Abstract
Inhibitors of topoisomerase I (Top1) that result in stalled Top1 cleavage complexes (Top1cc) are commonly employed against cancer. Combination chemotherapy with DNA repair inhibitors can potentially improve response to these widely used chemotherapeutics. One line of inquiry focuses on inhibitors of tyrosyl-DNA phosphodiesterase 1 (Tdp1), a repair enzyme for Top1cc. Tdp1 catalyzes the hydrolysis of DNA adducts covalently linked to the 3′-phosphate of DNA, including Top1-derived peptides and also 3′-phosphoglycolates. Tdp1 inhibitors should synergize not only with Top1-targeting drugs (camptothecins, indenoisoquinolines), but also with bleomycin, topoisomerase II (Top2) inhibitors (etoposide, doxorubicin) and DNA alkylating agents. Here, we summarize the structural-activity relationship obtained from the reported Tdp1 inhibitors. Better understanding of Top1cc repair in vivo coupled with detailed structural studies on Tdp1-inhibitor interaction will be crucial in guiding the rational design of Tdp1 inhibitors.
1. Introduction
Inhibitors of DNA topoisomerase I (Top1) are established effective anti-cancer chemotherapeutic agents (for reviews see1, 2). Camptothecin (CPT) and its derivatives reversibly bind and trap the Top1-DNA cleavage complex (Top1cc), which is normally a transient intermediate of the Top1 enzymatic reaction3. When replication or transcription machineries subsequently encounter the trapped Top1cc, DNA double-strand breaks (DSBs) are generated and cell death occurs as a consequence of DNA damage (Top1-DNA adducts and DSBs). In addition to the current Top1 inhibitors commercially available for cancer chemotherapy, several promising new derivatives with improved pharmacokinetic properties are currently in clinical trials1, 2, 4.
The repair mechanisms of Top1cc prove to be complex because several pathways are involved (for review see5). One of the pathways involves tyrosyl-DNA phosphodiesterase 1 (Tdp1), which catalyzes hydrolysis of the Top1 tyrosine residue covalently linked to the 3′-phosphate of DNA. The unique enzymatic activity of Tdp1 removes the covalent-linked Top1 from the DNA 3′-end after Top1 has been denatured or proteolysed6, 7. The discovery of a Tdp1 mutation responsible for the rare neurodegenerative disease, spinocerebellar ataxia with axonal neuropathy (SCAN1), further emphasizes the physiological importance of Tdp1 in central nervous system tissues8. While loss of Tdp1 does not appear to cause apparent cell dysfunctions, Tdp1 knockout mice and human cells deficient in Tdp1 or harboring the SCAN1 mutation all show hypersensitivity to CPT9-12. Conversely, human cells overexpressing Tdp1 show reduction in CPT- and etoposide-induced DNA damage13, 14. Therefore, chemotherapy combining Top1- or Top2-targeting drugs with Tdp1 inhibitor could potentially be more effective.
In addition to Tdp1, alternative pathways such as DNA repair (XPF/ERCC1, Mre11, CtIP), homologous recombination (BRCA1, BRCA2, CtIP, Mre11, Rad52) and cell cycle checkpoint signaling (Rad9, BRCA1, BRCA2, p53) are involved in the repair of CPT-induced DNA damage5. When these alternative pathways are inactivated, cells most likely rely more on the Tdp1-dependent pathway for repairing CPT-induced DNA damage15-25. Since many cancer cells are deficient in one or more DNA damage repair pathways, inhibiting Tdp1-dependent pathway have the potential to selectively sensitize cancer cells over normal cells to Top1 poisons2, 26.
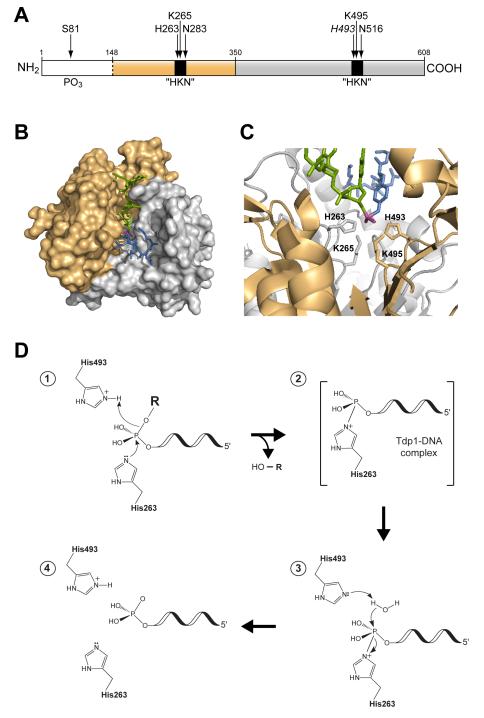
Mechanistic studies of Tdp1 activity have been enormously important in guiding the search for effective Tdp1 inhibitors. As a member of phospholipase D (PLD) superfamily, Tdp1 has two conserved HKD motifs located on different domains but clustering together in tertiary arrangement to form an active site27 (Figure. 1A and C). These two HKD motifs are responsible for two sequential hydrophilic attacks on the phosphodiester bond at the active site (Figure. 1D). Tdp1 can act on DNA substrates linked to a wide-range of 3′-blocking lesions via a phosphodiester linkage28, 29. The relative Km values of Tdp1 for different substrates should be helpful in rational design of Tdp1 inhibitors.
Figure 1.
A) Scheme of the domain structure of human Tdp1. The N-terminal and C-terminal domains correspond to residues 1-350 and 351-608, respectively. Positions of the “HKN” motifs and phosphorylation site (serine 81) are shown in black, with the position of the SCAN1 mutation (H493) shown in italics. B) Crystal structure of the quaternary complex consisting of truncated Tdp1 (Δ1-148), vanadate, a Top1 peptide, and single-strand DNA (PDB:1NOP). Shown as surface models, the N-terminal and C-terminal domains of Tdp1 are in light brown and gray, respectively [see (A)]. Shown in stick structures is the substrate transition-state mimic consisting of single-strand DNA in green, vanadate in magenta, and the peptide in blue. C) The active site residues of Tdp1 are shown in stick structures with the rest of the protein shown in ribbon diagram; the domain colors correspond to those shown in (A) and (B). The substrate transition-state mimic structures are in the same colors as in (B), seen here from the bottom of the binding cleft projecting outwards. For clarity, two loops in the N-terminal domain have been removed from the view. D) Proposed two-step catalytic mechanism of human Tdp1. (1) In the first step, His263 acts as a nucleophile, attacking the phosphorus atom in the phosphodiester bond between the 3′-lesion and the DNA 3′-oxygen. His493 donates a proton to the leaving group (HO-R). (2) A Tdp1-DNA intermediate forms wherein His263 is attached covalently to the 3′-end of the DNA via a phosphoamide bond. (3) A second nucleophilic attack by a water molecule activated by His493 hydrolyzes the phosphohistidine intermediate (4) The Tdp1 active site is regenerated and the 3′-phosphate DNA end is released.
Co-crystal structures of Tdp1 with a transition-state substrate mimic further illustrate the geometry of the Tdp1 active sites and the basis of Tdp1 catalytic activity (Figure. 1B and C)30. Assembled from vanadate, single-strand DNA, and a Top1-derived peptide, the three-part substrate mimic binds in a deep substrate-binding cleft30. The center of the cleft bottom is the active site formed by the two HKD motifs, covalently linked to the vanadate. On one side of the active site is a long and narrow cleft occupied by the single-strand DNA and on the opposite side a larger binding pocket occupied by the Top-derived peptide (Figure. 1B)30. The surface charge/contour of the substrate-binding site, as well as the conformation of the complex in transition-state revealed by the crystal structures should all be informative in the search for Tdp1 inhibitors.
Here we provide a brief summary of the available findings toward identifying specific Tdp1 inhibitors and address the issues that remain to be examined in this line of research.
2. Aminoglycoside Antibiotics and Ribosome Inhibitors
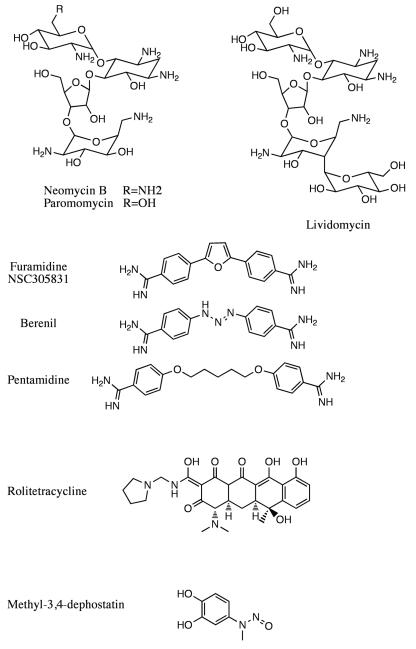
Because Tdp1 belongs to the PLD superfamily, known inhibitors for the other members of the family, such as neomycin (Figure 2), were tested for Tdp1 inhibitory activity. In vitro biochemical assays using recombinant Tdp1 enzyme demonstrate that neomycin inhibits Tdp1 but requires high concentrations (in the low millimolar range)31, 32. The inhibitory potency of neomycin is slightly weaker than that of vanadate, a known general inhibitor of phosphatases. Two other derivatives of neomycin tested, paromomycin and lividomycin (Figure 2), also inhibit Tdp1 activity at slightly higher concentrations31, 32. Given that these aminoglycoside compounds are established inhibitors of bacterial ribosomes, other bacterial ribosomal inhibitors were also screened using the same in vitro biochemical assay. All of the aminoglycoside and non-aminoglycoside antibiotics tested show weak inhibition against recombinant Tdp1 in the millimolar range31, 32. While the inhibitors identified in the initial screen were only weak Tdp1 inhibitors with uncharacterized mechanism, a reliable in vitro assay system was established for validation of other Tdp1 inhibitors.
Figure 2.
Chemical structures of the presented compounds
3. Furamidine
A high-throughput electrochemiluminescent assay was subsequently developed to screen the 2000-compound “diversity set” chemical library of the NCI-Developmental Therapeutics Program. Furamidine, a compound in phase III clinical trials against trypanosomiasis, a common parasitic disease prevalent in Africa, was identified as a positive hit in the screen (Figure 2). In vitro biochemical assays also confirmed that furamidine inhibits Tdp1 at low micromolar concentrations33, 34. Furamidine is known to bind to duplex DNA in the minor groove selectively at AT-rich sites, as well as intercalating between GC base pairs. Since no duplex DNA was present in the screen, the mode of action of furamidine is likely a novel one. Further investigation by surface plasmon resonance (SPR) analysis revealed that furamidine binds not only to DNA duplex, but also to single-stranded DNA, albeit to a lesser degree. Similar analysis also showed that furamidine interacts with Tdp1. These results raised the possibility of an inhibitory mode of action by combined interaction to the DNA substrate and to Tdp1, which is reminiscent of the interfacial inhibition of Top1cc by Top1 inhibitors 3, 35.
Two other diamidine compounds, berenil and pentamidine, are much less effective than furamidine at inhibiting Tdp1 (Figure 2)33, 34. Both compounds share the overall curved structure of furamidine, but substitute the furan ring linker in furamidine with other linking groups (Figure 2). This suggests that the furan linker is important in inhibiting Tdp1 activity, potentially by directly interacting with the DNA or Tdp1 or both, or by stabilizing the overall curvature of the compound.
4. Tetracyclines
A number of tetracycline compounds including rolitetracycline (Figure 2) were also identified as positive hits in two separate high-throughput screens for Tdp1 inhibitors. The preliminary results on tetracyclines showed micromolar inhibitory effect against Tdp136 but lacked apparent structure-activity relationship. Nevertheless, these compounds potentially warrant further investigation.
5. Phosphotyrosine Mimetics
The strongest Tdp1 inhibitors identified to date are Tdp1 phosphotyrosine substrate mimetics. Identified in two separate high through-put screens as positive hits, methyl-3,4-dephostatin37, 38 and the steroid compound NSC8891536, 39 both share similar structural characteristics as the phosphotyrosine substrate (Figure 2 & 3). Methyl-3,4-dephostatin is an aromatic amine derivative with a hydroxyl-substituted benzyl ring, bearing close resemblance to the phosphate group linked to the tyrosyl-moiety in the Tdp1 substrate (Figure 2). NSC88915 and its derivatives contain an aromatic sulfonyl ester group linked to a steroid, simulating the phosphate group covalently linked to the tyrosyl moiety on one side and the DNA oligonucleotide on the other (Figure 3).
Figure 3.
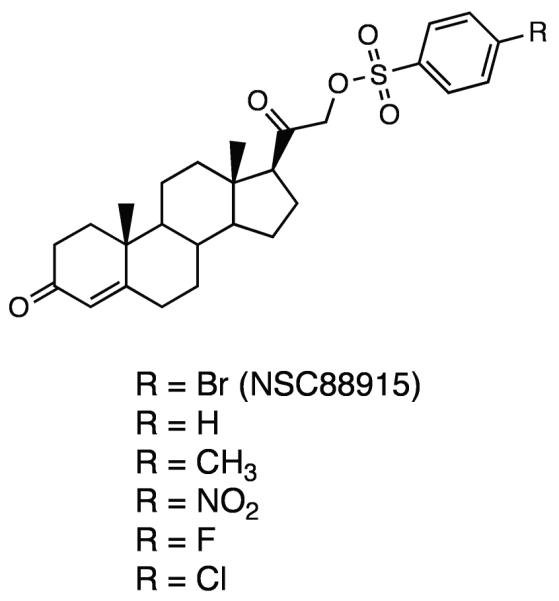
Chemical structures of the steroid derivatives
Close examination of the structure-activity relationship derived from methyl-3,4-dephostatin and NSC88915 and their respective derivatives is informative. For example, the only difference between dephostatin and methyl-3,4-dephostatin is the position of one hydroxyl substitution group on the aromatic ring (Figure 2). However, while methyl-3,4-dephostatin inhibits Tdp1 at sub-micromolar concentrations, dephostatin displays almost no activity in the same concentration range37, 38. Similarly, NSC88915 and its five derivatives only differ by the nature of the substitution at the para position of the aromatic ring (Figure 3). While the difference in activity for these six derivatives are within 10-fold of each other, another derivative lacking the aromatic ring all together lost its potency36, 39. Molecular dynamics of these seven steroid derivatives docking in the Tdp1 active site were simulated. Indeed the calculated free energy of binding for each compound closely correlates to its respective inhibitory activity against Tdp136, 39. All together, these results suggest that the aromatic ring linked to a non-hydrolysable phosphate mimic could potentially be active at inhibiting Tdp1. Furthermore, both the nature and position of the substitution groups could influence the potency of such inhibitors.
6. Expert Opinion
The effort to identify inhibitors of Tdp1 activity has led to development of a few high-throughput screen formats and several promising leads. The current efforts are largely on refining the structure-activity relationship of the existing inhibitors as well as searching for additional lead compounds. The biochemical activity of Tdp1 and various mutants is also an active area of research, which continues to guide the search for inhibitors of Tdp1. Further studies on the mode of actions of these lead inhibitors are warranted.
To better correlate inhibitor efficacies obtained from in vitro and in vivo screens, the expression and activity of Tdp1 in tumor tissues need to be further investigated. For example, non-small cell lung cancer tissues have been reported to show high level of Tdp1 expression and activity40. The relative contribution of the different DNA repair pathways in human cells also needs to be better understood. As discussed before, many cancer cells are deficient in one or more DNA damage response pathways. Thus, it will be important to develop clinical assays that can characterize tumors and identify deficiencies in specific DNA repair pathways. Tumors deficient in cell cycle signaling pathways (Chk2, ATM, p53), DNA repair (XPF, ERCC1, Mre11, Nbs1, CtIP) and/or homologous recombination (CtIP, BRCA1, BRCA2) should be prime candidates for combination chemotherapies with Top1 and Tdp1 inhibitors. Most notable examples might be colon cancers with Mre11 mutations41, 42 and lung cancers with ERCC1 mutations25, 43. It remains to be determined whether BRCA1- or BRCA2-deficient tumors will be selectively sensitized by Tdp1 inhibitors because, in budding yeast, Rad52 is epistatic with Tdp116, 20.
Tdp1 is involved in the repair of other types of lesions besides Top1cc, such as DNA strand breaks with phosphoglycolate adducts at 3′-ends28, 44, 45. The repair of those lesions by Tdp1 has been invoked to account for the sensitivity of Tdp1-deficient cells to bleomycin12. Moreover, the possible involvement of Tdp1 in the repair of Top2-induced DNA damage46 suggests that Tdp1 inhibitors may have value in wide range of combination therapies with DNA-targeting agents including, not only Top1 inhibitors but also bleomycin, Top2 inhibitors and potentially DNA alkylating agents.
The size of Top1cc-derived substrates in vivo remains elusive. While proteasome processing of Top1cc and poly(ADP-ribosylation) are required for the Tdp1-dependent repair pathway25, the extent of proteasome processing in vivo is not known7. The high-throughput screens to date employed a substrate that contains a single-tyrosine amino acid covalently linked to the 3′-phosphate of DNA. It should be noted that Tdp1 processes this particular substrate more efficiently than any substrates containing longer peptides, which are expected to be closer to the physiological Top1cc-derived substrates. The use of such an optimal substrate may underestimate the efficacy of inhibitors against more physiological substrates. Therefore, it is crucial to cross-validate any hits using both in vitro and in vivo systems. In vivo screening taking advantage of the differential sensitivity to Tdp1 inhibitors in Tdp1 knockout vs. wild-type cell lines are ongoing. Such efforts have the added benefits of potentially discovering prodrugs that would not be active in in vitro screens alone.
It is not surprising that the most potent Tdp1 inhibitors to date are non-hydrolysable Tdp1 substrate mimetics. Since the catalytic activity of Tdp1 is targeted towards a specific group of DNA lesions with well-defined structures, substrate mimetics potentially can inhibit Tdp1 with high degree of specificity. Another line of inquiry is to screen for small molecules that would act as interfacial inhibitors by stabilizing and trapping the Tdp1 intermediate covalently linked to DNA3, 47. Co-crystal structures of Tdp1 with lead compounds from either class or other detailed structural studies will provide valuable insights into their protein-drug interactions. Efforts on that front should pay handsome dividend to successful rational design of Tdp1 inhibitors.
Footnotes
Declaration of interest The authors declare no conflict of interest and have received no payment in preparation of this manuscript.
References
- 1.Pommier Y, Leo E, Zhang H, et al. DNA topoisomerases and their poisoning by anticancer and antibacterial drugs. Chem Biol. 2010 May 28;17(5):421–33. doi: 10.1016/j.chembiol.2010.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pommier Y. Topoisomerase I inhibitors: camptothecins and beyond. Nat Rev Cancer. 2006 Oct;6(10):789–802. doi: 10.1038/nrc1977. [DOI] [PubMed] [Google Scholar]
- 3.Marchand C, Antony S, Kohn KW, et al. A novel norindenoisoquinoline structure reveals a common interfacial inhibitor paradigm for ternary trapping of the topoisomerase I-DNA covalent complex. Mol Cancer Ther. 2006 Feb;5(2):287–95. doi: 10.1158/1535-7163.MCT-05-0456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Teicher BA. Next generation topoisomerase I inhibitors: Rationale and biomarker strategies. Biochem Pharmacol. 2008 Mar 15;75(6):1262–71. doi: 10.1016/j.bcp.2007.10.016. [DOI] [PubMed] [Google Scholar]
- **5.Pommier Y, Barcelo JM, Rao VA, et al. Repair of topoisomerase I-mediated DNA damage. Prog Nucleic Acid Res Mol Biol. 2006;81:179–229. doi: 10.1016/S0079-6603(06)81005-6. Comprehensive review on the repair of Top-1 mediated DNA damage
- 6.Debethune L, Kohlhagen G, Grandas A, et al. Processing of nucleopeptides mimicking the topoisomerase I-DNA covalent complex by tyrosyl-DNA phosphodiesterase. Nucleic Acids Res. 2002 Mar 1;30(5):1198–204. doi: 10.1093/nar/30.5.1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Interthal H, Champoux JJ. Effects of DNA and protein size on substrate cleavage by human tyrosyl-DNA phosphodiesterase (TDP1) Biochem J. 2011 Apr 4; doi: 10.1042/BJ20101841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *8.Takashima H, Boerkoel CF, John J, et al. Mutation of TDP1, encoding a topoisomerase I-dependent DNA damage repair enzyme, in spinocerebellar ataxia with axonal neuropathy. Nat Genet. 2002 Oct;32(2):267–72. doi: 10.1038/ng987. Study showing that a mutation in Tdp1 is responsible for the SCAN1 neurodegenerative disease
- 9.El-Khamisy SF, Katyal S, Patel P, et al. Synergistic decrease of DNA single-strand break repair rates in mouse neural cells lacking both Tdp1 and aprataxin. DNA Repair (Amst) 2009 Jun 4;8(6):760–6. doi: 10.1016/j.dnarep.2009.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Das BB, Antony S, Gupta S, et al. Optimal function of the DNA repair enzyme TDP1 requires its phosphorylation by ATM and/or DNA-PK. The EMBO Journal. 2009;28(23):3667–80. doi: 10.1038/emboj.2009.302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Katyal S, el-Khamisy SF, Russell HR, et al. TDP1 facilitates chromosomal single-strand break repair in neurons and is neuroprotective in vivo. EMBO J. 2007 Nov 14;26(22):4720–31. doi: 10.1038/sj.emboj.7601869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hirano R, Interthal H, Huang C, et al. Spinocerebellar ataxia with axonal neuropathy: consequence of a Tdp1 recessive neomorphic mutation? EMBO J. 2007 November 11;26(22):4732–43. doi: 10.1038/sj.emboj.7601885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *13.Barthelmes HU, Habermeyer M, Christensen MO, et al. TDP1 overexpression in human cells counteracts DNA damage mediated by topoisomerases I and II. J Biol Chem. 2004 Dec 31;279(53):55618–25. doi: 10.1074/jbc.M405042200. Study implicating Tdp1 in the repair of Top1 and Top2-mediated DNA lesions.
- 14.Nivens MC, Felder T, Galloway AH, et al. Engineered resistance to camptothecin and antifolates by retroviral coexpression of tyrosyl DNA phosphodiesterase-I and thymidylate synthase. Cancer Chemother Pharmacol. 2004 Feb;53(2):107–15. doi: 10.1007/s00280-003-0717-6. [DOI] [PubMed] [Google Scholar]
- *15.Pouliot JJ, Yao KC, Robertson CA, et al. Yeast gene for a Tyr-DNA phosphodiesterase that repairs topo I covalent complexes. Science. 1999;286:552–5. doi: 10.1126/science.286.5439.552. Discovery of the TDP1 gene.
- 16.Pouliot JJ, Robertson CA, Nash HA. Pathways for repair of topoisomerase I covalent complexes in Saccharomyces cerevisiae. Genes Cells. 2001;6(8):677–87. doi: 10.1046/j.1365-2443.2001.00452.x. [DOI] [PubMed] [Google Scholar]
- *17.Liu C, Pouliot JJ, Nash HA. Repair of topoisomerase I covalent complexes in the absence of the tyrosyl-DNA phosphodiesterase Tdp1. Proc Natl Acad Sci U S A. 2002 Nov 12;99(23):14970–5. doi: 10.1073/pnas.182557199. Genetic studies on alternative DNA repair pathways.
- 18.Eng WK, Faucette L, Johnson RK, et al. Evidence that DNA topoisomerase I is necessary for the cytotoxic effects of camptothecin. Mol Pharmacol. 1988;34:755–60. [PubMed] [Google Scholar]
- 19.Vance JR, Wilson TE. Repair of DNA strand breaks by the overlapping functions of lesion-specific and non-lesion-specific DNA 3′ phosphatases. Mol Cell Biol. 2001 Nov;21(21):7191–8. doi: 10.1128/MCB.21.21.7191-7198.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vance JR, Wilson TE. Yeast Tdp1 and Rad1-Rad10 function as redundant pathways for repairing Top1 replicative damage. Proc Natl Acad Sci U S A. 2002 Oct 15;99(21):13669–74. doi: 10.1073/pnas.202242599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Deng C, Brown JA, You D, et al. Multiple Endonucleases Function to Repair Covalent Topoisomerase I Complexes in Saccharomyces cerevisiae. Genetics. 2005 Apr 16;170:591–600. doi: 10.1534/genetics.104.028795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sartori AA, Lukas C, Coates J, et al. Human CtIP promotes DNA end resection. Nature. 2007 Nov 22;450(7169):509–14. doi: 10.1038/nature06337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nakamura K, Kogame T, Oshiumi H, et al. Collaborative action of Brca1 and CtIP in elimination of covalent modifications from double-strand breaks to facilitate subsequent break repair. PLoS Genet. 2010 Jan;6(1):e1000828. doi: 10.1371/journal.pgen.1000828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hartsuiker E, Neale MJ, Carr AM. Distinct Requirements for the Rad32(Mre11) Nuclease and Ctp1(CtIP) in the Removal of Covalently Bound Topoisomerase I and II from DNA. Mol Cell. 2009 Jan 16;33(1):117–23. doi: 10.1016/j.molcel.2008.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang YW, Regairaz M, Seiler JA, et al. Poly(ADP-ribose) polymerase and XPF-ERCC1 participate in distinct pathways for the repair of topoisomerase I-induced DNA damage in mammalian cells. Nucleic Acids Res. 2011 Jan 11; doi: 10.1093/nar/gkq1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **26.Dexheimer TS, Antony S, Marchand C, et al. Tyrosyl-DNA phosphodiesterase as a target for anticancer therapy. Anticancer Agents Med Chem. 2008 May;8(4):381–9. doi: 10.2174/187152008784220357. First review on Tdp1 as a target for anticancer therapy.
- 27.Davies DR, Interthal H, Champoux JJ, et al. Insights into substrate binding and catalytic mechanism of human tyrosyl-DNA phosphodiesterase (Tdp1) from vanadate and tungstate-inhibited structures. J Mol Biol. 2002 December 12;324(5):917–32. doi: 10.1016/s0022-2836(02)01154-3. [DOI] [PubMed] [Google Scholar]
- 28.Interthal H, Chen HJ, Champoux JJ. Human Tdp1 cleaves a broad spectrum of substrates, including phosphoamide linkages. J Biol Chem. 2005 October 10;280(43):36518–28. doi: 10.1074/jbc.M508898200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dexheimer TS, Stephen AG, Fivash MJ, et al. The DNA binding and 3′-end preferential activity of human tyrosyl-DNA phosphodiesterase. Nucleic Acids Res. 2010 April;38(7):2444–52. doi: 10.1093/nar/gkp1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *30.Davies DR, Interthal H, Champoux JJ, et al. Crystal structure of a transition state mimic for Tdp1 assembled from vanadate, DNA, and a topoisomerase I-derived peptide. Chem Biol. 2003 February;10(2):139–47. doi: 10.1016/s1074-5521(03)00021-8. Co-crystal structure of Tdp1-substrate complex
- *31.Liao Z, Thibaut L, Jobson A, et al. Inhibition of human tyrosyl-DNA phosphodiesterase by aminoglycoside antibiotics and ribosome inhibitors. Mol Pharmacol. 2006 Jul;70(1):366–72. doi: 10.1124/mol.105.021865. First report on Tdp1 inhibitors.
- 32.Pommier Y, inventor The Government of the United States of America, assignee Aminoglycosides and ribosome inhibitors as inhibitors of tyrosyl-DNA-phosphodiesterase. 60/661,306 USA patent. 2005
- 33.Antony S, Marchand C, Stephen AG, et al. Novel high-throughput electrochemiluminescent assay for identification of human tyrosyl-DNA phosphodiesterase (Tdp1) inhibitors and characterization of furamidine (NSC 305831) as an inhibitor of Tdp1. Nucleic Acids Res. 2007;35(13):4474–84. doi: 10.1093/nar/gkm463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pommier Y, Marchand C, Thibaut L, inventors; The Government of the United States of America, assignee Diamidine derivatives as inhibitors of human tyrosyl-DNA-phosphodiesterase (Tdp1) 60/786,604 USA patent. 2006
- 35.Pommier Y, Cherfils J. Interfacial protein inhibition: a nature’s paradigm for drug discovery. Trends Pharmacol Sci. 2005;28:136–45. doi: 10.1016/j.tips.2005.01.008. [DOI] [PubMed] [Google Scholar]
- 36.Pommier Y, Marchand C, Antony S, et al. inventors; The Government of the United States of America, assignee Steroid derivatives as inhibitors of human tyrosyl-DNA phosphodiesterase (Tdp1) 60/921,980 USA patent. 2007
- *37.Marchand C, Lea WA, Jadhav A, et al. Identification of phosphotyrosine mimetic inhibitors of human tyrosyl-DNA phosphodiesterase I by a novel AlphaScreen high-throughput assay. Mol Cancer Ther. 2009 Jan;8(1):240–8. doi: 10.1158/1535-7163.MCT-08-0878. Report on the first robotic high througput screen.
- 38.Pommier Y, Marchand C, Dexheimer T, et al. inventors; The Government of the United States of America, assignee Protein-Tyrosine Phosphatase Inhibitors as Inhibitors of TDP1 and Methods of Treating Disorders. 61/042,706 USA patent. 2008
- 39.Dexheimer TS, Gediya LK, Stephen AG, et al. 4-Pregnen-21-ol-3,20-dione-21-(4-bromobenzenesulfonate) (NSC 88915) and related novel steroid derivatives as tyrosyl-DNA phosphodiesterase (Tdp1) inhibitors. J Med Chem. 2009 Nov 26;52(22):7122–31. doi: 10.1021/jm901061s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu C, Zhou S, Begum S, et al. Increased expression and activity of repair genes TDP1 and XPF in non-small cell lung cancer. Lung Cancer. 2007 Mar;55(3):303–11. doi: 10.1016/j.lungcan.2006.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Giannini G, Rinaldi C, Ristori E, et al. Mutations of an intronic repeat induce impaired MRE11 expression in primary human cancer with microsatellite instability. Oncogene. 2004 Apr 8;23(15):2640–7. doi: 10.1038/sj.onc.1207409. [DOI] [PubMed] [Google Scholar]
- 42.Takemura H, Rao VA, Sordet O, et al. Defective Mre11-dependent Activation of Chk2 by Ataxia Telangiectasia Mutated in Colorectal Carcinoma Cells in Response to Replication-dependent DNA Double Strand Breaks. J Biol Chem. 2006 Oct 13;281(41):30814–23. doi: 10.1074/jbc.M603747200. [DOI] [PubMed] [Google Scholar]
- 43.Knez L, Sodja E, Kern I, et al. Predictive value of multidrug resistance proteins, topoisomerases II and ERCC1 in small cell lung cancer: A systematic review. Lung Cancer. 2011 Jun;72(3):271–9. doi: 10.1016/j.lungcan.2011.02.014. [DOI] [PubMed] [Google Scholar]
- 44.Zhou T, Akopiants K, Mohapatra S, et al. Tyrosyl-DNA phosphodiesterase and the repair of 3′-phosphoglycolate-terminated DNA double-strand breaks. DNA Repair (Amst) 2009 Jun 6;8(8):901–11. doi: 10.1016/j.dnarep.2009.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Das BB, Dexheimer TS, Maddali K, et al. Role of tyrosyl-DNA phosphodiesterase (TDP1) in mitochondria. Proc Natl Acad Sci U S A. 2010 Nov 16;107(46):19790–5. doi: 10.1073/pnas.1009814107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *46.Nitiss KC, Malik M, He X, et al. Tyrosyl-DNA phosphodiesterase (Tdp1) participates in the repair of Top2-mediated DNA damage. Proc Natl Acad Sci U S A. 2006 Jun 13;103(24):8953–8. doi: 10.1073/pnas.0603455103. Study implicating Tdp1 in the repair of Top2-mediated DNA lesions.
- 47.Pommier Y, Cherfils J. Interfacial inhibition of macromolecular interactions: nature’s paradigm for drug discovery. Trends Pharmacol Sci. 2005 Mar;26(3):138–45. doi: 10.1016/j.tips.2005.01.008. [DOI] [PubMed] [Google Scholar]