The temporal and spatial expression of proteins defines cell-specific activities and how a given cell responds to a local microenvironment. Thus, cells have evolved numerous integrated mechanisms to control which proteins are expressed and where they function at any given time. A small percentage of the human genome (<2%) encodes for messenger RNA (mRNA). A growing body of evidence suggests that noncoding RNAs (those that do not encode protein) constitute a substantial class of functional RNAs. Since the discovery that a small RNA molecule could control developmental transitions in Caenorhabditis elegans,1 microRNAs have been identified in numerous metazoans. In the past few years, human microRNAs (685 currently listed in the microRNA registry, version 11.0) have been shown to have a major role in complex gene regulatory networks and are estimated to contribute to the regulation of one third of all human genes. MicroRNAs, along with transcription factors, constitute the largest family of trans-acting, gene regulatory molecules, whose cell-specific expression directly and precisely regulate cellular functions. It is therefore not surprising that microRNAs are implicated in a wide variety of diseases, including cancer and diabetes mellitus. The role of microRNAs in the biology and pathobiology of GI tissues is now being unraveled. The purpose of this minireview is to provide current information on microRNA synthesis and processing, as well as microRNA-dependent gene regulation events that influence GI tissues in health and disease, and to highlight the potential of microRNAs for diagnosis and therapy.
MicroRNA Synthesis, Processing, and Function
The expression of a single microRNA potentially impacts the expression of a few to hundreds of proteins with a variety of cellular functions.2 Conversely, a single mRNA (and its translated protein) might be under stringent, but redundant, control of numerous microRNAs. To mechanistically address how cells control microRNA expression requires knowledge of their transcription and processing.
Synthesis and Processing
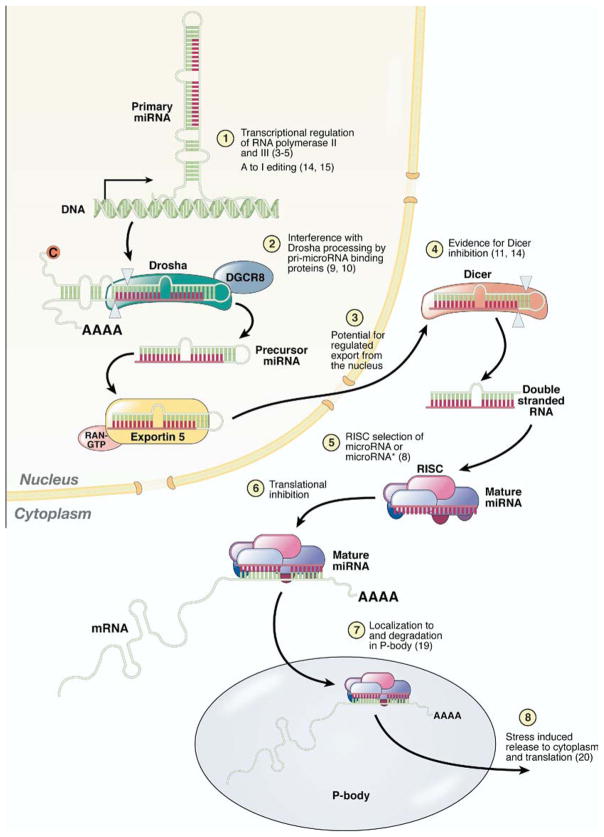
MicroRNAs are transcribed as mono- or polycistronic primary microRNAs (pri-microRNAs), which are processed to mature microRNAs (Figure 1). Human pri-microRNAs are often located between known protein encoding genes (intergenic microRNAs), but might also lie within an intron of a parental protein-coding gene (intronic microRNAs). Similar to protein-coding genes, intergenic microRNAs have promoters that interact with RNA polymerase II and transcription factors,3–5 permitting tissue-specific expression and signal-dependent regulation. Intronic microRNAs are cleaved from the primary mRNA transcript, suggesting mRNA promoter-dependent transcription of these microRNAs. The pri-microRNA molecule forms a hairpin structure recognized by the microprocessor complex containing the nuclear RNase III endonuclease, Drosha, and is rapidly processed to a 60- to 90-nucleotide stem loop RNA (~25–30 base pair stem). The resulting precursor microRNA (pre-microRNA) is actively transported from the nucleus through the nuclear export receptor, exportin 5. In the cytoplasm, the pre-microRNA is further processed by the RNAse III endonuclease, Dicer, which cleaves the precursor approximately 20 base pairs from the Drosha cut sites, thereby resulting in the formation of a short, double-stranded RNA molecule. The double-stranded microRNA associates with the RNA-induced silencing complex (RISC), where the RNA duplex is separated to yield a single-stranded mature microRNA, with strand selection based upon base-pair stability at the termini of the duplex.6,7 The “guide” strand typically loads into the RISC while the “passenger” strand (denoted by an asterisks; e.g., miR-199a*) is degraded. Recently, Kim et al8 have demonstrated that the microRNA* can load into the RISC and suppress translation of target mRNA, suggesting that specific spatial and temporal cellular environments may influence strand selection and, ultimately, protein expression.
Figure 1.
Biogenesis and processing of microRNAs. MicroRNAs are endogenous, non–protein-coding genes that are transcribed and ultimately processed into 20–22 nucleotide single-stranded RNA molecules. As indicated above and in the text, the production of a mature microRNA and ultimately translation inhibition is a highly regulated process.
In addition to acute and epigenetic transcriptional regulation, the potential exists for microRNA expression regulation at any of the posttranscriptional processing steps. For instance, posttranscriptional regulation has been demonstrated at the level of Drosha9,10 and/or Dicer,11 which enables the expression of either the primary or precursor microRNA, whereas expression of the mature, functional microRNA is inhibited.9 This regulated processing has been observed in embryonic tissues and human primary tumors.9 For example, Lin28, a developmentally regulated RNA binding protein that in conjunction with Nanog, Oct-4, and Sox2 can reprogram fibroblasts to pluripotency,12 selectively blocks the processing of primary let-7 microRNAs in embryonic cells.10 Interestingly, a Lin28 homolog, Lin28B, which also inhibits let-7 processing, is over-expressed in human hepatocellular carcinoma (HCC) as well as several cancer cell lines. Processing of the primary transcript can also include adenosine to inosine (A to I) editing by adenosine deaminases that act on RNA.13 This RNA editing might result in decreased cleavage by Dicer14 or, because inosine preferentially base pairs with guanosine, the modified nucleotide can alter target selection.15 Therefore, as with mRNAs, the functional expression of microRNAs is a complex, highly regulated process including both transcriptional and posttranscriptional processes.
Function
Frequently, there is dissociation between mRNA expression and cellular protein levels; microRNA-induced suppression of translation is a possible mechanism for this. MicroRNAs function to “fine tune” cellular expression of proteins. Unlike short interfering RNAs (siRNAs), which are designed to target mRNA with 100% complementarity and promote mRNA degradation, microRNA binding to mRNAs requires fewer complementary bases. Complementary base pairing between the microRNA seed region (nucleotides 2–8) and the 3′ untranslated region (UTR) of target mRNA is required for posttranscriptional gene silencing. However, recent reports have demonstrated the existence of microRNA targets within the coding sequence of several mRNAs.16,17 The degree of complementary base pairing over the entire length of the microRNA determines the mechanism of microRNA-induced mRNA translation suppression. Perfect or near-perfect complementarity between the microRNA and the mRNA results in mRNA degradation, similar to siRNA translational inhibition.18 More frequently, translational suppression occurs when the microRNA seed region binds target mRNA with less complementarity throughout the entire sequence. The precise mechanism of translational suppression by this pathway is not defined clearly, but most likely involves recruitment of mRNAs to processing bodies, intracellular organelles in which mRNAs could be sequestered or degraded.19 A recent report has described the microRNA-dependent sequestration of cationic amino acid transporter mRNA in hepatocyte processing bodies with subsequent release and rapid protein production during cellular stress.20 Therefore, microRNA-induced sequestration of mRNAs inhibits translation; conversely, protein levels can be rapidly increased under specific cellular conditions through release of the mRNA from the processing body.
Recently, reports have also defined a role for microRNAs in hepatitis C virus (HCV) replication.21 Specifically, miR-122 complementary sites were identified in both the 5′ and 3′ UTR of HCV RNA and were conserved among genotypes. The authors concluded that miR-122 does not affect viral RNA translation or stability but is likely recruited to the 5′ UTR for viral RNA replication. This study not only defines a novel role of animal microRNAs in viral replication, but also is the first to suggest that animal microRNAs contribute to gene transcription through recruitment to the 5′ UTR of transcripts.
Expression Profiling and Functional Analyses
Expression Profiling
Modifications of common molecular protocols allow for the analysis of mature microRNA expression. Microarray, cloning, and real-time polymerase chain reaction (PCR)-based approaches are commonly performed for microRNA expression analyses. Several microarray platforms are available; each address issues of sensitivity and specificity in a different manner. In general, the isolated or enriched microRNAs are labeled and hybridized to arrays spotted with microRNA-specific probes and scanned to obtain the relative expression in a given sample. For cloning analyses, small RNAs are enriched and cloned, followed by sequencing to identify microRNAs and their relative abundance.22,23 Large-scale, quantitative reverse transcriptase (RT) PCR is a technique frequently used to obtain the microRNA expression profile of a given sample. The small size of microRNAs precludes the use of standard RT PCR protocols; therefore, the reverse transcribed product must be extended before amplification. One way to achieve this is through the addition of a poly-A tail to mature microRNAs, which serves as a binding-site for oligo dT-primed reverse transcription. A 5′ extension on the oligo dT primer, including a universal primer binding site, extends the length of the synthesized first strand. Specific amplification thus depends on primers complementary to individual microRNAs. Whereas expression analyses are required to identify microRNAs with altered expression patterns in diseased tissues, functional analyses of the ability of these microRNAs to regulate expression of target mRNAs are essential to understand their impact on pathogenic pathways and processes.
Functional Analyses
Functional data exist for a small percentage of microRNAs. These studies are frequently performed by transfecting precursor microRNAs or antisense-inhibitor microRNAs (antagomirs) into cells and using immunoblot analyses to identify resulting changes in protein levels. One obvious disadvantage of this technique, as with the transfection of siRNAs, is the potential for off-target effects. Report assays (typically with firefly luciferase or green fluorescent protein) are utilized to show functional interactions between microRNAs and the 3′ UTR of a target mRNA. With this technique, the predicted target region of the mRNA is inserted into the 3′ UTR of the reporter construct or the entire 3′ UTR of the target mRNA is cloned at the 3′ end of the reporter coding sequence. The plasmid construct can then be transfected and the expression of the reporter, if targeted by the microRNA of interest, correlates inversely with the amount of the microRNA of interest.
Short hairpin RNAs (shRNAs), structural and functional homologs of pre-microRNAs encoded in a plasmid or viral vector, can be used to increase the expression of a microRNA in a specific cell type; this technique can generate continuous cell lines or transgenic animals with stable and heritable gene silencing. A drawback of this technique is the potential to inhibit endogenous microRNA function through saturation of a limiting level of 1 factors involved in microRNA biogenesis. Studies have demonstrated that over-expressed shRNAs saturated the activity of exportin 5, a factor required for pre-microRNA export from the nucleus, and interfered with endogenous microRNA processing.24,25 In vivo studies have also demonstrated the toxicity of this approach as shRNA expression in the livers of adult mice resulted in significant fatality.25 Furthermore, the expression of shRNAs can result in a nonspecific interferon response through RNA-activated protein kinase and interaction with Toll-like receptors (TLRs). Further optimization and regulation of shRNA expression,25 as well as nucleotide modifications that limit immune stimulation,26 could prove useful in moving this technology from bench to bedside.
Recently, locked nucleic acid (LNA) microRNA antagomirs were shown to be highly effective at modulating endogenous microRNA expression. LNA antagomirs, 1 type of modified antisense oligonucleotides (AMOs), are synthesized DNA molecules that contain modified ribose moieties in which the pentose sugar is constrained in the N-type conformation seen in A-form DNA. LNA-modified oligonucleotides have several advantages over standard antisense molecules. For example, the modified oligonucleotide increases thermal stability between the hybridized LNA molecule and target nucleotides, increasing binding affinity.27–29 The LNA modification also enhances mismatch discrimination, compared with un-modified antisense molecules, resulting in increased target recognition.30 Furthermore, LNA oligonucleotides exhibit high levels of stability (the LNA half-life in serum is reported to be ~15 hours, compared with ~12 hours for a 2′-O-methyl modified antisense molecule)29 and solubility in aqueous solutions.30 LNA-based microRNA probes are used commonly for in situ hybridization studies and array-based detection.
Several groups have developed microRNA knockout mice31–33 or transgenic animals that over-express microRNAs34–36 to study the roles of microRNAs in vivo. Zhao et al31 developed miR-1-2 knockout mice to study heart development and observed partial embryonic lethality owing to abnormal heart morphogenesis. Those mice that survived to adulthood demonstrated heart defects, including increased cardiomyocyte proliferation and electrophysiologic defects. Additionally, Johnnidis et al37 demonstrated that loss of function of a single miR-223 allele leads to an inflammatory lung pathology. The development of microRNA knockout mice is clearly a useful tool to improve our understanding of the relevance of microRNAs to GI function and disease.
MicroRNA Expression Profiles Are Altered in Malignant Diseases of the GI Tract
As with other complex gene regulatory networks, aberrant expression or processing of microRNAs could have profound effects on cellular function and contribute to disease states. Profiling studies have revealed that microRNA levels are dysregulated in hepatic,38–41 bile duct,42 colon,43,44 and pancreatic45–47 cancer cells. For example, members of the miR-17-92 cluster, as well as miR-21, miR-224, and miR-221/222, were typically expressed at higher levels in HCC tissues compared with normal tissues. Of interest, levels of let-7 family members, miR-29 family members and miR-103/107 were all dysregulated, but were either over-expressed or under-expressed in different tumor samples. These variations might have resulted from different methodologies or predisposing risk factors and etiologies of the HCCs studied. In colon cancer tissues, several microRNAs were expressed at altered levels in independent expression analysis studies. For example, miR-143 and miR-145 were consistently down-regulated,43,48,49 whereas miR-21 was consistently up-regulated43,48,49 in colorectal tumor samples versus normal tissues. Additionally, microRNA expression analysis of pancreatic tumors or cell lines derived from pancreatic tumors demonstrated altered expression compared with normal pancreatic tissue. Pancreatic tumors, regardless of histologic type, could be distinguished from normal pancreas by the over-expression of miR-103 and miR-109, and the lack of miR-155.46 However, conflicting expression data were reported from studies using both array45 and PCR-based approaches45,47 in that miR-155 was over-expressed in pancreatic ductal adenocarcinomas and cell lines derived from these tumors, which is consistent with solid tumors of other origins. The conflicting reports demonstrate the need to verify altered expression of individual microRNAs using several methodologies.
MicroRNAs Contribute to Cellular Processes Associated With GI Diseases
Important studies have demonstrated that microRNAs regulate tumor suppressors44,50 and oncogenes51–53 in a variety of tissues (Table 1). For instance, the let-7 family has been shown to regulate the Ras oncogene and the decreased let-7 expression observed in lung tumors is permissive for Ras over-expression.53 Therefore, decreased expression of this microRNA results in the over-expression of an oncogene, making let-7 a putative tumor suppressor. Specific examples of microRNA dys-regulation with functional implications for GI cancers include the frequent up-regulation of miR-21. Functional analysis has demonstrated that miR-21 contributes to regulation of the tumor suppressor PTEN in both cholangiocarcinoma and HCC54 and posttranscriptionally inhibits expression of the tumor suppressor PDCD4 in colorectal cell lines.50 Several studies also consistently reported miR-21 over-expression in pancreatic adenocarcinomas. Although the functional implications of miR-21 over-expression in this tissue has not been determined, it seems likely, based on studies from other tissues, that this microRNA regulates the expression of relevant tumor suppressor proteins. Furthermore, strong expression of this microRNA predicts limited survival in patients with node-negative pancreatic cancers.55
Table 1.
Examples of microRNAs with established targets in various organs within the gastrointestinal tract. Altered microRNA expression is frequently associated with known disease processes or conditions. The OMIM reference number for protein description is provided for each target protein (http://www.ncbi.nlm.nih.gov/sites/entrez?db=OMIM)
| microRNA | Established target (OMIM #) | Pathway/function | Disease/process | [Ref.] |
|---|---|---|---|---|
| Liver | ||||
| miR-122 | — | Cholesterol metabolism | [80] | |
| miR-122 | Viral RNA | Transcription | HCV | [21] |
| miR-122 | CAT-1 (104615) | Transport | — | [20] |
| miR-122 | Cyclin G1 (601578) | Cell cycle | Hepatocellular carcinoma | [59] |
| miR-21 | PTEN (601728) | Cell cycle | Hepatocellular carcinoma | [54] |
| miR-29 | MCL-1 (159552) | Apoptosis | Cholangiocarcinoma | [52] |
| let-7 | TLR4 (603030) | Infection/immune response | Cryptosporidiosis | [66] |
| miRNA-15a | CDC25A (116947) | Cell cycle | Cystic liver disease | [61] |
| miR-223 | STMN1 (151442) | Cell cycle/Signal transduction | Hepatocellular carcinoma | [84] |
| miR-148a | PXR (603065) | Nuclear receptor | Xenobiotic metabolism | [83] |
| Stomach | ||||
| miR-106b-25 cluster | E2F1 (189971) | Cell cycle | Gastric cancer | [82] |
| miR-15b/16 | BCL2 (151430) | Apoptosis | Gastric cancer | [85] |
| Pancreas | ||||
| miR-155 | TP53INP1 (606185) | Apoptosis | Ductal adenocarcinoma | [57] |
| Colon/Rectum | ||||
| let-7-a-1 | RAS (190070) | Signal transduction | Colorectal cancer | [53] |
| miR-143 | Erk5 (602521) | Signal transduction | Colorectal cancer | [62] |
| miR-21 | PDCD4 (608610) | Translation inhibition | Colorectal cancer | [50] |
| miR-126 | PI3K (603157) | Signal transduction | Colorectal cancer | [81] |
Apoptosis
Apoptosis is an important factor in the pathophysiology of several GI diseases. For instance, in primary liver cancers such as HCC and cholangiocarcinoma, malignant cells resist apoptotic signals, inappropriately surviving oncogenic transformation and often displaying resistance to chemotherapeutic agents. We reported that decreased miR-29b in the cultured malignant cholangiocarcinoma cell line (KMCH) increases Mcl-1 protein expression, compared with nonmalignant human cholangiocytes (H69), and contributes to survival of malignant cells.52 Datta et al56 recently reported the methylation-mediated silencing of miR-1-1 in both HCC cell lines and primary HCCs. Reintroduction of this microRNA to HCC cell lines promoted both cell-cycle arrest and apoptosis, possibly through the attenuated expression of the transcription factor FoxP1 and the receptor tyrosine kinase Met. miR-155 is over-expressed in pancreatic tumor samples; this microRNA targets and limits the expression of the stress-induced gene TP53INP1,57 which promotes p53-dependent apoptosis and is decreased in many pancreatic tumors. Animal models support the importance of microRNAs in apoptosis regulation. For example, Bim expression is increased in mice deficient in miR-17-92 and the mice die of pulmonary hypoplasia, possibly resulting from excessive cell death.58
Proliferation and Cell-Cycle Regulation
Increased proliferation accompanies dysregulated apoptosis in many cancers. MicroRNAs regulate the cell cycle and proliferation in many cell types. Gramantieri et al59 demonstrated that several microRNAs were differentially expressed between HCC and nonneoplastic liver tissues; miR-122 was down-regulated in HCC cells and its expression was inversely correlated with cyclin G1 expression. Furthermore, Shah et al60 demonstrated that agonist-activated peroxisome proliferator activated receptor-α, a member of the steroid hormone nuclear receptor superfamily, altered microRNA expression in a model of hepatocellular proliferation and tumorigenesis. One down-regulated microRNA, let-7c, targets the oncogenic transcription factor c-myc; let-7c down-regulation and the resulting increase in c-myc increased miR-17-92 expression and the proliferative capacity of hepatocytes. Cholangiocyte hyperproliferation was associated with cystogenesis in polycystic liver diseases, most notably with autosomal recessive polycystic kidney disease. Levels of miR-15a were significantly decreased in a cholangiocyte cell line derived from a rat model of autosomal recessive polycystic kidney disease compared with a normal cholangiocyte cell line; this microRNA targets the cell-cycle regulation gene Cdc25A. Transient over-expression of miR-15a ultimately led to decreased protein levels of Cdc25A and numbers of cells in the DNA synthesis phase of the cell cycle.61 MicroRNAs also contribute to intestinal epithelial cell hyperproliferation—transfection of the commonly dysregulated microRNAs miR-143 or miR-145 into colon cancer cell lines significantly inhibited cell proliferation. Furthermore, manipulation of cellular miR-143 levels altered the expression of ERK5, a mitogen-activated protein kinase that promotes proliferation.62
Differentiation
Using a cell culture model of intestinal cell differentiation, Hino et al63 found that miR-194 was induced and regulated by the transcription factor HNF1 during differentiation. Additionally, overlapping microRNA expression patterns exist between the embryonic colonic mucosa and colorectal cancer.64 The miR-17-92 cluster increased in embryonic colon mucosa and colorectal cancer tissues; this increase was associated with decreased levels of the transcription factor E2F1, which promotes apoptosis and regulates cell-cycle progression through the tumor suppressor protein Rb. Increased expression of the miR-17-92 cluster of microRNAs has been associated with colonic development, decreased apoptosis, increased proliferation, and cell-cycle progression in various tissues. The functional role of this cluster of microRNAs has been observed in transgenic mice that over-express the miR-17-92 cluster and develop a lymphoproliferative phenotype.36 Thus, it is intriguing to speculate that microRNAs have a similar role in human embryogenesis and neoplastic transformation.
Immune Regulation and Pathogen Recognition
MicroRNAs are implicated in the regulation of immune cell differentiation22,65 as well as innate66,67 and adaptive32,33 immune responses. Little is known about the mechanisms that control expression of proteins that contribute to chronic inflammatory diseases of the GI tract (ie, ulcerative colitis, Crohn’s disease, primary sclerosing cholangitis, or primary biliary cirrhosis). Although the contribution of microRNAs to chronic inflammatory conditions of the GI tract is unclear, their general contribution to the initiation and attenuation of inflammatory cascades is better understood. In an effort to find candidate microRNAs involved in host cell responses to microbial challenge, Taganov et al67 analyzed the microRNA expression profile of a monocytic leukemia cell line following TLR stimulation with ligands. Several microRNAs (miR-146a, miR-132, and miR-155) were up-regulated after stimulation with the bacterial endotoxin lipopolysaccharide. Functional analysis demonstrated that miR-146a expression is nuclear factor kappa B (NF-κB)–dependent and this microRNA targets interleukin-1 receptor-associated kinase and TRAF6, which each activate the TLR and pro-inflammatory cytokine signaling cascades. Therefore, expression of miR-146a has a negative effect on pro-inflammatory signaling in a leukemia cell line. Conversely, in a model of biliary cryptosporidiosis, infection of cultured human cholangiocytes with C parvum (a parasite that causes intestinal and biliary disease) decreased let-7i expression in a MyD88 and NF-κB–dependent manner. Decreased let-7i expression resulted in up-regulation of TLR4 in infected cells and increased NF-κB signaling.66 These data raise the possibility that microRNA-regulated posttranscriptional pathways contribute to host–cell responses to microbial infection by increasing inflammatory signaling in response to pathogens66 or attenuating the inflammatory response.67
Clinical Relevance
MicroRNAs for Diagnosis, Prognosis, and Therapy
Improved the understanding, detection, and delivery of microRNAs could have clinical value in approaches to GI diseases. Analysis of the microRNA profiles from diseased tissues might aide in diagnosis, allowing pathologists to distinguish between similar conditions, define the origin of a tumor of unknown primary, or distinguish benign from malignant lesions.38 Alternatively, microRNAs might provide evidence for underlying pathologies68 and improve de novo diagnosis; assays of sentinal microRNAs in blood, bile, or stool samples could be used to follow progression of a known condition. Additionally, prognostic information could be gained from knowing the expression level of individual microRNAs. Bloomston et al69 found that high expression levels of miR-196a-2 predicted poor survival of lymph node-positive pancreatic cancer patients and specific microRNA profiles have been associated with increased metastatic potential.12,70 This prognostic information may therefore act as a tool for disease management decisions. Further study is required to determine whether there are individual microRNAs or microRNA expression patterns associated with GI diseases that can be used in prognosis, such as determining progression of hepatitis or inflammatory bowel disease. This line of diagnostic and prognostic information will certainly mature as the phenotypic effects of individual microRNA alterations are better understood in the GI tract.
Therapeutically, it seems reasonable to predict that the microRNA profile of a tumor biopsy could be used to direct individualized therapy, based either on empiric correlations between microRNA expression profiles or on mechanism-based alterations in specific pathways caused by microRNA dysregulation. The identification of aberrantly expressed microRNAs and advances in our understanding of the pathways affected by microRNA dysregulation indicate the exciting possibility of therapeutic interventions involving the reintroduction of mature microRNAs (in instances where a microRNA is reduced or absent) or selective inhibition of over-expressed microRNAs (using AMOs). One advantage of microRNA-based therapy, compared with siRNA-based therapy, is that reintroduction of a microRNA results in expression of an endogenous regulator of physiologic processes. However, introducing antisense oligonucleotides to reduce the expression of a microRNA has the potential to generate undesirable off-target effects, although direct modulation of microRNA levels is not the only approach that could be used in microRNA-based therapy. Indeed, some cancers are marked by deficiencies in microRNA processing.9,71 In these instances, small molecule activators or repressors of endogenous microRNA processing or expression could affect a cellular phenotype to therapeutic gain.
MicroRNA-based therapies face many of the same considerable challenges as gene therapies, including selectivity, delivery, and efficacy. However, the approaches developed for in vivo delivery of siRNAs or shRNAs, including viral vectors, liposomes, and nanoparticles72–74 and specific nucleotide delivery through cell-surface receptor targeting,75–77 improve progress toward microRNA-based therapies. There are several examples of successful therapeutic application of RNA interference that use these technologies, typically in tissues that are easily accessible to delivery, such as the retina or lung epithelia.78,79
Although obstacles and concerns remain, successful application of microRNA therapies has been established in non-human primates, where antagonism of miR-122, through intraperitoneal delivery of an AMO, significantly lowered plasma cholesterol levels.80 Although other, more effective cholesterol-lowering therapeutics exist, this accomplishment highlights the opportunity to modulate organ function by controlling microRNA levels. Recently, Santaris Pharma (Denmark) initiated a clinical trial for treatment of HCV infection using SPC3649, an AMO that targets miR-122. Although SPC3649 is the only microRNA antagomir currently in clinical trials, other groups are likely to evaluate the therapeutic efficacy of other microRNA-based therapies. microRNAs have an important role in normal and pathologic cellular processes in the GI tract; their roles in effecting coordinated changes in expression of multiple proteins places these regulators in prime position for their utility in diagnosis, prognosis, and treatment of complex conditions.
Footnotes
The authors recognize the advances made by our colleagues in the field of microRNA biology. We regret that we cannot reference all of their significant work.
The authors disclose no conflicts.
References
- 1.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–54. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 2.Karginov FV, Conaco C, Xuan Z, et al. A biochemical approach to identifying microRNA targets. Proc Natl Acad Sci U S A. 2007;104:19291–6. doi: 10.1073/pnas.0709971104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–97. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 4.Zhao Y, Samal E, Srivastava D. Serum response factor regulates a muscle-specific microRNA that targets Hand2 during cardiogenesis. Nature. 2005;436:214–20. doi: 10.1038/nature03817. [DOI] [PubMed] [Google Scholar]
- 5.Borchert GM, Lanier W, Davidson BL. RNA polymerase III transcribes human microRNAs. Nat Struct Mol Biol. 2006;13:1097–101. doi: 10.1038/nsmb1167. [DOI] [PubMed] [Google Scholar]
- 6.Khvorova A, Reynolds A, Jayasena SD. Functional siRNAs and miRNAs exhibit strand bias. Cell. 2003;115:209–16. doi: 10.1016/s0092-8674(03)00801-8. [DOI] [PubMed] [Google Scholar]
- 7.Schwarz DS, Hutvagner G, Du T, et al. Asymmetry in the assembly of the RNAi enzyme complex. Cell. 2003;115:199–208. doi: 10.1016/s0092-8674(03)00759-1. [DOI] [PubMed] [Google Scholar]
- 8.Kim S, Lee UJ, Kim MN, et al. MicroRNA miR-199a* regulates the MET proto-oncogene and the downstream extracellular signal-regulated kinase 2 (ERK2) J Biol Chem. 2008;283:18158–66. doi: 10.1074/jbc.M800186200. [DOI] [PubMed] [Google Scholar]
- 9.Thomson JM, Newman M, Parker JS, et al. Extensive post-transcriptional regulation of microRNAs and its implications for cancer. Genes Dev. 2006;20:2202–7. doi: 10.1101/gad.1444406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Viswanathan SR, Daley GQ, Gregory RI. Selective blockade of microRNA processing by Lin28. Science. 2008;320:97–100. doi: 10.1126/science.1154040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Obernosterer G, Leuschner PJ, Alenius M, et al. Post-transcriptional regulation of microRNA expression. RNA. 2006;12:1161–7. doi: 10.1261/rna.2322506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yu J, Vodyanik MA, Smuga-Otto K, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–20. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 13.Luciano DJ, Mirsky H, Vendetti NJ, et al. RNA editing of a miRNA precursor. RNA. 2004;10:1174–7. doi: 10.1261/rna.7350304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kawahara Y, Zinshteyn B, Chendrimada TP, et al. RNA editing of the microRNA-151 precursor blocks cleavage by the Dicer-TRBP complex. EMBO Rep. 2007;8:763–9. doi: 10.1038/sj.embor.7401011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kawahara Y, Zinshteyn B, Sethupathy P, et al. Redirection of silencing targets by adenosine-to-inosine editing of miRNAs. Science. 2007;315:1137–40. doi: 10.1126/science.1138050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tay Y, Zhang J, Thomson AM, et al. MicroRNAs to Nanog, Oct4 and Sox2 coding regions modulate embryonic stem cell differentiation. Nature. 2008;455:1124–8. doi: 10.1038/nature07299. [DOI] [PubMed] [Google Scholar]
- 17.Forman JJ, Legesse-Miller A, Coller HA. A search for conserved sequences in coding regions reveals that the let-7 microRNA targets Dicer within its coding sequence. Proc Natl Acad Sci U S A. 2008;105:14879–84. doi: 10.1073/pnas.0803230105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bagga S, Bracht J, Hunter S, et al. Regulation by let-7 and lin-4 miRNAs results in target mRNA degradation. Cell. 2005;122:553–63. doi: 10.1016/j.cell.2005.07.031. [DOI] [PubMed] [Google Scholar]
- 19.Chan SP, Slack FJ. microRNA-mediated silencing inside P-bodies. RNA Biol. 2006;3:97–100. doi: 10.4161/rna.3.3.3499. [DOI] [PubMed] [Google Scholar]
- 20.Bhattacharyya SN, Habermacher R, Martine U, et al. Stress-induced reversal of microRNA repression and mRNA P-body localization in human cells. Cold Spring Harb Symp Quant Biol. 2006;71:513–21. doi: 10.1101/sqb.2006.71.038. [DOI] [PubMed] [Google Scholar]
- 21.Jopling CL, Yi M, Lancaster AM, et al. Modulation of hepatitis C virus RNA abundance by a liver-specific MicroRNA. Science. 2005;309:1577–81. doi: 10.1126/science.1113329. [DOI] [PubMed] [Google Scholar]
- 22.Wu H, Neilson JR, Kumar P, et al. miRNA profiling of naive, effector and memory CD8 T cells. PLoS ONE. 2007;2:e1020. doi: 10.1371/journal.pone.0001020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Landgraf P, Rusu M, Sheridan R, et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129:1401–14. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yi R, Doehle BP, Qin Y, et al. Overexpression of exportin 5 enhances RNA interference mediated by short hairpin RNAs and microRNAs. RNA. 2005;11:220–6. doi: 10.1261/rna.7233305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Grimm D, Streetz KL, Jopling CL, et al. Fatality in mice due to oversaturation of cellular microRNA/short hairpin RNA pathways. Nature. 2006;441:537–41. doi: 10.1038/nature04791. [DOI] [PubMed] [Google Scholar]
- 26.Gondai T, Yamaguchi K, Miyano-Kurosaki N, et al. Short-hairpin RNAs synthesized by T7 phage polymerase do not induce interferon. Nucleic Acids Res. 2008;36:e18. doi: 10.1093/nar/gkm1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Koshkin AA, Singh SK, Nielsen P, et al. LNA (locked nucleic acid): Synthesis of the adenine, cytosine, guanine, 5-methylcytosine, thymine and uracil bicyclonucleoside monomers, oligomerisation, and unprecedented nucleic acid recognition. Tetrahedron Letters. 1998;54:3607–30. [Google Scholar]
- 28.Braasch DA, Corey DR. Locked nucleic acid (LNA): fine-tuning the recognition of DNA and RNA. Chem Biol. 2001;8:1–7. doi: 10.1016/s1074-5521(00)00058-2. [DOI] [PubMed] [Google Scholar]
- 29.Kurreck J, Wyszko E, Gillen C, et al. Design of antisense oligonucleotides stabilized by locked nucleic acids. Nucleic Acids Res. 2002;30:1911–8. doi: 10.1093/nar/30.9.1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Grunweller A, Hartmann RK. Locked nucleic acid oligonucleotides: the next generation of antisense agents? BioDrugs. 2007;21:235–43. doi: 10.2165/00063030-200721040-00004. [DOI] [PubMed] [Google Scholar]
- 31.Zhao Y, Ransom JF, Li A, et al. Dysregulation of cardiogenesis, cardiac conduction, and cell cycle in mice lacking miRNA-1-2. Cell. 2007;129:303–17. doi: 10.1016/j.cell.2007.03.030. [DOI] [PubMed] [Google Scholar]
- 32.Rodriguez A, Vigorito E, Clare S, et al. Requirement of bic/microRNA-155 for normal immune function. Science. 2007;316:608–11. doi: 10.1126/science.1139253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thai TH, Calado DP, Casola S, et al. Regulation of the germinal center response by microRNA-155. Science. 2007;316:604–8. doi: 10.1126/science.1141229. [DOI] [PubMed] [Google Scholar]
- 34.Costinean S, Zanesi N, Pekarsky Y, et al. Pre-B cell proliferation and lymphoblastic leukemia/high-grade lymphoma in E(mu)-miR155 transgenic mice. Proc Natl Acad Sci U S A. 2006;103:7024–9. doi: 10.1073/pnas.0602266103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lu Y, Thomson JM, Wong HY, et al. Transgenic over-expression of the microRNA miR-17-92 cluster promotes proliferation and inhibits differentiation of lung epithelial progenitor cells. Dev Biol. 2007;310:442–53. doi: 10.1016/j.ydbio.2007.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xiao C, Srinivasan L, Calado DP, et al. Lymphoproliferative disease and autoimmunity in mice with increased miR-17-92 expression in lymphocytes. Nat Immunol. 2008;9:405–14. doi: 10.1038/ni1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Johnnidis JB, Harris MH, Wheeler RT, et al. Regulation of progenitor cell proliferation and granulocyte function by microRNA-223. Nature. 2008;451:1125–9. doi: 10.1038/nature06607. [DOI] [PubMed] [Google Scholar]
- 38.Ladeiro Y, Couchy G, Balabaud C, et al. MicroRNA profiling in hepatocellular tumors is associated with clinical features and oncogene/tumor suppressor gene mutations. Hepatology. 2008;47:1955–63. doi: 10.1002/hep.22256. [DOI] [PubMed] [Google Scholar]
- 39.Jiang J, Gusev Y, Aderca I, et al. Association of MicroRNA expression in hepatocellular carcinomas with hepatitis infection, cirrhosis, and patient survival. Clin Cancer Res. 2008;14:419–27. doi: 10.1158/1078-0432.CCR-07-0523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Huang YS, Dai Y, Yu XF, et al. Microarray analysis of microRNA expression in hepatocellular carcinoma and non-tumorous tissues without viral hepatitis. J Gastroenterol Hepatol. 2008;23:87–94. doi: 10.1111/j.1440-1746.2007.05223.x. [DOI] [PubMed] [Google Scholar]
- 41.Murakami Y, Yasuda T, Saigo K, et al. Comprehensive analysis of microRNA expression patterns in hepatocellular carcinoma and non-tumorous tissues. Oncogene. 2006;25:2537–45. doi: 10.1038/sj.onc.1209283. [DOI] [PubMed] [Google Scholar]
- 42.Meng F, Henson R, Lang M, et al. Involvement of human micro-RNA in growth and response to chemotherapy in human cholangiocarcinoma cell lines. Gastroenterology. 2006;130:2113–29. doi: 10.1053/j.gastro.2006.02.057. [DOI] [PubMed] [Google Scholar]
- 43.Cummins JM, He Y, Leary RJ, et al. The colorectal microRNAome. Proc Natl Acad Sci U S A. 2006;103:3687–92. doi: 10.1073/pnas.0511155103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Volinia S, Calin GA, Liu CG, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci U S A. 2006;103:2257–61. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Szafranska AE, Davison TS, John J, et al. MicroRNA expression alterations are linked to tumorigenesis and non-neoplastic processes in pancreatic ductal adenocarcinoma. Oncogene. 2007;26:4442–52. doi: 10.1038/sj.onc.1210228. [DOI] [PubMed] [Google Scholar]
- 46.Roldo C, Missiaglia E, Hagan JP, et al. MicroRNA expression abnormalities in pancreatic endocrine and acinar tumors are associated with distinctive pathologic features and clinical behavior. J Clin Oncol. 2006;24:4677–84. doi: 10.1200/JCO.2005.05.5194. [DOI] [PubMed] [Google Scholar]
- 47.Lee EJ, Gusev Y, Jiang J, et al. Expression profiling identifies microRNA signature in pancreatic cancer. Int J Cancer. 2007;120:1046–54. doi: 10.1002/ijc.22394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Michael MZ, O’Connor SM, van Holst Pellekaan NG, et al. Reduced accumulation of specific microRNAs in colorectal neoplasia. Mol Cancer Res. 2003;1:882–91. [PubMed] [Google Scholar]
- 49.Slaby O, Svoboda M, Fabian P, et al. Altered expression of miR-21, miR-31, miR-143 and miR-145 is related to clinicopathologic features of colorectal cancer. Oncology. 2007;72:397–402. doi: 10.1159/000113489. [DOI] [PubMed] [Google Scholar]
- 50.Asangani IA, Rasheed SA, Nikolova DA, et al. MicroRNA-21 (miR-21) post-transcriptionally downregulates tumor suppressor Pdcd4 and stimulates invasion, intravasation and metastasis in colorectal cancer. Oncogene. 2008;27:2128–36. doi: 10.1038/sj.onc.1210856. [DOI] [PubMed] [Google Scholar]
- 51.O’Donnell KA, Wentzel EA, Zeller KI, et al. c-Myc-regulated microRNAs modulate E2F1 expression. Nature. 2005;435:839–43. doi: 10.1038/nature03677. [DOI] [PubMed] [Google Scholar]
- 52.Mott JL, Kobayashi S, Bronk SF, et al. mir-29 regulates Mcl-1 protein expression and apoptosis. Oncogene. 2007;26:6133–40. doi: 10.1038/sj.onc.1210436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Johnson SM, Grosshans H, Shingara J, et al. RAS is regulated by the let-7 microRNA family. Cell. 2005;120:635–47. doi: 10.1016/j.cell.2005.01.014. [DOI] [PubMed] [Google Scholar]
- 54.Meng F, Henson R, Wehbe-Janek H, et al. MicroRNA-21 regulates expression of the PTEN tumor suppressor gene in human hepatocellular cancer. Gastroenterology. 2007;133:647–58. doi: 10.1053/j.gastro.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dillhoff M, Liu J, Frankel W, et al. MicroRNA-21 is Overexpressed in Pancreatic Cancer and a Potential Predictor of Survival. J Gastrointest Surg. 2008 Jul 19; doi: 10.1007/s11605-008-0584-x. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Datta J, Kutay H, Nasser MW, et al. Methylation mediated silencing of MicroRNA-1 gene and its role in hepatocellular carcinogenesis. Cancer Res. 2008;68:5049–58. doi: 10.1158/0008-5472.CAN-07-6655. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 57.Gironella M, Seux M, Xie MJ, et al. Tumor protein 53-induced nuclear protein 1 expression is repressed by miR-155, and its restoration inhibits pancreatic tumor development. Proc Natl Acad Sci U S A. 2007;104:16170–5. doi: 10.1073/pnas.0703942104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ventura A, Young AG, Winslow MM, et al. Targeted deletion reveals essential and overlapping functions of the miR-17 through 92 family of miRNA clusters. Cell. 2008;132:875–86. doi: 10.1016/j.cell.2008.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gramantieri L, Ferracin M, Fornari F, et al. Cyclin G1 is a target of miR-122a, a microRNA frequently down-regulated in human hepatocellular carcinoma. Cancer Res. 2007;67:6092–9. doi: 10.1158/0008-5472.CAN-06-4607. [DOI] [PubMed] [Google Scholar]
- 60.Shah YM, Morimura K, Yang Q, et al. Peroxisome proliferator-activated receptor alpha regulates a microRNA-mediated signaling cascade responsible for hepatocellular proliferation. Mol Cell Biol. 2007;27:4238–47. doi: 10.1128/MCB.00317-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lee SO, Masyuk T, Splinter P, et al. MicroRNA15a modulates expression of the cell-cycle regulator Cdc25A and affects hepatic cystogenesis in a rat model of polycystic kidney disease. J Clin Invest. 2008;118:3714–24. doi: 10.1172/JCI34922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Akao Y, Nakagawa Y, Naoe T. MicroRNAs 143 and 145 are possible common onco-microRNAs in human cancers. Oncol Rep. 2006;16:845–50. [PubMed] [Google Scholar]
- 63.Hino K, Tsuchiya K, Fukao T, et al. Inducible expression of microRNA-194 is regulated by HNF-1alpha during intestinal epithelial cell differentiation. RNA. 2008;14:1433–42. doi: 10.1261/rna.810208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Monzo M, Navarro A, Bandres E, et al. Overlapping expression of microRNAs in human embryonic colon and colorectal cancer. Cell Res. 2008;18:823–33. doi: 10.1038/cr.2008.81. [DOI] [PubMed] [Google Scholar]
- 65.Neilson JR, Zheng GX, Burge CB, et al. Dynamic regulation of miRNA expression in ordered stages of cellular development. Genes Dev. 2007;21:578–89. doi: 10.1101/gad.1522907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chen XM, Splinter PL, O’Hara SP, et al. A cellular micro-RNA, let-7i, regulates Toll-like receptor 4 expression and contributes to cholangiocyte immune responses against Cryptosporidium parvum infection. J Biol Chem. 2007;282:28929–38. doi: 10.1074/jbc.M702633200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Taganov KD, Boldin MP, Chang KJ, et al. NF-kappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc Natl Acad Sci U S A. 2006;103:12481–6. doi: 10.1073/pnas.0605298103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rosenfeld N, Aharonov R, Meiri E, et al. MicroRNAs accurately identify cancer tissue origin. Nat Biotechnol. 2008;26:462–9. doi: 10.1038/nbt1392. [DOI] [PubMed] [Google Scholar]
- 69.Bloomston M, Frankel WL, Petrocca F, et al. MicroRNA expression patterns to differentiate pancreatic adenocarcinoma from normal pancreas and chronic pancreatitis. JAMA. 2007;297:1901–8. doi: 10.1001/jama.297.17.1901. [DOI] [PubMed] [Google Scholar]
- 70.Budhu A, Jia HL, Forgues M, et al. Identification of metastasis-related microRNAs in hepatocellular carcinoma. Hepatology. 2008;47:897–907. doi: 10.1002/hep.22160. [DOI] [PubMed] [Google Scholar]
- 71.Karube Y, Tanaka H, Osada H, et al. Reduced expression of Dicer associated with poor prognosis in lung cancer patients. Cancer Sci. 2005;96:111–5. doi: 10.1111/j.1349-7006.2005.00015.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Abbas-Terki T, Blanco-Bose W, Deglon N, et al. Lentiviral-mediated RNA interference. Hum Gene Ther. 2002;13:2197–201. doi: 10.1089/104303402320987888. [DOI] [PubMed] [Google Scholar]
- 73.Lu PY, Xie F, Woodle MC. In vivo application of RNA interference: from functional genomics to therapeutics. Adv Genet. 2005;54:117–42. doi: 10.1016/S0065-2660(05)54006-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Schiffelers RM, Ansari A, Xu J, et al. Cancer siRNA therapy by tumor selective delivery with ligand-targeted sterically stabilized nanoparticle. Nucleic Acids Res. 2004;32:e149. doi: 10.1093/nar/gnh140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hu-Lieskovan S, Heidel JD, Bartlett DW, et al. Sequence-specific knockdown of EWS-FLI1 by targeted, nonviral delivery of small interfering RNA inhibits tumor growth in a murine model of metastatic Ewing’s sarcoma. Cancer Res. 2005;65:8984–92. doi: 10.1158/0008-5472.CAN-05-0565. [DOI] [PubMed] [Google Scholar]
- 76.McNamara JO, 2nd, Andrechek ER, Wang Y, et al. Cell type-specific delivery of siRNAs with aptamer-siRNA chimeras. Nat Biotechnol. 2006;24:1005–15. doi: 10.1038/nbt1223. [DOI] [PubMed] [Google Scholar]
- 77.Song E, Zhu P, Lee SK, et al. Antibody mediated in vivo delivery of small interfering RNAs via cell-surface receptors. Nat Biotechnol. 2005;23:709–17. doi: 10.1038/nbt1101. [DOI] [PubMed] [Google Scholar]
- 78.Bitko V, Musiyenko A, Shulyayeva O, et al. Inhibition of respiratory viruses by nasally administered siRNA. Nat Med. 2005;11:50–5. doi: 10.1038/nm1164. [DOI] [PubMed] [Google Scholar]
- 79.Shen J, Samul R, Silva RL, et al. Suppression of ocular neovascularization with siRNA targeting VEGF receptor 1. Gene Ther. 2006;13:225–34. doi: 10.1038/sj.gt.3302641. [DOI] [PubMed] [Google Scholar]
- 80.Esau C, Davis S, Murray SF, et al. miR-122 regulation of lipid metabolism revealed by in vivo antisense targeting. Cell Metab. 2006;3:87–98. doi: 10.1016/j.cmet.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 81.Guo C, Sah JF, Beard L, et al. The noncoding RNA, miR-126, suppresses the growth of neoplastic cells by targeting phosphatidylinositol 3-kinase signaling and is frequently lost in colon cancers. Genes Chromosomes Cancer. 2008;47:939–46. doi: 10.1002/gcc.20596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Petrocca F, Visone R, Onelli MR, et al. E2F1-regulated microRNAs impair TGFbeta-dependent cell-cycle arrest and apoptosis in gastric cancer. Cancer Cell. 2008;13:272–86. doi: 10.1016/j.ccr.2008.02.013. [DOI] [PubMed] [Google Scholar]
- 83.Takagi S, Nakajima M, Mohri T, et al. Post-transcriptional regulation of human pregnane X receptor by micro-RNA affects the expression of cytochrome P450 3A4. J Biol Chem. 2008;283:9674–80. doi: 10.1074/jbc.M709382200. [DOI] [PubMed] [Google Scholar]
- 84.Wong QW, Lung RW, Law PT, et al. MicroRNA-223 is commonly repressed in hepatocellular carcinoma and potentiates expression of Stathmin1. Gastroenterology. 2008;135:257–69. doi: 10.1053/j.gastro.2008.04.003. [DOI] [PubMed] [Google Scholar]
- 85.Xia L, Zhang D, Du R, et al. miR-15b and miR-16 modulate multidrug resistance by targeting BCL2 in human gastric cancer cells. Int J Cancer. 2008;123:372–9. doi: 10.1002/ijc.23501. [DOI] [PubMed] [Google Scholar]