Abstract
The extracellular matrix N-glycoprotein periostin is thought to enhance tumor invasion. In this study, the expression patterns of periostin and its splice isoforms were investigated in renal cell carcinoma (RCC). Periostin mRNA expression patterns were characterized in 30 fresh-frozen RCCs in normal fetal and adult renal tissues by both isoform-specific and nonspecific RT-PCR and by gene expression array analysis. Its protein expression was analyzed by immunohistochemistry, using tissue microarrays with tissue from 1007 RCC patients. Periostin mRNA in RCC was increased, as observed in both RT-PCR and gene microarray analyses, with significantly higher expression in the clear cell than in the papillary subtype. Four of eight periostin isoforms, identified in fetal kidney by direct sequencing, have not been described to date. Three isoforms could be detected in both RCC and matched non-neoplastic tissue, and one of them was expressed more frequently in RCC. Periostin protein was detected in both mesenchymal cells of the tumor stroma and epithelial tumor cells. Greater amounts of periostin in tumor epithelia correlated with the presence of sarcomatoid differentiation, higher tumor stage, lymph node metastases, and poor overall survival in the clear cell subtype. In conclusion, periostin expression in tumor epithelia may contribute to sarcomatoid differentiation and more aggressive behavior of RCC. The presence of a tumor-associated periostin isoform suggests splice-specific regulation in RCC tissue.
Renal cell carcinoma (RCC) is a heterogeneous epithelial tumor with frequent evidence of metastasis at the time of diagnosis. The major histologic subtypes are clear cell, papillary, and chromophobe RCC. Sarcomatoid differentiation is not its own entity and can be observed in all of these subtypes, where its presence is associated with a poor prognosis.1,2
Periostin, also termed osteoblast-specific factor 2, is a secreted matrix N-glycoprotein of 93 kDa.3 The N-terminal region contains four fasciclin-like domains as well as several glycosylation sites. This protein originally was identified in MC3T3-E1 osteoblast-like cells, where it promotes integrin-dependent cell adhesion and motility. It shares homology with the insect cell adhesion molecule fasciclin I, with human β IgH3, and is induced by transforming growth factor-β and bone morphogenic protein-2.3–5
Periostin is thought to promote epithelial-mesenchymal transition (EMT) of carcinoma cells by targeting the cross-talk between epidermal growth factor receptor and integrins at the plasma membrane with consecutive activation of the Akt/protein kinase B pathway.6 Through its binding to the integrins αvβ3, αvβ5, and αvβ6 periostin promotes cellular survival, angiogenesis, and resistance to hypoxia-induced cell death.7–11 Consequently, this protein is considered to be a prognostic marker. Furthermore, periostin was shown to be accessible by the blood stream after ex vivo perfusion and biotinylation of human kidney with tumor and therefore a potential target for stroma-directed tumor therapy.12,13
To date, six different splice isoforms of periostin have been reported, but only four of them (including the full-length variant) have been sequenced and annotated.13,14 Full-length and isoforms differ in their C-terminal sequences, and the N-terminus is responsible for integrin binding.3 All known isoforms are between 83 and 93 kDa in mass, and vary between 751 and 836 amino acids. The four known splice variants are characterized by an individual presence or absence of cassette exons 17 to 21. They are called isoform 1 (full-length variant with all exons), isoform 2 (absent exons: 17 and 18), isoform 3 (absent exons: 17 and 21), and isoform 4 (absent exons: 17, 18, and 21) with the following UniProtKB/Swiss-Prot accession numbers: D13666, D13665, AY140646, and AY918092, respectively.
Periostin is expressed mainly in collagen-rich and in fetal tissues such as periosteum, periodontal ligament, placenta, cardiac valves, and vasculature, and is up-regulated by mechanical stress during tissue generation and repair.3,15–21 Strong protein expression has been observed in lung, breast, ovarian, pancreatic, gastric, thyroid, colon, head and neck, prostate, liver, and other carcinomas, as well as in neuroblastoma, and was related to increased malignant behavior of those tumors.7–9,11,21–31 Consequently, the aim of this study was to characterize periostin expression patterns and to analyze periostin isoforms in RCC and in normal renal tissue.
Materials and Methods
Patients
First, we investigated a cohort of 30 surgically resected and fresh-frozen RCC tissue samples for periostin mRNA expression. Tumors were classified according to the 2004 World Health Organization classification of kidney tumors32 and the 2010 TNM edition (http://www.uicc.org/tnm). There were 23 clear-cell RCC (ccRCC) and 7 papillary RCC. The age range was 43 to 85 years and the tumor size range was 1.7 to 12.0 cm. Fourteen patients were stage pT1, 7 patients were stage pT2, and 9 patients were stage pT3/4. There were 4 Fuhrman grade 1, 15 Fuhrman grade 2, and 11 Fuhrman grade 3 cases. Frozen matched non-neoplastic tissue was available for 26 of these 30 cases. Selection criteria for the frozen samples were sufficient tissue quantity available and preserved morphology. All tumors were entirely reviewed on H&E sections.
Second, analysis of periostin gene expression levels was performed on an Affymetrix microarray (Affymetrix, Santa Clara, CA) of 102 RCCs. There were 6 normal renal tissues as control, 81 ccRCC, and 21 papillary RCC. Thirty-seven cases were diagnosed as pT1, 29 cases were diagnosed as pT2, and 36 cases were diagnosed as pT3. Ten cases were diagnosed as Fuhrman grade 1, 42 cases were diagnosed as Fuhrman grade 2, 44 cases were diagnosed as Fuhrman grade 3, and 6 cases were diagnosed as Fuhrman grade 4. These 102 RCCs spanned the years 1993 to 2007 at the University Hospital Zurich, and were reviewed on H&E by H.M. according to the World Health Organization classification. Total RNA from this cohort was reverse transcribed, labeled, and hybridized on HG-U133A high-throughput arrays (Affymetrix). The gene chips were read in a 96-format using an Affymetrix-HT scanner at the Broad Institute of the Massachusetts Institute of Technology and Harvard Medical School (Cambridge, MA). The Affymetrix raw data were normalized through GeneChip Robust Multi-array Average.33
Third, periostin protein expression was analyzed in a cohort of 1007 primary RCCs using TMA technology, as previously described.34 Tumors were collected at the University Hospital Zurich, the Kantonsspital St. Gallen, and the University Hospital Basel, all in Switzerland. All cases were reviewed by one pathologist (H.M.). The presence or absence of sarcomatoid differentiation was defined in all renal tumor subtypes according to previous publications.2,35 Information about sarcomatoid differentiation, lymphocyte infiltration, and necrosis was available only for the Zurich TMA. The median follow-up time was 39 months (range, 0 to 229 months). Survival time was available for 469 ccRCC patients. The study was approved by the local commission of ethics (reference number: StV 38-2005).
RNA Extraction and cDNA Synthesis
Total RNA from the 30 patients of the first cohort was isolated from 10 to 15 mg of fresh-frozen tissue using the RNeasy mini kit (Qiagen, Hombrechtikon, Switzerland). RT-PCR was performed using the high-capacity cDNA reverse-transcription kit (Applied Biosystems, Rotkreuz, Switzerland) following the manufacturer's protocol. RNA yield and purity were analyzed by NanoDrop 1000 (Thermo Scientific, Wohlen, Switzerland). RNA integrity was checked by capillary electrophoresis using a 2100 Agilent bioanalyzer (Agilent Technologies, Basel, Switzerland).
Quantitative RT-PCR Analysis
Quantitative RT-PCR was performed on an ABI 7900HT Fast RealTime PCR apparatus (Applied Biosystems) equipped with SDS 2.3 and RQ manager software (Applied Biosystems). cDNA, synthesized as described earlier,36 was used together with the following two PCR systems for gene expression assays (Applied Biosystems): Hs01566748 for the human periostin gene (GenBank accession no.: NM_006475) and Hs99999904 for human peptidylprolyl isomerase A (GenBank accession no.: NM_021130.3). The cycle threshold values for periostin were normalized first against peptidylprolyl isomerase A36 as endogenous control and second against normalized matched normal kidney tissue.
Semiquantitative RT-PCR Analysis
For semiquantitative RT-PCR we used primer pairs described by Castronovo et al13 that detect all isoform transcripts according to the Gene Bank accession number NM_006475 for the human periostin gene. The sequences are as follows: Per-F2, 5′-GCTGATAGATCTTATCCAGCAGACACACCTGTTG-3′; and Per-R2,' 5′-CGATTCAAGCTTAGTGATGGTGATGGTGATGCTGAGAACGACCTTCCCTTAATC-3′. Control glyceraldehyde-3-phosphate dehydrogenase primers are as follows: glyceraldehyde-3-phosphate dehydrogenase-F, 5′-TGGAAGGACTCATGACCACA-3′; and glyceraldehyde-3-phosphate dehydrogenase-R, 5′-TGCTGTAGCCAAATTCGTTG-3′.
Periostin PCR was performed with 35 cycles consisting of denaturation at 94°C for 30 seconds, annealing at 55°C for 30 seconds, and extension at 72°C for 50 seconds, followed by a final extension of 7 minutes. Glyceraldehyde-3-phosphate dehydrogenase PCR was performed with annealing at 50°C. The PCR products were separated by electrophoresis on 1.5% agarose gels.
Cloning and Sequencing of the Periostin Isoform Cassette (Exons 14 to 23)
Periostin isoforms were amplified by isoform-specific RT-PCR from cDNA libraries generated from fresh-frozen RCCs and normal fetal and adult kidney tissue, using the same primers as described earlier. The total RNA from human fetal kidney was purchased from Stratagene (La Jolla, CA). PCR products were cloned using the pGEM easy vector system (Promega, Wallisellen, Switzerland). Isolated clones were sequenced and analyzed.
Western Blot
The clear-cell RCC cell line 786−0, the human embryonic kidney cell line HEK293, the lung adenocarcinoma cell line A549, and the breast cancer cell line MCF7 were purchased and grown under conditions recommended by the American Type Culture Collection. The clear-cell RCC cell line RCC4 and the lung adenocarcinoma cell lines H23 and HCC827 were provided by Professor Wilhelm Krek (Department of Biology, ETH Zurich, Zurich, Switzerland) and by Dr. Roman Thomas (Max-Planck Institute for Neurological Research, Cologne, Germany), respectively.
Western blot of total cell lysates was performed as previously described,37 in reducing conditions using 10% (v/v) β-mercaptoethanol. Primary antibodies used were polyclonal anti-human periostin (dilution 1:5000; Biovendor Laboratory Medicine, Inc., Modrice, Czech Republique) and anti–β-actin (1:5000; Chemicon International, Temecula, CA). horseradish-peroxidase–conjugated anti-rabbit (1:2000; Millipore, Billerica, MA) and horseradish-peroxidase–conjugated anti-mouse (ab672, 1:2000; Abcam, Cambridge, UK) served as secondary antibodies. One hundred nanograms of purified recombinant periostin (R&D, Minneapolis, MN) served as positive control.
Immunohistochemistry
Both whole sections from the 30 RCC patients and TMA sections, 3-μm thick, were cut from representative paraffin and TMA blocks and mounted on superfrost slides (Menzel Gläser, Braunschweig, Germany). Periostin immunohistochemistry (IHC) was performed on the automated IHC platform BondMax (Leica Microsystems, Heerbrugg, Switzerland). Pretreatment was performed by heat-induced epitope retrieval, using the H2-Buffer (Leica Microsystems) for 30 minutes. The primary antibody anti-human periostin (Biovendor Laboratory Medicine, Inc.) was diluted 1:500. Immunoreactivity was detected with a corresponding secondary antibody included in the Refine-DAB (Leica Microsystems) detection kit. Scoring of protein expression was performed as recently described.22,25,31 The intensity in the tumor cells was scored as negative (0), weak (1+), moderate (2+), or strong (3+) by P.S. and G.K. After independent scoring, agreement was achieved between both observers. Periostin expression also was evaluated in the intratumoral stroma where it refers to the cytoplasm of stromal cells as well as to the stromal extracellular matrix. Elastica van Gieson and Alcian blue–periodic acid–Schiff stainings were manufactured to score the degree of collagen and elastin fibrillogenesis (Elastica van Gieson), as well as the amount of acid and neutral mucopolysaccharides (Alcian blue–periodic acid–Schiff) in the extracellular stromal matrix.
Statistics
Statistical analysis was performed using SPSS/PASW 18.0.0 software (IBM, Zurich, Switzerland) and Kendall's τ-b correlation tests for the 30-RCC cohort as well as χ2 association tests for the TMAs. Univariate survival analyses were performed by the Kaplan-Meier method and log-rank tests, using data dichotomized into a “periostin negative” (score = 0) and a “periostin positive” (score = 1 to 3) group. To evaluate the level of lymphocyte infiltration, each TMA core was analyzed for lymphocytes detectable in H&E staining. For statistical analysis, the level of lymphocyte infiltration was considered low if only single lymphocytes (<5) per punch were detected, and high when more lymphocytes were seen.
For the Affymetrix gene expression microarray, Significance Analysis of Microarrays was used to detect differential expression of the periostin gene between the two subtypes of papillary RCC and ccRCC.38
Statistical significance was assumed if the P value or the q-value was <0.05.
Results
Periostin mRNA Expression in RCC
The relative quantification of periostin mRNA levels was performed in the first cohort by TaqMan quantitative RT-PCR assay using a probe that binds a conserved region of the mRNA (boundary between exons 2 to 3), thus recognizing all known isoforms. The cycle threshold values were double normalized against the endogenous control peptidylprolyl isomerase A and against the matched normal control tissue. There was an average fivefold up-regulation of periostin mRNA in 15 (60%) of 26 frozen RCCs, whereas in the other 10 cases it was down-regulated with an average of 0.3-fold (Figure 1A). Periostin mRNA levels were higher in the clear-cell subtype compared with papillary RCC (P = 0.003).
Figure 1.
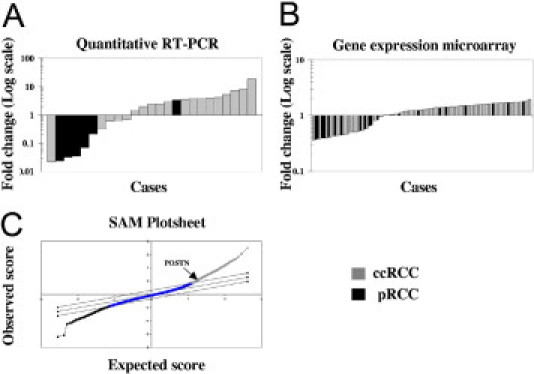
Periostin mRNA levels in RCC patients. A: Relative expression levels obtained from the quantitative RT-PCR analysis. B: Relative expression levels obtained from the Affymetrix microarray analysis. C: Significance Analysis of Microarrays (SAM) plotsheet of ccRCC versus papillary RCC (pRCC).
Gene expression levels of the periostin gene (POSTN_human) then were analyzed in the second cohort of 102 RCC patients using data from an Affymetrix gene expression microarray (Figure 1B). The data were processed using Significance Analysis of Microarrays to calculate differences in gene expression between ccRCC and papillary RCC (Figure 1C). Normalizing against normal renal tissue, 70% of the RCCs showed a 1.5-fold up-regulation of periostin mRNA, whereas the other 30% had a 0.5-fold decrease in the periostin mRNA levels. Higher periostin mRNA levels again were associated with the ccRCC subtype compared with papillary RCC (q = 0.02).
Isoform-Specific RT-PCR and Sequencing of the Periostin Isoform Cassette
To screen for periostin isoforms, total mRNA of the first cohort was reverse-transcribed. Because all described isoforms showed splice alterations in the C-terminus, we chose a PCR strategy covering the whole isoform cassette range from exon 14 to exon 23. Although we detected only five bands by agarose gel electrophoresis when analyzing the isoforms in fetal kidney (Figure 2a), sequence analysis from exons 14 to 23 revealed the presence of eight isoforms (Table 1). Four of them have been described previously and correspond to the accession numbers D13666, D13665, AY140646, and AY918092 in the UniProtKB/Swiss-Prot database (isoforms 1 to 4). The other 4 (isoforms 5 to 8) are new, with their missing exons presented in Table 1. The full-length periostin variant (isoform 1) as well as the isoforms 2, 5, 6, and 7 were detected only in fetal kidney but neither in RCC tissue nor in the corresponding normal control tissue. The other three isoforms were detected in both RCC and corresponding normal renal tissue (isoforms 3, 4, and 8) (Table 1). Two of these isoforms (3 and 4) have been described previously and correspond to variants AY140646 and AY918092 (Figure 2B). The third isoform (isoform 8) is an as yet unknown isoform of periostin in which the exons 17, 18, 19, and 21 are missing (Figure 2B).
Figure 2.
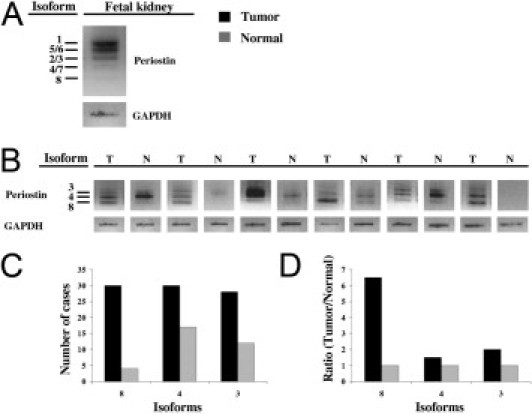
Periostin isoform expression in kidney tissue after semiquantitative PCR. Agarose gel bands from fetal kidney (A) and from RCC with matched normal kidney tissue (B). C: Isoform frequencies in RCC and matched normal tissue. D: Ratio between RCC and matched normal tissue for each isoform detected in this tissue. GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
Table 1.
Summary of Periostin Isoforms Detected in RCC and Fetal and Adult Normal Kidney
| Isoform | ID EMBL EBI | dbEST GenBank | Transcript (bp) | Amplicon (bp) | Peptide (aa) | Mass (kd) | Exons missing | Fetal | RCC | Normal |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | D13666 | 3213 | 624 | 836 | 93,314 | None | + | − | − | |
| 2 | D13665 | 3077 | 453 | 779 | 87,021 | 17,18 | + | − | − | |
| 3 | AY140646 | 2360 | 459 | 781 | 87,254 | 17,21 | + | + | + | |
| 4 | AY918092 | 2958 | 369 | 751 | 83,850 | 17,18,21 | + | + | + | |
| 5 | − | JG969042 | − | 540 | − | − | 21 | + | − | − |
| 6 | − | JG969043 | − | 534 | − | − | 18 | + | − | − |
| 7 | − | JG969044 | − | 363 | − | − | 17,18,19 | + | − | − |
| 8 | − | JG969045 | − | 279 | − | − | 17,18,19,21 | + | + | + |
In all patients of the first cohort, the frequency of isoforms 3 and 4 was similar in tumor and matched non-neoplastic tissue (Figure 2C). In contrast, isoform 8 was detected more frequently in tumor than in the corresponding normal tissue (ratio, 6.5:1; Figure 2, C and D). In four cases periostin isoform 8 expression was detectable in both normal and tumor tissue.
Periostin Protein Expression in Cell Lines
Western blot analysis of protein lysates using different periostin-positive cell lines showed the specificity of the polyclonal rabbit anti-human periostin antibody used for the immunohistochemical stainings. In fact, this antibody detected a band of about 93 kDa when using purified recombinant periostin and a band with a similar molecular weight from the cell lysates (Figure 3).
Figure 3.
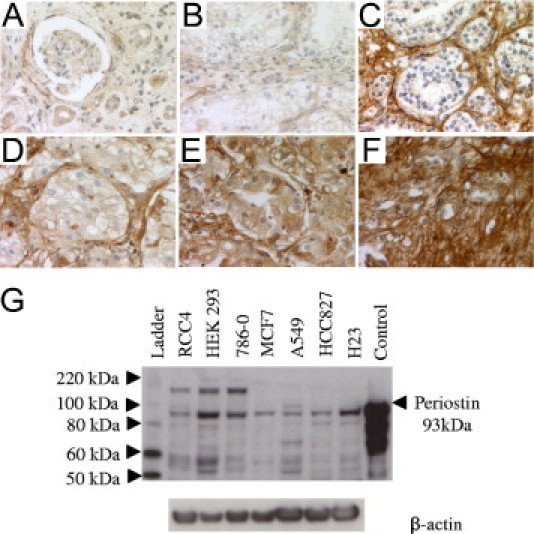
Periostin protein expression in malignant and normal kidney tissue. Weak periostin expression in normal renal tissue with renal tubules and glomeruli (A); ccRCC tissue with a negative epithelial periostin expression and a weak stromal expression (B); ccRCC tissue with a negative (C), weak (D), moderate (E), and strong (F) epithelial periostin expression and a strong stromal expression (C–F). Original magnifications of large sections, ×400 (A–F). G: Periostin immunoblot of different cell lines and purified recombinant periostin protein.
Periostin Protein Expression in RCC
The periostin immunoreactivity scores measured by IHC are presented in Table 2. In the first cohort of 30 RCC samples, protein expression was detected in the cytoplasm of tumor epithelia (indicated as “periostin tumor”), and in the cytoplasm of stromal fibroblasts and in the extracellular matrix (indicated together as “periostin stroma”). All 30 RCC cases were periostin stroma positive (any score of 1 to 3). The majority of the tumors (25 of 30) also were periostin tumor positive (any score of 1 to 3). Protein expression in the tumor epithelia significantly correlated with higher tumor grade (P = 0.03). Furthermore, stromal expression correlated with higher levels of collagen (P = 0.005) and acid mucopolysaccharides (P < 0.001), indicating that periostin is co-expressed together with collagen and acid mucopolysaccharides in the desmoplastic extracellular matrix. Representative stainings are depicted in Figure 3. The periostin protein was observed on some sections in which normal tissue was present next to the tumor. The epithelial cells of renal tubules had a weak to moderate periostin staining, whereas the surrounding stroma and the renal glomeruli were negative to weak.
Table 2.
Protein Expression Intensity Scores of Periostin, Collagen, Elastin, and Acid Mucopolysaccharides in the 30 RCCs
| Periostin |
Stroma |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Tumor |
Stroma |
Collagen |
Elastin |
Mucopol⁎ |
||||||
| Score | n | % | n | % | n | % | n | % | n | % |
| 0 | 5 | 16.7 | 0 | 0.0 | 0 | 0.0 | 12 | 40.0 | 1 | 3.3 |
| 1 | 21 | 70.0 | 1 | 3.3 | 8 | 26.7 | 10 | 33.3 | 14 | 46.7 |
| 2 | 3 | 10.0 | 10 | 33.3 | 8 | 26.7 | 4 | 13.3 | 5 | 16.7 |
| 3 | 1 | 3.3 | 19 | 63.3 | 14 | 46.7 | 4 | 13.3 | 10 | 33.3 |
| Total | 30 | 100.0 | 30 | 100.0 | 30 | 100.0 | 30 | 100.0 | 30 | 100.0 |
Alcian blue–positive acid mucopolysaccharides.
Finally, TMAs consisting of 1007 renal tumors and normal control kidney tissue were analyzed. Immunohistochemical analysis revealed periostin expression in the extracellular matrix in the majority of cases. This stromal expression was significantly higher in clear-cell than in papillary and chromophobe RCC (P < 0.001), and correlated with lower lymphocyte infiltration (P = 0.007) (see Supplemental Table S1 available at http://ajp.amjpathol.org) and lower periostin in tumor epithelia (P = 0.003), but no correlation was found with survival.
Therefore, we focused on periostin expression in tumor epithelia in the following analyses. Cytosolic protein expression correlated with the tumor type (P < 0.001). Among clear-cell RCC, high periostin tumor correlated with higher differentiation grade (P < 0.001), higher pT-category (P < 0.001), presence of lymph node metastases (P < 0.05), sarcomatoid differentiation (P < 0.001), presence of tumor necrosis (P < 0.001), and higher intratumoral lymphocyte infiltration (P < 0.001) (Table 3). Both high periostin in tumor epithelia and high lymphocyte infiltration correlated with decreased overall survival (P = 0.009 and P = 0.002, Figure 4, A and B, respectively).
Table 3.
Epithelial Periostin Expression in RCC and Clinicopathologic Parameters
| Total cases |
Periostin negative |
Periostin positive |
P values | ||||
|---|---|---|---|---|---|---|---|
| n | (%) | n | (%) | n | (%) | ||
| Subtype | |||||||
| ccRCC | 748 | 82.4 | 255 | 34 | 493 | 66 | <0.001 |
| pRCC | 112 | 12.3 | 9 | 8 | 103 | 92 | |
| chRCC | 48 | 5.3 | 1 | 2 | 47 | 98 | |
| Grade⁎ | |||||||
| 1 | 22 | 3.0 | 12 | 54 | 10 | 46 | <0.001 |
| 2 | 220 | 29.6 | 100 | 45 | 120 | 55 | |
| 3 | 317 | 42.6 | 102 | 32 | 215 | 68 | |
| 4 | 165 | 22.2 | 30 | 18 | 135 | 82 | |
| pT⁎ | |||||||
| 1 to 2 | 355 | 48.2 | 145 | 41 | 210 | 59 | <0.001 |
| 3 to 4 | 381 | 51.8 | 105 | 28 | 276 | 72 | |
| pN⁎ | |||||||
| 0 | 231 | 80.8 | 81 | 35 | 150 | 65 | 0.024 |
| 1 | 33 | 11.5 | 6 | 18 | 27 | 82 | |
| 2 | 22 | 7.7 | 4 | 18 | 18 | 82 | |
| Sarcomatoid differentiation⁎† | |||||||
| 0 | 179 | 72.5 | 90 | 50 | 89 | 50 | <0.001 |
| 1 | 68 | 27.5 | 12 | 18 | 56 | 82 | |
| Necrosis⁎† | |||||||
| 0 | 271 | 70.2 | 122 | 45 | 149 | 55 | <0.001 |
| 1 | 115 | 29.8 | 26 | 23 | 89 | 77 | |
| Lymphocyte infiltration⁎† | |||||||
| 0 | 45 | 18.1 | 27 | 60 | 18 | 40 | <0.001 |
| 1 | 127 | 51.2 | 57 | 45 | 70 | 55 | |
| 2 | 76 | 30.6 | 18 | 24 | 58 | 76 | |
ccRCC only.
Zurich TMA only.
Figure 4.
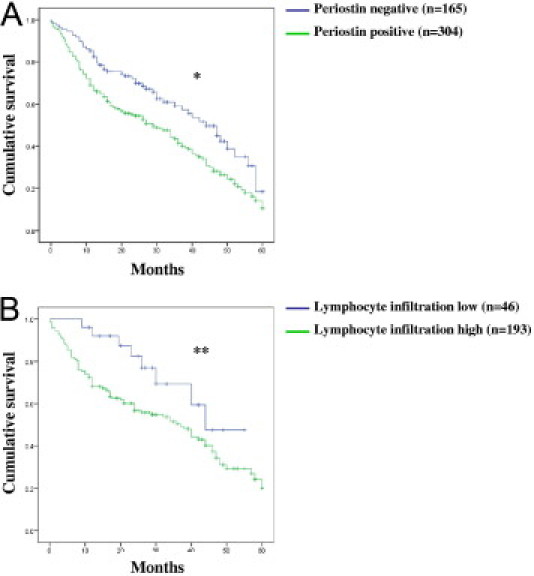
Cumulative univariate 5-year overall survival analysis. Kaplan-Meier curve (log-rank test) for periostin expression in the cytoplasm of tumor epithelial cells (*P = 0.009) (A) and for lymphocyte infiltration (**P = 0.002) (B). The median follow-up time was 39 months (range, 0 to 229 months). Periostin negative versus positive. Lymphocyte infiltration low versus high (for definition see text).
Discussion
In this study we provide evidence of periostin up-regulation at both the RNA and protein levels and of its significant association with the sarcomatoid differentiation in RCC. Moreover, we identified four new periostin splice isoforms, one of which is overexpressed significantly in RCC, compared with the surrounding non-neoplastic renal tissue.
We previously detected periostin in malignant pleural effusions of non-small cell lung cancer by shotgun mass spectrometry,24 suggesting that this extracellular matrix N-glycoprotein is secreted into body fluids. In a consecutive study, we developed a semiquantitative score for the evaluation of periostin protein expression in tumor epithelia and surrounding stroma of non-small cell lung cancer.25 This score was reproducible and permitted us to assess its expression in other cancer types.22,31,39,40 In non-small cell lung cancer, we found periostin to be a prognostic marker for decreased progression-free survival. Further, it correlated with other indicators of EMT such as vimentin.25 These findings underscored the important function of periostin in the regulation of EMT, which in turn is linked to increased invasive and thus metastatic properties of carcinoma cells.41
We now show a statistical correlation between periostin expression and sarcomatoid differentiation of RCC. Identification of sarcomatoid differentiation generally is of high importance for prognostic assessment of RCC patients. RCC is an extremely heterogeneous tumor and tumor cells with a spindle-like phenotype (sarcomatoid differentiation) may be identified only focally or be the main tumor component. In addition to different cytokeratins, these cells may co-express vimentin, another EMT marker. Vimentin expression is associated significantly with poor prognosis.42 Furthermore, we showed that the quantification of tumor areas with sarcomatoid differentiation in ccRCC is prognostically relevant.2,35 This finding was verified by others in larger tumor cohorts.1,43,44 We were able to confirm this in our TMA cohort, showing a close association between high expression of the EMT-related periostin protein with different indicators for poor patient prognosis, including higher tumor dedifferentiation grade, higher stage, and poor overall survival in ccRCC.
In our immunohistochemical analysis we examined periostin immunoreactivity in both cytoplasm of carcinoma cells and extracellular matrix of RCC. In the first cohort of 30 frozen samples of renal carcinoma cases, there were variations in the expression intensity of periostin in the extracellular matrix. In the TMA cohort, both stromal and epithelial periostin were related to lymphocyte infiltration. Strong epithelial periostin correlated with strong lymphocyte infiltration, whereas stromal periostin correlated with lower lymphocyte infiltration. These findings suggest that periostin might modulate the intratumoral lymphocyte infiltration. Thompson et al45 have indeed shown that interactions between immune cells and cancer cells may promote cancer progression by contributing to immune dysfunction in patients with RCC. They found a significant correlation between expression of programmed death-1 receptor on tumor-infiltrating T cells and the presence of sarcomatoid differentiation in RCC patients. In our study, high levels of periostin in the tumor epithelia correlated with higher lymphocyte infiltration and with sarcomatoid differentiation. Taken together, these data suggest that periostin levels in the tumor epithelia somehow might be related to immune dysfunction and more aggressive behavior in RCC. However, the molecular mechanism of interaction between periostin and tumor-infiltrating immune cells remains to be clarified.
It is of note that there were more periostin-negative cases in the TMA cohort compared with the smaller cohort of 30 RCC samples using whole tumor sections. This discrepancy may be a methodologic problem because the TMA technology uses large numbers of samples but with smaller tissue cores (0.6-mm diameter). The TMA analysis suggests that periostin expression in carcinoma cells is related more closely to RCC progression than stromal periostin. It is tempting to speculate that periostin expression in carcinoma cells is more important for invasion/metastasis of cancer cells than periostin expression in the extracellular matrix. Indeed, binding of periostin to the integrins at the plasma membrane leads to increased expression of genes that enhance invasion and metastasis of carcinoma cells,7–11 and this effect probably is exerted by an autocrine/paracrine loop at the close interface between tumor epithelia and surrounding stroma. Thus, periostin production by carcinoma cells may result in protein secretion at a site where it can exert its strongest effect, namely at the tumor invasion front.
According to our results, 60% of RCCs are characterized by an average fivefold up-regulation of periostin mRNA compared with normal tissue. This also was confirmed by IHC, showing increased protein levels in the tumor cells as well as in the stroma. A previous study was not able to find differences between periostin expression in tumor versus normal renal tissue. This was possibly owing to a relatively low case number, a high fluctuation between tissue samples, and diversity in cell type content.46 In our analysis of a gene expression microarray, periostin mRNA was 1.5-fold up-regulated in 70% of the RCCs in comparison with normal renal tissue. The difference in the average up-regulation between the results obtained from the quantitative RT-PCR analysis (average up-regulation of fivefold) and the gene expression microarray analysis (up-regulation of 1.5-fold) may be owing to, first, a different number of patients were enrolled in the two experiments, second, because we used matched normal renal tissues as a control in the quantitative RT-PCR whereas in the analysis of the gene expression microarray we used six nonmatched normal renal tissues as a control, and, third, because the RNA levels were detected with two different methods.
Our immunohistochemical analysis helps to clarify the localization of periostin. In fact, its localization has been a matter of controversy for a long time. Some studies showed the presence of periostin in stromal cells but not in cancer cells21,23,26,47 or both,29 whereas others found periostin mRNA only in cancer cells.9,11 The immunohistochemical analysis of our RCC cohorts provides evidence of protein presence in both tumor epithelia and stroma, which is in agreement with similar findings in lung, prostate, and liver carcinoma, as well as in malignant pleural mesothelioma.22,25,31,39,40 It remains to be clarified if the presence of periostin in the cytoplasm of tumor epithelia is caused by endogenous production or by an uptake mechanism from the surrounding extracellular matrix. We would rather favor endogenous production because periostin carries several N-glycomotifs in its N-terminal region, which is a typical feature of secreted proteins and mRNA was identified in the cytoplasm of breast and pancreatic cancer cells.9,11
In the desmoplastic stroma surrounding and intercalating the tumor, high levels of periostin correlated with increased amounts of collagen fibers as well as acid mucopolysaccharides. These results substantiate the concept that periostin isoforms exert their influence on dynamic stroma remodeling through their binding to collagen fibers48–50 in an acid mucopolysaccharide-rich environment. This is in line with the fact that periostin can bind the acid mucopolysaccharide heparin.51 Thus, next to collagen, periostin may be a major constituent of the desmoplastic microenvironment. However, there are no data about the exact stoichiometric relations.
Interestingly, four of the eight periostin isoforms identified in our analysis of fetal kidney tissue are unknown splice variants. The other four represent full-length periostin and three previously reported isoforms. The presence of variants in fetal kidney that are not expressed in RCC or adult kidney suggests a role of those proteins during embryogenesis of the kidney, including dynamic tissue remodeling, which comprises rearrangement of extracellular matrix fibers. Norris et al50 indeed showed that periostin binds collagen I and therefore is involved in stroma fibrillogenesis, influencing the biomechanical properties of fibrous connective tissues. Hoersch and Andrade-Navarro48 suggested that the C-terminal domain of periostin is responsible for binding to the matrix proteins such as collagen and fibronectin and that the isoforms may exert their influence on the extracellular matrix fibrillogenesis differently. Therefore, the cell type and the differentiation state specificity of periostin isoforms seems to reflect the differential modulation of periostin function through alternative splicing. This hypothesis is supported by the finding that overexpression of the isoform “variant I” (corresponding to isoform 4) led to increased invasiveness in in vitro and in vivo experiments using the human bladder cancer cell line SBT991 and the mouse malignant melanoma cell line B16F10.52 Furthermore, blocking the activity of periostin isoforms in the mouse osteoblast cell line MC3T3-E1 led to a significant reduction of osteoblast-specific differentiation markers.14
Among the three spliced periostin variants that were co-expressed in both RCC tissue and matched non-neoplastic tissue, two corresponded to the already described isoforms 3 and 4 (accession numbers: AY140646 and AY918092, UniProtKB/Swiss-Prot database), whereas, the third isoform represented an as yet unknown variant (named isoform 8). Interestingly, this new isoform was expressed more frequently in RCC than in matched non-neoplastic tissue. Even though the role of alternative splicing of periostin mRNA is not fully understood, these findings suggest that isoform 8 plays a role during tumor development, which is in line with the concept that some periostin variants may enhance tumor invasiveness. The fact that the already known full-length periostin (isoform 1), the isoform 2, and the newly identified isoforms 5 to 7 were detected neither in normal nor in tumor tissue of the kidney, leads to the hypothesis that the isoform regulation depends on the differentiation state and on the tissue type.
In summary, our study represents a comprehensive analysis of the EMT marker periostin in RCC, and indicates that this protein could be involved in the sarcomatoid differentiation of RCC, but further biomolecular studies are necessary to verify this hypothesis. Periostin splice isoforms were characterized in RCC and fetal and adult normal kidney. Furthermore, a novel isoform differentially expressed in normal kidney and RCC was identified.
Acknowledgments
We thank the Broad Institute Center for Genotyping and Analysis (MIT and Harvard) for performing gene expression array experiments; Susanne Dettwiler, Martina Storz, and Silvia Behnke for excellent technical support. Adriana von Teichmann is acknowledged for critical reading of the manuscript.
Footnotes
Supported by the Swiss National Science Foundation; the Cancer League, Zurich, Switzerland; and Helmut Horten Foundation, Agno, Switzerland.
Holger Moch and Alex Soltermann shared last authorship.
Supplemental material for this article can be found at http://ajp.amjpathol.org or at doi: 10.1016/j.ajpath.2011.05.035.
Supplementary data
References
- 1.Cheville J.C., Lohse C.M., Zincke H., Weaver A.L., Leibovich B.C., Frank I., Blute M.L. Sarcomatoid renal cell carcinoma: an examination of underlying histologic subtype and an analysis of associations with patient outcome. Am J Surg Pathol. 2004;28:435–441. doi: 10.1097/00000478-200404000-00002. [DOI] [PubMed] [Google Scholar]
- 2.Moch H., Gasser T., Amin M.B., Torhorst J., Sauter G., Mihatsch M.J. Prognostic utility of the recently recommended histologic classification and revised TNM staging system of renal cell carcinoma: a Swiss experience with 588 tumors. Cancer. 2000;89:604–614. [PubMed] [Google Scholar]
- 3.Takeshita S., Kikuno R., Tezuka K., Amann E. Osteoblast-specific factor 2: cloning of a putative bone adhesion protein with homology with the insect protein fasciclin I. Biochem J. 1993;294:271–278. doi: 10.1042/bj2940271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Horiuchi K., Amizuka N., Takeshita S., Takamatsu H., Katsuura M., Ozawa H., Toyama Y., Bonewald L.F., Kudo A. Identification and characterization of a novel protein, periostin, with restricted expression to periosteum and periodontal ligament and increased expression by transforming growth factor beta. J Bone Miner Res. 1999;14:1239–1249. doi: 10.1359/jbmr.1999.14.7.1239. [DOI] [PubMed] [Google Scholar]
- 5.Inai K., Norris R.A., Hoffman S., Markwald R.R., Sugi Y. BMP-2 induces cell migration and periostin expression during atrioventricular valvulogenesis. Dev Biol. 2008;315:383–396. doi: 10.1016/j.ydbio.2007.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yan W., Shao R. Transduction of a mesenchyme-specific gene periostin into 293T cells induces cell invasive activity through epithelial-mesenchymal transformation. J Biol Chem. 2006;281:19700–19708. doi: 10.1074/jbc.M601856200. [DOI] [PubMed] [Google Scholar]
- 7.Gillan L., Matei D., Fishman D.A., Gerbin C.S., Karlan B.Y., Chang D.D. Periostin secreted by epithelial ovarian carcinoma is a ligand for alpha(V)beta(3) and alpha(V)beta(5) integrins and promotes cell motility. Cancer Res. 2002;62:5358–5364. [PubMed] [Google Scholar]
- 8.Bao S., Ouyang G., Bai X., Huang Z., Ma C., Liu M., Shao R., Anderson R.M., Rich J.N., Wang X.F. Periostin potently promotes metastatic growth of colon cancer by augmenting cell survival via the Akt/PKB pathway. Cancer Cell. 2004;5:329–339. doi: 10.1016/s1535-6108(04)00081-9. [DOI] [PubMed] [Google Scholar]
- 9.Baril P., Gangeswaran R., Mahon P.C., Caulee K., Kocher H.M., Harada T., Zhu M., Kalthoff H., Crnogorac-Jurcevic T., Lemoine N.R. Periostin promotes invasiveness and resistance of pancreatic cancer cells to hypoxia-induced cell death: role of the beta4 integrin and the PI3k pathway. Oncogene. 2007;26:2082–2094. doi: 10.1038/sj.onc.1210009. [DOI] [PubMed] [Google Scholar]
- 10.Li P., Oparil S., Feng W., Chen Y.F. Hypoxia-responsive growth factors upregulate periostin and osteopontin expression via distinct signaling pathways in rat pulmonary arterial smooth muscle cells. J Appl Physiol. 2004;97:1550–1558. doi: 10.1152/japplphysiol.01311.2003. discussion, 1549. [DOI] [PubMed] [Google Scholar]
- 11.Shao R., Bao S., Bai X., Blanchette C., Anderson R.M., Dang T., Gishizky M.L., Marks J.R., Wang X.F. Acquired expression of periostin by human breast cancers promotes tumor angiogenesis through up-regulation of vascular endothelial growth factor receptor 2 expression. Mol Cell Biol. 2004;24:3992–4003. doi: 10.1128/MCB.24.9.3992-4003.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kudo Y., Siriwardena B.S., Hatano H., Ogawa I., Takata T. Periostin: novel diagnostic and therapeutic target for cancer. Histol Histopathol. 2007;22:1167–1174. doi: 10.14670/HH-22.1167. [DOI] [PubMed] [Google Scholar]
- 13.Castronovo V., Waltregny D., Kischel P., Roesli C., Elia G., Rybak J.N., Neri D. A chemical proteomics approach for the identification of accessible antigens expressed in human kidney cancer. Mol Cell Proteomics. 2006;5:2083–2091. doi: 10.1074/mcp.M600164-MCP200. [DOI] [PubMed] [Google Scholar]
- 14.Litvin J., Selim A.H., Montgomery M.O., Lehmann K., Rico M.C., Devlin H., Bednarik D.P., Safadi F.F. Expression and function of periostin-isoforms in bone. J Cell Biochem. 2004;92:1044–1061. doi: 10.1002/jcb.20115. [DOI] [PubMed] [Google Scholar]
- 15.Kruzynska-Frejtag A., Machnicki M., Rogers R., Markwald R.R., Conway S.J. Periostin (an osteoblast-specific factor) is expressed within the embryonic mouse heart during valve formation. Mech Dev. 2001;103:183–188. doi: 10.1016/s0925-4773(01)00356-2. [DOI] [PubMed] [Google Scholar]
- 16.Kruzynska-Frejtag A., Wang J., Maeda M., Rogers R., Krug E., Hoffman S., Markwald R.R., Conway S.J. Periostin is expressed within the developing teeth at the sites of epithelial-mesenchymal interaction. Dev Dyn. 2004;229:857–868. doi: 10.1002/dvdy.10453. [DOI] [PubMed] [Google Scholar]
- 17.Wilde J., Yokozeki M., Terai K., Kudo A., Moriyama K. The divergent expression of periostin mRNA in the periodontal ligament during experimental tooth movement. Cell Tissue Res. 2003;312:345–351. doi: 10.1007/s00441-002-0664-2. [DOI] [PubMed] [Google Scholar]
- 18.Nakazawa T., Nakajima A., Seki N., Okawa A., Kato M., Moriya H., Amizuka N., Einhorn T.A., Yamazaki M. Gene expression of periostin in the early stage of fracture healing detected by cDNA microarray analysis. J Orthop Res. 2004;22:520–525. doi: 10.1016/j.orthres.2003.10.007. [DOI] [PubMed] [Google Scholar]
- 19.Lindner V., Wang Q., Conley B.A., Friesel R.E., Vary C.P. Vascular injury induces expression of periostin: implications for vascular cell differentiation and migration. Arterioscler Thromb Vasc Biol. 2005;25:77–83. doi: 10.1161/01.ATV.0000149141.81230.c6. [DOI] [PubMed] [Google Scholar]
- 20.Shimazaki M., Kudo A. [Periostin, acting in regeneration of periodontal ligament, contributes to cardiac healing and tumor capsule formation] Fukuoka Igaku Zasshi. 2009;100:67–74. [PubMed] [Google Scholar]
- 21.Erkan M., Kleeff J., Gorbachevski A., Reiser C., Mitkus T., Esposito I., Giese T., Buchler M.W., Giese N.A., Friess H. Periostin creates a tumor-supportive microenvironment in the pancreas by sustaining fibrogenic stellate cell activity. Gastroenterology. 2007;132:1447–1464. doi: 10.1053/j.gastro.2007.01.031. [DOI] [PubMed] [Google Scholar]
- 22.Riener M.O., Fritzsche F.R., Soll C., Pestalozzi B.C., Probst-Hensch N., Clavien P.A., Jochum W., Soltermann A., Moch H., Kristiansen G. Expression of the extracellular matrix protein periostin in liver tumours and bile duct carcinomas. Histopathology. 2010;56:600–606. doi: 10.1111/j.1365-2559.2010.03527.x. [DOI] [PubMed] [Google Scholar]
- 23.Sasaki H., Dai M., Auclair D., Fukai I., Kiriyama M., Yamakawa Y., Fujii Y., Chen L.B. Serum level of the periostin, a homologue of an insect cell adhesion molecule, as a prognostic marker in nonsmall cell lung carcinomas. Cancer. 2001;92:843–848. doi: 10.1002/1097-0142(20010815)92:4<843::aid-cncr1391>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- 24.Soltermann A., Ossola R., Kilgus-Hawelski S., von Eckardstein A., Suter T., Aebersold R., Moch H. N-glycoprotein profiling of lung adenocarcinoma pleural effusions by shotgun proteomics. Cancer. 2008;114:124–133. doi: 10.1002/cncr.23349. [DOI] [PubMed] [Google Scholar]
- 25.Soltermann A., Tischler V., Arbogast S., Braun J., Probst-Hensch N., Weder W., Moch H., Kristiansen G. Prognostic significance of epithelial-mesenchymal and mesenchymal-epithelial transition protein expression in non-small cell lung cancer. Clin Cancer Res. 2008;14:7430–7437. doi: 10.1158/1078-0432.CCR-08-0935. [DOI] [PubMed] [Google Scholar]
- 26.Sasaki H., Yu C.Y., Dai M., Tam C., Loda M., Auclair D., Chen L.B., Elias A. Elevated serum periostin levels in patients with bone metastases from breast but not lung cancer. Breast Cancer Res Treat. 2003;77:245–252. doi: 10.1023/a:1021899904332. [DOI] [PubMed] [Google Scholar]
- 27.Kudo Y., Ogawa I., Kitajima S., Kitagawa M., Kawai H., Gaffney P.M., Miyauchi M., Takata T. Periostin promotes invasion and anchorage-independent growth in the metastatic process of head and neck cancer. Cancer Res. 2006;66:6928–6935. doi: 10.1158/0008-5472.CAN-05-4540. [DOI] [PubMed] [Google Scholar]
- 28.Puppin C., Fabbro D., Dima M., Di Loreto C., Puxeddu E., Filetti S., Russo D., Damante G. High periostin expression correlates with aggressiveness in papillary thyroid carcinomas. J Endocrinol. 2008;197:401–408. doi: 10.1677/JOE-07-0618. [DOI] [PubMed] [Google Scholar]
- 29.Li J.S., Sun G.W., Wei X.Y., Tang W.H. Expression of periostin and its clinicopathological relevance in gastric cancer. World J Gastroenterol. 2007;13:5261–5266. doi: 10.3748/wjg.v13.i39.5261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sasaki H., Sato Y., Kondo S., Fukai I., Kiriyama M., Yamakawa Y., Fuji Y. Expression of the periostin mRNA level in neuroblastoma. J Pediatr Surg. 2002;37:1293–1297. doi: 10.1053/jpsu.2002.34985. [DOI] [PubMed] [Google Scholar]
- 31.Tischler V., Fritzsche F.R., Wild P.J., Stephan C., Seifert H.H., Riener M.O., Hermanns T., Mortezavi A., Gerhardt J., Schraml P., Jung K., Moch H., Soltermann A., Kristiansen G. Periostin is up-regulated in high grade and high stage prostate cancer. BMC Cancer. 2010;10:273. doi: 10.1186/1471-2407-10-273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.WHO . Tumours of the urinary system and male genital organs: pathology and genetics. In: Eble John N., editor. IARC Press; Lyon, France: 2004. pp. 9–87. [Google Scholar]
- 33.Wu Z.J., Irizarry R.A., Gentleman R., Martinez-Murillo F., Spencer F. A model-based background adjustment for oligonucleotide expression arrays. J Am Stat Assoc. 2004;99:909–917. [Google Scholar]
- 34.Kononen J., Bubendorf L., Kallioniemi A., Barlund M., Schraml P., Leighton S., Torhorst J., Mihatsch M.J., Sauter G., Kallioniemi O.P. Tissue microarrays for high-throughput molecular profiling of tumor specimens. Nat Med. 1998;4:844–847. doi: 10.1038/nm0798-844. [DOI] [PubMed] [Google Scholar]
- 35.de Peralta-Venturina M., Moch H., Amin M., Tamboli P., Hailemariam S., Mihatsch M., Javidan J., Stricker H., Ro J.Y., Amin M.B. Sarcomatoid differentiation in renal cell carcinoma: a study of 101 cases. Am J Surg Pathol. 2001;25:275–284. doi: 10.1097/00000478-200103000-00001. [DOI] [PubMed] [Google Scholar]
- 36.Jung M., Ramankulov A., Roigas J., Johannsen M., Ringsdorf M., Kristiansen G., Jung K. In search of suitable reference genes for gene expression studies of human renal cell carcinoma by real-time PCR. BMC Mol Biol. 2007;8:47. doi: 10.1186/1471-2199-8-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Luu V.D., Boysen G., Struckmann K., Casagrande S., von Teichman A., Wild P.J., Sulser T., Schraml P., Moch H. Loss of VHL and hypoxia provokes PAX2 up-regulation in clear cell renal cell carcinoma. Clin Cancer Res. 2009;15:3297–3304. doi: 10.1158/1078-0432.CCR-08-2779. [DOI] [PubMed] [Google Scholar]
- 38.Tusher V.G., Tibshirani R., Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dahinden C., Ingold B., Wild P., Boysen G., Luu V.D., Montani M., Kristiansen G., Sulser T., Buhlmann P., Moch H., Schraml P. Mining tissue microarray data to uncover combinations of biomarker expression patterns that improve intermediate staging and grading of clear cell renal cell cancer. Clin Cancer Res. 2010;16:88–98. doi: 10.1158/1078-0432.CCR-09-0260. [DOI] [PubMed] [Google Scholar]
- 40.Schramm A., Opitz I., Thies S., Seifert B., Moch H., Weder W., Soltermann A. Prognostic significance of epithelial-mesenchymal transition in malignant pleural mesothelioma. Eur J Cardiothorac Surg. 2010;37:566–572. doi: 10.1016/j.ejcts.2009.08.027. [DOI] [PubMed] [Google Scholar]
- 41.Mimeault M., Batra S.K. Interplay of distinct growth factors during epithelial mesenchymal transition of cancer progenitor cells and molecular targeting as novel cancer therapies. Ann Oncol. 2007;18:1605–1619. doi: 10.1093/annonc/mdm070. [DOI] [PubMed] [Google Scholar]
- 42.Moch H., Schraml P., Bubendorf L., Mirlacher M., Kononen J., Gasser T., Mihatsch M.J., Kallioniemi O.P., Sauter G. High-throughput tissue microarray analysis to evaluate genes uncovered by cDNA microarray screening in renal cell carcinoma. Am J Pathol. 1999;154:981–986. doi: 10.1016/S0002-9440(10)65349-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kwak C., Park Y.H., Jeong C.W., Jeong H., Lee S.E., Moon K.C., Ku J.H. Sarcomatoid differentiation as a prognostic factor for immunotherapy in metastatic renal cell carcinoma. J Surg Oncol. 2007;95:317–323. doi: 10.1002/jso.20669. [DOI] [PubMed] [Google Scholar]
- 44.Mian B.M., Bhadkamkar N., Slaton J.W., Pisters P.W., Daliani D., Swanson D.A., Pisters L.L. Prognostic factors and survival of patients with sarcomatoid renal cell carcinoma. J Urol. 2002;167:65–70. [PubMed] [Google Scholar]
- 45.Thompson R.H., Dong H., Lohse C.M., Leibovich B.C., Blute M.L., Cheville J.C., Kwon E.D. PD-1 is expressed by tumor-infiltrating immune cells and is associated with poor outcome for patients with renal cell carcinoma. Clin Cancer Res. 2007;13:1757–1761. doi: 10.1158/1078-0432.CCR-06-2599. [DOI] [PubMed] [Google Scholar]
- 46.Tilman G., Mattiussi M., Brasseur F., van Baren N., Decottignies A. Human periostin gene expression in normal tissues, tumors and melanoma: evidences for periostin production by both stromal and melanoma cells. Mol Cancer. 2007;6:80. doi: 10.1186/1476-4598-6-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sasaki H., Lo K.M., Chen L.B., Auclair D., Nakashima Y., Moriyama S., Fukai I., Tam C., Loda M., Fujii Y. Expression of Periostin, homologous with an insect cell adhesion molecule, as a prognostic marker in non-small cell lung cancers. Jpn J Cancer Res. 2001;92:869–873. doi: 10.1111/j.1349-7006.2001.tb01174.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hoersch S., Andrade-Navarro M.A. Periostin shows increased evolutionary plasticity in its alternatively spliced region. BMC Evol Biol. 2010;10:30. doi: 10.1186/1471-2148-10-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sottile J., Shi F., Rublyevska I., Chiang H.Y., Lust J., Chandler J. Fibronectin-dependent collagen I deposition modulates the cell response to fibronectin. Am J Physiol Cell Physiol. 2007;293:C1934–C1946. doi: 10.1152/ajpcell.00130.2007. [DOI] [PubMed] [Google Scholar]
- 50.Norris R.A., Damon B., Mironov V., Kasyanov V., Ramamurthi A., Moreno-Rodriguez R., Trusk T., Potts J.D., Goodwin R.L., Davis J., Hoffman S., Wen X., Sugi Y., Kern C.B., Mjaatvedt C.H., Turner D.K., Oka T., Conway S.J., Molkentin J.D., Forgacs G., Markwald R.R. Periostin regulates collagen fibrillogenesis and the biomechanical properties of connective tissues. J Cell Biochem. 2007;101:695–711. doi: 10.1002/jcb.21224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Snider P., Hinton R.B., Moreno-Rodriguez R.A., Wang J., Rogers R., Lindsley A., Li F., Ingram D.A., Menick D., Field L., Firulli A.B., Molkentin J.D., Markwald R., Conway S.J. Periostin is required for maturation and extracellular matrix stabilization of noncardiomyocyte lineages of the heart. Circ Res. 2008;102:752–760. doi: 10.1161/CIRCRESAHA.107.159517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kim C.J., Isono T., Tambe Y., Chano T., Okabe H., Okada Y., Inoue H. Role of alternative splicing of periostin in human bladder carcinogenesis. Int J Oncol. 2008;32:161–169. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


