Abstract
Several studies have demonstrated an apparent link between positive selection on hematopoietic cells (HCs) and an “innate” T-cell phenotype. Whereas conventional CD8+ T cells are primarily selected on thymic epithelial cells (TECs) and certain innate T cells are exclusively selected on HCs, MHC class Ib-restricted CD8+ T cells appear to be selected on both TECs and HCs. However, whether TEC- and HC-selected T cells represent distinct lineages or whether the same T-cell precursors have the capacity to be selected on either cell type is unknown. Using an M3-restricted T-cell receptor transgenic mouse model, we demonstrate that not only are MHC class Ib-restricted CD8+ T cells capable of being selected on either cell type but that selecting cell type directly affects the phenotype of the resulting CD8+ T cells. M3-restricted CD8+ T cells selected on HCs acquire a more activated phenotype and possess more potent effector functions than those selected on TECs. Additionally, these two developmental pathways are active in the generation of the natural pool of M3-restricted CD8+ T cells. Our results suggest that these two distinct populations may allow MHC class Ib-restricted CD8+ T cells to occupy different immunological niches playing unique roles in immune responses to infection.
Keywords: CD8 T cells, H2-M3, innate lymphocytes, T-cell development
MHC class Ib genes, comprising the majority of the class I family, encode molecules that are expressed at lower levels than class Ia and have limited polymorphism. This large gene family includes but is not limited to MHC-linked H2-M3 (M3), Qa-1, and Qa-2 in mice; HLA-E, HLA-F, and HLA-G in humans; and MHC unlinked CD1 and MR1 molecules (1). CD1 molecules present lipid antigens to T cells, and CD1d, the only CD1 molecule found in mice, is required for the development a unique subset of T cells known as invariant natural killer T (iNKT) cells (2, 3). Unlike CD1 molecules, most MHC Ib molecules present peptide antigens to CD8+ T cells, and have been demonstrated to be involved in the immune response to a number of pathogens, including Mycobacterium tuberculosis (4–6), CMV (7), Salmonella enterica (8), Listeria monocytogenes (LM) (9–11), and Chlamydia pneumoniae (12).
M3 is one of the best-characterized MHC Ib molecules and preferentially binds N-formylated peptides (13). M3-restricted CD8+ T cells play a nonredundant role in antilisterial immunity, because M3-deficient (M3−/−) mice show increased susceptibility to listerial infection (14). Using T-cell receptor (TCR) transgenic (Tg) mice, we have shown that M3 is necessary and sufficient for the selection of M3-restricted T cells with no contribution from MHC Ia or other MHC Ib molecules (14, 15). Additionally, using a mouse model deficient in both MHC Ia and M3 molecules, we have shown that non-M3 MHC Ib-restricted CD8+ T cells are very similar to M3-restricted CD8+ T cells in terms of surface phenotype and expansion kinetics following LM infection (16). Based on this result, M3-restricted CD8+ T cells represent a reasonable model for the study of MHC Ib-restricted CD8+ T cells in general.
Most MHC Ib-restricted CD8+ T cells can be distinguished from MHC Ia-restricted T cells on the basis of a more “innate-like” phenotype. These cells express high levels of CD44 even in naive mice (17), are capable of rapidly producing cytokines after in vitro stimulation, and mount an immune response more quickly than MHC Ia-restricted CD8+ T cells following infection with LM (17, 18). Although the mechanisms responsible for the development of this innate-like phenotype are not fully understood, results from several studies suggest that the types of cells mediating selection of these T cells may contribute to their unique functional characteristics (1, 19).
Unlike MHC Ia-restricted T cells, which are positively selected by thymic epithelial cells (TECs) (20), iNKT cells are exclusively selected by CD1d-expressing thymocytes (21). Interestingly, a study using MHC Ia-deficient mice has demonstrated that MHC Ib-restricted CD8+ T cells, including M3-restricted CD8+ T cells, can be positively selected on hematopoietic cells (HCs) (19). However, it remains to be determined whether TEC- and HC-selected T cells represent two distinct T-cell lineages or whether certain T-cell precursors are capable of being selected by either pathway, expressing different phenotypes depending on how they are selected.
Using Tg mice expressing a TCR specific for the listerial peptide LemA presented by M3 (D7 Tg) (15), we show here that M3-restricted CD8+ T cells can be selected on both TECs and HCs. We demonstrate that D7 CD8+ T cells selected on HCs have more activated surface phenotypes and express increased amounts of Eomesodermin (Eomes), the T-box transcription factor shown to be up-regulated in a number of innate T-cell populations, including those found in mice deficient for IL-2–inducible T-cell kinase (22–24). Our results clearly demonstrate that M3-restricted CD8+ T cells can be generated from the same T-cell precursors selected on two distinct cell types and provide direct evidence that selecting cell type plays an important role in determining the phenotype and functional characteristics of these T cells.
Results
Phenotype and Effector Function of CD8+ T Cells in D7 TCR Tg Mice.
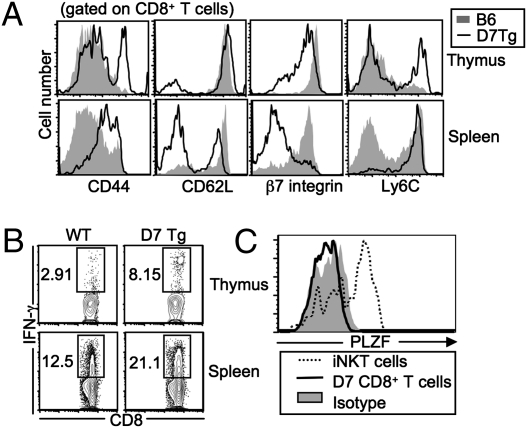
Previous studies have demonstrated that the majority of MHC Ib-restricted CD8+ T cells exhibit an activated phenotype in naive mice (17). We therefore examined the phenotype and effector functions of CD8+ T cells from D7 TCR Tg mice (D7 Tg: TCR Vα10 and Vβ5, specific for M3/LemA) (15). Similar to other MHC Ib-restricted CD8+ T cells, a significant proportion of CD8+ T cells from both the thymus and spleen of naive D7 Tg mice exhibited an activated phenotype (CD44hi, CD62Llo, β7 integrinlo, and Ly6C hi). In contrast, only a very small proportion of CD8+ T cells from age-matched WT B6 mice had an activated phenotype (Fig1A). Consistent with their surface phenotype, D7 CD8+ T cells demonstrated a more potent IFN-γ response following in vitro stimulation with phorbol 12-myristate 13-acetate (PMA) and ionomycin compared with CD8+ T cells from B6 mice (Fig. 1B). Several recent studies have identified the transcription factor promyelocytic leukemia zinc finger (PLZF) as a key regulator in the development of a number of innate T-cell populations (25–27). However, we found that PLZF expression levels in D7 CD8+ T cells were much lower than in iNKT cells and more similar to the levels in conventional T cells (Fig. 1C).
Fig. 1.
Characterization of CD8+ T cells in D7 Tg mice. (A) Splenocytes and thymocytes from D7+Rag−/− and B6 mice were stained with mAb against various lymphocyte activation markers. Data shown are representative of three to six mice in each group. (B) Ex vivo stimulation of CD8+ T cells from D7+Rag−/− and B6 mice. CD8+ T cells were stimulated with PMA and ionomycin and then subjected to intracellular IFN-γ staining. Data shown are representative of two independent experiments with two mice in each group. (C) PLZF expression on D7 thymocytes, iNKT cells (TCRβ+CD1d/α-galactosylceramide tetramer+), and conventional CD8+ T cells from B6 mice. The expression of PLZF on conventional CD8+ T cells was indistinguishable from isotype control. Data shown are representative of two independent experiments.
D7 TCR CD8+ T Cells Can Be Selected by Either HCs or TECs.
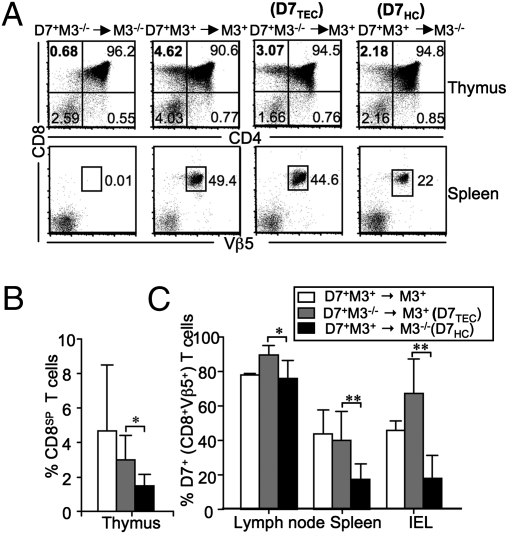
Unlike conventional CD8+ T cells, which are selected on MHC Ia-expressing TECs (20), it is well established that some MHC Ib-restricted CD8+ T cells can be selected on HCs (19). To determine whether M3-restricted CD8+ T cells are selected exclusively on HCs or can be selected on both HCs and TECs, we generated chimeric mice by adoptive transfer of bone marrow (BM) cells from D7 Tg+M3−/−Rag−/− (D7+M3−/−) and D7 Tg+M3+Rag−/− (D7+M3+) donors into irradiated M3−/−Rag−/− (M3−/−) and M3+Rag−/− (M3+) recipients. Mice in the Rag-deficient background were used in this study to avoid complications associated with the expression of unrelated TCRs. Selection was evaluated 8–10 wk after adoptive transfer by measuring the proportion of CD8 single positive thymocytes and Vβ5+CD8+ T cells in the thymus and spleens of chimeric animals, respectively. Expression of M3 on either TECs (D7+M3−/−→M3+) or HCs (D7+M3+→M3−/−) was sufficient to support selection of D7 CD8+ T cells. However, no selection of D7 CD8+ T cells was observed in D7+M3−/−→M3−/− chimeric mice that completely lacked M3 expression (Fig. 2 A and B). It is worth noting that although both TECs and HCs are capable of selecting D7 CD8+ T cells, TEC-mediated selection appears to be the more efficient pathway, because we consistently observed more D7 CD8+ T cells in D7+M3−/−→ M3+ chimera than in D7+M3+→M3−/− chimera in all examined lymphoid tissues (Fig. 2C). In addition, TEC-selected CD8+ T cells appear to be more efficiently recruited to the small intestine than HC-selected cells, an outcome most likely attributable to phenotypic differences described below.
Fig. 2.
Selection of M3-restricted CD8+ T cells on HCs and TECs. Thymocytes and splenocytes from indicated chimeric mice were stained with antibodies specific for CD8β, CD4, and Vβ5. (A) (Upper) Numbers represent percentages of total thymocyte population within each quadrant. (Lower) Percentages of CD8+Vβ5+ cells within the total splenocyte population. Because both donor and recipient mice are on the Rag−/− background, all Vβ5+ cells in chimeric mice are D7 T cells. (B) Percentage of CD8 single positive cells within the total thymocyte population from various BM chimeras. (C) Mean percentage of CD8+Vβ5+ cells in the total lymphocyte populations from the lymph node, spleen, and small intestine ± SEM. *P < 0.05; **P < 0.01. Data shown are representative of four independent experiments with two to three mice per group. IEL, intestinal epithelial lymphocytes.
D7 CD8+ T Cells Selected on HCs Are More Activated Than Those Selected on TECs.
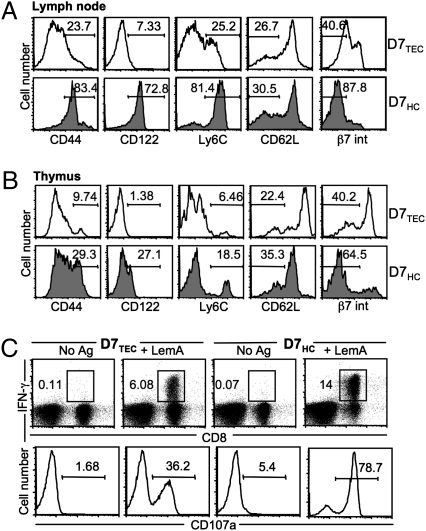
Because D7 CD8+ T cells have a noticeably different phenotype from conventional CD8+ T cells, we were interested in any phenotypic differences that might exist between TEC- and HC-selected D7 CD8+ T cells. We found that although the majority of HC-selected D7 CD8+ T cells (D7HCs) in the periphery were CD44hi, Ly6Chi, CD122+, and CD62Llo, TEC-selected D7 CD8+ T cells (D7TECs) exhibited a somewhat less activated phenotype (Fig. 3A). β7 integrin is a cell adhesion molecule that is important in mediating efficient homing and retention of lymphocytes into the gut (28). Higher expression of β7 integrin on the surface of D7TECs agrees with our earlier observation that D7TECs traffic more efficiently to the gut than D7HCs, suggesting that these two phenotypically distinct T-cell subsets might play distinct immunological roles. Interestingly, mature D7HC and D7TEC thymocytes also demonstrated similar (although less striking) differences in activation marker expression (Fig. 3B), indicating that the phenotypic differences are at least partially acquired as a result of interactions occurring during thymic selection and perhaps further amplified by peripheral activation.
Fig. 3.
D7 CD8+ T cells selected on HCs are more activated and possess more potent effector function than those selected on TECs. Expression of activation markers on lymphocytes isolated from chimeric mice. The percentages of CD8+Vβ5+ cells (A; lymph node) and HSAloCD8+ cells (B; thymus) displaying each activation marker are shown. Data shown are representative of four independent experiments with two mice per group. (C) Splenocytes from D7TECs and D7HCs were stimulated with LemA. Intracellular IFN-γ production and CD107a expression were analyzed. Data shown are representative of two independent experiments with three mice per group.
D7 CD8+ T Cells Selected on HCs Are Capable of More Potent Effector Function.
To assess differences in effector function between these two subsets of D7 CD8+ T cells, splenocytes from chimeric mice were stimulated in vitro with LemA and the levels of intracellular IFN-γ and extent of degranulation (as measured by CD107a expression) were determined by flow cytometry. Although both D7HCs and D7TECs responded to LemA stimulation, consistent with their surface phenotypes, D7HCs mounted a significantly more robust response (Fig. 3C). D7HCs also demonstrated higher proliferative capacity (measured by carboxyfluorescein succinimidyl ester dilution) following stimulation with LemA (Fig. S1).
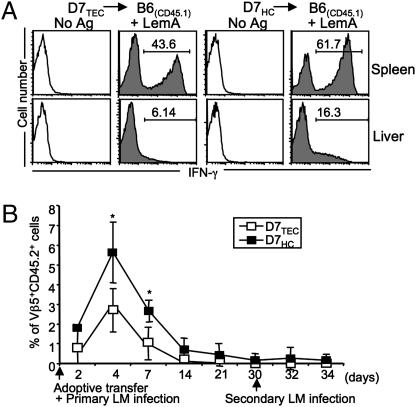
To compare the functional properties of D7HCs and D7TECs in vivo further, we adoptively transferred each subset of T cells into naive B6 congenic (CD45.1+) mice and examined their effector functions following infection with LM. Seven days postinfection, splenocytes and hepatic leukocytes were harvested from infected mice and stimulated in vitro with LemA, and their IFN-γ responses were determined. Similar to our earlier results, although both groups of T cells responded in an antigen-specific manner, D7HCs mounted a more potent response (Fig. 4A).
Fig. 4.
D7 T cells selected on TECs and HCs respond with similar kinetics but different magnitudes during LM infection. Sorted D7TECs and D7HCs (CD45.2+) were adoptively transferred into CD45.1 B6 congenic mice and recipient mice infected with LM. (A) Seven days postinfection, splenocytes and hepatic leukocytes from recipient mice were stimulated with LemA. The percentages of cells producing IFN-γ within the CD45.2+CD8+ LemA-specific D7TEC or D7HC population are shown. (B) Percentages of Vβ5+CD45.2+ cells detected in the blood of recipient mice at different time points following primary and secondary LM infection. *P < 0.05. Data shown are representative of two independent experiments with three to four mice per group.
During LM infection, M3-restricted T cells have been shown to expand faster than conventional CD8+ T cells but are incapable of mounting a robust secondary response on rechallenge (29). Because conventional CD8+ T cells are almost exclusively selected on TECs, we were interested in comparing the kinetics of D7TEC and D7HC expansion after infection. Somewhat surprisingly, although the magnitude of the D7TEC response was lower than the D7HC response, the two T-cell populations exhibited identical kinetics in response to LM infection and neither group was capable of significant expansion following secondary infection (Fig. 4B). Taken together, these results demonstrate that although D7 CD8+ T cells develop distinct surface and effector phenotypes depending on what cells mediate their selection, certain characteristics that distinguish MHC Ib-restricted T cells from conventional T cells are unaffected by selection pathway.
D7 CD8+ T Cells Selected on HCs Up-Regulate a Specific Transcription Factor Involved in Innate T-Cell Development.
The T-box transcription factor Eomes is significantly up-regulated in innate CD8+ T cells compared with conventional CD8+ T cells (23). It is therefore possible that Eomes plays a role in determining the phenotype of M3-restricted CD8+ T cells. In addition to comparing Eomes expression between D7TECs and D7HCs, we were interested in comparing the expression levels of CD122 (the receptor specifying IL-15 responsiveness), which, similar to CD44, is expressed at a high level on innate T cells under naive conditions (30).
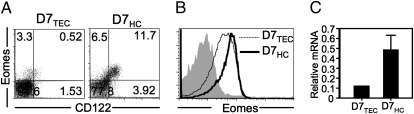
We harvested liver leukocytes and thymocytes from chimeric mice; depleted immature thymocytes; and, in addition to assessing CD122 expression, determined the relative amounts of intracellular Eomes via intracellular staining (Fig. 5 A and B). We also isolated total RNA from both sets of D7 CD8+ T cells and analyzed Eomes expression by real-time RT-PCR (Fig. 5C). Although a significant proportion of D7HC thymocytes are CD122+Eomes+, this population is almost absent in the thymus of D7TEC mice. Our finding that D7HCs express higher levels of Eomes suggests a potential role for the transcription factor in determining the innate-like phenotype of M3-restricted CD8+ T cells selected on HCs.
Fig. 5.
D7 T cells selected on HCs express higher levels of Eomes than D7 T cells selected on TECs. (A) Expression of CD122 and Eomes on heat stable antigenhi-depleted thymocytes from D7TEC and D7HC chimeric mice. (B) Expression of Eomes on hepatic leukocytes from D7TEC and D7HC chimeric mice, compared with isotypye control (gray filled). Histograms are gated on CD8+ cells. Data shown are representative of two separate experiments with four mice per group. (C) Real-time RT-PCR of Eomes mRNA from D7 T cells sorted from D7TEC and D7HC chimeric mice. Bar graph represents the mean ± SEM of triplicate determinants of Eomes expression normalized to GAPDH expression levels.
Natural Pool of H2-M3–Restricted CD8+ T Cells Consists of T Cells Selected by Both HC- and TEC-Mediated Pathways.
Unlike CD1d-restricted iNKT cells, which can be easily detected in naive mice using CD1d/α-galactosylceramide (αGalCer) tetramer, low frequency and a diverse TCR repertoire complicate tetramer-based detection of M3-restricted CD8+ T cells in naive mice. Although the D7 Tg mouse is useful, it was important to determine the phenotype of M3-restricted CD8+ T cells in naive B6 mice and to identify what cells support selection of these T cells under more accurate physiological conditions.
To characterize the naive M3-restricted LemA-specific T-cell population, we used a tetramer-based enrichment approach that has been developed recently to detect low-frequency antigen-specific T cells in naive mice (31). Whereas conventional CD8+ T cells were predominantly CD44lo, Ly6Clo, and CD62Lhi, a significant proportion of M3-restricted LemA-specific CD8+ T cells in naive B6 mice appeared to possess an innate-like phenotype. These M3/LemA+CD8+ T cells (CD44hi, Ly6Chi, and CD62Llo) exhibited a surface marker expression profile that closely resembled the pattern observed in D7 Tg mice (Fig. 6A).
Fig. 6.
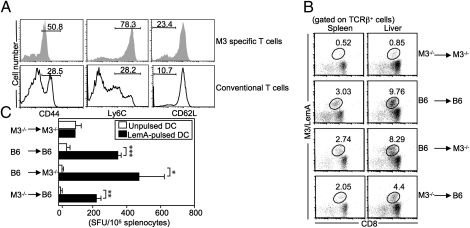
Natural (polyclonal) pool of M3-restricted CD8+ T cells can be selected by both HC- and TEC-mediated pathways. (A) Surface phenotypes of M3-restricted and conventional CD8+ T cells in naive B6 mice. (B and C) BM chimeric mice were immunized with LemA-pulsed BMDCs. Seven days later, the M3-restricted LemA-specific T-cell response was analyzed. (B) Percentages of CD8+M3/LemA tetramer+ cells in DC-immunized chimeric mice. (C) Splenocytes from day 7 immunized chimeric mice were cultured with unpulsed or LemA-pulsed BMDCs isolated from B6 mice. IFN-γ production was quantified via IFN-γ enzyme-linked immunospot assay. SFU, spot forming unit. *P < 0.05; **P < 0.01; ***P < 0.001. Results are representative of three independent experiments with two to three mice per group.
To determine which cell types mediate selection of M3-restricted CD8+ T cells under polyclonal conditions, we generated BM chimeric mice by transferring either B6 or M3−/− BM into irradiated B6 or M3−/− recipients. Because significant numbers of antigen-specific M3-restricted CD8+ T cells are only detectable after antigen-induced cellular expansion, BM chimeric mice were immunized with LemA-pulsed BM-derived dendritic cells (BMDCs) 10 wk following adoptive transfer. Seven days postimmunization, a significant number of M3/LemA tetramer+CD8+ T cells were detected in the spleen and liver of both B6→M3−/− (HC-selected) and M3−/−→B6 (TEC-selected) chimeric mice but not in the negative control group (M3−/−→M3−/−). In agreement with our D7 Tg mouse model, HC-selected cells appeared to mount a more robust response than TEC-selected cells, most noticeably in the liver of immunized animals (Fig. 6B).
To examine the functional properties of these two types of M3-restricted CD8+ T cells further, splenocytes isolated from each group of chimeric mice were stimulated with LemA-pulsed antigen presenting cells (APCs), and the number of IFN-γ–producing cells was measured by enzyme-linked immunospot assay. Similar to results from the D7 Tg mouse, both HC- and TEC-selected M3-restricted CD8+ T cells were capable of producing IFN-γ in an antigen-specific manner (Fig. 6C). These results demonstrate that the natural pool of M3-restricted CD8+ T cells consists of two phenotypically distinct populations resulting from selection by either TEC- or HC-mediated pathways.
Discussion
The results presented here provide direct experimental evidence that selecting cell type plays an important role in determining the phenotype of M3-restricted CD8+ T cells. Using M3/LemA-specific TCR Tg mice, we demonstrate that M3-restricted CD8+ T cells with identical TCRs can be selected on either TECs (D7TECs) or HCs (D7HCs), giving rise to two phenotypically and functionally divergent populations. Interestingly, recent studies have suggested a model in which IL-4–producing PLZF+ T cells are responsible for regulating the innate phenotype of MHC Ia-restricted CD8+ T cells found in KLF2- and Id3-deficient animals (32, 33). We were, however, unable to detect PLZF-expressing T cells in the thymus of D7 Tg mice, suggesting that differences in selecting cell type are sufficient to regulate the development of an innate-like phenotype in M3-restricted CD8+ T cells. To corroborate our findings on the polyclonal level, we characterized the M3-restricted LemA-specific CD8+ T-cell population in naive B6 mice, demonstrating that this population appeared to consist of two phenotypically distinct subsets. Further studies using BM chimeras showed conclusively that selection of M3-restricted CD8+ T cells under polyclonal conditions occurs efficiently on either TECs or HCs.
Although the unique phenotypic characteristics of M3-restricted CD8+ T cells are dependent on their selecting cell types, the question remains as to why MHC Ib-restricted CD8+ T cells can be selected on both TECs and HCs, whereas conventional T cells and iNKT cells are only efficiently selected on TECs and HCs, respectively (20, 21, 34). Although conventional T-cell selection is thought to be dependent on certain specialized cell-surface molecules and cytokines provided by TECs (35), there is some evidence to suggest that cells with high affinities for self-MHC molecules may be less dependent on these TEC-provided signals and, as such, are capable of being selected on HCs (19, 36). The fact that iNKT cells are solely selected on HCs and are thought to have higher affinities for selecting ligands than conventional T cells (2, 37, 38) appears to support the notion that TCR affinity plays an important role in determining which cells are capable of being selected on HCs.
To test if this hypothesis could explain M3-restricted T-cell selection, we compared the TCR affinities of iNKT cells with D7 Tg T cells. Using tetramer binding and decay experiments, we determined that D7 CD8+ T cells bind M3–LemA complexes with lower affinity than iNKT cells; however, based on comparisons with previously published reports (37, 38), D7 CD8+ T cells still have significantly higher affinity for peptide–MHC complexes than conventional CD8+ T cells (Fig. S2). This intermediate TCR affinity (most likely reflective of the limited repertoire of N-formylated “self”-peptides necessary for positive selection) might explain why M3-restricted CD8+ T cells are efficiently selected on both TECs and HCs.
Our results suggest a basic model whereby MHC Ib-restricted CD8+ T cells initiate a specific developmental program based on specific signals received from the selecting cells. In the case of cells selected on HCs, this signaling cascade results in the up-regulation of transcription factors, including but not necessarily limited to Eomes, resulting in the development of a more innate preactivated phenotype. Another transcription factor associated with CD8+ T-cell effector function, T-bet, was similarly up-regulated in D7HCs, in contrast to PLZF, which was barely detectable in either group of cells (Fig. S3). These cells up-regulate activation markers, such as CD122 and CD44, and rapidly produce IFN-γ on stimulation. Such HC-dependent selection may involve homotypic interactions between members of the signalling lymphocyte activation molecule (SLAM) family of cell-surface receptors, exclusively expressed on thymocytes and other HCs. Mice deficient in the SLAM-associated protein (SAP) have a severe defect in iNKT cell numbers (39), and SAP appears to be the main signaling pathway responsible for the development of innate CD4+ T cells selected on HCs [class II major histocompatibility complex transactivator (CIITA) Tg] (40). Conversely, cells that are selected on TECs initiate a different developmental program resulting in a more naive phenotype.
In describing this model, we must acknowledge that this developmental “programming” may not occur in all MHC Ib-restricted CD8+ T cells. A previously published report using Tg mice that express a TCR specific for an insulin-derived peptide presented by Qa-1 (41) showed that although these Qa-1–restricted CD8+ T cells are capable of being positively selected on either cell type, there are no phenotypic differences between the two T-cell populations. Unlike the majority of class Ib-restricted CD8+ T cells, however, these T cells do not exhibit an activated phenotype in naive mice (42). Because the majority of CD8+ T cells in both MHC Ia−/− (17) and MHC Ia−/−M3−/− animals (16) express an activated phenotype, the D7 Tg model appears to represent the majority of MHC Ib-restricted CD8+ T cells better.
Although our results do show that the innate-like phenotype of M3-restricted CD8+ T cells is attributable, in part, to selection on HCs, we noticed that a small proportion of D7 T cells selected on TECs expressed an activated phenotype. This finding agrees with previously published work suggesting that some aspects of the innate-like phenotype are intrinsic to MHC Ib-restricted CD8+ T cells (19) and independent of selecting cell type. Interestingly, the proportion of activated D7TECs was significantly higher in the periphery than in the thymus. There is some evidence that commensal bacteria might play a role in the peripheral activation of M3-restricted CD8+ T cells (43), and it is possible that exposure to certain products derived from the normal microbial flora might also contribute to the activated phenotype of these T cells in naive mice. In fact, D7 Tg mice but not histocompatibility-Y antigen (H-Y) Tg mice treated from birth with antibiotic-containing water have lower numbers of activated CD8+ T cells than untreated Tg mice (Fig. S4).
Based on our results, the unique phenotype of M3-restricted CD8+ T cells is most likely attributable to a combination of T-cell intrinsic factors, signals from selecting cells, and stimulation by microbial antigens. Of particular note is the remarkable ability of these T cells to be “programmed” depending on the cell types that mediate their positive selection. Our data not only confirm the role played by HCs in determining the phenotype of MHC Ib-restricted CD8+ T cells but further suggest that both selection pathways may have physiologically significant roles. Specifically, M3-restricted CD8+ T cells selected on TECs have higher levels of β7 integrin expression and, as a result, traffic more efficiently to the gut. It is therefore possible that these two different selecting pathways allow M3-restricted CD8+ T cells, and possibly other MHC Ib-restricted CD8+ T cells, to occupy a number of different immunological niches possibly playing unique roles in mediating immune function.
Materials and Methods
Mice.
C57BL/6 (B6), CD45.1 congenic B6, and Rag-2–deficient mice (Rag−/−) were purchased from Jackson Laboratories. D7 Tg (15) and M3−/− (14) mice were generated in our laboratory and were backcrossed at least 10 times to the B6 background. D7 Tg mice were crossed onto the Rag−/− background and further crossed with M3−/− mice for these studies. All animal work was approved by the Institutional Animal Care and Use Committee.
Antibodies and Tetramers.
FITC-conjugated anti-CD8β, CD44, Ly6C, CD24, Vβ5, CD45.1, CD45.2, and CD107; phycoerythrin (PE)-conjugated anti-CD8α, B220, CD122, and Eomes; peridinin chlorophyll protein complex (PerCP)-conjugated anti-CD4, B220, and Ly6C; allophycocyanin-conjugated IFN-γ, and CD11c; PerCP Cy5.5-conjugated anti–TCR-β; pacific blue-conjugated anti-B220, CD11b, and CD11c; and biotin-conjugated anti-CD62L, CD45.1, and CD45.2 were purchased from BD Biosciences or eBioscience. Anti-PLZF was purchased from Santa Cruz. CD1d/α-GalCer tetramers were generated in our laboratory, and H2-M3/LemA tetramers were provided by the National Institutes of Health tetramer core facility.
Flow Cytometry.
Single-cell suspensions were prepared by standard procedures and stained with the appropriate combinations of mAbs. PLZF expression was analyzed via intracellular staining using the FoxP3 staining buffer set (eBioscience) with 4 μg/mL anti-PLZF mAb. For Eomes staining, thymocytes were first depleted of immature cells using anti-CD24 mAb and a magnetic affinity cell sorting separation column. Cells were fixed, permeabilized, and then stained with anti-Eomes mAb. Flow cytometry was performed with a FACSCanto II (BD Biosciences) and analyzed using FlowJo software (Tree Star Inc.).
BM Chimeras.
Donor BM cells were depleted of mature T cells using anti-Thy1.2 mAb (AT83.A-6) and rabbit complement. A total of 1 × 107 T cell-depleted BM cells were injected i.v. per irradiated recipient. Chimeras were analyzed by flow cytometry or used for immunization 8–10 wk later.
BMDC Generation and DC Immunization.
BMDCs were prepared as described previously (16). DCs were pulsed with 1 μM LemA peptide (fMIGWII) for 6 h and injected i.v. into BM chimeric mice (1 × 106 peptide-coated DCs per mouse).
Bacteria and LM Infection.
The recombinant LM strain rLM-ovalbumin (OVA) was grown in brain-heart infusion broth supplemented with 5 μg/mL erythromycin. For primary infections, mice were infected i.v. with 5 × 104 cfu rLM-OVA. When necessary, secondary infection was performed 1 mo after primary infection with 1 × 106 cfu rLM-OVA.
Intracellular Cytokine Staining and Degranulation Assay.
Splenocytes were stimulated with PMA (20 ng/mL)/ionomycin (1 μM) or 5 μM LemA peptide for 3–5 h in the presence of 10 μM monensin and, in relevant experiments, anti-CD107a mAb. Cells were washed and stained for cell surface markers. After fixation with 4% paraformaldehyde and permeabilization with 0.15% saponin, cells were stained with anti–IFN-γ mAb.
Enzyme-Linked Immunospot Assay.
Multiscreen-IP filter plates (Millipore) were coated with anti–IFN-γ mAb, washed, and blocked with RPMI-10. Splenocytes from various BM chimeras were cultured with LemA-pulsed or unpulsed BMDCs. Plates were incubated for 18 h, and IFN-γ–producing cells were quantified with an ImmunoSpot reader (Cellular Technology).
RNA Extraction and Quantitative Real-Time PCR.
D7 T cells (CD8+Vβ5+ cells) from BM chimeras were sorted via MoFlo (Beckman Coulter Inc.). Total RNA was isolated from purified D7TECs or D7HCs using an RNeasy kit (Qiagen) and reverse-transcribed using SuperScript II RT (Invitrogen). Real-time PCR was performed on an i-cycler (BioRad) using SYBR Green Master Mix (BioRad). Transcripts for murine Eomes were quantified with primers (forward primer: 5′-TGAATGAACCTTCCAAGACTCAGA-3′; reverse primer: 5′-TGAATGAACCTTCCAAGACTCAGA-3′) and normalized to GAPDH (forward primer: 5′-TTCACCACCATGGAGAAGGC-3′; reverse primer, 5′-GGCATGGACTGTGGTCATGA-3′).
Tetramer Enrichment.
Splenocytes were stained with PE-M3/LemA tetramer, washed, and incubated with anti-PE microbeads (Miltenyi Biotec) for 30 min at 4 °C. Cells were purified using a magnetized LS column (Miltenyi Biotec) and stained with allophycocyanin-M3/LemA tetramer, anti-CD3 (FITC), anti-CD8 (V500), anti-CD44 (Alexa Fluor 700), and PerCP-conjugated anti-Ly6C and anti-CD62L mAb. Cells were also stained with Pacific Blue-conjugated anti-B220, CD11b, and CD11c (dump gate).
Statistical Analysis.
Statistical analyses were performed using PRISM software (GraphPad). Statistical significance of differences was calculated using Student t tests. P values <0.05 were considered significant (*P < 0.05; **P < 0.01; ***P < 0.001).
Supplementary Material
Acknowledgments
We thank National Institutes of Health tetramer core facility for M3/LemA tetramers; the Northwestern University flow cytometry core facility for cell sorting services; Jessica Rojas, Sharmila Shanmuganad, Stephen Wood, and Chunting Yang for technical assistance; and Dr. L. Lefrancois for providing rLM-Ova. This work is supported by National Institutes of Health Grant R01 AI40301 (to C-R.W.) and Cancer Center Support Grant NCI CA060553 to The Robert H. Lurie Comprehensive Cancer Center at Northwestern University.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1105118108/-/DCSupplemental.
References
- 1.Rodgers JR, Cook RG. MHC class Ib molecules bridge innate and acquired immunity. Nat Rev Immunol. 2005;5:459–471. doi: 10.1038/nri1635. [DOI] [PubMed] [Google Scholar]
- 2.Bendelac A, Rivera MN, Park SH, Roark JH. Mouse CD1-specific NK1 T cells: Ddevelopment, specificity, and function. Annu Rev Immunol. 1997;15:535–562. doi: 10.1146/annurev.immunol.15.1.535. [DOI] [PubMed] [Google Scholar]
- 3.Chen YH, Chiu NM, Mandal M, Wang N, Wang CR. Impaired NK1+ T cell development and early IL-4 production in CD1-deficient mice. Immunity. 1997;6:459–467. doi: 10.1016/s1074-7613(00)80289-7. [DOI] [PubMed] [Google Scholar]
- 4.Heinzel AS, et al. HLA-E-dependent presentation of Mtb-derived antigen to human CD8+ T cells. J Exp Med. 2002;196:1473–1481. doi: 10.1084/jem.20020609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Joosten SA, et al. Mycobacterium tuberculosis peptides presented by HLA-E molecules are targets for human CD8 T-cells with cytotoxic as well as regulatory activity. PLoS Pathog. 2010;6:e1000782. doi: 10.1371/journal.ppat.1000782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chun T, et al. Induction of M3-restricted cytotoxic T lymphocyte responses by N-formylated peptides derived from Mycobacterium tuberculosis. J Exp Med. 2001;193:1213–1220. doi: 10.1084/jem.193.10.1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pietra G, et al. HLA-E-restricted recognition of cytomegalovirus-derived peptides by human CD8+ cytolytic T lymphocytes. Proc Natl Acad Sci USA. 2003;100:10896–10901. doi: 10.1073/pnas.1834449100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Salerno-Gonçalves R, Fernandez-Viña M, Lewinsohn DM, Sztein MB. Identification of a human HLA-E-restricted CD8+ T cell subset in volunteers immunized with Salmonella enterica serovar Typhi strain Ty21a typhoid vaccine. J Immunol. 2004;173:5852–5862. doi: 10.4049/jimmunol.173.9.5852. [DOI] [PubMed] [Google Scholar]
- 9.Pamer EG, Wang CR, Flaherty L, Lindahl KF, Bevan MJ. H-2M3 presents a Listeria monocytogenes peptide to cytotoxic T lymphocytes. Cell. 1992;70:215–223. doi: 10.1016/0092-8674(92)90097-v. [DOI] [PubMed] [Google Scholar]
- 10.Gulden PH, et al. A Listeria monocytogenes pentapeptide is presented to cytolytic T lymphocytes by the H2-M3 MHC class Ib molecule. Immunity. 1996;5:73–79. doi: 10.1016/s1074-7613(00)80311-8. [DOI] [PubMed] [Google Scholar]
- 11.Princiotta MF, Lenz LL, Bevan MJ, Staerz UD. H2-M3 restricted presentation of a Listeria-derived leader peptide. J Exp Med. 1998;187:1711–1719. doi: 10.1084/jem.187.10.1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tvinnereim A, Wizel B. CD8+ T cell protective immunity against Chlamydia pneumoniae includes an H2-M3-restricted response that is largely CD4+ T cell-independent. J Immunol. 2007;179:3947–3957. doi: 10.4049/jimmunol.179.6.3947. [DOI] [PubMed] [Google Scholar]
- 13.Loveland B, Wang CR, Yonekawa H, Hermel E, Lindahl KF. Maternally transmitted histocompatibility antigen of mice: A hydrophobic peptide of a mitochondrially encoded protein. Cell. 1990;60:971–980. doi: 10.1016/0092-8674(90)90345-f. [DOI] [PubMed] [Google Scholar]
- 14.Xu H, Chun T, Choi HJ, Wang B, Wang CR. Impaired response to Listeria in H2-M3-deficient mice reveals a nonredundant role of MHC class Ib-specific T cells in host defense. J Exp Med. 2006;203:449–459. doi: 10.1084/jem.20051866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chiu NM, et al. The selection of M3-restricted T cells is dependent on M3 expression and presentation of N-formylated peptides in the thymus. J Exp Med. 1999;190:1869–1878. doi: 10.1084/jem.190.12.1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cho H, Choi HJ, Xu H, Felio K, Wang CR. Nonconventional CD8+ T cell responses to Listeria infection in mice lacking MHC class Ia and H2-M3. J Immunol. 2011;186:489–498. doi: 10.4049/jimmunol.1002639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Seaman MS, Wang CR, Forman J. MHC class Ib-restricted CTL provide protection against primary and secondary Listeria monocytogenes infection. J Immunol. 2000;165:5192–5201. doi: 10.4049/jimmunol.165.9.5192. [DOI] [PubMed] [Google Scholar]
- 18.Ploss A, Leiner I, Pamer EG. Distinct regulation of H2-M3-restricted memory T cell responses in lymph node and spleen. J Immunol. 2005;175:5998–6005. doi: 10.4049/jimmunol.175.9.5998. [DOI] [PubMed] [Google Scholar]
- 19.Urdahl KB, Sun JC, Bevan MJ. Positive selection of MHC class Ib-restricted CD8(+) T cells on hematopoietic cells. Nat Immunol. 2002;3:772–779. doi: 10.1038/ni814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Anderson G, Owen JJ, Moore NC, Jenkinson EJ. Thymic epithelial cells provide unique signals for positive selection of CD4+CD8+ thymocytes in vitro. J Exp Med. 1994;179:2027–2031. doi: 10.1084/jem.179.6.2027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bendelac A. Positive selection of mouse NK1+ T cells by CD1-expressing cortical thymocytes. J Exp Med. 1995;182:2091–2096. doi: 10.1084/jem.182.6.2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Atherly LO, et al. The Tec family tyrosine kinases Itk and Rlk regulate the development of conventional CD8+ T cells. Immunity. 2006;25:79–91. doi: 10.1016/j.immuni.2006.05.012. [DOI] [PubMed] [Google Scholar]
- 23.Horai R, et al. Requirements for selection of conventional and innate T lymphocyte lineages. Immunity. 2007;27:775–785. doi: 10.1016/j.immuni.2007.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Broussard C, et al. Altered development of CD8+ T cell lineages in mice deficient for the Tec kinases Itk and Rlk. Immunity. 2006;25:93–104. doi: 10.1016/j.immuni.2006.05.011. [DOI] [PubMed] [Google Scholar]
- 25.Alonzo ES, Sant’Angelo DB. Development of PLZF-expressing innate T cells. Curr Opin Immunol. 2011;23:220–227. doi: 10.1016/j.coi.2010.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kovalovsky D, et al. The BTB-zinc finger transcriptional regulator PLZF controls the development of invariant natural killer T cell effector functions. Nat Immunol. 2008;9:1055–1064. doi: 10.1038/ni.1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Savage AK, et al. The transcription factor PLZF directs the effector program of the NKT cell lineage. Immunity. 2008;29:391–403. doi: 10.1016/j.immuni.2008.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gorfu G, Rivera-Nieves J, Ley K. Role of beta7 integrins in intestinal lymphocyte homing and retention. Curr Mol Med. 2009;9:836–850. doi: 10.2174/156652409789105525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kerksiek KM, Busch DH, Pilip IM, Allen SE, Pamer EG. H2-M3-restricted T cells in bacterial infection: Rapid primary but diminished memory responses. J Exp Med. 1999;190:195–204. doi: 10.1084/jem.190.2.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dubois S, Waldmann TA, Müller JR. ITK and IL-15 support two distinct subsets of CD8+ T cells. Proc Natl Acad Sci USA. 2006;103:12075–12080. doi: 10.1073/pnas.0605212103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moon JJ, et al. Tracking epitope-specific T cells. Nat Protoc. 2009;4:565–581. doi: 10.1038/nprot.2009.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Weinreich MA, Odumade OA, Jameson SC, Hogquist KA. T cells expressing the transcription factor PLZF regulate the development of memory-like CD8+ T cells. Nat Immunol. 2010;11:709–716. doi: 10.1038/ni.1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Verykokakis M, Boos MD, Bendelac A, Kee BL. SAP protein-dependent natural killer T-like cells regulate the development of CD8(+) T cells with innate lymphocyte characteristics. Immunity. 2010;33:203–215. doi: 10.1016/j.immuni.2010.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ohteki T, MacDonald HR. Major histocompatibility complex class I related molecules control the development of CD4+8− and CD4−8− subsets of natural killer 1.1+ T cell receptor-alpha/beta+ cells in the liver of mice. J Exp Med. 1994;180:699–704. doi: 10.1084/jem.180.2.699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fink PJ, Bevan MJ. Positive selection of thymocytes. Adv Immunol. 1995;59:99–133. doi: 10.1016/s0065-2776(08)60630-6. [DOI] [PubMed] [Google Scholar]
- 36.Zerrahn J, et al. Class I MHC molecules on hematopoietic cells can support intrathymic positive selection of T cell receptor transgenic T cells. Proc Natl Acad Sci USA. 1999;96:11470–11475. doi: 10.1073/pnas.96.20.11470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kalergis AM, et al. Efficient T cell activation requires an optimal dwell-time of interaction between the TCR and the pMHC complex. Nat Immunol. 2001;2:229–234. doi: 10.1038/85286. [DOI] [PubMed] [Google Scholar]
- 38.Savage PA, Boniface JJ, Davis MM. A kinetic basis for T cell receptor repertoire selection during an immune response. Immunity. 1999;10:485–492. doi: 10.1016/s1074-7613(00)80048-5. [DOI] [PubMed] [Google Scholar]
- 39.Nichols KE, et al. Regulation of NKT cell development by SAP, the protein defective in XLP. Nat Med. 2005;11:340–345. doi: 10.1038/nm1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li W, et al. The SLAM-associated protein signaling pathway is required for development of CD4+ T cells selected by homotypic thymocyte interaction. Immunity. 2007;27:763–774. doi: 10.1016/j.immuni.2007.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sullivan BA, Kraj P, Weber DA, Ignatowicz L, Jensen PE. Positive selection of a Qa-1-restricted T cell receptor with specificity for insulin. Immunity. 2002;17:95–105. doi: 10.1016/s1074-7613(02)00343-6. [DOI] [PubMed] [Google Scholar]
- 42.Sullivan BA, Reed-Loisel LM, Kersh GJ, Jensen PE. Homeostatic proliferation of a Qa-1b-restricted T cell: A distinction between the ligands required for positive selection and for proliferation in lymphopenic hosts. J Immunol. 2004;173:6065–6071. doi: 10.4049/jimmunol.173.10.6065. [DOI] [PubMed] [Google Scholar]
- 43.Lenz LL, Dere B, Bevan MJ. Identification of an H2-M3-restricted Listeria epitope: Implications for antigen presentation by M3. Immunity. 1996;5:63–72. doi: 10.1016/s1074-7613(00)80310-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.