Abstract
Here we describe the first detailed catalogue of gene expression in the developing lower urinary tract (LUT), including epithelial and mesenchymal portions of the developing bladder, urogenital sinus, urethra and genital tubercle (GT) at E13 and E14. Top compartment-specific genes implicated by the microarray data were validated using wholemount in situ hybridization (ISH) over the entire LUT. To demonstrate the potential of this resource to implicate developmentally critical features, we focused on gene expression patterns and pathways in the sexually indeterminate, androgen-independent GT. GT expression patterns reinforced the proposed similarities between development of GT, limb and craniofacial prominences. Comparison of spatial expression patterns predicted a network of Wnt7a-associated GT-enriched epithelial genes, including Gjb2, Dsc3, Krt5 and Sostdc1. Known from other contexts, these genes are associated with normal epidermal differentiation, with disruptions in Dsc3 and Gjb2 showing palmo-plantar keratoderma in the limb. We propose that this gene network contributes to normal foreskin, scrotum and labial development. As several of these are known regulated by, or contain cis elements responsive to retinoic acid, estrogen, or androgen, this implicates this pathway in the later androgen-dependent development of the GT.
Keywords: genital tubercle development, lower urinary tract development, gene expression, Wnt, appendage development
Introduction
Congenital anomalies of the urogenital tract are the third most common birth defects, with abnormalities of the external genitalia being the most prevalent manifestation. Cryptorchidism (undescended testes) and hypospadias (in which the urethral orifice is located on the underside of the penis or at the perineum) each occur in as many as 1 in 125 males born (Kurzrock et al, 1999; Cox et al, 2008) and the prevalence of these two conditions appears to be rising, potentially due to environmental exposure to anti-androgenic compounds (Miyagawa et al, 2002; Brouwers et al, 2007; Wang and Baskin, 2008). These and other less common anomalies of the external genitalia (diphallia, micropenis) create social as well as reproductive challenges for the affected individuals. The lower urinary tract (LUT) is defined as extending from the bladder to the meatal opening of the urethra. Congenital anomalies of the kidney and urinary tract (CAKUT) include such defects as vesicoureteric reflux (VUR) (improper insertion of the ureters into the bladder) and posterior urethral valves (fine outgrowths of urethral tissue that result in bladder outlet obstruction) (Kerecuk et al, 2008). Such conditions are quite common (VUR presents in 1–2% of all children (Kerecuk et al, 2008)), with thesequelae of most LUT problems presenting later in life (urge incontinence, stress incontinence, cystitis, neurogenic bladder, overactive bladder, erectile dysfunction) and being aetiologically poorly understood.
Despite a physiological relationship with the mesodermally-derived kidney and ureters, the LUT and GT have distinct embryological origins. The LUT develops from the endodermal cloaca, which becomes separated into the urogenital sinus and rectum by the downgrowth of the urorectal septum around E10.5 in mouse. The urogenital sinus gives rise to the internal urethra and bladder. While both become lined with a water-tight urothelial layer, these two regions differentiate into distinct structures with respect to musculature, stroma, epithelial layers and innervation. The role of the LUT is storage and voiding. Regulation of appropriate micturition requires complex neural connections and, unlike most internal organs, control of storage and voiding by the LUT involves voluntary regulation. Locally, this involves sympathetic and parasympathetic innervation of the bladder and somatic motor nerves to regulate contraction of the urethral sphincter (Fowler et al, 2008). As well as providing a barrier, the urothelial cells of the bladder also act as mechanosensors and are actively involved in triggering neural responses in response to bladder expansion. The importance of the urothelium in this process was recently highlighted by the presence of urothelial hyperplasia and non-voiding contractions in bladders from mice defective for either uroplakin II or IIIa (Aboushwareb et al, 2009). The genital tubercle (GT) gives rise to the male and female external genitalia (penis, clitoris, scrotum, labia, foreskin) and represents the intersection between the reproductive and urinary tracts. The primary swellings of the genital tubercle form the penis and clitoris, while the preputial swellings give rise to the foreskin in males and the labia minora in females. A second set of swellings, the labioscrotal swellings, arise later in development and give rise to the scrotum and labia majora in males and females, respectively. The LUT and GT physically intersect as the urethra passes through the GT. The GT is proposed to be derived from all three germ layers. The urethral plate (UP) is derived from urethral endoderm (Siefert et al, 2008). The GT surface other than the urethral plate is derived from surface ectoderm and the core of the extending appendage is derived from lateral plate mesoderm (Perriton et al, 2002). Murine GT development commences at approximately E10.75, with the appearance of paired swellings ventral to the cloacal membrane (Perriton et al, 2002). Without a urethral plate, the GT will fail to form. The urethral epithelium of the GT does not grow out from the urethra but forms in situ along the GT itself (Pennington and Hutson, 2002). GT development involves an early androgen-independent phase and a later androgen-dependent phase, after which sexual dimorphism is evident. In mouse, this dimorphism is not evident until approximately E16, before which there has already been significant outgrowth and patterning during an androgen-independent phase.
The genital tubercle develops as an out-budding appendage, hence it has been proposed that there is congruence in the genetic basis of GT patterning with other out-budding appendages, including the limb and craniofacial prominences. Of these three appendages, limb patterning has been most highly studied. The limb has three key axes of development, the anterior-posterior axis (thumb to little finger), proximal-distal axis (arm to finger tips) and dorsal-ventral or plantar-palmar (back of hand to palm) axis. Major growth factor pathways involved in limb development include the hedgehog, fibroblast growth factor (FGF), bone morphogenetic protein (BMP) pathways and Wnt pathways (Goodman et al, 2000; Ahn et al, 2001; Hill et al, 2006; Geetha-Loganathan et al, 2008; Witte et al, 2009). Sonic hedgehog (Shh) is expressed in a posterior zone of polarizing activity (ZPA) and is critical for limb anterior-posterior (A-P) patterning. Duplication of this region on the anterior side of the limb leads to a mirror image duplication of digits (polydactyly). Extension in the distal direction involves signals from the apical ectodermal ridge (AER), notably Fgf8, but also Fgf4, Fgf9 and Fgf17 (Mariani et al, 2008). These same signaling pathways are involved in GT development. Here, Shh is expressed in the endodermal UP along the ventral surface of the GT (Haraguchi et al, 2001; Perriton et al, 2002). The UP has been proposed to act as an organizing centre similar to the ZPA in the limb. Transplantation of this region into the anterior limb of the chick induces digit duplication, however it also results in the dimpling of the skin and formation of a furrow similar in structure to the UP (Haraguchi et al, 2001; Perriton et al, 2002). The UP expression of Shh influences the development of the adjacent mesenchyme, eliciting extension to form the GT. Shh was proposed to regulate the expression of Fgf8 and Bmp7, which are also expressed in the UP and distal urethral epithelium (DUE), and Wnt5a, Bmp2, Bmp4 and Fgf10, which are expressed in the mesenchyme surrounding the urethral plate (Yamaguchi et al, 1999; Haraguchi et al, 1999; Perriton et al, 2002). Expression of Shh is required throughout the period of androgen-independent GT extension. By removing smoothened from different layers of the developing GT, it was also demonstated that the primary target of Shh is the GT mesenchyme (Lin et al, 2009). Shh knockout mice show a loss of GT accompanied by a loss of expression of Bmp4 and Fgf10, suggesting that Shh is upstream of both of these growth factor pathways. As for the limb, the GT undergoes considerable proliferative extension prior to structural differentiation. In the limb, this outgrowth is driven by the expression of Fgf8 in the AER. The expression of Fgf8 in the DUE was similarly presumed to be required for GT extension and outgrowth (Morgan et al, 2003; Yamada et al, 2006). Certainly the presumptive region of Fgf8 expression is lost when Shh is deleted. Surprisingly, the conditional removal of Fgf8 from the cloacal and urethral epithelium did not affect GT outgrowth, even though there was a loss of expression of putative downstream targets of Fgf8 (Bmp4, Wnt5a, Hoxd13) and no evidence of a redundant role for other members of the Fgf family (Siefert et al, 2009). Wnt5a has been shown to be involved in outgrowth of GT and limb (Yamaguchi et al, 1999). The loss of Shh also reduces canonical Wnt signaling demonstrating that anterioposterior extension due to mesenchymal proliferation is likely require Wnt signaling from the DUE (Miyagawa et al, 2009). Indeed, concommitent loss of Shh, Wnt5a and axin2 expression is seen in two models of disrupted GT and anorectal development, Danforth’s short tail (Sd) and all-trans RA treated mice (Nakata et al, 2009).
GT expression of Shh also regulates Hoxd13 and Hoxa13 expression, although independently to Wnt and Fgf signalling. (Lin et al, 2009). The homeodomain genes Hoxa13 and Hoxd13 are also expressed in the UP, whereas Msx1 and the Shh receptor, Ptch1, are expressed in the surrounding mesenchyme. Here again, there is a similarity to limb where Hoxa13 and Hoxd13 are required for the development and A-P patterning of the autopods (digits) (Cobb and Duboule, 2005). Homozygote knockouts of these genes result in loss of the limbs and phallus, whereas heterozygotes display disrupted patterning in both locations (reviewed in Perriton et al, 2002). In humans, mutations in these genes result in hand-foot-genital syndrome (Goodman et al, 2000). In the posterior embryo, these homeodomain genes also control formation of the terminal vertebrae and the anus. The UP is therefore also thought to be critical for both dorsal-ventral patterning and proximal-distal extension of the GT.
As noted, congenital GT defects are very common. Based on observations in mice, hypospadias may result from disruptions in Fgfr2, Fgf8, Fgf10 and Bmp7, all of which are involved in UP patterning (Beleza-Meireles et al, 2007). In humans, hypospadias has been observed in Opitz syndrome (MID1 mutation), Pallister-Hall syndrome (GLI3 mutation), Reiger syndrome, type 1 (PITX2 mutation), Hand-foot-genital syndrome (HOXA13, HOXD13 mutations) and Split hand/split-foot malformation type 1 and type 4 (mutations in Dlx5, Dlx6 and Dss1) (Yamada et al, 2006; Suzuki et al, 2008). Micropenis occurs in Pallister-Hall syndrome, Robinow syndrome (ROR2 mutation), Ulnar mammary syndrome (TBX3 mutation) and X-linked lissencephaly (ARX mutation) (reviewed in Yamada et al, 2006). Mutations in CXorf6 have also been identified in isolated cases of non-syndromic hypospadias (Kalfa et al, 2008).
Despite the clinical relevance and prevalence of defects, there have been almost no systems biology applied to the molecular basis of LUT/GT development. Li et al (2006) performed expression analysis of mouse GT (E14, E15, E16 and E17), but focused on those genes expressed during the later androgen-dependent phase of development. In this study, we have addressed this gap by creating a comprehensive dataset of microarray and in situ gene expression of the E13 and E14 LUT/GT. In order to test the utility of this dataset, we analysed in more depth gene expression in the GT. Ontological analysis of GT-enriched genes reaffirmed the importance of Wnt signaling and defined an epidermal, oestrogen-responsive, Wnt7a-associated gene network potentially involved in foreskin/labial development and possibly defective in hypospadias.
Materials and methods
At E13.5, a pregnant female FVB/N mouse was sacrificed by CO2 asphyxiation and, following a midline abdominal incision, the gravid uteri were removed and placed in PBS on ice. Embryos were dissected under magnification. Bladder and urethra were dissected free of surrounding tissues and then divided at the level of ureteral insertion. Bladder segments were incubated in 20 mM EDTA in Tyrode’s solution for 20 minutes at 37°C. Urethral segments were treated with Trypsin (1 mg/ml in PBS) for 20 minutes (37°C). In each instance, epithelium was separated from mesenchyme by rimming with a fine needle. Tissues were then placed in RLT Buffer containing ß-ME, according to manufacturer’s instructions (RNeasy Micro Kit, Qiagen Inc., Valencia, CA) and rapidly frozen in dry ice prior to storage at −80°C for subsequent RNA purification. This was achieved using a spin-column system (RNeasy Micro Kit, Qiagen Inc., Valencia, CA), according to manufacturer’s directions, and stored at −80°C prior to amplification.
At E14, a pregnant female FVB/N mouse carrying the SMGA/EGFP transgene (Szucsik et al. 2004) was sacrificed by CO2 asphyxiation and following a midline abdominal incision the gravid uteri were removed and placed in PBS on ice. Embryos were dissected under GFP illumination. The lower urinary tract was trisected into bladder (EGFP positive dome), UGS (EGFP negative) and urethra (EGFP positive). A separate, nontransgenic pregnant FVB/N female was used to obtain samples of the genital tubercle at E14. Tissues were rapidly frozen in dry ice and stored at −80°C for subsequent RNA purification using a spin-column system (RNeasy Micro Kit, Qiagen Inc., Valencia, CA), according to manufacturer’s directions, and stored at −80°C prior to amplification.
Expression profiling, data normalisation and analysis
Purified RNA was analyzed for concentration and integrity using the Agilent RNA 6000 Pico Kit (Agilent Technologies Inc., Santa Clara, CA) and then 0.5–1.5 ng (in a volume of 1 μl) underwent two-round amplification (Epicenter Biotechnologies, Madison, WI) according to manufacturer’s directions except for the addition of Full SpectrumTm multistart primers (SBI System Biosciences, Mountain View, CA) per the manufacturer’s directions. The resultant product was biotinylated and hybridized to Affymetrix Mouse MOE 430 2.0 microarray chips (Affymetrix Inc., Santa Clara, CA). CEL files were generated using Expression Console v1.1.1 (Affymetrix) and subjected to RMA normalization using Affymetrix CDF probeset definitions and GeneSpringGX v7.3.1 (Agilent). The relative expression of each probeset in each sample was calculated as the ratio relative to the median of its values as measured across the entire GUDMAP MOE 430 2.0 dataset that contains developing and postnatal tissues from kidney, lower urinary tract, and gonadal tissues. Probesets to be analyzed for differential expression across the developing lower urinary tract samples were then selected by filtering for those with RMA expression intensity greater than 6.2 in at least two replicates from any sample type. A general ANOVA was used to identify probesets that are differentially expressed between sample types with less than 5% false discovery using Benjamini Hochberg p-value correction. From the resulting 18K probesets, those with highest relative expression in the Genital Tubercle were identified based on normalized expression as referenced to the diverse panel of other developing and postnatal genitourinary samples. GeneSet enrichment analysis was performed using ToppGene (Chen et al, 2009). Genes that exhibited shared properties with respect to geneset enrichment features were transformed into network representations using CytoScape as previously described (Brunskill et al. 2008) using the ToppCluster server (http://toppcluster.cchmc.org/; Kaimal et al., submitted).
Collection of material for in situ hybridisation
For in situ hybridisation, outbred CD1 embryos were collected from two time points, TS17 (10.5 dpc) and TS21 (13.5 dpc), where 0.5dpc was defined as noon of the day on which mating plug was observed. Embryos were dissected in ice-cold PBS under a dissecting microscope and fixed in fresh 4% paraformaldehyde (PFA) at 4° C overnight. Tissues were then washed in PBTX and dehydrated through a PBTX/methanol series (25%, 50%, 75%) and stored in 100% methanol at −20° C until usage. TS17 whole embryos were collected with a puncture in the back of the head to prevent subsequent stain trapping in the brain. Tail somites were counted for TS17 litters to confirm embryos are within the range of 35–39 somites, as described by Kaufman (1994). The lower urogenital system plus genital tubercle were collected from TS21 and sex was determined by gonadal inspection.
Riboprobe Synthesis
The complete protocol for digoxigenin (Dig)-labelled riboprobe synthesis is available and described in detail on the GUDMAP gene expression database, (http://www.gudmap.org/Research/Protocols/Little.html). A brief overview, with minor changes to the protocol, is described here. Primers were ordered from Invitrogen according to the microarray output and were designed to amplify a 3′ UTR region of a gene between 500–800 bp. Riboprobes were amplified from either the Fantom II plasmid pool or the combination of both 12.5 and 15.5 dpc whole embryonic mouse cDNA. The 3′ primers were tagged with a T7 polymerase sequence which allows in vitro transcription of Dig-labelled riboprobes using T7 RNA polymerase. (Roche) Riboprobes were then purified with lithium chloride precipitation. A master mix was made with each riboprobe consisting of: 4M LiCl (10 μl), 0.2M EDTA, pH 7 (8 μl), 100% Ethanol (300μl), and water (100 μl) and stored at −20° C overnight. Samples were then spun for 20 minutes at 13,000 rpm at 4° C with supernatant discarded after the spin. Samples were gently washed with 1ml of chilled 70% ethanol, then spun for 5 minutes at 13000rpm at 4° C. Supernatant were discarded and samples dried for 10 minutes at room temperature. Pellets from each sample were resuspended with 25ul of water and stored at −70° C.
Whole mount in situ hybridisation and imaging of WISH
The complete protocol for whole mount in situ hybridisation is available and described in detail on the GUDMAP gene expression database. (http://www.gudmap.org/Research/Protocols/Little.html). In brief, whole mount in situ hybridisation was performed by using the BioLane HTI robot with digoxigenin–labelled antisense riboprobes. Tissues were rehydrated by a series of methanol/PBTX washes, followed by a short digestion of Proteinase K, and re-fixed by 0.2% gluteraldehyde/4% PFA before incubation in pre-hybridisation solution at 65° C for 2 hours. Tissues were later transferred into pre-hybridisation solution containing 0.2 μg/ml of riboprobe, incubated at 65° C overnight. Following post-hybridisation washes, a 2 hours incubation in pre-block solution followed by incubation in pre-adsorbed anti-DIG/pre-block solution. After the post-antibody washes, to detect the hybridised alkaline phosphatase activity, chromogenic substrates NBT/BCIP were used. Time for colour development varies between 15 minutes up to 300 minutes depending on the specificity of the riboprobe. Once the tissue has reached its optimal intensity, the tissues were washed with 1% Triton X -100/PBS at 4° C for a duration up to 2 days, later fixed with 4% PFA and stored in PBS at 4° C. All whole mount tissues were photographed manually on a 1.5% agarose gel dish, using Nikon SMZ1500 stereomicroscope system with a Nikon DXM1200f, 12 megapixel digital camera, ACT-2U Image Application Software and Adobe Photoshop CS2. For imaging of bisected genital tubercles, this region was dissected away from the rest of the lower urinary tissues in PBS on a Petri dish, and each sample was bisected either longitudinally or transversely with the aid of a 30G needle and number 11 surgical blade.
Pseudo colored gene expression overlays
To compare multiple gene expression domains Adobe Photoshop CS2 software was used to produce pseudo colored gene expression images of overlaid, stage-matched tubercles at 13.5dpc. Experimental image data was first outlined to obtain the structural outer edge of the tubercle and inverted to produce a black and white image. These were then aligned with up to two other genes using the RGB image format.
Results
Tissue was collected for RNA extraction from E13 urethra and bladder in which the epithelial and mesenchymal layers were separated from each other using trypsin digestion or EDTA treatment, respectively (Figure 1A). Tissue was also collected from E14 lower urinary tract (LUT) micro-dissected into bladder, urogenital sinus (UGS; includes the junction between the urethra, bladder and ureters), urethra and GT as illustrated in Figure 1B. All data has been submitted to GEO and is available from the GUDMAP database (www.gudmap.org), from which the expression level of all genes present on the microarray can be queried individually.
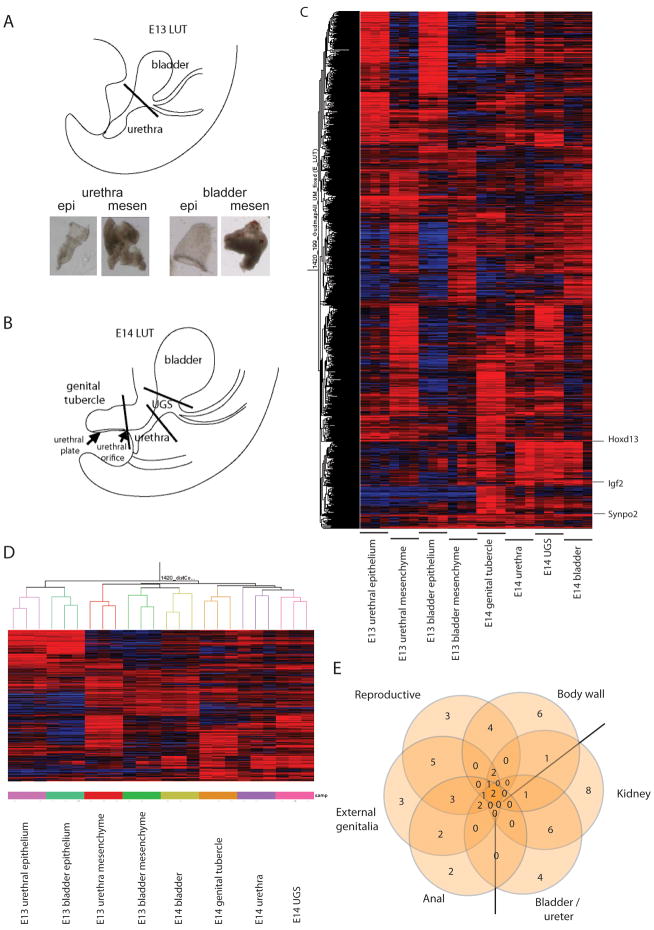
Figure 1. Defining the gene expression profile of the developing lower urinary tract.
A) Diagram of the E13 LUT indicating the point of dissection to separate E13 urethra from E13 bladder and the resultant separation of epithelium from mesenchyme achieved using trypsin digestion and EDTA dissociation, respectively. B) Diagram of the E14 LUT indicating the point of dissection to delineate four spatial regions; genital tubercle (GT), urethra, UGS and bladder. C) Heat map of the 1420 most highly expressed genes across all eight compartments of the developing LUT. D) Cluster diagram highlighting the hierarchical relationships between all samples analysed (eight compartments in triplicate). Note that all replicates correctly clustered with their compartment of origin. E) Venn diagram indicating the degree of gene overlap between the processes of reproductive, external genital, anal, body wall, kidney and bladder/ureter development identified in the GT dataset.
Creation of an overall view of differential gene expression across the developing lower urinary tract
In order to identify sets of genes that are either highly expressed or comparatively over-expressed in one or more of the sampled individual LUT/GT compartments during development, probesets were first filtered to identify those genes expressed at least at the 40th percentile of raw RMA normalized intensity in at least two replicates of each sample type. From these, probes were identified that exhibited the highest ranked relative expression as compared to the globally referenced dataset, i.e. their having normalized expression in the top 2% per each of the LUT sample types (~ 250–400 probesets per each of the 8 compartments). The combination of all these probesets were hierarchically clustered and Figure 1C shows a heat map of these 1420 probesets across all compartments. These are presented after hierarchical clustering to display the most highly expressed genes across the different LUT sample types. A full gene list and heatmap of the expression profiles manifested by these transcripts is provided in Supplementary Table 1. The expression of previously known compartment markers, including Tbx20 and Tbx18 in the bladder and the strong expression of Hoxd13 in all compartments, supports the accuracy of the data (Figure 1C). To determine potential functional roles for these genes, the 1420 probeset list was enriched with public database annotations (e.g. GO, Jax Mouse, OMIM, TFBS, Drug Bank, TargetScan, MSigDB, etc.) using Toppgene (Chen et al, 2009). The results of these analyses are shown in Supplementary Table 2. This revealed expression of a large number of DNA binding transcription factors, particularly homeobox (Hoxd13, Msx1, Six1, Irx3, Irx5, Msx2, Isl1, Pitx1, Pitx2, Hoxa10, Zfhx4, Dlx5, Zfhx3, Mkx, Satb2, Prrx1, Prrx2, Alx3, Alx1, Cux1) and forkhead genes (Foxf2, Foxd1, Foxa1, Foxn3, Foxq1); fibronectin III (FnIII) domain encoding genes (Ptprj, Ptpro, Flrt3, Flrt2, Cntn3, Dcc, Crlf1, Epha5, Epha4, Epha7, Robo1) and genes encoding cell-cell adhesion molecules. The most active biological processes included cell-cell adhesion with 21/104 cadherin domain containing proteins being expressed in the developing LUT (Pcdh10, Pcdh18, Cdh11, Pcdha10, Pcdha11, Pcdha13, Dsg2, Frem1, Dsc2, Pcdha6, Pcdha7, Pcdha9, Pcdha2, Pcdha3, Pcdha4, Pcdha5, Pcdha1, Pcdhb7, Pcdhc2, Pcdhac1, Pcdhgb2). The most active pathways were the Wnt signaling (Wnt5A, Gng4, Myh3, Pcdh10, Pcsh18, Nkd2, Wnt7A, Cdh11, Pcdha10, Pcdha11, Pcdha13, FrzB, Pcdha6, Pcdha7, Pcdha9, Pcdha2, Pcdha3, Pcdha4, Pcdha5, Pcdha1, Pcdhb7, Bcl9, Fzd6, Ankrd6, Wnt6, Myst4, Pcdhac2, Pcdhac1, Axin2, Lef1, Pcdhgb2, Dkk2, Dkk1, Dkk3, Axin2, Wif1, Nfat5), hedgehog signaling (Shh, Bmp5, Gli3, Wnt6, Wnt5A, Wnt7A) and cadherin signaling pathways. Toppgene analysis identified many genes involved in limb/face/appendage and skeletal development. In addition, there were many genes previously implicated in neural development and ectodermal/epidermal/epithelial development genes networks, notably cadherins, collagens and keratins. Examining the data for genes linked to human or mouse phenotypes involving the urogenital tract revealed genes known to be critical for reproductive, genital, bladder/ureter and kidney development. This set of genes overlapped genes involved in anal development, presumably reflecting the common origin of the bladder and urethra from the cloaca, and body wall development. The strongest associations were seen between genes affecting external genitalia/reproductive tract, anal canal and body wall development (TP63, Sall1, Sall4, Gli3, Pitx2, Tbx3) (Figure 1E, Supplementary Table 3). Sall1 and p63 are involved in the development of all of these organs/tissues, Sall4 is involved in all but body wall and Six1 is involved in body wall, kidney and ureter development (Supplementary Table 3).
Defining the most active networks in each compartment
In Figure 1C there is both considerable overlap of gene expression between all compartments of the LUT as well as clusters of gene expression more active in specific compartments. Heat maps for each compartment are presented in Supplementary Figure 1 and complete gene lists for each compartment in Supplementary Table 1. To understand the major processes occurring in each compartment, Toppgene analysis of each compartment was performed to identify the most represented molecular functions, biological processes, cellular components, protein domains, pathways and phenotypes. A summary of this analysis is presented in Table 2. A complete analysis of gene ontology for each compartment-enriched gene set is presented in Supplementary Table 2. This analysis reinforced the prevalence of cadherin and Wnt signaling, the predominance of transcription factor activity and the association of LUT compartments with genes involved in appendage, skeletal, neural, epithelial and urogenital development. Cluster analysis suggested strong alignment between E13 bladder epithelium and E13 urethra epithelium, but less of a relationship between the bladder and urethral mesenchyme of the same timepoint (Figure 1D). E13 bladder mesenchyme was most similar to E14 total bladder due to the expression of muscle genes (Tnnt2, Kcnq5, Actg2, Myh11). These two bladder samples were also outliers in their lack of association with cadherin or Wnt signalling. E14 UGS and urethra clustered together, whereas GT was distinct aside from a cluster of genes co-expressed in E13 urethra mesenchyme (Fgf9, Alx3, Foxd1, Msx1, Wnt5a). These genes have all previously been reported in GT development, reflecting the fact that the E13 urethra sample would have included GT. E13 urethral mesenchyme and UGS showed significant overlap (Myrip, Mapk10, Prph, Phox2b, Sthm2, Sthm2, Gdap1) with enrichment for genes involved in neurogenesis and neural crest, including the GDNF/Ret pathway (Gfra1, Gfra2).
Table 2. Observed gene expression patterns for genital tubercle-enriched genes.
Affy ID, Affymetrix ID; DGTE, distal genital tubercle epithelium; DGTM, distal genital tubercle mesenchyme; DUE, distal urethral epithelium; PUE, proximal urethral epithelium; UPM, mesenchyme underlying the urethral plate; PPSM, preputial swelling mesenchyme; PPSE, preputial swelling epithelium; PPG, preputial gland.
| Symbol | Cluster | Affy ID | Distal GT | Urethral epithelium | UPM | Preputial swelling | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| DGTM | DGTE | DUE | PUE | PPSM | PPSE | PPG | ||||
| Col17a1 | GT epithelium - enriched | 1418799_a_at | - | + | + | - | - | - | + | - |
| Gjb2 | GT epithelium - enriched | 1423271_at | - | + | - | - | - | - | + | - |
| Krt2-6a | GT epithelium - enriched | 1422784_at | - | - | + | + | - | - | - | - |
| Krt2-5 | GT epithelium - enriched | 1424096_at | - | + | + | + | - | - | + | - |
| Dlx5 | GT epithelium - enriched | 1449863_a_at | + | - | - | - | + | - | - | - |
| Dsc3 | GT epithelium - enriched | 1434534_at | + | + | + | - | - | - | + | - |
| Wnt7a | GT epithelium - enriched | 1423367_at | - | + | + | + | - | - | + | - |
| Serpinb3b | GT epithelium - enriched | 1422940_x_at | + | - | - | + | + | - | - | - |
| Cxcl14 | GT epithelium - enriched | 1418457_at | - | + | + | - | - | - | + | - |
| Shox2 | GT mesenchyme - enriched | 1438042_at | - | - | - | - | - | + | - | - |
| Lef1 | GT mesenchyme - enriched | 1454734_at | + | - | - | - | + | + | - | + |
| Syt13 | GT mesenchyme - enriched | 1434470_at | + | - | - | - | - | - | - | - |
| Prdm1 | GT mesenchyme - enriched | 1420425_at | + | - | - | - | + | + | - | - |
| Dkk1 | GT mesenchyme - enriched | 1420360_at | + | - | + | - | + | - | - | - |
| Tcfap2b | GT mesenchyme - enriched | 1435670_at | + | - | - | - | + | - | + | - |
| Hs3st3b1 | GT mesenchyme - enriched | 1433977_at | + | - | - | - | + | + | - | - |
| Wnt5a | GT mesenchyme - enriched | 1436791_at | + | - | + | - | + | - | - | - |
| Kcnab1 | GT mesenchyme - enriched | 1448468_a_at | + | - | - | - | + | + | - | - |
| Apcdd1 | GT mesenchyme - enriched | 1449070_x_at | + | - | - | - | + | + | - | - |
| GT mesenchyme - enriched | 1440630_at | + | - | - | + | + | - | - | - | |
| Unc5d | GT mesenchyme - enriched | 1440484_at | + | - | - | + | + | - | - | - |
| Kcna4 | GT mesenchyme - enriched | 1438613_at | + | - | - | + | - | + | - | - |
| D930002L09Rik | GT mesenchyme - enriched | 1444452_at | + | - | - | + | - | - | - | - |
| Satb2 | GT mesenchyme - enriched | 1427017_at | + | - | - | + | + | - | - | - |
| Frzb | GT mesenchyme - enriched | 1416658_at | + | - | - | - | + | - | - | - |
| Mup1 | E14 GT restricted | 1420465_s_at | + | - | - | - | - | - | - | - |
| Dlx6os1 | E14 GT restricted | 1438799_at | + | - | - | - | + | - | - | - |
| Trpm1 | E14 GT restricted | 1437445_at | + | - | - | - | - | - | - | - |
| Msx1 | E14 GT restricted | 1448601_s_at | + | - | - | - | + | - | - | - |
| Col19a1 | E14 GT restricted | 1456953_at | + | - | - | + | + | - | - | - |
| Col19a1 | E14 GT restricted | 1421698_a_at | + | - | - | + | + | - | - | - |
| Ptgfr | E14 GT restricted | 1420349_at | + | - | - | + | + | - | - | - |
| E14 GT restricted | 1445804_at | + | - | - | + | + | - | - | - | |
| Tle4 | E14 GT restricted | 1430384_at | - | - | - | + | - | - | - | - |
| Dlx6os2 | E14 GT restricted | 1440797_at | + | - | - | + | + | - | - | - |
| Sostdc1 | E14 GT restricted | 1449340_at | - | + | + | - | - | - | + | - |
| Wif1 | E13 urethra and GT | 1425425_a_at | + | - | - | - | + | + | - | - |
| Dlx1 | E13 urethra and GT | 1449470_at | - | + | + | - | + | - | - | - |
| Gja1 | E13 urethra and GT | 1415801_at | + | - | + | - | + | + | - | + |
| Osr2 | E13 urethra and GT | 1426155_a_at | + | - | - | - | + | + | - | - |
| Msx2 | E13 bladder and GT | 1449559_at | + | + | + | - | + | - | - | - |
| Tcfap2c | E13 bladder and GT | 1448977_at | + | - | - | - | + | - | - | + |
| Cpm | E13 bladder and GT | 1429413_at | + | - | + | - | + | - | - | - |
LUT compartment-enriched gene expression highlights innervation and vascularisation
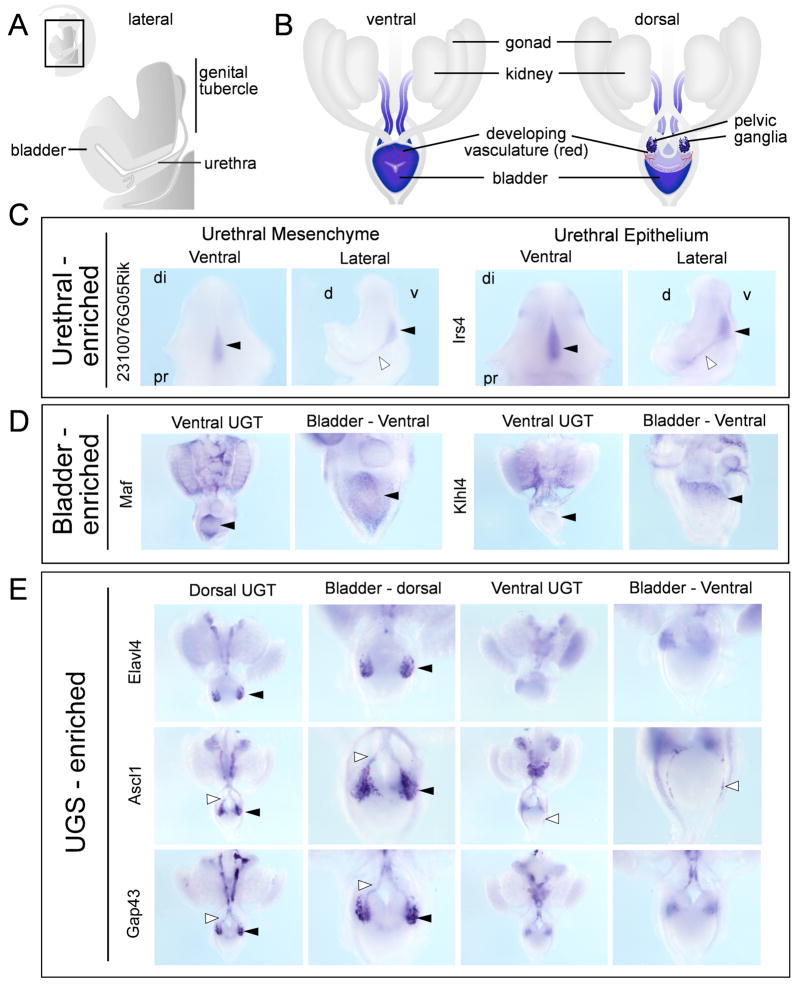
The expression profiling was then reanalyzed so as to specifically identify differences in gene expression between the compartments being assessed. The intention of identifying compartment-enriched genes was to identify unique markers of each compartment. Supplementary Table 4 lists all genes displaying compartment-enriched expression for each compartment. Figure 2 shows a heat map highlighting gene clusters with urethra-enriched or GT-enriched gene expression. Within the heatmap it is possible to identify E14 urethra-enriched genes that are likely to mark epithelium versus mesenchyme by examining their co-expression in E13 urethra and bladder epithelium. Wholemount in situ hybridisation (WISH) was used to validate enrichment of gene expression in each compartment. Riboprobes were generated for a total of 94 genes (Supplementary Table 5) and all WISH results were fully annotated for gene expression and submitted to the Genitourinary Development Molecular Anatomy Project website (www.gudmap.org; McMahon et al, 2008) and riboprobe sequences are shown in Supplementary Table 5). WISH was performed on E13.5 embryos for 11 urethra-specific genes (Figure 3C), including Irs4 and 2310076G05Rik, seventeen E14 UGS-enriched genes, including Elavl4, Gap43 and Ascl1 (Figure 3D), and eight E14 bladder-enriched genes, including Maf and Klhl4 (Figure 3E). Quite a number of UGS-enriched genes were expressed in the pelvic ganglia. Bladder-enriched genes highlighted bladder innervation and vasculature as well as markers specific to the dome of the bladder (Figure 3E). Schematic representations of these regions of the LUT are shown in Figure 3A and B.
Figure 2. Heat map of gene expression across E14 urethral-enriched and genital tubercle-enriched genes.
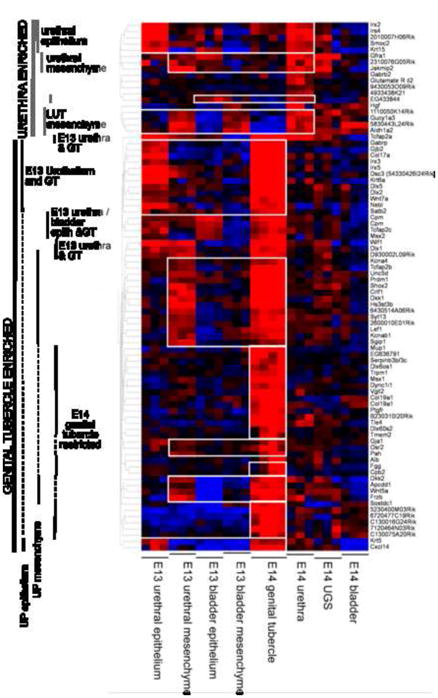
Urethral-enriched genes can be subdivided into epithelial versus mesenchymal based on the expression in E13 urethra. Genital tubercle-enriched genes were subdivided into urethral plate (UP) epithelium (expressed in genital tubercle and E13 urethral plate epithelium but not E14 urethra), UP mesenchyme (expressed in genital tubercle and E13 urethral mesenchyme but not E14 urethra) and genital tubercle-specific genes (expressed only in GT).
Figure 3. Validation of E14 urethral, bladder and UGS-enriched genes.
A) Schematic diagram of a bisected LUT/GT, lateral view, illustrating the bladder, UGS and urethra. up, urethral plate; due, distal urethral epithelium B) Schematic diagram of whole urogenital tracts, ventral and dorsal views. C) Urethral-enriched genes with epithelial (lrs4) and mesenchymal (2310076G05Rik) expression displayed as ventral (left) and lateral (right) views. Expression can be seen along the UP (solid arrowheads) and in the urethra itself (open arrowheads). D) E13 bladder-enriched genes Maf and Klhl4 showing expression in vasculature and differential expression in the dome of the bladder (solid arrowheads, rostral end). E) UGS-enriched genes Elav4, Ascl1 and Gap43 revealing specific expression in migrating neural crest cells (open arrowheads) and developing ganglia (solid arrowheads) at the base of the dorsal surface of the bladder. di, distal; pr, proximal; d, dorsal; v, ventral.
Analysis of androgen-independent genital tubercle development
To validate the utility of the dataset, we focused on the gene expression observed in the E14 GT given that this was the most unique set of compartment-enriched sets. As previous studies have reported expression of genes from within the BMP, FGF, Wnt and hedgehog pathways in GT development, we examined the expression of all members of these pathways present on the Affymetrix chips (Supplementary Table 6). This confirmed the expression of many pathway members known to be expressed in the GT although, as expected, few of these genes showed GT-enriched or -specific gene expression. Genes from the FGF/Hedgehog/Wnt pathways showing expression in E13 LUT and E14 GT included Fgf9 (mesenchyme), Pik3ca, Npc1, Fgfr3, Fgfbp1, Wnt4, Wnt5a, Wnt7a, Wnt7b and Wnt10a. A smaller number of genes from the BMP pathway showed expression in the E13 LUT and E14 GT, including Bmp2, Bmp4, Bmp7, Fstl1 and Gsk3b. Bmp5 showed GT-restricted gene expression at E14, but expression levels were low. The genes selected as being GT-enriched were analysed for pathway, disease, or GO term associations (Supplementary Table 7). As for the analysis of the most active pathways in this compartment, this revealed a strong link to Wnt pathway signaling. It also reinforced the previously proposed association between GT-enriched genes and genes involved in limb and craniofacial development (Supplementary Tables 2 and 7, Table 1).
Table 1. Overview of Toppgene ontological analysis of the most active genes in each compartment of the developing LUT examined.
For each category, features that are common between different compartments are highlighted in distinct colours to assist in visual assessment of similiarities between compartments. No pathways were strongly evident in E13 urethral mesenchyme, E13 bladder mesenchyme or E14 bladder. No human phenotypes were linked to genes expressed in the E13 bladder mesenchyme.
| E13 Ureth Epith | E13 Ureth Mesen | E13 Bladder Epith | E13 Bladder Mesen | E14GT | E14 Urethra | E14 UGS | E14 Bladder | |
|---|---|---|---|---|---|---|---|---|
| Molecular Function | TF activity | TF activity | TF activity | VEGFR binding | TF activity | TF activity | calcium ion binding | TFactivity |
| structural molecules | calcium ion binding | cytoskeleton | DNA binding | calcium ion binding | calcium ion binding | TF activity | TGF beta binding | |
| calcium ion binding | ephrin R activity | Cl channel activity | calmodulin binding | chromatin binding | chemorepellant | receptor protein kinase | transferase activity | |
|
| ||||||||
| Biological Process | epithelial/epidermal devel | neural development | epithelial/epidermal dev | axon guidance | limb/appendage devel | neural development | neural development | lung development |
| cell adhesion | cell adhesion | regulation of transcription | cell motion/localisation | neural development | cell-cell adhesion | cell adhesion | epithelial development | |
| regulation of transcription | axonogenesis | autonomic NS/neural crest | cell adhesion | cell adhesion | tube development | axonogenesis | reguation of transcription | |
|
| ||||||||
| Cellular Component | apicolateral junction | neuron projection | apicolateral junction | extracellular matrix | plasma membrane | extracellular space | neuron projection | extracellular matrix |
| cell junction | synapse | cell-cell junction | chromosome passenger complex | cell-cell junction | plasma membrane | intrinsic to plasma memb | extracellular space | |
| intercalated disc | plasma membrane | basolaternal membrane | extracellular space | intercalated disc | extracellular matrix | nucleosome | plasma membrane | |
|
| ||||||||
| Domain | cadherin | cadherin | plectin | bZIP | cadherin | cadherin | cadherin | ETS domain 3 |
| bZIP | nuclear orpan receptor | marvel | TSP1 | homeobox | forkhead | stathmin | ZnF GATA | |
| Plectin | stathmin | LDLa | HELP | OAR | thyroglobulin | Fnlll | Fnlll | |
|
| ||||||||
| Pathway | cadherin signaling | breast cancer E2 signaling | wnt signaling | cadherin signaling | cadherin signaling | |||
| wnt signaling | cell adhesion | cadherin signaling | wnt signaling | wnt signaling | ||||
| breast cancer E2 signaling | tight junction | basal cell carcinoma | axon guidance | SLE | ||||
|
| ||||||||
| Human Phenotype | abnormal keratinisation | Hirschsprung disease | abnormality of the uterus | onset in 1 st or 2nd decade | abnormal phalanges/limb | abnormality of face | renal dysplasia/VUR | cardiovascular dilatation |
| abnormalities of skin | Anal stenosis | renal dysplasia | hyperkeratosis | external genital abnorm | genital abnormality | muscular hypotonia | ||
| abnormal mouth/teeth | Abnormal limb musc | cutaneous syndactyly | abnormality of the head/face | abnormality of phalanges | Polydactyly | molluscoid pseudotumors | ||
|
| ||||||||
| Mouse Phenotype | abnormal skin morphology | abnormal neurons | abnormal mesonephros morpholc | impaired smooth muscle | abnormal limb | perinatal lethality | abnormal neuron | absent tracheal rings |
| abnormal vagus ganglion | abnormal musculature | abnormal epidermis morph | abnormal umbilical artery | abnormal face/mouth/head | abnormal oral/facial | neonatal lethality | abnormal ischium | |
| abnormal esophagus morph | perinatal lethality | thickened epidermis | abnormal skeleton | abnormal skeleton | abnormal digestive sys | conductive hearing loss | abnormal vertebrae | |
|
| ||||||||
| Disease | Epidermolysis Bullosa | Hirschsprung Disease | renal adysplasia | autistic disorder | autistic disorder | salivary gland neoplasms | salivary gland neoplasms | Autistic Disorder |
| Keratoderma, Palmoplantar | Autistic Disorder | nonsyndromic cleft lip +/−palate | hypertrophic neuropathy | Branchio-Oto-Renal syn | Branchio-Oto-Renal syn | Branchio-Oto-Renal syn | Ehlers-Danlos Type 2 | |
| neoplasia | Waardenburg Shah syn | prostate cancer | oral submucous fibrosis | Epidermolysis Bullosa | autonomic control loss | Williams Syndrome | oral submucous fibrosis | |
| insulin resistance | protein QT loci | infertility, male | intervertebral disc disease | female UG disease | head/neck Neoplasms | Autistic Disorder | reperfusion Injury | |
| infertility, male | Pheochromocytoma | prostatic neoplasms | pulmonary function traits | Keratoderma, Palmoplantar | neoplasm metastasis | Hirschprung disease | Obesity | |
VEGFR: vascular endothelial growth factor receptor; SLE: systemic lupus erythematosis; VUR: vesicoureteric reflux; QT: quantitative trait; TGF: transforming growth factor; CI: chloride
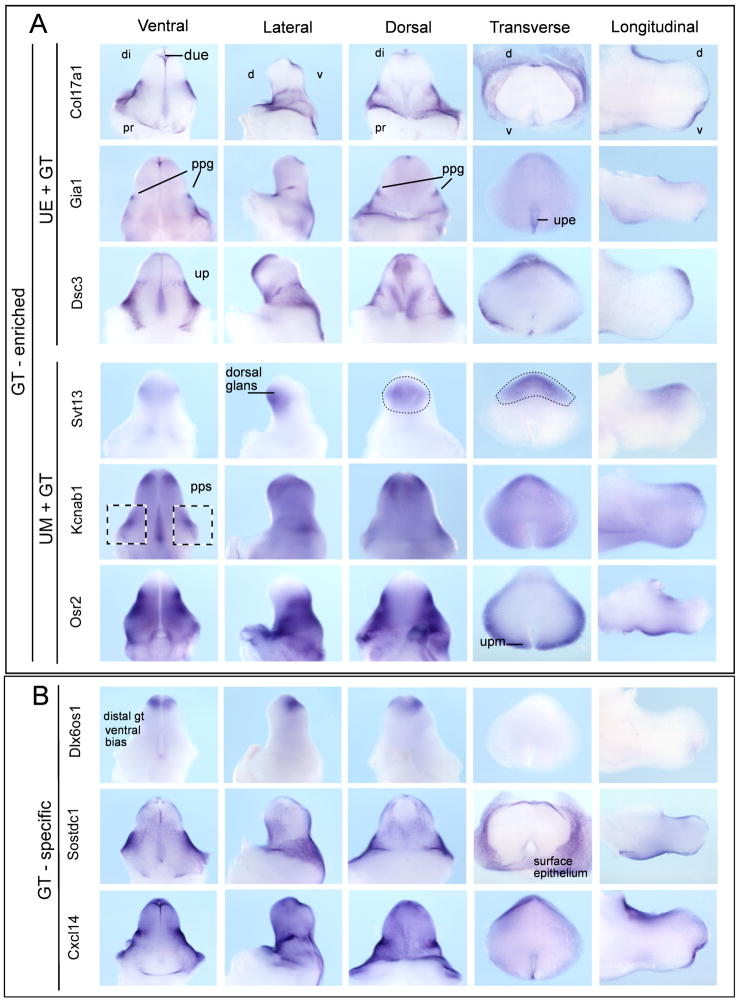
WISH of E13.5 GT was performed to validate the microarray data and investigate the spatial relationship of the GT epithelial-enriched (“on” in GT and E13 urethral epithelium), GT mesenchyme-enriched (“on” in GT and E13 urethral mesenchyme) and GT-specific genes (only observed in E14 GT). As GT was not separated from LUT at E13, genes expressed in E13 urethral epithelium or mesenchyme, but not expressed in E14 urethra, were likely to represent early GT genes, possibly enriched for those expressed along the urethral plate. Those restricted to the epithelium at E13 were most likely to be expressed in the epithelium of urethral plate of the E14 GT, while those expressed in the mesenchyme at E13 were likely to be in the mesenchyme along the urethral plate in the E14 GT. WISH validated these assumptions. In order to fully annotate the gene expression patterns observed, we modified the previous genitourinary tract ontology (Little et al, 2006) to increase the level of anatomical resolution for the TS21 genital tubercle (Supplementary Figure 2). Figure 4 demonstrates examples of GT-enriched and GT-restricted gene expression patterns. A schematic representation of the features of the developing GT can be seen at http://www.gudmap.org/Internal/Consortium/Work_In_Progress/Organ_Summaries/index.html (currently requires username: Gudmap_pr, password: 888gudmap). In total, validation of GT-enriched or restricted expression was observed for 43/58 genes examined by WISH (74% validation of array data). Table 2 summarises the annotated expression patterns these genes in the developing GT. For those where validation was not observed, signal was either absent/very weak or expression was seen in additional sites. The former is likely to result from the reduced sensitivity of the ISH in comparison to microarray (negatives). The latter may result from the riboprobe detecting an isoform of the gene not identified by the Affymetrix oligonucleotide (additional sites of expression).
Figure 4. Validation of E14 genital tubercle-enriched gene expression.
A) Genital tubercle-enriched genes predicted to be expressed in UP epithelium and GT (Col17a, Gja1, Dsc3) or UP mesenchyme and GT (Syt13, Kcnab1, Osr2). Expression can also be seen in preputial gland (Gja1), distal urethral epithelium (Col17a, Gja1, Dsc3), surface epithelium of glans and preputial swelling (Col17a, Gja1, Dsc3) and preputial swelling mesenchyme (Kcnab1, Osr2). B) Genital tubercle-restricted genes (Dlx6os1, Sostdc1, Cxcl14) showed expression in preputial swelling epithelium (Sostdc1, Cxcl14), distinct domains across the dorsal GT (Dlx6os1, Sostdc1, Cxcl14) and distal genital tubercle (glans) (Dlx6os1). Material is presented from left to right as wholemounts of ventral GT, lateral GT (dorsal to the left side), dorsal GT, and transverse and longitudinal sections through wholemount specimens. di, distal; pr, proximal; d, dorsal; v, ventral; due, distal urethral epithelium; up, urethral plate; upe, up epithelium; upm, up mesenchyme; ppg, preputial gland; pps, preputial swelling.
Novel gene expression domains on the dorsal surface of the presumptive glans
The dorsal side of the extending genital tubercle is longer than the ventral surface such that there is an overhanging dorsal lip and the orifice of the DUE is on the ventral side. At E13.5, there is a discernable glans that is most prominent from the dorsal side. Analysis of the gene expression patterns seen in E13.5 GT revealed some distinct patterns of expression as yet unreported. Of particular interest were very tightly delineated domains of expression on the dorsal surface of the genital tubercle. These genes did not show dorsally-restricted expression, but did reveal specific dorsal patterns of expression around the tip of the extending glans that allowed the subdivision of distal genital tubercle into a number of expression patterns not previously reported. Dlx6os1, Cpm, Mup1, Sty13 and Hs3st3b1 each showed distinct but non-overlapping expression in specific regions of the distal GT (some epithelial, some mesenchymal) other than the DUE (Figure 4, 6 and www.gudmap.org). Distinct gene expression patterns were also seen on the preputial swellings (e.g. Sostdc1, Dsc3, Figure 4) and preputial glands (e.g. Gja1, Figure 4).
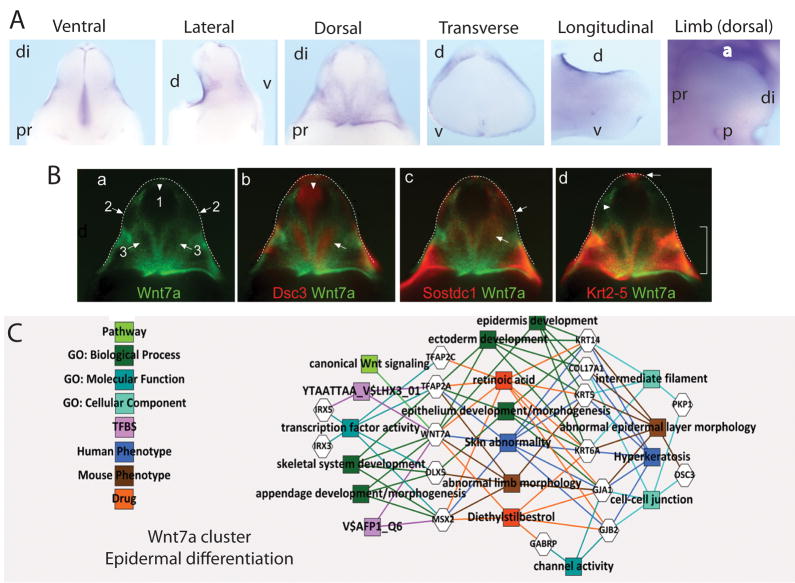
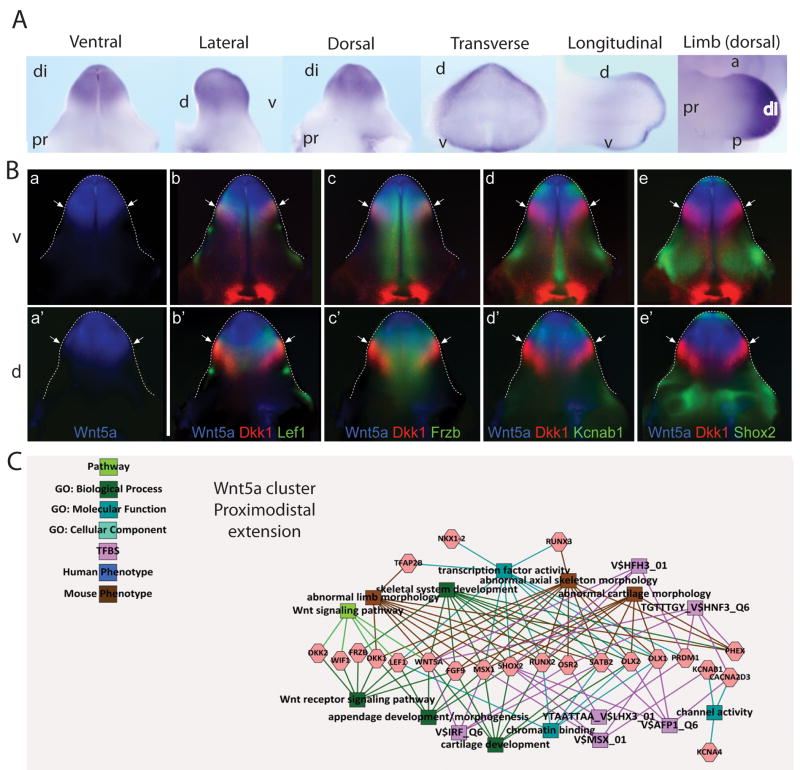
Figure 6. Identification of Wnt7a associated gene networks in GT development.
A. Wholemount ISH of Wnt7a in the developing E13.5 GT showing ventral, lateral, dorsal, transverse and longitudinal views, as well as Wnt5a expression in the limb (dorsal view). B. Pseudo-colored expression pattern overlays of known and candidate genes of a Wnt7a network. Experimental WISH images of stage-matched genital tubercles at 13.5dpc were pseudo-colored and overlaid with Wnt7a (a–d). Dorsal (d; a–d) views are shown and the edge of the GT is outlined with a dotted line. Wnt7a network genes include Wnt7a alone (a), Dsc3/Wnt7a (b), Sostdc1/Wnt7a (c) and Krt5/Wnt7a (d). (a) Arrows indicate the main regions (1–3) on the dorsal side of the GT where Wnt7a was not expressed. (b) Dsc3 showed a complementary pattern to Wnt7a with strong, tightly restricted expression in regions 1 and 3 (arrows), but was expressed in the epithelium. (c,d) Sostdc1 and Krt5 showed overlapping expression with Wnt7a. Sostdc1 was expressed more broadly than Wnt7a and extended into regions 2 and 3 (arrows), however like Wnt7a, expression was absent from the central region 1. (d) Krt5 expression closely resembled Wnt7a with strong expression in the proximal Wnt7a domains. In the distal Wnt7a domains, only weak Krt5 expression was seen (arrowhead), except for a small region of strong expression at the distal GT tip (arrow). C. GT epithelium-enriched gene cluster associated via hierarchical clustering decorated with GO terms and known transcription factor binding sites and known human and mouse phenotype associations. This cluster shows strong links to retinoic acid and diethylstilbestrol regulation as well as epithelium and epidermal differentiation, notably in the limbs.
Validation of Wnt5a signaling in GT mesenchyme and identification of an associated gene network
The WISH-confirmed GT-enriched genes contained many Wnt signaling pathways members (see Supplementary Figure 3). As previously reported, Wnt5a was expressed in the mesenchyme of the distal genital tubercle (Yamaguchi et al, 1999; Suzuki et al, 2003) (Figure 5A). In order to predict genes interacting with Wnt5a within this compartment, we generated pseudo-colored overlays of WISH results from all GT-enriched genes. This allowed the direct alignment of up to three genes to examine in detail their spatial relationships (Figure 5B). Wnt5a expression overlapped with the distal GT mesenchymal expression of Frizzled b (Frzb), a secreted frizzled protein that acts as an inhibitor of the pathway (Figure 5Bc, c′). The proximal edges of the Wnt5a expression domain were also marked by a unique and distinct expression pattern of Dkk1, an inhibitor of canonical Wnt signaling (Figure 5Bb–e, b′–e′). This combination of ligand and inhibitor appears to delineate the GT equivalent of a progress zone in the glans/distal GT. Comparison of this domain of expression with other genes suggests a possible interaction with several other genes from within the GT mesenchyme-enriched cluster. Kcnab1 shows overlapping expression with Wnt5a in the glans, but is expressed in a wider region of the GT mesenchyme, notably not overlapping the expression of Dkk1 (Figure 5Bd, d′). The Wnt downstream signaling molecule Lef1 also shows partial synexpression with Wnt5a (Figure 5Bb, b′). In contrast, the short stature homeobox 2 gene (Shox2) shows a complementary pattern of expression (Figure 5Be, e′). These visual comparisons of spatial expression patterns suggest an interaction between these genes. To both validate these predictions and define potentially Wnt5a-interacting genes, the initial GT-enriched dataset was re-analyzed to seek genes displaying synexpression across the LUT/GT with Wnt5a using K-means and hierarchical clustering. This identified a cluster of genes with moderate to high expression in the E13 urethral mesenchyme (Wnt5a, Dkk2, Frzb, Wif1, Lef1, Phex, Runx2, Dlx2, Msx1, Dlx1, Shox1, Satb2, Osr2, Runx5, Cacna2d3, Prdm1, Kcnab1, Tfap2b, Nkx1.2, Kcna4). Toppgene analysis of this cluster identified enrichment for genes involved in skeletal and cartilage development/morphology (Figure 5C). For example, Runx2 has previously been shown to be regulated by Shox2 in limb chondrogenesis prior to long bone formation (Cobb et al, 2006). These genes often displayed differential anterior-posterior limb and dorsal-ventral GT expression, reinforcing congruence in the development of these two appendicular structures. Many also showed differential proximal-distal patterning in both structures, as is the case for Wnt5a. This suggests possible interactions between these genes in proximal-distal extension of the GT.
Figure 5. Identification of Wnt5a associated gene networks in GT development.
A. Wholemount ISH of Wnt5a in the developing E13.5 GT showing ventral, lateral, dorsal, transverse and longitudinal views, as well as Wnt5a expression in the limb (dorsal view). di, distal; pr, proximal; d, dorsal; v, ventral; a, anterior; p, posterior. B. Pseudo-colored expression pattern overlays of known and candidate genes of a Wnt5a network. Experimental WISH images of stage-matched genital tubercles at 13.5dpc were pseudo-colored and overlaid with either Wnt5a (a–e, a′–e′). Ventral (v; a–e) and dorsal (d; a′–e′) views are shown and the edge of the GT is outlined with a dotted line. Wnt5a network genes include Wnt5a alone (a, a′), Wnt5a/Dkk1/Lef1 (b, b′), Wnt5a/Dkk1/Frzb (c, c′), Wnt5a/Dkk1/Kcnab1 (d, d′) and Wnt5a/Dkk1/Shox2 (e, e′). Arrows indicate the border of the Wnt5a distal GT expression domain. All genes showed some degree of overlap with the Wnt5a domain. Dkk1 showed overlapping expression with Lef1 (b, b′) and Frzb (c, c′), in contrast to Kcnab1 (d, d′) and Shox2 (e, e′), which did not overlap with the distal Dkk1 domain. C. GT mesenchyme-enriched gene cluster associated via hierarchical clustering decorated with GO terms and known transcription factor binding sites and known human and mouse phenotype associations. This cluster shows strong links to limb morphogenesis, particularly of mesodermal elements including cartilage and bone.
Evidence for a Wnt7a-associated gene network in the epidermis of the GT
In contrast to Wnt5a, Wnt7a was expressed in the surface epithelium of the GT with stronger expression on the dorsal surface (Figure 6A). Little attention has been paid to expression in the dorsal GT. Wnt7a was also expressed on the surface epithelium of the preputial swellings (Figure 6A). Pseudo-coloured overlays suggested co-expression of Wnt7a, the BMP antagonist sclerostin domain containing-1 (Sostdc1) and keratin 2–5 (Krt5) in tightly defined regions of the dorsal epithelial layer of the GT. Expression of these genes was also seen in the preputial swellings, but these genes were excluded from the dorsal distal glans other than at the DUE (Figure 6Bcd). Also epithelial in expression were several genes with complementary expression patterns on the dorsal distal glans. These included desmocollin 3 (Dsc3) (Figure 6Bb) and Gjb2 (Figure 4), both genes previously associated with skin development. K-means and heirarchical clustering supported the association of these and other genes as representing a synexpression group in E13 urethral epithelium, therefore potentially restricted to surface epithelium (Wnt7a, Dlx5, Msx2, Tfap2c, Krt2–5, Gja1, Irx3, Irx5, Tfap2a, Gabrp, Krt6a, Krt14, Col17a, Dsc3 and Pkp1). Toppgene analysis of this Wnt7a-associated cluster showed a strong link with epithelial and epidermis development/morphology, including clear links to skin abnormalities (including hyperkeratosis) in humans (Figure 6C). Genes within this cluster are involved in cell-cell junction and intermediate filament biology and some (Gjb2, Dsc3 and Krt5) have previously been reported to be involved in plantar/palmar expression in the developing limb, as has Wnt7a. Of particular note, many genes within this cluster are regulated by diethylstilbestrol (Wnt7a, Msx2, Gja1, Gjb2, Tfap2c, Gabrp, Krt6a) and/or retinoic acid (Krt14, Krt6a, Wnt7a, Tfap2a, Krt5, Gja1, Gjb2, Tfap2c). A final cluster of genes was identified using K means and hierarchical clustering. This represented GT-specific genes, many of which showed distinct proximal-distal restriction of expression in the GT (Supplementary Figure 4). Heatmaps of each of these clusters (GT mesenchyme (Wnt5a) associated, GT epithelium (Wnt7a) associated and GT specific) are shown in Supplmentary Figure 5.
A comparison of gene expression during appendage formation
Congruence of molecular processes in appendicular outgrowth was first observed between limb and branchial arch development with the observation of a key role for Fgf8 in the patterning of the first branchial arch (Trumpp et al, 1999). As for the limb, Fgf8 expression is seen in the overlying ectoderm of the first branchial arch is required for the survival and migration of the underlying mesenchyme. A similar congruence was then observed for genital tubercle development (Haraguchi et al, 2000; Yamada et al, 2006) with hedgehog and Fgf signaling showing considerable similarities in all three tissues. This began to explain the coincidence of defects within these tissues in many birth defects in humans (Schneider et al, 1999). Our analysis of the sites of expression of genes within the Wnt5a associated network in the genital tubercle would support a role for this cluster in appendage outgrowth. WISH was also performed for these genes in whole embryo to examine limb and craniofacial prominence gene expression. This showed very tight concordance in spatial expression patterns between limb and GT, as previously hypothesized (Trumpp et al, 1999; Schneider et al, 1999). Supplementary Figure 6 schematically represents this congruence in gene expression between two of these appendicular structures; limb and GT. To further examine the hypothesis that there are similarities in GT patterning with the other appendages in the embryo, we performed WISH on E10.5 and E11.5 embryos examining expression in limb, GT and branchial arches. In total, 74% (23/31) of GT-confirmed genes showed expression in limb and 71% (22/31) showed expression in the brnachial arches/craniofacial prominences (Supplementary Table 8). In almost all cases, no other strong sites of gene expression were observed, supporting the notion of congruence in regulation of appendage development. As previously described, there was also a congruence the axis of expression between dorsal-ventral GT and anterior-posterior limb (Figure 7; Shox2, Tfap2b, Msx1, Tfap2c). However, a careful analysis of all GT gene expression patterns in comparison with limb expression patterns indicated that this was not always the case (Figure 7).
Figure 7. Examination of the congruence between gene expression polarity in genital tubercle versus limb.
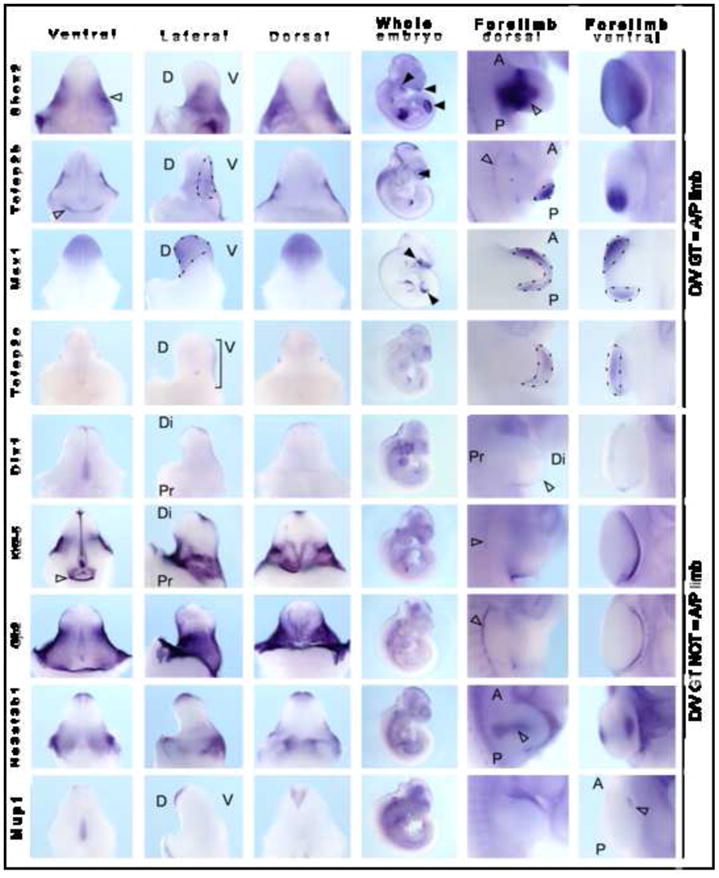
Wholemount ISH was performed on E13.5 genital tubercle, E10.5 whole embryo and E10.5 forelimb. Specimens are presented from left to right as ventral GT, lateral GT, dorsal GT, E10.5 whole embryo, E11.5 dorsal limb and apical/ventral forelimb limb views. Expression of Shox2, Tcfap2b and Msx1 showed congruence of dorsal/ventral GT and anterior/posterior limb (lateral GT = DV limb). Shox2 was expressed in the mesenchyme of the paired lateral preputial swellings (open arrowhead) and was also expressed in central mesenchyme of the developing limb. Tcfap2b showed preputial swelling expression. D, dorsal; V, ventral; A, anterior; P, posterior; Di, distal; Pr, proximal.
Discussion
This study describes the creation and analysis of the first comprehensive catalogue of gene expression profiles describing the development of the early murine lower urinary tract and genital tubercle. Li et al (2006) previously performed microarray analysis of GT development, however the data presented only listed genes that increased in E16 and E17 GT versus E14. In addition, this data was collected from cDNA arrays and not Affymetrix. Hence a comparison cannot be made with this data. A comparison of these analyses with previous literature, together with our extensive wholemount ISH analysis supports the validity of the data. Hence, this will be a valuable resource for investigating the molecular basis of morphogenesis in this organ system and to investigate correlations between development and disease. By way of example, mutations in the renin-angiotensin system, Robo2, HNF1β (TCF2), PAX2 (renal-coloboma syndrome), EYA1 and SIX1 (branchio-oto-renal syndrome (BORS)) and SALL1 (Townes-Brocks syndrome (TBS)) have been reported in CAKUT patients (Weber et al, 2008). Although not previously associated with bladder, urethra or genital defects, strong and compartment-enriched expression in the developing LUT/GT was identified for a number of these CAKUT-associated genes showed. SALL1 mutations in TBS result in renal dysplasia and VUR. Our data shows that Sall1 is clearly expressed in the urethra mesenchyme and GT, but not in the bladder. Only a single case report exists of a TBS patient with hypospadias (Salerno et al, 2000), but our data suggest that Sall1 may be more commonly involved in GT defects. Sall1 showed strong synexpression with Sox11, suggesting that mutations in this gene may also cause similar defects. Expression of SIX1 and EYA1, both mutated in BOR, showed highly similar gene expression patterns in the LUT. Neither was expressed in the bladder, but both were expressed in the E13 urethral mesenchyme, E14 GT, urethra and UGS. There have been no links between these genes and genital defects, but both are associated with defects in the development of other appendages (limbs, craniofacial structures) suggesting a possible involvement in GT as well.
As a further example of the validity and potential of this dataset, Toppgene analysis of the this dataset also annotated six genes as being involved in anal, body wall, reproductive and genital development (TP63, Sall1, Gli3, Sall4, Pitx2, Tbx3). Patients with anorectal malformations show high levels of genital (13.63%) or urologic (11.36%) anomalies (Saha et al, 2005), suggesting the need to carefully examine the urogenital tracts of newborns with anorectal anomalies. Nakata et al (2009) have recently also reported an association between Wnt5a expression in mesenchyme close to the cloacal membrane and normal anorectal development. The link between the body wall has been less noted, although it is known that prune belly syndrome, a male-associated anomaly in the musculature of the abdominal body wall, is also associated with cryptorchidism and urinary tract anomalies (Bogart et al, 2006). Prune belly syndrome has also been linked to posterior urethral valves (Weber et al, 2005) and loci on human chromosomes 1q and 11p, as well as occurring in patients with Six1 mutations and renal defects (BOR syndrome).
To test the utility of this catalogue in investigating the development of an individual compartment, we focused on the genital tubercle and have validated and fully described the spatial expression pattern of 31 genes likely to be critical for GT extension and differentiation. GT development has been proposed to be similar to the development of other appendages, including the limb and face. Toppgene analysis confirmed this suggestion although the most enriched pathway in the GT was Wnt signaling, a growth factor family known to be involved in the limb (Witte et al, 2009). In the limb, Wnt5a is involved in proximal-distal limb extension as loss of Wnt5a reduces proliferation within the mesenchymal progress or undifferentiated zone below the AER, resulting in limb shortening and loss of more distal structures (digits) (Yamaguchi et al, 1999). This can be mimicked by inactivation of mesenchymal β-catenin (Hill et al, 2006), suggesting that this process involves canonical Wnt signaling (Geetha-Loganathan et al, 2008). The canonical Wnt inhibitor, Dkk1, is expressed in the AER and loss of this gene widens the AER but does not disturb plantar-palmar patterning (Adamska et al, 2003), linking Dkk1 with Wnt5a rather than Wnt7a activity. Similarly in the GT, loss of Wnt5a is also known to disrupt proximal-distal extension (Yamaguchi et al, 1999). Wnt5a expression has been reported to be expressed in the GT mesenchyme and appears to be regulated, along with Msx2, Lef1 and Bmp4, by signals from the distal urethral epithelium (Lin et al, 2008). Our data on Wnt5a agrees with the literature in that expression was seen in the GT mesenchyme. Our clustering and expression analyses implied interaction between Wnt5a and other known Wnt pathway members, including Frzb (secreted frizzled related protein 3, Sfrp3), Dkk1, Dkk2, Msx1, Tcfap2b and Lef1. An association between Wnt5a and Frzb in digit formation has already been described (Chimal-Monray et al, 2002), supporting the validity of this cluster. Alignment of the expression patterns of these genes showed a distinct band of Dkk1 expression delineating the boundary of the Wnt5a expression in the emerging glans region, presumably delimiting the boundary of Wnt5a-driven canonical Wnt signaling.
Differentiation between the plantar and palmar (dorsal and ventral) surface of the limb requires dorsal Wnt7a signaling accompanied by repression of that signal on the ventral surface by the transcription factor, engailed-1 (En-1) (Geetha-Loganathan et al, 2008). Dorsal Lmx1b expression is also thought to be regulated by Wnt7a (Chen and Johnson, 2002; Geetha-Loganathan et al, 2008). Rare occasions have been reported where there has been a duplication of the palmar surface due to mutation in Wnt7a (Al-Qattan et al, 2009). The role of Wnt signaling in the developing ectoderm of the GT has not previously been investigated. Unlike the limb, Wnt7a expression was not restricted to the dorsal surface of the GT although in this region it did show a specific pattern of expression. Alignment of ISH and gene cluster analysis identified other genes expressed in the GT surface epithelium (Sostdc1, Gjb2, Dsc3 and Krt5), most of which have known roles in epidermal differentiation in other tissues. Gjb2 encodes a gap junction protein also called connexin 26. Mutations in connexin 26 result in deafness as a result of cornification of the epithelium within the cochlea of the ear (Richard, 2000; Houang et al, 2002). Connexin 26 and connexin 31 (Gjb3) are crucial for normal epidermal development and, as well as deafness, connexin 26 mutations can result in palmo-plantar keratoderma (Richard, 2000; de Zwart-Storm et al, 2008). Dsc3 is a cadherin-like protein present in desmosomes and previously reported to be expressed in the epidermis of the foreskin (King et al, 1995). Dsc3 is upregulated along with β-catenin in squamous cell carcinoma (Wang et al, 2007) and disruptions of squamous and stratified epithelium. This includes palmo-plantar keratoderma and immunobullous disease in a patient with circulating antibodies against Dsc3 (Bolling et al, 2007). This patient presented hyperkeratotic ridges on their palms as well as intra-epidermal pustules. Based on these prior associations, we can predict that this cluster of genes is involved in differentiation of the epidermis of the GT, but not necessarily the epithelium of the UP.
Genes within the predicted Wnt7a-associated network are known to be modulated by retinoic acid (RA) and diethylstilbestrol (DES). Sexual dimorphism of the external genitalia is dependent on steroid hormones and there have been many reports linking imbalance between estrogen and androgen signaling to hypospadias, including the use of DES during pregnancy (Brouwers et al, 2007). This is thought to affect development during the androgen-sensitive sexually dimorphic stage of GT development (>E16) rather than during early development. An association between these epidermal GT genes and DES implies either a previously unreported early oestrogen-sensitive period or involvement in both the androgen-independent and subsequent androgen-sensitive phase of GT development. DES exposure during development also results in metaplasia of the normally simple columnar epithelium of the uterus, thereby preventing embryo implantation (Huang et al, 2005). Here Msx2 is required for normal Wnt7a expression and DES can repress the expression of both Msx2 and Wnt7a in turn (Huang et al, 2005). This effect is mediated via the estrogen receptor, since ER-KO mice treated with DES do not show a reduction in Wnt7a (Couse et al, 2004). Hence, Wnt7a and other DES-responsive genes in this network may also be involved in female reproductive tract development. Sexual dimorphism of the external genitalia is also thought to involve RA signaling due to the dynamic expression of RARs, RXRs, RALDH2 and CYP26 (Ogino et al, 2001). Administration of RA to mouse embryos as young as E9 does not affect genital tubercle extension, but does disrupt the formation of the urethral plate. Our re-examination of these data revealed undescribed defects in the development of the preputial swellings and the shape of the distal glans (Ogino et al, 2001). Studies on the effect of an absence of RA on limb development in the quail embryo showed that while all axes were disrupted, this particularly affected limb dorsal-ventral patterning genes, including Wnt7a and En-1 (Stratford et al, 1999). While this does not eliminate a role for Wnt7a in other axes of GT development, it is suggestive of limb/GT congruence and strengthens the association between the genes in this cluster.
Overall, our observations suggest that Wnt7a may play a role in epidermal differentiation of the GT in collaboration with genes known to play roles in plantar-palmar epidermal dimorphism. The expression of Wnt7a-synexpressed genes in the epithelium of the preputial swelling may implicate this network in the differentiation of the foreskin and labia rather than in extension of the glans. This division of expression may serve to demarcate the development of the smooth skin of the penis and clitoris and the epidermis that will give rise to the foreskin, labia minora and scrotum (Figure 6A, B). The link to androgen regulation may also suggest that at least some forms of hypospadias or other congenital glans malformations may involve disruptions to epidermal development. Indeed, a review of 137 non-diphallic non-hypospadiac glans defects noted dermatological lesions in 15% of cases (Papali et al, 2008).
In this study, gene ontology, pathway and network analysis of the microarray data has reinforced the concept of molecular congruence in appendage development. However, while initially arising as paired genital swellings, the glans in both sexes (penis and clitoris) becomes a single midline structure, unlike the limbs. Significant morphological differences exist between the dorsal and ventral surfaces of the developing limb, both externally with respect to skin, hair and nail formation, and internally with respect to extensor and flexor muscle positioning. The same is true for the anterior-posterior axis of the limbs. This is not as prominent in the external genitalia. While duplication of the Shh-expressing ZPA on the posterior surface of the limb (duplication of the A-P axis) gives rise to a duplication of digits, a dorsal duplication of the Shh-expressing urethral plate never occurs. Duplication of the urethral plate can occur if there is an underlying duplication of the urethral endoderm, but this duplication occurs ventrally and gives rise to a total GT duplication, referred to as diphallia in males (Kaufman et al, 1990; Gryftopoulos et al, 2002; Mughal et al, 2003). This is usually accompanied by urethral or bladder duplication and imperforate anus. Such phallic duplications almost always occur in the lateral plane such that the two appendages are positioned side by side. Rare cases of duplication apparently in the dorsal-ventral axis usually involve incomplete or glans-only duplication with no accompanying urethral or bladder duplication (Melekos et al, 1986; Gavali et al, 2002). If the limb A-P axis is equivalent to GT D-V axis, lateral diphallia would be equivalent to a duplication of the hand in the D-V axis, which has never been reported. We therefore conclude that while early congruences in patterning exist between different appendages, particularly in relation to extension, there remain significant and anatomically obvious differences between these structures and hence the molecular regulation of these processes.
Supplementary Material
Heatmaps of the most active genes for each of the eight compartments of the LUT examined.
Changes to anatomical ontology of the urogential tract with respect to the developing genital tubercle.
Microarray analysis revealed a cluster of members of the Wnt signaling pathway, with expression enriched in E14 genital tubercle. These included Wnt ligands (Wnt5a, Wnt7a), signaling molecules (Lef1), inhibitors (Dkk1, Frzb, Wif1) and targets (Sostdc1). Wholemount ISH of E13.5 GT displayed (from left to right) as ventral, lateral and dorsal wholemounts, and transverse and longitudinal sections demonstrate the expression of these Wnt pathway genes. Expression in E11.5 limb is also shown. Di, distal; Pr, proximal; D, dorsal; V, ventral; A, anterior; P, posterior.
Cluster association diagrams for genital tubercle-enriched genes
Heatmaps of genital tubercle-enriched and genital-tubercle-specific clusters 1–3.
Diagrammatic representations of congruence between spatial gene expression patterns in the extension of genital tubercle and limb. Congruence applies between ventral GT and posterior limb, dorsal GT and anterior limb and lateral GT and dorsal limb. Signaling for extension comes from the UP in the GT and the ZPA in the limb. GT, genital tubercle; AER, apical ectodermal ridge; ZPA, zone of polarizing activity; UP, urethral plate; UM, urethral mesenchyme; DGT, distal genital tubercle (glans).
List of the 1420 most highly active genes in the developing LUT incorporating individual lists for each compartment.
Toppgene analyses for the most highly active genes in each compartment of the developing LUT
LUT active genes associated with reproductive, genital, anal, body wall, kidney/ureter and bladder development
Gene lists for the most compartment-enriched genes in each compartment of the developing LUT
Lists of enriched genes for all E14 LUT compartments (GT, bladder, UGS, urethra) including Affymetrix identification numbers and primers used for the generation of riboprobes.
Heat map and gene list for BMP, FGF, Wnt and hedgehog pathway expression across the developing LUT
Toppgene analysis of the 98 probesets identified as enriched in or specific to E14 GT
Summary of expression in GT, limb and craniofacial arch for 37 confirmed GT-enriched or specific genes.
Acknowledgments
We thank Emmanuelle Lesieur for assistance in material collection and Jane Brennan, Jane Armstrong, Chris Armit, Jamie Davies, Derek Houghton, Sue Lloyd-MacGilp, Mehran Sharghi, Xinguin Pi, Yogmatee Roochun, Ying Cheng, Simon Harding, Jamie Davies and Duncan Davidson for GUDMAP editorial and database support. We also thank Carol Wicking for scientific opinion on limb and facial development. ML is a Principal Research Fellow of the National Health and Medical Research Council. This work was supported by the National Institutes of Health via awards DK070136 (MHL) and DK070219 (JLL).
Grant information: Grant sponsor National Institute of Health; Grant numbers DK070136 (MHL) and DK070219 (JLL).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aboushwareb T, Zhou G, Deng FM, Turner C, Andersson KE, Tar M, Zhao W, Melman A, D’Agostino R, Jr, Sun TT, Christ GJ. Alterations in bladder function associated with urothelial defects in uroplakin II and IIIa knockout mice. Neurourol Urodyn. 2009 doi: 10.1002/nau.20688. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adamska M, MacDonald BT, Meisler MH. Doubleridge, a mouse mutant with defective compaction of the apical ectodermal ridge and normal dorsal-ventral patterning of the limb. Dev Biol. 2003;255:350–362. doi: 10.1016/s0012-1606(02)00114-8. [DOI] [PubMed] [Google Scholar]
- Ahn K, Mishina Y, Hanks MC, Behringer RR, Crenshaw EB., III BMPR-1A signaling is required for the formation of the apical ectodermal ridge and dorsal-ventral patterning of the limb. Development. 2001;128:4449–4451. doi: 10.1242/dev.128.22.4449. [DOI] [PubMed] [Google Scholar]
- Al-Qattan MM, Al-Balwi M, Eyaid W, Al-Abdulkarim I, Al-Turki S. Congenital duplication of the palm syndrome: gene analysis and the molecular basis of its clinical features. J Hand Surgery. 2009;34E:247–251. doi: 10.1177/1753193408099828. [DOI] [PubMed] [Google Scholar]
- Beleza-Meireles A, Lundberg F, Lagerstedt K, Zhou X, Omrani D, Frisen L, Norddenskjold A. FGFR2, FGF8, FGF10 and BMP7 as candidate genes for hypospadias. Eur J Hum Genetics. 2007;15:405–410. doi: 10.1038/sj.ejhg.5201777. [DOI] [PubMed] [Google Scholar]
- Bogart MM, Arnold HE, Greer KE. Prune-belly syndrome in two children and review of the literature. Pediatr Dermatol. 2006;23:342–345. doi: 10.1111/j.1525-1470.2006.00265.x. [DOI] [PubMed] [Google Scholar]
- Bolling MC, Mekkes JR, Goldschmidt WF, van Noesel CJ, Jonkman MF, Pas HH. Acquired palmoplantar keratoderma and immunobullous disease associated with antibodies to desmocollin 3. Br J Dermatol. 2007;157:168–173. doi: 10.1111/j.1365-2133.2007.07920.x. [DOI] [PubMed] [Google Scholar]
- Brouwers MM, Feitz WF, Roelofs LA, Kiemeney LA, de Gier RP, Roeleveld N. Risk factor for hypospadias. Eur J Pediatr. 2007;166:671–678. doi: 10.1007/s00431-006-0304-z. [DOI] [PubMed] [Google Scholar]
- Brunskill EW, Aronow BJ, Georgas K, Valerius MT, Aronow JE, Kaimal V, Jegga AG, Grimmond S, McMahon AP, Patterson L, Little MH, Potter SS. Atlas of Gene Expression in Developing Kidney at Microanatomic Resolution. Developmental Cell. 2008;15:781–791. doi: 10.1016/j.devcel.2008.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Johnson RL. Interactions between dorsal-ventral patterning genes Imx1b, engrailed-1 and wnt-7a in the vertebrate limb. Int J Dev Biol. 2002;46:937–941. [PubMed] [Google Scholar]
- Chen J, Bardes EE, Aronow BJ, Jegga AG. ToppGene Suite for gene list enrichment analysis and candidate gene prioritization. Nucleic Acids Res. 2009;37(Web Server issue):W305–311. doi: 10.1093/nar/gkp427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chimal-Monroy J, Montero JA, Gañan Y, Macias D, Garcia-Porrero JA, Hurle JM. Comparative analysis of the expression and regulation of Wnt5a, Fz4, and Frzb1 during digit formation and in micromass cultures. Dev Dyn. 2002;224(3):314–320. doi: 10.1002/dvdy.10110. [DOI] [PubMed] [Google Scholar]
- Cobb J, Duboule D. Comparative analysis of genes downstream of the Hoxd cluster in developing digits and external genitalia. Development. 2005;132:3055–3067. doi: 10.1242/dev.01885. [DOI] [PubMed] [Google Scholar]
- Cobb J, Dierich A, Huss-Garcia Y, Duboule D. A mouse model for human short-stature syndromes identifies Shox2 as an upstream regulator of Runx2 during long-bone development. Proc Natl Acad Sci U S A. 2006;103:4511–4515. doi: 10.1073/pnas.0510544103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couse JF, Korach KS. Estrogen receptor-alpha mediates the detrimental effects of neonatal diethylstilbestrol (DES) exposure in the murine reproductive tract. Toxicology. 2004;205:55–63. doi: 10.1016/j.tox.2004.06.046. [DOI] [PubMed] [Google Scholar]
- Cox MJ, Coplen DE, Austin PF. The incidence of disorders of sexual differentiation and chromosomal abnormalities of cryptorchidism and hypspadias stratified by meatal location. J Urol. 2008;180:2649–2652. doi: 10.1016/j.juro.2008.08.058. [DOI] [PubMed] [Google Scholar]
- De Zwart-Storm EA, van Geel M, van Neer PA, Steijlen PM, Martin PE, van Steensel MA. A novel missense mutations in the second extracellular domain of GFB2, p. Ser183Phe, causes a syndrome of focal palmoplantar keratoderma with deafness. Am J Pathol. 2008;173(4):1113–1119. doi: 10.2353/ajpath.2008.080049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler CJ, Griffiths D, de Groat WC. The neural control of micturition. Nature Reviews. 2008;9:453–466. doi: 10.1038/nrn2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavali S, Deshpande AV, Sanghani HH, Hirugade ST, Talpallikar MC, Borwankar SS. Glanular diphallus with urethral stricture. Pediatr Surg Int. 2002;18:70–71. doi: 10.1007/s003830200018. [DOI] [PubMed] [Google Scholar]
- Geetha-Loganathan P, Nimmagadda S, Scaal M. Wnt signaling in limb organogenesis. Organogenesis. 2008;4(2):109–115. doi: 10.4161/org.4.2.5857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman FR, Bacchelli C, Brady AF, Brueton LA, Fryns JP, Mortlock DP, Innis JW, Holmes LB, Donnenfeld AE, Feingold M, Beemer FA, Hennekam RC, Scambler PJ. Novel HOXA13 mutations and the phenotypic syndrome of hand-foot-genital syndrome. Am J Hum Genet. 2000;67:197–202. doi: 10.1086/302961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gyftopoulos K, Wolffenbuttel KP, Nijman RJ. Clinical and embryologic aspects of penile duplication and associated anomalies. Urology. 2002;60(4):675–679. doi: 10.1016/s0090-4295(02)01874-5. [DOI] [PubMed] [Google Scholar]
- Haraguchi R, Suzuki K, Murakami R, Sakai M, Kamikawa M, Kengaku M, Sekine K, Kawano H, Kato S, Ueno N, Yamada G. Molecular analysis of external genitalia formation: the role of fibroblast growth factor (Fgf) genes during genital tubercle formation. Development. 2000;127(11):2471–2479. doi: 10.1242/dev.127.11.2471. [DOI] [PubMed] [Google Scholar]
- Haraguchi R, Mo R, Hui C, Motoyama J, Makino S, Shiroishi T, Gaffield W, Yamada G. Unique functions of Sonic hedgehog signaling during external genitalia development. Development. 2001;128(21):4241–4250. doi: 10.1242/dev.128.21.4241. [DOI] [PubMed] [Google Scholar]
- Haraguchi R, Kotoyama J, Sasaki H, Satoh Y, Miyagawa S, Nakagata N, Moon A, Yamada G. Molecular analysis of coordinated bladder and urogenital organ formation by hedgehog signaling. Development. 2007;134:525–533. doi: 10.1242/dev.02736. [DOI] [PubMed] [Google Scholar]
- Houang M, Gourmelen M, Moatti L, Le BY, Garabédian EN, Denoyelle F. Hypogonadotrophic hypogonadism associated with prelingual deafness due to a connexin 26 gene mutation. J Pediatr Endocrinol Metab. 2002;15(2):219–223. doi: 10.1515/jpem.2002.15.2.219. [DOI] [PubMed] [Google Scholar]
- Hill TP, Taketo MM, Birchmeier W, Hartmann C. Multiple roles of mesenchymal beta-catenin during limb patterning. Development. 2006;133:1219–1229. doi: 10.1242/dev.02298. [DOI] [PubMed] [Google Scholar]
- Huang WW, Yin Y, Bi Q, Chiang TC, Vuoristo J, McLachlan JA, Ma L. Developmental diethylstilbestrol exposure alters genetic pathways of uterine cytodifferentiation. Mol Endocrinol. 2005;19:669–682. doi: 10.1210/me.2004-0155. [DOI] [PubMed] [Google Scholar]
- Kalfa N, Liu B, Ophir K, Audran F, Wang MH, Mei C, Sulton C, Baskin LS. Mutations of CXorf6 are associated with a range of severities of hypospadias. Eur J Endocrinol. 2008;159:452–458. doi: 10.1530/EJE-08-0085. [DOI] [PubMed] [Google Scholar]
- Kaufman A, Guia R, Davila H, Kaufman A. Diphallus with third urethra. Urology. 1990;35(3):257–260. doi: 10.1016/0090-4295(90)80045-o. [DOI] [PubMed] [Google Scholar]
- Kerecuk L, Schreuder MF, Woolf AS. Renal tract malformations: perspectives for nephrologists. Nature Clin Prac. 2008;4:312–325. doi: 10.1038/ncpneph0807. [DOI] [PubMed] [Google Scholar]
- King IA, Sullivan KH, Bennett R, Jr, Buxton RS. The desmocollins of human foreskin epidermis: identification and chromosomal assignment of a third gene and expression patterns of the three isoforms. J Invest Dermatol. 1995;105(3):314–321. doi: 10.1111/1523-1747.ep12319935. [DOI] [PubMed] [Google Scholar]
- Kurzrock EA, Baskin LS, Cunha GR. Ontogeny of the male urethra: theory of endodermal differentiation. Differentiation. 1999;64:115–122. doi: 10.1046/j.1432-0436.1999.6420115.x. [DOI] [PubMed] [Google Scholar]
- Li J, Willingham E, Baskin LS. Gene expression profiles in mouse urethral development. BJU International. 2006;98:880–885. doi: 10.1111/j.1464-410X.2006.06435.x. [DOI] [PubMed] [Google Scholar]
- Lin C, Yin Y, Long F, Ma L. Tissue-specific requirements of β-catenin in external genitalia development. Development. 2008;135:2815–2825. doi: 10.1242/dev.020586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C, Yin Y, Veith GM, Fisher AV, Long F, Ma L. Temporal and spatial dissection of Shh signaling in genital tubercle development. Development. 2009;136:3959–3967. doi: 10.1242/dev.039768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Little MH, Brennan J, Georgas K, Davies J, Davidson D, et al. A high-resolution anatomical ontology of the developing murine genitourinary tract. Gene Expr Patterns. 2007;7(6):680–699. doi: 10.1016/j.modgep.2007.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariani FV, Ahn CP, Martin GR. Genetic evidence that FGFs have an instructive role in limb proximal-distal patterning. Nature. 2008;453(7193):401–405. doi: 10.1038/nature06876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon AP, Aronow BJ, Davidson DR, Davies JA, Gaido KW, Grimmond S, Lessard JL, Little MH, Potter SS, Wilder EL, Zhang P. GUDMAP: the genitourinary developmental molecular anatomy project. J Amer Society Nephrol. 2008;19(4):667–671. doi: 10.1681/ASN.2007101078. [DOI] [PubMed] [Google Scholar]
- Melekos MD, Barbalias GA, Asbach HW. Penile duplication. Urology. 1986;27(3):258–259. doi: 10.1016/0090-4295(86)90285-2. [DOI] [PubMed] [Google Scholar]
- Miyagawa S, Buchanan DL, Sato T, Ohta Y, Nishina Y, Iguchi T. Characterization of diethylstilbestrol-induced hypospadias in female mice. Anat Rec. 2002;266:43–50. doi: 10.1002/ar.10033. [DOI] [PubMed] [Google Scholar]
- Miyagawa S, Moon A, Haraguchi R, Inoue C, Harada M, Nakahara C, Suzuki K, Matsumaru D, Kaneko T, Matsuo I, Yang L, Taketo MM, Iguchi T, Evans SM, Yamada G. Dosage-depenent hedgehog signals integrated with Wnt/b-catenin signaling regulate external genitalia formation as an appendicular program. Development. 2009;136:3969–3978. doi: 10.1242/dev.039438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan EA, Nguyen SB, Scott V, Stadler HS. Loss of Bmp7 and Fgf8 signaling in Hoxd13-mutant mice causes hypospadias. Development. 2003;130:3095–3109. doi: 10.1242/dev.00530. [DOI] [PubMed] [Google Scholar]
- Mughal SA, Soomro S, Shaikh JM. Double phallus. J Coll Physicians Surg Pak. 2003;13(9):534–535. [PubMed] [Google Scholar]
- Nakata M, Takada Y, Hishiki T, Saito T, Terui K, Sato Y, Koseki H, Yoshida H. Induction of Wnt5a-expressing mesenchymal cells adjacent to the cloacal plate is an essential process for its proximodistal elongation and subsequent anorectal development. Pediatr Res. 2009;66(2):149–154. doi: 10.1203/PDR.0b013e3181aa304a. [DOI] [PubMed] [Google Scholar]
- Ogino Y, Suzuki K, Haraguchi R, Satoh Y, Dolle P, Yamada G. External genitalia formation: role of fibroblast growth factor, retinoic acid and distal urethral epithelium. Ann NY Acad Sci. 2001;948:13–31. [PubMed] [Google Scholar]
- Papali AC, Alpert SA, Edmondson JD, Maizels M, Yerkes E, Hagerty J, Chaviano A, Kaplan WE. A review of pediatric glans malformations: a handy clinical reference. J Urol. 2008;180:1737–1742. doi: 10.1016/j.juro.2008.04.079. [DOI] [PubMed] [Google Scholar]
- Pennington EC, Hutson JM. The urethral plate – does it grow into the genital tubercle or within it? BJU International. 2002;89:733–739. doi: 10.1046/j.1464-410x.2002.02656.x. [DOI] [PubMed] [Google Scholar]
- Perriton CL, Powles N, Chiang C, Machonochie MK, Cohn MJ. Sonic hedgehog signaling from the urethral epithelium controls external genital development. Dev Biol. 2002;247(1):26–46. doi: 10.1006/dbio.2002.0668. [DOI] [PubMed] [Google Scholar]
- Richard G. Connexins: a connection with the skin. Exp Dermatol. 2000;9(2):77–96. doi: 10.1034/j.1600-0625.2000.009002077.x. [DOI] [PubMed] [Google Scholar]
- Saha SR, Roy AK, Saha S. Incidence of associated congenital anomalies in anorectal malformations. J Indian Med Assoc. 2005;103:690–691. [PubMed] [Google Scholar]
- Salerno A, Kohlhase J, Kaplan BS. Townes-Brocks syndrome and renal dysplasia: a novel mutation in the SALL1 gene. Pediatr Nephrol. 2000;14:25–28. doi: 10.1007/s004670050006. [DOI] [PubMed] [Google Scholar]
- Schneider RA, Hu D, Helms JA. From head to toe: conservation of molecular signals regulating limb and craniofacial morphogenesis. Cell Tissue Res. 1999;296(1):103–109. doi: 10.1007/s004410051271. [DOI] [PubMed] [Google Scholar]
- Siefert AW, Harfe BD, Cohn MJ. Cell lineage analysis demonstrated an endodermal origin of the distal urethra and perineum. Dev Biol. 2008;318:143–152. doi: 10.1016/j.ydbio.2008.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siefert AW, Yamaguchi T, Cohn MJ. Functional and phylogenetic analysis shows that Fgf8 is a marker of genital induction in mammals but is not required for external genital development. Development. 2009;136:2643–2651. doi: 10.1242/dev.036830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stratford T, Logan C, Zile M, Maden M. Abnormal anteroposterior and dorsoventral patterning of the limb bud in the absence of retinoids. Mech Dev. 1999;81:115–125. doi: 10.1016/s0925-4773(98)00231-7. [DOI] [PubMed] [Google Scholar]
- Suzuki K, Bachiller D, Chen YP, Kamikawa M, Ogi H, Haraguchi R, Ogino Y, Minami Y, Mishina Y, Ahn K, Crenshaw EB, 3rd, Yamada G. Regulation of outgrowth and apoptosis for the terminal appendage: external genitalia development by concerted actions of BMP signaling. Development. 2003;130(25):6209–6220. doi: 10.1242/dev.00846. [DOI] [PubMed] [Google Scholar]
- Suzuki K, Haraguchi R, Ogata T, Barbieri O, Alegria O, Vieux-Rochas M, Nakagata N, Ito M, Mills AA, Kurita T, Levi G, Yamada G. Abnormal urethra formation in mouse models of split-hand/split-foot malformation type 1 and type 4. Eur J Human Genet. 2008;16:36–44. doi: 10.1038/sj.ejhg.5201925. [DOI] [PubMed] [Google Scholar]
- Szucsik JC, Lewis AG, Marmer DJ, Lessard JL. Urogenital tract expression of enhanced green fluorescent protein in transgenic mice driven by a smooth muscle gamma-actin promoter. J Urol. 2004;171(2 Pt 1):944–949. doi: 10.1097/01.ju.0000099168.25976.9a. [DOI] [PubMed] [Google Scholar]
- Trumpp A, Depew MJ, Rubenstein JLR, Bishap JM, Martin GR. Cre-mdeiated gene inactivation demonstrates that FGF8 is required for cell survival and patterning of the first branchial arch. Genes & Dev. 1999;13:3136–3148. doi: 10.1101/gad.13.23.3136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang MH, Baskin LS. Endocrine disruptors, genital development and hypospadias. J Androl. 2008;29:499–505. doi: 10.2164/jandrol.108.004945. [DOI] [PubMed] [Google Scholar]
- Wang L, Liu T, Wang Y, Cao L, Nishioka M, Aguirre RL, Ishikawa A, Geng L, Okada N. Altered expresson of desmocollin 3, desmoglein 3 and beta-catenin in oral squamous cell carcinoma: correlation with lymph node metastasis and cell proliferation. Virchows Arch. 2007;451(5):959–966. doi: 10.1007/s00428-007-0485-5. [DOI] [PubMed] [Google Scholar]
- Weber S, Mir S, Schlingmann KP, Nurnberg G, Becker C, Kara PE, Ozkayin N, Konrad M, Nurnberg P, Schaefer F. Gene locus ambiguity in posterior urethral valves/prune belly syndrome. Pediatr Nephrol. 2005;20:1036–1042. doi: 10.1007/s00467-005-1977-7. [DOI] [PubMed] [Google Scholar]
- Weber S, Moriniere V, Knüppel T, Charbit M, Dusek J, Ghiggeri GM, Jankauskiené A, Mir S, Montini G, Peco-Antic A, Wühl E, Zurowska AM, Mehls O, Antignac C, Schaefer F, Salomon R. Prevalence of mutations in renal developmental genes in children with renal hypodysplasia: results of the ESCAPE study. J Am Soc Nephrol. 2006;17:2864–2870. doi: 10.1681/ASN.2006030277. [DOI] [PubMed] [Google Scholar]
- Witte F, Dokas J, Neuendorf F, Mundlos S, Stricker S. Comprehensive expression analysis of all Wnt genes and their major secreted antagonists during mouse limb development and cartilage differentiation. Gene Exp Patterns. 2009;2:215–223. doi: 10.1016/j.gep.2008.12.009. [DOI] [PubMed] [Google Scholar]
- Yamaguchi TP, Bradley A, McMahon AP, Jones SA. Wnt5a pathway underlies outgrowth of multiple structures in the vertebrate embryo. Development. 1999;126:1211–1223. doi: 10.1242/dev.126.6.1211. [DOI] [PubMed] [Google Scholar]
- Yamada G, Suzuki K, Haraguchi R, Miyagawa S, Satoh Y, Kamimura M, Nakagata N, Kataoka H, Kuroiwa A, Chen Y. Molecular genetic cascades for external genitalia formation: an emerging organogenesis program. Dev Dyn. 2006;235:1738–1752. doi: 10.1002/dvdy.20807. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Heatmaps of the most active genes for each of the eight compartments of the LUT examined.
Changes to anatomical ontology of the urogential tract with respect to the developing genital tubercle.
Microarray analysis revealed a cluster of members of the Wnt signaling pathway, with expression enriched in E14 genital tubercle. These included Wnt ligands (Wnt5a, Wnt7a), signaling molecules (Lef1), inhibitors (Dkk1, Frzb, Wif1) and targets (Sostdc1). Wholemount ISH of E13.5 GT displayed (from left to right) as ventral, lateral and dorsal wholemounts, and transverse and longitudinal sections demonstrate the expression of these Wnt pathway genes. Expression in E11.5 limb is also shown. Di, distal; Pr, proximal; D, dorsal; V, ventral; A, anterior; P, posterior.
Cluster association diagrams for genital tubercle-enriched genes
Heatmaps of genital tubercle-enriched and genital-tubercle-specific clusters 1–3.
Diagrammatic representations of congruence between spatial gene expression patterns in the extension of genital tubercle and limb. Congruence applies between ventral GT and posterior limb, dorsal GT and anterior limb and lateral GT and dorsal limb. Signaling for extension comes from the UP in the GT and the ZPA in the limb. GT, genital tubercle; AER, apical ectodermal ridge; ZPA, zone of polarizing activity; UP, urethral plate; UM, urethral mesenchyme; DGT, distal genital tubercle (glans).
List of the 1420 most highly active genes in the developing LUT incorporating individual lists for each compartment.
Toppgene analyses for the most highly active genes in each compartment of the developing LUT
LUT active genes associated with reproductive, genital, anal, body wall, kidney/ureter and bladder development
Gene lists for the most compartment-enriched genes in each compartment of the developing LUT
Lists of enriched genes for all E14 LUT compartments (GT, bladder, UGS, urethra) including Affymetrix identification numbers and primers used for the generation of riboprobes.
Heat map and gene list for BMP, FGF, Wnt and hedgehog pathway expression across the developing LUT
Toppgene analysis of the 98 probesets identified as enriched in or specific to E14 GT
Summary of expression in GT, limb and craniofacial arch for 37 confirmed GT-enriched or specific genes.