more RNA degradation. Antibodies against SRSF2 can immunoprecipitate cdk9 and in the absence of SRSF2 polII association with P-TEFb is dramatically reduced, showing that SRSF2 can enhance transcriptional elongation and facilitate the interaction between polII and pTEFb (107). Thus, these studies demonstrate a requirement for SR proteins in connecting splicing to transcription.
The kinetic model
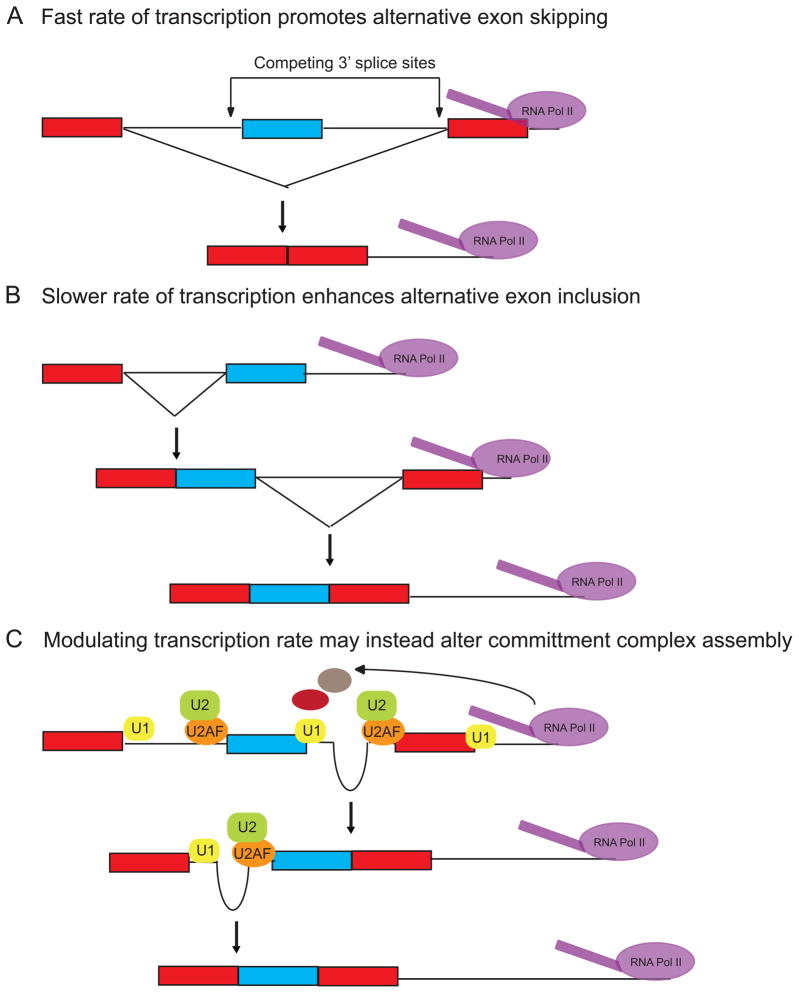
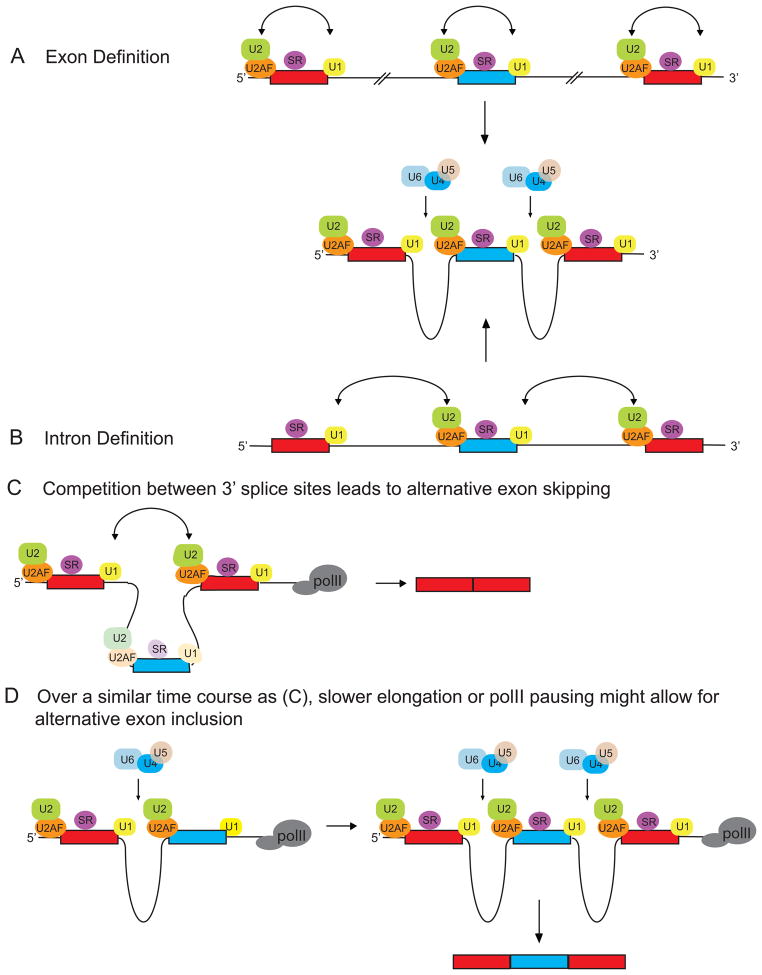
The ‘kinetic model’ of splicing regulation posits that alternative exon use can be regulated through adjustments to the polII elongation rate (22)(Figure 3A, 3B). A prediction of this model is that synthesis of an intron downstream of an alternative exon will lag that of the upstream one when transcribed by a slower polymerase (Figure 3B). This lag should afford the spliceosome more time to assemble upon and excise the upstream intron without competition from splice sites further downstream. A slower transcriptional rate will thus promote increased exon inclusion while faster transcriptional rates will result in near simultaneous presentation of both flanking introns to the splicing machinery. According to the model, the presence of a stronger, more rapidly assembling downstream 3′splice site would favor skipping of the alternative exon with a weaker 3′ splice site (Figure 3A). In this way, any factor that modulates the rate of transcription can conceivably change regulated splicing outcomes as well.
Figure 3. A slower rate of transcriptional elongation promotes alternative exon inclusion.
(a) Polymerases working at a relatively fast rate of transcription will rapidly transcribe the alternative region, such that the3′splice site of the alternative and downstream constitutive exons are both available for pairing with the 5′ss of the upstream constitutive exon. Competition between the stronger constitutive and weaker alternative 3′ splice sites will favor use of the downstream constitutive 3′ss and promote alternative exon skipping.
(b) The kinetic model posits that a slower transcriptional elongation rate will result in a longer time lapse between synthesis of the upstream alternative exon and the downstream constitutive one than if transcribed by a faster polymerase. This lag will afford the spliceosome more time to recognize the weaker 3′ss of the alternative exon without competition from the downstream constitutive exon, resulting in increased alternative exon inclusion. Pausing of the polymerase within the intron downstream of the alternative exon could similarly permit increased splicing of the regulated exon.
(c) In the case of FN alternative exon 33, the downstream intron is excised prior to the upstream one. In this case, it is thought that the rate of transcription alters commitment complex assembly on the pre-mRNA rather than affecting the order of intron excision per se. In this case, splicing choices would be decided prior to intron excision.
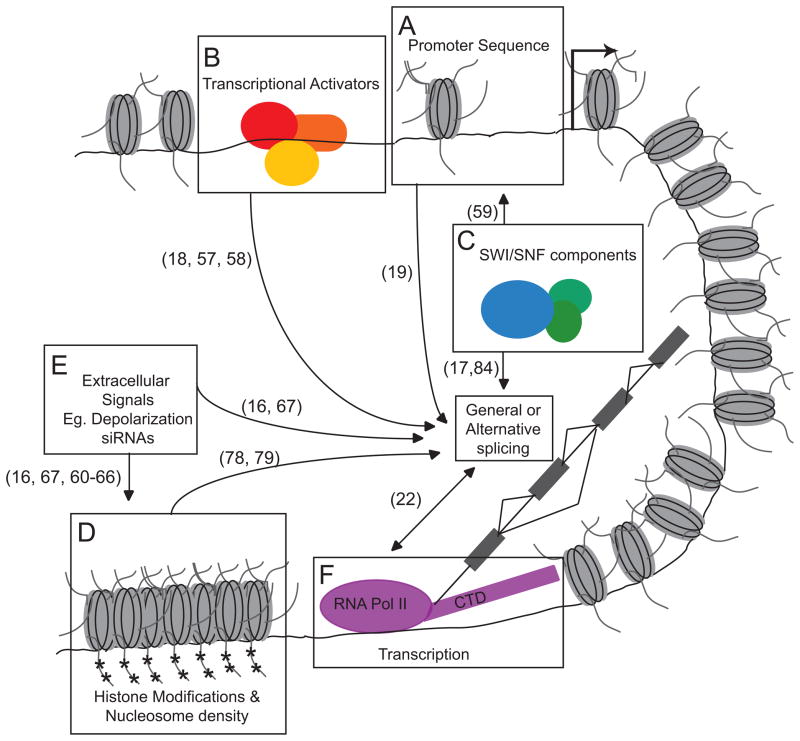
As described above, promoter sequences, transcriptional activators and chromatin remodeling enzymes are factors with established roles in transcriptional regulation that also influence the inclusion level of some alternative exons (17–19)(Figure 4). The TGS-like mechanism of chromatin condensation around the endogenous FN exon 33 region is thought to decrease DNA accessibility to the polymerase, thereby hindering its forward progression and forcing transcription to proceed more slowly. The kinetic model predicts that this will result in increased alternative exon inclusion, which is indeed the observed outcome in this case. It follows that a more open chromatin state should permit faster polII elongation and a commensurate rise in exon skipping, and is seen upon depolarization of neuronal N2A cells with KCl. That chromatin structure can seemingly influence splicing choices through modulating the rate of transcription has leant support to the idea that other variables similarly control the rate of transcription and thereby influence splicing of regulated exons. However, it is also possible that transacting factors might influence splicing by interfering with spliceosome assembly independently of their role in transcription (84).
Figure 4. Alternative splicing choices are affected by transcriptional-and chromatin-associated parameters.
The boxed regions and arrowheads represent the different aspects of chromatin structure and transcriptional dynamics that are known to affect splicing choices. The annotation(s) next to each arrowhead indicate the reference number for the studies that describe each effect. (A) FN alternative exon 33 inclusion levels differ when transcribed from different promoter sequences (B) Transcriptional activators that bind to promoter sequences, such as the CAPER proteins, can alter the splicing patterns of mRNAs produced from the downstream transcriptional unit. It is not known whether this effect is direct or through changes in the rate of transcription. (C) Nucleosome remodeling factors can influence regulated splicing choices. The SWI/SNF subunit Brm1 can bind to the U1 and U5 snRNPs and alter splicing patters of RNAs transcribed from genes that require SWI/SNF remodeling at the promoter for activation. (D) Dense chromatin structures are predicted to hinder polII accessibility to the DNA and possibly slow transcriptional rates. The kinetic model posits that this will increase alternative exon inclusion rates. Conversely, relaxed chromatin is thought to enhance transcription and thus, favor regulated exon skipping. (E) SiRNA’s targeted to intronic regions upstream of alternative FN exon 33 induce chromatin condensation and increase inclusion of FN exon 33. Depolarizing stimuli leads to a more open chromatin state and a decrease in inclusion of the NCAM exon 18. (F) Chromatin structure is implicated in regulating transcriptional rate, which in turn might effect changes in alternative splicing choices as posited by the kinetic model. Alternatively, some of these variables may interfere with spliceosome assembly to influence splicing choice.
Current definitions of the relationship between transcription rate and the effect on splicing are qualitative. In an effort to try and quantify splicing under coupled conditions, steady state measurements of nascent RNA have shown that the intron downstream of FN exon 33 is removed prior to the upstream one, when the regulated exon is included (14, 39). Interestingly, the relative abundance of each intermediate (the species in which only one of the two flanking introns has been removed) is not affected by a decrease in elongation rate (39). These findings are difficult to reconcile with the kinetic model as it is described above and it has been suggested that the transcriptional rate may instead affect the assembly of commitment complexes across the alternative region, rather than intron excision per se (39)(Figure 3C). However, until the rates of spliceosome assembly and catalysis can be measured relative to the rate of RNA synthesis, it will be difficult to test many aspects of this model.
THE SIGNIFICANCE OF COUPLING SPLICING TO TRANSCRIPTION ON EXON DEFINITION
Alternative exon definition
Nearly all alternative splicing patterns involve a competition between splice sites, and any process that alters the relative kinetics of splicing assembly can modulate splice site selection and pairing (109). Under conditions where introns are relatively short, splice sites bookending an intron pair up to bring the exons into juxtaposition in a model known as intron-definition (110)(Figure 5B). In metazoans, introns are generally much longer than exons, and data supports a model in which the two splice sites flanking each exon are recognized as an independent unit prior to pairing of the exons across the intervening intron (56)(Figure 5A). This exon definition process requires that that the two exons flanking an intron be fully synthesized prior to spliceosomal assembly (110).
Figure 5. Endogenous pre-mRNA’s assemble ‘exon defined’ rather than ‘intron defined’ complexes.
(A) In metazoans, where introns are much longer than exons, the U1 snRNP bound at a 5′ss stimulates binding of U2AF at the upstream 3′ss to form a cross-exon E complex. In this manner, each exon is recognized as an independent unit prior to pairing of two exons across an intron, and is known as ‘exon definition’.
(B) RNAs containing short introns, such as those used for invitro splicing, assemble spliceosomes via intron definition. Under intron-defined conditions, U1 bound at a 5′ss will form a cross intron E complex with U2AF bound at the downstream 3′ss.
(C) To accommodate skipping of an alternative exon, a prediction of the exon definition model is that transcription of the entire alternative region be complete prior to splicing. Under these conditions, the stronger, more rapidly assembling 3′ splice site of the downstream constitutive exon is predicated to out compete the weaker 3′ss of the alternative exon for pairing with the upstream constitutive exon. This would result in regulated exon skipping.
(D) Under conditions of slower transcriptional elongation or polymerase pausing in the downstream intron, alternative exon definition and pairing can proceed without competition from the stronger constitutive downstream exon, thus providing a potential mechanism by which enhanced inclusion of the regulated exon can be achieved while still adhering to an exon-defined mode of splicing.
In vivo, where splicing occurs upon nascent transcripts with long introns, the exon-definition model nicely explains pairing of consecutive constitutive exons (110). In the case of regulated cassette exon splicing, the exon-definition model demands that transcription of the alternative region completes prior to splicing in order to accommodate exclusion of the regulated exon. Under these conditions, it is not entirely clear how exon inclusion is achieved, because competition between the downstream constitutive exon and alternative exon, which usually has weaker splice sites, is thought to favor exon skipping (Figure 5C). The revised kinetic model posits that a slower rate of transcription permits recognition and pairing of the alternative exon with the upstream constitutive exon prior to complete synthesis of the downstream constitutive exon, resulting in increased exon inclusion. Pausing of the polymerase downstream of the regulated exon would similarly allow for increased splicing of the alternative exon. Thus, coupling of splicing to transcription provides a potential explanation for how alternative exon inclusion might be achieved during transcription, while still adhering to an exon-defined mode of spliceosome assembly(Figure 5D).
Terminal exon definition
The first and last exons in a pre-mRNA each have only one flanking splice site and therefore must be recognized differently than internal exons. In the case of first exon definition, this apparent problem has an elegant solution. After initiation of transcription, the first exon undergoes rapid 7-methylguanosine (7mG) cap addition as described above (88–91). The newly formed cap is bound by the nuclear cap binding complex which aids U1 assembly at the cap-proximal 5′ss (111). Thus, co-transcriptional cap addition acts in lieu of an upstream splice site to enable identification of the first exon within a pre-mRNA transcript.
Mechanisms regulating definition of the 3′ terminal exon are more complex. The 3′ terminus of the last exon is bounded by the poly(A) signal, which is composed of a conserved AAUAAA hexamer and a U or GU-rich tract 20–50nt downstream (Figure 6). These two elements flank the cleavage and polyadenylation (CPA) site. Cleavage and Polyadenylation Specificity Factor (CPSF) is a complex of four proteins that binds to the AAUAAA element, of which the CPSF-73 subunit has been identified as the endonuclease responsible for cleavage at the CPA site (112) (Figure 6). The U/GU tract is bound by the Cleavage stimulation Factor (CstF). Cleavage Factors (CF) I and II and symplekin are also required for the cleavage reaction and addition of the poly(A) tail is catalyzed by Poly(A) polymerase (PAP). These factors are essential for correct cleavage and polyadenylation at the majority of poly(A) sites (For a complete review see (113)and (114))
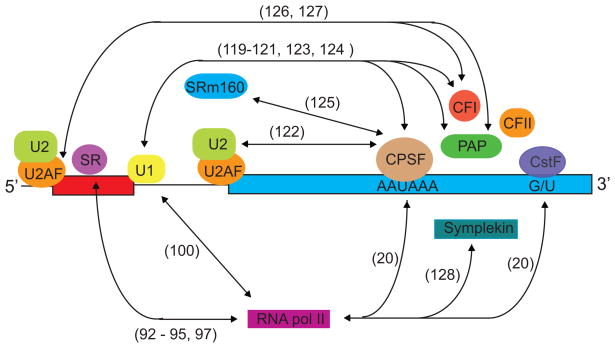
Figure 6. Factors involved in 3′ end processing interact with members of the splicing apparatus.
Double arrowheads indicate known interactions (direct and indirect) between splicing factors and the cleavage and polyadenylation machinery. For clarity, interactions that occur between spliceosomal factors during spliceosomal assembly, or between the cleavage and polyadenylation factors during 3′ end processing have been omitted. The annotation(s) next to each arrowhead indicate the citation number for the studies that describe the indicated interactions.
Cellular extracts that support both splicing and 3′ end processing show that poly(A) site mutations inhibit splicing across the terminal 3′ intron, but do not affect splicing of upstream introns (115). This result implies that 3′ terminal exon definition is dependent on cleavage at the poly(A) site. Moreover, mutations within the 5′ss, 3′ss or polypyrimidine tract of the last intron similarly impede processing at the CPA site(116–118), and demonstrate that concerted recognition of both the 3′ terminal intron and the poly(A) site are required for efficient processing of the 3′ terminal exon.
A number of reports document interactions between splicing proteins and 3′ end processing factors that have interesting functional consequences. One example involves the U1 snRNP specific-U1A protein which autoregulates it’s own transcript by interfering with polyadenylation (119). The 3′ untranslated region of the U1A transcript contains two binding sites for the U1A protein. When bound to the RNA, U1A interacts with PAP to inhibit cleavage and polyadenylation of the U1A transcript (120). The domain of U1A that is inhibitory to PAP also prevents splicing of an adenoviral template when added to an in vitro system supporting splicing and CPA (121). Thus, the U1A splicing factor can affect processing of it’s own transcript by interfering with cleavage and polyadenylation.
The CPSF complex is an essential 3′ end-processing factor and it co-purifies with the U2 snRNP-specific SF3b proteins in fractionated cellular extract. Direct interactions between CPSF subunits and SF3b49 and 130 occur in vitro (122). Extracts depleted of CPSF100 fail to splice a MINX pre-mRNA containing a strong SV40 polyadenylation signal (MINXSVL), although depletion of CPSF100 has no effect on the splicing of a MINX pre-mRNA lacking a poly(A) site, suggesting that the CPSF-SF3b interactions are of consequence (122). These two examples lend credence to the idea that the splicing machinery assembled across the terminal intron functionally interacts with the 3′ end processing apparatus to define the 3′ terminal exon. Additional contacts between 3′ end processing components and spliceosomal factors have been described in the literature and are depicted in Figure 6 (123–128). However, little is known regarding the significance of most of these interactions.
The mechanisms regulating definition of the first and last exons are clearly different to those employed to recognize internal ones. The 5′ end of the first exon is defined by the capping reaction, which also couples transcriptional initiation and early elongation to splicing of the first intron in a pre-mRNA (89–91, 111). While the mechanisms controlling 3′ terminal exon definition are not yet clear, the current data invoke a situation in which the spliceosome that assembles across the 3′ terminal intron makes a series of concerted contacts with the 3′ end processing machinery that functionally links splicing to cleavage and polyadenylation during transcription.
Conclusions
It has been 25 years since splicing was first observed to occur during transcription (13). It is now clear that these two processes are functionally coupled and do not act in isolation. Promoter sequences (19), the factors that bind them (18, 57), chromatin-remodeling enzymes (17)and changes in chromatin structure (16, 67)can all modify splicing outcomes, but understanding how these variables exert their influence on splicing will require further analysis of their mechanism(s) of action.
Direct recruitment of the capping enzymes by the CTD is well documented and provides credence to the recruitment model as a means to couple transcription to RNA processing(89–91). However, since evidence for binding of bona fide splicing factors to the CTD is lacking, the role of polII in bringing splicing factors to the nascent RNA remains unclear. Further development of tractable coupled systems that support regulated splicing is imperative to understanding whether the recruitment and kinetic models accurately explain at least some aspects of splicing regulation at it occurs during transcription.
Sensitive fluorescent assays, including single molecule fluorescence resonance energy transfer (smFRET) that function in vitro and in live cells (129–131), are being developed. These techniques hold much promise for revealing the kinetics of spliceosome assembly both in vitro and in vivo, and may provide clues to in situ protein-protein and protein-RNA interactions that could potentially functionally couple transcription to capping, splicing and 3′ end processing of pre-mRNA transcripts.
Acknowledgments
I thank Erik Anderson, Douglas Black, Lauren Gehman, Timothy Nilsen, Karla Neugebauer and James Manley for their constructive comments on the manuscript. I apologize to colleagues whose valuable work I have been unable to include due to space limitations. My work has been supported in part through fellowships from the American Association of University Women, the Warsaw family foundation and the UCLA Dissertation year committee as well as grants awarded to Douglas. L. Black by the National Institutes of Health.
References
- 1.Wahl MC, Will CL, Luhrmann R. The spliceosome: design principles of a dynamic RNP machine. Cell. 2009;136(4):701–718. doi: 10.1016/j.cell.2009.02.009. [DOI] [PubMed] [Google Scholar]
- 2.Tycowski KT, Kolev NG, Conrad NK, Fok V, Steitz JA. The Ever-Growing World of Small Nuclear Ribonucleoproteins. In: Gesteland RF, Cech TR, Atkins JF, editors. The RNA World. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2006. [Google Scholar]
- 3.Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, et al. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456(7221):470–476. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nilsen TW, Graveley BR. Expansion of the eukaryotic proteome by alternative splicing. Nature. 2010;463(7280):457–463. doi: 10.1038/nature08909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Black DL. Mechanisms of alternative pre-messenger RNA splicing. Annu Rev Biochem. 2003;72:291–336. doi: 10.1146/annurev.biochem.72.121801.161720. [DOI] [PubMed] [Google Scholar]
- 6.Chen M, Manley JL. Mechanisms of alternative splicing regulation: insights from molecular and genomics approaches. Nat Rev Mol Cell Biol. 2009;10(11):741–754. doi: 10.1038/nrm2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shepard PJ, Hertel KJ. The SR protein family. Genome Biol. 2009;10(10):242. doi: 10.1186/gb-2009-10-10-242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Manley JL, Krainer AR. A rational nomenclature for serine/arginine-rich protein splicing factors (SR proteins) Genes Dev. 2010;24(11):1073–1074. doi: 10.1101/gad.1934910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Padgett RA, Hardy SF, Sharp PA. Splicing of adenovirus RNA in a cell-free transcription system. Proc Natl Acad Sci U S A. 1983;80(17):5230–5234. doi: 10.1073/pnas.80.17.5230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Krainer AR, Maniatis T, Ruskin B, Green MR. Normal and mutant human beta-globin pre-mRNAs are faithfully and efficiently spliced in vitro. Cell. 1984;36(4):993–1005. doi: 10.1016/0092-8674(84)90049-7. [DOI] [PubMed] [Google Scholar]
- 11.Hernandez N, Keller W. Splicing of in vitro synthesized messenger RNA precursors in HeLa cell extracts. Cell. 1983;35(1):89–99. doi: 10.1016/0092-8674(83)90211-8. [DOI] [PubMed] [Google Scholar]
- 12.Green MR, Maniatis T, Melton DA. Human beta-globin pre-mRNA synthesized in vitro is accurately spliced in Xenopus oocyte nuclei. Cell. 1983;32(3):681–694. doi: 10.1016/0092-8674(83)90054-5. [DOI] [PubMed] [Google Scholar]
- 13.Osheim YN, Miller OL, Jr, Beyer AL. RNP particles at splice junction sequences on Drosophila chorion transcripts. Cell. 1985;43(1):143–151. doi: 10.1016/0092-8674(85)90019-4. [DOI] [PubMed] [Google Scholar]
- 14.Pandya-Jones A, Black DL. Co-transcriptional splicing of constitutive and alternative exons. RNA. 2009;15(10):1896–1908. doi: 10.1261/rna.1714509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Listerman I, Sapra AK, Neugebauer KM. Cotranscriptional coupling of splicing factor recruitment and precursor messenger RNA splicing in mammalian cells. Nat Struct Mol Biol. 2006;13(9):815–822. doi: 10.1038/nsmb1135. [DOI] [PubMed] [Google Scholar]
- 16.Allo M, Buggiano V, Fededa JP, Petrillo E, Schor I, et al. Control of alternative splicing through siRNA-mediated transcriptional gene silencing. Nat Struct Mol Biol. 2009;16(7):717–724. doi: 10.1038/nsmb.1620. [DOI] [PubMed] [Google Scholar]
- 17.Batsche E, Yaniv M, Muchardt C. The human SWI/SNF subunit Brm is a regulator of alternative splicing. Nat Struct Mol Biol. 2006;13(1):22–29. doi: 10.1038/nsmb1030. [DOI] [PubMed] [Google Scholar]
- 18.Dowhan DH, Hong EP, Auboeuf D, Dennis AP, Wilson MM, et al. Steroid hormone receptor coactivation and alternative RNA splicing by U2AF65-related proteins CAPERalpha and CAPERbeta. Mol Cell. 2005;17(3):429–439. doi: 10.1016/j.molcel.2004.12.025. [DOI] [PubMed] [Google Scholar]
- 19.Cramer P, Pesce CG, Baralle FE, Kornblihtt AR. Functional association between promoter structure and transcript alternative splicing. Proc Natl Acad Sci U S A. 1997;94(21):11456–11460. doi: 10.1073/pnas.94.21.11456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McCracken S, Fong N, Yankulov K, Ballantyne S, Pan G, et al. The C-terminal domain of RNA polymerase II couples mRNA processing to transcription. Nature. 1997;385(6614):357–361. doi: 10.1038/385357a0. [DOI] [PubMed] [Google Scholar]
- 21.Munoz MJ, Perez Santangelo MS, Paronetto MP, de la Mata M, Pelisch F, et al. DNA damage regulates alternative splicing through inhibition of RNA polymerase II elongation. Cell. 2009;137(4):708–720. doi: 10.1016/j.cell.2009.03.010. [DOI] [PubMed] [Google Scholar]
- 22.de la Mata M, Alonso CR, Kadener S, Fededa JP, Blaustein M, et al. A slow RNA polymerase II affects alternative splicing in vivo. Mol Cell. 2003;12(2):525–532. doi: 10.1016/j.molcel.2003.08.001. [DOI] [PubMed] [Google Scholar]
- 23.Carrillo Oesterreich F, Preibisch S, Neugebauer KM. Global analysis of nascent RNA reveals transcriptional pausing in terminal exons. Mol Cell. 2010;40(4):571–581. doi: 10.1016/j.molcel.2010.11.004. [DOI] [PubMed] [Google Scholar]
- 24.Lacadie SA, Tardiff DF, Kadener S, Rosbash M. In vivo commitment to yeast cotranscriptional splicing is sensitive to transcription elongation mutants. Genes Dev. 2006;20(15):2055–2066. doi: 10.1101/gad.1434706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gunderson FQ, Johnson TL. Acetylation by the transcriptional coactivator Gcn5 plays a novel role in co-transcriptional spliceosome assembly. PLoS Genet. 2009;5(10):e1000682. doi: 10.1371/journal.pgen.1000682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Alexander RD, Innocente SA, Barrass JD, Beggs JD. Splicing-dependent RNA polymerase pausing in yeast. Mol Cell. 2010;40(4):582–593. doi: 10.1016/j.molcel.2010.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Beyer AL, Osheim YN. Splice site selection, rate of splicing, and alternative splicing on nascent transcripts. Genes Dev. 1988;2(6):754–765. doi: 10.1101/gad.2.6.754. [DOI] [PubMed] [Google Scholar]
- 28.Kiseleva E, Wurtz T, Visa N, Daneholt B. Assembly and disassembly of spliceosomes along a specific pre-messenger RNP fiber. EMBO J. 1994;13(24):6052–6061. doi: 10.1002/j.1460-2075.1994.tb06952.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sass H, Pederson T. Transcription-dependent localization of U1 and U2 small nuclear ribonucleoproteins at major sites of gene activity in polytene chromosomes. J Mol Biol. 1984;180(4):911–926. doi: 10.1016/0022-2836(84)90263-8. [DOI] [PubMed] [Google Scholar]
- 30.Alzhanova-Ericsson AT, Sun X, Visa N, Kiseleva E, Wurtz T, Daneholt B. A protein of the SR family of splicing factors binds extensively to exonic Balbiani ring pre-mRNA and accompanies the RNA from the gene to the nuclear pore. Genes Dev. 1996;10(22):2881–2893. doi: 10.1101/gad.10.22.2881. [DOI] [PubMed] [Google Scholar]
- 31.Zeng C, Kim E, Warren SL, Berget SM. Dynamic relocation of transcription and splicing factors dependent upon transcriptional activity. EMBO J. 1997;16(6):1401–1412. doi: 10.1093/emboj/16.6.1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Neugebauer KM, Roth MB. Distribution of pre-mRNA splicing factors at sites of RNA polymerase II transcription. Genes Dev. 1997;11(9):1148–1159. doi: 10.1101/gad.11.9.1148. [DOI] [PubMed] [Google Scholar]
- 33.Misteli T, Caceres JF, Spector DL. The dynamics of a pre-mRNA splicing factor in living cells. Nature. 1997;387(6632):523–527. doi: 10.1038/387523a0. [DOI] [PubMed] [Google Scholar]
- 34.Sass H. Features of in vitro puffing and RNA synthesis in polytene chromosomes of Chironomus. Chromosoma. 1980;78(1):33–78. doi: 10.1007/BF00291908. [DOI] [PubMed] [Google Scholar]
- 35.Bauren G, Belikov S, Wieslander L. Transcriptional termination in the Balbiani ring 1 gene is closely coupled to 3′-end formation and excision of the 3′-terminal intron. Genes Dev. 1998;12(17):2759–2769. doi: 10.1101/gad.12.17.2759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wetterberg I, Bauren G, Wieslander L. The intranuclear site of excision of each intron in Balbiani ring 3 pre-mRNA is influenced by the time remaining to transcription termination and different excision efficiencies for the various introns. RNA. 1996;2(7):641–651. [PMC free article] [PubMed] [Google Scholar]
- 37.Tennyson CN, Klamut HJ, Worton RG. The human dystrophin gene requires 16 hours to be transcribed and is cotranscriptionally spliced. Nat Genet. 1995;9(2):184–190. doi: 10.1038/ng0295-184. [DOI] [PubMed] [Google Scholar]
- 38.Wuarin J, Schibler U. Physical isolation of nascent RNA chains transcribed by RNA polymerase II: evidence for cotranscriptional splicing. Mol Cell Biol. 1994;14(11):7219–7225. doi: 10.1128/mcb.14.11.7219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.de la Mata M, Lafaille C, Kornblihtt AR. First come, first served revisited: factors affecting the same alternative splicing event have different effects on the relative rates of intron removal. RNA. 2010;16(5):904–912. doi: 10.1261/rna.1993510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ule J, Stefani G, Mele A, Ruggiu M, Wang X, et al. An RNA map predicting Nova-dependent splicing regulation. Nature. 2006;444(7119):580–586. doi: 10.1038/nature05304. [DOI] [PubMed] [Google Scholar]
- 41.Marshall NF, Price DH. Purification of P-TEFb, a transcription factor required for the transition into productive elongation. J Biol Chem. 1995;270(21):12335–12338. doi: 10.1074/jbc.270.21.12335. [DOI] [PubMed] [Google Scholar]
- 42.Singh J, Padgett RA. Rates of in situ transcription and splicing in large human genes. Nat Struct Mol Biol. 2009;16(11):1128–1133. doi: 10.1038/nsmb.1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Huranova M, Ivani I, Benda A, Poser I, Brody Y, et al. The differential interaction of snRNPs with pre-mRNA reveals splicing kinetics in living cells. J Cell Biol. 2010;191(1):75–86. doi: 10.1083/jcb.201004030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cramer P, Bushnell DA, Fu J, Gnatt AL, Maier-Davis B, et al. Architecture of RNA polymerase II and implications for the transcription mechanism. Science. 2000;288(5466):640–649. doi: 10.1126/science.288.5466.640. [DOI] [PubMed] [Google Scholar]
- 45.Corden JL, Cadena DL, Ahearn JM, Jr, Dahmus ME. A unique structure at the carboxyl terminus of the largest subunit of eukaryotic RNA polymerase II. Proc Natl Acad Sci U S A. 1985;82(23):7934–7938. doi: 10.1073/pnas.82.23.7934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xu YX, Manley JL. Pin1 modulates RNA polymerase II activity during the transcription cycle. Genes Dev. 2007;21(22):2950–2962. doi: 10.1101/gad.1592807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Noble CG, Hollingworth D, Martin SR, Ennis-Adeniran V, Smerdon SJ, et al. Key features of the interaction between Pcf11 CID and RNA polymerase II CTD. Nat Struct Mol Biol. 2005;12(2):144–151. doi: 10.1038/nsmb887. [DOI] [PubMed] [Google Scholar]
- 48.Komarnitsky P, Cho EJ, Buratowski S. Different phosphorylated forms of RNA polymerase II and associated mRNA processing factors during transcription. Genes Dev. 2000;14(19):2452–2460. doi: 10.1101/gad.824700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kelly WG, Dahmus ME, Hart GW. RNA polymerase II is a glycoprotein. Modification of the COOH-terminal domain by O-GlcNAc. J Biol Chem. 1993;268(14):10416–10424. [PubMed] [Google Scholar]
- 50.Chapman RD, Heidemann M, Albert TK, Mailhammer R, Flatley A, et al. Transcribing RNA polymerase II is phosphorylated at CTD residue serine-7. Science. 2007;318(5857):1780–1782. doi: 10.1126/science.1145977. [DOI] [PubMed] [Google Scholar]
- 51.Misteli T, Spector DL. RNA polymerase II targets pre-mRNA splicing factors to transcription sites in vivo. Mol Cell. 1999;3(6):697–705. doi: 10.1016/s1097-2765(01)80002-2. [DOI] [PubMed] [Google Scholar]
- 52.Meininghaus M, Chapman RD, Horndasch M, Eick D. Conditional expression of RNA polymerase II in mammalian cells. Deletion of the carboxyl-terminal domain of the large subunit affects early steps in transcription. J Biol Chem. 2000;275(32):24375–24382. doi: 10.1074/jbc.M001883200. [DOI] [PubMed] [Google Scholar]
- 53.Hirose Y, Tacke R, Manley JL. Phosphorylated RNA polymerase II stimulates pre-mRNA splicing. Genes Dev. 1999;13(10):1234–1239. doi: 10.1101/gad.13.10.1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Natalizio BJ, Robson-Dixon ND, Garcia-Blanco MA. The Carboxyl-terminal Domain of RNA Polymerase II Is Not Sufficient to Enhance the Efficiency of Pre-mRNA Capping or Splicing in the Context of a Different Polymerase. J Biol Chem. 2009;284(13):8692–8702. doi: 10.1074/jbc.M806919200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Aebi M, Hornig H, Padgett RA, Reiser J, Weissmann C. Sequence requirements for splicing of higher eukaryotic nuclear pre-mRNA. Cell. 1986;47(4):555–565. doi: 10.1016/0092-8674(86)90620-3. [DOI] [PubMed] [Google Scholar]
- 56.Talerico M, Berget SM. Effect of 5′ splice site mutations on splicing of the preceding intron. Mol Cell Biol. 1990;10(12):6299–6305. doi: 10.1128/mcb.10.12.6299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rosonina E, Bakowski MA, McCracken S, Blencowe BJ. Transcriptional activators control splicing and 3′-end cleavage levels. J Biol Chem. 2003;278(44):43034–43040. doi: 10.1074/jbc.M307289200. [DOI] [PubMed] [Google Scholar]
- 58.Rosonina E, Ip JY, Calarco JA, Bakowski MA, Emili A, et al. Role for PSF in mediating transcriptional activator-dependent stimulation of pre-mRNA processing in vivo. Mol Cell Biol. 2005;25(15):6734–6746. doi: 10.1128/MCB.25.15.6734-6746.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kwon H, Imbalzano AN, Khavari PA, Kingston RE, Green MR. Nucleosome disruption and enhancement of activator binding by a human SW1/SNF complex. Nature. 1994;370(6489):477–481. doi: 10.1038/370477a0. [DOI] [PubMed] [Google Scholar]
- 60.Pal-Bhadra M, Bhadra U, Birchler JA. Cosuppression of nonhomologous transgenes in Drosophila involves mutually related endogenous sequences. Cell. 1999;99(1):35–46. doi: 10.1016/s0092-8674(00)80060-4. [DOI] [PubMed] [Google Scholar]
- 61.Mette MF, van der Winden J, Matzke MA, Matzke AJ. Production of aberrant promoter transcripts contributes to methylation and silencing of unlinked homologous promoters in trans. EMBO J. 1999;18(1):241–248. doi: 10.1093/emboj/18.1.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Grishok A, Sinskey JL, Sharp PA. Transcriptional silencing of a transgene by RNAi in the soma of C. elegans. Genes Dev. 2005;19(6):683–696. doi: 10.1101/gad.1247705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Reinhart BJ, Bartel DP. Small RNAs correspond to centromere heterochromatic repeats. Science. 2002;297(5588):1831. doi: 10.1126/science.1077183. [DOI] [PubMed] [Google Scholar]
- 64.Mette MF, Aufsatz W, van der Winden J, Matzke MA, Matzke AJ. Transcriptional silencing and promoter methylation triggered by double-stranded RNA. EMBO J. 2000;19(19):5194–5201. doi: 10.1093/emboj/19.19.5194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pal-Bhadra M, Leibovitch BA, Gandhi SG, Rao M, Bhadra U, et al. Heterochromatic silencing and HP1 localization in Drosophila are dependent on the RNAi machinery. Science. 2004;303(5658):669–672. doi: 10.1126/science.1092653. [DOI] [PubMed] [Google Scholar]
- 66.Volpe TA, Kidner C, Hall IM, Teng G, Grewal SI, Martienssen RA. Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science. 2002;297(5588):1833–1837. doi: 10.1126/science.1074973. [DOI] [PubMed] [Google Scholar]
- 67.Schor IE, Rascovan N, Pelisch F, Allo M, Kornblihtt AR. Neuronal cell depolarization induces intragenic chromatin modifications affecting NCAM alternative splicing. Proc Natl Acad Sci U S A. 2009;106(11):4325–4330. doi: 10.1073/pnas.0810666106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Xie J, Black DL. A CaMK IV responsive RNA element mediates depolarization-induced alternative splicing of ion channels. Nature. 2001;410(6831):936–939. doi: 10.1038/35073593. [DOI] [PubMed] [Google Scholar]
- 69.Schones DE, Cui K, Cuddapah S, Roh TY, Barski A, et al. Dynamic regulation of nucleosome positioning in the human genome. Cell. 2008;132(5):887–898. doi: 10.1016/j.cell.2008.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Spies N, Nielsen CB, Padgett RA, Burge CB. Biased chromatin signatures around polyadenylation sites and exons. Mol Cell. 2009;36(2):245–254. doi: 10.1016/j.molcel.2009.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Schwartz S, Meshorer E, Ast G. Chromatin organization marks exon-intron structure. Nat Struct Mol Biol. 2009;16(9):990–995. doi: 10.1038/nsmb.1659. [DOI] [PubMed] [Google Scholar]
- 72.Andersson R, Enroth S, Rada-Iglesias A, Wadelius C, Komorowski J. Nucleosomes are well positioned in exons and carry characteristic histone modifications. Genome Res. 2009;19(10):1732–1741. doi: 10.1101/gr.092353.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chen W, Luo L, Zhang L. The organization of nucleosomes around splice sites. Nucleic Acids Res. 2010;38(9):2788–2798. doi: 10.1093/nar/gkq007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hon G, Wang W, Ren B. Discovery and annotation of functional chromatin signatures in the human genome. PLoS Comput Biol. 2009;5(11):e1000566. doi: 10.1371/journal.pcbi.1000566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Nahkuri S, Taft RJ, Mattick JS. Nucleosomes are preferentially positioned at exons in somatic and sperm cells. Cell Cycle. 2009;8(20):3420–3424. doi: 10.4161/cc.8.20.9916. [DOI] [PubMed] [Google Scholar]
- 76.Tilgner H, Nikolaou C, Althammer S, Sammeth M, Beato M, et al. Nucleosome positioning as a determinant of exon recognition. Nat Struct Mol Biol. 2009;16(9):996–1001. doi: 10.1038/nsmb.1658. [DOI] [PubMed] [Google Scholar]
- 77.Huff JT, Plocik AM, Guthrie C, Yamamoto KR. Reciprocal intronic and exonic histone modification regions in humans. Nat Struct Mol Biol. 2010;17(12):1495–1499. doi: 10.1038/nsmb.1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sims RJ, 3rd, Millhouse S, Chen CF, Lewis BA, Erdjument-Bromage H, et al. Recognition of trimethylated histone H3 lysine 4 facilitates the recruitment of transcription postinitiation factors and pre-mRNA splicing. Mol Cell. 2007;28(4):665–676. doi: 10.1016/j.molcel.2007.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Luco RF, Pan Q, Tominaga K, Blencowe BJ, Pereira-Smith OM, Misteli T. Regulation of alternative splicing by histone modifications. Science. 2010;327(5968):996–1000. doi: 10.1126/science.1184208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Natalizio BJ, Garcia-Blanco MA. In vitro coupled transcription splicing. Methods. 2005;37(4):314–322. doi: 10.1016/j.ymeth.2005.07.013. [DOI] [PubMed] [Google Scholar]
- 81.Hicks MJ, Yang CR, Kotlajich MV, Hertel KJ. Linking splicing to Pol II transcription stabilizes pre-mRNAs and influences splicing patterns. PLoS Biol. 2006;4(6):e147. doi: 10.1371/journal.pbio.0040147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Yu Y, Das R, Folco EG, Reed R. A model in vitro system for co-transcriptional splicing. Nucleic Acids Res. 2010 doi: 10.1093/nar/gkq620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lazarev D, Manley JL. Concurrent splicing and transcription are not sufficient to enhance splicing efficiency. RNA. 2007;13(9):1546–1557. doi: 10.1261/rna.595907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Tyagi A, Ryme J, Brodin D, Ostlund Farrants AK, Visa N. SWI/SNF associates with nascent pre-mRNPs and regulates alternative pre-mRNA processing. PLoS Genet. 2009;5(5):e1000470. doi: 10.1371/journal.pgen.1000470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Stenesh J. Biochemistry. Plenum; New York: 1998. [Google Scholar]
- 86.Egloff S, O’Reilly D, Chapman RD, Taylor A, Tanzhaus K, et al. Serine-7 of the RNA polymerase II CTD is specifically required for snRNA gene expression. Science. 2007;318(5857):1777–1779. doi: 10.1126/science.1145989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lu H, Zawel L, Fisher L, Egly JM, Reinberg D. Human general transcription factor IIH phosphorylates the C-terminal domain of RNA polymerase II. Nature. 1992;358(6388):641–645. doi: 10.1038/358641a0. [DOI] [PubMed] [Google Scholar]
- 88.McCracken S, Fong N, Rosonina E, Yankulov K, Brothers G, et al. 5′-Capping enzymes are targeted to pre-mRNA by binding to the phosphorylated carboxy-terminal domain of RNA polymerase II. Genes Dev. 1997;11(24):3306–3318. doi: 10.1101/gad.11.24.3306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ho CK, Sriskanda V, McCracken S, Bentley D, Schwer B, Shuman S. The guanylyltransferase domain of mammalian mRNA capping enzyme binds to the phosphorylated carboxyl-terminal domain of RNA polymerase II. J Biol Chem. 1998;273(16):9577–9585. doi: 10.1074/jbc.273.16.9577. [DOI] [PubMed] [Google Scholar]
- 90.Cho EJ, Takagi T, Moore CR, Buratowski S. mRNA capping enzyme is recruited to the transcription complex by phosphorylation of the RNA polymerase II carboxy-terminal domain. Genes Dev. 1997;11(24):3319–3326. doi: 10.1101/gad.11.24.3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Schroeder SC, Schwer B, Shuman S, Bentley D. Dynamic association of capping enzymes with transcribing RNA polymerase II. Genes Dev. 2000;14(19):2435–2440. doi: 10.1101/gad.836300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kim E, Du L, Bregman DB, Warren SL. Splicing factors associate with hyperphosphorylated RNA polymerase II in the absence of pre-mRNA. J Cell Biol. 1997;136(1):19–28. doi: 10.1083/jcb.136.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Robert F, Blanchette M, Maes O, Chabot B, Coulombe B. A human RNA polymerase II-containing complex associated with factors necessary for spliceosome assembly. J Biol Chem. 2002;277(11):9302–9306. doi: 10.1074/jbc.M110516200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Tanner S, Stagljar I, Georgiev O, Schaffner W, Bourquin JP. A novel SR-related protein specifically interacts with the carboxy-terminal domain (CTD) of RNA polymerase II through a conserved interaction domain. Biol Chem. 1997;378(6):565–571. doi: 10.1515/bchm.1997.378.6.565. [DOI] [PubMed] [Google Scholar]
- 95.Bourquin JP, Stagljar I, Meier P, Moosmann P, Silke J, et al. A serine/arginine-rich nuclear matrix cyclophilin interacts with the C-terminal domain of RNA polymerase II. Nucleic Acids Res. 1997;25(11):2055–2061. doi: 10.1093/nar/25.11.2055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ahn SH, Kim M, Buratowski S. Phosphorylation of serine 2 within the RNA polymerase II C-terminal domain couples transcription and 3′ end processing. Mol Cell. 2004;13(1):67–76. doi: 10.1016/s1097-2765(03)00492-1. [DOI] [PubMed] [Google Scholar]
- 97.Yuryev A, Patturajan M, Litingtung Y, Joshi RV, Gentile C, et al. The C-terminal domain of the largest subunit of RNA polymerase II interacts with a novel set of serine/arginine-rich proteins. Proc Natl Acad Sci U S A. 1996;93(14):6975–6980. doi: 10.1073/pnas.93.14.6975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.de la Mata M, Kornblihtt AR. RNA polymerase II C-terminal domain mediates regulation of alternative splicing by SRp20. Nat Struct Mol Biol. 2006;13(11):973–980. doi: 10.1038/nsmb1155. [DOI] [PubMed] [Google Scholar]
- 99.Sapra AK, Anko ML, Grishina I, Lorenz M, Pabis M, et al. SR protein family members display diverse activities in the formation of nascent and mature mRNPs in vivo. Mol Cell. 2009;34(2):179–190. doi: 10.1016/j.molcel.2009.02.031. [DOI] [PubMed] [Google Scholar]
- 100.Das R, Yu J, Zhang Z, Gygi MP, Krainer AR, et al. SR proteins function in coupling RNAP II transcription to pre-mRNA splicing. Mol Cell. 2007;26(6):867–881. doi: 10.1016/j.molcel.2007.05.036. [DOI] [PubMed] [Google Scholar]
- 101.Mili S, Steitz JA. Evidence for reassociation of RNA-binding proteins after cell lysis: implications for the interpretation of immunoprecipitation analyses. RNA. 2004;10(11):1692–1694. doi: 10.1261/rna.7151404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Neugebauer KM. On the importance of being co-transcriptional. J Cell Sci. 2002;115(Pt 20):3865–3871. doi: 10.1242/jcs.00073. [DOI] [PubMed] [Google Scholar]
- 103.Chew SL, Liu HX, Mayeda A, Krainer AR. Evidence for the function of an exonic splicing enhancer after the first catalytic step of pre-mRNA splicing. Proc Natl Acad Sci U S A. 1999;96(19):10655–10660. doi: 10.1073/pnas.96.19.10655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Cao W, Jamison SF, Garcia-Blanco MA. Both phosphorylation and dephosphorylation of ASF/SF2 are required for pre-mRNA splicing in vitro. RNA. 1997;3(12):1456–1467. [PMC free article] [PubMed] [Google Scholar]
- 105.Harper JE, Manley JL. Multiple activities of the human splicing factor ASF. Gene Expr. 1992;2(1):19–29. [PMC free article] [PubMed] [Google Scholar]
- 106.Xu X, Yang D, Ding JH, Wang W, Chu PH, et al. ASF/SF2-regulated CaMKIIdelta alternative splicing temporally reprograms excitation-contraction coupling in cardiac muscle. Cell. 2005;120(1):59–72. doi: 10.1016/j.cell.2004.11.036. [DOI] [PubMed] [Google Scholar]
- 107.Lin S, Coutinho-Mansfield G, Wang D, Pandit S, Fu XD. The splicing factor SC35 has an active role in transcriptional elongation. Nat Struct Mol Biol. 2008;15(8):819–826. doi: 10.1038/nsmb.1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Kwon YS, Garcia-Bassets I, Hutt KR, Cheng CS, Jin M, et al. Sensitive ChIP-DSL technology reveals an extensive estrogen receptor alpha-binding program on human gene promoters. Proc Natl Acad Sci U S A. 2007;104(12):4852–4857. doi: 10.1073/pnas.0700715104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Reed R, Maniatis T. A role for exon sequences and splice-site proximity in splice-site selection. Cell. 1986;46(5):681–690. doi: 10.1016/0092-8674(86)90343-0. [DOI] [PubMed] [Google Scholar]
- 110.Berget SM. Exon recognition in vertebrate splicing. J Biol Chem. 1995;270(6):2411–2414. doi: 10.1074/jbc.270.6.2411. [DOI] [PubMed] [Google Scholar]
- 111.Lewis JD, Izaurralde E, Jarmolowski A, McGuigan C, Mattaj IW. A nuclear cap-binding complex facilitates association of U1 snRNP with the cap-proximal 5′ splice site. Genes Dev. 1996;10(13):1683–1698. doi: 10.1101/gad.10.13.1683. [DOI] [PubMed] [Google Scholar]
- 112.Mandel CR, Kaneko S, Zhang H, Gebauer D, Vethantham V, et al. Polyadenylation factor CPSF-73 is the pre-mRNA 3′-end-processing endonuclease. Nature. 2006;444(7121):953–956. doi: 10.1038/nature05363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Millevoi S, Vagner S. Molecular mechanisms of eukaryotic pre-mRNA 3′ end processing regulation. Nucleic Acids Res. 2010;38(9):2757–2774. doi: 10.1093/nar/gkp1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Mandel CR, Bai Y, Tong L. Protein factors in pre-mRNA 3′-end processing. Cell Mol Life Sci. 2008;65(7–8):1099–1122. doi: 10.1007/s00018-007-7474-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Niwa M, Berget SM. Mutation of the AAUAAA polyadenylation signal depresses in vitro splicing of proximal butnot distal introns. Genes Dev. 1991;5(11):2086–2095. doi: 10.1101/gad.5.11.2086. [DOI] [PubMed] [Google Scholar]
- 116.Dye MJ, Proudfoot NJ. Terminal exon definition occurs cotranscriptionally and promotes termination of RNA polymerase II. Mol Cell. 1999;3(3):371–378. doi: 10.1016/s1097-2765(00)80464-5. [DOI] [PubMed] [Google Scholar]
- 117.Cooke C, Hans H, Alwine JC. Utilization of splicing elements and polyadenylation signal elements in the coupling of polyadenylation and last-intron removal. Mol Cell Biol. 1999;19(7):4971–4979. doi: 10.1128/mcb.19.7.4971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Rigo F, Kazerouninia A, Nag A, Martinson HG. The RNA tether from the poly(A) signal to the polymerase mediates coupling of transcription to cleavage and polyadenylation. Mol Cell. 2005;20(5):733–745. doi: 10.1016/j.molcel.2005.09.026. [DOI] [PubMed] [Google Scholar]
- 119.Boelens WC, Jansen EJ, van Venrooij WJ, Stripecke R, Mattaj IW, Gunderson SI. The human U1 snRNP-specific U1A protein inhibits polyadenylation of its own pre-mRNA. Cell. 1993;72(6):881–892. doi: 10.1016/0092-8674(93)90577-d. [DOI] [PubMed] [Google Scholar]
- 120.van Gelder CW, Gunderson SI, Jansen EJ, Boelens WC, Polycarpou-Schwarz M, et al. A complex secondary structure in U1A pre-mRNA that binds two molecules of U1A protein is required for regulation of polyadenylation. EMBO J. 1993;12(13):5191–5200. doi: 10.1002/j.1460-2075.1993.tb06214.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Gunderson SI, Vagner S, Polycarpou-Schwarz M, Mattaj IW. Involvement of the carboxyl terminus of vertebrate poly(A) polymerase in U1A autoregulation and in the coupling of splicing and polyadenylation. Genes Dev. 1997;11(6):761–773. doi: 10.1101/gad.11.6.761. [DOI] [PubMed] [Google Scholar]
- 122.Kyburz A, Friedlein A, Langen H, Keller W. Direct interactions between subunits of CPSF and the U2 snRNP contribute to the coupling of pre-mRNA 3′ end processing and splicing. Mol Cell. 2006;23(2):195–205. doi: 10.1016/j.molcel.2006.05.037. [DOI] [PubMed] [Google Scholar]
- 123.Awasthi S, Alwine JC. Association of polyadenylation cleavage factor I with U1 snRNP. RNA. 2003;9(11):1400–1409. doi: 10.1261/rna.5104603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Lutz CS, Murthy KG, Schek N, O’Connor JP, Manley JL, Alwine JC. Interaction between the U1 snRNP-A protein and the 160-kD subunit of cleavage-polyadenylation specificity factor increases polyadenylation efficiency in vitro. Genes Dev. 1996;10(3):325–337. doi: 10.1101/gad.10.3.325. [DOI] [PubMed] [Google Scholar]
- 125.McCracken S, Lambermon M, Blencowe BJ. SRm160 splicing coactivator promotes transcript 3′-end cleavage. Mol Cell Biol. 2002;22(1):148–160. doi: 10.1128/MCB.22.1.148-160.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Vagner S, Vagner C, Mattaj IW. The carboxyl terminus of vertebrate poly(A) polymerase interacts with U2AF 65 to couple 3′-end processing and splicing. Genes Dev. 2000;14(4):403–413. [PMC free article] [PubMed] [Google Scholar]
- 127.Millevoi S, Loulergue C, Dettwiler S, Karaa SZ, Keller W, et al. An interaction between U2AF 65 and CF I(m) links the splicing and 3′ end processing machineries. EMBO J. 2006;25(20):4854–4864. doi: 10.1038/sj.emboj.7601331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Xiang K, Nagaike T, Xiang S, Kilic T, Beh MM, et al. Crystal structure of the human symplekin-Ssu72-CTD phosphopeptide complex. Nature. 2010;467(7316):729–733. doi: 10.1038/nature09391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Crawford DJ, Hoskins AA, Friedman LJ, Gelles J, Moore MJ. Visualizing the splicing of single pre-mRNA molecules in whole cell extract. RNA. 2008;14(1):170–179. doi: 10.1261/rna.794808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Abelson J, Blanco M, Ditzler MA, Fuller F, Aravamudhan P, et al. Conformational dynamics of single pre-mRNA molecules during in vitro splicing. Nat Struct Mol Biol. 2010;17(4):504–512. doi: 10.1038/nsmb.1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Sakon JJ, Weninger KR. Detecting the conformation of individual proteins in live cells. Nat Methods. 2010;7(3):203–205. doi: 10.1038/nmeth.1421. [DOI] [PMC free article] [PubMed] [Google Scholar]