Abstract
Publicly available molecular datasets can be used for independent verification or investigative repurposing, but depends on the presence, consistency and quality of descriptive annotations. Annotation and indexing of molecular datasets using well-defined controlled vocabularies or ontologies enables accurate and systematic data discovery, yet the majority of molecular datasets available through public data repositories lack such annotations. A number of automated annotation methods have been developed; however few systematic evaluations of the quality of annotations supplied by application of these methods have been performed using annotations from standing public data repositories. Here, we compared manually-assigned Medical Subject Heading (MeSH) annotations associated with experiments by data submitters in the PRoteomics IDEntification (PRIDE) proteomics data repository to automated MeSH annotations derived through the National Center for Biomedical Ontology Annotator and National Library of Medicine MetaMap programs. These programs were applied to free-text annotations for experiments in PRIDE. As many submitted datasets were referenced in publications, we used the manually curated MeSH annotations of those linked publications in MEDLINE as “gold standard”. Annotator and MetaMap exhibited recall performance 3-fold greater than that of the manual annotations. We connected PRIDE experiments in a network topology according to shared MeSH annotations and found 373 distinct clusters, many of which were found to be biologically coherent by network analysis. The results of this study suggest that both Annotator and MetaMap are capable of annotating public molecular datasets with a quality comparable, and often exceeding, that of the actual data submitters, highlighting a continuous need to improve and apply automated methods to molecular datasets in public data repositories to maximize their value and utility.
Keywords: Proteomics, Annotations, Ontologies, Concept Identification, Natural Language Processing, MEDLINE
1 Introduction
Advances in high-throughput molecular measurement technologies continued to dramatically increase; and due to this, the demand for the sharing of data and results from these technologies has intensified [1, 2]. Many journals now require the public availability of such data [3], and the National Institutes of Health requires grant applicants to include a section on data sharing if their proposals exceed a threshold amount.
As a result of these policies, the amount of data in international repositories has grown exponentially. One such example is NCBI GEO, an international repository for gene expression data developed and maintained by the NLM [4]. As of this writing, GEO holds 531,381 samples (i.e., gene expression microarray experiments) and has been doubling in size every year or two. Similarly, the ArrayExpress repository maintained by the EBI contains 153,233 independent samples [5]. Recently, international repositories for proteomic experimental datasets have also been instituted. PeptideAtlas, supported by the Institute for Systems Biology, is an example of one such repository [6]. As of this writing, PeptideAtlas holds the raw and processed mass spectra for 366 experiments. Similarly, the PRIDE database at the EBI currently holds over 13 thousand experiments, 4 million protein identifications, and 100 million spectra [7].
One major barrier to the wider usage of this public molecular data is poor indexing using controlled vocabularies and ontologies [8, 9]. The contents of most of these repositories provide basic search functionality across the free-text annotations associated with the data and do not leverage the biomedical knowledge inherent in the annotations. The primary means of data access is typically the unique identifier assigned to the experiment by the repository (e.g., GEO accession number), which is typically provided in publications describing experimental results. But the potential for these and other repositories is more than just serving as a reference for those who wish to verify and validate published findings. We and others have shown that primary research can be performed using data stored in these repositories [10–14].
GEO, ArrayExpress, PeptideAtlas, and PRIDE are all successful databases and possess either fully integrated or advance search interface to retrieve relevant datasets. For example, GEO appears to be indexed with MeSH and free text queries are usually translated to MeSH term searches. PRIDE can be browsed using a variety of biomedical ontologies but no semantic expansion is performed on free text searches. Surprisingly, most repositories, even GEO or ArrayExpress that are compliant with MIAME and MAGE-ML standard formats [3, 15], do not provide tools to the dataset submitters to annotate their own submitted data using controlled vocabularies or ontologies. Consequently, contextual annotations are still represented by unstructured narrative text and determining the phenotypes, diseases, and environmental contexts studied by these experiments is no longer a tractable manual process.
We have previously proposed that the free-text annotations associated with experimental datasets can be processed using concept identification programs to yield a high-content set of contextual details [16]. While it is possible to use automated methods to extract concepts related to phenotypes, diseases, and drugs studied in an experiment, it may still be argued that the original dataset submitter of a dataset might better be able to assign concepts to their data in a more sensitive and specific manner than text parsing. Numerous errors in automated concept identification from the annotations of molecular datasets have been described [13]. It has been suggested that the best long-term strategy to deal with the mapping problem will be to ask dataset submitters of data to label their own data with concepts chosen from an ontology relevant to their experimentation.
However, it is not a foregone conclusion that annotations supplied by data submitters will outperform annotations applied to the same data by automated annotation methods. Annotating free text without support of an integrated annotation service is a non-trivial task and a burden to the submitters that do not necessarily have the knowledge and/or time to provide high quality annotation during the submission process. The EBI PRIDE repository, described above, is unique in that it has asked, but not enforced, dataset submitters to select and submit MeSH terms along with their proteomic datasets. Thus, in this work, we compared these submitter-assigned MeSH terms with the concepts found by applying concept identification tools to free-text annotations in PRIDE.
We compared both submitter-assigned and concept-identified MeSH against a gold standard consisting of the MeSH terms assigned to those MEDLINE publications associated with PRIDE entries. MeSH terms that are later assigned by NLM indexers to publications are obviously not available earlier to dataset submitters, and we suspect that the terms used by dataset submitters in the submission of data to EBI PRIDE are not otherwise easily available or queried by the NLM indexers in annotating publications. Thus, we feel these annotations are reasonably independent and can enable a valuable comparison. We compared the output of two automated concept identification methods, the NCBO Annotator [17, 18] and the NLM MetaMap [19]. Our goal was to answer which concept assignment method (manual or automated) is most accurate, and determine whether dataset submitters are educated enough about concept-assignment, that their assignments can sufficiently enable the search and integration of datasets.
2 Material and methods
2.1 PRIDE database
We downloaded all 11694 PRIDE data files representing 9317 unique PRIDE entries from the EBI FTP server on July 27, 2010. Each PRIDE entry has several free-text fields, and we specifically extracted the following: “Title”, “ShortLabel”, “sampleDescription”, “ProtocolName”, “sampleName” and “additional” (Figure 1). We also extracted the PubMed IDs and MeSH terms associated with these datasets.
Figure 1.
Example of PRIDE entry raw XML file displaying the free-text fields used by NLM MetaMap and NCBO Annotator.
2.2 MEDLINE gold standard
Only 2452 PRIDE entries were associated to one or more scientific publications. Of those we retrieved 88 unique PubMed IDs corresponding to publications. Numerous PRIDE entries were annotated with the same PubMed ID. Through MEDLINE, we retrieved overall 298 unique MeSH terms that we associated back to their respective PRIDE entry to build our gold standard set of annotations. We used all MEDLINE MeSH terms, and did not focus just on primary MeSH terms. Trained NLM indexers assign these MeSH annotations by contrast to PRIDE dataset submitters, who may not be very familiar with MeSH structure. To test our hypothesis, we also collected the MeSH annotation entered by the dataset submitter. We found 203 PRIDE entries were annotated with 32 distinct MeSH terms
2.3 Natural language processing programs
We evaluated the performance of two concept identification software systems on annotating the PRIDE free-text fields using MeSH terms. The MetaMap developed by the NLM, represents the state of the art in automated biomedical text indexing [19]. MetaMap is a well-established software system under development and evaluation at the NLM for indexing MEDLINE. MetaMap indexes free-text with concepts from the Unified Medical Language System (UMLS) Metathesaurus. The UMLS contain multiple controlled vocabularies and ontologies such as Gene Ontology, SNOMED-CT and MeSH. The 2010 release of the program was used.
The NCBO Annotator tool was chosen as a promising newer alternative to MetaMap, considering its overall higher precision and faster execution time obtained across different biomedical resources [17, 18]. The Annotator service is a Representational State Transfer (REST)-based web service. Annotator has the capability of matching free-text against a variety of standardized vocabularies and ontologies, as well as in the UMLS Metathesaurus. In this study, only the MeSH structure was used to annotate PRIDE entries. Precision (1) and recall (2) for newly mapped annotations were calculated as follow:
| (1) |
| (2) |
2.4 Network representation
Annotator annotated 9311 PRIDE entries that grouped into 373 clusters sharing the exact same group of MeSH annotation. We build a network from these clusters based on the shared MeSH annotation between clusters. Clusters were displayed using Cytoscape [20]. By setting a threshold over the minimum number of MeSH terms to be shared between clusters we discovered highly connected subset of clusters.
3 Results
3.1 Automated annotation results
Using MetaMap and Annotator, we were able to successfully annotate 9315 and 9311 of the 9317 PRIDE entries, respectively (Table 1). MetaMap assigned 862 unique MeSH terms compared to 504 for Annotator. This represents a much greater number than the current state of MeSH annotations in PRIDE with only 32 different concepts over 203 PRIDE entries.
Table 1.
Precision and recall obtained using MEDLINE annotation’s gold standard for Annotator and MetaMap programs and original dataset submitter annotations.
| Method | Precision (%) | Recall (%) | Annotated entries (%) | Average MeSH annotation per entry |
|---|---|---|---|---|
| Annotator | 32.96 | 1.83 | 9311 (99.9%) | 5.3 |
| MetaMap | 35.37 | 2.45 | 9315 (99.9%) | 8.1 |
| User Submitted | 97.58 | 0.59 | 203 (2.2%) | 1.5 |
3.2 Precision and recall using MEDLINE gold standard
We compared the precision and recall obtained with MetaMap and Annotator with the ones obtained by dataset submitter using our MEDLINE gold standard (Table 1). We found that the precision achieved by both concept identification programs were similarly high, with scores of 32.96% and 35.37% for Annotator and MetaMap respectively. Dataset submitters scored almost perfect precision (97.58%) in tagging their experiments with MeSH concepts. However, dataset submitters scored very low on recall, at 0.59%. Automated concept identification programs outperformed the recall from dataset submitters by more than three fold. These results have to be contrasted with the number of PRIDE entry annotated by both automated concept identification programs reaching above 99.9% coverage of the entire database compared to only 2.2% for dataset submitters.
3.3 User submitted annotation as gold standard
To place the precision and recall from Table 1 in context, we compared MetaMap and Annotator tagging of PRIDE entries with MeSH terms assigned by dataset submitters to their own data (Table 2).
Table 2.
Comparison of precision and recall of annotations between for Annotator and MetaMap when considering user submitted MeSH annotation as gold standard.
| Method | Precision (%) | Recall (%) |
|---|---|---|
| Annotator | 20.97 | 79.48 |
| MetaMap | 15.66 | 79.44 |
We observed that both MetaMap and Annotator were able to retrieve most of the dataset submitters MeSH terms with a recall of 79.44% and 79.48%, respectively. The higher precision observed for Annotator is most likely explained by MetaMap tagging datasets with a much greater number of MeSH terms than Annotator (see Table 1), which consequently lowered MetaMap precision.
3.4 Automated annotations group biologically related samples
To further validate the relevance of the annotation retrieved using automated concept identification programs, we displayed the newly generated annotations in a network. We chose the annotations from the Annotator as a proof-of-concept here; however, an identical approach can be undertaken using MetaMap annotations. We found that all 9311 PRIDE IDs annotated using the Annotator could be grouped into 373 clusters sharing exactly identical sets of MeSH annotations between them. The largest PRIDE cluster contained 4090 unique PRIDE entries sharing three identical MeSH terms. All the entries in this cluster belonged to the same project: “Quantitative Proteomics Analysis of the Secretory Pathway”. Similarly the second largest cluster of 916 PRIDE entries represented the single project: “GPMDB Submission: Protein complexes in Saccharomyces cerevisiae”. We then determined the level similarity between these 373 clusters through the number of shared MeSH terms between each cluster.
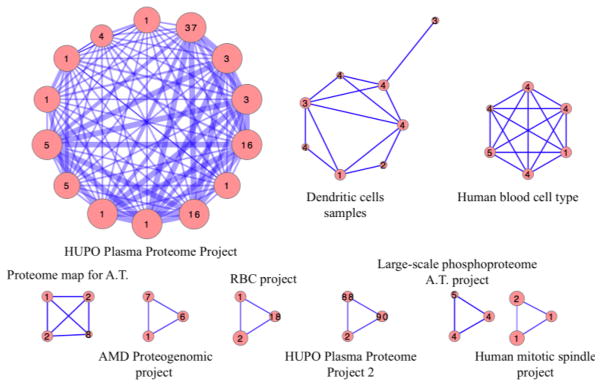
Figure 2 shows PRIDE clusters having at least 10 MeSH annotations in common. We found that highly connected clusters are grouped mainly according to PRIDE projects as well as biological context. For example, all 95 experiments from the “HUPO Plasma Proteome Project” were found to share high number of MeSH annotation. More interestingly, the second cluster represent 25 individual experiments related to dendritic cells (a sub-type of immune cells specialized in antigen presenting) but from two different projects.
Figure 2.
PRIDE entries clusters sharing 10 or more common MeSH annotation organized according to experiments and tissues of origin. Numbers displayed in nodes are number of PRIDE entries in cluster. Width of the edges is proportional to number of MeSH term in common between clusters. HUPO = Human Proteome Organization; AMD = Acid Mine Drainage; RBC = Red Blood Cells; A.T. = Arabidopsis Thaliana.
4 Discussion
Secondary use of the exponentially growing numbers of publicly-available molecular measurements depends on our ability to discover and identify them within international repositories. We found that dataset submitters, not otherwise trained specifically in standardized vocabularies, can submit relevant MeSH terms. This finding is encouraging for public repositories and should engage them in including controlled vocabularies into their submission workflow.
However, while dataset submitters are precise in using terms that NLM indexers use on their resulting papers, we found that dataset submitters do not annotate their dataset in great depth, using on an average only 1.5 MeSH terms per PRIDE experiment, thus leading to low recall when compared against our MEDLINE gold standard. This behavior of the data submitters might be explained in part by the current non-enforcement of the MeSH annotation step in PRIDE and also by the lack of awareness within the community of the benefit of annotation with standardized terms. Additionally, ontologies are often large and finding all relevant concepts for the experiment at hand can be laborious and time consuming. Lastly, there are many ontologies that frequently overlap and are regularly updated and expanded.
Interestingly, the automated concept-identification methods we tested exhibited a higher recall than the dataset submitters in capturing the terms used by NLM indexers, but at the same time, automated concept-identification methods demonstrated low precision against the specific MeSH terms selected by dataset submitters. This suggests while dataset submitters actually type in good terms in the free text annotations of their datasets, they are not judging these as relevant enough to put them into their list of MeSH terms. Future hybrid systems suggesting to dataset submitters several candidate terms from their own free text might be an ideal solution to the annotation problem. Additionally, excluding specific categories (also called: semantic types) of MeSH terms, such as “Geographic Area” or “Professional Society”, could reduce noise and increase precision of automated annotations. MetaMap and Annotator demonstrated an excellent ability to retrieve the dataset submitter MeSH terms with almost 80% of recall. In our hands, MetaMap exhibited a lower precision (5%) than Annotator for user submitted annotation. We do not imply here that one automated concept-identification method is better than the other; instead we used both to ensure our results were not an artifact of one particular concept identification tool. Improvement of the indexing quality performed by Annotator and MetaMap are possible by tuning the numerous parameters within these programs.
MEDLINE indexing includes a very large spectrum of annotations where not all MeSH terms relevant to the research papers are also relevant for the associated PRIDE experiments. This could explain the overall low recall rates for automated and submitter supplied annotation when using MEDLINE indexing as gold standard; as in some case it is virtually impossible to capture from the free text available through PRIDE the MeSH terms from MEDLINE. Thus, we are likely underestimating the true recall that could have been achieved if MEDLINE MeSH annotation could be filtered for annotation relevant to the PRIDE dataset only. Despite this, we feel this gold standard serves as a good metric for comparing different methods, since user submitted annotation displayed 97.58% precision indicating that MEDLINE indexers annotate the published work with the same concept independently from the dataset submitters. Additionally, MeSH annotations from MEDLINE are widely accepted and used throughout the scientific community. We and others have used publication-based MeSH concepts to annotate and find experimental datasets through MEDLINE relations [21].
Network analysis revealed meaningful annotation of PRIDE entries grouping biologically relevant experiments. Clusters grouping experiments from the same projects were found predominantly as they shared almost identical titles, descriptions, sample names and protocol descriptions.
5 Conclusion
We showed that automated concept identification methods could reach a higher recall of MeSH annotations than human dataset submitters who are not specifically trained in annotating their own experiments with MeSH concepts. Human submitters achieved the highest precision when annotating their experiments, but we found they are also using too few terms to annotate their experiments. Additionally, we showed that automated concept identification programs could successfully map proteomic data from PRIDE given only the free text fields provided by datasets submitters. However, having dataset submitters select their own MeSH terms without assisted term selection, does not necessarily add much value beyond using the MeSH terms provided through MEDLINE or found with automated concept identification programs.
Our results were established numerically and graphically, and both suggest that our method is a realistic approach to annotating the PRIDE database, and by extension other molecular databases. Concept identification programs, such as the NLM MetaMap or NCBO Annotator, are freely available and perhaps could be used by submitters to scan their abstract for relevant MeSH terms prior to submission. Enabling dataset submitters to scan their free text to attain better MeSH annotations would add little, if any, burden on the submitters, but could provide an enormous benefit in the areas of data mining, verification of research results, secondary use of data, and overall accessibility of high-throughput molecular data to the public.
Acknowledgments
We acknowledge funding and support from the National Library of Medicine (R01 LM009719), the Stanford Summer Research Program (SSRP), the Hewlett Packard Foundation, and the Lucile Packard Foundation for Children's Health. We acknowledge Dr. Clement Jonquet for helpful discussions.
Abbreviations
- NCBO
National Center for Biomedical Ontology
- MeSH
Medical Subject Headings
- GEO
Gene Expression Omnibus
- PRIDE
PRoteomics IDEntifications
- NLM
National Library of Medicine
- UMLS
Unified Medical Language System
- NCBI
National Center for Biotechnology Information
- EBI
European Bioinformatics Institute
- FTP
File Transfer Protocol
Footnotes
Conflict of Interest
The authors declare that there are no conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Perou CM. Show me the data! Nat Genet. 2001;29:373. doi: 10.1038/ng1201-373. [DOI] [PubMed] [Google Scholar]
- 2.Lussier YA, Butte AJ, Hunter L. Current methodologies for translational bioinformatics. J Biomed Inform. 2010;43:355–7. doi: 10.1016/j.jbi.2010.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brazma A, Hingamp P, Quackenbush J, Sherlock G, Spellman P, Stoeckert C, Aach J, Ansorge W, Ball CA, Causton HC, Gaasterland T, Glenisson P, Holstege FC, Kim IF, Markowitz V, Matese JC, Parkinson H, Robinson A, Sarkans U, Schulze-Kremer S, Stewart J, Taylor R, Vilo J, Vingron M. Minimum information about a microarray experiment (MIAME)-toward standards for microarray data. Nat Genet. 2001;29:365–71. doi: 10.1038/ng1201-365. [DOI] [PubMed] [Google Scholar]
- 4.Barrett T, Suzek TO, Troup DB, Wilhite SE, Ngau WC, Ledoux P, Rudnev D, Lash AE, Fujibuchi W, Edgar R. NCBI GEO: mining millions of expression profiles--database and tools. Nucleic Acids Res. 2005;33:D562–6. doi: 10.1093/nar/gki022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Parkinson H, Kapushesky M, Shojatalab M, Abeygunawardena N, Coulson R, Farne A, Holloway E, Kolesnykov N, Lilja P, Lukk M, Mani R, Rayner T, Sharma A, William E, Sarkans U, Brazma A. ArrayExpress--a public database of microarray experiments and gene expression profiles. Nucleic Acids Res. 2007;35:D747–50. doi: 10.1093/nar/gkl995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Deutsch EW. The PeptideAtlas Project. Methods Mol Biol. 2010;604:285–96. doi: 10.1007/978-1-60761-444-9_19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jones P, Cote RG, Martens L, Quinn AF, Taylor CF, Derache W, Hermjakob H, Apweiler R. PRIDE: a public repository of protein and peptide identifications for the proteomics community. Nucleic Acids Res. 2006;34:D659–63. doi: 10.1093/nar/gkj138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dudley J, Butte AJ. Enabling integrative genomic analysis of high-impact human diseases through text mining. Pac Symp Biocomput. 2008:580–91. [PMC free article] [PubMed] [Google Scholar]
- 9.Shah NH, Jonquet C, Chiang AP, Butte AJ, Chen R, Musen MA. Ontology-driven indexing of public datasets for translational bioinformatics. BMC Bioinformatics. 2009;10(Suppl 2):S1. doi: 10.1186/1471-2105-10-S2-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stuart JM, Segal E, Koller D, Kim SK. A gene-coexpression network for global discovery of conserved genetic modules. Science. 2003;302:249–55. doi: 10.1126/science.1087447. [DOI] [PubMed] [Google Scholar]
- 11.Shen-Orr SS, Tibshirani R, Khatri P, Bodian DL, Staedtler F, Perry NM, Hastie T, Sarwal MM, Davis MM, Butte AJ. Cell type-specific gene expression differences in complex tissues. Nature methods. 7:287–9. doi: 10.1038/nmeth.1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wolfe CJ, Kohane IS, Butte AJ. Systematic survey reveals general applicability of “guilt-by-association” within gene coexpression networks. BMC Bioinformatics. 2005;6:227. doi: 10.1186/1471-2105-6-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Butte AJ, Kohane IS. Creation and implications of a phenome-genome network. Nat Biotechnol. 2006;24:55–62. doi: 10.1038/nbt1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Patel CJ, Bhattacharya J, Butte AJ. An Environment-Wide Association study (EWAS) on type 2 diabetes mellitus. PLoS ONE. 2010;5:e10746. doi: 10.1371/journal.pone.0010746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Spellman PT, Miller M, Stewart J, Troup C, Sarkans U, Chervitz S, Bernhart D, Sherlock G, Ball C, Lepage M, Swiatek M, Marks WL, Goncalves J, Markel S, Iordan D, Shojatalab M, Pizarro A, White J, Hubley R, Deutsch E, Senger M, Aronow BJ, Robinson A, Bassett D, Stoeckert CJ, Jr, Brazma A. Design and implementation of microarray gene expression markup language (MAGE-ML) Genome Biol. 2002;3:46. doi: 10.1186/gb-2002-3-9-research0046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Butte AJ, Chen R. Finding disease-related genomic experiments within an international repository: first steps in translational bioinformatics. AMIA ... Annual Symposium proceedings/AMIA Symposium; 2006. pp. 106–10. [PMC free article] [PubMed] [Google Scholar]
- 17.Jonquet C, Shah NH, Musen MA. The open biomedical annotator. AMIA Summit on Translational Informatics. 2009 [PMC free article] [PubMed] [Google Scholar]
- 18.Shah NH, Bhatia N, Jonquet C, Rubin D, Chiang AP, Musen MA. Comparison of concept recognizers for building the Open Biomedical Annotator. BMC Bioinformatics. 2009;10(Suppl 9):S14. doi: 10.1186/1471-2105-10-S9-S14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Aronson AR. Effective mapping of biomedical text to the UMLS Metathesaurus: the MetaMap program. Proc AMIA Symp. 2001:17–21. [PMC free article] [PubMed] [Google Scholar]
- 20.Cline MS, Smoot M, Cerami E, Kuchinsky A, Landys N, Workman C, Christmas R, Avila-Campilo I, Creech M, Gross B, Hanspers K, Isserlin R, Kelley R, Killcoyne S, Lotia S, Maere S, Morris J, Ono K, Pavlovic V, Pico AR, Vailaya A, Wang PL, Adler A, Conklin BR, Hood L, Kuiper M, Sander C, Schmulevich I, Schwikowski B, Warner GJ, Ideker T, Bader GD. Integration of biological networks and gene expression data using Cytoscape. Nat Protoc. 2007;2:2366–82. doi: 10.1038/nprot.2007.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dudley JT, Tibshirani R, Deshpande T, Butte AJ. Disease signatures are robust across tissues and experiments. Molecular systems biology. 2009;5:307. doi: 10.1038/msb.2009.66. [DOI] [PMC free article] [PubMed] [Google Scholar]